In November 2021, the Omicron variant (B.1.1.529) emerged and was designated a variant of concern (VOC) by the World Health Organization. Recently, Omicron was reported to extensively escape neutralizing antibodies elicited by COVID-19 vaccination or natural infection [1–3]. However, whether Omicron evades the T cell immunity elicited by COVID-19 vaccination or natural infection remains to be elucidated. To address this issue, we analyzed the amino acid sequences of T cell epitopes identified from the original SARS-CoV-2 strain (Wuhan-Hu-1) in the Omicron variant (hCoV-19/South Africa/CERI-KRISP-K032284/2021).
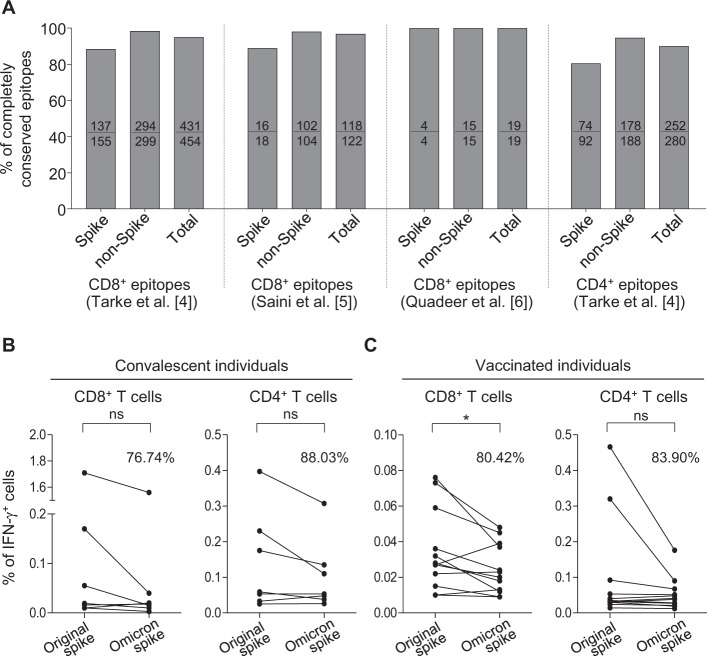
First, we examined 454 major histocompatibility complex (MHC) class I-restricted CD8+ T cell epitopes that were previously identified by activation-induced marker (AIM) assays [4]. In the Omicron variant, 88.4% (137/155) of epitopes from the spike protein and 98.3% (294/299) of epitopes from nonspike proteins were fully conserved (Fig. 1A), and 94.9% (431/454) of CD8+ T cell epitopes were completely conserved. We performed a similar analysis with 122 CD8+ T cell epitopes that were previously identified by a systematic analysis using peptide-MHC class I complex multimers [5]. In the Omicron variant, 88.9% (16/18) and 98.1% (102/104) of epitopes from the spike and nonspike proteins, respectively, were fully conserved, and 96.7% (118/122) of CD8+ T cell epitopes were completely conserved. We identified 19 dominant epitopes by a meta-analysis of CD8+ T cell epitope data from 18 studies with 852 COVID-19 convalescents [6], and all 19 CD8+ T cell epitopes were completely conserved in the Omicron variant. We also examined 280 MHC class II-restricted CD4+ T cell epitopes that were previously identified by AIM assays [4]. In the Omicron variant, 80.4% (74/92) and 94.7% (178/188) of epitopes from the spike and nonspike proteins, respectively, were fully conserved, and 90.0% (252/280) of CD4+ T cell epitopes were completely conserved. These results indicate that the majority of T cell epitopes are considerably conserved in the Omicron variant.
Fig. 1.
T cell analysis against the Omicron variant. A Amino acid sequences of T cell epitopes identified from the original SARS-CoV-2 strain (NC_045512.2) were examined in the Omicron variant (EPI_ISL_6699770, GISAID). Four different sets of previously identified T cell epitopes were used for comparisons. Percentages of completely conserved epitopes are presented as bar graphs. B, C IFN-γ intracellular staining was performed using PBMCs from COVID-19 convalescents (N = 7; 45–125 days after symptom onset) and BNT162b2-vaccinated individuals (N = 12; 3 months after the second vaccination). PBMCs were stimulated with OLP pools to spike the original strain and Omicron. The ratio of the frequency of IFN-γ+ cells after stimulation with the Omicron spike OLPs to the frequency after stimulation with the original spike OLPs is presented as a percentage. Statistical analysis was performed using paired t tests. *p < 0.05; ns not significant
Next, we performed IFN-γ intracellular cytokine staining (ICS) assays by stimulating peripheral blood mononuclear cells (PBMCs) using overlapping peptide (OLP) pools for the spike protein of the original SARS-CoV-2 strain and Omicron. PBMCs were obtained from individuals who had recovered from infection with the original SARS-CoV-2 and individuals vaccinated with BNT162b2. Among convalescent individuals, the frequency of IFN-γ+ cells after stimulation with the Omicron spike OLPs was 76.74% and 88.03% of the frequency after stimulation with the original spike OLPs in CD8+ and CD4+ T cells, respectively, without significant differences (Fig. 1B). Among vaccinated individuals, the frequency of IFN-γ+ cells after stimulation with the Omicron spike OLPs was 80.42% and 83.90% of the frequency after stimulation with the original spike OLPs in CD8+ and CD4+ T cells, respectively (Fig. 1B). The decrease in the CD8+ T cell response was significant (P = 0.03), but the change in the CD4+ T cell response was not. These results demonstrate that considerable proportions (>75%) of SARS-CoV-2 spike-specific memory CD8+ and CD4+ T cells elicited by COVID-19 vaccination or natural infection recognize and respond to the Omicron spike.
