Abstract
The burden of coronavirus disease 2019 (COVID-19) in children represents a fraction of cases worldwide, yet a subset of those infected are at risk for severe disease. We measured plasma severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) RNA in a cohort of 103 children hospitalized with COVID-19 with diverse clinical manifestations. SARS-CoV-2 RNAemia was detected in 27 (26%) of these children, lasted for a median of 6 (interquartile range, 2–9) days, and was associated with higher rates of oxygen administration, admission to the intensive care unit, and longer hospitalization.
Keywords: children, COVID-19, disease severity, infants, outcomes, RNAemia, SARS-CoV-2, viremia
In a cohort of 103 children and adolescents hospitalized with COVID-19 with diverse clinical manifestations, SARS-CoV-2 RNAemia was detected in 26% of them, lasted for 6 (interquartile range, 2–9) days, and was associated with worse clinical outcomes.
The coronavirus disease 2019 (COVID-19) pandemic remains an enormous challenge worldwide due to the high infectivity of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the causative agent, and the tremendous variability in clinical presentations [1]. Children are generally less likely to experience severe COVID-19 compared to adults; however, young children—particularly those <1 year of age—and adolescents are at a greater risk for serious illness [2–4]. Additionally, children and young adolescents are at risk of developing a rare but serious condition after SARS-CoV-2 infection that has been termed multisystem inflammatory syndrome in children (MIS-C) [5].
Previous studies conducted in adults indicated that detection of SARS-CoV-2 RNA in plasma occurred more frequently in patients with severe disease and was associated with worse clinical outcomes including mortality [6]. The frequency of SARS-CoV-2 RNAemia and its prognostic implications in children with COVID-19 remain poorly defined.
In this study, we analyzed the frequency of plasma SARS-CoV-2 RNA detection in children hospitalized with COVID-19, and its association with various clinical characteristics and disease severity parameters.
METHODS
A convenience sample of children and adolescents <21 years of age hospitalized with COVID-19 were prospectively enrolled at Nationwide Children’s Hospital (NCH) in Columbus, Ohio. Blood and nasopharyngeal (NP) research samples were obtained within a median of 21 (interquartile range [IQR], 13–39) hours of hospitalization for SARS-CoV-2 quantitation by real-time polymerase chain reaction (PCR). Additional blood research samples were collected every 3 (IQR, 2–5) days during the acute hospitalization until patient discharge. We excluded from the analysis children in whom paired NP and blood samples were not obtained, children with MIS-C, or those in whom confirmatory NP SARS-CoV-2 PCR was negative.
At enrollment, we collected demographic and clinical information including duration of illness on all study patients. Other parameters of disease severity were also collected including presence and duration of fever, oxygen administration, pediatric intensive care unit (PICU) admission, duration of hospitalization, and administration of COVID-19–directed therapies including remdesivir, tocilizumab, bamlanivimab, or convalescent plasma/intravenous immunoglobulin, as well as systemic steroids and anticoagulation medications. This information was collected using standardized questionnaires designed for the study using portable tablets, and information was automatically transferred to a secure database (REDCap). During the study period all children hospitalized at NCH, irrespective of the presence of symptoms, underwent NP SARS-CoV-2 testing per standard of care using a PCR assay (GeneXpert Xpress SARS-CoV-2; Cepheid, Sunnyvale, California); the US Centers for Disease Control and Prevention 2019-nCoV_N1 primers and probes [6]; or the FilmArray Respiratory Viral Panel (BioFire, Salt Lake City, Utah). Plasma SARS-CoV-2 RNAemia was assessed as part of the research study.
