Abstract
Fluorescent amplified-fragment length polymorphism (FAFLP), a genotyping technique with phylogenetic significance, was applied to 123 isolates of Neisseria meningitidis. Nine of these were from an outbreak in a British university; 9 were from a recent outbreak in Pontypridd, Glamorgan; 15 were from sporadic cases of meningococcal disease; 26 were from the National Collection of Type Cultures; 58 were carrier isolates from Ironville, Derbyshire; 1 was a disease isolate from Ironville; and five were representatives of invasive clones of N. meningitidis. FAFLP analysis results were compared with previously published multilocus sequence typing (MLST) and pulsed-field gel electrophoresis (PFGE) results. FAFLP was able to identify hypervirulent, hyperendemic lineages (invasive clones) of N. meningitidis as well as did MLST. PFGE did not discriminate between two strains from the outbreak that were classified as similar but distinct by FAFLP. The results suggest that high resolution of N. meningitidis for outbreak and other epidemiological analyses is more cost efficient by FAFLP than by sequencing procedures.
Neisseria meningitidis is an obligate human pathogen of worldwide significance whose effects range from asymptomatic carriage to lethal systemic infection. Its ability to cause outbreaks with significant mortality is the focus of considerable public health concern. Meningococci are carried asymptomatically in the nasopharynxes of up to 15% of the population, and all age groups, but especially children, may contract meningococcal septicemia or meningitis. Fatality is high even with antibiotic and supportive therapy.
The meningococcus is an antigenically complex bacterium with multiple genetic mechanisms for initiating changes to its cell surface to evade host immune defenses. Thirteen serogroups are recognized on the basis of capsular polysaccharide antigens, five of which (A, B, C, Y, and W-135) are commonly associated with disease. Antigenic diversity in the PorB and PorA outer membrane proteins defines serotypes and serosubtypes, respectively. Meningococci are transformable, and there is frequent lateral transfer of antigen-encoding genes (capsular switching).
The population genetics of N. meningitidis have been analyzed by multilocus enzyme electrophoresis (MLEE) (4) and, more recently, by multilocus sequence typing (MLST) (16). MLST-based measurement of the selectively neutral variation that slowly accumulates in the meningococcal population shows that the species has a very complex population structure. It is largely panmictic, i.e., nonclonal, but it contains transient clones of variable stability. Meningococcal strains with an increased attack rate tend to arise by random assortment and horizontal gene transfer of the alleles that determine disease-causing propensity (15, 22). MLEE has identified several of these hypervirulent, hyperendemic electropherotypes (ETs) or complexes of related N. meningitidis strains associated with disease. These include ET37, ET5, and the A4 complex.
Sensitive and reproducible meningococcal typing methods are required not only for population genetic and epidemiological investigations but also for vaccine-related studies. It is necessary to identify outbreaks associated with particular serogroups (as vaccines are serogroup specific), to demonstrate epidemiological links between cases or between cases and carriers in an outbreak, to monitor the changing epidemiology of disease, and to evaluate new vaccines. Phenotypic typing methods used to examine isolates for characteristics below the species level, such as serogrouping, suffer from several problems, including antigenic variability, poor expression or masking of surface antigens, the inability to subtype all isolates, and the need to continually enlarge the reagent panel. Molecular typing methods for meningococci such as MLEE, MLST based solely on housekeeping genes (seven-locus MLST), and pulsed-field gel electrophoresis (PFGE) achieve discrimination in different ways. MLEE and seven-locus MLST are methods based on variations that accumulate very slowly and are suitable for long-term and global epidemiology. PFGE and other methods based on the selection of highly variable regions of the genome which include appropriate restriction enzymes or PCR priming sites identify the microvariation that is required to distinguish between strains circulating within a geographical location. For meningococci, PFGE provides greater discriminatory power than does serology for epidemiological investigation (2, 3). Furthermore, MLST which includes sequences of two variable antigen genes (9-locus MLST) is able to distinguish between strains identical by other molecular methods such as PFGE (7).
Amplified fragment length polymorphism (AFLP) analysis is a PCR-based genome sampling technique that reproducibly generates a specific profile for each bacterial clone. First described by Vos et al. (21), AFLP is emerging as a convenient tool for the study of genetic diversity (6, 10–14). In the fluorescent AFLP (FAFLP) format for the MLEE-defined EcoR reference collection of 72 Escherichia coli strains, AFLP yields groupings nearly identical to those of MLEE (and, by implication, those of MLST) (1).
