ABSTRACT
The dynamics of SARS-CoV-2 infection in COVID-19 patients are highly variable, with a subset of patients demonstrating prolonged virus shedding, which poses a significant challenge for disease management and transmission control. In this study, the long-term dynamics of SARS-CoV-2 infection were investigated using a human well-differentiated nasal epithelial cell (NEC) model of infection. NECs were observed to release SARS-CoV-2 virus onto the apical surface for up to 28 days postinfection (dpi), further corroborated by viral antigen staining. Single-cell transcriptome sequencing (sc-seq) was utilized to explore the host response from infected NECs after short-term (3-dpi) and long-term (28-dpi) infection. We identified a unique population of cells harboring high viral loads present at both 3 and 28 dpi, characterized by expression of cell stress-related genes DDIT3 and ATF3 and enriched for genes involved in tumor necrosis factor alpha (TNF-α) signaling and apoptosis. Remarkably, this sc-seq analysis revealed an antiviral gene signature within all NEC cell types even at 28 dpi. We demonstrate increased replication of basal cells, absence of widespread cell death within the epithelial monolayer, and the ability of SARS-CoV-2 to replicate despite a continuous interferon response as factors likely contributing to SARS-CoV-2 persistence. This study provides a model system for development of therapeutics aimed at improving viral clearance in immunocompromised patients and implies a crucial role for immune cells in mediating viral clearance from infected epithelia.
KEYWORDS: COVID-19, nasal epithelial cells, SARS-CoV-2, single-cell sequencing, viral persistence
INTRODUCTION
SARS-CoV-2 is a positive-stranded RNA virus in the family Coronaviridae and the causative agent of the COVID-19 pandemic. Efforts at containing the pandemic are hampered by the ease of viral transmission, asymptomatic viral shedding, a variable incubation period, and the emergence of viral variants (1–4). In human hosts, the primary site of SARS-CoV-2 infection is postulated to be the upper airways, including the nasal tissue (5). This is facilitated by higher expression of the ACE2 entry receptor on proximal airway tissue relative to the distal airways (6). Several groups, including ours, have utilized ex vivo differentiated airway epithelial cells (AECs) as a model system for dissecting SARS-CoV-2-mediated pathogenesis (6–10). Differentiated AECs cultured at the air-liquid interface (ALI) better recapitulate the cellular diversity, spatial arrangement, and polarized nature of the airway epithelium compared to cell lines (11, 12).
Differentiated AECs are composed of a heterogeneous collection of cell types, derived from progenitor/stem cells which are initially isolated and expanded from primary human tissue (13–15). Subsequent differentiation at the air-liquid interface results in the generation of a pseudostratified mucociliary epithelium. Recent studies, notably from those utilizing single-cell RNA sequencing (sc-seq) technology, have shed light on the differentiation trajectories giving rise to these various cell types (14, 16, 17). In differentiated AECs derived from human upper airway tissue, basal cells possess multipotency and replenish the epithelial structure (16, 18). Basal cells differentiate into club cells via an intermediate suprabasal cell type (14). Club cells in turn can give rise to goblet cells responsible for mucus production and multiciliated cells (MCCs) which possess motile cilia (14). Deuterosomal cells, enriched for centriole-related genes, have been described as a precursor of MCCs originating downstream of club cells (14). A minor population of ionocytes, expressing the CFTR channel protein and responsible for regulating airway surface physiology, have also been identified within AECs, likely arising directly from basal cells (15, 19). Other rare cell types within AECs include tuft cells, neuroendocrine cells and mucous-multiciliated cells (15, 20, 21). Uncovering differentiation hierarchies of these epithelial cell types is still an ongoing area of research, and further complexities and revisions to their cellular ontology are likely to emerge.
COVID-19 patients can exhibit broad variability in the time required for resolution of symptoms, but infectious virus shedding is usually not detected beyond 2 weeks post-symptom onset (22, 23). However, persistent infections have been observed in immunocompromised patients with infectious virus isolated for several months after initial infection (24, 25). In addition, reverse transcriptase PCR (RT-PCR) positive respiratory samples have been detected beyond 14 days post-acute infection in a subsection of immunocompetent patients, though with the absence of infectious virus shedding (26, 27). Lastly, a varying proportion of patients exhibit “long-COVID” or post-acute COVID syndrome, where symptoms are observed 12 weeks or more after acute infection and after discharge from hospital (28–31). Epidemiological data as well as mechanistic explanations for the above-described observations are incomplete and currently the subject of intense research. Here, we explore a specific facet relevant to this discussion—viral persistence within host cells. We provide evidence from virus titers, viral antigen staining, viral transcripts in sc-RNA seq data sets, and the presence of a continuous immune response to demonstrate persistent infection of airway epithelial cells by SARS-CoV-2 in vitro.
RESULTS
Prolonged release of SARS-CoV-2 from infected airway epithelial cells.
We previously demonstrated that differentiated nasal epithelial cells (NECs) are readily susceptible to SARS-CoV-2 infection, and viral kinetics and host responses were studied up to 3 days postinfection (dpi) (7). To determine the long-term kinetics of virus release from infected airway epithelial cells, NECS were infected with SARS-CoV-2, and virus titers were assayed from the apical side at various time points up to 28 dpi. We observed, on average, SARS-CoV-2 virus titers of 1.6 × 105 to 9.3 × 106 50% tissue culture infective dose (TCID50)/mL at each endpoint (Fig. 1A). Importantly, viral titers at 14, 21, and 28 dpi were not significantly lower than that at 3 dpi, indicating the presence of live virus in apical secretions from infected epithelial cells for an extended period of time. We observed similar data with 2 donor-derived bronchial epithelial cells (BECs) (Fig. 1A, orange data points). With this single-endpoint sampling experimental setup, it is difficult to determine if virus particles were released throughout the course of infection or were produced during an early time point and then persisted on the mucus-rich apical secretions. We therefore performed a series of experiments incorporating repeated sampling of the same infected AECs from the apical side at 3, 7, 14, and 21 dpi. At each time point, the apical surface is rinsed with phosphate-buffered saline (PBS) (used for virus titration; refer to Materials and Methods for more details), which significantly removes residual extracellular virus. It was observed that apical viral titers from dynamic sampling were generally lower than the viral titers from endpoint sampling at corresponding time points (Fig. 1A and B), indicating that at least a portion of the viral loads detected at late time points were derived from residual virus released early during the infection and surviving in the extracellular milieu. This observation is consistent with prior studies reporting a half-life for SARS-CoV-2 of several days in human mucus (32) and liquid media (33). Virus release was nevertheless observed at each of the time points assayed during dynamic sampling (Fig. 1B). Crucially, average virus loads at 28 dpi (6.5 × 105 TCID50/mL, n = 5) were observed to be similar to that at 3 dpi (6.0 × 105 TCID50/mL, n = 5), despite 4 rounds of repeated sampling and rinsing of the apical surface. Taken together, these findings demonstrate that infected AECs can release significant amounts of SARS-CoV-2 for a prolonged duration postinfection.
FIG 1.
SARS-CoV-2 replication kinetics in AECs. Apical viral titers from infected NECs (blue) and BECs (orange) at each of the indicated time points (n = 4 to 8), as detected via (A) single-endpoint sampling or (B) repeated sampling of the same apical surface for each time point. Each dot represents a different human-donor-derived AEC. Data are represented as the mean ± standard error of the mean (SEM). P values indicated are derived from Student's t test.
