Abstract
Chagas disease, caused by Trypanosoma cruzi, can reactivate and cause severe acute disease in immunocompromised patients such as those infected with human immunodeficiency virus (HIV). We conducted amplicon deep sequencing of a 327-bp fragment of the tcscd5 gene using an Ion Torrent PGM directly from clinical samples from HIV patients with high parasitemia. We describe the within-host diversity, both characterizing the discrete typing unit of the infections and confirming the presence of multistrain infections, directly from clinical samples. This method can rapidly provide information on the genetic diversity of T. cruzi infection, which can have direct impacts on clinical disease.
Keywords: Trypanosoma cruzi, Chagas disease, HIV, genetic diversity, discrete typing unit, multiclonality, amplicon deep sequencing, tcsc5d
Amplicon deep sequencing can be used to characterize multiplicity of infection and genotype Trypanosoma cruzi infections in patient coinfected with human immunodeficiency virus.
Trypanosoma cruzi, the cause of Chagas disease, infects 5–6 million people worldwide, with >38 000 new infections occurring yearly [1]. Trypanosoma cruzi is diploid, with a clonal population structure. However, the species is genetically diverse, with 7 discrete typing units (DTUs) that infect humans [2]. Individual DTUs have distinct, but often overlapping, geographic distributions, with TcI predominating in human infections north of the Amazon and TcII, TcV, and TcVI being most common in the Southern Cone of South America, including Bolivia [3]. TcIII and TcIV are primarily sylvatic DTUs found mainly in the Amazon forest [3]. DTUs isolated from human infections usually reflect common local strains, confounding attempts to definitively correlate DTU with clinical outcomes, but some studies have found associations between DTU and gastrointestinal symptoms, progression to cardiomyopathy, and acute orally transmitted disease, as summarized in Messenger et al [2].
There is a paucity of data, however, about DTU-related clinical differences in human immunodeficiency virus (HIV) coinfections, though multiple DTUs have been reported in HIV-infected patients [2]. In patients infected with HIV, reactivation of chronic T. cruzi infection can cause severe neurological syndromes with mortality as high as 80% [4]. Bolivia has the highest prevalence of Chagas disease in the world at about 6% [1] and exceeding 50% in some villages in the Bolivian Chaco [5]. In a study of acutely ill HIV-infected patients in Santa Cruz, Bolivia, we found that 25% were also seropositive for T. cruzi (unpublished data).
Next-generation sequencing (NGS) is a powerful tool to understand the genetics and epidemiology of infections that has been rarely utilized to study the epidemiology or pathogenesis of Chagas disease. In this study, we demonstrate the utility of amplicon deep sequencing to characterize the diversity of chronic T. cruzi infection in immunocompromised persons. In contrast to chronically infected immunocompetent persons, HIV/T. cruzi–coinfected persons frequently exhibit very high levels of parasitemia due to immunosuppression, which provides sufficient genetic material for sequencing without first culturing the parasite. Culture of T. cruzi can select for certain strains more adapted to growth in vitro, which can lead to underestimation of parasite diversity and mismeasurement of haplotype frequencies [6]. Sequencing directly from clinical samples can provide a more accurate representation of the diversity and relative frequencies of parasite populations in human infection. Amplicon deep sequencing from clinical samples is sensitive enough to detect mixed-genotype infections and avoids biases introduced by culture. We targeted the tcsc5d gene, which putatively encodes a C-5 sterol desaturase (lathosterol/episterol oxidase) on chromosome 22, because it is a single-copy nuclear gene that contains single-nucleotide polymorphisms (SNPs) without significant insertions or deletions, allowing us to estimate multiplicity of infection. Additionally, it can be used to distinguish between different DTUs [7]. Here we show that deep sequencing of T. cruzi directly from clinical samples is possible with sufficient levels of parasitemia.
MATERIALS AND METHODS
Blood samples were obtained from HIV-positive subjects from Cochabamba (HIV167) and Santa Cruz (HIV70, HIV76, HIV141), Bolivia. T. cruzi infection was confirmed by at least 2 serological tests and by quantitative polymerase chain reaction (qPCR) as described below [8]. Human blood samples were also examined by microscopy. DNA from stock strains (maintained by the Miles laboratory, London School of Tropical Medicine and Hygiene) of each DTU maintained in culture were analyzed in parallel for reference (Supplementary Table) with the exception of the reference strain from DTU IV, which was degraded and did not amplify; however, TcIV is primarily a sylvatic strain that very rarely infects humans.