As FAFLP has also been used successfully for the investigation of outbreaks of Streptococcus pyogenes and Staphylococcus aureus (5, 9), it appears that FAFLP might be of general use for the study of micro- and macrovariation between bacterial strains, including effectiveness in outbreak investigations and studies of the population genetics of N. meningitidis. We have therefore determined the congruence of FAFLP with other molecular typing methods, namely, MLST and PFGE, for N. meningitidis and compared them for efficiency. We have analyzed strains from two outbreaks of meningitis previously characterized by MLST and PFGE and 58 N. meningitidis isolates from carriers in the village of Ironville, Derbyshire, United Kingdom. This village, population 1,600, has recently (between August 1997 and August 1999) experienced a protracted outbreak of invasive meningococcal disease, with five confirmed and seven probable meningitis cases. The patients were between 3 and 9 years old.
MATERIALS AND METHODS
Strains.
A total of 123 isolates of N. meningitidis were examined. Thirty-three were disease causing or disease associated (from asymptomatic patient contacts). They included nine from the 1997 Southampton University outbreak in the south of England (S1 and S3 to S10) and nine from the 1999 Pontypridd outbreak in Wales (P1 and P3 to P10); 15 were from epidemiologically unrelated meningococcal disease (prefixed X) and were isolated between 1990 and 1993. Twenty-six strains, dating from 1934 to 1987, were from the National Collection of Type Cultures (NCTC; prefixed N). Fifty-eight carriage isolates were from throat swabs of healthy individuals from Ironville (no prefix). Screening of the population had been undertaken in May 1998 to ascertain the serogroup, serotype, and serosubtype of carrier strains, and 116 people in Ironville (approximately 7.25% of the population) were found to carry N. meningitidis asymptomatically. Five strains of known MLEE type 37 were also analyzed. These were one disease isolate from Ironville, isolated in 1997 (Ironville); one from the north of England, isolated in 1999 (ET37); one from Mali, isolated in 1989 (ET37a); one from Israel, isolated in 1988 (ET37b); and one from Scotland, isolated in 1990 (ET37c). One known MLEE type five (ET5) strain, isolated in Norway in 1982, was also analyzed.
Serotyping.
Isolates were serogrouped and serotyped by the Meningococcal Reference Unit (Manchester Public Health Laboratory) using a standard reference collection of monoclonal antibodies (8).
Genomic DNA extraction.
Bacteria were grown for 24 h on chocolate agar at 37°C in a 5% CO2 atmosphere. Stock cultures were maintained in 16% (vol/vol) glycerol broth on Preserver Beads (Technical Services Consultants, Cambridgeshire, United Kingdom) at −70°C. Genomic DNA was extracted from plate cultures as follows. One-half loopful (10 μl) of bacterial growth was removed from a chocolate agar plate by sweeping an inoculating loop over the agar surface. Cells were suspended in Tris-EDTA-glucose, centrifuged at 7,500 × g, resuspended in 100 μl of lysozyme, and incubated at 37°C for 1 h. Cell wall debris, polysaccharides, and proteins were selectively precipitated with cetyltrimethylammonium bromide, and DNA was recovered from the supernatant by isopropanol precipitation.
FAFLP analysis.
Genomic DNAs were purified by ethanol precipitation using standard methods, and the pellet was then resuspended in 15.8 μl of water.
Genomic DNAs were digested in a 20-μl (total volume) mixture containing 500 ng of DNA, 4 U of MseI (New England Biolabs [NEB]), 2 μl of 10× MseI buffer (NEB), 0.2 μl of bovine serum albumin (NEB), and 1 μl of RNase A (10 mg/ml) that was incubated for 2 h at 37°C. Five units of EcoRI (Life Technologies) was then added, along with 1.68 μl of Tris buffer (pH 7.6) and 2.1 μl of NaCl (0.5 M) to give a final volume of 25 μl and incubated at 37°C for 2 h. Five microliters of EcoRI adapter (2 μM), 5 μl of MseI adapter (20 μM), 5 μl of ligase buffer (NEB), and 0.1 μl of T4 DNA ligase (40 U/μl of NEB) were added in a final volume of 50 μl, and the mixture was incubated at 12°C overnight. The enzyme was denatured at 65°C for 20 min, and the reaction mixture was stored at −20°C until use. The sequence of the EcoRI adapter was: 5′-CTCGTAGACTGCGTACC CATCTGACGCATGGTTAA-5′
The sequence of the MseI adapter was: 5′-TACTCAGGACTCATC GAGTCCTGAGTAGCAG-5′
PCR was performed in a final volume of 20 μl, consisting of 2 μl of 10× PCR buffer, (Life technologies), 1.5 mM MgCl2, 0.5 U of Taq polymerase, 200 μM deoxynucleoside triphosphates, 0.25 μM Mse-O primer, 0.1 μM Eco-T primer, and 2 μl of stored postligation mix. The primer sequences were as follows: Mse-0, 5′-GATGAGTCCTGAGTAA-3′; Eco-T, 5′-GACTGCGTACCAATTCT-3′. The Eco-T primer (MWG Biotech) was fluorescently labeled with 5-carboxyfluorescein. Amplification was performed on a PE9600 with predenaturation at 94°C for 2 min, followed by 30 cycles of denaturation at 94°C for 20 s, a 30-s annealing step, and extension at 72°C for 2 min. The annealing temperature for the first cycle, 66°C, was subsequently reduced by 1°C for the next nine cycles, with the remaining 20 cycles at an annealing temperature of 56°C. This was followed by a 30-min incubation at 60°C. PCR amplicons (FAFLP profiles) were separated on a 377 automated sequencer (PE Biosystems) using 36-cm plates and a 5% SinGel (FMC) at 3 kV and 51°C for 2.5 h with an internal molecular size marker (ROX 500; PE Biosystems).
