Abstract
The adult neurogenic niches are complex multicellular systems, receiving regulatory input from a multitude of intracellular, juxtacrine, and paracrine signals and biological pathways. Within the niches, adult neural stem cells (aNSCs) generate astrocytic and neuronal progeny, with the latter predominating in physiological conditions. The new neurons generated from this neurogenic process are functionally linked to memory, cognition, and mood regulation, while much less is known about the functional contribution of aNSC-derived newborn astrocytes and adult-born oligodendrocytes. Accumulating evidence suggests that the deregulation of aNSCs and their progeny can impact, or can be impacted by, aging and several brain pathologies, including neurodevelopmental and mood disorders, neurodegenerative diseases, and also by insults, such as epileptic seizures, stroke, or traumatic brain injury. Hence, understanding the regulatory underpinnings of aNSC activation, differentiation, and fate commitment could help identify novel therapeutic avenues for a series of pathological conditions. Over the last two decades, small non-coding RNAs (sncRNAs) have emerged as key regulators of NSC fate determination in the adult neurogenic niches. In this review, we synthesize prior knowledge on how sncRNAs, such as microRNAs (miRNAs) and piwi-interacting RNAs (piRNAs), may impact NSC fate determination in the adult brain and we critically assess the functional significance of these events. We discuss the concepts that emerge from these examples and how they could be used to provide a framework for considering aNSC (de)regulation in the pathogenesis and treatment of neurological diseases.
Keywords: adult hippocampal neurogenesis, neurodegeneration, neural stem cells, small non-coding RNA, microRNA, piRNA
Introduction
After birth, most of the mammalian brain presents a cellular environment that is refractory to the generation of new neurons under physiological conditions (Kempermann, 2015). However, adult neural stem cells (aNSCs), residing within spatially restricted microenvironments of the adult brain, referred to as “niches” (Schofield, 1978; Lim et al., 2007), maintain the capability to self-renew, proliferate and generate differentiated progeny, mainly neurons, and, infrequently, astrocytes (Bottes et al., 2021). The subventricular zone (SVZ) of the lateral ventricles and the subgranular zone (SGZ) of the dentate gyrus (DG) in the hippocampus (Gage, 2019) are two of the main neurogenic niches in the adult mammalian brain. Here, we will focus on the adult hippocampal niche, because of the functional relevance of the hippocampus to memory and cognition, its link to neurodevelopmental and mood disorders and neurodegenerative diseases, and its sensitivity to insults such as epileptic seizures, stroke, or traumatic brain injury (for systematic reviews on the functional significance of the adult hippocampal neurogenic niche, see Kempermann et al., 2018; Duque and Spector, 2019; Snyder, 2019; Lucassen et al., 2020; Babcock et al., 2021; Denoth-Lippuner and Jessberger, 2021).
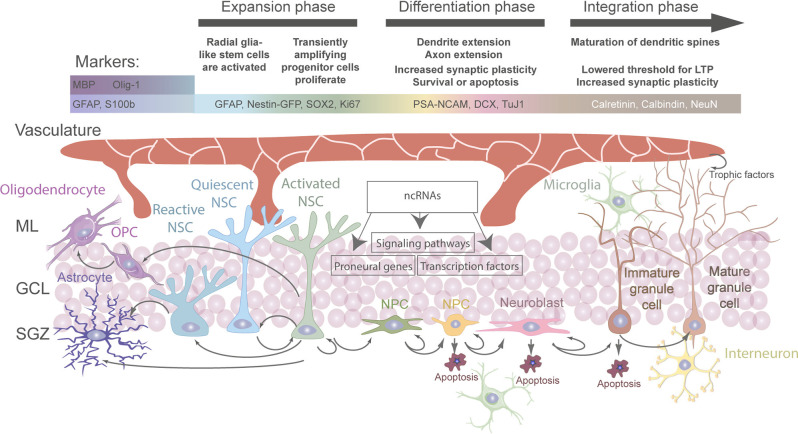
Generally, stem cell niches are characterized by the ability to anatomically host stem cells and functionally control their development in vivo in order to adapt to organismal and environmental requirements (Alonso, 2009; Li and Guo, 2021). At the adult hippocampal neurogenic niche, this is achieved by a complex crosstalk between aNSCs and other cell types present (Figure 1; Vicidomini et al., 2020). More specifically, the milieu of the adult hippocampal neurogenic niche is tightly regulated by intracellular and extracellular signals (e.g. aging, stress, physical exercise, environmental enrichment, learning, pathological alterations, neurotrophins, growth factors, signaling pathways, extracellular matrix mediators and extracellular vesicles, vasculature-secreted factors and local circuit activitiy; for systematic reviews, see Balu and Lucki, 2009; Aimone et al., 2014; Baptista and Andrade, 2018; Bonafina et al., 2020; Losurdo and Grilli, 2020; Vicidomini et al., 2020). Thereby, a regulated balance between aNSC quiescence, proliferation, self-renewal, differentiation, and fate choice contributes to lifelong hippocampal plasticity and fitness, while preventing neurodegeneration and other pathologies (Faigle and Song, 2013; Fitzsimons et al., 2014; Yao et al., 2016; Lopatina et al., 2019; Toda et al., 2019; Vicidomini et al., 2020; Kim et al., 2021; Li and Guo, 2021). Hence, understanding the regulation of aNSCs and their crosstalk with other cell types within the hippocampal neurogenic niche is indispensable to fully map the mechanisms governing their fate determination under physiological conditions and in the diseased brain.
Figure 1.
Regulation and fate of aNSC in the adult hippocampal stem cell niche. Overview of all cell types involved directly or indirectly in the regulation of stem cell activation and fate determination in the adult hippocampus. Cell-intrinsic and extrinsic signals regulate the different stages of aNSC quiescence, activation, neurogenesis, astrogliogenesis, and oligodendrogliogenesis in the adult dentate gyrus (DG).
A considerable part of the regulatory complexity required for proper aNSC homeostasis in the adult hippocampal niche is executed by non-protein-coding RNAs (ncRNAs), which comprise a significant proportion of the transcriptional output of the eukaryotic genome, contributing to both housekeeping and key regulatory processes (Carninci et al., 2005; Dunham et al., 2012; Salta and De Strooper, 2012, 2017b; Salvatori et al., 2020). These functions are regarded as particularly relevant for cells such as aNSCs, which require fast adaptation of transcriptome and protein abundance, especially when transitioning through cellular states while undergoing fate decisions (Encinas and Fitzsimons, 2017). However, given that the majority of ncRNA species are still functionally uncharacterized, it remains unclear how the diverse ncRNA-mediated pathways regulate fate determination in aNSCs. A deeper understanding of how ncRNA functions impact aNSCs may provide a better mechanistic appreciation of physiological brain development, its plasticity, and the disorders arising from the deregulation of aNSCs. Such insights may also prove pivotal for the next generation of RNA-based therapeutics targeting abnormal gene expression in aNSCs, in order to prevent, counteract or correct disruption of their homeostasis, activation and differentiation.
Here, we review recent discoveries reporting the regulation of cell intrinsic programs by small ncRNAs (sncRNAs) in several immature cell types of the adult hippocampal neurogenic niche, we discuss the mechanisms by which they impact fate determination of these cell populations and how, sometimes, sncRNAs may interact with each other to act as intercellular signals and regulate neighboring cells within the hippocampal neurogenic niche.
Adult Neural Stem Cells in The Hippocampus: Complexity and Regulation
In the adult hippocampus, aNSCs occupy a narrow area between the hilus and the granular cell layer of the DG, known as SGZ (Kempermann et al., 2004), which forms part of the neurogenic niche (Figure 1). ANSCs and their immature progeny in the adult hippocampal neurogenic niche have often been classified using divergent nomenclature, which we summarize in Table 1. ANSCs are also known as “radial glia-like cells” (RGLs), due to their similarity to embryonic radial glial NSCs, and “quiescent neural stem cells” or “type 1 cells” (Table 1; Kempermann et al., 2004; Encinas et al., 2011). ANSCs are maintained in a reversible cell cycle arrest state, termed quiescence. Daughter cells generated by initial mitotic divisions of activated aNSCs exhibit reduced self-renewal and increased lineage restriction and proliferative potential (Berg et al., 2018). These highly proliferative progeny are functionally classified as “(transiently) amplifying (neural) progenitors” (herein collectively termed neural progenitor cells, “NPCs”), “intermediate progenitors”, or based on marker expression, as “type 2a cells” with a glial phenotype, and “type 2b cells” with a neuronal phenotype (Table 1). NPCs generate immature neuroblasts, also termed “type 3 cells,” which exit the cell cycle to become immature neurons first and, over a period of several weeks, newborn mature dentate granule neurons (“granule cells”, DGCs; Table 1; Encinas et al., 2011; Kempermann, 2015; Berg et al., 2018). Oligodendrocyte precursor cells (OPCs) are also present in the adult DG, yet to a significantly lesser extent, and not found to be restricted to the subgranular zone (Encinas et al., 2011). In addition, neuron-glial antigen 2 (NG2, a chondroitin sulfate proteoglycan)-expressing progenitor cells are present and continue to proliferate in the adult CNS (Ffrench-Constant and Raff, 1986; Wolswijk and Noble, 1989), and may represent a distinct class of proliferating glial cells in the adult hippocampus (Klempin et al., 2011; Schouten et al., 2020). Yet, their intrinsic stem cell potential and functional contribution are not well-characterized, with some studies reporting that NG2-expressing cells are multipotent progenitors that generate astrocytes and mature neurons in the postnatal hippocampus (Belachew et al., 2003), while others suggest that they may be OPCs (Dawson et al., 2000). Of note, single-cell transcriptomic profiling studies in the mouse DG have demonstrated that cells of the neurogenic lineage may co-exist in a heterogeneous continuum of cell states (Shin et al., 2015; Artegiani et al., 2017). A similar heterogeneity has been reported for oligodendrocytic and astrocytic precursors in several brain regions, including, among others, cortical, subcortical, and hippocampal subfields (Steindler and Laywell, 2003; Marques et al., 2016).
Table 1.
Terminology used to describe aNSCs and their progeny.
| Terminology | Others | Cellular state | Reference(s)Our review |
|---|---|---|---|
| Adult neural stem cells (aNSCs) | Radial glia-like cells Quiescent neural stem cells Type-1 cells | Quiescent/Activated/Proliferative | Kempermann et al. (2004) and Encinas et al. (2011) |
| Neural progenitor cells (NPCs) | Intermediate progenitors Transiently amplifying neural progenitors Type 2a cells (glial phenotype) Type 2b cells (neuronal phenotype) | Proliferative | Kempermann et al. (2004), Encinas et al. (2011), and Berg et al. (2018) |
| Oligodendrocyte precursor cells | NG2-positive cells | Proliferative | Dawson et al. (2000) and Belachew et al. (2003) |
| Neuroblasts | Type 3 cells | Largely postmitotic | Kempermann et al. (2004), Encinas et al. (2011), and Berg et al. (2018) |
Besides aNSCs and their progeny, the adult hippocampal niche is a dynamic structure containing multiple other cell types (Figure 1). The functional intercellular crosstalk among the niche-resident cells allows for proper maintenance of the aNSC pool, progenitor proliferation, differentiation, survival, and maturation (Vicidomini et al., 2020). Of note, aNSCs and NPCs can regulate their own homeostasis, as well as the differentiation and maturation of their progeny, through autocrine and paracrine signals (Schultheiß et al., 2013; Vicidomini et al., 2020; Shariq et al., 2021). Similarly, hippocampal niche-resident astrocytes have been shown to provide both structural and functional support to aNSCs and their progeny, and to control cell proliferation, differentiation, migration, and synaptic integration of adult-born granule neurons (Bonafina et al., 2020). Moreover, microglia, the brain-resident immune cells, actively regulate multiple phases of neurogenesis (Butovsky et al., 2006; Ekdahl et al., 2009; Woodbury et al., 2015; Pérez-Rodríguez et al., 2021). In addition, blood vessels localize within the neurogenic niche and their capillary loops are found in close proximity to aNSCs and dividing NPCs (Licht and Keshet, 2015). As such, the vascular compartment can impact neurogenesis through direct contact with, and paracrine signaling from, endothelial and mural cells that make up blood vessels, or indirectly, via transporting systemic signals (in the form of soluble factors) from blood circulation into the neurogenic niche (Fitzsimons et al., 2013; Lee et al., 2013; Villeda et al., 2014; Licht and Keshet, 2015; Smith et al., 2015; Bátiz et al., 2016; Schouten et al., 2020). Last, mature hippocampal DGCs can modulate aNSC homeostasis via cell autonomously secreted factors and by recruiting circuit mechanisms involving, among others, local inhibitory interneurons and mossy cells (Gobeske et al., 2009; Ma et al., 2009; Jang et al., 2013; Brooker et al., 2017).
How this melting pot of cells and signals is orchestrated to eventually direct aNSCs and other niche-resident progenitors towards quiescence, neurogenesis, astrogliogenesis, and oligodendrogliogenesis is of particular importance for understanding the functional relevance of these processes in both the healthy and the diseased adult brain. In the next sections, we will focus on the pivotal regulatory role of sncRNAs as cell-intrinsic or -extrinsic signals and will critically discuss the emerging evidence supporting distinct key functions for these small but mighty regulators in the adult hippocampal niche.
Small Non-Coding RNAs
NcRNAs are RNA molecules not translated into proteins. They are traditionally classified based on their average size into small (<200 nucleotides) and long (>200 nucleotides) species (Hombach and Kretz, 2016; Salvatori et al., 2020). These molecules are highly versatile and exhibit a very diverse mechanistic repertoire, ranging from regulation of chromatin state, transcription, epigenetic events, alternative splicing, and RNA interference (RNAi)-mediated gene silencing, to effects on mRNA stability, translational regulation, and sequestration of RNA and proteins from their natural targets in the intracellular space (Schouten et al., 2012; Salta and De Strooper, 2017b; Figure 2). In this review, we focus on regulatory sncRNAs (Figure 2), and in particular on microRNAs (miRNAs), which are the most studied sncRNA subclass with well-documented and emerging roles in the regulation of aNSCs and their progeny. We also discuss the functions of PIWI-interacting RNAs (piRNAs), which are rapidly emerging as important regulators of somatic stem cells. For further information on long ncRNAs in the central nervous system (CNS), readers are invited to consider several previous reviews that cover the topic in more detail (Salta and De Strooper, 2017b; Zhang et al., 2019b).
Figure 2.
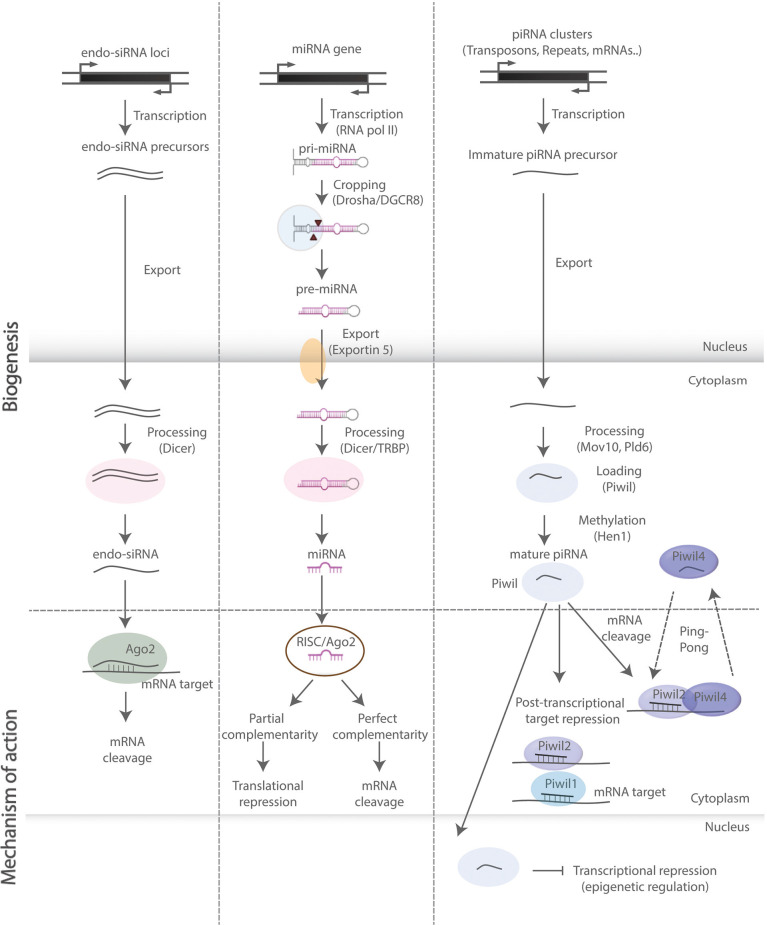
SncRNA biogenesis and mode of action. Schematic representation of biosynthetic and regulatory pathways of the sncRNA species discussed in this review. Endo-siRNAs are short double-stranded RNAs, processed by Dicer and incorporated into the RNA-induced ribonucleoprotein silencing complex (RISC), upon which they bind onto target mRNAs and induce mRNA cleavage. MiRNAs are primarily transcribed by RNA polymerase II and subsequently processed by—among others—Drosha and Dicer into double-stranded miRNA precursor molecules. The guide strand is then incorporated into RISC leading to translational repression and mRNA degradation upon base pairing with complementary sites on mRNA targets. PiRNAs are processed in a Dicer-independent manner and act through binding with PIWI-clade proteins exerting both transcriptional and post-transcriptional regulatory effects (“ping-pong” amplification loop).
