Abstract
To prescribe effective treatment schemes for patients with tuberculosis, more-efficient susceptibility testing techniques for Mycobacterium tuberculosis are needed, especially in regions with multidrug resistance. Etest (AB BIODISK, Solna, Sweden) is a simple technique that provides quantitative drug susceptibility results for M. tuberculosis in 5 to 10 days from a culture grown at low cost. The performance of Etest was compared to that of the reference proportion method, using 95 M. tuberculosis clinical isolates of which 42.1% (40 of 95) were resistant to at least one antibiotic by the reference method. Overall agreement between Etest and the reference method was 98.9% (94 of 95) for detection of multidrug resistance; for resistance to individual drugs, agreement was 97.9% (93 of 95) for rifampin, 96.0% (92 of 95) for ethambutol, 94.7% (90 of 95) for isoniazid, and 85.3% (81 of 95) for streptomycin. This study supports the utility of Etest for timely detection of drug resistance in M. tuberculosis and for use in tuberculosis control programs.
Tuberculosis (TB) is a growing global health problem, both in terms of disease burden and in terms of resistance to conventional chemotherapy (7). The regions where TB is more prevalent lack the resources to implement appropriate measures to control the disease (13); hence, it is likely that the problem will increase further. The standard treatment of TB as recommended by the World Health Organization (WHO) is a multidrug regimen that includes four antibiotics (rifampin [RIF], isoniazid [INH], pyrazinamide, and streptomycin [STR] or ethambutol [EMB]). This treatment scheme is usually effective against Mycobacterium tuberculosis (5, 21). However, in settings with a high frequency of drug resistance, this regimen is ineffective and results in lower cure rates (1). The continued use of the standard treatment when the cure rate is low maintains or increases the rates of resistance (13).
The bactericidal activity of both RIF and INH in killing M. tuberculosis (30) makes these drugs most effective for standard treatment of TB. When an M. tuberculosis strain is resistant to at least these two antibiotics, the effectiveness of the standard treatment is diminished by 15 to 77% (13). Therefore, resistance to these two drugs (34) defines multidrug resistance (MDR) with significant clinical impact.
The prevalence of TB in any region is influenced by biological, behavioral, and socioeconomic factors (15). These factors also affect the appearance of MDR TB, which is a manmade phenomenon that originates principally through inadequate chemotherapy (14). A declining public health infrastructure associated with increasing levels of MDR TB (18, 19) can occur in any country on a focal basis. The global and widespread emergence of drug-resistant TB is supported by the fact that MDR prevalences are 1.6% in the United States, 1.1% in the United Kingdom, 4.6% in Argentina, 6.6% in the Dominican Republic, and 14.4% in Latvia (23).
Early recognition and appropriate treatment have been proven to be one of the most effective strategies to control MDR TB (14) even in human immunodeficiency virus (HIV)-infected populations (31). Knowledge of the drug susceptibility pattern of the MDR clinical isolate is necessary to design and prescribe an appropriate treatment for the patient. Susceptibility testing can prevent treatment failures and thereby diminish the number of secondary cases of MDR TB (3).
The method recommended by the NCCLS for susceptibility testing of M. tuberculosis is the modified agar proportion. The BACTEC system (Becton Dickinson) is also widely used. The proportion method is an inexpensive and relatively simple technique, which provides results in 3 weeks from a cultured isolate. The BACTEC system provides results in only 5 days but requires expensive equipment and reagents and technical expertise. Molecular techniques such as PCR and DNA hybridization assays provide results in 24 h, but they require specialized equipment and highly skilled personnel, and they have not yet been developed for all known mutations and antimycobacterial drugs (30). Cost-effective techniques that do not depend on prior identification of the molecular mechanisms of resistance are needed for the rapid diagnosis of MDR, wherever it may prevail.