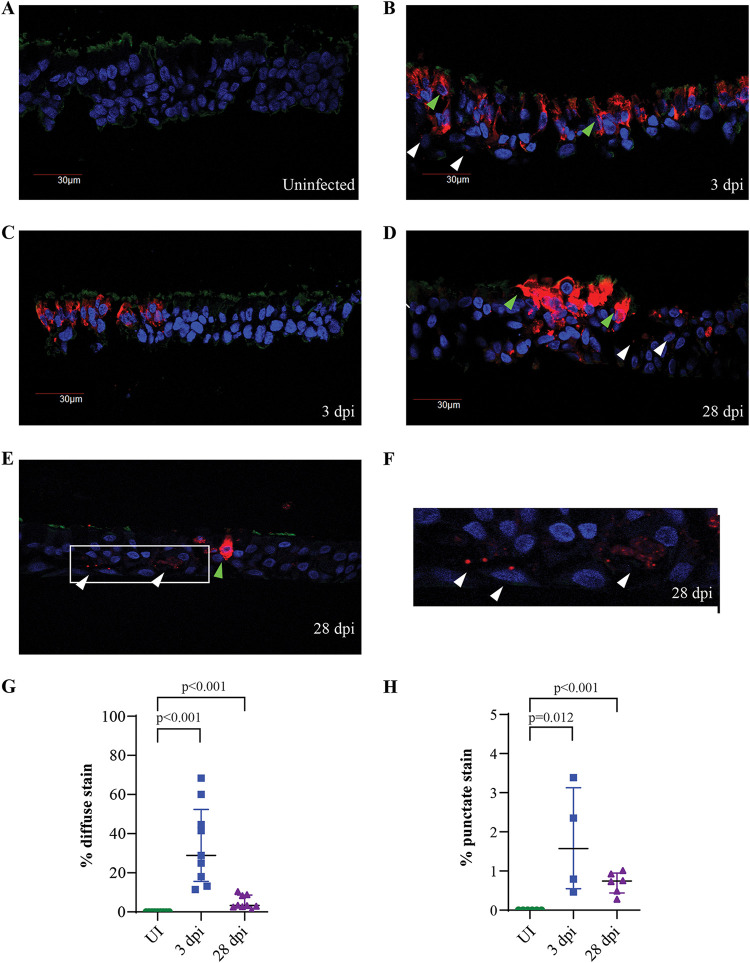
We next performed immunofluorescence (IF) staining on paraformaldehyde (PFA)-fixed NECs from uninfected, 3-dpi, and 28-dpi samples. Viral antigen was not detected from uninfected NECs, indicating the absence of background staining (Fig. 2A). Two distinct staining patterns for the SARS-CoV-2 nucleoprotein were observed within infected samples—intense viral staining diffused throughout the extranuclear compartment (green arrows, Fig. 2B, D, and E) and localized punctate staining originating from a few foci within a cell (white arrows, Fig. 2B and D to F). Both viral antigen staining patterns were observed at 3 and 28 dpi, supporting the previous observation of a persistent infection within NECs. However, both staining patterns were more abundant at 3 dpi than at to 28 dpi (Fig. 2G and H), indicating that a relatively small number of infected cells at 28 dpi are sufficient for sustaining virus production at similar levels to that observed during the early phases of the infection (Fig. 1A). We observed limited aberrations to the epithelial barrier in some regions of infected NECs which overlapped with heavily infected clusters of cells (Fig. 2D). However, at both 3 and 28 dpi, we did not observe regions with clear loss of cellular content despite abundant cells staining positive for viral antigen (Fig. 2C to E).
FIG 2.
Detection of SARS-CoV-2 viral antigen from infected NECs. (A to F) IF staining for SARS-CoV-2 nucleoprotein (red), DAPI nuclear stain (blue), and βIV-tubulin (green) from (A) uninfected, (B and C) 3-dpi, and (D to F) 28-dpi infected NECs. Green arrows indicate examples of cells with diffuse viral antigen staining, and white arrows indicate cells with punctate staining for viral antigen. Panel F is zoomed in from the indicated image region in panel E to better illustrate punctate focal staining for viral antigen. Representative images obtained from at least 3 different donor-derived NEC inserts processed for IF staining are shown. (G and H) The percentage of cells within NEC structures displaying (G) diffuse viral antigen staining and (H) punctate viral antigen-positive foci at each of the indicated time points. Each dot represents data obtained from a different human donor-derived NEC cross-section. Data are represented as the mean ± SEM. The indicated P values are derived from Student's t test.
Delineating cell type heterogeneity in NECs and changes to cell type proportions upon infection.
To interrogate changes in cellular composition and transcriptional responses associated with short-term (3-dpi) and long-term (28-dpi) SARS-CoV-2 infection, sc-seq was carried out with pooled samples derived from uninfected, 3-dpi, and 28-dpi NECs. After preprocessing for quality control and removal of debris and cell doublets, 13,986 cell transcriptomes were de novo clustered using the Uniform Manifold Approximation and Projection (UMAP) algorithm, followed by examination of key marker genes from each cluster to assign cell type identities (Fig. 3A and B). Basal cells were identified as KRT15+ KRT5+ cells enriched for DLK2 and TP63 expression (Fig. 3B). Goblet cells strongly expressed genes encoding mucins and other secretory proteins (TFF3, SCGB1A1) and were observed to form two separate clusters. Goblet 1 cells had higher expression of mucin genes MUC5AC and MUC5B, while goblet 2 cells had elevated expression of the adhesion protein CEACAM6 and lower expression of MUC5AC and MUC5B (Fig. 3A and B). Dividing cells (MKI67), deuterosomal cells (PLK4), MCCs (CCDC153, MLF1), and ionocytes (CFTR, FOXI1) were identified by expression of their respective marker genes (Fig. 3B). Suprabasal and club cells were observed as intermediates on a differentiation continuum from basal cells to goblet and ciliated cells, consistent with their reported ontology. As such, significant overlap in gene expression was observed between basal and suprabasal cells and between club and goblet cells. Suprabasal cells were identified as KRT5+ cells which poorly expressed basal markers (DLK2 low, TP63low), while club cells were KRT5low and expressed SCGB1A1 in common with goblet cells, but not the mucins MUC5AC and MUC5B (Fig. 3B). Lastly, we observed a distinct cell cluster which did not uniquely express any of the canonical markers associated with major airway epithelial cell types but highly expressed cell-stress-related genes DDIT3 (DNA damage inducible transcript 3), a transcription factor involved in the endoplasmic reticulum (ER) stress response, and ATF3 (activating transcription factor 3), a common stress-response transcription factor (Fig. 3C). This cluster was annotated as DDIT3high, was observed to increase in proportion upon infection (Fig. 3D), and will be discussed in more detail in the subsequent section (Fig. 4). All of the major airway epithelial cell types were conserved in sc-seq transcriptomes derived from uninfected, 3-dpi, and 28-dpi NECs (Fig. S1A and B). Dividing cells showed a marked increase in proportion at 28 dpi compared to uninfected or 3-dpi samples, indicating increased cell proliferation is associated with prolonged infection (Fig. 3B). IF staining for KI67, a specific marker of dividing cells, corroborated these observations with KI67+ cells observed to be significantly higher at 28 dpi compared to uninfected (UI) and 3-dpi NEC samples (Fig. S2A to D). In conclusion, we delineate all major airway epithelial cell types in the sc-seq data sets generated in this study and observe that these populations are conserved between libraries generated from uninfected, 3-dpi, and 28-dpi NECs, with two notable changes—the DDIT3high cluster increasing after infection and dividing cell population increasing at 28 dpi.
FIG 3.
Clustering of sc-seq transcriptomes identified 10 airway epithelial cell types within NECs. (A) Pooled sc-seq transcriptomes from uninfected and 3- and 28-dpi NECs clustered and annotated as indicated on the figure. (B) Violin plots illustrating expression of key cell-type markers across the 10 epithelial cell types. (C) Specific expression of cell stress-related genes, DDIT3 and ATF3, from the DDIT3high cell cluster. (D) The percentage change in the proportion of each cell type at 3 and 28 dpi relative to the uninfected NEC samples.
FIG 4.
SARS-CoV-2 viral reads detected across multiple epithelial cell types; the DDIT3high cluster consists of heavily infected cells. (A) SARS-CoV-2 viral reads overlaid with clustering of sc-seq transcriptomes derived from uninfected, 3- and 28-dpi NECs. (B) Histograms of viral readsper cell from 3- and 28-dpi NECs. Blue lines indicate the cutoff for demarcating infected versus bystander cells (4 reads per cell), as determined by Otsu’s thresholding. (C) Percentage of cell transcriptomes with >4 and >50 viral reads across each of the epithelial cell types. (D) Sc-seq transcriptomes from the DDIT3high cluster reannotated with transcriptional signatures from the other 9 epithelial cell types in this study and the resulting annotations quantified and displayed as a bar chart on the right panel.
Distribution and abundance of airway epithelial cell types in infected versus uninfected sc-seq libraries. (A) Clustering of sc-seq transcriptomes from uninfected (top panel), 3-dpi (middle panel), and 28-dpi (bottom panel) NECs. Each color-coded cluster corresponds to the same cell type annotation displayed for the pooled UMAP in Fig. 3A. (B) Number of cell transcriptomes from each of the airway epithelial cell types in the uninfected, 3-dpi, and 28-dpi libraries. Download FIG S1, TIF file, 2.7 MB (2.7MB, tif) .