The output gel file was analyzed and the bands were sized using the software package Genescan (PE Biosystems). For analysis of fragments, the presence or absence of which was confirmed by eye, files were exported to Genotyper (PE Biosystems). Eight isolates were used as a control for both digestion-ligation and PCR. One hundred samples were analyzed more than once, and the profiles were compared to test reproducibility. Data were translated into binary in Excel, showing the presence or absence of a band, and exported into a custom-designed software application that calculated Dice coefficients between pairs of isolates (17). These data were exported into Neighbor, a part of the Phylip package which uses the unweighted pair group method with arithmetic averages (UPGMA) to construct a tree by successive clustering using an average-linkage method of clustering.
RESULTS
FAFLP reproducibility.
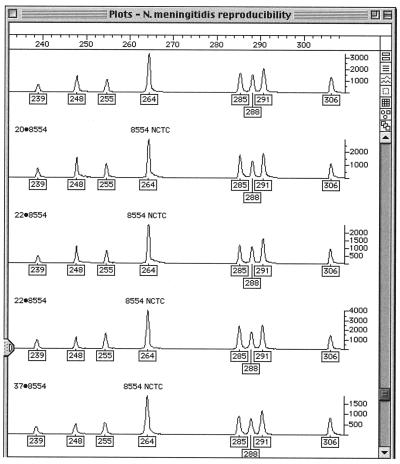
One hundred of the isolates were examined twice. All duplicates gave FAFLP profiles identical to those of the originals. Gel and amplification controls always gave identical profiles. Figure 1 shows an example of the reproducibility of the FAFLP analysis of meningococcus where five FAFLP replicates of strain NCTC 8554 yield identical FAFLP profiles.
FIG. 1.
Genotyper output of FAFLP analysis of replicates of NCTC type strain 8554 in the range of 210- to 310-bp fragments. The five traces represent separate FAFLP reactions on the same DNA extract, run on different gels. The boxed numbers under the peaks of the traces are the fragment sizes in base pairs assigned by comparison with the standard curve generated with the internal size standard.
FAFLP fragment amplification patterns.
A total of 131 distinct DNA fragments were amplified from the genomes of 123 meningococcal isolates analyzed in this study. Fragments in the size range of 100 to 306 bp were used for analysis as a small number of isolates gave suitable signal strength for analysis only within this size range. The FAFLP profiles of individual isolates contained between 18 and 39 bands. For every isolate, three distinct DNA fragments were always amplified; i.e., they were present in every FAFLP profile. The FAFLP profiles yielded the tree shown in Fig. 2 and 3. Tree-building programs using algorithms different from UPGMA, including parsimony, gave trees with the same topology (data not shown).