Etest (AB BIODISK, Solna, Sweden) is a recent innovation for quantitative antibiotic susceptibility testing of a wide variety of microorganisms (24). Preliminary studies of its application to M. tuberculosis have shown good agreement with the reference agar proportion method and BACTEC (100, 97.5, 91.3, and 98.7% agreement for RIF, INH, EMB, and STR, respectively) using a sample of clinical isolates with a low frequency of resistance (4.9%; 4 of 81) (12). These results, and the feasibility of obtaining quantitative MIC results within 5 days at a modest cost without specialized equipment, prompted further evaluation of Etest for the detection of drug-resistant TB. In this study, the performance of Etest relative to the proportion method was investigated using clinical isolates of M. tuberculosis from patients in a population with a high prevalence of MDR TB (4, 18, 19). To evaluate the ability of Etest to detect MDR TB, our comparison to the “gold standard” with isolates having a higher frequency of resistance would be desirable (2, 25). This is the first study in which Etest is validated with clinical isolates of M. tuberculosis with a high prevalence of drug resistance (45.3% [43 of 95] of the isolates studied were resistant to at least one drug).
MATERIALS AND METHODS
Clinical isolates.
A total of 95 clinical isolates of M. tuberculosis were obtained from 95 patients from Buenaventura, Colombia (19). Patients were selected according to their history of receiving at least 1 month of TB treatment; 37 had records of previous treatment(s) (acquired-resistance group) and 58 did not (primary-resistance group). The clinical isolates were grown on modified Ogawa-Kudoh slants (26) for use in drug susceptibility testing.
M. tuberculosis reference strains.
M. tuberculosis H37Rv (ATCC 27294) (susceptible to all antituberculous agents) and M. tuberculosis AWC (resistant to INH and STR) (32) were used for quality control. Both reference strains were used as controls in every Etest evaluation.
Susceptibility testing.
Susceptibilities to INH, RIF, STR, and EMB were determined in a double-blind manner by the proportion method and Etest. Testing by the modified agar proportion method was performed at the Centers for Disease Control and Prevention (CDC, Atlanta, Ga.) as recommended by Kent and Kubica (17). The Etest was performed at the Centro Internacional de Entrenamiento e Investigaciones Médicas (CIDEIM) as follows. Colonies of M. tuberculosis from Ogawa-Kudoh slant cultures incubated for 3 to 4 weeks were suspended in Middlebrook 7H9 broth (Difco, Detroit, Mich.) using 3-mm glass beads to achieve a turbidity equivalent to a McFarland standard of 3.0 (∼9 × 108 CFU/ml). Middlebrook medium was prepared and stored for up to 30 days at 4°C and was protected from light (either sterile or during incubation). Five Middlebrook 7H11 agar plates (100 mm) supplemented with 10% oleic acid, albumin, dextrose, and catalase (OADC) (Difco) were inoculated by swabbing the mycobacterial suspension onto the agar surface. One Etest strip was placed on each plate after 24 h of preincubation of the inoculated plate at 37°C with 5% CO2. No strip was placed on the fifth plate, which served as a growth control. The plates were incubated under the same conditions for 5 to 10 days, after which the MIC was read. The MIC was the value where the growth inhibition ellipse intersected the strip or as specified in the AB BIODISK Etest technical guide no. 6 for M. tuberculosis (1997). As a control, each experiment included susceptibility testing of the reference strains. For the interpretation of susceptibility categories, the following susceptibility breakpoints were utilized: ≤1.0, 0.2, 5.0, and 2.0 μg/ml for RIF, INH, STR, and EMB, respectively (22). Isolates with discordant categorical results were reassessed by Etest and the proportion method, unless the isolate was no longer viable.
To determine the reproducibility of Etest, a set of 24 strains (chosen at random) was evaluated at least twice in separate experiments for the four drugs. For cases where paired results were not available due to contamination, the complete drug set was reevaluated and the values were included in the reproducibility analysis.
Statistical analyses.