Copyright © 2022 Gamage et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Increased KI67+ cells at 28 dpi. (A to C) IF staining for KI67 (red), DAPI nuclear stain (blue), and P63 (green) from (A) uninfected (UI), (B) 3-dpi, and (C) 28-dpi infected NECs. Arrows indicate cells staining positive for KI67. Representative images obtained from at least 3 different donor-derived NEC cross-sections processed for IF staining are shown. (D) Percentage of KI67+cells within NEC structures at each of the indicated time points. Each dot represents data obtained from a different human donor-derived NEC cross-section. Data are represented as the mean ± SEM. The indicated P values are derived from Student's t test. Download FIG S2, TIF file, 2.7 MB (2.7MB, tif) .
Copyright © 2022 Gamage et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
High expression of SARS-CoV-2 genes in the DDIT3high cell cluster.
SARS-CoV-2 viral reads were detected in the sc-seq transcriptomes of both 3- and 28-dpi samples (Fig. 4A and B), although it was observed that the viral reads per cell from 28-dpi NECs were approximately 3-fold lower than at 3 dpi (Fig. S3A). The uninfected sample had only 22 total viral reads, while 3- and 28-dpi samples had 415,285 and 27,262 total viral reads, respectively, indicating high specificity of the algorithm for parsing total transcripts and identifying those mapping to SARS-CoV-2. Viral reads mapped primarily to the 3′ untranscribed region (UTR), consistent with the polyA targeting of the gene expression profiling primers (Fig. S3B). Upon transposing viral reads to individual cell transcriptomes, it was observed that at both 3 and 28 dpi, SARS-CoV-2 transcripts broadly originated from all cell types (Fig. 4A and C). At 3 and 28 dpi, we detected 3,499/5,016 (69.8%) and 357/4984 (7.16%) cell transcriptomes positive for at least four viral reads, respectively (Fig. 4B). However, heavily infected cells (>50 viral transcripts per cell) originated primarily from the DDIT3high cluster (Fig. 4C). To further delineate the identity of these cells, this cluster was reannotated using transcriptional signatures derived from the remaining 9 clusters from the NEC data sets generated in this study (Fig. 4D). DDIT3high cells were composed of multiple individual cell types, including suprabasal, club, goblet, and ciliated cells (Fig. 4D). Taking these findings together, we demonstrate that SARS-CoV-2 is capable of infecting multiple NEC subsets at both 3 and 28 dpi and that a subset of infected cells harbor very high viral loads and a sufficiently different host transcriptional profile, causing them to cluster separately from their native cell type on a UMAP projection.
SARS-CoV-2 viral reads detected from 3- and 28-dpi NECs. (A) Viral reads per cell detected in cell transcriptomes derived from uninfected (D0), 3-dpi (D3), and 28-dpi (D28) NEC samples. (B) Coverage plot of pooled SARS-CoV-2 reads from 3-dpi and 28-dpi NEC samples, overlaid with the SARS-CoV-2 genome, demonstrating that the majority of viral reads map to the 3′ genome terminus. Download FIG S3, TIF file, 1.1 MB (793.7KB, tif) .
Copyright © 2022 Gamage et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The DDIT3high cell cluster has reduced IFN signaling and higher TNF-α signaling gene-set enrichment compared to other cell types.
Analysis of key gene expression pathways induced within NECs at 3 dpi identified a dominant interferon alpha (IFN-α) and IFN-γ response across all airway epithelial cell types (Fig. 5A). The DDIT3high cluster was uniquely enriched for “TNF-α signaling via NFκb” and “apoptosis” pathways, indicating that high levels of virus replication within NECs can trigger cell death pathways, likely via the cell extrinsic death pathway which can be mediated by tumor necrosis factor (TNF) signaling. Interestingly, it was observed that although the DDIT3high cluster was significantly enriched for IFN-α and -γ responses, this enrichment was the smallest across all cell types, an unexpected observation, as higher virus replication would be expected to correlate with greater stimulation of pathogen recognition receptors (Fig. 5A). Specific interferon-stimulated genes (ISGs) examined, including ISG15, IFIT3, RSAD2, and OAS1 were strongly induced across multiple cell types compared to the uninfected sample, while still exhibiting a notably reduced induction from DDIT3high cells (Fig. 5B). Similarly, gene expression heat map analysis revealed that while the DDIT3high cluster had the highest expression of SARS-CoV-2 genes and TNF-α signaling genes (Fig. 5C, upper-middle panel), these cells had subdued expression of key IFN-α signaling genes (Fig. 5C, lower panel). These observations are consistent with SARS-CoV-2 subverting host IFN signaling within heavily infected cells. In contrast, the goblet 2 cluster exhibited strong expression of IFN-α signaling genes, indicating a key role for this cell type in antiviral signaling during SARS-CoV-2 infection.
FIG 5.
The DDIT3high cluster has reduced IFN response, and higher TNF-σ signaling gene set enrichment. (A) Bubble-plot of the top enriched Hallmark gene sets from differentially expressed genes (DEGs) at 3 dpi compared to uninfected NECs (D3/D0). The color and size of each bubble is proportional to the adjusted P value and the percentage of enriched genes from each gene set, respectively. Only gene-sets with an adjusted P value < 0.05 are shown. (B) Split violin plots illustrating upregulation of key antiviral genes across epithelial cell types at 3 dpi compared to uninfected NECs. The DDIT3high cluster has a relatively lower upregulation of antiviral genes relative to the other cell types. (C) Heat map of the most highly expressed genes in the “TNF-α signaling” (in blue) and “IFN-α response” (in red) Hallmark gene sets across cell transcriptomes derived from 3-dpi NEC samples, arranged according to cell type clusters and sorted for SARS-CoV-2 viral reads (top panel).
Diverse inflammatory mediators produced from SARS-CoV-2 infected NECs.
Next, to identify cell type-specific contributions to the inflammatory milieu produced during SARS-CoV-2 infection of airways, we visualized the expression patterns of inflammatory mediators (by retrieving all genes within KEGG pathway hsa04060 for cytokine-cytokine receptor interaction) that were significantly induced (log2 fold change, >1.5) at 3 dpi compared to the uninfected sample and expressed in at least 5% of any one cell type. Chemokines and members of the TNF ligand superfamily constituted the majority of this gene list (Fig. 6A). All cell types, and in particular, goblet 2 cells, upregulated TNFSF13B, which encodes a potent B cell activator, driving the proliferation and differentiation of B cells (34, 35). Infection also resulted in the increased expression of CXCL10 and CXCL11 by multiple cell subsets, including goblet 2 cells. Both CXCL10 and CXCL11 signal via the CXCR3 receptor to mediate chemotaxis of activated T cells (36). Interleukin-6 (IL-6) was induced in a minor population of DDIT3high cells after infection. However, genes encoding common proinflammatory cytokines such as TNF-α, IL-1β, IL-12 (p35 and p40 genes), and IFN-γ were not observed to be expressed in any of the cell types to a significant degree and proportion, indicating that immune cells are likely the core contributors of these cytokines in COVID-19 patients. Surprisingly, we also did not observe induction of IFN genes in the above-described analysis, although we were capable of detecting a clear IFN response from all cell types. As this prior analysis required at least 5% of the cells in a cluster to express the gene of interest and a log2 fold change > 1.5, we reanalyzed the expression of type I, II, and III IFNs between 3 dpi and uninfected controls without this threshold criteria. IFN-λ1 and IFN-β were weakly upregulated from some clusters upon infection (Fig. 6B). Further analysis indicated sparse but detectable expression of both cytokines from cell transcriptomes specifically upon infection notably the expression of IFN-λ1 from goblet 2 cells (Fig. 6C). In conclusion, secretory factors produced by various epithelial cell types in response to SARS-CoV-2 infection mediate the recruitment and development of an adaptive immune response, while the production of IFN-λ1 and IFN-β from a very limited number of cells are responsible for maintaining an antiviral state across the NEC structure upon infection.
FIG 6.
A limited number of cells are responsible for producing type I and type III IFN from infected NECs at 3 dpi. (A) Diverse inflammatory mediators produced from various epithelial cell types from 3-dpi infected NECs. (B) Induction of type I and type III IFN genes across epithelial cell types from infected NECs at 3 dpi relative to uninfected NECs (D3/D0). Only IFN genes with detectable expression across the pooled sc-seq data set were included in this analysis. (C) Expression of IFN-β1 and IFN-λ1 across cell transcriptomes from uninfected and 3-dpi infected NEC samples, indicating positive expression of IFN from a very limited subset of cells upon infection.