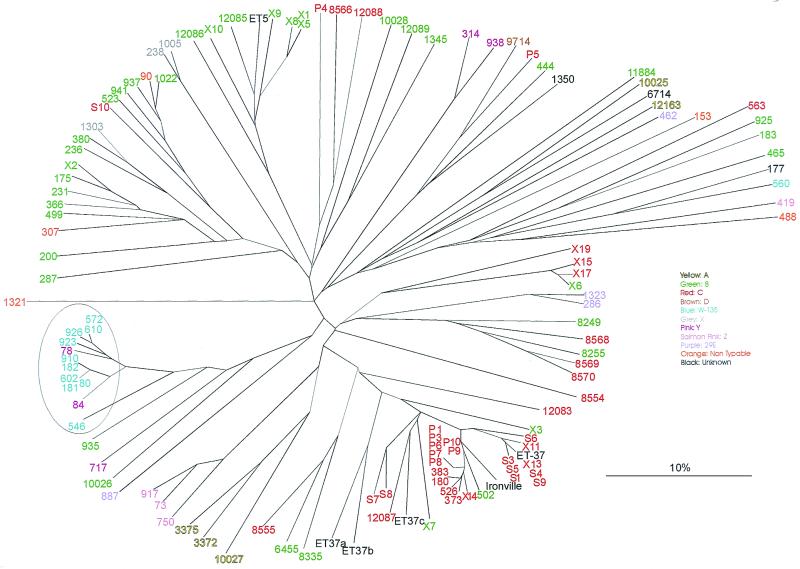
FIG. 2.
Tree representing FAFLP data for 119 N. meningitidis isolates with the serogroup, where known, indicated for each isolate (see key). One serogroup, W-135, clustered closely by FAFLP (circled), an exception being Ironville isolate 560.
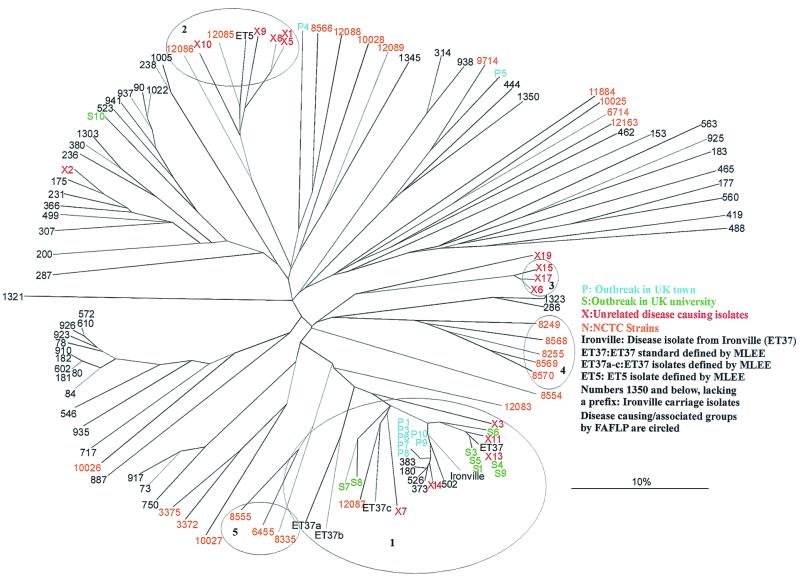
FIG. 3.
Tree identical to that in Fig. 2 except that the invasive disease-causing isolates and the carriage isolates are indicated. FAFLP separated the disease-causing strains into five clusters (circled). The largest of these, cluster 1, consisted of the majority of Southampton and Pontypridd, United Kingdom (UK), outbreak strains.
FAFLP association with serogroup profiles.
Figure 2 shows the relationship of serogroup with genotype. Only one serogroup, W-135 (circled in Fig. 2), clustered closely by FAFLP, an exception being Ironville isolate 560, placed at the 2 o'clock position among carriage strains of various serogroups.
Isolates of serogroups B and C were found throughout the tree. There were certain regions where serogroup B isolates (in green) were more prevalent—for example, at 9 to 12 o'clock in Fig. 2. Serogroup C isolates (in red) were most prevalent at the 3 to 6 o'clock position.
FAFLP of disease-causing strains.
The disease-causing isolates of N. meningitidis formed five distinct clusters, circled in Fig. 3. (Figures 2 and 3 are identical in form and content except for color coding to show serogroup and epidemiological context, respectively.)
Cluster 1 in Fig. 3 consists of the majority of the disease strains. Five are apparently epidemiologically unrelated disease isolates (prefixed X). Eight are from the Southampton University outbreak (S1 and S3 to S9, from patients and carriers. Seven are from the Pontypridd outbreak (P1 and P3 to P10, from patients and carriers). One is a disease-associated NCTC strain (12087) deposited in 1987. Five strains are of a known MLEE type defined as being part of the ET37 complex of N. meningitidis (Ironville and ET37 to ET37c). Also found in cluster 1 are five isolates from the Ironville carriage study (180, 373, 383, 502, and 526). All of these Ironville isolates belonged to serogroup C except 502, which belonged to serogroup B. All of the strains and isolates in cluster 1 belong to serogroup B or C. The remainder of the Ironville carriage isolates are distributed throughout the tree, with the majority of isolates having unique profiles.