Previously known VOCs, including the Beta (B.1.351) and Delta (B.1.617.2) variants, reduce the neutralizing activities of antibodies induced by COVID-19 vaccination or natural infection [7]. However, such VOCs rarely evade memory T cell responses elicited by COVID-19 vaccination or natural infection [8]. In principle, VOCs can hardly evade T cell responses because multiple T cell epitopes are scattered across structural and nonstructural proteins.
COVID-19 vaccination or natural infection induces not only neutralizing antibodies but also memory T cells. Virus-specific CD8+ and CD4+ T cells exert antiviral functions by eliminating virus-infected cells and producing effector cytokines, leading to rapid control of viral infection and reducing disease severity [9]. Although the Omicron variant escapes neutralizing antibodies induced by COVID-19 vaccination or natural infection [1–3], our current analysis demonstrates that T cell epitopes are considerably conserved in the Omicron variant and that substantial proportions of memory T cells elicited by COVID-19 vaccination or natural infection respond to the Omicron spike. These results indicate that memory T cells may provide protective immunity during reinfection or breakthrough infection with the Omicron variant.
Supplementary information
Acknowledgements
This study was supported by the National Research Foundation Grant NRF-2018M3A9D3079498 and the Institute for Basic Science (IBS), Korea, under project code IBS-R801-D2.
Author contributions
SJC, JYN, SK, S-HP, and E-CS designed the research. HWJ collected the clinical specimens. D-UK performed the experiments. SJC, D-UK, S-HP, HWJ, and E-CS analyzed the results. SJC, JYN, and E-CS wrote the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
These authors contributed equally: Seong Jin Choi, Dong-Uk Kim.
Supplementary information
The online version contains supplementary material available at 10.1038/s41423-022-00838-5.
References
- 1.Liu L, Iketani S, Guo Y, Chan JFW, Wang M, Liu L, et al. Striking antibody evasion manifested by the Omicron variant of SARS-CoV-2. Nature. 2021. 10.1038/d41586-021-03826-3. [DOI] [PubMed]
- 2.Planas D, Saunders N, Schwartz O Considerable escape of SARS-CoV-2 Omicron to antibody neutralization. Nature. 2021. 10.1038/d41586-021-03827-2. [DOI] [PubMed]
- 3.Cele S, Jackson L, Sigal A Omicron extensively but incompletely escapes Pfizer BNT162b2 neutralization. Nature. 2021. 10.1038/d41586-021-03824-5. [DOI] [PMC free article] [PubMed]
- 4.Tarke A, Sidney J, Kidd CK, Dan JM, Ramirez SI, Yu ED, et al. Comprehensive analysis of T cell immunodominance and immunoprevalence of SARS-CoV-2 epitopes in COVID-19 cases. Cell Rep Med. 2021;2:100204. doi: 10.1016/j.xcrm.2021.100204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Saini SK, Hersby DS, Tamhane T, Povlsen HR, Hernandez SPA, Nielsen M, et al. SARS-CoV-2 genome-wide T cell epitope mapping reveals immunodominance and substantial CD8+ T cell activation in COVID-19 patients. Sci Immunol. 2021;6:eabf7550. doi: 10.1126/sciimmunol.abf7550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Quadeer AA, Ahmed SF, McKay MR. Landscape of epitopes targeted by T cells in 852 individuals recovered from COVID-19: meta-analysis, immunoprevalence, and web platform. Cell Rep Med. 2021;2:100312. doi: 10.1016/j.xcrm.2021.100312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Planas D, Veyer D, Baidaliuk A, Staropoli I, Guivel-Benhassine F, Rajah MM, et al. Reduced sensitivity of SARS-CoV-2 variant Delta to antibody neutralization. Nature. 2021;596:276–80. doi: 10.1038/s41586-021-03777-9. [DOI] [PubMed] [Google Scholar]
- 8.Tarke A. Impact of SARS-CoV-2 variants on the total CD4+ and CD8+ T cell reactivity in infected or vaccinated individuals. Cell Rep Med. 2021;2:100355. doi: 10.1016/j.xcrm.2021.100355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Noh JY, Jeong HW, Kim JH, Shin EC. T cell-oriented strategies for controlling the COVID-19 pandemic. Nat Rev Immunol. 2021;21:687–8. doi: 10.1038/s41577-021-00625-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.