Abstract
Florfenicol, a veterinary fluorinated analog of thiamphenicol, is approved for treatment of bovine respiratory pathogens in the United States. However, florfenicol resistance has recently emerged among veterinary Escherichia coli isolates incriminated in bovine diarrhea. The flo gene, which confers resistance to florfenicol and chloramphenicol, has previously been identified in Photobacterium piscicida and Salmonella enterica serovar Typhimurium DT104. The flo gene product is closely related to the CmlA protein identified in Pseudomonas aeruginosa. The cmlA gene confers nonenzymatic chloramphenicol resistance via an efflux mechanism. Forty-eight E. coli isolates recovered from calves with diarrhea, including 41 that were both chloramphenicol and florfenicol resistant, were assayed for the presence of both flo and cmlA genes. Forty-two of the 44 isolates for which florfenicol MICs were ≥16 μg/ml were positive via PCR for the flo gene. All E. coli isolates for which florfenicol MICs were ≤8 μg/ml were negative for the flo gene (n = 4). Twelve E. coli isolates were positive for cmlA, and chloramphenicol MICs for all 12 were ≥32 μg/ml. Additionally, eight isolates were positive for both flo and cmlA, and both florfenicol and chloramphenicol MICs for these isolates were ≥64 μg/ml. DNA sequence analysis of the E. coli flo gene demonstrated 98% identity to the published GenBank sequences of both serovar Typhimurium floSt and P. piscicida pp-flo. The flo gene was identified on high-molecular-weight plasmids of approximately 225 kb among the majority of florfenicol-resistant E. coli isolates. However, not all of the florfenicol-resistant E. coli isolates tested contained the large flo-positive plasmids. This suggests that several of the E. coli isolates may possess a chromosomal flo gene. The E. coli flo gene specifies nonenzymatic cross-resistance to both florfenicol and chloramphenicol, and its presence among bovine E. coli isolates of diverse genetic backgrounds indicates a distribution much wider than previously thought.
Currently, there is increased public and scientific interest in the use of therapeutic and subtherapeutic antimicrobials in animals. This is due primarily to the possible emergence and dissemination of multiple-drug-resistant zoonotic bacterial pathogens (1, 8, 9, 22, 27, 32). Antimicrobial drug-resistant bacterial pathogens in animals pose a risk not only to animal health, but possibly to humans via transmission as food-borne pathogens (8, 9, 22, 27, 32).
Infections caused by antibiotic-resistant bacteria are a severe and costly animal health problem. These infections prolong illness and, if not treated in time with more costly, alternative antimicrobial agents, can lead to increased morbidity and mortality. One of the more challenging diseases facing cattle veterinarians and producers today is colibacillosis, a severe form of diarrhea, which causes significant financial loss to cow and calf producers (10, 37). There are multiple infectious etiological agents that can cause colibacillosis in cattle; however, Escherichia coli is recognized as the single most important bacterial cause (31, 36, 37). Numerous E. coli serotypes have been incriminated, but the majority are enterotoxigenic E. coli (ETEC) strains. These strains usually possess the K99 (F5) adhesion and produce heat-stable (STa or STb) and/or heat-labile (LT) enterotoxins (36, 37). The most important aspect of treating E. coli-related colibacillosis is to correct the accompanying electrolyte loss, dehydration, and acidosis. However, antimicrobial therapy is often initiated at the same time in an attempt to eliminate the pathogenic E. coli.
Florfenicol [d-threo-3-fluoro-2-dichloroacetamido-1-(4-methylsulfonylphenyl)-1-propanol] is a fluorinated structural analog of thiamphenicol and chloramphenicol approved by the Food and Drug Administration (FDA) in 1996 for treatment of bovine respiratory pathogens such as Pasteurella spp. However, it is not currently approved for treatment of E. coli-related cattle enteric diseases in the United States. Additionally, there are no approved NCCLS breakpoints for E. coli currently available; however, the resistance breakpoint for bovine respiratory pathogens (i.e., Pasteurella multocida) is ≥8 μg/ml (25).
Florfenicol is not approved for human use; however, it is related to chloramphenicol and can select for cross-resistance among bacterial pathogens. Florfenicol has been shown to have a spectrum of activity similar to that of chloramphenicol, except that it is active at lower concentrations than chloramphenicol against a variety of clinical bacterial isolates, including chloramphenicol-resistant bacteria (16, 26, 33). Florfenicol's mechanism of action is directed at disrupting bacterial protein synthesis by binding to the 50S subunit of the bacterial ribosome and is generally considered to be bacteriostatic (12, 23, 26, 33).