In the Southampton University outbreak, S1, S3, and S5 had the same FAFLP profile, as did isolates S4 and S9. Isolates S6, S7, and S8 had unique profiles. All of these isolates were in FAFLP cluster 1. S10, by contrast, was very dissimilar by FAFLP analysis, being among carriage strains positioned just to the left of cluster 2. The nine isolates from the Southampton University outbreak included four meningitis isolates (S1, S3, S4, and S5), a contact of the patient who was the source of S4 (S9), and four asymptomatic-carriage isolates (S6, S7, S8, and S10). Table 1 compares FAFLP with other genotypic and phenotypic methods used to analyze the Southampton University isolates, as determined by Feavers et al. (7). As noted above, FAFLP analysis generated two profiles among the five meningitis isolates. Isolates S1, S3, and S5 shared a profile, and this had a single 223-bp fragment difference from the profile of meningitis isolate S4 and patient contact isolate S9. This fragment difference corresponded to observed differences between these two profiles with respect to phenotype (C:NT:P1.5 versus C:2a:NT), the porB allele (2-37 versus 2-36), and the porA allele (5a,10d versus 5,2).
TABLE 1.
Genotypes and phenotypes of Southampton outbreak isolates
| Isolate no. | FAFLP | MLSTb | Variable allele sequence typea
|
PFGEc | Phenotype | Isolate source | ||
|---|---|---|---|---|---|---|---|---|
| porA allele | porB allele | tpbB allele | ||||||
| 1 | Ia | 50 | 5a,10d | 2-37 | 2 | A1 | C:NT:P1.5 | Patient |
| 3 | Ia | 50 | 5a,10d | 2-37 | 2 | A1 | C:NT:P1.5 | Patient |
| 4 | Ib | 11 | 5,2d | 2-36 | 2 | A1 | C:2a:NT | Patient |
| 5 | Ia | 50 | 5a,10d | 2-37 | 2 | A1 | C:NT:P1.5 | Patient |
| 6 | II | 51 | 19d,15 | 2-38 | 1 | C | C:2a:P1.5 | Carrier |
| 7 | III | 11 | 5,2 | 2-39 | 1 | B1 | C:NT:P1.5,2 | Carrier |
| 8 | IV | 52 | NA | 2-2 | 1 | B21 | C:2a:NT | Carrier |
| 9 | Ib | 11 | 5,2d | 2-36 | 2 | A1 | C:2a:NT | Patient contact |
| 10 | V | NDe | ND | ND | ND | D | B:21:P1.1 | Carrier |
MLST 11 and 50 to 52 are ET37-like isolates.
Results obtained from Feavers et al. (7).
PFGE data, kindly supplied by the Meningococcal Reference Unit (Manchester Public Health Laboratory) were obtained by the methods described by Feavers et al. (7) using SpeI, NheI, and SfiI.
This allele contained insertion sequence IS1301.
ND, not determined.
Analysis of the Pontypridd outbreak (Fig. 3, cluster 1) showed that isolates P1, P3, P6, P7, P8, P9, and P10, all causing invasive disease, were identical by FAFLP, although the signal intensity of some fragments varied between isolates. By contrast, P4, from a patient suspected of being infected, and P5, from a carrier, were dissimilar from each other and from the other Pontypridd isolates. Table 2 compares FAFLP with other genotypic and phenotypic methods for the Pontypridd isolates. Six disease isolates from this outbreak had the same MLST profile (profile 11; Table 2), and all except P4 were of serotype C:2a:P1.5. Asymptomatic-carriage isolates P3 and P8, which are included in this FAFLP cluster 1, were also of this serotype. Pontypridd outbreak isolate P4 was of serotype C:1:P1.15 and had a unique MLST profile. Carriage isolate P5 was of serotype C:NT:P1.14 and had a unique MLST profile. By PFGE, all but two (P9 and P10) of these Pontypridd isolates had identical macrorestriction profiles. Two of the three distinct FAFLP profiles found for the Pontypridd isolates corresponded to the distinct phenotype of P5, and the other was found for disease isolate P4. P4 and P5 had 10 and 13 amplified fragment differences, respectively, from the FAFLP profile shared by the other seven isolates.
TABLE 2.