Persistent IFN response observed across all cell types at 28 dpi.
Lastly, we investigated gene expression pathways induced at 28 dpi, compared to the uninfected sample. Remarkably, genes involved in the IFN-α and IFN-γ response pathways were still significantly enriched across all cell types (Fig. 7A). Within the goblet 2 cluster, which was observed to mount a robust antiviral response at 3 dpi (Fig. 5C), IFN response genes were consistently the most upregulated genes compared to the uninfected sample (Fig. 7B). Similarly, key ISGs, including ISG15, MX1, and IFITM1, were significantly upregulated across all clusters at 28 dpi compared to the uninfected control (Fig. 7C), although this expression was less than that observed at 3 dpi (Fig. 7D), consistent with the reduced viral transcripts detected at 28 dpi relative to 3 dpi. The chemokine CXCL10 was significantly elevated in the basolateral media from infected NECs at 7, 14, 21, and 28 dpi compared to uninfected controls, further validating the ISG induction observed at the transcriptional level at 28 dpi and demonstrating a persistent immune response from infected NECs (Fig. 8).
FIG 7.
Broad IFN response observed across cell types from infected NECs at 28 dpi. (A) Bubble plot of IFN-γ and IFN-α response gene set enrichment in DEGs at 28 dpi compared to uninfected NECs (D28/D0). The color and size of each bubble is proportional to the adjusted P value and the percentage of enriched genes from each gene set, respectively. Only gene sets with an adjusted P value of <0.05 are shown. (B) Volcano plot of genes within the goblet 2 cell cluster from infected NECs at 28 dpi compared to uninfected NECs (D28/D0). Significant DEGs with an adjusted P value of <0.05 and log2 fold change (FC) of at least 1.5 are indicated as maroon dots; all other DEGs are indicated as gray dots. The top 15 upregulated genes by log FC are annotated, with genes in the “Hallmark Interferon Alpha response” list annotated in red. (C) Split violin plots illustrating upregulation of key antiviral genes across epithelial cell types at 28 dpi (D3) compared to uninfected NECs (D0), and (D) 3 dpi (D3) compared to 28 dpi (D28) infected NECs.
FIG 8.

Elevated CXCL10 levels are observed in basolateral media from infected NECs for up to 28 days postinfection. CXCL10 cytokine concentrations (pg/mL, log10) were assayed from the basolateral media at each of the indicated postinfected time points. Data are shown as the mean ± SEM. Each dot represents a different human-donor-derived NEC. Indicated P values are derived from Student's t test.
DISCUSSION
Pathogens display various adaptations to prolong the duration of infection and dissemination. Here, we demonstrate that SARS-CoV-2 is capable of productively infecting airway epithelia for up to 28 days postinfection. We further identify three aspects of this host-pathogen interaction which likely contribute to persistent infection—the ability of SARS-CoV-2 to continually infect host cells while evading a tissue-wide antiviral response, limited cell death within the infected epithelium, and proliferating basal cells replacing cells lost during infection. These findings are consistent with recent reports on persistent SARS-CoV-2 infection observed with other in vitro models. Apical release of SARS-CoV-2 was observed at 51 days postinfection from human BECs (37), and progeny virus release into the media was reported up to 23 days from human intestinal epithelial cells (38). Strikingly, infectious virus shedding has been observed for several months after initial infection in immunocompromised patients (24, 25, 39–41). In immunocompetent, asymptomatic COVID-19 patients, antigen persistence and viral RNA have been detected in the small intestine 4 months after infection (42). Another report observed antigen persistence, viral RNA, and inflammation in the olfactory mucosa of patients with prolonged or recurrent olfactory function loss multiple months after first onset of COVID-19 symptoms (43). Taken together, these observations support the capacity for prolonged SARS-CoV-2 replication within infected host tissue, particularly in the absence of competent humoral and cell-mediated immune responses.
Host-transcriptional responses observed upon short-term (3 dpi) SARS-CoV-2 infection in this study are consistent with previously published sc-seq analysis of SARS-CoV-2-infected AECs (8, 9). Ravindra et al. also observed type I and type III IFN expression and the broad induction of ISGs across all epithelial cell types upon infection of HBECs. Bystander versus infected cell analysis revealed the specific upregulation of genes involved in apoptosis and inflammation (e.g., NFKBIA and NFKBIZ) from infected cells, similarly to our observations (9). We identified a DDIT3high cell cluster consisting of heavily infected cells, which was determined to be of a heterogeneous cell type composition. Transcriptional signatures corresponding to MCCs, goblet, basal, and other cell types were observed from cell transcriptomes in this cluster. Therefore, although there is an emerging consensus that SARS-CoV-2 establishes initial infection via ciliated cells (6, 44), broad susceptibility of multiple epithelial cell types to SARS-CoV-2 infection is evident, particularly at later stages of infection. We further demonstrate that heavily infected cells display a lower upregulation of ISGs relative to other cell types. IFN expression (IFN-λ1 and IFN-β) was also detected in only a small fraction of infected cells, despite widespread detection of viral transcripts across cell transcriptomes at 3 dpi. Fiege et al. also observed distinct clusters of heavily infected cells composed of a variety of cell phenotypes upon sc-seq analysis of SARS-CoV-2-infected tracheobronchial epithelial cells. Similarly, significantly lower expression of ISGs was observed in cells with the highest levels of viral RNA (8). Multiple SARS-CoV-2 proteins, including NSP1, ORF3b, and ORF6, are capable of antagonizing IFN signaling (45–48). Reduced IFN signaling from infected cells could dampen induction of an antiviral state in neighboring tissue, enabling the establishment of a persistently infected state.
We acknowledge specific limitations of our work. Apical viral loads were observed to be comparable between 3 and 28 dpi, but sc-seq-derived viral reads from 28-dpi NECs were significantly lower than those from 3-dpi NECs. This discrepancy is likely explained by the potential disproportionate loss of virus harboring apoptotic cells during single-cell dissociation and library preparation between time points. The stronger immune response elicited from infected cells at 3 dpi could also limit the packaging and release of infectious virus particles despite the higher viral transcripts detected at this time point. However, despite differences in the magnitude of virus quantification between assays, our analysis demonstrated that a clear population of cell transcriptomes at both 3 and 28 dpi contain high (>50) viral reads along with the upregulation of cell stress-related genes. We further presented multiple corroborating lines of evidence for persistent infection, via live virus titration from apical samples up to 28 dpi despite repeated washes, viral antigen staining at 28 dpi by IF, and CXCL10 chemokine secretion observed up to 28 dpi, each of which was observed in every donor-derived NEC assayed in this study (n ≥ 3). Our work is also limited to work with one SARS-CoV-2 virus lineage. Further work needs to be done with more recently emerged variants, such as the beta and delta variants, reported to have increased syncytial activity (49), which could play a further role in viral persistence within host tissue.
Importantly, this work highlights that specialized immune cells are required to fully clear SARS-CoV-2 infection from infected epithelial tissue. Chemokines and immune activating factors upregulated from infected NECs such as CXCL10, CXCL11, and TNFSF13B (BAFF) are capable of mediating the infiltration and maturation of these specialized immune cells. Effective viral clearance can be mediated by immunoglobulins which neutralize extracellular virus, and effector T cells which lyse and kill infected cells displaying viral antigens on the cell surface. However, immunocompromised patients remain vulnerable to persistent SARS-CoV-2 infections, a significant concern given the emerging endemicity of SARS-CoV-2 and the observation that treatment with remdesivir and intravenous immunoglobulin or plasma therapies have not been fully curative in some case reports (25, 40). Viral and immune response persistence observed in vitro in this work may also be relevant to the study of long-COVID pathophysiology, in at least a subsection of patients with incomplete viral clearance where “hidden” reservoirs of the virus may contribute to prolonged inflammatory activation. Lastly, persistent SARS-CoV-2 infection in AEC cultures can provide an important model system for evaluating novel therapeutics which improve viral clearance. In particular, it enables screening therapeutics which can control virus replication and release from the apical surface of AECs upon drug delivery to the basolateral compartment, as the inability to cross the epithelial barrier can impede therapeutic efficacy in vivo. As such, we expect the experimental setup and findings described here to serve as the basis for further investigating mechanisms of SARS-CoV-2 persistence and as a platform for evaluation of novel therapeutics.