SARS-CoV-2 Viral Loads
Nasopharyngeal swabs were collected at enrollment, placed in viral transport media, transported immediately to the laboratory, aliquoted, and stored at –80°C. Similarly, blood samples were collected in ethylenediaminetetraacetic acid tubes (BD Vacutainer, Franklin Lakes, New Jersey) and centrifuged, and aliquots of plasma were stored at –80°C until processed in batches. Viral RNA was extracted from 200 µL of NP or plasma samples using the QIAcube HT instrument (Qiagen, Germantown, Maryland) and eluted into 100 µL volume. In brief, SARS-CoV-2 viral loads were measured using a 2-step reverse-transcription (RT) quantitative PCR assay targeting the conserved region of the N1 gene using described primers and probes [6]. A standard curve was generated from a dilution series of a known concentration (107copies/mL) of the N1 gene of SARS-CoV-2. Standards and negative controls were included and tested with each PCR assay. The lower limit of detection of the assay was 1–2 copies/reaction (~200 copies/mL).
Statistical Analysis
Data are reported using descriptive statistics, with frequency and percentage for categorical variables and mean (standard deviation) or median (IQR, 25%–75%) for continuous variables according to data distribution. Categorical data were analyzed by χ2 or Fisher exact test, and continuous variables by Mann–Whitney test. Analyses were conducted using GraphPad Prism version 9 (GraphPad Software, San Diego, California), with a 2-sided P value < .05 considered statistically significant.
Ethical Considerations
This study was approved by the Institutional Review Board (IRB) at Nationwide Children’s Hospital (IRB number STUDY00000921), classified as a level 1 risk clinical study—no greater than minimal risk (pursuant under 45 Code of Federal Regulations [CFR] 46.404 and 21 CFR 50.51). Informed consent procedures followed Nationwide Children’s Research Responsible Conduct Guidelines.
RESULTS
From March 2020 to May 2021, 685 patients with COVID-19 were hospitalized at NCH. Of those, we enrolled a convenience sample 157 patients, and excluded 54 from analyses for the reasons outlined in Supplementary Figure 1, leaving a total of 103 children and adolescents <21 years of age (median [IQR] age, 9.2 [0.6–16.1] years) with paired NP and blood samples. Of the 103 patients, 23 (22%) were identified by screening on admission, and 80 (78%) were hospitalized with symptomatic COVID-19. The most common diagnoses in the 23 patients identified by screening were appendicitis (n = 7), hospitalization for a traumatic injury or presurgical evaluation (n = 7), and other infectious disease diagnoses (n = 7).
Plasma SARS-CoV-2 RNA was detected in 26% (27/103) of patients, 26 with symptomatic COVID-19, and in a 13-year-old female who presented with ruptured appendicitis. Thus, the frequency of plasma SARS-CoV-2 detection was 4% (1/23) in children identified by screening compared with 33% (26/80) in the symptomatic COVID-19 cohort (P = .006). The demographic, clinical characteristics, laboratory parameters, and clinical outcomes of study patients according to the presence of RNAemia are included in Table 1. Median age of patients with SARS-CoV-2 RNAemia was 1.6 years compared to 11.1 years in the non-RNAemia group, although this difference did not reach statistical significance. Also, 44% (12/27) of patients with SARS-CoV-2 RNAemia were <1 year of age compared with 24% (18/76) in the non-RNAemia cohort (P = .05). Sex, race, ethnicity, and presence of underlying conditions were not different between children with and without SARS-CoV-2 RNAemia.
Table 1.