MiRNAs are a class of small (19–22 nucleotides in length) single-stranded ncRNAs. AGO proteins associate with miRNAs and other sncRNAs acting in RNA-based silencing mechanisms by inhibiting protein synthesis and altering RNA stability (Hutvagner and Simard, 2008; Figure 2). Their functions are particularly important for the fine-tuning of gene expression in brain development, physiology, and homeostasis (Saba and Schratt, 2010; Barca-Mayo and De Pietri Tonelli, 2014; Zhang et al., 2019b). Of note, miRNAs may repress individual targets only to a limited extent (Baek et al., 2008; Selbach et al., 2008). Thereby, their regulatory potential may often rely on the synergistic action of several miRNAs over distinct sets of transcripts, which in aNSCs often encode proteins that share similar or converging biological functions (Barca-Mayo and De Pietri Tonelli, 2014; Schouten et al., 2015; Pons-Espinal et al., 2017).
PiRNAs are a class of animal-specific single-stranded sncRNAs defined by their ability to associate with PIWI proteins, a subclass of Dicer-binding AGO proteins (Doyle et al., 2012). PiRNAs are longer (21–35 nucleotides in length) than miRNAs and endogenous siRNAs, and unlike the former classes, they are processed in a Dicer-independent manner, bear 2′-O-methyl-modified 3′ termini and only function through binding with PIWI-clade proteins, rather than AGO-clade proteins (Figure 2). Although much less studied than miRNAs, piRNAs are present in the mammalian CNS, while nearly 30,000 neuronal piRNAs have been identified in the mouse brain throughout development, suggesting that they are expressed and dynamically regulated during brain development (Ghoshes et al., 2016). In addition, several studies have shown that piRNAs might regulate neuronal function and synaptic plasticity in the mammalian brain (Wakisaka and Imai, 2019).
The disruption of miRNA and piRNA expression has been linked to a multitude of brain pathologies, including neurodegenerative diseases (Salta and De Strooper, 2012, 2017b; Wakisaka and Imai, 2019; Wu and Kuo, 2020; Huang and Wong, 2021), psychiatric (Yoshino and Dwivedi, 2020) and neurodevelopmental disorders (Barca-Mayo and De Pietri Tonelli, 2014; Zhang et al., 2019d). In agreement with their general involvement in health and disease, accumulating evidence links sncRNAs to aNSC regulation under both physiological (Figure 3) and pathological conditions (Table 2). In the following sections, we discuss the existing evidence demonstrating how sncRNA species, particularly miRNAs and piRNAs, can impact aNSC maintenance, fate determination, differentiation, and maturation of their progeny.
Figure 3.
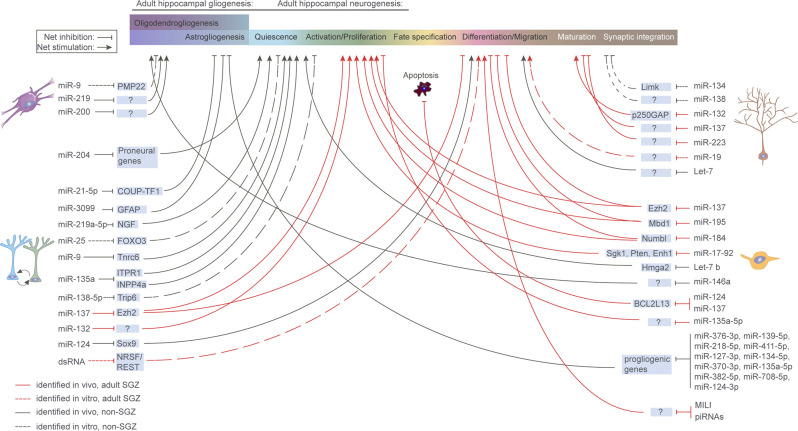
microRNA regulation of aNSCs and their progeny. Overview of miRNAs known to be functionally implicated in aNSC quiescence, activation, fate determination, and progeny maturation under physiological conditions in the murine hippocampus. Continuous red line: in vivo study, refers to adult SGZ; Dashed red line: in vitro study, refers to adult SGZ; Continuous black line: in vivo study, refers to systems other than SGZ; Dashed black line: in vitro study, refers to systems other than SGZ.
Table 2.
MicroRNAs implicated in the regulation of aNSCs and their progeny in neurological diseases.
| Neurogenesis | |||
|---|---|---|---|
| Non-coding RNA(s) | Disease | Functional relevance | References |
| miR-132 | Alzheimer’s disease | -Decreased expression in AD hippocampus. -Exerts proneurogenic effects in aNSCs and their progeny. | Walgrave et al. (2021a) |
| miR-124 | Parkinson’s disease | Increases the number of neuroblasts reaching the granular cell layer of the olfactory bulb (SVZ). | Saraiva et al. (2016) |
| miR-30 family | Depression | - Decreased expression in the mouse model of depression. - Mediates chronic stress-induced depression-like phenotype by altering hippocampal neurogenesis and neuroplasticity via controlling epigenetic and transcription regulators, such as Mll3 and Runx1; and cell signaling regulators, like Socs3, Ppp3r1, Gpr125, and Nrp1. | Khandelwal et al. (2019) |
| miR-19 | Schizophrenia | Upregulated in SGZ-NPCs. | Han et al. (2016) |
| miR-124 and miR-137 | Epilepsy | Concomitant knockdown of the two prevents hippocampal aNSC loss upon non-convulsive seizures. | Bielefeld et al. (2017) and Bielefeld et al. (2019) |
| Astrogliogenesis/Reactive astrogliosis | |||
| Non-coding RNA(s) | Disease | Functional relevance | References |
| miR-132 | Epilepsy | - Increased expression in reactive astrocytes in epileptogenic human and rat hippocampus. - Reduces pro-epileptogenic factors in cultured astrocytes. | Korotkov et al. (2020) |
| miR-146a | Epilepsy | - Increased expression in reactive astrocytes in epileptogenic human and rat hippocampus. | Aronica et al. (2010) |
| Down syndrome | - Increased expression in reactive astrocytes in the brain of individuals with Down syndrome. | Arena et al. (2017) | |
| miR-181a | Ischemia | Its inhibition induces neurogenesis partly via astrocytic dedifferentiation in the hippocampus. | Griffiths et al. (2019) |
| miR-302/367 | Alzheimer’s disease | Induce conversion of reactive astrocytes to neurons in DG. | Ghasemi-Kasman et al. (2018) |
| Oligodendrogliogenesis | |||
| Non-coding RNA(s) | Disease | Functional relevance | References |
| miR-146a | Stroke | Induces remyelination by increasing differentiation of OPCs to oligodendrocytes. | Santra et al. (2014) and Liu et al. (2017) |
| Multiple sclerosis | Enhances myelination and increased OPC differentiation. | Zhang et al. (2019a) | |
| miR-23a, miR-219a, and miR-338 | Demyelinating insult | Promote oligodendrocytic differentiation and remyelination. | Santos et al. (2020) |
microRNAs
Regulation of aNSC Quiescence
Effects in Physiological Conditions
aNSCs in the adult brain are typically quiescent (Doetsch et al., 1999), allowing for reactivation and cell cycle re-entry to generate neuronal and gliogenic progeny upon distinct environmental stimuli (Delgado et al., 2021). Maintaining the quiescent state of aNSCs is thought to be critical for preserving the NSC pool during aging (Encinas et al., 2011), as overactivation of aNSCs may deplete the aNSC pool (Sierra et al., 2015). Thus, this intricate balance between aNSC quiescence and activation drives neuro- and gliogenesis, while safeguarding a resident aNSC population from decay (Harris et al., 2021). Over the recent years, extensive studies have highlighted several cell-intrinsic and extrinsic cues, as well as systemic factors, that play pivotal roles in the regulation of aNSC quiescence, both in health and disease (Figure 1).
Key signaling pathways acting as regulators of NSC quiescence include, among others, Notch (Ehm et al., 2010; Imayoshi et al., 2010; Lugert et al., 2010), ephrins in endothelial cells (Ottone et al., 2014), cadherins in ependymal cells (Porlan et al., 2014), BMP, WNT (Li and Clevers, 2010; Mira et al., 2010), GABAergic (Song et al., 2012; Giachino et al., 2014), and glutamatergic signaling (Lugert et al., 2010; Sierra et al., 2015; Bielefeld et al., 2019). For an extensive overview of all signaling pathways and proteins involved in maintaining aNSC quiescence, we refer to an excellent recent review (Urbán et al., 2019).
Although all these molecular cascades provide potential targets for ncRNA control, only a few sncRNAs have been identified that regulate aNSC quiescence, all of them being miRNAs (Figure 3). A key miRNA controlling NSC quiescence is miR-9. As will be discussed later, miR-9 is classically known to be a regulator of the NPC proliferation/differentiation balance when localized to the cytoplasm through Tlx, Foxo1 and Notch (Zhao et al., 2009; Kim et al., 2015). However, subsets of aNSCs are extensively enriched for miR-9. When localized to the nucleus, miR-9 acts as a rheostat for aNSC activation/quiescence, through its target TNRC6 and the Notch pathway (Katz et al., 2016).
MiR-204 has been shown to be transferred from the choroid plexus to the cerebrospinal fluid, allowing long-distance regulatory control of aNSCs residing in the SVZ. Silencing miR-204 results in significant activation of quiescent aNSCs, through the upregulation of a plethora of neurogenic target genes (Lepko et al., 2019).
Another example of systemic regulation of aNSC quiescence under the control of sncRNAs is Foxo3, a downstream factor of the IGF signaling pathway. Foxo3 is required to maintain NSC quiescence in both the SVZ and SGZ (Paik et al., 2009; Renault et al., 2009), and forms a feedback loop with the miR-106b-25 cluster, which is encoded in an intronic region of a Foxo3 target gene (Mcm7; Brett et al., 2011). The Foxo3 mRNA is one of the predicted targets of the transcribed miR-25, creating a negative feedback loop controlling aNSC quiescence. Yet, the involvement of this feedback loop in the regulation of NSC quiescence has not been experimentally validated in vivo.
Effects in Pathological Conditions
ANSC activation is linked to several neurological pathologies, of which epileptic seizures have been widely validated in animal models (Lugert et al., 2010; Sierra et al., 2015). Overactivation of aNSCs under pathological conditions results in a rapid depletion of the aNSC pool and a concomitant decrease in the neurogenic capacity in the DG (Sierra et al., 2015), suggesting an association with the cognitive deficits (Cho et al., 2015). Several studies have mapped the ncRNA response to seizures in both mice and humans (Schouten et al., 2015; Bencurova et al., 2017; Bielefeld et al., 2017; Brennan and Henshall, 2020). Interestingly, the expression of both the miR-106b-25 cluster and miR-9 in the hippocampus are altered at different stages after seizure onset (McKiernan et al., 2012; Risbud and Porter, 2013; Wang et al., 2015). However, none of these alterations or their potential role in the regulation of NSC activation have been experimentally validated. Of note, a lack of aNSC activation is thought to be involved in neuropsychiatric disorders, of which depression is the most extensively studied. As antidepressant drugs increase neurogenesis, and electroconvulsive shock therapy induces aNSC activation, it has been suggested that a lack of aNSC activation may underlie the development of depression (Encinas et al., 2006; Hanson et al., 2011). However, clear evidence for a potential role of sncRNAs in this imbalance of aNSC activation has not been reported yet.
Regulation of Neurogenesis From aNSCs
A first demonstration of the dependency of NSCs on miRNA regulation comes from studies in which one of the essential miRNA-processing enzymes, Dicer, was deleted in adult or embryonic NSCs, precluding global miRNA generation (Saurat et al., 2013; Pons-Espinal et al., 2017; Xu et al., 2020). ANSC-specific ablation of Dicer in mouse DG and rescue with mature miRNAs confirmed that miRNAs, but not miRNA-independent Dicer functions, are required for the generation of adult-born DGCs (Pons-Espinal et al., 2017). However, in the absence of reliable cell type-specific markers for the selective genetic targeting of aNSCs, differentiating between NSC-specific and non-specific effects remains challenging. Another interesting observation is that miRNAs respond to some of the extrinsic stimuli that act as triggers of AHN in DG. More specifically, miRNAs were shown to mediate some of the enhancing effects of both environmental enrichment and physical exercise on AHN (Barak et al., 2013; Pons-Espinal et al., 2019; Walgrave et al., 2021a), possibly placing miRNAs upstream of signaling pathways and molecular regulators that induce AHN. In addition, miRNAs frequently function in concerted action with transcription factors and chromatin modifiers, as we discuss in more detail in the following sections. Thereby, miRNAs provide an additional layer to ensure the robustness of gene regulatory programs dictating the fate transitions along the neurogenic trajectory (Encinas and Fitzsimons, 2017; Stappert et al., 2018).
Effects in Physiological Conditions: Early Neurogenic Events
Besides evidence derived from global inhibition of miRNA biogenesis, individual miRNAs, or miRNA networks can also induce either widespread effects on the adult neurogenic process or preferentially regulate distinct stages along the neurogenic trajectory from aNSCs to adult-born neurons. The miRNA impact on the net outcome of neuronal differentiation can be attributed to the regulation of upstream processes, direct targeting of differentiation effectors, or both. However, non-cell-autonomous effects mediated by other niche-resident cells cannot be ruled out, given the lack of aNSC/NPC targeting specificity in the transgenic mice generation strategy. Lentiviral knockdown of miR-132, primarily in aNSCs in the mouse DG, blocked the running-induced increase in NPC proliferation and neuronal differentiation, suggesting that miR-132 is required for the running-mediated effects on AHN (Walgrave et al., 2021a). Although systematic validation of the pertinent miR-132 targetome in aNSCs is still pending, miR-132 overexpression induced broad effects in genes involved in cell cycle regulation and neuronal differentiation, as assessed by single-cell RNA-sequencing in Nestin-positive niche-resident cells. Functioning as a gatekeeper of the balance between proliferation and differentiation, miR-138-5p acts through inactivation of the TRIP6, a molecule that is necessary and sufficient to maintain aNSC proliferation and self-renewal capacity. Downregulation of miR-138-5p in vitro promotes aNSC proliferation and inhibits neuronal differentiation, whereas miR-138-5p overexpression exerts opposite effects (Wang et al., 2017b). While such shifts are easier to monitor in in vitro aNSC mono-cultures, differentiating between effects on distinct stages of aNSC self-renewal and NPC proliferation in vivo would require further systematic fate mapping.
Interestingly, miR-137, a miRNA epigenetically repressed by MeCP2, a DNA methyl-CpG-binding protein also regulating AHN, was shown to induce NPC proliferation in SGZ at the expense of neuronal differentiation, upon retroviral overexpression in the adult DG (Szulwach et al., 2010). In a feedback regulatory loop back onto chromatin, Ezh2, a histone methyltransferase and polycomb group protein, was identified as a direct target of miR-137, mediating its effects on AHN and exemplifying the complex miRNA interactomes in the adult hippocampal neurogenic niche. A similar interplay between miRNA and epigenetic regulation was reported for miR-184 and miR-195, which regulate the balance between aNSC activation/proliferation and differentiation in the adult SGZ niche by forming a regulatory network with their epigenetic repressor, Mbd1. MiR-184 then targets Numbl, an important player during brain development (Liu et al., 2010), whereas miR-195 signals back to Mbd1 (Liu et al., 2013). In the same vein, let-7b regulates the expression of Hmga2, a small chromatin-associated protein that can modulate transcription by altering chromatin structure (Nishini et al., 2008). Aging-associated Hmga2 deficiency in adult mouse SVZ, which is partially mediated by increased let-7b expression, reduces the self-renewal potential of aNSC in the aged brain. Apart from a pivotal role as a rheostat between aNSC quiescence and activation (Katz et al., 2016), as discussed earlier, miR-9 was also reported to suppress aNSC proliferation and accelerate neuronal differentiation in the embryonic mouse SVZ, via a negative feedback regulatory loop with its target, TLX (Zhao et al., 2009). These observations were confirmed in primary NSC/NPC cultures derived from neonatal mouse brain, where miR-9 was shown to positively regulate neuronal differentiation via a regulatory cascade involving Foxo1 and Notch signaling (Kim et al., 2015). Although suggestive of a potent miR-9-dependent gene regulatory network fine-tuning NPC proliferation and neuronal differentiation during embryonic and early postnatal stages, a similar role for miR-9 in the adult brain has not yet been reported. Another miRNA with crucial functions in mammalian CNS is miR-124, the most abundant miRNA in the adult brain (Lagos-Quintana et al., 2002). MiR-124 was reported to be a key determinant of neuronal fate in aNSCs in mouse SVZ (Åkerblom et al., 2012), where miR-124 overexpression induces neuronal differentiation by directly targeting Sox9, a key transcription factor for adult neurogenesis (Cheng et al., 2009). Of note, miR-124 also regulates and is regulated by REST, a primary transcriptional repressor responsible for inhibiting the expression of neuronal genes in non-neuronal cells (Conaco et al., 2006). Whether the miR-124/REST regulatory loop is also involved in AHN remains to be assessed.