Neither chloramphenicol acetyltransferase (CAT), the enzyme responsible for most of the plasmid-mediated resistance to chloramphenicol (12), nor the known nonenzymatic chloramphenicol resistance gene (cmlA) confers resistance to florfenicol (14, 18, 23). Until recently, no genes conferring resistance to fluorinated derivatives of chloramphenicol or thiamphenicol were known. However, in 1996 researchers in Japan identified a novel plasmid-encoded gene (pp-flo) from Photobacterium piscicida that encoded resistance to both chloramphenicol and florfenicol (19). More recently, Bolton et al. described a gene with 97% homology to the pp-flo gene among Salmonella enterica serovar Typhimurium DT104 isolates, which they termed floSt (9). Salmonella isolates possessing the floSt gene also exhibited dual resistance to chloramphenicol and florfenicol (4, 9). The flo gene has also been identified recently among chloramphenicol-resistant Salmonella enterica serovar Agona isolates recovered from poultry in Belgium and florfenicol-resistant E. coli from poultry in the United States (13, 18).
The present study involves 48 antimicrobial-resistant strains of E. coli isolated from diarrheic calves and submitted to the North Dakota Veterinary Diagnostic Laboratory (ND-VDL) from 1997 to 1998. Since there was limited information regarding florfenicol resistance in the literature and virtually none concerning E. coli, a study was initiated to determine the mechanism of resistance among these bovine E. coli isolates.
MATERIALS AND METHODS
Bacterial isolates.
The present study focuses on 48 E. coli strains recovered from calf diarrhea cases submitted to the ND-VDL from 1997 to 1998. All calves were under 2 weeks of age, and several of the case histories indicated that florfenicol was used in an extra-label manner in an attempt to cure the diarrhea. E. coli bacteria were isolated from necropsy or fecal specimens from calves suffering diarrhea. The specimens were plated on blood and MacConkey agar plates. Indole and oxidase tests were performed on lactose-positive colonies. API 20E test strips (bioMérieux Vitek, Hazelwood, Mo.) were also used to confirm E. coli identification. Bovine E. coli isolates displaying decreased susceptibilities to florfenicol were subsequently collected for further analysis to determine the mechanism of resistance. Isolates were stored as 10% glycerol stocks at −80°C until analysis.
Antimicrobial susceptibility determination.
Antimicrobial MICs for E. coli isolates were determined using the Sensititre automated antimicrobial susceptibility system (Trek Diagnostic Systems, Westlake, Ohio) and interpreted according to the NCCLS guidelines for broth microdilution methods (24, 25). Sensititre susceptibility testing was performed according to the manufacturer's instructions. The following antimicrobials were assayed: amikacin, amoxicillin-clavulanic acid, ampicillin, apramycin, ceftiofur, ceftriaxone, cephalothin, chloramphenicol, ciprofloxacin, florfenicol, gentamicin, kanamycin, nalidixic acid, streptomycin, sulfamethoxazole, tetracycline, and trimethoprim-sulfamethoxazole. E. coli ATCC 25922, E. coli ATCC 35218, and Pseudomonas aeruginosa ATCC 27853 were used as controls in MIC determinations.
PCR, isolation, and sequencing of the E. coli flo and cmlA genes.
Oligonucleotides were synthesized by Biosynthesis (Lewisville, Tex.). The PCR-derived flo probe was obtained using previously published primers (9) and yielded a 215-bp amplicon within the coding region. Two additional PCR primers were created flanking the entire pp-flo gene. E. coli Flo-F corresponded to nucleotides 981 to 1004 (TTGTTGTTGCGGCGCTCTGTAAGG), and E. coli Flo-R corresponded to nucleotides 2405 to 2384 (CGGCGACGGCGATGAACTGAAC) of the published sequence (GenBank accession no. D37826), yielding a predicted amplicon of 1,424 bp. The PCR-derived cmlA probe was amplified using previously published primers (18) and yielded a 698-bp amplicon. PCRs were carried out in a thermocycler 2400 (Perkin-Elmer Applied Biosystems, Norwalk, Conn.). Two hundred microliters of each PCR product was separated by 1.5% agarose gel electrophoresis at 75 V for 2 h. The appropriate bands were excised and purified utilizing GenElute Spin columns (Supelco, Bellefonte, Pa.). The purified PCR amplicons were sequenced at the University of Minnesota Advanced Genetic Analysis Center, St. Paul, Minn. Sequence comparisons were made using the NCBI-BLAST program (2).
PFGE.
Five-milliliter broth cultures of each isolate were grown overnight at 37°C. The cell pellet was collected, embedded in agarose plugs, and lysed as previously described (5, 30). Agarose plugs were digested with 10 U of XbaI (Promega, Madison, Wis.) overnight at 37°C. Digested DNA was separated on a 1.2% agarose gel using the CHEF-DRII pulsed-field gel electrophoresis (PFGE) system (Bio-Rad, Hercules, Calif.). Electrophoresis was carried out for 25 h at 6 V with a ramped pulse time of 2 to 40 s in 0.5× Tris-borate-EDTA (TBE) buffer (14°C). Molecular weight standards were Saccharomyces cerevisiae YPH 755 chromosomes (Roche Biochemicals, Indianapolis, Ind.). Gel documentation and phylogenetic analysis of the E. coli isolates were performed using the RFLPScan and TREECON programs as previously described (21).