DNA was extracted from either clotted blood (all samples) or whole blood with ethylenediaminetetraacetic acid and guanidine (HIV167) using a Qiacube automated system with Qiagen reagents (Qiagen N.V., Hilden, Germany). Presence of T. cruzi was detected using qPCR with the Cruzi 1 and Cruzi 2 primers and Cruzi 3 probe using FastStart Universal Probe Master (Rox) reagents from Roche Diagnostics according to published methods [8]. Samples were considered positive if the cycle threshold (Ct) was less than the cutoff determined using a standard curve.
A fragment of the tcsc5d gene (GenBank: HQ586986 [CL-Brener] and JN050564–JN050587; https://tritrypdb.org/tritrypdb/app/record/gene/TcCLB.507853.10) was amplified using hemi-nested PCR with outer primers forward 5′-GGACGTGGCGTTTGATTTAT and reverse 5′-TCCCATCTTCTTCGTTGACT-3′ using published conditions [7]. The second reaction was performed using inner forward primer 5′-CCTTGTGATGGATTGGTCA-3′ with the same reverse primer to obtain a 326-bp-long fragment (see Supplementary Table 1 for reaction conditions). A 10-base barcode tag was included at the 5′ end of the inner forward primer so amplicons could be pooled during library preparation. Hemi-nested PCR was performed using Phusion Taq (NEB, Ipswich, Massachusetts) for the outer PCR and Roche Diagnostics FastStart High Fidelity enzyme blend (Hoffman-La Roche, Basel, Switzerland) to generate the inner PCR fragment. Amplicon libraries were generated from pooled amplicons using the Ion Plus Fragment Library Kit and Ion Xpress Barcode Adapters 1–16 kit following manufacturer instructions. Sequencing was performed on the Ion Torrent platform using a 318-size chip by the University of North Carolina High-Throughput Sequencing Facility. Samples were amplified and sequenced in duplicate.
Sequence extraction, clustering, and comparison of clusters from replicates were all done using SeekDeep version 2.1.1 (https://seekdeep.brown.edu/) [9]. Data were first demultiplexed according to barcode and then replicate PCRs were compared to find haplotypes. The demultiplexed sequences were matched to barcodes and primers and both were removed. The quality filtering, also using Seekdeep, was done using a sliding window of 50 bases, moving by 5 bases, requiring an average quality score of 20 or above setting a minimum length of 300 bp. Replicates from resulting filtered sequences were then clustered to compared to find haplotypes. Only haplotypes that were found in both sample replicates were retained. Replicates where prevalence of any given haplotype was absolutely different by >15% were rejected. Samples that had <200 reads in any replicate were rejected. Sequences were aligned using MEGA version 10.0 [10]. Data were visualized using SeekDeep [9].
Ethical Considerations
This analysis was exempt from Institutional Review Board approval at UNC because only de-identified samples were used. The studies that served as sources of human samples were approved by the institutional review boards at John Hopkins Bloomberg School of Public Health (Baltimore, Maryland); the London School of Tropical Medicine and Hygiene (London, United Kingdom); Universidad Católica Boliviana (Santa Cruz, Bolivia); Hospital Clínico Viedma (Cochabamba, Bolivia); Instituto de Desarrollo Humano (Cochabamba, Bolivia); Colectivo de Estudios Aplicados, Desarrollo Social, Salud y Medio Ambiente (Cochabamba, Bolivia); Asociación Benéfica PRISMA (Lima, Peru); and Universidad Peruana Cayetano Heredia (Lima, Peru).