Demographic, Clinical Characteristics, Laboratory Parameters, and Clinical Outcomes of All Study Patients
| Characteristic | SARS-CoV-2 RNAemia (n = 27) | No SARS-CoV-2 RNAemia (n = 76) | P Value |
|---|---|---|---|
| Demographic characteristics | |||
| Age, y | 1.6 (0.1–13.7) | 11.1 (1.4–16.1) | .16 |
| Sex (male/female) | 16 (59)/11 (41) | 42 (55)/34 (45) | .82 |
| Race | .46 | ||
| White | 12 (44) | 40 (53) | |
| Black | 11 (41) | 18 (24) | |
| Multiracial | 3 (11) | 10 (13) | |
| Asian | 1 (4) | 6 (8) | |
| Other | 0 | 2 (2) | |
| Ethnicity | .76 | ||
| Hispanic or Latino | 5 (18) | 13 (17) | |
| Not Hispanic or Latino | 22 (78) | 63 (83) | |
| Not reported | 1 (4) | 0 (0) | |
| Underlying conditions | 15 (56) | 33 (43) | .37 |
| Obesity | 4 (15) | 17 (22) | |
| Immunocompromiseda | 4 (15) | 3 (4) | |
| Respiratory (CLD/asthma) | 3 (11) | 9 (12) | |
| Otherb | 4 (15) | 4 (5) | |
| Clinical and laboratory parameters | |||
| Days of symptoms | 2 (1–3) | 4 (2–7) | .005 |
| Presence of fever | 22 (81) | 42 (55) | .02 |
| Duration of fever, d | 2.5 (1.7–7.5) | 3 (2–6) | .82 |
| ALCc | |||
| Infants | 4120 (1292–5512) | 4171 (2424–5800) | .51 |
| Children >1 y | 778 (337–1067) | 1725 (1113–2518) | .001 |
| C-reactive protein, mg/dL | 1.5 (0.5–3.7) | 1.7 (0.6–4.5) | .28 |
| Medications used | |||
| Anti–COVID-19 medicationsd | 9 (33) | 10 (13) | .04 |
| Anticoagulant | 6 (22) | 10 (13) | .37 |
| Systemic steroids | 10 (37) | 19 (25) | .31 |
| Outcomes of care | |||
| PICU admission | 11 (41) | 15 (20) | .04 |
| O2 administration at admission | 10 (37) | 12 (16) | .03 |
| Hospital length of stay, d | 2.6 (1.7–14.0) | 2.2 (1.4–4.7) | .07 |
| Viral load data | |||
| NP SARS-CoV-2 load, log10 copies/mL | 8.2 (5.1–9.1) | 4.7 (3.5–7.1) | <.0001 |
| NP SARS-CoV-2 load, Ct value | 20.9 (17.9–30.6) | 31.9 (25.6–36.8) | <.0001 |
Categorical data are expressed as frequencies (%) and analyzed using Fisher exact test or χ2 test. Continuous variables are expressed as median (interquartile range) and analyzed using Mann–Whitney rank test. Values in bold indicate significant P values.
Abbreviations: ALC, absolute lymphocyte count; CI, confidence interval; CLD, chronic lung disease; COVID-19, coronavirus disease 2019; NP, nasopharyngeal; PICU, pediatric intensive care unit; O2, oxygen; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
Immunocompromised host includes congenital immunodeficiency (n = 1), sickle cell disease with asplenia (n = 2), solid organ transplant (n = 1), and acute lymphocytic leukemia (n = 3).
Other includes congenital heart disease (n = 3), diabetes (n = 3), cerebral palsy (n = 1), and prematurity (n = 1).
Six of 30 infants (20%) and 17 of 73 (23%) children did not have ALC data available.
Anti–COVID-19 medications include remdesivir (n = 14), convalescent plasma/intravenous immunoglobulin (n = 3), and bamlanivimab (n = 2).
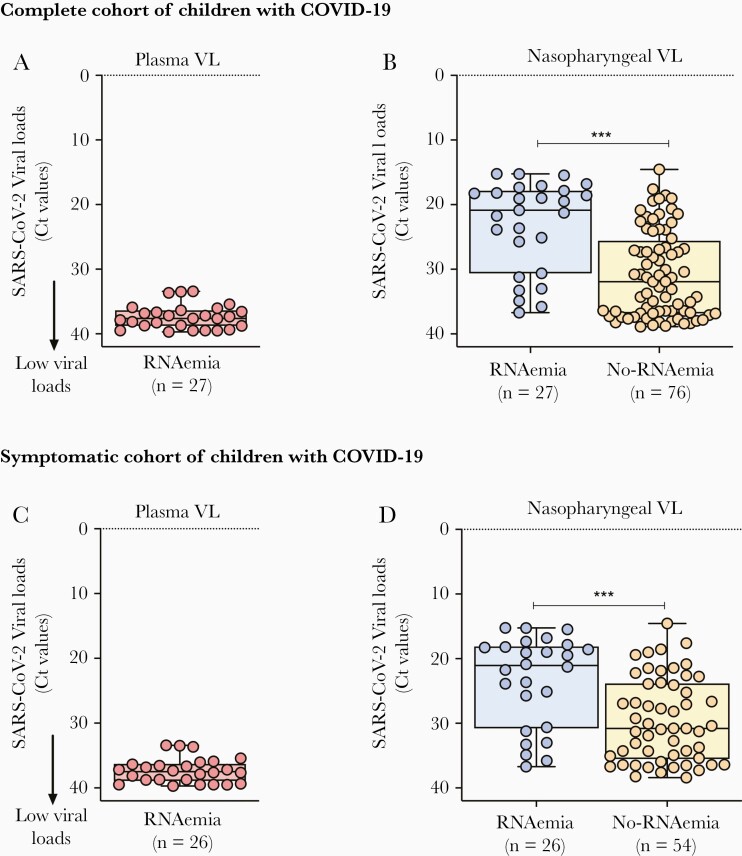