Data were analyzed with the statistical package SPSS for Windows, release 7.5 (SPSS, Inc.). The reproducibility of the categorical results was described by percent agreement and the Kappa statistic, a measure of the percent agreement beyond that expected by chance (9). The reproducibilities of Etest MICs were evaluated by determining the reliability coefficient after converting the values to log2 MIC (6) (the Pearson regression test was not used because tests were repeated more than twice). The sensitivity, specificity, and positive and negative predictive values (16) of Etest were determined using the proportion method as the reference method. The positive and negative predictive values were determined for several theoretical prevalences using Bayes' theorem (16). McNemar's test with a level of α = 0.05 was used to determine if the difference between the two methods was significant.
RESULTS
The reference agar proportion method identified 43.2% (41 of 95) of the isolates as resistant to at least one of the four antibiotics. Resistance to INH was the highest at 38.9% (37 of 95), while resistances to STR, RIF, and EMB were 22.1% (21 of 95), 22.1% (21 of 95), and 7.4% (7 of 95), respectively. Resistance to one, two, three, and four antibiotics was observed in 13.7% (13 of 95), 15.7% (15 of 95), 9.5% (9 of 95), and 4.2% (4 of 95) of the isolates, respectively. MDR was found in 21.1% (20 of 95) of the isolates. Resistance to RIF was strongly associated with resistance to INH (20 of the 21 RIF-resistant isolates were also resistant to INH).
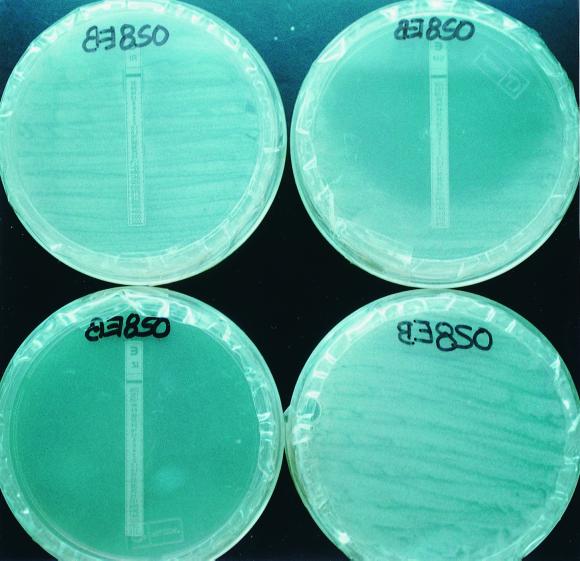
Etest results were easily read and interpreted (Fig. 1) (no differences were observed between Etest MIC interpretations by independent readers). The percentages of agreement, sensitivity, specificity, and positive and negative predictive values for Etest were high with regard to the individual drugs and MDR criteria (Table 1). Agreement with the proportion method was greater for RIF, INH, and EMB than for STR.
FIG. 1.
Etest results for a clinical isolate of M. tuberculosis after 10 days of incubation. Resistance to RIF (MIC, >32 μg/ml) is evidenced by the presence of growth along the strip (top left); susceptibility to INH (MIC, <0.016 μg/ml) is evidenced by a lack of growth in the whole plate (bottom left); and an inhibition ellipse is observed in the STR plate (top right), in which a MIC of 0.25 μg/ml (susceptible) is read. No Etest strip was placed on the control plate (bottom right). The EMB plate is not shown here.
TABLE 1.