MATERIALS AND METHODS
Collection and culture of air-liquid interface (ALI) in vitro-differentiated upper airway human nasal epithelial cells (NECs) and lower airway human bronchial epithelial cells (BECs).
This study was approved by the National Healthcare Group Domain-Specific Board of Singapore (DSRB ref. D/11/228) and the institutional review board of the National University of Singapore (IRB ref. NUS-IRB-2020-33). To generate NECs in ALI culture, nasal biopsy specimens were obtained from inferior turbinate of healthy donors undergoing septal deviation surgery who gave prior written informed consent. All subjects were free of symptoms of upper respiratory tract infection (URTI) and did not take any forms of glucocorticoids (GC) or antibiotics within 3 months before the study, at the time of collection. The human nasal epithelial stem/progenitor cells (hNESPCs) were isolated and expanded from the tissue biopsy specimens according to previously standardized protocols (7, 50). Briefly, upper airway primary cells were subjected to isolation to select for hNESPCs, which were then expanded using Dulbecco’s modified Eagle medium nutrient mixture F-12 (DMEM/F12) (Gibco-Invitrogen) containing 10 ng/mL of human epithelial growth factor (EGF; Gibco-Invitrogen), 5 μg/mL of insulin (Sigma), 0.1 nM cholera toxin (Sigma), 0.5 μg/mL of hydrocortisone (Sigma), 2 nM/mL of 3, 3′, 5-triiodo-l-thyronine (T3) (Sigma), 10 μL/mL of N-2 supplement (Gibco-Invitrogen), and 100 IU/mL of antibiotic-antimycotic (Gibco-Invitrogen). The enriched hNESPCs were then transferred onto 12-well 0.4-μm transwell inserts (Corning) upon reaching sufficient confluence. Once the cells on the transwell are confluent, growth medium from both apical and basal chamber was discarded and replaced with 700 μL of PneumaCult-ALI medium with inducer supplements (STEMCELL Technologies, Inc.) in the basal chamber only to establish ALI conditions. The cells were cultured in ALI culture for 4 weeks, with media replacement every 2 to 3 days. Fully differentiated NECs after 3 to 4 weeks of differentiation were used for subsequent SARS-CoV-2 infection experiments. To generate BECs, primary bronchial cells were obtained from commercial sources (EpitheliX SARL, Switzerland) for expansion following the NEC expansion and differentiation protocol.
AEC infection and virus titration.
AEC infection with SARS-CoV-2 and virus titration were performed as described previously (7). In brief, the apical surfaces of AECs were washed once in Dulbecco’s phosphate-buffered saline (dPBS), and then incubated with virus inoculum at a multiplicity of infection (MOI) of 0.1 for 60 min in a 37°C incubator, after which the inoculum was removed, and the apical surface was washed twice in dPBS before incubating for the indicated period of time. Basolateral medium was replaced with fresh medium every 3 to 4 days. For quantifying virus release at each of the indicated time points, for both single endpoint sampling and repeated sampling, 100 μL of dPBS was added to the apical surface, incubated for 10 min, and then aspirated and used for virus titration.
Cross-section of Transwell NEC and BEC preparation.
Transwells with infected NECs and BECs were treated with 4% paraformaldehyde for > 48 h, followed by immersion in 70% ethanol overnight prior to transfer out of the biosafety level 3 (BSL-3) facility. The infected NECs and BECs on the transwells were paraffin embedded for processing and subsequent IF staining. Briefly, the transwell inserts were dehydrated in ascending grades of ethanol followed by treatment with 2× xylene for 10 min. Following this, the membrane was removed from the transwell and placed into cassettes for subsequent treatment with 2× liquid paraffin for 30 min each. The solidified membranes were embedded in a paraffin boat taking into account orientation for sectioning. Then, 4- to 5-μM sections were cut according to the standard procedures, mounted on glass slides, and allowed to dry for subsequent processing.
Immunofluorescence (IF) staining.
Expression of viral nucleocapsid (NC), βIV-tubulin, P63, and KI67 was assayed via IF staining of the NEC transwell cross-sections. Rabbit monoclonal against SARS-CoV-2 nucleocapsid (Invitrogen, Waltham, MA, USA) and KI67 (Abcam, Cambridge, UK) were used at dilutions of 1:1,000 and 1:400, respectively. Mouse monoclonal antibodies against βIV-tubulin, and P63 (Abcam, Cambridge UK) were used at dilutions of 1:1,000 and 1:80, respectively. Paraffin-embedded sections were dewaxed, rehydrated, and antigen retrieved prior to IF staining. All sections were permeabilized using 0.1% TritonX-100 for 10 min at room temperature, followed by three 1× PBS washes. Cross-sections were blocked with 10% goat serum for 30 min at room temperature and incubated with a primary antibody solution (diluted with 1% goat serum) overnight at 4°C. After overnight incubation, sections were then incubated for 1 h with Alexa Fluor 488- or Alexa Fluor 594-conjugated secondary antibodies in the dark at room temperature, after which, coverslips were mounted onto the slides using SlowFade Gold antifade reagent with 4′,6-diamidino-2-phenylindole (DAPI) (Life Technologies). The slides were analyzed using both fluorescence microscopy (Olympus IX51), a TissueFAXS analyzer, and confocal microscopy.
Quantification of viral and cellular marker staining in NECs.
Transwell sections were scanned with the TissueFAXS slide scanner (TissueGnostics). The resulting region overviews of the merged and individual channels (DAPI, FITC, Texas red) were exported as a .tiff file and subsequently processed using the ImageJ cell counter plugin. The transwell section was scanned from end to end, marking each respective positive stain with a marker according to the cell type. The total number of cells was determined by manually counting the total number of individual nuclei from the DAPI region overview.
10× Genomics single-cell sequencing library preparation and sequencing.
NECs (n = 2) and BECs (n = 2) derived from different donors each were differentiated under ALI culture conditions as described above. They were then infected with SARS-CoV-2 for 28 days (28-dpi sample), cultured at ALI for 25 days, and then infected for 3 days (3-dpi sample) or mock infected with PBS for 28 days (UI sample) before being processed for sc-seq on the same day. As a result, we ensured that all NEC samples from the infected (3- and 28-dpi) and uninfected controls were of the same in vitro culture duration and were processed for sc-seq in the same batch. Cells were dissociated by treatment with trypsin-EDTA (Gibco) and pelleted by centrifugation at 400 × g for 5 min, and the cell pellet was washed twice with PBS + 0.04% bovine serum albumin (BSA) before enumeration with a Scepter cell-counter (Merck Millipore). AECs from each of the 3 infection conditions were processed as three separate reactions using a Chromium Next GEM single cell 3′ kit and chromium controller (10x Genomics). Each reaction contained a pooled mix of 2 NEC and 2 BEC samples from a separate infection condition (UI, 3-dpi, or 28-dpi). GEM generation and barcoding were performed within the ABSL3 facility according to manufacturer’s instructions (Chromium single cell 3′ reagent kits v3 guide, 10x Genomics), with an additional heat inactivation step included during GEM RT-incubation (step 1: 53°C for 45 min, additional heat inactivation step as step 2: 60°C for 30 min, step 3: 85°C for 5 min, before the hold step at 4°C). Subsequent post-GEM-RT cleanup and cDNA amplification, and 3′ gene expression library construction were performed under BSL2+ conditions according to the manufacturer’s instructions. The resulting three libraries were pooled and sequenced in three lanes of a HiSeq4000 (Novogene), generating 110 Gb of raw data for each library. Raw fastq files were demultiplexed and aligned using Cellranger 5.0 (10x Genomics), using GRCh38 as reference.
Genotype sequencing.
For each donor, genomic DNA was extracted from uninfected NECs using a QIAamp DNA minikit (Qiagen). The isolated genomic DNA was used for genotyping to aid sample demultiplexing. Genotyping was performed on an Illumina global screening array-24-MD v3.0 bead chip (Illumina; cat no. 20030773) according to the manufacturer’s protocol. The GenomeStudio2.0 platform was utilized to visualize and analyze genotyping data. Genotype data were strand-corrected (https://github.com/seasky002002/Strandscript), filtered (minor allele frequency [MAF] = 0.01, minor allele count [MAC] = 1), and exon filtered. Subsequently, the vcf file was phased and imputed using Beagle over a 1000 Genomes reference. It was lastly filtered with dosage R-squared (DR2) < 0.9 before use.
Sc-seq analysis.