Median viral loads in the 27 patients in whom SARS-CoV-2 was detected in plasma were 3.1 (IQR, 2.9–3.6) log10 copies/mL (or 37.6 [IQR, 36.3–38.8] RT-PCR cycle threshold [Ct] values), and RNAemia lasted for 6 (IQR, 2–9) days in the 11 symptomatic patients in whom sequential samples were obtained during the hospitalization. The plasma SARS-CoV-2 load in the asymptomatic patient was 2.9 log10 copies/mL (38.2 Ct values) and was measured only once at study enrollment. Patients with SARS-CoV-2 RNAemia also had significantly higher viral loads in nasopharyngeal samples compared with patients without RNAemia (20.9 vs 31.9 Ct values; P < .0001) (Figure 1A and 1B). Similar results were obtained when SARS-CoV-2 viral loads were exclusively analyzed in the symptomatic COVID-19 cohort (Figure 1C and 1D).
Figure 1.
Nasopharyngeal and plasma severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) viral load (VL) in children and adolescents with coronavirus disease 2019 (COVID-19). A and C, The y-axis represents the reverse-transcription polymerase chain reaction cycle threshold (Ct) SARS-CoV-2 values in plasma samples from all children and adolescents with SARS-CoV-2 RNAemia (red dots) (A) and in those with symptomatic COVID-19 (C). B and D, Nasopharyngeal SARS-CoV-2 loads in all children and adolescents with RNAemia (blue dots) and without RNAemia (yellow dots) (B), and exclusively in children with symptomatic COVID-19 (D). Data represent median (interquartile range). Comparisons by Mann–Whitney rank test; ∗∗∗P < .001; ∗∗∗∗P < .0001.
At study enrollment, patients with SARS-CoV-2 RNAemia had shorter duration of symptoms (2 vs 4 days, respectively; P = .005) and had fever more frequently (81% vs 55%, respectively; P = .02). In addition, lymphopenia, defined as an absolute lymphocyte count of <4500 cells/µL in infants and <1500 cells/µL in children >1 year of age [7], was significantly more common in children with SARS-CoV-2 RNAemia than in those without (72% vs 43%; P = .01). A higher proportion of patients with SARS-CoV-2 RNAemia were treated with anti–COVID-19–directed therapies during their hospitalization (33% vs 13%; P = .04) Table 1. In addition, patients with SARS-CoV-2 RNAemia compared with children without RNAemia had greater disease severity in the different clinical outcomes evaluated including supplemental oxygen administration (37% vs 16%; P = .03), need for PICU admission (41% vs 20%; P = .04), and a trend toward longer duration of hospitalization that was not significantly different (2.6 vs 2.2 days; P = .07). There was no mortality in any of the enrolled patients.
Sensitivity analyses, excluding patients in whom SARS-CoV-2 was identified by screening, confirmed that detection of plasma SARS-CoV-2 RNAemia was associated with worse disease severity as defined by longer duration of hospitalization (median, 3.1 [IQR, 1.7–14.7] vs 1.9 [IQR, 1.2–4.1] days for the RNAemia and non-RNAemia groups, respectively; P = .02), and higher odds of PICU admission (odds ratio, 2.67 [95% confidence interval, .97–6.64]; P = .07) (Supplementary Table 1).