Effects in Physiological Conditions: Late Neurogenic Events
Newly born immature neurons in the SVZ and SGZ need to migrate over distinct distances that depend on the neurogenic niche in which they developed. Let-7 and miR-19 regulate the migration of adult-born neurons in the olfactory bulb and the DG, respectively (Han et al., 2016; Petri et al., 2017). The regulation of the maturation of newborn neurons at late neurogenic stages has also been documented for several miRNAs. As most of these studies employed either in vitro or embryonic systems, it is possible that these regulatory mechanisms are not applicable in the adult brain, and even if they were, they may represent generalized miRNA functions, and not adult-born neuron-specific phenomena. These limitations further complicate conclusions regarding AHN-specific miRNA-dependent effects on memory formation and cognition (for a review on miRNA roles in memory formation, see Saab and Mansuy, 2014). Along these lines, an additional putative role for miR-124 in the neurogenic process is the induction of both dendritic arborization and axonal growth via repression of RhoG, a small GTPase involved in neuritogenesis (Franke et al., 2012). However, these effects have only been monitored in cultured hippocampal neurons. Reversely, miR-134 and miR-138, a brain-specific and a brain-enriched miRNA, respectively, are localized to the synapto-dendritic compartment and were shown to negatively regulate the development of dendritic spines in hippocampal neurons in vitro (Schratt et al., 2006; Siegel et al., 2009). Interestingly, miR-134 acts by targeting Limk1 in response to neuronal stimulation by BDNF, a key modulator of neuronal plasticity (Schratt et al., 2006). Another neuronal activity- and CREB-regulated miRNA, miR-132, was also previously shown to promote neurite outgrowth, both in cultured cortical neurons via its direct target, p250GAP (Vo et al., 2005), and in vivo (Magill et al., 2010; Luikart et al., 2011; Pathania et al., 2012; Walgrave et al., 2021a). Of note, miR-132 is emerging as a key component of structural plasticity networks regulating dendritic arborization, survival, and synaptic integration, both in postnatal neurogenesis at the SVZ (Pathania et al., 2012) and cell-autonomously, in adult-born neurons in DG (Magill et al., 2010; Luikart et al., 2011; Walgrave et al., 2021a). Conversely, both miR-137 and miR-223 negatively impact dendritic outgrowth in newly born GCs in the adult mouse brain upon retroviral targeting of adult NPCs (Smrt et al., 2010; Harraz et al., 2014). Taken together, these findings strongly support a multilayered role for miRNAs in the neurogenic process in general, and in AHN in particular. Considering the contextualization component of many miRNA regulatory effects, validation studies in the (aging) brain are required to assess the conservation of similar miRNA-mediated mechanisms.
Effects in Pathological Conditions
AHN has been shown to be impaired by several pathologies, including neurodegenerative diseases, mood disorders, epilepsy, and substance use disorders. Given the pivotal role of miRNAs in controlling critical steps of the neurogenic process discussed before, the link between miRNA deregulation and these pathologies in the context of neurogenesis has been investigated on several occasions, although direct functional links have mainly been drawn from in vitro studies and rodent models.
Neurodegenerative Disorders
Alterations in adult neurogenesis are a common phenomenon in several neurodegenerative disorders, which could potentially contribute to their pathophysiology. For example, aberrant AHN has been reported in Alzheimer’s disease (AD) patients and mouse models (Babcock et al., 2021), and recent findings suggest that in AD patients, the number and the maturation of newly born GCs progressively decline as AD pathology advances (Moreno-Jiménez et al., 2019; Tobin et al., 2019). Restoring adult hippocampal neurogenesis deficits in several AD mouse models was shown to improve AHN-related memory (Choi et al., 2018; Kim et al., 2019; Walgrave et al., 2021a). Intriguingly, miR-132 is consistently downregulated in the hippocampus of human AD patients (Salta and De Strooper, 2017a) and has been recently shown to be a potent regulator of AHN, exerting cell-autonomous proneurogenic effects in aNSCs and their progeny. Furthermore, it was shown that miR-132 replacement in adult mouse AD hippocampus restores AHN and relevant memory deficits, opening up novel avenues for possible therapeutic approaches targeting miR-132 in neurodegeneration (Walgrave et al., 2021b).
Another neurodegenerative disease associated with impaired AHN is Parkinson’s disease (PD; reviewed in Lim et al., 2018). Signaling pathways implicated in the neuroinflammatory response and autophagy are crucial in PD pathogenesis and could potentially be orchestrated by different miRNAs, among other regulators (Nuzziello and Liguori, 2019). However, to date, the only evidence linking miRNAs, PD, and adult neurogenesis involves the SVZ, where neurogenesis is found markedly impaired in the PD brain. Assessing the neurogenic and migratory potential of SVZ-derived neuroblasts, both in physiological conditions and in a 6-OHDA-mouse model of PD, showed that single administration of nanoparticle-encapsulated miR-124, was able to promote migration of neuroblasts reaching the granular cell layer of the olfactory bulb and ameliorating motor symptoms (Saraiva et al., 2016).
Mood and Psychiatric Disorders
In addition to neurodegenerative diseases, mood and psychiatric disorders have also been linked to AHN. Aberrant AHN has been associated with depression, schizophrenia, and anxiety (Kang et al., 2016). Evidence shows that antidepressants increase AHN and that enhanced AHN is sufficient to promote resilience to anxiety and depression-related behaviors in a mouse model of stress (Hill et al., 2015; Kang et al., 2016; Planchez et al., 2021). Recent studies suggested once again a putative role for miRNAs in the interplay between AHN and mood disorders. The miR-30 family was shown to mediate a chronic stress-induced depression-like phenotype by altering hippocampal neurogenesis and neuroplasticity via controlling epigenetic, transcription, and cell signaling regulators (Khandelwal et al., 2019).
Abnormal migration of adult-born hippocampal neurons has been described in schizophrenia (Duan et al., 2007; Kim et al., 2009), and rare inherited copy number variants of RAPGEF2 have been associated with familiar schizophrenia (Xu et al., 2009). In a study investigating the role of miR-19 and RAPGEF2 in schizophrenia using patient-derived hippocampal NPCs (SZ-NPCs), miR-19 was found upregulated in SZ-NPCs, and conservation of the regulation of RAPGEF2 by miR-19 was demonstrated in human embryonic stem cell-derived hippocampal NPCs (Han et al., 2016). Of note, miRNA-19 belongs to the family of polycistronic miR-17 clusters, including also miR-17-92 and miR-106b-25b, which, as discussed above, have been shown to be important for aNSC proliferation (Brett et al., 2011; Jin et al., 2016). Hence, it would be interesting to assess whether the other members of the miR-17 clusters are also altered in schizophrenia and if they contribute to pathology.
Epilepsy
Temporal lobe epilepsy (TLE), characterized by seizures originating in many cases from the hippocampal region, is the most common form of epilepsy. AHN is particularly sensitive to seizures. Pioneering studies showed that epileptic seizures cause a significant increase in the short-term proliferation rate of NSCs, while diminishing long-term proliferation (Parent et al., 1997), and that newborn neurons in the epileptic brain also display several morphological and functional alterations, suggesting a possible role for aberrant AHN in epilepsy (Scharfman et al., 2000; Dashtipour et al., 2001; Murphy et al., 2011, 2012; Wood et al., 2011; Bielefeld et al., 2014). Over the recent years, it has been shown that many miRNAs known to regulate AHN are also deregulated during acute seizure events or chronic epilepsy, particularly in the DG (Schouten et al., 2015), providing further insights into the role of AHN in the context of epilepsy (Bielefeld et al., 2017). Of note, concomitant knockdown of miR-124 and miR-137 has been shown to prevent hippocampal aNSC loss upon non-convulsive seizures (Bielefeld et al., 2019).
Regulation of Adult Astrogliogenesis
Effects in Physiological Conditions
New astrocytes are continuously generated at low levels alongside the new neurons in the adult hippocampus (Kuhn et al., 1996; Suh et al., 2007; Bottes et al., 2021). However, there is still not a clear consensus as to whether astrocytes are generated by multipotent NSCs or by a distinct type of stem cells, which have some characteristics of differentiated astrocytes (Alvarez-Buylla et al., 2001; Song et al., 2002; Steiner et al., 2004). On the molecular level, two transcriptional mechanisms controlling neuron-astroglia fate decisions in the adult hippocampus have been recently described, the Notch2-Id4 axis, possibly in synergy with transforming growth factor-β (TGF-β)/bone morphogenetic protein (BMP) pathway (Zhang et al., 2019c), and the COUP-TFI(Nr2f1) pathway (Bonzano et al., 2018).
Despite several studies indicating that miRNAs are essential for the developmental switch from neurogenesis to gliogenesis in mice (Zheng et al., 2010; Shenoy et al., 2015; Tsuyama et al., 2015), currently, understanding of their role in the control of adult astrogliogenesis is missing. As mentioned earlier, a study addressing this question showed that loss of Dicer-dependent miRNA impairs neurogenesis but not astrogliogenesis in the adult hippocampus in vivo (Pons-Espinal et al., 2017). However, different mechanisms could account for this observation. Firstly, it is possible that different subtypes of neural and glial progenitor cells exist in the adult hippocampal niche (Pilz et al., 2018; Bottes et al., 2021), which differentially respond to miRNA depletion. Secondly, astrocytes in glia-like hippocampal aNSCs might represent a “default” developmental path rather than a true fate choice (Encinas et al., 2011; Kempermann, 2011), and thus, their generation could be less dependent on miRNAs. Finally, it is also possible that miRNAs need to suppress glial fates in order to sustain neurogenesis. This hypothesis would be consistent with the evidence in mouse cortical development, where COUP-TF1 is actively suppressed by miR-21-5p (Terrigno et al., 2018) and GFAP by miR-3099 (Zainal Abidin et al., 2019), and in the adult hippocampus, where Drosha functions as a molecular barrier, which, by preventing oligodendrogliogenesis, restricts the multi-lineage potential of aNSCs (Rolando et al., 2016).
Effects in Pathological Conditions
The discovery of astrocyte-like NSCs in the hippocampus has also shed new light on the role of sncRNAs in reactive gliosis, which is increasingly viewed as an important aspect to address in order to better understand the pathological and/or age-dependent loss of neurogenesis, as well as to explore the potential of RNA therapeutics in various brain diseases. Indeed, altered expression of several miRNAs, particularly miR-146a, miR-155, and miR-132 has been reported in reactive astrocytes in the postnatal hippocampus of various neurodegenerative and ischemia models, or following status epilepticus (Aronica et al., 2010; Arena et al., 2017; Korotkov et al., 2020). Moreover, manipulation of specific miRNAs, such as miR-181a or miR-302/367, in reactive hippocampal astrocytes could ameliorate some of the neural deficits in models of focal ischemia or AD (Ghasemi-Kasman et al., 2018; Griffiths et al., 2019).
Regulation of Adult Oligodendrogliogenesis
Effects in Physiological Conditions
Most knowledge on the regulation of oligodendrogliogenesis by miRNAs originates from research in embryonic development. During the embryonic development of the CNS, inhibition of miRNA formation by conditional Dicer knockout in the spinal cord resulted in a disruption of both astrogliogenesis and oligodendrogliogenesis, without affecting neurogenesis, indicating that miRNAs are essential for the initiation of gliogenesis (Zheng et al., 2010).
Similarly, the importance of miRNAs in oligodendrogliogenesis has been unveiled using Dicer ablation in OPCs. Dicer mutant mice display a lack of differentiated OPCs resulting in demyelination defects, and OPCs isolated from these mice are unable to properly differentiate in vitro (Dugas et al., 2010). When comparing the expression of miRNAs in OPCs and mature oligodendrocytes, a 10 to 100-fold increase of miR-219, miR-338, and miR-138 was observed. The most strongly induced miRNA among these, miR-219, directly targets genes involved in maintaining OPC proliferation, and increasing miR-219 levels stimulate OPCs to exit proliferation and start differentiation (Dugas et al., 2010; Li et al., 2019). Furthermore, transplantation of OPCs overexpressing miR-219 into a cuprizone-induced demyelinated mouse model promoted remyelination and improved cognitive function, highlighting the therapeutic relevance of miR-129-mediated regulation of oligodendrogliogenesis (Fan et al., 2017). Of interest, postnatal oligodendrocytic cell lines possess a characteristic miRNA signature composed of 43 miRNAs dynamically expressed during oligodendrogliogenesis. Within this group of 43 miRNAs, miR-9 seems of particular importance for the physiological regulation of oligodendrogliogenesis, as its expression inversely correlates with the expression of its target, the peripheral myelin protein PMP22 (Lau et al., 2008). Additionally, this study demonstrated that particular miRNAs are expressed at specific stages during OPC differentiation to mature oligodendrocytes (Lau et al., 2008). Interestingly, oligodendrogliogenesis provides a physio/pathologically relevant system for the study of functional synergy between multiple miRNAs. MiR-219 cooperates with miR-338 in regulating myelination in the CNS (Zhao et al., 2010; Wang et al., 2017a). Moreover, the increased levels of mature oligodendrocytes observed in the presence of the receptor for advanced glycation end products (RAGE) antagonist, FPS-ZM1, correlated with increased expression of miR-23a, miR-219a, and miR-338, three miRNAs associated with oligodendrocytic differentiation and remyelination (Santos et al., 2020).
While under physiological conditions aNSCs mostly produce neurons and astrocytes and not oligodendrocytes (Suh et al., 2007; Jessberger et al., 2008), hippocampal aNSCs can redirect their fate to the oligodendrocyte lineage upon genetic manipulation. The first example was provided by overexpression of the transcription factors Ascl1, Olig2, or Sox10 in vivo, which resulted in a switch toward the oligodendrocytic lineage (Jessberger et al., 2008; Braun et al., 2015). Follow-up studies have shown that Drosha deletion in hippocampal aNSCs impairs neurogenesis and instead induces oligodendrocytic commitment and production. Even though Drosha is part of the microRNA microprocessor, it can also directly destabilize mRNAs, suggesting that the observed regulation of aNSC fate towards oligodendrocytes may be independent of miRNAs and dependent on Drosha-mediated repression of Neurofibromin 1 (Rolando et al., 2016). Indeed, Neurofibromin 1 inactivation in vivo revealed a latent oligodendrocytic lineage potential of hippocampal aNSCs (Sun et al., 2015). Nevertheless, recent studies have shown that neutralization of the neurotrophin nerve growth factor promotes oligodendrogliogenesis from hippocampal neurospheres by increasing miR-219a-5p levels, highlighting the role of miRNA-dependent mechanisms in this process (Brandi et al., 2021).
Effects in Pathological Conditions
Many of the studies addressing the role of miRNAs in OPCs have used animal models of neurological injuries, such as stroke-induced cerebral ischemia and traumatic brain injury. In these models, a positive neurological outcome has been associated with increased oligodendrogliogenesis, which contributes to remyelination (Santra et al., 2014; Xiong et al., 2015). One of the best-characterized consequences of CNS injury is the induction of inflammation. In particular, the Toll-like receptor (TLR) proinflammatory signaling pathway regulates tissue injury (Symons et al., 2006). MiR-146(a) is an inflammation-associated miRNA that downregulates proinflammatory cytokines and TLR-mediated pathways (Taganov et al., 2006; Nahid et al., 2011). In OPCs, miR-146a is upregulated by thymosin β4, resulting in the suppression of TLR-mediated proinflammatory pathways (Santra et al., 2014). Further studies have demonstrated that thymosin β4 upregulated miR-200a in NPCs in the peri-infarct area in a model of middle cerebral artery occlusion (Santra et al., 2016). In this model, thymosin β4-mediated miR-200 upregulation resulted in increased expression of the oligodendrocyte marker myelin basic protein (MBP; Santra et al., 2016). In agreement with these studies, overexpression of miR-146a in NPCs differentiated them into OPCs, and increased the expression of myelin proteins in OPCs, indicating that miRNAs mediate some of the effects of stroke on oligodendrogliogenesis (Liu et al., 2017). Similar results were obtained in a mouse model of multiple sclerosis, where miR-146a promoted OPC differentiation and enhanced remyelination (Zhang et al., 2019a), further highlighting the involvement of sncRNAs in myelination-related disorders.