Mapping of the flo gene in bovine E. coli isolates.
The flo gene was initially identified in a transferable R plasmid in the fish pathogen P. piscicida (18). A flo homolog was recently mapped within a multidrug resistance locus that is flanked by two class 1 integrons in serovar Typhimurium DT104 and serovar Agona (3, 11, 13). PCR primers were developed to determine if the E. coli flo gene had a genetic organization similar to that observed in either serovar Typhimurium DT104 or P. piscidida. The first PCR primers were designed to amplify a portion of flo and genes mapping downstream (tetR) within the Salmonella multidrug resistance (MDR) locus (GenBank accession no. AF071555). The second PCR set was targeted to flo and the broad-host-range plasmid RSF1010 replicon identified in P. piscicida (GenBank accession no. D37826). Primers flomap2F (GGGATCGGCGAAACTTTAC) and flomap2R (TGTGGTCGGTTCCGTTCTC) anneal at nucleotide positions 5330 of flo and 6073 of tetR within the serovar Typhimurium DT104 (GenBank accession no. AF07155), resulting in a 744-bp amplicon. Primers flomap2F (GGGATCGGCGAAACTTTAC) and pp-floMapR (TCCGCGTCCTTGCAATAC) anneal to nucleotide positions 1953 and 2723 of the P. piscicida plasmid-mediated flo sequence (GenBank accession no. D37826), resulting in a 771-bp amplicon. The PCR conditions used have been previously described (17).
Detection of high-molecular-weight plasmids by S1 nuclease treatment and PFGE.
Due to their structure, large circular plasmids are not resolved by PFGE. This problem can be overcome by using S1 nuclease digestion prior to PFGE. S1 nuclease creates breaks in the plasmids, which allow them to enter the agarose gel during PFGE. Agarose plugs prepared for PFGE were treated with S1 nuclease (Roche Biochemicals) as previously described (6, 22). Plasmids were separated on a 1.5% agarose gel using the CHEF-DRII PFGE system (Bio-Rad). Plasmids were electrophoresed initially for 14 h at 6 V with a pulse time of 45 s, followed by a program of 6 h at 6 V with a pulse time of 25 s. Molecular weight standards were S. cerevisiae YPH 755 chromosomes (Roche Biochemicals).
Southern hybridization analysis.
Megabase DNA was blotted to nylon membranes using the Bio-Rad procedure. Briefly, DNA in ethidium bromide-stained gels was nicked using a UV Transilluminator (Fisher Scientific) at 60 mJ of energy. Gels were soaked in 0.4 N NaOH–1.5 M NaCl for 15 min, and then DNA was transferred for 4 h to Magnagraph nylon (MSI, Westborough, Mass.) using the model 785 Vacuum Blotter (Bio-Rad) with NaOH as the buffer. After transfer, nylon membranes were washed briefly in 0.5 M Tris, pH 7.6, followed by 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) (29). Membranes were blocked and probed with a PCR-amplified portion of the florfenicol resistance gene probe as previously described (18).
Nucleotide sequence accession number.
The sequence of the bovine E. coli flo gene has been assigned GenBank accession no. AF252855.
RESULTS AND DISCUSSION
Antimicrobial resistance patterns in bovine E. coli.
Forty-eight E. coli isolates recovered from calves with diarrhea were tested for their resistance to antimicrobial agents of human and veterinary significance according to NCCLS broth microdilution methods and guidelines (Table 1). Interestingly, 90% of bovine E. coli isolates were resistant to chloramphenicol (Table 1), an antibiotic that has been banned from veterinary use in food animals in the United States since the 1980s (15). Ninety-two percent of E. coli isolates were also resistant to florfenicol based on the NCCLS breakpoint for bovine respiratory pathogens, ≥8 μg/ml (25). In addition to chloramphenicol and florfenicol resistance, all bovine isolates were resistant to at least four antimicrobials and 37 (77%) isolates were resistant to at least nine antimicrobials. The most common antimicrobial resistance pattern observed in the bovine E. coli isolates included resistance to chloramphenicol, florfenicol, amoxicillin, ampicillin, ceftiofur, cephalothin, gentamicin, kanamycin, streptomycin, sulfamethoxazole, tetracycline, and trimethoprim-sulfamethoxazole (n = 14).
TABLE 1.