Genotypes and phenotypes of Pontypridd outbreak isolates
| Isolate no. | FAFLP | MLST | PFGEa | Phenotype | Isolate source |
|---|---|---|---|---|---|
| 1 | VI | 11b | A | C:2a:P1.5 | Patient |
| 3 | VI | 11 | A | C:2a:P1.5 | Carrier |
| 4 | VII | Unique | A | C:1:P1.15 | Suspected infection |
| 5 | VIII | Unique | A | C:NT:P1.14 | Carrier |
| 6 | VI | 11 | A | C:2a:P1.5 | Patient |
| 7 | VI | 11 | A | C:2a:P1.5 | Patient |
| 8 | VI | 11 | A | C:2a:P1.5 | Carrier |
| 9 | VI | 11 | A2(B) | C:2a:P1.5 | Patient |
| 10 | VI | 11 | A1 | C:2a:P1.5 | Patient |
FAFLP cluster 2 (Fig. 3) contained five epidemiologically unrelated meningitis isolates (prefixed X) and NCTC strains 12085 and 12086. It also contained a representative of the hypervirulent and hyperendemic ET5 clone complex isolated in Norway in 1982 and initially identified by MLEE. All but one of the isolates in cluster 2 was known to belong to serogroup B, except for ET5, whose serogroup was unknown.
FAFLP cluster 3 consisted of three isolates from the epidemiologically unrelated disease-causing (X) collection of isolates (X6, X15, and X17). Of these three, two were of serogroup C and one was of serogroup B. Clusters 4 and 5 consisted entirely of disease-causing NCTC isolates dating from 1934 to 1987.
Epidemiologically unrelated disease-causing isolates were found in clusters 1, 2, and 3. One of these isolates, X13, had the same FAFLP profile as isolates S4 and S9 from the Southampton University outbreak.
DISCUSSION
Congruence of FAFLP with MLST.
The development of 9-locus MLST (7), which includes sequences from two variable antigen genes together with sequences from selectively neutral housekeeping genes has, for the first time, enabled both the macro- and microvariation of N. meningitidis to be analyzed using the same method. Previously, global epidemiology (macrovariation) of isolates has been studied using MLEE (and subsequently MLST based on housekeeping genes) while outbreak investigation (microvariation) of isolates has been done using a method such as PFGE. A further advantage of MLST over other methods is its portability between laboratories (16).
Data from this study show that FAFLP has the same high level of resolution for the Southampton University outbreak investigation as 9-locus MLST as determined by Feavers et al. (7). FAFLP also clusters together hypervirulent lineages such as ET37 and ET5 in the same way as MLST. All of the Southampton University outbreak isolates except S10 were in a single FAFLP cluster (1) and were identified as belonging to the ET37 complex by MLST (7). Similarly, all of the Pontypridd outbreak isolates except P4 and P5 were in FAFLP cluster 1 and the ET37 complex by MLST (data not shown). Two isolates related to, but not part of, FAFLP cluster 1 (N12083 and N8554) grouped away from cluster 1 and were not assigned to the ET37 complex by MLST (data not shown).
Thus, insofar as FAFLP cluster 1 corresponds to ET37, FAFLP (a technique that samples throughout the genome, examining both variable and nonvariable regions, as demonstrated by predictive modeling experiments with a known genome sequence [1]), has been shown to be capable of identifying an MLST-MLEE-defined clone. Further evidence that FAFLP can identify significant MLST-MLEE clones is provided by data from FAFLP cluster 2. This cluster consisted of five epidemiologically unrelated disease isolates, two recent disease-causing NCTC strains isolated in the 1980s, and one isolate obtained in Norway in 1982 that is representative of ET5, another hypervirulent lineage. These findings on meningococcal MLEE clones agree with our previously published FAFLP analysis of the EcoR reference collection, which represents the genetic diversity of 72 E. coli strains. In that study, FAFLP was fully congruent with the five MLEE-defined phylogenetic groups of Escherichia coli for that collection of strains (1).
Olive et al. recently published costs for DNA-based typing of microbial organisms (18). Using their calculations, the estimated cost for one FAFLP reaction is $20 whereas the cost for carrying out 9-locus MLST on a single isolate, which in our study gave the same amount of information as a single FAFLP reaction, is $360. Thus, FAFLP is much more cost efficient than 9-locus MLST. The setup costs for FAFLP and MLST ($45,000 to $130,000) are the same (18).
Congruence of FAFLP with PFGE.
The data from both FAFLP and PFGE for the two outbreak investigations studied here were comparable, but PFGE was unable to discriminate between two similar but distinct strains from the Southampton University outbreak. FAFLP had greater resolution than PFGE for the Pontypridd outbreak. The increased degree of resolution achieved by FAFLP can be attributed to the double restriction enzyme digestion employed by FAFLP, as opposed to the single enzyme digestion used in PFGE, and the consequently higher number of DNA fragments analyzed by FAFLP (23). The cost of a single PFGE reaction is estimated at $22, compared with $20 for FAFLP. The setup costs for PFGE are less, only $10,000 to $20,000 (18).