Regulation of the Crosstalk Between NSCs and Microglia
DG microglia display distinct gene expression profiles compared to microglia found in other hippocampal regions (St-Pierre et al., 2020), suggesting that they may be specialized for crosstalk with niche cells, like aNSCs (Kreisel et al., 2019). In fact, aNSCs express a multitude of receptors for immune molecules and cytokines, allowing their regulation by microglial immune mediators (Ekdahl et al., 2009). As such, microglia may be involved in different phases of neurogenesis (Ekdahl et al., 2009), through different functions, such as initiating the inflammatory response and phagocytosis (Sierra et al., 2010, 2019; Paolicelli et al., 2011; Abiega et al., 2016).
Regulation of inflammation is one of the best characterized microglial functions and, thus, inflammatory mediators have also been studied in the context of the microglia-aNSC crosstalk (Ekdahl et al., 2003; Monje et al., 2003). However, the outcome in vivo is dependent on the balance of pro- and anti-inflammatory mediators in the neurogenic niche (Monje et al., 2003; Butovsky et al., 2006; Woodbury et al., 2015; Diaz-Aparicio et al., 2020). The role of microglial miRNAs in neuroinflammatory modulation has been widely documented. MiRNAs regulate the expression of many cytokines and the function of several pro-inflammatory signaling pathways in microglia, as previously reviewed (Wang and Wang, 2018; Guo et al., 2019). Microglial miRNAs such as miR-206, miR-155, and miR-32-5p, favor a pro-inflammatory microglial phenotype (Butovsky et al., 2015; Xing et al., 2016; Yan et al., 2018), while others, like miR-93, miR-367, miR-26a, miR-124, miR-199b, and miR-27a, inhibit pro-inflammatory signaling (Kumar et al., 2015; Wang et al., 2015; Louw et al., 2016; Zhou et al., 2016; Lv et al., 2017; Tian et al., 2017; Guo et al., 2019). Many of these miRNAs have an impact on the expression of cytokines that have been directly linked to aNSC fate regulation, thus, potentially regulating important aNSC features, such as proliferation and cell fate. However, there is an overall scarcity of reports directly assessing the impact of the microglial inflammation-related miRNAs on the regulation of aNSCs. One of the few studies addressing this, showed that knocking out microglial miR-155 promotes neurogenesis in NSCs cultured with LPS-activated microglia (Woodbury et al., 2015). Taken together, these observations suggest that microglial inflammation-related miRNAs may play a key role in the modulation of aNSC fate.
Phagocytosis is another of the main microglial functions that has a direct impact on aNSCs (Diaz-Aparicio et al., 2020). Microglial miRNAs also regulate microglial phagocytosis, thereby potentially altering aNSC fate. MiR-146a is mainly overexpressed by microglia in prion disease mouse brain (Saba et al., 2012). Microglial miR-146a increases by TLR2 stimulation in vitro, disrupting inflammatory signaling pathways and predictably targeting two phagocytic mediators of the oxidative burst (Saba et al., 2012; Guo et al., 2019). Upregulation of microglial miRNA-34a expression has been linked to decreased expression of triggering receptor expressed in myeloid/microglial cells-2 (TREM2) in microglia in vitro (Alexandrov et al., 2013), which has been linked to a decrease in microglial phagocytosis of Aβ42 in vitro (Bhattacharjee et al., 2016). Induced overexpression of miR-124, a miRNA shown to inhibit neuroinflammation and to drive macrophage differentiation towards microglia (Ponomarev et al., 2011), has also been shown to inhibit the phagocytosis of apoptotic cells in the optic tectum of zebrafish larvae (Svahn et al., 2016). Although there are no studies linking miRNA regulation of microglial phagocytosis to aNSC fate, the aforementioned miRNA-driven changes in phagocytosis could potentially regulate aNSC function and fate through changes in the microglial phagocytic secretome, as previously described (Diaz-Aparicio et al., 2020). Overall, the extent, complexity, and implications of the miRNA-driven crosstalk between microglia and aNSCs remain to be fully elucidated.
Piwi-Interacting RNAs and Other Classes of sncRNAs
Mounting evidence has shown that some of the PIWI proteins and Piwi-interacting RNAs (piRNAs; Figure 2) are expressed in the CNS of different animal models, and participate in the processes of neurodevelopment, neurotransmission, transient focal ischemia and neurodegeneration (Kaur et al., 2018; Kim, 2019; Huang and Wong, 2021). In particular, PIWIL1 (MIWI in the mouse) and piRNAs have been linked to neuronal injury and axonal regeneration in rodents (Phay et al., 2018; Sohn et al., 2019) and C. elegans (Kim et al., 2018), but the underlying mechanisms are still obscure. In the mouse developing cerebral cortex, the knockdown of MIWI resulted in a multipolar morphology of neurons and a larger number of primary neurites, suggesting that the piRNA pathway can regulate neuronal migration and maturation during development (Zhao et al., 2016). The expression of PIWIL2 (MILI in mouse) protein has also been reported in the adult mouse brain, with hypomethylation of transposons and behavioral defects observed in MILI knockout mice (Nandi et al., 2016). A more recent study has described behavioral impairment in mice upon hippocampal MILI and/or MIWI depletion. Knockdown of MILI results in hyperactivity, whereas knockdown of both MILI and MIWI leads to enhanced contextual fear memory without affecting anxiety (Leighton et al., 2019).
The aforementioned studies and the majority of those which investigated the possible roles of the piRNA pathway in mammalian CNS, have focused on neurons (Dharap et al., 2011; Lee et al., 2011; Ghoshes et al., 2016; Nandi et al., 2016; Zhao et al., 2016; Leighton et al., 2019). However, as piRNA levels in these cells are low compared to germline cells, the proposed function of the piRNA pathway in synaptic plasticity and memory control (if any) is debated.
Interestingly, the hippocampus shows the highest number of unique piRNA sequences and piRNA transcripts (5494 piRNAs) in the adult brain (Perera et al., 2019). In agreement with this observation, it was recently demonstrated that the piRNA pathway has a crucial role in maintaining the homeostasis and the fate of hippocampal NSCs (Gasperini et al., 2021). In particular, the expression of MILI and piRNAs was found to be enriched in NPCs of the postnatal mouse hippocampus, compared to their differentiated progeny, and piRNA functions were shown to be essential for neurogenesis. Moreover, inhibition of the piRNA pathway in adult NPCs leads to senescence-associated inflammation and an increase in reactive astrogliogenesis (Gasperini et al., 2021), a condition which has been linked to age-related impairment of neurogenesis and neurodegeneration.
Despite the growing interest in brain-related piRNAs, the unavailability of proper methods and tools to investigate the piRNA pathway outside gonads, and therefore the lack of a fully characterized pathway in neuronal tissues, complicates research in this field. Indeed, most of the existing studies did not provide conclusive evidence that piRNA-dependent, rather than piRNA-independent, functions of these proteins drove the various phenotypes. Functions of PIWI proteins were shown to regulate chromatin structure/stability, histone/DNA methylation, DNA repair transcription, and RNA turnover and translation in gonads (Czech et al., 2018; Ozata et al., 2019), but these mechanisms are poorly investigated in CNS. Because PIWI protein functions have been attributed roles in several CNS diseases, including neurodevelopmental and neurodegenerative diseases and various cancers, a better understanding of these processes in aNSCs entails important translational implications.
With respect to additional classes of sncRNAs, at present very little is known about their involvement in the regulation of aNSC homeostasis, activation, and fate determination. Interestingly, a double-stranded RNA (dsRNA) molecule (Figure 2) expressed at the SGZ of DG, was shown to regulate fate determination of hippocampal aNSCs in vitro (Kuwabara et al., 2004). This dsRNA bares a recognition motif for NRSF/REST, a major negative transcriptional regulator of neuronal differentiation. Interaction between the dsRNA and the NRSF/REST machinery results in the transition from aNSCs to a neuronal (but not glial) fate, while the effects seem to be mediated by a dsRNA/protein interplay (Figure 3).
Discussion
The complexity of the biological processes involved in aNSC homeostasis, including activation, self-renewal, and fate determination, requires tight control of numerous multilayered regulatory programs. SncRNAs, as other ncRNA species, regulate gene expression at the transcriptional and post-transcriptional level and are additionally considered to be the main players in epigenetic regulation.
In this review, we have summarized evidence indicating that sncRNAs, and primarily microRNAs, play major regulatory roles at virtually all stages of aNSC homeostasis in both health and disease. We have discussed in detail the role of individual miRNAs, in the regulation of key targets crucial for aNSC quiescence, and for their activation, which can eventually result in neurogenesis, astrogliogenesis or oligodendrogliogenesis. Moreover, we surveyed miRNA roles in the functional crosstalk between aNSCs and other relevant cell types in the adult hippocampal neurogenic niche, in particular microglia. However, with the advent of novel technologies, we have only recently begun to unveil previously hidden layers of the biology of sncRNAs. For instance, tRNA-derived fragments are a new class of sncRNAs with noncanonical biological functions unrelated to protein synthesis (Torres et al., 2019); they are produced from tRNAs by specific cleavage events catalyzed by ribonucleases, such as Dicer; and have been shown to increase during disease, where they seem to regulate stress responses, proteostasis, and neuronal survival (Fagan et al., 2021). Interestingly, tRNA-derived piRNAs, putatively requiring PIWI protein function in somatic cells, have also been identified (Keam et al., 2014). How these and other newly identified sncRNAs may independently or in synergy (with miRNAs, piRNAs, or other ncRNA species) impact phenomena related to aNSCs remains elusive.
Further contributing to our understanding of the complexity of sncRNA-mediated regulation, the functional crosstalk across and within different classes of sncRNAs has been shown to enable precise regulation of several brain functions (Mehta et al., 2021). Functional pleiotropy (each miRNA may simultaneously regulate multiple targets) and convergence onto shared targets within distinct pathways (multiple miRNAs may simultaneously regulate the same target) are two key elements of miRNA biology (Barca-Mayo and De Pietri Tonelli, 2014; Walgrave et al., 2021b) with functional relevance in AHN (for a review, see Stappert et al., 2018). Systematic interrogation of the intricate targetomes of single miRNAs or of synergistic miRNA networks in the context of aNSC regulation will enable a more accurate profiling of their regulatory repertoire and, hence, of their potential therapeutic use as mediators of regeneration of distinct cell types in the adult brain. Even though these interactions may also be critical for maintaining the heterogeneity of the adult hippocampal niche by determining the balance between neurogenesis and astrogliogenesis (Pons-Espinal et al., 2017), the majority of such mechanistic aspects have not been experimentally addressed in the context of AHN, apart from a few exceptions (Schouten et al., 2015; Jin et al., 2016; Pons-Espinal et al., 2017; Wang et al., 2017b; Bielefeld et al., 2019; Pan et al., 2019).
As an additional layer of intercellular communication, aNSCs and other cell types in the niche are capable of releasing and taking up extracellular vesicles like many other cell types in the mammalian brain (Lopez-Verrilli and Court, 2013; Luarte et al., 2016; Vogel et al., 2018). Extracellular vesicles can be divided into several categories depending on size, among which exosomes have been mostly implicated in the regulation of the aNSC niche. Exosomal content varies greatly based on the cell type from which they are released and the (patho)physiological state of the cell. In general, exosomes contain a plethora of non-coding RNAs, including miRNAs, piRNAs, tRNAs, long non-coding RNAs (lncRNAs), as well as mRNAs, lipids, and transcription factors (Valadi et al., 2007; O’Brien et al., 2020). Exosomes in the neurogenic niche have been implicated in all stages of neurogenesis, ranging from the regulation of aNSC activation (Cheng et al., 2021) to synaptic pruning of mature neuronal spines through microglia (Bahrini et al., 2015). An example of exosome-mediated intercellular signaling and synergy between multiple miRNAs is the regulation of neuroplasticity and functional recovery after stroke by the miR-17-92 cluster. Exosomes extracted from multipotent mesenchymal stromal cells and artificially enriched in miRNAs from the miR-17-92 cluster, increased oligodendrogliogenesis, neurogenesis, neurite remodeling, and neuronal dendrite plasticity in the ischemic boundary zone after stroke, possibly by activating the PI3K/protein kinase B/mechanistic target of rapamycin/glycogen synthase kinase 3β signaling pathway (Xin et al., 2017). In addition, intercellular communication via exosomal miRNAs has been shown to impact brain physiology and a diverse range of pathologies (Xia et al., 2019). More recently, endogenous sncRNAs, termed “glycoRNAs”, were found to bear glycan conjugations and to primarily localize on the cell membrane, putatively acting as signals in the intercellular communication and further contributing to intricate cellular interaction pathways (Flynn et al., 2021).
An issue confounding the current understanding of hippocampal aNSC biology is that a big part of our knowledge on the sncRNA-mediated regulation of aNSCs is derived from in vitro systems and from studies in embryogenesis or in the adult SVZ niche. Yet, evidence from heterotopical transplantation experiments demonstrated the significance of the niche microenvironment in fate choice heterogeneity of aNSCs (Beyer et al., 2020), suggesting that findings not derived from the adult SGZ should not be extrapolated to the adult DG without further experimental validation. In addition, given the context-specificity of several miRNA regulatory networks, extrapolating such observations to putative effects in AHN should be done with caution.
Another pivotal limitation is the vast knowledge gap with respect to whether the coding and non-coding mechanisms regulating aNSC quiescence, activation, and fate choice in the adult rodent brain are also conserved in humans. Studies in human systems in vitro and high-resolution cellular and molecular profiling of the adult hippocampal neurogenic niche in the human brain will shed more light on the existence of aNSCs, their putative heterogeneity, and their regulation in health and disease.
In conclusion, the intricate sncRNA landscape of mammalian cells has only recently started to be appreciated. Understanding the complex sncRNA-mediated regulatory mechanisms involved in aNSC homeostasis, including self-renewal, activation, and fate commitment will be key to decoding the regulatory determinants that can be used to leverage aNSC biology in brain diseases. Further studies focusing on the adult hippocampal neurogenic niche and considering the elaborate sncRNA functional communication both among them and with target pathways will be critical to profile the full regulatory potential of sncRNAs in this regard, as well as their future applications in the next generation of (RNA) therapies, particularly targeting age-related pathologies.
Author Contributions
CF and ES conceived the article. AP, GT, OA, PB, CG, DD, CF, and ES discussed, wrote and corrected the manuscript. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We apologize to all our colleagues, whose original contributions we have not included in this review, due to space limitations.
Funding
AP, GT, and ES received funding from Alzheimer Nederland (WE.03-2020-04) and Health Holland (LSHM20095). CF, PB, and OA were financed by the European Commission co-fund ERA-NET-NEURON grant EJTC 2016 to CF, Netherlands Organization for Scientific Research (NWO) and Alzheimer Nederland (WE.03-2020-07). OA was financed by a postdoctoral fellowship from the Government of the Basque Country. DD was partly financed by the AIRC-IG 2017 # 20106 grant.