Antimicrobial resistance phenotypes of bovine E. coli isolates
| Class and/or antimicrobial | % Resistant strainsa(n = 48) |
|---|---|
| Phenicols | |
| Florfenicolb | 92 |
| Chloramphenicol | 90 |
| Penicillins | |
| Ampicillin | 88 |
| Amoxicillin-clavulanic acid | 69 |
| Cephalosporins | |
| Cephalothin | 77 |
| Ceftiofur | 69 |
| Ceftriaxone | 15 |
| Tetracycline | 100 |
| Aminoglycosides | |
| Amikacin | 0 |
| Apramycin | 8 |
| Gentamicin | 63 |
| Kanamycin | 92 |
| Streptomycin | 100 |
| Sulfonamides and potentiated sulfonamides | |
| Sulfamethoxazole | 100 |
| Trimethoprim-sulfamethoxazole | 69 |
| Quinolones and fluoroquinolones | |
| Nalidixic acid | 8 |
| Ciprofloxacin | 0 |
With multiple antibiotic resistance phenotypes so common among the bovine E. coli isolates, the question then becomes, what is the driving selection pressure that is maintaining chloramphenicol resistance in the absence of its use? One possibility is the use of florfenicol, a fluorinated derivative of chloramphenicol that was approved for treatment of bovine respiratory disease in 1996. Bacterial cross-resistance to chloramphenicol and florfenicol is being increasingly reported and has been attributed to a homolog of the chloramphenicol resistance efflux gene, cmlA (4, 9, 13, 18) and not the traditional CAT genes (7, 12, 14, 26, 33, 34, 35). This cmlA analog, termed flo, was first described in the fish pathogen P. piscicida, isolated in Japan, where florfenicol is routinely used in aquaculture (19). This gene has since been found in other bacteria including serovar Typhimurium DT104 (4, 9), serovar Agona (13), and avian E. coli (18). We investigated the possibility that the emerging chloramphenicol resistance phenotype observed in bovine E. coli isolates actually was due to the dissemination of this florfenicol resistance gene.
Identification of the flo resistance gene in bovine E. coli.
PCR primers, based on the published pp-flo nucleotide sequence, amplified a 1.4-kb PCR product from chloramphenicol- and florfenicol-resistant bovine E. coli isolates. DNA sequence analysis of the E. coli flo gene demonstrated 98% identity to the published GenBank sequences of both serovar Typhimurium floSt and P. piscicida pp-flo (AF118107 and D37826, respectively). DNA sequence analysis using the NCBI-BLAST program (2) predicted an open reading frame (ORF) of 1,215 bp, demonstrating the greatest identity (98%) with the pp-flo gene from P. piscicida (19), the cmlA-like gene described in the DT104 MDR locus (11), and the floSt gene described by Bolton et al. (9). This 1,215-bp ORF translates into a hypothetical protein of 404 amino acids. Using BLAST-p (2), it was determined that this hypothetical protein demonstrated 98 and 89% amino acid sequence homology with the previously described floSt and pp-flo gene products, respectively. There was also 49% identity between the E. coli hypothetical Flo protein and the previously described nonenzymatic chloramphenicol resistance gene product CmlA.
Evidence that the flo resistance gene is widely disseminated among bovine E. coli isolates.
Forty-one (85%) of the E. coli isolates assayed were resistant to both chloramphenicol (MIC, ≥32 μg/ml) and florfenicol (MIC, ≥16 μg/ml). For 36 of these E. coli isolates, florfenicol MICs were ≥128 μg/ml, higher than those previously reported for florfenicol-resistant E. coli (9) but comparable to MICs observed for serovar Typhimurium DT104 (18). These isolates were further analyzed for the presence of both flo and cmlA efflux genes. Forty-two of 44 isolates that displayed decreased susceptibilities to florfenicol (MIC, ≥16 μg/ml) were positive by PCR for the flo gene (Table 2). All E. coli isolates for which florfenicol MICs were ≤8 μg/ml were negative for the flo gene (n = 4).
TABLE 2.
Prevalence of flo and cmlA genes in chloramphenicol-resistant bovine E. coli
Twelve E. coli isolates were positive by PCR for the cmlA gene. Additionally, eight isolates were positive for both flo and cmlA genes, and both florfenicol and chloramphenicol MICs for these isolates were elevated (≥64 μg/ml) (Table 2). Four isolates were negative for flo but positive for cmlA. Interestingly, for two of these four isolates, florfenicol MICs were elevated (≥16 μg/ml). The putative chloramphenicol efflux gene cmlA has previously been shown not to confer resistance to florfenicol and has been further identified on a transposable element in P. aeruginosa (7, 14, 18). However, there is very little information regarding the presence of cmlA in E. coli and resulting resistance phenotypes. It is possible that the cmlA genes in the two florfenicol-resistant bovine E. coli isolates that lack flo accumulated mutations resulting in florfenicol being recognized as an efflux substrate. Further research on these two strains is certainly warranted.