FAFLP association with serogroup profiles.
It is known that ET37 complex strains are mainly of serogroup C, but serogroup B isolates of this complex are increasingly being isolated from patients, and recent studies have demonstrated that meningococci are capable of capsular switching among serogroups B, C, and W-135 in vivo while retaining hypervirulence capabilities (19, 20). Evidence for possible capsular switching in this study is best illustrated by the very closely related Ironville ET37 strains, of which four belong to serogroup C and one belongs to serogroup B. As no suitable serogroup B vaccine is available, it is important to monitor the present national vaccination campaign in the United Kingdom for capsular switching of hypervirulence complexes to serogroup B due to selective pressure. Further evidence of recent capsular switching is shown by serogroup W-135, which is fairly tightly clustered by FAFLP, apart from one strain quite distinct from the rest.
In summary, to distinguish outbreak isolates from unrelated but temporally and geographically proximate cases, rapid and accurate characterization of meningococcal isolates is needed. We believe that this need can be met by FAFLP analysis, shown here to cost effectively establish the identity of strains and their genetic relationships. In the present study, FAFLP was congruent with the established genotypic method of MLEE or MLST and distinguished between carriage and invasive isolates, as MLST did. The technique has the potential to identify hypervirulent lineages defined by ET or MLST. Thus, in addition to identifying microvariation that will distinguish strains circulating within a geographic location in a more efficient manner than MLST, FAFLP can simultaneously contribute to the wider understanding of the population genetics and global epidemiology of the meningococcus.
ACKNOWLEDGMENT
We thank the Meningococcal Reference Unit (Manchester Public Health Laboratory), especially Steve Gray, for kindly supplying PFGE data.
REFERENCES
- 1.Arnold C, Metherell L, Willshaw G, Maggs A, Stanley J. Predictive fluorescent amplified fragment length polymorphism analysis of Escherichia coli: a high resolution typing method with phylogenetic significance. J Clin Microbiol. 1999;37:1274–1279. doi: 10.1128/jcm.37.5.1274-1279.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berron S, De La Fuente L, Martin E, Vazquez J A. Increasing incidence of meningococcal diseases in Spain associated with a new variant of serogroup C. Eur J Clin Microbiol Infect Dis. 1998;17:85–89. doi: 10.1007/BF01682161. [DOI] [PubMed] [Google Scholar]
- 3.Bevanger L, Bergh K, Gisnas G, Caugant D A, Froholm L O. Identification of nasopharyngeal carriage of an outbreak strain of Neisseria meningitidis by pulsed-field gel electrophoresis versus phenotypic methods. J Med Microbiol. 1998;47:993–998. doi: 10.1099/00222615-47-11-993. [DOI] [PubMed] [Google Scholar]
- 4.Caugant D A, Bovre K, Gaustad P, Bryn K, Holten E, Hoiby A, Froholm L O. Multilocus genotypes determined by enzyme electrophoresis of Neisseria meningitidis isolated from patients with systemic disease and from healthy carriers. J Gen Microbiol. 1985;132:641–652. doi: 10.1099/00221287-132-3-641. [DOI] [PubMed] [Google Scholar]
- 5.Desai M, Tanna A, Wall R, Efstratiou A, George R, Stanley J. Fluorescent amplified-fragment length polymorphism analysis of an outbreak of group A streptococcal invasive disease. J Clin Microbiol. 1998;36:3133–3137. doi: 10.1128/jcm.36.11.3133-3137.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dijkshoorn L, Aucken H, Gerner-Smidt P, Janssen P, Kaufmann M E, Garaizar J, Ursing J, Pitt T L. Comparison of outbreak and nonoutbreak Acinetobacter baumanni strains by genotypic and phenotypic methods. J Clin Microbiol. 1996;34:1519–1525. doi: 10.1128/jcm.34.6.1519-1525.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Feavers I M, Gray S J, Urwin R, Russell J E, Bygraves J A, Kaczmarski E B, Maiden M C. Multilocus sequence typing and antigen gene sequencing in the investigation of a meningococcal disease outbreak. J Clin Microbiol. 1999;37:3883–3887. doi: 10.1128/jcm.37.12.3883-3887.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frasch C E, Zollinger W D, Poolman J T. Serotype antigens of Neisseria meningitidis and a proposal scheme for designation of serotypes. Rev Infect Dis. 1985;7:504–510. doi: 10.1093/clinids/7.4.504. [DOI] [PubMed] [Google Scholar]