References
- Abiega O., Beccari S., Diaz-Aparicio I., Nadjar A., Layé S., Leyrolle Q., et al. (2016). Neuronal hyperactivity disturbs ATP microgradients, impairs microglial motility and reduces phagocytic receptor expression triggering apoptosis/microglial phagocytosis uncoupling. PLoS Biol. 14:e1002466. 10.1371/journal.pbio.1002466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aimone J., Li Y., Lee S., Clemenson G., Deng W., Gage F. (2014). Regulation and function of adult neurogenesis: from genes to cognition. Physiol. Rev. 94, 991–1026. 10.1152/physrev.00004.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Åkerblom A., Sachdeva A., Barde A., Verp A., Gentner A., Trono A., et al. (2012). MicroRNA-124 is a subventricular zone neuronal fate determinant. J. Neurosci. 32, 8879–8889. 10.1523/JNEUROSCI.0558-12.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandrov P., Zhao Y., Jones B., Bhattacharjee S., Lukiw W. (2013). Expression of the phagocytosis-essential protein TREM2 is down-regulated by an aluminum-induced miRNA-34a in a murine microglial cell line. J. Inorg. Biochem. 128, 267–269. 10.1016/j.jinorgbio.2013.05.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso M. (2009). “Neurogenic Niche,” in Encyclopedia of Neuroscience, eds Binder M. D., Hirokawa N., Windhorst U. (Berlin, Heidelberg: Springer Berlin Heidelberg; ), 2682–2685. [Google Scholar]
- Alvarez-Buylla A., García-Verdugo J., Tramontin A. (2001). A unified hypothesis on the lineage of neural stem cells. Nat. Rev. Neurosci. 2, 287–293. 10.1038/35067582 [DOI] [PubMed] [Google Scholar]
- Arena A., Iyer A. M., Milenkovic I., Kovacs G. G., Ferrer I., Perluigi M., et al. (2017). Developmental expression and dysregulation of miR-146a and miR-155 in Down’s syndrome and mouse models of Down’s syndrome and Alzheimer’s disease. Curr. Alzheimer Res. 14, 1305–1317. 10.2174/1567205014666170706112701 [DOI] [PubMed] [Google Scholar]
- Aronica E., Fluiter K., Iyer A., Zurolo E., Vreijling J., van Vliet E., et al. (2010). Expression pattern of miR-146a, an inflammation-associated microRNA, in experimental and human temporal lobe epilepsy. Eur. J. Neurosci. 31, 1100–1107. 10.1111/j.1460-9568.2010.07122.x [DOI] [PubMed] [Google Scholar]
- Artegiani B., Lyubimova A., Muraro M., van Es J., van Oudenaarden A., Clevers H. (2017). A single-cell RNA sequencing study reveals cellular and molecular dynamics of the hippocampal neurogenic niche. Cell Rep. 21, 3271–3284. 10.1016/j.celrep.2017.11.050 [DOI] [PubMed] [Google Scholar]
- Babcock K., Page J., Fallon J., Webb A. (2021). Adult hippocampal neurogenesis in aging and Alzheimer’s disease. Stem Cell Rep. 16, 681–693. 10.1016/j.stemcr.2021.01.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek D., Villén J., Shin C., Camargo F., Gygi S., Bartel D. (2008). The impact of microRNAs on protein output. Nature 455, 64–71. 10.1038/nature07242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahrini I., Song J., Diez D., Hanayama R. (2015). Neuronal exosomes facilitate synaptic pruning by up-regulating complement factors in microglia. Sci. Rep. 5:7989. 10.1038/srep07989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balu D., Lucki I. (2009). Adult hippocampal neurogenesis: regulation, functional implications and contribution to disease pathology. Neurosci. Biobehav. Rev. 33, 232–252. 10.1016/j.neubiorev.2008.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baptista P., Andrade J. (2018). Adult hippocampal neurogenesis: regulation and possible functional and clinical correlates. Front. Neuroanat. 12:44. 10.3389/fnana.2018.00044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barak B., Shvarts-Serebro I., Modai S., Gilam A., Okun E., Michaelson D., et al. (2013). Opposing actions of environmental enrichment and Alzheimer’s disease on the expression of hippocampal microRNAs in mouse models. Transl. Psychiatry 3:e304. 10.1038/tp.2013.77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barca-Mayo O., De Pietri Tonelli D. (2014). Convergent microRNA actions coordinate neocortical development. Cell. Mol. Life Sci. 71, 2975–2995. 10.1007/s00018-014-1576-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bátiz L. F., Castro M., Burgos P., Velásquez Z., Muñoz R., Lafourcade C., et al. (2016). Exosomes as novel regulators of adult neurogenic niches. Front. Cell. Neurosci. 9:501. 10.3389/fncel.2015.00501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belachew S., Chittajallu R., Aguirre A., Yuan X., Kirby M., Anderson S., et al. (2003). Postnatal NG2 proteoglycan-expressing progenitor cells are intrinsically multipotent and generate functional neurons. J. Cell Biol. 161, 169–186. 10.1083/jcb.200210110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bencurova P., Baloun J., Musilova K., Radova L., Tichy B., Pail M., et al. (2017). MicroRNA and mesial temporal lobe epilepsy with hippocampal sclerosis: whole miRNome profiling of human hippocampus. Epilepsia 58, 1782–1793. 10.1111/epi.13870 [DOI] [PubMed] [Google Scholar]
- Berg D., Bond A., Ming G., Song H. (2018). Radial glial cells in the adult dentate gyrus: what are they and where do they come from? F1000Res. 7:277. 10.12688/f1000research.12684.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyer F., Jadasz J., Samper Agrelo I., Schira-Heinen J., Groh J., Manousi A., et al. (2020). Heterogeneous fate choice of genetically modulated adult neural stem cells in gray and white matter of the central nervous system. Glia 68, 393–406. 10.1002/glia.23724 [DOI] [PubMed] [Google Scholar]
- Bhattacharjee S., Zhao Y., Dua P., Rogaev E., Lukiw W. (2016). microRNA-34a-mediated down-regulation of the microglial-enriched triggering receptor and phagocytosis-sensor TREM2 in age-related macular degeneration. PLoS One 11:e0150211. 10.1371/journal.pone.0150211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bielefeld P., Mooney C., Henshall D., Fitzsimons C. (2017). miRNA-mediated regulation of adult hippocampal neurogenesis; implications for epilepsy. Brain Plast. 3, 43–59. 10.3233/BPL-160036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bielefeld P., Schouten M., Meijer G., Breuk M., Geijtenbeek K., Karayel S., et al. (2019). Co-administration of anti microRNA-124 and -137 oligonucleotides prevents hippocampal neural stem cell loss upon non-convulsive seizures. Front. Mol. Neurosci. 12:31. 10.3389/fnmol.2019.00031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bielefeld P., van Vliet E., Gorter J., Lucassen P., Fitzsimons C. (2014). Different subsets of newborn granule cells: a possible role in epileptogenesis? Eur. J. Neurosci. 39, 1–11. 10.1111/ejn.12387 [DOI] [PubMed] [Google Scholar]
- Bonafina A., Paratcha G., Ledda F. (2020). Deciphering new players in the neurogenic adult hippocampal niche. Front. Cell Dev. Biol. 8:548. 10.3389/fcell.2020.00548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonzano S., Crisci I., Podlesny-Drabiniok A., Rolando C., Krezel W., Studer M., et al. (2018). Neuron-astroglia cell fate decision in the adult mouse hippocampal neurogenic niche is cell-intrinsically controlled by COUP-TFI in vivo. Cell Rep. 24, 329–341. 10.1016/j.celrep.2018.06.044 [DOI] [PubMed] [Google Scholar]
- Bottes S., Jaeger B. N., Pilz G.-A., Jörg D. J., Cole J. D., Kruse M., et al. (2021). Long-term self-renewing stem cells in the adult mouse hippocampus identified by intravital imaging. Nat. Neurosci. 24, 225–233. 10.1038/s41593-020-00759-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandi R., Fabiano M., Giorgi C., Arisi I., La Regina F., Malerba F., et al. (2021). Nerve growth factor neutralization promotes oligodendrogenesis by increasing miR-219a-5p levels. Cells 10:405. 10.3390/cells10020405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun S., Pilz G., Machado R., Moss J., Becher B., Toni N., et al. (2015). Programming hippocampal neural stem/progenitor cells into oligodendrocytes enhances remyelination in the adult brain after injury. Cell Rep. 11, 1679–1685. 10.1016/j.celrep.2015.05.024 [DOI] [PubMed] [Google Scholar]
- Brennan G., Henshall D. (2020). MicroRNAs as regulators of brain function and targets for treatment of epilepsy. Nat. Rev. Neurol. 16, 506–519. 10.1038/s41582-020-0369-8 [DOI] [PubMed] [Google Scholar]
- Brett J., Renault V., Rafalski V., Webb A., Brunet A. (2011). The microRNA cluster miR-106b~25 regulates adult neural stem/progenitor cell proliferation and neuronal differentiation. Aging (Albany. NY) 3, 108–124. 10.18632/aging.100285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooker S., Gobeske K., Chen J., Peng C., Kessler J. (2017). Hippocampal bone morphogenetic protein signaling mediates behavioral effects of antidepressant treatment. Mol. Psychiatry 22, 910–919. 10.1038/mp.2016.160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butovsky O., Jedrychowski M., Cialic R., Krasemann S., Murugaiyan G., Fanek Z., et al. (2015). Targeting miR-155 restores abnormal microglia and attenuates disease in SOD1 mice. Ann. Neurol. 77, 75–99. 10.1002/ana.24304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butovsky O., Ziv Y., Schwartz A., Landa G., Talpalar A., Pluchino S., et al. (2006). Microglia activated by IL-4 or IFN-gamma differentially induce neurogenesis and oligodendrogenesis from adult stem/progenitor cells. Mol. Cell. Neurosci. 31, 149–160. 10.1016/j.mcn.2005.10.006 [DOI] [PubMed] [Google Scholar]
- Carninci P., Kasukawa T., Katayama S., Gough J., Frith M. C., Maeda N., et al. (2005). The transcriptional landscape of the mammalian genome. Science 309, 1559–1563. 10.1126/science.1112014 [DOI] [PubMed] [Google Scholar]
- Cheng X., Li W., Zhao R., Li H., Qin J., Tian M., et al. (2021). The role of hippocampal niche exosomes in rat hippocampal neurogenesis after fimbria-fornix transection. J. Biol. Chem. 296:100188. 10.1074/jbc.RA120.015561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng L., Pastrana E., Tavazoie M., Doetsch F. (2009). miR-124 regulates adult neurogenesis in the subventricular zone stem cell niche. Nat. Neurosci. 12, 399–408. 10.1038/nn.2294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho K. O., Lybrand Z. R., Ito N., Brulet R., Tafacory F., Zhang L., et al. (2015). Aberrant hippocampal neurogenesis contributes to epilepsy and associated cognitive decline. Nat. Commun. 6:6606. 10.1038/ncomms7606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi S., Bylykbashi E., Chatila Z., Lee S., Pulli B., Clemenson G., et al. (2018). Combined adult neurogenesis and BDNF mimic exercise effects on cognition in an Alzheimer’s mouse model. Science 361:eaan8821. 10.1126/science.aan8821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conaco C., Otto S., Han J., Mandel G. (2006). Reciprocal actions of REST and a microRNA promote neuronal identity. Proc. Natl. Acad. Sci. U. S. A. 103, 2422–2427. 10.1073/pnas.0511041103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czech B., Munafò M., Ciabrelli F., Eastwood E., Fabry M., Kneuss E., et al. (2018). piRNA-guided genome defense: from biogenesis to silencing. Annu. Rev. Genet. 52, 131–157. 10.1146/annurev-genet-120417-031441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dashtipour K., Tran P., Okazaki M., Nadler J., Ribak C. (2001). Ultrastructural features and synaptic connections of hilar ectopic granule cells in the rat dentate gyrus are different from those of granule cells in the granule cell layer. Brain Res. 890, 261–271. 10.1016/s0006-8993(00)03119-x [DOI] [PubMed] [Google Scholar]
- Dawson M., Levine J., Reynolds R. (2000). NG2-expressing cells in the central nervous system: are they oligodendroglial progenitors? J. Neurosci. Res. 61, 471–479. [DOI] [PubMed] [Google Scholar]
- Delgado A., Maldonado-Soto A., Silva-Vargas V., Mizrak D., von Känel T., Tan K., et al. (2021). Release of stem cells from quiescence reveals gliogenic domains in the adult mouse brain. Science 372, 1205–1209. 10.1126/science.abg8467 [DOI] [PubMed] [Google Scholar]
- Denoth-Lippuner A., Jessberger S. (2021). Formation and integration of new neurons in the adult hippocampus. Nat. Rev. Neurosci. 22, 223–236. 10.1038/s41583-021-00433-z [DOI] [PubMed] [Google Scholar]
- Dharap A., Nakka C., Vemuganti R. (2011). Altered expression of PIWI RNA in the rat brain after transient focal ischemia. Stroke 42, 1105–1109. 10.1161/STROKEAHA.110.598391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-Aparicio I., Paris I., Sierra-Torre V., Plaza-Zabala A., Rodríguez-Iglesias N., Márquez-Ropero Maletic-Savatic M., et al. (2020). Microglia Actively Remodel Adult Hippocampal Neurogenesis through the Phagocytosis Secretome. J. Neurosci. 40, 1453–1482. 10.1523/JNEUROSCI.0993-19.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doetsch F., García-Verdugo J., Alvarez-Buylla A. (1999). Regeneration of a germinal layer in the adult mammalian brain. Proc. Natl. Acad. Sci. U S A 96, 11619–11624. 10.1073/pnas.96.20.11619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle M., Jaskiewicz L., Filipowicz W. (2012). Dicer proteins and their role in gene silencing pathways. Enzymes 32, 1–35. 10.1016/B978-0-12-404741-9.00001-5 [DOI] [Google Scholar]
- Duan X., Chang J., Ge S., Faulkner R., Kim J., Kitabatake Y., et al. (2007). Disrupted-In-Schizophrenia 1 regulates integration of newly generated neurons in the adult brain. Cell 130, 1146–1158. 10.1016/j.cell.2007.07.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dugas J., Cuellar T., Scholze A., Ason B., Ibrahim A., Emery B., et al. (2010). Dicer1 and miR-219 Are required for normal oligodendrocyte differentiation and myelination. Neuron 65, 597–611. 10.1016/j.neuron.2010.01.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunham I., Kundaje A., Aldred S. F., Collins P. J., Davis C. A., Doyle F., et al. (2012). An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74. 10.1038/nature11247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duque A., Spector R. (2019). A balanced evaluation of the evidence for adult neurogenesis in humans: implication for neuropsychiatric disorders. Brain Struct. Funct. 224, 2281–2295. 10.1007/s00429-019-01917-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehm O., Göritz C., Covic M., Schäffner I., Schwarz T. J., Karaca E., et al. (2010). RBPJκ-dependent signaling is essential for long-term maintenance of neural stem cells in the adult hippocampus. J. Neurosci. 30, 13794–13807. 10.1523/JNEUROSCI.1567-10.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekdahl C., Claasen J., Bonde S., Kokaia Z., Lindvall O. (2003). Inflammation is detrimental for neurogenesis in adult brain. Proc. Natl. Acad. Sci. U. S. A. 100, 13632–13637. 10.1073/pnas.2234031100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekdahl C., Kokaia Z., Lindvall O. (2009). Brain inflammation and adult neurogenesis: the dual role of microglia. Neuroscience 158, 1021–1029. 10.1016/j.neuroscience.2008.06.052 [DOI] [PubMed] [Google Scholar]
- Encinas J., Fitzsimons C. (2017). Gene regulation in adult neural stem cells. Current challenges and possible applications. Adv. Drug Deliv. Rev. 120, 118–132. 10.1016/j.addr.2017.07.016 [DOI] [PubMed] [Google Scholar]
- Encinas J., Michurina T., Peunova N., Park J., Tordo J., Peterson D., et al. (2011). Division-coupled astrocytic differentiation and age-related depletion of neural stem cells in the adult hippocampus. Cell Stem Cell 8, 566–579. 10.1016/j.stem.2011.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Encinas J. M., Vaahtokari A., Enikolopov G. (2006). Fluoxetine targets early progenitor cells in the adult brain. Proc. Natl. Acad. Sci. U S A 103, 8233–8238. 10.1073/pnas.0601992103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagan S., Helm M., Prehn J. (2021). tRNA-derived fragments: A new class of non-coding RNA with key roles in nervous system function and dysfunction. Prog. Neurobiol. 205:102118. 10.1016/j.pneurobio.2021.102118 [DOI] [PubMed] [Google Scholar]