DISCUSSION
In this cohort of pediatric patients with COVID-19, more than one-fourth had detectable SARS-CoV-2 RNA in their plasma, and plasma RNAemia was associated with worse clinical outcomes, defined as supplemental oxygen administration, requirement of PICU care, and duration of hospitalization.
The presence of underlying conditions and age have been identified as risk factors for severe COVID-19 in adults, but there still exists a substantial knowledge gap concerning the factors that influence disease severity among children with SARS-CoV-2 infection [8]. Viremia associated with other respiratory viruses that commonly affect children, such as rhinovirus or influenza virus, has been identified as a risk factor for symptomatic and/or severe disease [9, 10]. The relationship between SARS-CoV-2 viremia and COVID-19 severity in children, however, is not completely understood. In this cohort of children with COVID-19, the frequency of SARS-CoV-2 RNAemia was similar to that previously described in adults, and although no mortality was documented, detection of plasma SARS-CoV-2 RNA lasted for almost 1 week and was associated with enhanced disease severity [6, 11]. Studies conducted in pediatric patients suggested that children with COVID-19 have higher viral loads in NP samples compared with adults, but low or negligible rates of viremia [12, 13]. Those findings are in contrast to our study, which could be attributed in part to the different populations included, as in those studies the majority of children were of older age. In our study, although age ranges overlapped between groups, the median age of children in whom SARS-CoV-2 was detected in plasma was lower than that of children without RNAemia; 44% of viremic children were <1 year old and 52% were <2 years of age. Thus, we present additional evidence that younger children—especially those <1 year of age—might be at increased risk for severe illness from SARS-CoV-2 infection.
Our work identifies a relationship between RNAemia and severe presentation of COVID-19 in children, but overall favorable outcomes as no fatalities were documented. The underlying mechanisms leading to SARS-CoV-2 viremia are not well understood, but other studies conducted in adults correlated SARS-CoV-2 RNAemia with higher levels of chemokines and biomarkers indicative of a systemic inflammatory response, especially interleukin 6, ferritin, lactate dehydrogenase, and C-reactive protein [14, 15]. While those markers were not systematically measured in our patients, lymphopenia was significantly more frequent in children with SARS-CoV-2 RNAemia. Nevertheless, identification of plasma SARS-CoV-2 RNAemia could be used by clinicians to identify COVID-19 patients that may be at risk for severe disease and might benefit from early and targeted interventions with antivirals or other immunomodulatory therapies.
There are limitations to our study. Since viral load changes rapidly during the infection, the time between symptom onset and sampling is an important variable. Some patients with severe disease may delay entry to care, missing the peak period of plasma SARS-CoV-2 RNA detection, suggesting that the rates described in this study might have underestimated the real frequency and degree of SARS-CoV-2 RNAemia. Additionally, the patients enrolled in this study represent a convenience sample of hospitalized children, with limited numbers of sequential samples, and it did not include outpatients. Including outpatients with COVID-19 into the study would have allowed us to ascertain the broader role of SARS-CoV-2 RNAemia as a predictor of severity. Nevertheless, significant effort was made to obtain NP swabs and blood from every enrolled subject, which should compensate for any bias created from the sampling method. Additionally, we did not confirm whether RNA detected in plasma was indicative of infectious virions or simply shedding from the respiratory tract in the more severe forms of the disease.
In summary, we report an association between the detection of SARS-CoV-2 RNA in plasma samples and severity of presentation in hospitalized children and adolescents with COVID-19. Although the mechanisms by which RNAemia is associated with worse clinical outcomes in children with COVID-19 needs to be explored, larger prospective studies are needed to validate the value of SARS-CoV-2 RNAemia as a predictor of disease severity and outcomes in children.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Supplementary materials consist of data provided by the author that are published to benefit the reader. The posted materials are not copyedited. The contents of all supplementary data are the sole responsibility of the authors. Questions or messages regarding errors should be addressed to the author.
Notes
Acknowledgments. We thank all members of the clinical research team at Nationwide Children’s Hospital for their extraordinary efforts in helping to enroll our patients; Desiree Jones for her help with patient and data coordination; and, especially, our patients and their families for their participation in the study.