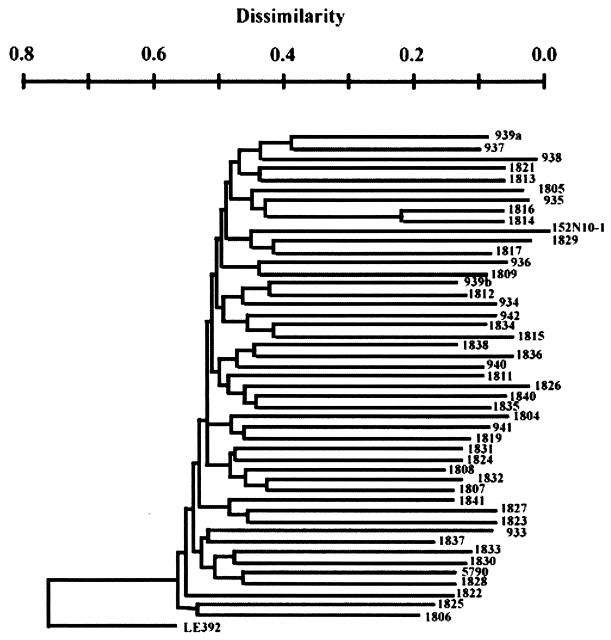
To determine whether florfenicol-resistant bovine E. coli isolates represented dissemination of a clonal strain, 45 of the 48 isolates were analyzed by PFGE. Using the neighbor-joining method and rooting the tree against E. coli strain LE392 (K-12), 44 different branches were observed (Fig. 1). Only two isolates, CVM1814 and CVM1816, displayed the same PFGE pattern. This extreme genetic diversity suggests that florfenicol resistance is not limited to a particular chromosomal background and is most likely due to dissemination of the flo gene via mobile transposons and/or a plasmid(s).
FIG. 1.
Phylogenetic tree of florfenicol-resistant bovine E. coli. Total DNA was digested with XbaI and separated by PFGE. Molecular weight markers were S. cerevisiae strain YPH 755 chromosomes. Similarities among E. coli PFGE patterns were identified by cluster analysis using the neighbor-joining method to draw a phylogenetic tree (21). Phylogenetic analysis identified 44 PFGE patterns in the 45 florfenicol-resistant bovine E. coli isolates assayed.
The flo resistance gene maps to high-molecular-weight plasmids in bovine E. coli.
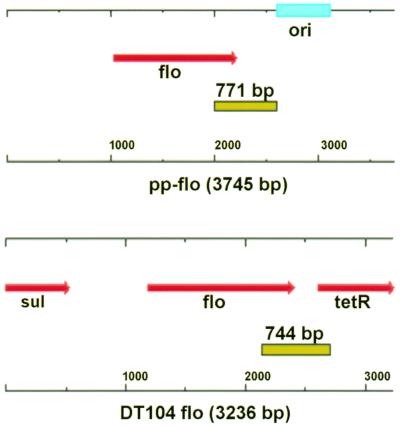
Recently, Briggs and Fratamico characterized the MDR locus of the penta-antibiotic-resistant serovar Typhimurium DT104 (11). This locus contains the florfenicol resistance gene flo, which is flanked on one side by a class 1 integron containing the streptomycin/spectinomycin resistance gene aadA2 and on the other by the tetracycline resistance gene, tetA (3, 11, 13). To determine whether flo-positive bovine E. coli isolates have a similar genetic organization, PCR primers targeted to flo and genes downstream in the MDR locus of serovar Typhimurium DT104 were designed (Fig. 2). PCR analysis of a serovar Typhimurium DT104 isolate resulted in a PCR product of approximately 750 bp, corresponding in size to the predicted amplicon between flo and tetR (Fig. 2). No PCR amplicons were observed for primers anchored in DNA sequences downstream of the DT104 MDR locus and flo in the florfenicol-resistant bovine E. coli isolates. However, several florfenicol-resistant bovine E. coli isolates yielded 770-bp amplicons using PCR primers within flo and downstream within the P. piscicida R plasmid RSF1010 replicon (Fig. 2). This suggests horizontal transfer of the E. coli flo gene via acquisition of large broad-host-range plasmids.
FIG. 2.
Comparison of the location of the flo gene in serovar Typhimurium DT104, P. piscicida, and E. coli. PCR primers were designed to amplify a portion of flo and genes mapping downstream (tetR) within the Salmonella MDR locus or to amplify flo and the RSF1010 replicon identified in P. piscicida. Primers flomap2F and pp-floMapR anneal to nucleotide positions 1953 and 2723 of the P. piscicida plasmid-mediated flo sequence, resulting in a 771-bp amplicon. Primers flomap2F and flomap2R anneal at nucleotide positions 5330 of flo and 6073 of tetR within the serovar Typhimurium DT104 MDR locus, resulting in a 744-bp amplicon.