- Faigle R., Song H. (2013). Signaling mechanisms regulating adult neural stem cells and neurogenesis. Biochim. Biophys. Acta 1830, 2435–2448. 10.1016/j.bbagen.2012.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan H., Chen L., Qu X., Ren C., Wu X., Dong F., et al. (2017). Transplanted miR-219-overexpressing oligodendrocyte precursor cells promoted remyelination and improved functional recovery in a chronic demyelinated model. Sci. Rep. 7:41407. 10.1038/srep41407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ffrench-Constant C., Raff M. (1986). Proliferating bipotential glial progenitor cells in adult rat optic nerve. Nature 319, 499–502. 10.1038/319499a0 [DOI] [PubMed] [Google Scholar]
- Fitzsimons C., van Bodegraven E., Schouten M., Lardenoije R., Kompotis K., Kenis G., et al. (2014). Epigenetic regulation of adult neural stem cells: implications for Alzheimer’s disease. Mol. Neurodegener. 9:25. 10.1186/1750-1326-9-25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzsimons C. P., van Hooijdonk L. W. A., Schouten M., Zalachoras I., Brinks V., Zheng T., et al. (2013). Knockdown of the glucocorticoid receptor alters functional integration of newborn neurons in the adult hippocampus and impairs fear-motivated behavior. Mol. Psychiatry 18, 993–1005. 10.1038/mp.2012.123 [DOI] [PubMed] [Google Scholar]
- Flynn R., Pedram K., Malaker S., Batista P., Smith B., Johnson A., et al. (2021). Small RNAs are modified with N-glycans and displayed on the surface of living cells. Cell 184, 3109–3124.e22. 10.1016/j.cell.2021.04.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke K., Otto W., Johannes S., Baumgart J., Nitsch R., Schumacher S. (2012). miR-124-regulated RhoG reduces neuronal process complexity via ELMO/Dock180/Rac1 and Cdc42 signalling. EMBO J. 31, 2908–2921. 10.1038/emboj.2012.130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gage F. (2019). Adult neurogenesis in mammals. Science 364, 827–828. 10.1126/science.aav6885 [DOI] [PubMed] [Google Scholar]
- Gasperini C., Tuntevski K., Pelizzoli R., Van A. L., Mangoni D., Cossu R. M., et al. (2021). The piRNA pathway sustains adult neurogenesis by reducing protein synthesis and cellular senescence. bioRxiv [Preprint]. 10.1101/2020.09.15.297739 [DOI] [Google Scholar]
- Ghasemi-Kasman M., Shojaei A., Gol M., Moghadamnia A., Baharvand H., Javan M. (2018). miR-302/367-induced neurons reduce behavioral impairment in an experimental model of Alzheimer’s disease. Mol. Cell. Neurosci. 86, 50–57. 10.1016/j.mcn.2017.11.012 [DOI] [PubMed] [Google Scholar]
- Ghoshes Y., Seridis L., Ravasis T., Takahas H., Orlands V., Carnins P., et al. (2016). Characterization of piRNAs across postnatal development in mouse brain. Sci. Rep. 6:25039. 10.1038/srep25039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giachino C., Barz M., Tchorz J., Tome M., Gassmann M., Bischofberger J., et al. (2014). GABA suppresses neurogenesis in the adult hippocampus through GABAB receptors. Development 141, 83–90. 10.1242/dev.102608 [DOI] [PubMed] [Google Scholar]
- Gobeske K., Das S., Bonaguidi M., Weiss C., Radulovic J., Disterhoft J., et al. (2009). BMP signaling mediates effects of exercise on hippocampal neurogenesis and cognition in mice. PLoS One 4:e7506. 10.1371/journal.pone.0007506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths B., Ouyang Y., Xu L., Sun X., Giffard R., Stary C. (2019). Postinjury inhibition of miR-181a promotes restoration of hippocampal CA1 neurons after transient forebrain ischemia in rats. eNeuro 6:ENEURO.0002-19.2019. 10.1523/ENEURO.0002-19.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y., Hong W., Wang X., Zhang P., Körner H., Tu J., et al. (2019). MicroRNAs in microglia: how do microRNAs affect activation, inflammation, polarization of microglia and mediate the interaction between microglia and glioma? Front. Mol. Neurosci. 12:125. 10.3389/fnmol.2019.00125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J., Kim H., Schafer S., Paquola A., Clemenson G., Toda T., et al. (2016). Functional implications of miR-19 in the migration of newborn neurons in the adult brain. Neuron 91, 79–89. 10.1016/j.neuron.2016.05.034 [DOI] [PubMed] [Google Scholar]
- Hanson N., Owens M., Nemeroff C. (2011). Depression, antidepressants and neurogenesis: a critical reappraisal. Neuropsychopharmacology 36, 2589–2602. 10.1038/npp.2011.220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harraz M., Xu J., Guiberson N., Dawson T., Dawson V. (2014). MiR-223 regulates the differentiation of immature neurons. Mol. Cell. Ther. 2:18. 10.1186/2052-8426-2-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris L., Rigo P., Stiehl T., Gaber Z. B., Austin S. H. L., Masdeu M. D. M., et al. (2021). Coordinated changes in cellular behavior ensure the lifelong maintenance of the hippocampal stem cell population. Cell Stem Cell 28, 863–876.e6. 10.1016/j.stem.2021.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill A., Sahay A., Hen R. (2015). Increasing adult hippocampal neurogenesis is sufficient to reduce anxiety and depression-like behaviors. Neuropsychopharmacology 40, 2368–2378. 10.1038/npp.2015.85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hombach S., Kretz M. (2016). Non-coding RNAs: classification, biology and functioning. Adv. Exp. Med. Biol. 937, 3–17. 10.1007/978-3-319-42059-2_1 [DOI] [PubMed] [Google Scholar]
- Huang X., Wong G. (2021). An old weapon with a new function: PIWI-interacting RNAs in neurodegenerative diseases. Transl. Neurodegener. 10:9. 10.1186/s40035-021-00233-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutvagner G., Simard M. (2008). Argonaute proteins: key players in RNA silencing. Nat. Rev. Mol. Cell Biol. 9, 22–32. 10.1038/nrm2321 [DOI] [PubMed] [Google Scholar]
- Imayoshi I., Sakamoto M., Yamaguchi M., Mori K., Kageyama R. (2010). Essential roles of Notch signaling in maintenance of neural stem cells in developing and adult brains. J. Neurosci. 30, 3489–3498. 10.1523/JNEUROSCI.4987-09.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang M. H., Bonaguidi M. A., Kitabatake Y., Sun J., Song J., Kang E., et al. (2013). Secreted frizzled-related protein 3 regulates activity-dependent adult hippocampal neurogenesis. Cell Stem Cell 12, 215–223. 10.1016/j.stem.2012.11.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessberger S., Toni N., Clemenson G., Ray J., Gage F. (2008). Directed differentiation of hippocampal stem/progenitor cells in the adult brain. Nat. Neurosci. 11, 888–893. 10.1038/nn.2148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin J., Kim S., Liu X., Zhang H., Zhang C., Seo J., et al. (2016). miR-17–92 cluster regulates adult hippocampal neurogenesis, anxiety and depression. Cell Rep. 16, 1653–1663. 10.1016/j.celrep.2016.06.101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang E., Wen Z., Song H., Christian K., Ming G. (2016). Adult neurogenesis and psychiatric disorders. Cold Spring Harb. Perspect. Biol. 8:a019026. 10.1101/cshperspect.a019026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz S., Cussigh D., Urbán N., Blomfield I., Guillemot F., Bally-Cuif L., et al. (2016). A nuclear role for miR-9 and argonaute proteins in balancing quiescent and activated neural stem cell states. Cell Rep. 17, 1383–1398. 10.1016/j.celrep.2016.09.088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur H., Sarmah D., Saraf J., Vats K., Kalia K., Borah A., et al. (2018). Noncoding RNAs in ischemic stroke: time to translate. Ann. N. Y. Acad. Sci. 1421, 19–36. 10.1111/nyas.13612 [DOI] [PubMed] [Google Scholar]
- Keam S., Young P., McCorkindale A., Dang T., Clancy J., Humphreys D., et al. (2014). The human Piwi protein Hiwi2 associates with tRNA-derived piRNAs in somatic cells. Nucleic Acids Res. 42, 8984–8995. 10.1093/nar/gku620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kempermann G. (2011). The pessimist’s and optimist’s views of adult neurogenesis. Cell 145, 1009–1011. 10.1016/j.cell.2011.06.011 [DOI] [PubMed] [Google Scholar]
- Kempermann G. (2015). Adult neurogenesis: an evolutionary perspective. Cold Spring Harb. Perspect. Biol. 8:a018986. 10.1101/cshperspect.a018986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kempermann G., Gage F., Aigner L., Song H., Curtis M., Thuret S., et al. (2018). Human adult neurogenesis: evidence and remaining questions. Cell Stem Cell 23, 25–30. 10.1016/j.stem.2018.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kempermann G., Jessberger S., Steiner B., Kronenberg G. (2004). Milestones of neuronal development in the adult hippocampus. Trends Neurosci. 27, 447–452. 10.1016/j.tins.2004.05.013 [DOI] [PubMed] [Google Scholar]
- Khandelwal N., Dey S., Chakravarty S., Kumar A. (2019). miR-30 family miRNAs mediate the effect of chronic social defeat stress on hippocampal neurogenesis in mouse depression model. Front. Mol. Neurosci. 12:188. 10.3389/fnmol.2019.00188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K. (2019). PIWI proteins and piRNAs in the nervous system. Mol. Cells 42, 828–835. 10.14348/molcells.2019.0241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T., Chen L., Ge S. (2021). The interplay of neurovasculature and adult hippocampal neurogenesis. Neurosci. Lett. 760:136071. 10.1016/j.neulet.2021.136071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Cho J., Kang H. (2019). Protective effect of exercise training against the progression of Alzheimer’s disease in 3xTg-AD mice. Behav. Brain Res. 374:112105. 10.1016/j.bbr.2019.112105 [DOI] [PubMed] [Google Scholar]
- Kim J., Duan X., Liu C., Jang M., Guo J., Pow-anpongkul N., et al. (2009). DISC1 regulates new neuron development in the adult brain via modulation of AKT-mTOR signaling through KIAA1212. Neuron 63, 761–773. 10.1016/j.neuron.2009.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Hwang I., Muller F., Paik J. (2015). Functional regulation of FoxO1 in neural stem cell differentiation. Cell Death Differ. 22, 2034–2045. 10.1038/cdd.2015.123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K., Tang N., Andrusiak M., Wu Z., Chisholm A., Jin Y. (2018). A neuronal piRNA pathway inhibits axon regeneration in C. elegans. Neuron 97, 511–519.e6. 10.1016/j.neuron.2018.01.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klempin F., Kronenberg G., Cheung G., Kettenmann H., Kempermann G. (2011). Properties of doublecortin-(DCX)-expressing cells in the piriform cortex compared to the neurogenic dentate gyrus of adult mice. PLoS One 6:e25760. 10.1371/journal.pone.0025760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korotkov A., Broekaart D., Banchaewa L., Pustjens B., van Scheppingen J., Anink J., et al. (2020). microRNA-132 is overexpressed in glia in temporal lobe epilepsy and reduces the expression of pro-epileptogenic factors in human cultured astrocytes. Glia 68, 60–75. 10.1002/glia.23700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreisel T., Wolf B., Keshet E., Licht T. (2019). Unique role for dentate gyrus microglia in neuroblast survival and in VEGF-induced activation. Glia 67, 594–618. 10.1002/glia.23505 [DOI] [PubMed] [Google Scholar]
- Kuhn H., Dickinson-Anson H., Gage F. (1996). Neurogenesis in the dentate gyrus of the adult rat: age-related decrease of neuronal progenitor proliferation. J. Neurosci. 16, 2027–2033. 10.1523/JNEUROSCI.16-06-02027.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A., Bhatia H., de Oliveira A., Fiebich B. (2015). microRNA-26a modulates inflammatory response induced by toll-like receptor 4 stimulation in microglia. J. Neurochem. 135, 1189–1202. 10.1111/jnc.13364 [DOI] [PubMed] [Google Scholar]
- Kuwabara T., Hsieh J., Nakashima K., Taira K., Gage F. (2004). A small modulatory dsRNA specifies the fate of adult neural stem cells. Cell 116, 779–793. 10.1016/s0092-8674(04)00248-x [DOI] [PubMed] [Google Scholar]
- Lagos-Quintana M., Rauhut R., Yalcin A., Meyer J., Lendeckel W., Tuschl T. (2002). Identification of tissue-specific microRNAs from mouse. Curr. Biol. 12, 735–739. 10.1016/s0960-9822(02)00809-6 [DOI] [PubMed] [Google Scholar]
- Lau P., Verrier J., Nielsen J., Johnson K., Notterpek L., Hudson L. (2008). Identification of dynamically regulated microRNA and mRNA networks in developing oligodendrocytes. J. Neurosci. 28, 11720–11730. 10.1523/JNEUROSCI.1932-08.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee E., Banerjee S., Zhou H., Jammalamadaka A., Arcila M., Manjunath B., et al. (2011). Identification of piRNAs in the central nervous system. RNA 17, 1090–1099. 10.1261/rna.2565011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. W., Haditsch U., Cord B., Guzman R., Kim S., Boettcher C., et al. (2013). Absence of CCL2 is sufficient to restore hippocampal neurogenesis following cranial irradiation. Brain. Behav. Immun. 30, 33–44. 10.1016/j.bbi.2012.09.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leighton L., Wei W., Marshall P., Ratnu V., Li X., Zajaczkowski E., et al. (2019). Disrupting the hippocampal Piwi pathway enhances contextual fear memory in mice. Neurobiol. Learn. Mem. 161, 202–209. 10.1016/j.nlm.2019.04.002 [DOI] [PubMed] [Google Scholar]
- Lepko T., Pusch M., Müller T., Schulte D., Ehses J., Kiebler M., et al. (2019). Choroid plexus-derived miR-204 regulates the number of quiescent neural stem cells in the adult brain. EMBO J. 38:e100481. 10.15252/embj.2018100481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Clevers H. (2010). Coexistence of quiescent and active adult stem cells in mammals. Science 327, 542–545. 10.1126/science.1180794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Guo W. (2021). Neural stem cell niche and adult neurogenesis. Neuroscientist 27, 235–245. 10.1177/1073858420939034 [DOI] [PubMed] [Google Scholar]
- Li F., Zhou M., Liu N., Yang Y., Xing H., Lu Y., et al. (2019). MicroRNA-219 inhibits proliferation and induces differentiation of oligodendrocyte precursor cells after contusion spinal cord injury in rats. Neural Plast. 2019:9610687. 10.1155/2019/9610687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licht T., Keshet E. (2015). The vascular niche in adult neurogenesis. Mech. Dev. 138, 56–62. 10.1016/j.mod.2015.06.001 [DOI] [PubMed] [Google Scholar]
- Lim J., Bang Y., Choi H. (2018). Abnormal hippocampal neurogenesis in Parkinson’s disease: relevance to a new therapeutic target for depression with Parkinson’s disease. Arch. Pharm. Res. 41, 943–954. 10.1007/s12272-018-1063-x [DOI] [PubMed] [Google Scholar]
- Lim D. A., Huang Y. C., Alvarez-Buylla A. (2007). The adult neural stem cell niche: lessons for future neural cell replacement strategies. Neurosurg. Clin. N. Am. 18, 81–92. 10.1016/j.nec.2006.10.002 [DOI] [PubMed] [Google Scholar]
- Liu X., Chopp M., Pan W., Wang X., Fan B., Zhang Y., et al. (2017). MicroRNA-146a promotes oligodendrogenesis in stroke. Mol. Neurobiol. 54, 227–237. 10.1007/s12035-015-9655-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C., Teng Z., McQuate A., Jobe E., Christ C., von Hoyningen-Huene S., et al. (2013). An epigenetic feedback regulatory loop involving microRNA-195 and MBD1 governs neural stem cell differentiation. PLoS One 8:e51436. 10.1371/journal.pone.0051436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C., Teng Z., Santistevan N., Szulwach K., Guo W., Jin P., et al. (2010). Epigenetic regulation of miR-184 by MBD1 governs neural stem cell proliferation and differentiation. Cell Stem Cell 6, 433–444. 10.1016/j.stem.2010.02.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopatina O. L., Malinovskaya N. A., Komleva Y. K., Gorina Y. V., Shuvaev A. N., Olovyannikova R. Y., et al. (2019). Excitation/inhibition imbalance and impaired neurogenesis in neurodevelopmental and neurodegenerative disorders. Rev. Neurosci. 30, 807–820. 10.1515/revneuro-2019-0014 [DOI] [PubMed] [Google Scholar]
- Lopez-Verrilli M., Court F. (2013). Exosomes: mediators of communication in eukaryotes. Biol. Res. 46, 5–11. 10.4067/S0716-97602013000100001 [DOI] [PubMed] [Google Scholar]
- Losurdo M., Grilli M. (2020). Extracellular vesicles, influential players of intercellular communication within adult neurogenic niches. Int. J. Mol. Sci. 21:8819. 10.3390/ijms21228819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louw A., Kolar M., Novikova L., Kingham P., Wiberg M., Kjems J., et al. (2016). Chitosan polyplex mediated delivery of miRNA-124 reduces activation of microglial cells in vitro and in rat models of spinal cord injury. Nanomedicine 12, 643–653. 10.1016/j.nano.2015.10.011 [DOI] [PubMed] [Google Scholar]