RESULTS
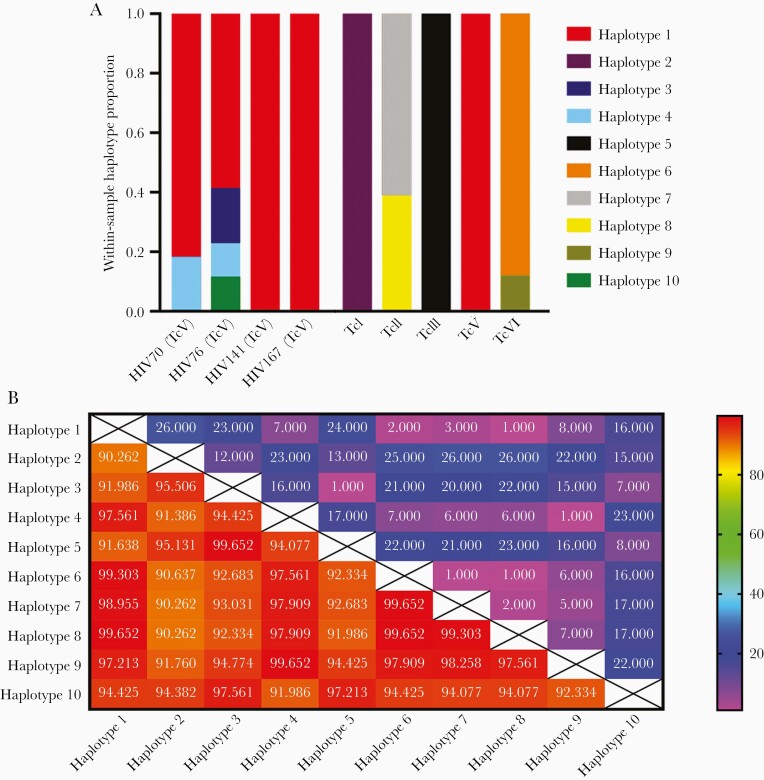
We amplified and sequenced the target amplicon from 4 samples from HIV-infected patients (Table 1) as well as cultures of reference DTUs. After quality filters were applied, we identified 10 unique haplotypes (Figure 1A and Supplementary Table 2). The predominant haplotype in the 4 human samples was identical to the TcV reference DTU. The distribution of SNPs across the amplicon varied, with some areas showing high levels of diversity and others not (Supplementary Figures 1 and 2).
Table 1.
Patient Characteristics
| Code | Age | Sex | CD4 Count | HIV Viral Load (copies/ml) | Micromethod Result | Parasite Load (parasites/ml) | Clinical Description |
|---|---|---|---|---|---|---|---|
| HIV70 | 41 | Male | 179 | 83 653 | Positive | 556 726 (clot) | Diagnosed with HIV 1 year prior to enrollment; sensory and motor deficit below T5; hypotensive (BP 80/60); LP with protein 130, glucose 18, 2 TNCs; rapidly declined and developed respiratory failure and died 22 days after enrollment |
| HIV76 | 43 | Male | NA | NA | Positive | 228 424 (clot) | Newly diagnosed HIV; diarrhea, pneumonia, malnutrition, GCS 12; died 6 days after enrollment |
| HIV141 | 57 | Male | NA | NA | Positive | 134 254 (clot) | Newly diagnosed HIV; gastroenteritis, dehydration, oropharyngeal thrush; died 3 months after enrollment |
| HIV167 | 27 | Male | 15 | 6705 | Positive | 9951 (clot) 21 462 (EDTA) |
Newly diagnosed HIV; pneumonia, gastroenteritis, oropharyngeal thrush, malnutrition (weight 42 kg); mild bilateral tremor; LAFB on EKG; normal MMSE |
Abbreviations: BP, blood pressure; EDTA, ethylenediaminetetraacetic acid; EKG, electrocardiogram; GCS, Glasgow Coma Score; HIV, human immunodeficiency virus; LAFB, left anterior fascicular block; LP, lumbar puncture; MMSE, Mini-Mental State Examination; NA, not available; TNC, total nucleated cell.
Figure 1.
A, tcsc5d haplotype distribution in human clinical samples, cultured reference strains, and a dog from rural Bolivia. B, Heatmap showing the number of single-nucleotide polymorphism differences (blue and pink) and percentage sequence identity (red and orange) between haplotypes.
At least 1 (HIV76) and possibly 2 (HIV70) samples showed evidence of infection by at least 2 strains of T. cruzi and potentially as many as 4 based on the number of haplotypes identified. Trypanosoma cruzi is a diploid organism, making it impossible to distinguish between infection by 2 unique strains or a strain heterozygous at the target gene; however, the widely different haplotype frequencies in both samples, with haplotype 1 representing the majority of reads with <20% abundance for any other haplotype, suggest that these represent polyclonal infections rather than heterozygosity.
Haplotype sequences included several SNPs useful for genotyping that were consistent with DTU TcV [7]. the most common in Bolivian human infections. Blood from patient HIV70 was cultured in Santa Cruz, Bolivia. The culture was extracted and resulting DNA was analyzed using PCR-restriction fragment length polymorphism analysis with the restriction enzymes hsp60 and gpi (Supplementary Figure 3). The PCR–restriction fragment length polymorphism pattern was most consistent with DTU TcV or VI, concordant with our sequencing results. The dominant haplotype in HIV70 is identical to TcV, as expected. Haplotype 4, the minority population found in this patient, differs by only 1 nucleotide from one of the haplotypes (haplotype 9) sequenced from the TcVI reference strain (Figure 1B), also a relatively frequent cause of human infections in Bolivia [2].