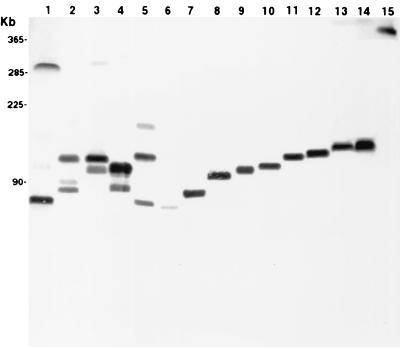
Since PCR mapping indicated linkage between flo and the RSF1010 replicon sequence, we preceded to confirm the position of flo on a plasmid(s) in bovine E. coli. The flo gene was identified on high-molecular-weight plasmids by S1 nuclease digestion and PFGE. The flo resistance gene mapped to bovine E. coli plasmids of approximately 225 kb. These plasmids were larger than previously described flo-containing plasmids in avian E. coli (18). The presence of such large plasmids is not unprecedented in E. coli. Mitsuda et al. previously reported the presence of a 170-kb plasmid in an ETEC isolate (22). Additionally, the original report of florfenicol resistance among P. piscicida strains attributed it to a high-molecular-weight transferable R plasmid (19). Not all of the flo-positive bovine E. coli isolates contained the large flo-positive plasmids, indicating that some of the genes are chromosomally located. Like serovar Typhimurium DT104 and serovar Agona, the flo gene in these bovine E. coli isolates appears to be chromosomally located. However, the flo genes mapped to a variety of different XbaI DNA fragments in bovine E. coli (Fig. 3). This is in contrast to the common 10-kb XbaI DNA fragment containing flo in serovar Typhimurium DT104 (11), and may be due to chromosomal integration of flo containing DNA mobile elements.
FIG. 3.
Location of the flo resistance gene in bovine E. coli genomes by PFGE and Southern analysis. Total E. coli genomic DNA, plasmid and chromosome, was cut with XbaI, separated by PFGE, and probed with labeled flo. Lanes 2 to 15, bovine E. coli isolates. Avian E. coli strain 5790 served as the positive control for flo (lane 1).
Is the widespread dissemination of florfenicol resistance in bovine E. coli due to a common flo-containing plasmid?
A common 225-kb flo-containing plasmid was identified in several genetically distinct bovine E. coli isolates. Southern analysis was used to determine if the florfenicol resistance gene, flo, mapped to a common XbaI DNA fragment. This would indicate that florfenicol resistance emerged in bovine E. coli due to dissemination of a common resistance plasmid containing flo. On the contrary, we found that flo mapped to a diverse array of XbaI DNA fragments in bovine E. coli, ranging in size from 72 to 529 kb (Fig. 3, lanes 2 to 15). The flo gene mapped to 14 distinct XbaI DNA fragments in 32 bovine E. coli isolates that were positive for flo. A common 124-kb XbaI DNA fragment containing flo was identified among nine genetically distinct bovine E. coli isolates. However, the 124-kb XbaI fragment was not located on a common plasmid in those isolates. The flo probe also hybridized to two XbaI fragments in four of the E. coli isolates, indicating that multiple copies were present (Fig. 3, lanes 2 to 5). These data suggest that the dissemination of florfenicol resistance is not attributed to one common plasmid.
In conclusion, the E. coli flo gene specifies nonenzymatic cross-resistance to both florfenicol and chloramphenicol, and its presence among bovine E. coli isolates of diverse genetic backgrounds indicates a distribution much wider than previously thought. Florfenicol and chloramphenicol resistance was not associated with one particular E. coli chromosomal genotype and is most likely due to the dissemination of the flo gene via high-molecular-weight plasmids and/or a mobile transposon(s). The bovine E. coli florfenicol resistance gene flo does not appear to be part of the serovar Typhimurium DT104 MDR locus. However, flanking DNA surrounding the E. coli flo gene is similar to the R plasmid RSF1010 sequence from P. piscicida. This suggests the possibility that florfenicol resistance in E. coli arose from intraspecies transfer of broad-host-range plasmids, which has been observed for other antimicrobial resistance phenotypes (20, 28). We also report the first occurrence of the cmlA gene among pathogenic E. coli isolates and the possibility that there are yet-to-be-characterized mechanisms of resistance to florfenicol. The emergence and dissemination of florfenicol resistance among potentially pathogenic bovine E. coli isolates will limit the use of this antimicrobial as an extra-label alternative for treating calf diarrhea. Research is currently under way to further characterize the putative efflux mechanism attributed to the expression of the flo and cmlA genes in florfenicol- and chloramphenicol-resistant E. coli isolates.
ACKNOWLEDGMENT
This work was supported by Public Health Service grant supplement AI22383 from the National Institutes of Health.