- Luarte A., Bátiz L., Wyneken U., Lafourcade C. (2016). Potential therapies by stem cell-derived exosomes in CNS diseases: focusing on the neurogenic niche. Stem Cells Int. 2016:5736059. 10.1155/2016/5736059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucassen P., Fitzsimons C., Salta E., Maletic-Savatic M. (2020). Adult neurogenesis, human after all (again): classic, optimized and future approaches. Behav. Brain Res. 381:112458. 10.1016/j.bbr.2019.112458 [DOI] [PubMed] [Google Scholar]
- Lugert S., Basak O., Knuckles P., Haussler U., Fabel K., Götz M., et al. (2010). Quiescent and active hippocampal neural stem cells with distinct morphologies respond selectively to physiological and pathological stimuli and aging. Cell Stem Cell 6, 445–456. 10.1016/j.stem.2010.03.017 [DOI] [PubMed] [Google Scholar]
- Luikart B., Bensen A., Washburn E., Perederiy J., Su K., Li Y., et al. (2011). miR-132 mediates the integration of newborn neurons into the adult dentate gyrus. PLoS One 6:e19077. 10.1371/journal.pone.0019077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv Y., Ou-Yang A., Fu L. (2017). MicroRNA-27a negatively modulates the inflammatory response in lipopolysaccharide-stimulated microglia by targeting TLR4 and IRAK4. Cell. Mol. Neurobiol. 37, 195–210. 10.1007/s10571-016-0361-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma D., Jang M., Guo J., Kitabatake Y., Chang M., Pow-Anpongkul N., et al. (2009). Neuronal activity-induced Gadd45b promotes epigenetic DNA demethylation and adult neurogenesis. Science 323, 1074–1077. 10.1126/science.1166859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magill S., Cambronne X., Luikart B., Lioy D., Leighton B., Westbrook G., et al. (2010). microRNA-132 regulates dendritic growth and arborization of newborn neurons in the adult hippocampus. Proc. Natl. Acad. Sci. U S A 107, 20382–20387. 10.1073/pnas.1015691107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques S., Zeisel A., Codeluppi S., van Bruggen D., Mendanha Falcão A., Xiao L., et al. (2016). Oligodendrocyte heterogeneity in the mouse juvenile and adult central nervous system. Science 352, 1326–1329. 10.1126/science.aaf6463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKiernan R. C., Jimenez-Mateos E. M., Bray I., Engel T., Brennan G. P., Sano T., et al. (2012). Reduced mature MicroRNA levels in association with dicer loss in human temporal lobe epilepsy with hippocampal sclerosis. PLoS One 7:35921. 10.1371/journal.pone.0035921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta S., Chokkalla A., Vemuganti R. (2021). Noncoding RNA crosstalk in brain health and diseases. Neurochem. Int. 149:105139. 10.1016/j.neuint.2021.105139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mira H., Andreu Z., Suh H., Chichung Lie D., Jessberger S., Consiglio A., et al. (2010). Signaling through BMPR-IA regulates quiescence and long-term activity of neural stem cells in the adult hippocampus. Cell Stem Cell 7, 78–89. 10.1016/j.stem.2010.04.016 [DOI] [PubMed] [Google Scholar]
- Monje M., Toda H., Palmer T. (2003). Inflammatory blockade restores adult hippocampal neurogenesis. Science 302, 1760–1765. 10.1126/science.1088417 [DOI] [PubMed] [Google Scholar]
- Moreno-Jiménez E., Flor-García M., Terreros-Roncal J., Rábano A., Cafini F., Pallas-Bazarra N., et al. (2019). Adult hippocampal neurogenesis is abundant in neurologically healthy subjects and drops sharply in patients with Alzheimer’s disease. Nat. Med. 25, 554–560. 10.1038/s41591-019-0375-9 [DOI] [PubMed] [Google Scholar]
- Murphy B., Hofacer R., Faulkner C., Loepke A., Danzer S. (2012). Abnormalities of granule cell dendritic structure are a prominent feature of the intrahippocampal kainic acid model of epilepsy despite reduced postinjury neurogenesis. Epilepsia 53, 908–921. 10.1111/j.1528-1167.2012.03463.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy B., Pun R., Yin H., Faulkner C., Loepke A., Danzer S. (2011). Heterogeneous integration of adult-generated granule cells into the epileptic brain. J. Neurosci. 31, 105–117. 10.1523/JNEUROSCI.2728-10.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandi S., Chandramohan D., Fioriti L., Melnick A., Hébert J., Mason C., et al. (2016). Roles for small noncoding RNAs in silencing of retrotransposons in the mammalian brain. Proc. Natl. Acad. Sci. U S A 113, 12697–12707. 10.1073/pnas.1609287113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nahid M., Satoh M., Chan E. (2011). Mechanistic role of microRNA-146a in endotoxin-induced differential cross-regulation of TLR signaling. J. Immunol. 186, 1723–1734. 10.4049/jimmunol.1002311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishini J., Kii I., Chadi K., Morrii S. (2008). Hmga2 promotes neural stem cell self-renewal in young but not old mice by reducing p16Ink4a and p19Arf Expression. Cell 135, 227–239. 10.1016/j.cell.2008.09.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuzziello N., Liguori M. (2019). The MicroRNA centrism in the orchestration of neuroinflammation in neurodegenerative diseases. Cells 8:1193. 10.3390/cells8101193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien K., Breyne K., Ughetto S., Laurent L., Breakefield X. (2020). RNA delivery by extracellular vesicles in mammalian cells and its applications. Nat. Rev. Mol. Cell Biol. 21, 585–606. 10.1016/j.jmbbm.2021.105020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottone C., Krusche B., Whitby A., Clements M., Quadrato G., Pitulescu M., et al. (2014). Direct cell-cell contact with the vascular niche maintains quiescent neural stem cells. Nat. Cell Biol. 16, 1045–1056. 10.1038/ncb3045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozata D., Gainetdinov I., Zoch A., O’Carroll D., Zamore P. (2019). PIWI-interacting RNAs: small RNAs with big functions. Nat. Rev. Genet. 20, 89–108. 10.1038/s41576-018-0073-3 [DOI] [PubMed] [Google Scholar]
- Paik J., Ding Z., Narurkar R., Ramkissoon S., Muller F., Kamoun W., et al. (2009). FoxOs cooperatively regulate diverse pathways governing neural stem cell homeostasis. Cell Stem Cell 5, 540–553. 10.1016/j.stem.2009.09.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan W., Chopp M., Fan B., Zhang R., Wang X., Hu J., et al. (2019). Ablation of the microRNA-17–92 cluster in neural stem cells diminishes adult hippocampal neurogenesis and cognitive function. FASEB J. 33, 5257–5267. 10.1096/fj.201801019R [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paolicelli R., Bolasco G., Pagani F., Maggi L., Scianni M., Panzanelli P., et al. (2011). Synaptic pruning by microglia is necessary for normal brain development. Science 333, 1456–1458. 10.1126/science.1202529 [DOI] [PubMed] [Google Scholar]
- Parent J., Yu T., Leibowitz R., Geschwind D., Sloviter R., Lowenstein D. (1997). Dentate granule cell neurogenesis is increased by seizures and contributes to aberrant network reorganization in the adult rat hippocampus. J. Neurosci. 17, 3727–3738. 10.1523/JNEUROSCI.17-10-03727.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathania M., Torres-Reveron J., Yan L., Kimura T., Lin T., Gordon V., et al. (2012). miR-132 enhances dendritic morphogenesis, spine density, synaptic integration and survival of newborn olfactory bulb neurons. PLoS One 7:e38174. 10.1371/journal.pone.0038174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perera B., Tsai Z., Colwell M., Jones T., Goodrich J., Wang K., et al. (2019). Somatic expression of piRNA and associated machinery in the mouse identifies short, tissue-specific piRNA. Epigenetics 14, 504–521. 10.1080/15592294.2019.1600389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Rodríguez D., Blanco-Luquin I., Mendioroz M. (2021). The participation of microglia in neurogenesis: a review. Brain Sci. 11:658. 10.3390/brainsci11050658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petri R., Pircs K., Jönsson M., Åkerblom M., Brattås P., Klussendorf T., et al. (2017). let-7 regulates radial migration of new-born neurons through positive regulation of autophagy. EMBO J. 36, 1379–1391. 10.15252/embj.201695235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phay M., Kim H., Yoo S. (2018). Analysis of piRNA-like small non-coding RNAs present in axons of adult sensory neurons. Mol. Neurobiol. 55, 483–494. 10.1007/s12035-016-0340-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilz G., Bottes S., Betizeau M., Jörg D., Carta S., Simons B., et al. (2018). Live imaging of neurogenesis in the adult mouse hippocampus. Science 359, 658–662. 10.1126/science.aao5056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Planchez B., Lagunas N., Le Guisquet A., Legrand M., Surget A., Hen R., et al. (2021). Increasing adult hippocampal neurogenesis promotes resilience in a mouse model of depression. Cells 10:972. 10.3390/cells10050972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponomarev E. D., Veremeyko T., Barteneva N., Krichevsky A. M., Weiner H. L. (2011). MicroRNA-124 promotes microglia quiescence and suppresses EAE by deactivating macrophages via the C/EBP-α-PU.1 pathway. Nat. Med. 17, 64–70. 10.1038/nm.2266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pons-Espinal M., de Luca E., Marzi M., Beckervordersandforth R., Armirotti A., Nicassio F., et al. (2017). Synergic functions of miRNAs determine neuronal fate of adult neural stem cells. Stem Cell Rep. 8, 1046–1061. 10.1016/j.stemcr.2017.02.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pons-Espinal M., Gasperini C., Marzi M., Braccia C., Armirotti A., Pötzsch A., et al. (2019). MiR-135a-5p is critical for exercise-induced adult neurogenesis. Stem Cell Rep. 12, 1298–1312. 10.1016/j.stemcr.2019.04.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porlan E., Martí-Prado B., Morante-Redolat J., Consiglio A., Delgado A., Kypta R., et al. (2014). MT5-MMP regulates adult neural stem cell functional quiescence through the cleavage of N-cadherin. Nat. Cell Biol. 16, 629–638. 10.1038/ncb2993 [DOI] [PubMed] [Google Scholar]
- Renault V., Rafalski V., Morgan A., Salih D., Brett J., Webb A., et al. (2009). FoxO3 regulates neural stem cell homeostasis. Cell Stem Cell 5, 527–539. 10.1016/j.stem.2009.09.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risbud R. M., Porter B. E. (2013). Changes in MicroRNA expression in the whole hippocampus and hippocampal synaptoneurosome fraction following pilocarpine induced status epilepticus. PLoS One 8:e53464. 10.1371/journal.pone.0053464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolando C., Erni Engler A., Grison A., Beattie R., Erni Engler A., Gokhale P., et al. (2016). Multipotency of adult hippocampal nscs in vivo is restricted by drosha/NFIB. Cell Stem Cell 19, 653–662. 10.1016/j.stem.2016.07.003 [DOI] [PubMed] [Google Scholar]
- Saba R., Gushue S., Huzarewich R., Manguiat K., Medina S., Robertson C., et al. (2012). MicroRNA 146a (miR-146a) is over-expressed during prion disease and modulates the innate immune response and the microglial activation state. PLoS One 7:e30832. 10.1371/journal.pone.0030832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saab B., Mansuy I. (2014). Neuroepigenetics of memory formation and impairment: the role of microRNAs. Neuropharmacology 80, 61–69. 10.1016/j.neuropharm.2014.01.026 [DOI] [PubMed] [Google Scholar]
- Saba R., Schratt G. (2010). MicroRNAs in neuronal development, function and dysfunction. Brain Res. 1338, 3–13. 10.1016/j.brainres.2010.03.107 [DOI] [PubMed] [Google Scholar]
- Salta E., De Strooper B. (2012). Non-coding RNAs with essential roles in neurodegenerative disorders. Lancet Neurol. 11, 189–200. 10.1016/S1474-4422(11)70286-1 [DOI] [PubMed] [Google Scholar]
- Salta E., De Strooper B. (2017a). microRNA-132: a key noncoding RNA operating in the cellular phase of Alzheimer’s disease. FASEB J. 31, 424–433. 10.1096/fj.201601308 [DOI] [PubMed] [Google Scholar]
- Salta E., De Strooper B. (2017b). Noncoding RNAs in neurodegeneration. Nat. Rev. Neurosci. 18, 627–640. 10.1038/nrn.2017.90 [DOI] [PubMed] [Google Scholar]
- Salvatori B., Biscarini S., Morlando M. (2020). Non-coding RNAs in nervous system development and disease. Front. Cell Dev. Biol. 8:273. 10.3389/fcell.2020.00273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos G., Barateiro A., Brites D., Fernandes A. (2020). S100B impairs oligodendrogenesis and myelin repair following demyelination through RAGE engagement. Front. Cell. Neurosci. 14:279. 10.3389/fncel.2020.00279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santra M., Chopp M., Santra S., Nallani A., Vyas S., Zhang Z., et al. (2016). Thymosin beta 4 up-regulates miR-200a expression and induces differentiation and survival of rat brain progenitor cells. J. Neurochem. 136, 118–132. 10.1111/jnc.13394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santra M., Zhang Z., Yang J., Santra S., Santra S., Chopp M., et al. (2014). Thymosin β4 up-regulation of microRNA-146a promotes oligodendrocyte differentiation and suppression of the toll-like proinflammatory pathway. J. Biol. Chem. 289, 19508–19518. 10.1074/jbc.M113.529966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saraiva C., Paiva J., Santos T., Ferreira L., Bernardino L. (2016). MicroRNA-124 loaded nanoparticles enhance brain repair in Parkinson’s disease. J. Control. Release 235, 291–305. 10.1016/j.jconrel.2016.06.005 [DOI] [PubMed] [Google Scholar]
- Saurat N., Andersson T., Vasistha N., Molnár Z., Livesey F. (2013). Dicer is required for neural stem cell multipotency and lineage progression during cerebral cortex development. Neural. Dev. 8:14. 10.1186/1749-8104-8-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharfman H., Goodman J., Sollas A. (2000). Granule-like neurons at the hilar/CA3 border after status epilepticus and their synchrony with area CA3 pyramidal cells: functional implications of seizure-induced neurogenesis. J. Neurosci. 20, 6144–6158. 10.1523/JNEUROSCI.20-16-06144.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schofield R. (1978). The relationship between the spleen colony-forming cell and the haemopoietic stem cell. a hypothesis. Blood Cells 4, 7–25. [PubMed] [Google Scholar]
- Schouten M., Bielefeld P., Garcia-Corzo L., Passchier E., Gradari S., Jungenitz T., et al. (2020). Circadian glucocorticoid oscillations preserve a population of adult hippocampal neural stem cells in the aging brain. Mol. Psychiatry 25, 1382–1405. 10.1038/s41380-019-0440-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schouten M., Buijink M., Lucassen P., Fitzsimons C. (2012). New neurons in aging brains: molecular control by small non-coding RNAs. Front. Neurosci. 6:25. 10.3389/fnins.2012.00025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schouten M., Fratantoni S., Hubens C., Piersma S., Pham T., Bielefeld P., et al. (2015). MicroRNA-124 and -137 cooperativity controls caspase-3 activity through BCL2L13 in hippocampal neural stem cells. Sci. Rep. 5:12448. 10.1038/srep12448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schratt G., Tuebing F., Nigh E., Kane C., Sabatini M., Kiebler M., et al. (2006). A brain-specific microRNA regulates dendritic spine development. Nature 439, 283–289. 10.1038/nature04367 [DOI] [PubMed] [Google Scholar]
- Schultheiß C., Abe P., Hoffmann F., Mueller W., Kreuder A.-E., Schütz D., et al. (2013). CXCR4 prevents dispersion of granule neuron precursors in the adult dentate gyrus. Hippocampus 23, 1345–1358. 10.1002/hipo.22180 [DOI] [PubMed] [Google Scholar]
- Selbach M., Schwanhäusser B., Thierfelder N., Fang Z., Khanin R., Rajewsky N. (2008). Widespread changes in protein synthesis induced by microRNAs. Nature 455, 58–63. 10.1038/nature07228 [DOI] [PubMed] [Google Scholar]