Comparison between Etest and the reference agar proportion method
| Antibiotic | Agreementa | Sensitivityb | Specificityc | PPVd | NPVe | Agreementa upon retesting |
|---|---|---|---|---|---|---|
| RIF | 97.9 (93/95) | 100.0 (21/21); 80.8–100.0 | 97.3 (72/74); 89.7–99.5 | 91.3 (21/23); 70.5–98.5 | 100.0 (72/72); 93.7–100 | 100.0 (95/95) |
| INH | 94.7 (90/95) | 94.6 (35/37); 80.5–99.1 | 94.8 (55/58); 84.7–98.7 | 92.1 (35/38); 77.5–97.9 | 96.5 (55/57); 86.8–99.4 | 97.9 (93/95) |
| EMB | 96.0 (92/95) | 85.7 (6/7); 42.0–99.2 | 97.7 (86/88); 91.3–99.6 | 75.0 (6/8); 35.6–95.5 | 98.9 (86/87); 92.9–99.9 | 100.0 (95/95) |
| STR | 85.3 (81/95) | 85.7 (18/21); 62.6–96.2 | 85.1 (63/74); 74.5–92.0 | 62.1 (18/29); 42.4–78.7 | 95.5 (63/66); 86.4–98.8 | 94.7 (90/95) |
| MDR (RIF and INH) | 98.9 (94/95) | 100.0 (21/21); 80.8–100.0 | 98.6 (73/74); 91.7–99.9 | 95.5 (21/22); 75.1–99.8 | 100.0 (73/73); 93.8–100.0 | 100.0 (95/95) |
| Any resistance (RIF, INH, EMB, or STR) | 93.7 (356/380) | 93.0 (80/86); 84.9–97.1 | 93.9 (276/295); 90.3–96.2 | 81.6 (80/98); 72.3–88.5 | 97.9 (276/282); 95.2–99.1 | 98.2 (373/380) |
Percent (number of concordant results/total results).
Ability of Etest to detect resistance, expressed as percent (number of isolates resistant by both methods/number resistant by the proportion method); 95% confidence interval.
Ability of Etest to detect susceptibility, expressed as percent (number of isolates susceptible by both methods/number susceptible by the proportion method); 95% confidence interval.
Positive predictive value, expressed as percent (number of isolates resistant by both methods/number resistant by Etest); 95% confidence interval.
Negative predictive value, expressed as percent (number of isolates susceptible by both methods/number susceptible by Etest); 95% confidence interval.
In 22 isolates, 24 of 88 initial comparisons between the proportion method and Etest were discordant. After blinded retesting by both methods with these 22 isolates, only seven discrepancies remained (Table 2). Upon retesting, the reference proportion method yielded a different categorical result for nine of the discrepancies, while Etest yielded a different result for eight. Thirteen of these 17 changes were from susceptible to resistant. The retest results showed higher percentages of agreement than the original values (Table 1).
TABLE 2.
Isolates with discordant results between Etest and agar proportion method
| Isolate | Antibiotic | Proportion method resulta (initial/retest) | Etest result (MIC, in μg/ml) for initial testing/retesting | Status after retesting |
|---|---|---|---|---|
| 044EB | INH | R/R | S (0.016)/S (0.064) | Discordant |
| 032EB | INH | S/S | R (12.0)/R (48.0) | Discordant |
| 032EB | STR | S/S | R (128.0)/R (256.0) | Discordant |
| 015EB | STR | R/R | S (1.5)/ND | Discordant |
| 020EBS | STR | S/S | R (4.0)/R (96.0) | Discordant |
| 085VT | STR | S/S | R (4.0)/ND | Discordant |
| 091VT | STR | S/S | R (3.0)/ND | Discordant |
| 002EB | INH | S/ND | R (0.25)/S (0.016) | Concordant |
| 043EB | INH | S/R | R (32)/R (0.5) | Concordant |
| 090VT | INH | R/R | S (<0.016)/R (8) | Concordant |
| 19EB | STR | S/ND | R (4.0)/S (0.94) | Concordant |
| 090VT | STR | R/R | S (<0.016)/R (>256) | Concordant |
| 049EB | STR | S/R | R (6.0)/R (96) | Concordant |
| 03EBS | STR | S/R | R (8.0)/R (>256) | Concordant |
| 05EBS | STR | S/R | R (16)/R (>256) | Concordant |
| 06EBS | STR | R/R | S (0.016)/R (>256) | Concordant |
| 07EBS | STR | R/R | R (3.0)/S (0.125) | Concordant |
| 08EBS | STR | S/ND | R (3.0)/S (0.5) | Concordant |
| 092VT | STR | S/R | R (96)/R (96) | Concordant |
| 10EBS | EMB | S/R | R (>256)/R (>256) | Concordant |
| 132VT | EMB | R/R | S (0.125)/R (8) | Concordant |
| 034VT | EMB | S/R | R (>256)/R (8) | Concordant |
| 086VT | RIF | S/R | R (1.0)/R (1.5) | Concordant |
| 23EBS | RIF | S/R | R (32)/R (1.0) | Concordant |
R, resistant; S, sensitive; ND, not done (isolate was nonviable or contaminated at the time of retesting).