REFERENCES
- 1.Adesiyun A A, Kaminjolo J S. Susceptibility to antibiotics of Escherichia coli strains isolated from diarrhoeic and non-diarrhoeic livestock in Trinidad. Rev Elev Med Vet Pays Trop. 1992;45:260–262. [PubMed] [Google Scholar]
- 2.Altschul S F, Madden T L, Schäffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arcangioli M A, Leroy-Sétrin S, Martel J L, Chaslus-Dancla E. A new chloramphenicol and florfenicol resistance gene flanked by two integron structures in Salmonella typhimurium DT104. FEMS Microbiol Lett. 1999;174:327–332. doi: 10.1111/j.1574-6968.1999.tb13586.x. [DOI] [PubMed] [Google Scholar]
- 4.Arcangioli M A, Leroy-Sétrin S, Martel J L, Chaslus-Dancla E. Evolution of chloramphenicol resistance, with emergence of cross-resistance to florfenicol, in bovine Salmonella Typhimurium strains implicates definitive phage type (DT) 104. J Med Microbiol. 2000;49:103–110. doi: 10.1099/0022-1317-49-1-103. [DOI] [PubMed] [Google Scholar]
- 5.Barrett T J, Lior H, Green J H, Khakhria R, Wells J G, Bell B P, Greene K D, Lewis J, Griffin P M. Laboratory investigation of a multistate food-borne outbreak of Escherichia coli O157:H7 by using pulsed-field gel electrophoresis and phage typing. J Clin Microbiol. 1994;32:3013–3017. doi: 10.1128/jcm.32.12.3013-3017.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barton B M, Harding G P, Zuccarelli A J. A general method for detecting and sizing large plasmids. Anal Biochem. 1995;226:235–240. doi: 10.1006/abio.1995.1220. [DOI] [PubMed] [Google Scholar]
- 7.Bissonnette L, Champetier S, Buisson J P, Roy P H. Characterization of the nonenzymatic chloramphenicol resistance (cmlA) gene of the In4 integron of Tn1696: similarity of the product to transmembrane transport proteins. J Bacteriol. 1991;173:4493–4502. doi: 10.1128/jb.173.14.4493-4502.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blanco J E, Blanco M, Mora A, Blanco J. Prevalence of bacterial resistance to quinolones and other antimicrobials among avian Escherichia coli strains isolated from septicemic and healthy chickens in Spain. J Clin Microbiol. 1997;35:2184–2185. doi: 10.1128/jcm.35.8.2184-2185.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bolton L F, Kelly L, Lee M D, Cray P F, Maurer J J. Detection of multidrug-resistant Salmonella enterica serotype Typhimurium DT104 based on a gene which confers cross-resistance to florfenicol and chloramphenicol. J Clin Microbiol. 1999;37:1348–1351. doi: 10.1128/jcm.37.5.1348-1351.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bradford P A, Petersen P J, Fingerman I M, White D G. Emergence of expanded-spectrum cephalosporin resistance in E. coli isolates associated with bovine calf diarrheal disease. J Antimicrob Chemother. 1999;44:607–610. doi: 10.1093/jac/44.5.607. [DOI] [PubMed] [Google Scholar]
- 11.Briggs C E, Fratamico P M. Molecular characterization of an antibiotic resistance gene cluster of Salmonella typhimurium DT104. Antimicrob Agents Chemother. 1999;43:846–849. doi: 10.1128/aac.43.4.846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cannon M, Harford S, Davies J A. A comparative study on the inhibitory actions of chloramphenicol, thiamphenicol and some fluorinated derivatives. J Antimicrob Chemother. 1990;26:307–317. doi: 10.1093/jac/26.3.307. [DOI] [PubMed] [Google Scholar]
- 13.Cloeckaert A, Boumedine K S, Flaujac G, Imberechts H, D'Hooghe I, Chaslus-Dancla E. Occurrence of a Salmonella enterica serovar Typhimurium DT104-like antibiotic resistance gene cluster including the floR gene in S. enterica serovar Agona. Antimicrob Agents Chemother. 2000;44:1359–1361. doi: 10.1128/aac.44.5.1359-1361.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dorman C J, Foster T J. Nonenzymatic chloramphenicol resistance determinants specified by plasmids R26 and R55–1 in Escherichia coli K-12 do not confer high-level resistance to fluorinated analogues. Antimicrob Agents Chemother. 1982;22:912–914. doi: 10.1128/aac.22.5.912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gilmore A. Chloramphenicol and the politics of health. Can Med Assoc J. 1996;134:423–435. [PMC free article] [PubMed] [Google Scholar]
- 16.Graham R, Palmer D, Pratt B C, Hart C A. In vitro activity of florphenicol. Eur J Clin Microbiol Infect Dis. 1998;7:691–694. doi: 10.1007/BF01964257. [DOI] [PubMed] [Google Scholar]
- 17.Hudson C R, Quist C, Lee M D, Keyes K, Dodson S V, Morales C, Sanchez S, White D G, Maurer J J. Genetic relatedness of Salmonella isolates from nondomestic birds in the Southeastern United States. J Clin Microbiol. 2000;38:1860–1865. doi: 10.1128/jcm.38.5.1860-1865.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Keyes K, Hudson C, Maurer J J, Thayer S, White D G, Lee M D. Detection of florfenicol resistance genes in Escherichia coli isolated from sick chickens. Antimicrob Agents Chemother. 2000;44:421–424. doi: 10.1128/aac.44.2.421-424.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim E, Aoki T. Sequence analysis of the florfenicol resistance gene encoded in the transferable R-plasmid of a fish pathogen, Pasteurella piscicida. Microbiol Immunol. 1996;9:665–669. doi: 10.1111/j.1348-0421.1996.tb01125.x. [DOI] [PubMed] [Google Scholar]