DISCUSSION
Though polyclonal infections have been characterized from culture and using other modalities [11, 12], to our knowledge no one has used deep sequencing to detect polyclonal T. cruzi infections directly from clinical samples. Multiplicity of infection does occur in chronic Chagas disease, and here we show that it can be seen in reactivation, defined by microscopically detectable parasitemia, in HIV-infected patients. HIV/T. cruzi–coinfected persons often exhibit high-level parasitemia that provides the opportunity to study multiplicity of infection [13]. Here we show that deep sequencing can be done directly from clinical samples, though sensitivity is currently limited to qPCR Ct values of <25–28 (approximately 0.0005–0.00005 ng parasite DNA/μL) by the primers we used. Though we only present data from 4 patients here, we provide proof-of-concept for use of this technique. To our knowledge, this work represents the first use of this genotyping scheme directly from human samples (without passage through culture) and the first use of deep sequencing to characterize T. cruzi populations in HIV-infected patients.
One reason for limited sensitivity is the use of a single-copy nuclear gene like tcsc5d, which distinguishes between different copies within a single genome and multiple infection within a host, in contrast to other schemes that target multicopy genes [11]. The tcsc5d gene is useful for DTU-typing [7], though our truncated amplicon could not definitively distinguish all 6 compared to sequencing the full gene; however, the predominant haplotype in our clinical samples was identical to the TcV reference strain, the most common DTU in human infections in this area of Bolivia [3]. We included results from the reference strains to demonstrate the robustness of our methods. The TcII strain contained 2 haplotypes (7 and 8), which could reflect T. cruzi’s diploid genome, though it could also represent PCR or sequencing artifact, as the haplotypes differed by only 2 SNPs. TcV is a hybrid of TcII and TcII, but we isolated only 1 haplotype. TcVI, also a TcII/TcIII hybrid, contained 2 haplotypes that differed by 6 SNPs [14]. When we aligned the haplotypes against reference tcsc5d sequences (Supplementary Figure 2) [7], we identified a number of conserved heterozygous SNPs in TcV and TcVI. The levels of heterozygosity observed in this short fragment for TcV and TcVI are consistent with other T. cruzi housekeeping genes [15]. Though similar across this fragment, there is a SNP at position 111 that can distinguish between TcV and TcVI, although we acknowledge that gene conversion events in other TcV samples could potentially confound this.
In conclusion, this work demonstrates the feasibility of amplicon deep sequencing for T. cruzi directly from clinical samples, without need for passage through culture or xenodiagnosis. Culture from blood or by xenodiagnosis is laborious and inefficient, requiring up to several weeks to grow sufficient parasite quantities; furthermore, culture can select for culture-adapted strains, impose genetic bottlenecks reducing diversity in multiclonal populations, and introduce bias into sequencing results [6]. The tcsc5d gene is an ideal sequencing target because it is a single-copy nuclear gene, allowing more accurate determination of multiplicity of infection, and it is sufficiently variable to detect inter- and intra-DTU variation. While other genotyping algorithms compatible with deep sequencing exist, notably use of the mini-exon region [12], they are unable to differentiate all 7 DTUs. This is the first time tcsc5d has been used to characterize parasite genetic diversity in human infections directly from extracted blood samples using deep sequencing technology. We hope this technique can be extended to investigate intrahost parasite dynamics, parasite evolution, and transmission patterns.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Acknowledgments. The Working Group on Chagas Disease in Bolivia and Peru includes Daniel Clark, Jorge Flores, Roni Colanzi, Jeong Choi, Gerson Galdos, Mauricio Dorn, Omar Gandarilla, Enzo Fortuny, Anne Palumbo, Lisbeth Ferrufino, Monica Pajuelo, Melissa Reimer, and Sandra Mendoza Guerrero . We thank the Michael Miles group (London School of Tropical Medicine and Hygiene) for the reference strains. We acknowledge Steven R. Meshnick’s (now deceased) contributions as well.
Financial support. This work was supported by the National Institute of Allergy and Infectious Diseases (grant numbers K23 AI113197 and P30 AI50410 to N. M. B. and K24 AI13499 to J. J. J.); the Burroughs Wellcome Fund/American Society of Tropical Medicine and Hygiene (to N. M. B.); and the Fogarty International Center (grant number R25 TW009340 to N. M. B.).