For the reproducibility analysis, paired results with all 24 isolates were obtained for STR only; for the other drugs only 20 of 24 were available due to contamination. In order to analyze the reproducibility of Etest, the Kappa statistic was determined using the categorical results, and the reliability coefficient was determined using the MICs converted to log2 MIC. The percent agreement was high, ranging from 79 to 95%. The Kappa statistics were all above 0.4, indicating that the Etest results are reproducible for the four antibiotics. Likewise, the reliability of the MICs was substantiated by reliability coefficients between 0.88 and 0.96 (Table 3).
TABLE 3.
Reproducibility of Etest
To establish the theoretical positive and negative predictive values for Etest in different scenarios of drug resistance, Bayes' theorem (16) was applied using a range of resistance prevalences of 0 to 42.4% as reported in 28 countries for each antibiotic or its combinations (23). The values obtained (data not shown) indicate that the negative predictive values varied between 95 and 100.0% in all cases, while the positive predictive values varied between 0 and 91%.
DISCUSSION
In this study of M. tuberculosis isolates with a high frequency of drug resistance, Etest provided reproducible categorical and quantitative results as previously reported (32). Etest had an average of 93.0 to 93.9% sensitivity, specificity, and agreement with the proportion method, supporting the reliability of the method for the four drugs evaluated.
The Etest was proven to yield reproducible results using a sample of isolates chosen randomly. However, it was observed that when the discrepancies between Etest and the proportion method were reevaluated, the result varied in 8 out of 24 tests by Etest, and in 9 out of 24 tests by the proportion method. Thirteen of 17 discrepancies varied from susceptible to resistant, which might have been due to an in vitro selection of a resistant subpopulation during subculturing. In optimal conditions, the same subculture would have been used to perform the drug susceptibility testing by both methods, but this was not feasible, and different subcultures were used (i.e., subcultures were performed to send the isolates to CDC), which might in part explain the variation in the categorical results. The variation in the MICs observed between repeats (e.g., isolate 90VT or 06EBS in Table 2), could be explained by poor growth in the initial testing. The retest results were not used to recalculate the specificity, sensitivity, and negative and positive predictive values of the Etest, because such discrepant analysis is not recommended for diagnostic techniques due to frequent overestimation of performance (20).
In the case of EMB, the small number of resistant cases (7.3% [7 of 95]) limited the stability of the sensitivity estimation, while the low prevalence of resistance decreased the positive predictive value (Table 1).
To effectively control TB, it is preferable to overdiagnose resistance than to miss patients with resistant strains of M. tuberculosis. The positive and negative predictive values of Etest indicate that the technique is able to accurately detect resistance and susceptibility to the four antibiotics and their combinations. Etest can positively impact TB control as a tool for individual patient testing and surveillance programs in settings with high resistance prevalences, in addition to its advantages in shorter turnaround time, simplicity of performance, and lower cost.
Good performance for RIF, as previously reported for Etest (12, 26, 32), was also observed in this study, where agreement was 97.9%. The proposal that testing of RIF alone may be adequate to diagnose MDR is applicable to Etest. Using RIF resistance as a surrogate marker for MDR (8, 11, 29, 33) was supported by our findings, where 95.2% (20 of 21) of the RIF-resistant M. tuberculosis isolates were MDR. Etest could be used as a screen for MDR by testing RIF. The RIF-resistant isolates detected should be further confirmed by Etest and/or reference methods to determine MDR status by performing complete susceptibility testing (including second- and third-line drugs) to select an appropriate treatment scheme. Patients who have RIF-sensitive isolates can be treated with the standard regimen recommended by WHO.