Cell identities were first demultiplexed using demuxlet (https://github.com/statgen/demuxlet), removing doublets of different samples. DoubletFinder was then used downstream to further remove heterotypic cell type doublets. Homotypic doublets were removed based on the spread of “nUMI” and “nGenes” parameters for each cell barcode. Cells with more than 20% mitochondrial genes were excluded, a relatively lenient threshold to avoid excluding heavily infected cells which are apoptotic, as these are specifically of interest for this study. There were totals of 14,627 and 1,829 cell transcriptomes passing quality control (QC) from the NEC and BEC samples, respectively. Due to the lower number of BEC transcriptomes post-QC, subsequent sc-seq analysis was performed only using the NEC data set. The cleaned count matrix was then imported into Seurat (v4.0) for principal-component analysis (PCA), uniform manifold approximation and projection (UMAP), and graph-based clustering. Numbers of clusters were defined using the clustree package (v0.4.3), and cluster annotations were assigned based on expression of marker genes consistent with the literature and previously published sc-seq annotations for AECs. A debris cluster was identified and removed from the analysis based on absence of cell-type-specific markers, low reads per cell (nCount_RNA), and low genes per cell (nFeauture_RNA). Differential gene expression analysis was performed using edgeR (v3.32.1). For reassigning cell-type identity to transcriptomes within the DDIT3high cluster, reference-based cell type annotation was performed using SingleR (v1.4.1), with count tables from the other 9 clusters pooled across all 3 libraries used as a reference. Gene set enrichment analysis was performed using fgsea (v1.16), as previously described (7). For plotting pathways enriched upon SARS-CoV-2 infection in 3-dpi compared to uninfected NECs (Fig. 5A), significant pathways (adjusted P value < 0.05) with a positive normalized enrichment score (NES) were used for the bubble-plot generation. For plotting IFN signaling pathways enriched upon SARS-CoV-2 infection in 28-dpi compared to uninfected NECs (Fig. 5A), clusters with significant enrichment (adjusted P value < 0.05) and a positive normalized enrichment score (NES) for Hallmark pathways “interferon gamma response” and “interferon alpha response” were displayed as a bubble plot. Graphics were created using GraphPad Prism 8, ggplot2 (v3.2.1), and pheatmap (v1.0.12).
Viral gene expression.
Viral reads were extracted from the raw single-cell fastq libraries with the viral-track pipeline (https://github.com/PierreBSC/Viral-Track) (51). The viral reference genome was the WuHu1 strain extracted from NCBI. Due to the 3′ end biased sequencing of the 10x Genomics pipeline, we additionally included the 3′ UTR in the virus annotation together with the predefined open reading frames (ORFs). Viral read cutoffs to delineate infected versus bystander cells were estimated by applying Otsu’s thresholding (in-house Python script), after log transformation of pooled viral-track output from all 3 libraries (D0, D3, and D28 data sets). Cell barcodes with less than 4 viral reads in libraries derived from 3-dpi and 28-dpi NECs were classified as bystander cells, and cell barcodes with more than 4 viral reads were classified as infected cells.
Cytokine quantification.
Basolateral media from uninfected or infected NECs at each of the indicated time points were assayed for 48 human cytokines using the Bio-Plex Pro human cytokine screening panel (Bio-Rad) as previously described (7). For each of the indicated time points, for both the uninfected and infected NEC samples, the basolateral medium was changed 3 days prior to each basolateral sampling time point, so that the cytokines secreted by NECs during the preceding 72 h were captured for each time point.
Data availability.
All sc-seq data generated from uninfected and SARS-CoV-2-infected NECs for this study have been deposited in NCBI’s Gene Expression Omnibus and are accessible through GEO series accession number GSE182475.
ACKNOWLEDGMENTS
We thank the staff and surgeon team in the Department of Otolaryngology, Head and Neck Surgery, National University Health System (NUH), for help with acquiring human biopsy samples and the operations team at the Duke-NUS Medical School ABSL3 facility for logistics management. The schematic in Fig. 1A was created with Biorender.
This research was funded by the National Medical Research Council, Singapore no. MOH-COVID19RF2-0001 (to R.T.P.L., D.Y.W., and L.-F.W.), COVID19RF-003 (to L.-F.W.), and MOH-OFYIRG19May-0007 (to K.S.T.); K.S.T. is a recipient of support from the European Allergy and Clinical Immunology (EAACI) Research Fellowship 2019.
We declare no conflict of interest.
Conceptualization: A.M.G., K.S.T., J.L. R.T.P.L., D.Y.W., L.-F.W. Investigation and methodology: A.M.G., K.S.T., Z.Z.R.L., J.L., C.W.T., L.M.T., P.N.V. Formal analysis: A.M.G., K.S.T., W.O.Y.C. Software (bioinformatics pipelines): W.O.Y.C., D.R., Q.X.X.L., S.P. Resources (human nasal biopsy samples): Y.K.O. and M.T. Writing: A.M.G., K.S.T., W.O.Y.C., J.L., Z.Z.R.L., D.Y.W., L.-F.W. Funding acquisition and supervision: R.T.P.L., D.Y.W., L.-F.W. All authors have read and agreed to the final version of the manuscript.
Contributor Information
De Yun Wang, Email: entwdy@nus.edu.sg.
Lin-Fa Wang, Email: linfa.wang@duke-nus.edu.sg.
Kanta Subbarao, The Peter Doherty Institute for Infection and Immunity.
REFERENCES
- 1.Qiu X, Nergiz AI, Maraolo AE, Bogoch II, Low N, Cevik M. 2021. The role of asymptomatic and pre-symptomatic infection in SARS-CoV-2 transmission-a living systematic review. Clin Microbiol Infect 27:511–519. doi: 10.1016/j.cmi.2021.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Meyerowitz EA, Richterman A, Bogoch II, Low N, Cevik M. 2021. Towards an accurate and systematic characterisation of persistently asymptomatic infection with SARS-CoV-2. Lancet Infect Dis 21:e163–e169. doi: 10.1016/S1473-3099(20)30837-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cevik M, Tate M, Lloyd O, Maraolo AE, Schafers J, Ho A. 2021. SARS-CoV-2, SARS-CoV, and MERS-CoV viral load dynamics, duration of viral shedding, and infectiousness: a systematic review and meta-analysis. Lancet Microbe 2:e13–e22. doi: 10.1016/S2666-5247(20)30172-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Harvey WT, Carabelli AM, Jackson B, Gupta RK, Thomson EC, Harrison EM, Ludden C, Reeve R, Rambaut A, Consortium C-GU, Peacock SJ, Robertson DL, COVID-19 Genomics UK (COG-UK) Consortium . 2021. SARS-CoV-2 variants, spike mutations and immune escape. Nat Rev Microbiol 19:409–424. doi: 10.1038/s41579-021-00573-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gallo O, Locatello LG, Mazzoni A, Novelli L, Annunziato F. 2021. The central role of the nasal microenvironment in the transmission, modulation, and clinical progression of SARS-CoV-2 infection. Mucosal Immunol 14:305–316. doi: 10.1038/s41385-020-00359-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hou YJ, Okuda K, Edwards CE, Martinez DR, Asakura T, Dinnon KH, 3rd, Kato T, Lee RE, Yount BL, Mascenik TM, Chen G, Olivier KN, Ghio A, Tse LV, Leist SR, Gralinski LE, Schafer A, Dang H, Gilmore R, Nakano S, Sun L, Fulcher ML, Livraghi-Butrico A, Nicely NI, Cameron M, Cameron C, Kelvin DJ, de Silva A, Margolis DM, Markmann A, Bartelt L, Zumwalt R, Martinez FJ, Salvatore SP, Borczuk A, Tata PR, Sontake V, Kimple A, Jaspers I, O'Neal WK, Randell SH, Boucher RC, Baric RS. 2020. SARS-CoV-2 reverse genetics reveals a variable infection gradient in the respiratory tract. Cell 182:429–446.e414. doi: 10.1016/j.cell.2020.