Disclaimer. The content of this work is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health (NIH).
Financial support. This study was supported in part by the National Institutes of Health (grant number AI131386) and by Nationwide Children’s Hospital intramural funds.
Potential conflicts of interest. O. R. and A. M. have received research grants from Janssen and Merck. A. M. has received fees for participation in advisory boards from Janssen, Roche, Sanofi, and Merck and fees for lectures from Sanofi. O. R. has received fees for participation in advisory boards from Adagio, Sanofi, Lilly, Merck, and Pfizer and fees for lectures from Pfizer. M. E. P. has received grants from the Cystic Fibrosis Foundation, Janssen, and Pfizer. P. J. S. has received a research grant from Merck, unrelated to the present work. All other authors report no potential conflicts of interest.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Guan WJ, Ni ZY, Hu Y, et al. Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med 2020; 382:1708–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Leidman E, Duca LM, Omura JD, Proia K, Stephens JW, Sauber-Schatz EK.. COVID-19 trends among persons aged 0–24 years—United States, March 1–December 12, 2020. MMWR Morb Mortal Wkly Rep 2021; 70:88–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Shekerdemian LS, Mahmood NR, Wolfe KK, et al. Characteristics and outcomes of children with coronavirus disease 2019 (COVID-19) infection admitted to US and Canadian pediatric intensive care units. JAMA Pediatr 2020; 174:868–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. DeBiasi RL, Song X, Delaney M, et al. Severe coronavirus disease-2019 in children and young adults in the Washington, DC, Metropolitan Region. J Pediatr 2020; 223:199–203.e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Son MBF, Murray N, Friedman K, et al. ; Overcoming COVID-19 Investigators. Multisystem inflammatory syndrome in children—initial therapy and outcomes. N Engl J Med 2021; 385:23–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Fajnzylber J, Regan J, Coxen K, et al. ; Massachusetts Consortium for Pathogen Readiness. SARS-CoV-2 viral load is associated with increased disease severity and mortality. Nat Commun 2020; 11:5493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Régent A, Kluger N, Bérezné A, Lassoued K, Mouthon L.. Lymphocytopenia: aetiology and diagnosis, when to think about idiopathic CD4(+) lymphocytopenia? [in French]. Rev Med Interne 2012; 33:628–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wu Z, McGoogan JM.. Characteristics of and important lessons from the coronavirus disease 2019 (COVID-19) outbreak in China: summary of a report of 72314 cases from the Chinese Center for Disease Control and Prevention. JAMA 2020; 323:1239–42. [DOI] [PubMed] [Google Scholar]
- 9. Baillie VL, Moore DP, Mathunjwa A, Morailane P, Simões EAF, Madhi SA.. A prospective case-control study on the association of rhinovirus nasopharyngeal viral load and viremia in South African children hospitalized with severe pneumonia. J Clin Virol 2020; 125:104288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tse H, To KK, Wen X, et al. Clinical and virological factors associated with viremia in pandemic influenza A/H1N1/2009 virus infection. PLoS One 2011; 6:e22534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hogan CA, Stevens BA, Sahoo MK, et al. High frequency of SARS-CoV-2 RNAemia and association with severe disease. Clin Infect Dis 2021; 72:e291–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yonker LM, Neilan AM, Bartsch Y, et al. Pediatric severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2): clinical presentation, infectivity, and immune responses. J Pediatr 2020; 227:45–52.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pourakbari B, Mahmoudi S, Mahmoudieh Y, et al. SARS-CoV-2 RNAaemia in children: an Iranian referral hospital-based study. J Med Virol 2021; 93:5452–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bermejo-Martin JF, González-Rivera M, Almansa R, et al. Viral RNA load in plasma is associated with critical illness and a dysregulated host response in COVID-19. Crit Care 2020; 24:691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Li Y, Schneider AM, Mehta A, et al. SARS-CoV-2 viremia is associated with distinct proteomic pathways and predicts COVID-19 outcomes. J Clin Invest 2021; 131:e148635. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.