In settings like the Colombian port city of Buenaventura, where resources are severely limited and the basic needs of 52.2% of the rural population are unmet (28), no routine susceptibility testing is performed despite a high frequency of MDR TB cases. Under such circumstances, screening for MDR TB patients using Etest for RIF could strengthen the TB control program. This practice would be cost-effective for several reasons. Etest could be more easily implemented and sustained, because it reduces the number of operator-dependent variations in the preparation and dilution of each antibiotic. Screening for RIF resistance would reduce the number of multidrug susceptibility tests. This strategy would allow the TB program to diagnose MDR earlier and reduce the costly management of MDR cases by reducing the number of contacts with MDR TB. Equally important would be the reduction in ineffective prescriptions for standard TB treatment for MDR TB, reducing the selection of new MDR TB strains in the population.
The Etest offers decision makers an economically and technologically feasible means of drug susceptibility testing that may not be possible with the proportion method. Even in situations where resources are not the limiting factor, the Etest offers an important advantage in time and simplicity. The results obtained with four of the first-line antibiotics in these drug-resistant isolates encourage the development of Etest for other first-, second-, and third-line antibiotics. The availability of the Etest for the spectrum of antibiotics used to manage MDR cases would facilitate the treatment and control of MDR TB.
In conclusion, Etest was found to be a robust, sensitive, and specific tool for the timely detection of drug resistance in M. tuberculosis. These features support its use in TB control programs, especially in settings with high levels of drug resistance.
ACKNOWLEDGMENTS
This work was supported by the Secretaría de Salud del Valle del Cauca; the Division of AIDS, STD, and Tuberculosis Laboratory Research, CDC; and CIDEIM. The Young Investigator program of COLCIENCIAS supported M. S. Orozco.
We thank S. Brim and B. Metchock, from the Division of AIDS, STD, and Tuberculosis Laboratory Research, CDC, for performing the proportion method testing; N. Saravia for assistance in preparing this report; E. Jaramillo, H. Hernández, and L. Osorio for critical reading of the text; J. Robledo for evaluation by the proportion method of the first set of clinical isolates in the preliminary stages of the study; and A. M. Benítez for the isolation and propagation of M. tuberculosis from patient samples. We also thank the personnel of the Matías Lumumba Hospital in Buenaventura for the clinical sample collection and dispatching and D. J. McFadden for collaboration in the introduction of the Etest at CIDEIM.
Footnotes
Dedicated to the memory of Luis Ernesto Giraldo.
REFERENCES
- 1.Bloch A, Simone P, McRay E. Preventing multi-drug resistant tuberculosis. JAMA. 1996;275:487–489. [PubMed] [Google Scholar]
- 2.Browner W S, Newman T B, Cummings S R. Designing a new study: diagnostic tests. In: Hulley S B, Cummings S R, editors. Designing clinical research. An epidemiological approach. Baltimore, Md: Williams & Wilkins; 1988. pp. 88–97. [Google Scholar]
- 3.Centers for Disease Control and Prevention. Initial therapy for tuberculosis in the era of multidrug resistance. Recommendations of the Advisory Council for the Elimination of Tuberculosis. Morb Mortal Wkly Rep. 1993;42:RR-7. [PubMed] [Google Scholar]