- 20.Liebert C A, Hall R M, Summers A O. Transposon Tn21, flagship of the floating genome. Microbiol Mol Biol Rev. 1999;63:507–522. doi: 10.1128/mmbr.63.3.507-522.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maurer J J, Lee M D, Lobsinger C, Brown T, Maier M, Thayer S G. Molecular typing of avian Escherichia coli isolates by random amplification of polymorphic DNA. Avian Dis. 1998;42:431–451. [PubMed] [Google Scholar]
- 22.Mitsuda T, Muto T, Yamada M, Kobayashi N, Toba M, Aihara Y, Ito A, Yokota S. Epidemiological study of a food-borne outbreak of enterotoxigenic Escherichia coli O25:NM by pulsed-field gel electrophoresis and randomly amplified polymorphic DNA analysis. J Clin Microbiol. 1998;36:652–656. doi: 10.1128/jcm.36.3.652-656.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nagabhushan T L, Kandasamy D, Tsai H, Turner W N, Miller G H. Novel class of chloramphenicol analogues with activity against chloramphenicol-resistant and chloramphenicol-susceptible organisms. In: Nelson J D, Grassi C, editors. Current chemotherapy and infectious disease. Washington, D.C.: American Society for Microbiology; 1980. pp. 442–443. [Google Scholar]
- 24.National Committee for Clinical Laboratory Standards. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, 4th ed. Approved standard M7–A4. Villanova, Pa: National Committee for Clinical Laboratory Standards; 1997. [Google Scholar]
- 25.National Committee for Clinical Laboratory Standards. Performance standards for antimicrobial disk and dilution susceptibility tests for bacteria isolated from animals. Approved standard M31-A. Villanova, Pa: National Committee for Clinical Laboratory Standards; 1999. [Google Scholar]
- 26.Neu H C, Fu K P. In vitro activity of chloramphenicol and thiamphenicol analogues. Antimicrob Agents Chemother. 1980;18:311–316. doi: 10.1128/aac.18.2.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Piddock L J V. Does the use of antimicrobial agents in veterinary medicine and animal husbandry select antibiotic-resistant bacteria that infect man and compromise antimicrobial chemotherapy? J Antimicrob Chemother. 1996;38:1–3. doi: 10.1093/jac/38.1.1. [DOI] [PubMed] [Google Scholar]
- 28.Salyers A A, Amabile-Cuevas C F. Why are antibiotic resistance genes so resistant to elimination? Antimicrob Agents Chemother. 1997;41:2321–2325. doi: 10.1128/aac.41.11.2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 30.Schwartz D C, Cantor C R. Separation of yeast chromosome-sized DNAs by pulsed field gradient gel electrophoresis. Cell. 1984;37:67–75. doi: 10.1016/0092-8674(84)90301-5. [DOI] [PubMed] [Google Scholar]
- 31.Smith C J, Marron M, Smith S G J. Fimbriae of Escherichia coli. In: Gyles C L, editor. Escherichia coli in domestic animals and humans. Oxon, United Kingdom: CAB International; 1994. pp. 399–435. [Google Scholar]
- 32.Smith K E, Besser J M, Hedberg C W, Leano F T, Bender J B, Wicklund J H, Johnson B P, Moore K A, Osterholm M T. Quinolone-resistant Campylobacter jejuni infections in Minnesota, 1992–1998. New Engl J Med. 1999;340:1525–1532. doi: 10.1056/NEJM199905203402001. [DOI] [PubMed] [Google Scholar]
- 33.Syriopoulou V P, Harding A L, Goldmann D A, Smith A L. In vitro antibacterial activity of fluorinated analogs of chloramphenicol and thiamphenicol. Antimicrob Agents Chemother. 1981;19:294–297. doi: 10.1128/aac.19.2.294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Trieu-Cuot P, De Cespedes G, Bentorcha F, Delbos F, Gaspar E, Horaud T. Study of heterogeneity of chloramphenicol acetyltransferase (CAT) genes in streptococci and enterococci by polymerase chain reaction: characterization of a new CAT determinant. Antimicrob Agents Chemother. 1993;37:2593–2598. doi: 10.1128/aac.37.12.2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vassort-Bruneau, C., M. C. Lesage-Descauses, J. L. Martel, J. P. Lafont, and E. Chaslus-Dancla. CAT III chloramphenicol resistance in Pasteurella haemolytica and Pasteurella multocida isolated from calves. J. Antimicrob. Chemother. 38:205–213. [DOI] [PubMed]
- 36.Wolf M K. Occurrence, distribution, and associations of O and H serogroups, colonization factor antigens, and toxins of enterotoxigenic Escherichia coli. Clin Microbiol Rev. 1997;10:569–584. doi: 10.1128/cmr.10.4.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wray C, McLaren I M, Carroll P J. Escherichia coli isolated from farm animals in England and Wales between 1986 and 1991. Vet Rec. 1993;133:439–442. doi: 10.1136/vr.133.18.439. [DOI] [PubMed] [Google Scholar]