- 9.Grady R, Desai M, O'Neill G, Cookson B, Stanley J. Genotyping of epidemic methicillin-resistant Staphylococcus aureus phage-type 15 isolates by fluorescent amplified-fragment length polymorphism. J Clin Microbiol. 1999;37:3198–3203. doi: 10.1128/jcm.37.10.3198-3203.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huys G, Coopman R, Janssen P, Kersters K. High-resolution genotypic analysis of the genus Aeromonas by AFLP fingerprinting. Int J Syst Bacteriol. 1996;46:572–580. doi: 10.1099/00207713-46-2-572. [DOI] [PubMed] [Google Scholar]
- 11.Huys G, Kersters I, Coopman R, Janssen P, Kersters K. Genotypic diversity among Aeromonas isolates recovered from drinking water production plants as revealed by AFLPTM analysis. Syst Appl Microbiol. 1996;19:428–435. [Google Scholar]
- 12.Janssen P, Coopman R, Huys G, Swings J, Bleeker M, Vos P, Zabeau M, Kersters K. Evaluation of the DNA fingerprinting method AFLP as a new tool in bacterial taxonomy. Microbiology. 1996;142:1881–1893. doi: 10.1099/13500872-142-7-1881. [DOI] [PubMed] [Google Scholar]
- 13.Janssen P, Dijkshoorn L. High resolution DNA fingerprinting of Acinetobacter outbreak strains. FEMS Microbiol Lett. 1996;142:191–194. doi: 10.1111/j.1574-6968.1996.tb08429.x. [DOI] [PubMed] [Google Scholar]
- 14.Kiem P, Kalif A, Schupp J, Hill K, Travis S E, Richmond K, Adair D M, Hugh-Jones M, Kuske C R, Jackson P. Molecular evolution and diversity in Bacillus anthracis as detected by amplified length polymorphism markers. J Bacteriol. 1997;179:818–824. doi: 10.1128/jb.179.3.818-824.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maiden M C, Feavers I M. Population genetics and global epidemiology of the human pathogen Neisseria meningitidis. In: Baumberg S, Young J P W, Wellington E M H, Saunders J R, editors. Population genetics of bacteria. Cambridge, United Kingdom: University Press; 1995. pp. 269–293. [Google Scholar]
- 16.Maiden M C J, Bygraves J A, Feil E, Morelli G, Russell J E, Urwin R, Zhang Q, Zhou J, Zurth K, Caugant D A, Feavers I M, Achtman M, Spratt B. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Microbiology. 1998;95:3140–3145. doi: 10.1073/pnas.95.6.3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nei M, Li W-H. Mathematical model for studying genetic variations in terms of restriction endonucleases. Proc Natl Acad Sci USA. 1979;76:5269–5273. doi: 10.1073/pnas.76.10.5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Olive D M, Bean P. Principles and applications of methods for DNA-based typing of microbial organisms. J Clin Microbiol. 1999;37:1661–1669. doi: 10.1128/jcm.37.6.1661-1669.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Popovic T, Sacchi C T, Reeves M W, Whitney A M, Mayer L W, Noble C A, Ajello G W, Mostashari F, Bendana N, Lingappa J, Hajjeh R, Rosenstein N E. Neisseria meningitidis serogroup W135 isolates associated with the ET-37 complex. Emerg Infect Dis. 2000;6:428–429. doi: 10.3201/eid0604.000423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Swartley J S, Marfin A A, Edupuganti S, Liu L-J, Cieslak P, Wenger J D, Stephens D S. Capsule switching of Neisseria meningitidis. Proc Natl Acad Sci USA. 1997;94:271–276. doi: 10.1073/pnas.94.1.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vos P, Hogers R, Bleeker M, Reijans M, van de Lee T, Hornes M, Frijters A, Pot J, Peleman J, Kulper M, Zabeau M. AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res. 1995;23:4407–4414. doi: 10.1093/nar/23.21.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yakubu D E, Abadi F J R, Pennington T H. Molecular typing methods for Neisseria meningitidis. J Med Microbiol. 1999;48:1055–1064. doi: 10.1099/00222615-48-12-1055. [DOI] [PubMed] [Google Scholar]
- 23.Zhao S, Mitchell S E, Meng J, Kresovich S, Doyle M P, Dean R E, Casa A M, Weller J W. Genomic typing of Escherichia coli O157:H7 by semi-automated fluorescent AFLP analysis. Microbes Infect. 2000;2:107–113. doi: 10.1016/s1286-4579(00)00278-1. [DOI] [PubMed] [Google Scholar]