- 4.Centers for Disease Control and Prevention. Acquired multi-drug resistant TB—Buenaventura, Colombia. Morb Mortal Wkly Rep. 1998;47:759–761. [PubMed] [Google Scholar]
- 5.Crofton J, Chaulet P, Maher D. Guidelines for the management of drug-resistant tuberculosis. WHO/TB/96.210. Geneva, Switzerland: World Health Organization; 1997. [Google Scholar]
- 6.DeVellis R F. Applied social research methods series. Vol. 26. Newbury Park, Calif: SAGE Publications, Inc.; 1991. Scale of development. Theory and applications; pp. 25–42. [Google Scholar]
- 7.Dye C, Scheele S, Dolin P, Pathania V, Raviglione M C. Global burden of tuberculosis. Estimated incidence, prevalence, and mortality by country. JAMA. 1999;282:677–686. doi: 10.1001/jama.282.7.677. [DOI] [PubMed] [Google Scholar]
- 8.Eltringham I J, Drobniewski F A, Mangan J A, Butcher P D, Wilson S M. Evaluation of reverse transcription-PCR and a bacteriophage-based assay for rapid phenotypic detection of rifampin resistance in clinical isolates of Mycobacterium tuberculosis. J Clin Microbiol. 1999;37:3524–3527. doi: 10.1128/jcm.37.11.3524-3527.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fleiss J L. Statistical methods for rates and proportions. New York, N.Y: John Wiley & Sons, Inc.; 1981. pp. 217–220. [Google Scholar]
- 10.Frieden T R, Sterling T, Pablos-Méndez A, Kilburn J O, Cauthen G M, Dooley S W. The emergence of drug-resistant tuberculosis in New York City. N Engl J Med. 1993;328:521–527. doi: 10.1056/NEJM199302253280801. [DOI] [PubMed] [Google Scholar]
- 11.Gamboa F, Cardona P J, Manterola J M, Lonca J, Matas L, Padilla E, Manzano J R, Ausina V. Evaluation of a commercial probe assay for detection of rifampin resistance in Mycobacterium tuberculosis directly from respiratory and nonrespiratory clinical samples. Eur J Clin Microbiol Infect Dis. 1998;17:189–192. doi: 10.1007/BF01691116. [DOI] [PubMed] [Google Scholar]
- 12.Hausdorfer J, Sompek E, Allerberge F, Dietrich M P, Rusch-Gerdes S. Etest for susceptibility of Mycobacterium tuberculosis. Int J Tuber Lung Dis. 1998;2:751–755. [PubMed] [Google Scholar]
- 13.Heymann S J, Brewer T F, Wilson M E, Fineberg H V. The need for global action against multidrug-resistant tuberculosis. JAMA. 1999;281:2138–2141. doi: 10.1001/jama.281.22.2138. [DOI] [PubMed] [Google Scholar]
- 14.Iseman M D. Treatment and implications of multidrug-resistant tuberculosis for the 21st century. Chemotherapy. 1999;45(Suppl. 2):34–40. doi: 10.1159/000048480. [DOI] [PubMed] [Google Scholar]
- 15.Jaramillo E. Encompassing treatment with prevention: the path for lasting control of tuberculosis. Soc Sci Med. 1999;49:393–404. doi: 10.1016/s0277-9536(99)00114-8. [DOI] [PubMed] [Google Scholar]
- 16.Jekel J F, Elomore J G, Katz D L. Saunders text and review series. Philadelphia, Pa: W. B. Saunders Company; 1996. Epidemiology, biostatistics and preventive medicine; pp. 89–102. [Google Scholar]
- 17.Kent P T, Kubica G P. Public health mycobacteriology: a guide for the level III laboratory. Atlanta, Ga: Centers for Disease Control; 1985. pp. 71–120. [Google Scholar]
- 18.Laserson K F, Osorio L, Hernández H, Benítez A M, Villegas M V, Rodríguez E, Binkin N J. Risk factors for chronic, drug resistant tuberculosis, Buenaventura, Colombia. Int J Tuber Lung Dis. 1998;2:S311. [PubMed] [Google Scholar]
- 19.Laserson K F, Osorio L, Sheppard J D, Hernández H, Benítez A M, Brim S, Woodley C L, Hazbón M H, Villegas M V, Castaño M C, Henríquez N, Rodríguez E, Metchok B, Binkin N J. Clinical mismanagement or community outbreak? An analysis of chronic, drug-resistant tuberculosis in Buenaventura, Colombia, 1999. Int J Tuber Lung Dis. 2000;4:673–683. [PubMed] [Google Scholar]