- Shariq M., Sahasrabuddhe V., Krishna S., Radha S., Nruthyathi B. R., Bellampalli R., et al. (2021). Adult neural stem cells have latent inflammatory potential that is kept suppressed by Tcf4 to facilitate adult neurogenesis. Sci. Adv. 7:eabf5606. 10.1126/sciadv.abf5606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shenoy A., Danial M., Blelloch R. (2015). Let-7 and miR-125 cooperate to prime progenitors for astrogliogenesis. EMBO J. 34, 1180–1194. 10.15252/embj.201489504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J., Berg D., Zhu Y., Shin J., Song J., Bonaguidi M., et al. (2015). Single-cell RNA-Seq with waterfall reveals molecular cascades underlying adult neurogenesis. Cell Stem Cell 17, 360–372. 10.1016/j.stem.2015.07.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel G., Obernosterer G., Fiore R., Oehmen M., Bicker S., Christensen M., et al. (2009). A functional screen implicates microRNA-138-dependent regulation of the depalmitoylation enzyme APT1 in dendritic spine morphogenesis. Nat. Cell Biol. 11, 705–716. 10.1038/ncb1876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sierra A., Encinas J., Deudero J., Chancey J., Enikolopov G., Overstreet-Wadiche L., et al. (2010). Microglia shape adult hippocampal neurogenesis through apoptosis-coupled phagocytosis. Cell Stem Cell 7, 483–495. 10.1016/j.stem.2010.08.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sierra A., Martín-Suárez S., Valcárcel-Martín R., Pascual-Brazo J., Aelvoet S., Abiega O., et al. (2015). Neuronal hyperactivity accelerates depletion of neural stem cells and impairs hippocampal neurogenesis. Cell Stem Cell 16, 488–503. 10.1016/j.stem.2015.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sierra A., Paolicelli R., Kettenmann H. (2019). Cien Años de microglía: milestones in a century of microglial research. Trends Neurosci. 42, 778–792. 10.1016/j.tins.2019.09.004 [DOI] [PubMed] [Google Scholar]
- Smith L. K., He Y., Park J.-S., Bieri G., Snethlage C. E., Lin K., et al. (2015). β2-microglobulin is a systemic pro-aging factor that impairs cognitive function and neurogenesis. Nat. Med. 21, 932–937. 10.1038/nm.3898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smrt R., Szulwach K., Pfeiffer R., Li X., Guo W., Pathania M., et al. (2010). MicroRNA miR-137 regulates neuronal maturation by targeting ubiquitin ligase mind bomb-1. Stem Cells 28, 1060–1070. 10.1002/stem.431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder J. (2019). Recalibrating the relevance of adult neurogenesis. Trends Neurosci. 42, 164–178. 10.1016/j.tins.2018.12.001 [DOI] [PubMed] [Google Scholar]
- Sohn E., Jo Y., Park H. (2019). Downregulation MIWI-piRNA regulates the migration of Schwann cells in peripheral nerve injury. Biochem. Biophys. Res. Commun. 519, 605–612. 10.1016/j.bbrc.2019.09.008 [DOI] [PubMed] [Google Scholar]
- Song H., Stevens C., Gage F. (2002). Astroglia induce neurogenesis from adult neural stem cells. Nature 417, 39–44. 10.1038/417039a [DOI] [PubMed] [Google Scholar]
- Song J., Zhong C., Bonaguidi M. A., Sun G. J., Hsu D., Gu Y., et al. (2012). Neuronal circuitry mechanism regulating adult quiescent neural stem-cell fate decision. Nature 489, 150–154. 10.1038/nature11306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stappert L., Klaus F., Brüstle O. (2018). MicroRNAs engage in complex circuits regulating adult neurogenesis. Front. Neurosci. 12:707. 10.3389/fnins.2018.00707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steindler D., Laywell E. (2003). Astrocytes as stem cells: nomenclature, phenotype and translation. Glia 43, 62–69. 10.1002/glia.10242 [DOI] [PubMed] [Google Scholar]
- Steiner B., Kronenberg G., Jessberger S., Brandt M., Reuter K., Kempermann G. (2004). Differential regulation of gliogenesis in the context of adult hippocampal neurogenesis in mice. Glia 46, 41–52. 10.1002/glia.10337 [DOI] [PubMed] [Google Scholar]
- St-Pierre M., Šimončičová E., Bögi E., Tremblay M. (2020). Shedding light on the dark side of the microglia. ASN Neuro 12:1759091420925335. 10.1177/1759091420925335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh H., Consiglio A., Ray J., Sawai T., D’Amour K., Gage F. (2007). in vivo fate analysis reveals the multipotent and self-renewal capacities of Sox2+ neural stem cells in the adult hippocampus. Cell Stem Cell 1, 515–528. 10.1016/j.stem.2007.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun G., Zhou Y., Ito S., Bonaguidi M., Stein-O’Brien G., Kawasaki N., et al. (2015). Latent tri-lineage potential of adult hippocampal neural stem cells revealed by Nf1 inactivation. Nat. Neurosci. 18, 1722–1724. 10.1038/nn.4159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svahn A., Giacomotto J., Graeber M., Rinkwitz S., Becker T. (2016). miR-124 Contributes to the functional maturity of microglia. Dev. Neurobiol. 76, 507–518. 10.1002/dneu.22328 [DOI] [PubMed] [Google Scholar]
- Symons A., Beinke S., Ley S. (2006). MAP kinase kinase kinases and innate immunity. Trends Immunol. 27, 40–48. 10.1016/j.it.2005.11.007 [DOI] [PubMed] [Google Scholar]
- Szulwach K., Li X., Smrt R., Li Y., Luo Y., Lin L., et al. (2010). Cross talk between microRNA and epigenetic regulation in adult neurogenesis. J. Cell Biol. 189, 127–141. 10.1083/jcb.200908151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taganov K., Boldin M., Chang K., Baltimore D. (2006). NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc. Natl. Acad. Sci. U S A 103, 12481–12486. 10.1073/pnas.0605298103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terrigno M., Bertacchi M., Pandolfini L., Baumgart M., Calvello M., Cellerino A., et al. (2018). The microRNA miR-21 is a mediator of FGF8 action on cortical COUP-TFI translation. Stem Cell Rep. 11, 756–769. 10.1016/j.stemcr.2018.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian F., Yuan C., Hu L., Shan S. (2017). MicroRNA-93 inhibits inflammatory responses and cell apoptosis after cerebral ischemia reperfusion by targeting interleukin-1 receptor-associated kinase 4. Exp. Ther. Med. 14, 2903–2910. 10.3892/etm.2017.4874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobin M., Musaraca K., Disouky A., Shetti A., Bheri A., Honer W., et al. (2019). Human hippocampal neurogenesis persists in aged adults and Alzheimer’s disease patients. Cell Stem Cell 24, 974–982.e3. 10.1016/j.stem.2019.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toda T., Parylak S. L., Linker S. B., Gage F. H. (2019). The role of adult hippocampal neurogenesis in brain health and disease. Mol. Psychiatry 24, 67–87. 10.1038/s41380-018-0036-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres A., Reina O., Stephan-Otto Attolini C., Ribas de Pouplana L. (2019). Differential expression of human tRNA genes drives the abundance of tRNA-derived fragments. Proc. Natl. Acad. Sci. U S A 116, 8451–8456. 10.1073/pnas.1821120116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuyama J., Bunt J., Richards L., Iwanari H., Mochizuki Y., Hamakubo T., et al. (2015). MicroRNA-153 regulates the acquisition of gliogenic competence by neural stem cells. Stem Cell Rep. 5, 365–377. 10.1016/j.stemcr.2015.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbán N., Blomfield I., Guillemot F. (2019). Quiescence of adult mammalian neural stem cells: a highly regulated rest. Neuron 104, 834–848. 10.1016/j.micpath.2021.105341 [DOI] [PubMed] [Google Scholar]
- Valadi H., Ekström K., Bossios A., Sjöstrand M., Lee J., Lötvall J. (2007). Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 9, 654–659. 10.1038/ncb1596 [DOI] [PubMed] [Google Scholar]
- Vicidomini C., Guo N., Sahay A. (2020). Communication, cross talk and signal integration in the adult hippocampal neurogenic niche. Neuron 105, 220–235. 10.1016/j.neuron.2019.11.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villeda S. A., Plambeck K. E., Middeldorp J., Castellano J. M., Mosher K. I., Luo J., et al. (2014). Young blood reverses age-related impairments in cognitive function and synaptic plasticity in mice. Nat. Med. 20, 659–663. 10.1038/nm.3569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vo N., Klein M., Varlamova O., Keller D., Yamamoto T., Goodman R., et al. (2005). A cAMP-response element binding protein-induced microRNA regulates neuronal morphogenesis. Proc. Natl. Acad. Sci. U S A 102, 16426–16431. 10.1073/pnas.0508448102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel A., Upadhya R., Shetty A. (2018). Neural stem cell derived extracellular vesicles: attributes and prospects for treating neurodegenerative disorders. EBioMedicine 38, 273–282. 10.1016/j.ebiom.2018.11.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakisaka K., Imai Y. (2019). The dawn of pirna research in various neuronal disorders. Front. Biosci. (Landmark Ed). 24, 1440–1451. 10.2741/4789 [DOI] [PubMed] [Google Scholar]
- Walgrave H., Balusu S., Snoeck S., Vanden Eynden E., Craessaerts K., Thrupp N., et al. (2021a). Restoring miR-132 expression rescues adult hippocampal neurogenesis and memory deficits in Alzheimer’s disease. Cell Stem Cell 28, 1805–1821.e8. 10.1016/j.stem.2021.05.001 [DOI] [PubMed] [Google Scholar]
- Walgrave H., Zhou L., Strooper B. D., Salta E. (2021b). The promise of microRNA-based therapies in Alzheimer’s disease: challenges and perspectives. Mol. Neurodegener. 16:76. 10.1186/s13024-021-00496-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Moyano A., Ma Z., Deng Y., Lin Y., Zhao C., et al. (2017a). miR-219 cooperates with miR-338 in myelination and promotes myelin repair in the CNS. Dev. Cell 40, 566–582.e5. 10.1016/j.devcel.2017.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Li J., Yang J., Zhang L., Gao S., Jiao F., et al. (2017b). MicroRNA-138-5p regulates neural stem cell proliferation and differentiation in vitro by targeting TRIP6 expression. Mol. Med. Rep. 16, 7261–7266. 10.3892/mmr.2017.7504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Wang T. (2018). MicroRNAs of microglia: wrestling with central nervous system disease. Neural Regen. Res. 13, 2067–2072. 10.4103/1673-5374.241444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Yu J.-T., Tan L., Tian Y., Ma J., Tan C.-C., et al. (2015). Genome-wide circulating microRNA expression profiling indicates biomarkers for epilepsy. Sci. Rep. 5:9522. 10.1038/srep09522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolswijk G., Noble M. (1989). Identification of an adult-specific glial progenitor cell. Development 105, 387–400. [DOI] [PubMed] [Google Scholar]
- Wood J., Jackson J., Jakubs K., Chapman K., Ekdahl C., Kokaia Z., et al. (2011). Functional integration of new hippocampal neurons following insults to the adult brain is determined by characteristics of pathological environment. Exp. Neurol. 229, 484–493. 10.1016/j.expneurol.2011.03.019 [DOI] [PubMed] [Google Scholar]
- Woodbury M., Freilich R., Cheng C., Asai H., Ikezu S., Boucher J., et al. (2015). miR-155 is essential for inflammation-induced hippocampal neurogenic dysfunction. J. Neurosci. 35, 9764–9781. 10.1523/JNEUROSCI.4790-14.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Kuo H. (2020). Functional roles and networks of non-coding RNAs in the pathogenesis of neurodegenerative diseases. J. Biomed. Sci. 29:49. 10.1186/s12929-020-00636-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia X., Wang Y., Huang Y., Zhang H., Lu H., Zheng J. (2019). Exosomal miRNAs in central nervous system diseases: biomarkers, pathological mediators, protective factors and therapeutic agents. Prog. Neurobiol. 183:101694. 10.1016/j.pneurobio.2019.101694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin H., Katakowski M., Wang F., Qian J.-Y., Liu X. S., Ali M. M., et al. (2017). Correction to: microRNA-17–92 cluster in exosomes enhance neuroplasticity and functional recovery after stroke in rats. Stroke 48:e137. 10.1161/STR.0000000000000130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing H., Guo S., Zhang Y., Zheng Z., Wang H. (2016). Upregulation of microRNA-206 enhances lipopolysaccharide-induced inflammation and release of amyloid-β by targeting insulin-like growth factor 1 in microglia. Mol. Med. Rep. 14, 1357–1364. 10.3892/mmr.2016.5369 [DOI] [PubMed] [Google Scholar]
- Xiong Y., Zhang Y., Mahmood A., Chopp M. (2015). Investigational agents for treatment of traumatic brain injury. Expert Opin. Investig. Drugs 24, 743–760. 10.1517/13543784.2015.1021919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y., Hajdukiewicz K., Tiwari A., Przybyś J., Parkitna J., Novak M., et al. (2020). MicroRNAs are indispensable for the proliferation and differentiation of adult neural progenitor cells in mice. Biochem. Biophys. Res. Commun. 530, 209–214. 10.1016/j.bbrc.2020.06.143 [DOI] [PubMed] [Google Scholar]
- Xu B., Woodroffe A., Rodriguez-Murillo L., Roos J., van Rensburg E., Abecasis G., et al. (2009). Elucidating the genetic architecture of familial schizophrenia using rare copy number variant and linkage scans. Proc. Natl. Acad. Sci. U S A 106, 16746–16751. 10.1073/pnas.0908584106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan T., Zhang F., Sun C., Sun J., Wang Y., Xu X., et al. (2018). miR-32>–5p-mediated Dusp5 downregulation contributes to neuropathic pain. Biochem. Biophys. Res. Commun. 495, 506–511. 10.1016/j.bbrc.2017.11.013 [DOI] [PubMed] [Google Scholar]
- Yao B., Christian K., He C., Jin P., Ming G., Song H. (2016). Epigenetic mechanisms in neurogenesis. Nat. Rev. Neurosci. 17, 537–549. 10.1038/nrn.2016.70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshino Y., Dwivedi Y. (2020). Non-coding RNAs in psychiatric disorders and suicidal behavior. Front. Psychiatry 11:543893. 10.3389/fpsyt.2020.543893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zainal Abidin S., Fam S., Chong C., Abdullah S., Cheah P., Nordin N., et al. (2019). miR-3099 promotes neurogenesis and inhibits astrogliogenesis during murine neural development. Gene 697, 201–212. 10.1016/j.gene.2019.02.014 [DOI] [PubMed] [Google Scholar]
- Zhang J., Zhang Z., Lu M., Zhang Y., Shang X., Chopp M. (2019a). MiR-146a promotes oligodendrocyte progenitor cell differentiation and enhances remyelination in a model of experimental autoimmune encephalomyelitis. Neurobiol. Dis. 125, 154–162. 10.1016/j.nbd.2019.01.019 [DOI] [PubMed] [Google Scholar]
- Zhang P., Wu W., Chen Q., Chen M. (2019b). Non-Coding RNAs and their Integrated Networks. J. Integr. Bioinform. 16:20190027. 10.1515/jib-2019-0027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R., Boareto M., Engler A., Louvi A., Giachino C., Iber D., et al. (2019c). Id4 downstream of notch2 maintains neural stem cell quiescence in the adult hippocampus. Cell Rep. 28, 1485–1498.e6. 10.1016/j.celrep.2019.07.014 [DOI] [PubMed] [Google Scholar]
- Zhang S., Gao J., Liu C. (2019d). The role of non-coding RNAs in neurodevelopmental disorders. Front. Genet. 10:1033. 10.3389/fgene.2019.01033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X., He X., Han X., Yu Y., Ye F., Chen Y., et al. (2010). MicroRNA-mediated control of oligodendrocyte differentiation. Neuron 65, 612–626. 10.1016/j.neuron.2010.02.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C., Sun G., Li S., Shi Y. (2009). A feedback regulatory loop involving microRNA-9 and nuclear receptor TLX in neural stem cell fate determination. Nat. Struct. Mol. Biol. 16, 365–371. 10.1038/nsmb.1576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao P., Yao M., Chang S., Gou L., Liu M., Qiu Z., et al. (2016). Erratum to: novel function of PIWIL1 in neuronal polarization and migration via regulation of microtubule-associated proteins. Mol. Brain 9:21. 10.1186/s13041-016-0201-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng K., Li H., Zhu Y., Zhu Q., Qiu M. (2010). MicroRNAs are essential for the developmental switch from neurogenesis to gliogenesis in the developing spinal cord. J. Neurosci. 30, 8245–8250. 10.1523/JNEUROSCI.1169-10.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H., Wang L., Xu Q., Fan Z., Zhu Y., Jiang H., et al. (2016). Downregulation of miR-199b promotes the acute spinal cord injury through IKKβ-NF-κB signaling pathway activating microglial cells. Exp. Cell Res. 349, 60–67. 10.1016/j.yexcr.2016.09.020 [DOI] [PubMed] [Google Scholar]