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gamage AM, Tan KS, Chan WOY, Liu J, Tan CW, Ong YK, Thong M, Andiappan AK, Anderson DE, Wang DY, Wang L-F. 2020. Infection of human Nasal Epithelial Cells with SARS-CoV-2 and a 382-nt deletion isolate lacking ORF8 reveals similar viral kinetics and host transcriptional profiles. PLoS Pathog 16:e1009130. doi: 10.1371/journal.ppat.1009130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fiege JK, Thiede JM, Nanda HA, Matchett WE, Moore PJ, Montanari NR, Thielen BK, Daniel J, Stanley E, Hunter RC, Menachery VD, Shen SS, Bold TD, Langlois RA. 2021. Single cell resolution of SARS-CoV-2 tropism, antiviral responses, and susceptibility to therapies in primary human airway epithelium. PLoS Pathog 17:e1009292. doi: 10.1371/journal.ppat.1009292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ravindra NG, Alfajaro MM, Gasque V, Huston NC, Wan H, Szigeti-Buck K, Yasumoto Y, Greaney AM, Habet V, Chow RD, Chen JS, Wei J, Filler RB, Wang B, Wang G, Niklason LE, Montgomery RR, Eisenbarth SC, Chen S, Williams A, Iwasaki A, Horvath TL, Foxman EF, Pierce RW, Pyle AM, van Dijk D, Wilen CB. 2021. Single-cell longitudinal analysis of SARS-CoV-2 infection in human airway epithelium identifies target cells, alterations in gene expression, and cell state changes. PLoS Biol 19:e3001143. doi: 10.1371/journal.pbio.3001143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pizzorno A, Padey B, Julien T, Trouillet-Assant S, Traversier A, Errazuriz-Cerda E, Fouret J, Dubois J, Gaymard A, Lescure X, Dulière V, Brun P, Constant S, Poissy J, Lina B, Yazdanpanah Y, Terrier O, Rosa-Calatrava M. 2020. Characterization and treatment of SARS-CoV-2 in nasal and bronchial human airway epithelia. bioRxiv doi: 10.1016/j.xcrm.2020.100059. [DOI] [PMC free article] [PubMed]
- 11.Zarkoob H, Allue-Guardia A, Chen YC, Jung O, Garcia-Vilanova A, Song MJ, Park JG, Oladunni F, Miller J, Tung YT, Kosik I, Schultz D, Yewdell J, Torrelles JB, Martinez-Sobrido L, Cherry S, Ferrer M, Lee EM. 2021. Modeling SARS-CoV-2 and influenza infections and antiviral treatments in human lung epithelial tissue equivalents. bioRxiv doi: 10.1101/2021.05.11.443693. [DOI] [PMC free article] [PubMed]
- 12.Heinen N, Klohn M, Steinmann E, Pfaender S. 2021. In vitro lung models and their application to study SARS-CoV-2 Pathogenesis and disease. Viruses 13:792. doi: 10.3390/v13050792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fulcher ML, Gabriel S, Burns KA, Yankaskas JR, Randell SH. 2005. Well-differentiated human airway epithelial cell cultures. Methods Mol Med 107:183–206. doi: 10.1385/1-59259-861-7:183. [DOI] [PubMed] [Google Scholar]
- 14.Ruiz Garcia S, Deprez M, Lebrigand K, Cavard A, Paquet A, Arguel MJ, Magnone V, Truchi M, Caballero I, Leroy S, Marquette CH, Marcet B, Barbry P, Zaragosi LE. 2019. Novel dynamics of human mucociliary differentiation revealed by single-cell RNA sequencing of nasal epithelial cultures. Development 146:978–990. doi: 10.1242/dev.177428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Davis JD, Wypych TP. 2021. Cellular and functional heterogeneity of the airway epithelium. Mucosal Immunol 14:dev177428. doi: 10.1038/s41385-020-00370-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bukowy-Bieryllo Z. 2021. Long-term differentiating primary human airway epithelial cell cultures: how far are we? Cell Commun Signal 19:63. doi: 10.1186/s12964-021-00740-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Plasschaert LW, Zilionis R, Choo-Wing R, Savova V, Knehr J, Roma G, Klein AM, Jaffe AB. 2018. A single-cell atlas of the airway epithelium reveals the CFTR-rich pulmonary ionocyte. Nature 560:377–381. doi: 10.1038/s41586-018-0394-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rock JR, Onaitis MW, Rawlins EL, Lu Y, Clark CP, Xue Y, Randell SH, Hogan BLM. 2009. Basal cells as stem cells of the mouse trachea and human airway epithelium. Proc Natl Acad Sci USA 106:12771–12775. doi: 10.1073/pnas.0906850106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Montoro DT, Haber AL, Biton M, Vinarsky V, Lin B, Birket SE, Yuan F, Chen S, Leung HM, Villoria J, Rogel N, Burgin G, Tsankov AM, Waghray A, Slyper M, Waldman J, Nguyen L, Dionne D, Rozenblatt-Rosen O, Tata PR, Mou H, Shivaraju M, Bihler H, Mense M, Tearney GJ, Rowe SM, Engelhardt JF, Regev A, Rajagopal J. 2018. A revised airway epithelial hierarchy includes CFTR-expressing ionocytes. Nature 560:319–324. doi: 10.1038/s41586-018-0393-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zaragosi LE, Deprez M, Barbry P. 2020. Using single-cell RNA sequencing to unravel cell lineage relationships in the respiratory tract. Biochem Soc Trans 48:327–336. doi: 10.1042/BST20191010. [DOI] [PubMed] [Google Scholar]
- 21.Vieira Braga FA, Kar G, Berg M, Carpaij OA, Polanski K, Simon LM, Brouwer S, Gomes T, Hesse L, Jiang J, Fasouli ES, Efremova M, Vento-Tormo R, Talavera-Lopez C, Jonker MR, Affleck K, Palit S, Strzelecka PM, Firth HV, Mahbubani KT, Cvejic A, Meyer KB, Saeb-Parsy K, Luinge M, Brandsma CA, Timens W, Angelidis I, Strunz M, Koppelman GH, van Oosterhout AJ, Schiller HB, Theis FJ, van den Berge M, Nawijn MC, Teichmann SA. 2019. A cellular census of human lungs identifies novel cell states in health and in asthma. Nat Med 25:1153–1163. doi: 10.1038/s41591-019-0468-5. [DOI] [PubMed] [Google Scholar]
- 22.van Kampen JJA, van de Vijver D, Fraaij PLA, Haagmans BL, Lamers MM, Okba N, van den Akker JPC, Endeman H, Gommers D, Cornelissen JJ, Hoek RAS, van der Eerden MM, Hesselink DA, Metselaar HJ, Verbon A, de Steenwinkel JEM, Aron GI, van Gorp ECM, van Boheemen S, Voermans JC, Boucher CAB, Molenkamp R, Koopmans MPG, Geurtsvankessel C, van der Eijk AA. 2021. Duration and key determinants of infectious virus shedding in hospitalized patients with coronavirus disease-2019 (COVID-19). Nat Commun 12:267. doi: 10.1038/s41467-020-20568-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Young BE, Ong SWX, Ng LFP, Anderson DE, Chia WN, Chia PY, Ang LW, Mak TM, Kalimuddin S, Chai LYA, Pada S, Tan SY, Sun L, Parthasarathy P, Fong SW, Chan YH, Tan CW, Lee B, Rotzschke O, Ding Y, Tambyah P, Low JGH, Cui L, Barkham T, Lin RTP, Leo YS, Renia L, Wang LF, Lye DC, Singapore Novel Coronavirus Outbreak Research Team . 2020. Viral dynamics and immune correlates of COVID-19 disease severity. Clin Infect Dis doi: 10.1093/cid/ciaa1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aydillo T, Gonzalez-Reiche AS, Aslam S, van de Guchte A, Khan Z, Obla A, Dutta J, van Bakel H, Aberg J, Garcia-Sastre A, Shah G, Hohl T, Papanicolaou G, Perales MA, Sepkowitz K, Babady NE, Kamboj M. 2020. Shedding of viable SARS-CoV-2 after immunosuppressive therapy for cancer. N Engl J Med 383:2586–2588. doi: 10.1056/NEJMc2031670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Choi B, Choudhary MC, Regan J, Sparks JA, Padera RF, Qiu X, Solomon IH, Kuo HH, Boucau J, Bowman K, Adhikari UD, Winkler ML, Mueller AA, Hsu TY, Desjardins M, Baden LR, Chan BT, Walker BD, Lichterfeld M, Brigl M, Kwon DS, Kanjilal S, Richardson ET, Jonsson AH, Alter G, Barczak AK, Hanage WP, Yu XG, Gaiha GD, Seaman MS, Cernadas M, Li JZ. 2020. Persistence and evolution of SARS-CoV-2 in an immunocompromised host. N Engl J Med 383:2291–2293. doi: 10.1056/NEJMc2031364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tan GSE, Ding Y, Cui L, Mak TM, Mok CK, Kurup A, Parthasarathy P, Chia WN, Wang LF, Lin RTP, Leo YS, Vasoo S. 2021. Positive RT-PCR detected in patients recovered from COVID-19. Ann Acad Med Singap 50:191–194. doi: 10.47102/annals-acadmedsg.2020387. [DOI] [PubMed] [Google Scholar]