- 20.Miller W C. Can we do better than discrepant analysis for new diagnostic test evaluation? Clin Infect Dis. 1998;27:1186–1193. doi: 10.1086/514996. [DOI] [PubMed] [Google Scholar]
- 21.Mushtaque A, Chowdhuri R. Success with the DOTS strategy. Lancet. 1999;353:969–973. doi: 10.1016/s0140-6736(99)02119-4. [DOI] [PubMed] [Google Scholar]
- 22.National Committee for Clinical Laboratory Standards. Antimycobacterial susceptibility testing for Mycobacterium tuberculosis. Tentative standard M24 T. Villanova, Pa: National Committee for Clinical Laboratory Standards; 1995. [Google Scholar]
- 23.Pablos-Méndez A, Raviglione M C, Laszlo A. Global surveillance for antituberculous-drug resistance, 1994–1997. N Engl J Med. 1998;338:1641–1649. doi: 10.1056/NEJM199806043382301. [DOI] [PubMed] [Google Scholar]
- 24.Pfaller M A, Jones R N, Doern G V, Salazar J C. Multicenter evaluation of antimicrobial resistance to six broad-spectrum beta-lactams in Colombia: comparison of data from 1997 and 1998 using the Etest method. The Colombian Antimicrobial Resistance Study Group. Diagn Microbiol Infect Dis. 1999;35:235–241. doi: 10.1016/s0732-8893(99)00077-2. [DOI] [PubMed] [Google Scholar]
- 25.Riegelman R K, Hirsch R P. Cómo estudiar un estudio y probar una prueba: lectura crítica de la literatura médica. 2nd ed. 1998. pp. 112–122. . Publicación científíca no. 531. Organización Panamericana de la Salud, Washington, D. C. [PubMed] [Google Scholar]
- 26.Saito H, Hiramíne S, Watanabe T. Isolation of tubercle bacteria using Ogawa egg medium modified by addition of Tween 80. Zentbl Bakteriol Orig A. 1978;242:132–136. [PubMed] [Google Scholar]
- 27.Sánchez L, Londoño D, Arango A I, Mattar S. In vitro activity of antituberculous agents against Mycobacterium tuberculosis isolates from Bogota, DC (Colombia) evaluated by the Etest. Diagn Microbiol Infect Dis. 1999;35:109–112. doi: 10.1016/s0732-8893(99)00068-1. [DOI] [PubMed] [Google Scholar]
- 28.Secretaría de Salud Departamental del valle del Cauca. Estadísticas básicas en salud del Valle del Cauca. Cali, Colombia: Secretaría de Salud del Valle del Cauca; 1993. [Google Scholar]
- 29.Siddiqi N, Shamim M, Jain N K, Rattan A, Amin A, Katoch V M, Sharma S K, Hasnain S E. Molecular genetic analysis of multi-drug resistance in Indian isolates of Mycobacterium tuberculosis. Mem Inst Oswaldo Cruz. 1998;93:589–594. doi: 10.1590/s0074-02761998000500006. [DOI] [PubMed] [Google Scholar]
- 30.Telenti A. Genetics of drug resistance in tuberculosis. Clin Chest Med. 1997;18:55–64. doi: 10.1016/s0272-5231(05)70355-5. [DOI] [PubMed] [Google Scholar]
- 31.Turret G S, Telzak E E, Torian L V, Blum S, Assand D, Weisfuse I, Fazal B A. Improved outcomes for patients with multi-drug resistant tuberculosis. Clin Infect Dis. 1995;21:1238–1244. doi: 10.1093/clinids/21.5.1238. [DOI] [PubMed] [Google Scholar]
- 32.Wanger A, Mills K. Testing Mycobacterium tuberculosis susceptibility to ethambutol, isoniazid, rifampin, and streptomycin by using Etest. J Clin Microbiol. 1996;364:1672–1676. doi: 10.1128/jcm.34.7.1672-1676.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Watterson S A, Wilson S M, Yates M D, Drobniewski F A. Comparison of three molecular assays for rapid detection of rifampin resistance in Mycobacterium tuberculosis. J Clin Microbiol. 1998;36:1969–1973. doi: 10.1128/jcm.36.7.1969-1973.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.World Health Organization. Tuberculosis programme: framework for effective tuberculosis control. WHO/TB/94.179. Geneva, Switzerland: World Health Organization; 1994. [Google Scholar]