Potential conflicts of interest. All authors: No reported conflicts of interest.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
Presented in part: Annual meeting of the American Society of Tropical Medicine and Hygiene, October 2015, Philadelphia, Pennsylvania. Poster 1085.
Contributor Information
Working Group on Chagas Disease in Bolivia and Peru:
Daniel Clark, Jorge Flores, Roni Colanzi, Jeong Choi, Gerson Galdos, Mauricio Dorn, Omar Gandarilla, Enzo Fortuny, Anne Palumbo, Lisbeth Ferrufino, Monica Pajuelo, Melissa Reimer, and Sandra Mendoza Guerrero
References
- 1. World Health Organization. Chagas disease in Latin America: an epidemiological update based on 2010 estimates. Wkly Epidemiol Rec 2015; 90:33–44. [PubMed] [Google Scholar]
- 2. Messenger LA, Miles MA, Bern C. Between a bug and a hard place: Trypanosoma cruzi genetic diversity and the clinical outcomes of Chagas disease. Expert Rev Anti Infect Ther 2015; 13:995–1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Brenière SF, Waleckx E, Barnabé C. Over six thousand Trypanosoma cruzi strains classified into discrete typing units (DTUs): attempt at an inventory. PLoS Negl Trop Dis 2016; 10:e0004792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. de Almeida EA, Ramos Júnior AN, Correia D, Shikanai-Yasuda MA. Co-infection Trypanosoma cruzi/HIV: systematic review (1980–2010). Rev Soc Bras Med Trop 2011; 44:762–70. [DOI] [PubMed] [Google Scholar]
- 5. Samuels AM, Clark EH, Galdos-Cardenas G, et al. ; Working Group on Chagas Disease in Bolivia and Peru . Epidemiology of and impact of insecticide spraying on Chagas disease in communities in the Bolivian Chaco. PLoS Negl Trop Dis 2013; 7:e2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Devera R, Fernandes O, Coura JR. Should Trypanosoma cruzi be called “cruzi” complex? A review of the parasite diversity and the potential of selecting population after in vitro culturing and mice infection. Mem Inst Oswaldo Cruz 2003; 98:1–12. [DOI] [PubMed] [Google Scholar]
- 7. Cosentino RO, Agüero F. A simple strain typing assay for Trypanosoma cruzi: discrimination of major evolutionary lineages from a single amplification product. PLoS Negl Trop Dis 2012; 6:e1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Piron M, Fisa R, Casamitjana N, et al. Development of a real-time PCR assay for Trypanosoma cruzi detection in blood samples. Acta Trop 2007; 103:195–200. [DOI] [PubMed] [Google Scholar]
- 9. Hathaway NJ, Parobek CM, Juliano JJ, Bailey JA. SeekDeep: single-base resolution de novo clustering for amplicon deep sequencing. Nucleic Acids Res 2018; 46:e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 2018; 35:1547–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Llewellyn MS, Messenger LA, Luquetti AO, et al. Deep sequencing of the Trypanosoma cruzi GP63 surface proteases reveals diversity and diversifying selection among chronic and congenital Chagas disease patients. PLoS Negl Trop Dis 2015; 9:e0003458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Villanueva-Lizama L, Teh-Poot C, Majeau A, Herrera C, Dumonteil E. Molecular genotyping of Trypanosoma cruzi by next-generation sequencing of the mini-exon gene reveals infections with multiple parasite discrete typing units in chagasic patients from Yucatan, Mexico. J Infect Dis 2019; 219:1980–8. [DOI] [PubMed] [Google Scholar]
- 13. de Freitas V, da Silva S, Sartori A, et al. Real-time PCR in HIV/Trypanosoma cruzi coinfection with and without Chagas disease reactivation: association with HIV viral load and CD4 level. PLoS Negl Trop Dis 2011; 5:e1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lewis MD, Llewellyn MS, Yeo M, Acosta N, Gaunt MW, Miles MA. Recent, independent and anthropogenic origins of Trypanosoma cruzi hybrids. PLoS Negl Trop Dis 2011; 5:e1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yeo M, Mauricio IL, Messenger LA, et al. Multilocus sequence typing (MLST) for lineage assignment and high resolution diversity studies in Trypanosoma cruzi. PLoS Negl Trop Dis 2011; 5:e1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.