- 27.Owusu D, Pomeroy MA, Lewis NM, Wadhwa A, Yousaf AR, Whitaker B, Dietrich E, Hall AJ, Chu V, Thornburg N, Christensen K, Kiphibane T, Willardson S, Westergaard R, Dasu T, Pray IW, Bhattacharyya S, Dunn A, Tate JE, Kirking HL, Matanock A, Household Transmission Study Team . 2021. Persistent SARS-CoV-2 RNA shedding without evidence of infectiousness: a cohort study of individuals with COVID-19. J Infect Dis doi: 10.1093/infdis/jiab107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang C, Huang L, Wang Y, Li X, Ren L, Gu X, Kang L, Guo L, Liu M, Zhou X, Luo J, Huang Z, Tu S, Zhao Y, Chen L, Xu D, Li Y, Li C, Peng L, Li Y, Xie W, Cui D, Shang L, Fan G, Xu J, Wang G, Wang Y, Zhong J, Wang C, Wang J, Zhang D, Cao B. 2021. 6-month consequences of COVID-19 in patients discharged from hospital: a cohort study. Lancet 397:220–232. doi: 10.1016/S0140-6736(20)32656-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carfi A, Bernabei R, Landi F, Gemelli Against COVID-19 Post-Acute Care Study Group . 2020. Persistent symptoms in patients after acute COVID-19. JAMA 324:603–605. doi: 10.1001/jama.2020.12603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sudre CH, Murray B, Varsavsky T, Graham MS, Penfold RS, Bowyer RC, Pujol JC, Klaser K, Antonelli M, Canas LS, Molteni E, Modat M, Jorge Cardoso M, May A, Ganesh S, Davies R, Nguyen LH, Drew DA, Astley CM, Joshi AD, Merino J, Tsereteli N, Fall T, Gomez MF, Duncan EL, Menni C, Williams FMK, Franks PW, Chan AT, Wolf J, Ourselin S, Spector T, Steves CJ. 2021. Attributes and predictors of long COVID. Nat Med 27:626–631. doi: 10.1038/s41591-021-01292-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nalbandian A, Sehgal K, Gupta A, Madhavan MV, McGroder C, Stevens JS, Cook JR, Nordvig AS, Shalev D, Sehrawat TS, Ahluwalia N, Bikdeli B, Dietz D, Der-Nigoghossian C, Liyanage-Don N, Rosner GF, Bernstein EJ, Mohan S, Beckley AA, Seres DS, Choueiri TK, Uriel N, Ausiello JC, Accili D, Freedberg DE, Baldwin M, Schwartz A, Brodie D, Garcia CK, Elkind MSV, Connors JM, Bilezikian JP, Landry DW, Wan EY. 2021. Post-acute COVID-19 syndrome. Nat Med 27:601–615. doi: 10.1038/s41591-021-01283-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kwon T, Gaudreault NN, Richt JA. 2021. Seasonal stability of SARS-CoV-2 in biological fluids. bioRxiv doi: 10.1101/2021.04.07.438866:2021.2004.2007.438866. [DOI] [PMC free article] [PubMed]
- 33.Sun Z, Cai X, Gu C, Zhang R, Han W, Qian Y, Wang Y, Xu W, Wu Y, Cheng X, Yuan Z, Xie Y, Qu D. 2020. Survival of SARS-COV-2 under liquid medium, dry filter paper and acidic conditions. Cell Discov 6:57. doi: 10.1038/s41421-020-00191-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schneider P, MacKay F, Steiner V, Hofmann K, Bodmer JL, Holler N, Ambrose C, Lawton P, Bixler S, Acha-Orbea H, Valmori D, Romero P, Werner-Favre C, Zubler RH, Browning JL, Tschopp J. 1999. BAFF, a novel ligand of the tumor necrosis factor family, stimulates B cell growth. J Exp Med 189:1747–1756. doi: 10.1084/jem.189.11.1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mackay F, Browning JL. 2002. BAFF: a fundamental survival factor for B cells. Nat Rev Immunol 2:465–475. doi: 10.1038/nri844. [DOI] [PubMed] [Google Scholar]
- 36.Groom JR, Luster AD. 2011. CXCR3 ligands: redundant, collaborative and antagonistic functions. Immunol Cell Biol 89:207–215. doi: 10.1038/icb.2010.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hao S, Ning K, Kuz CA, Vorhies K, Yan Z, Qiu J. 2020. Long-term modeling of SARS-CoV-2 infection of in vitro cultured polarized human airway epithelium. mBio 11:e02852-20. doi: 10.1128/mBio.02852-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee S, Yoon GY, Myoung J, Kim SJ, Ahn DG. 2020. Robust and persistent SARS-CoV-2 infection in the human intestinal brush border expressing cells. Emerg Microbes Infect 9:2169–2179. doi: 10.1080/22221751.2020.1827985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reuken PA, Stallmach A, Pletz MW, Brandt C, Andreas N, Hahnfeld S, Loffler B, Baumgart S, Kamradt T, Bauer M. 2021. Severe clinical relapse in an immunocompromised host with persistent SARS-CoV-2 infection. Leukemia 35:920–923. doi: 10.1038/s41375-021-01175-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kemp SA, Collier DA, Datir RP, Ferreira I, Gayed S, Jahun A, Hosmillo M, Rees-Spear C, Mlcochova P, Lumb IU, Roberts DJ, Chandra A, Temperton N, COVID-19 Genomics UK (COG-UK) Consortium, Sharrocks K, Blane E, Modis Y, Leigh KE, Briggs JAG, van Gils MJ, Smith KGC, Bradley JR, Smith C, Doffinger R, Ceron-Gutierrez L, Barcenas-Morales G, Pollock DD, Goldstein RA, Smielewska A, Skittrall JP, Gouliouris T, Goodfellow IG, Gkrania-Klotsas E, Illingworth CJR, McCoy LE, Gupta RK. 2021. SARS-CoV-2 evolution during treatment of chronic infection. Nature 592:277–282. doi: 10.1038/s41586-021-03291-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tarhini H, Recoing A, Bridier-Nahmias A, Rahi M, Lambert C, Martres P, Lucet JC, Rioux C, Bouzid D, Lebourgeois S, Descamps D, Yazdanpanah Y, Le Hingrat Q, Lescure FX, Visseaux B. 2021. Long-term severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infectiousness among three immunocompromised patients: from prolonged viral shedding to SARS-CoV-2 superinfection. J Infect Dis 223:1522–1527. doi: 10.1093/infdis/jiab075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gaebler C, Wang Z, Lorenzi JCC, Muecksch F, Finkin S, Tokuyama M, Cho A, Jankovic M, Schaefer-Babajew D, Oliveira TY, Cipolla M, Viant C, Barnes CO, Bram Y, Breton G, Hagglof T, Mendoza P, Hurley A, Turroja M, Gordon K, Millard KG, Ramos V, Schmidt F, Weisblum Y, Jha D, Tankelevich M, Martinez-Delgado G, Yee J, Patel R, Dizon J, Unson-O'Brien C, Shimeliovich I, Robbiani DF, Zhao Z, Gazumyan A, Schwartz RE, Hatziioannou T, Bjorkman PJ, Mehandru S, Bieniasz PD, Caskey M, Nussenzweig MC. 2021. Evolution of antibody immunity to SARS-CoV-2. Nature 591:639–644. doi: 10.1038/s41586-021-03207-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.de Melo GD, Lazarini F, Levallois S, Hautefort C, Michel V, Larrous F, Verillaud B, Aparicio C, Wagner S, Gheusi G, Kergoat L, Kornobis E, Donati F, Cokelaer T, Hervochon R, Madec Y, Roze E, Salmon D, Bourhy H, Lecuit M, Lledo PM. 2021. COVID-19-related anosmia is associated with viral persistence and inflammation in human olfactory epithelium and brain infection in hamsters. Sci Transl Med 13:eabf8396. doi: 10.1126/scitranslmed.abf8396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ahn JH, Kim J, Hong SP, Choi SY, Yang MJ, Ju YS, Kim YT, Kim HM, Rahman MDT, Chung MK, Hong SD, Bae H, Lee CS, Koh GY. 2021. Nasal ciliated cells are primary targets for SARS-CoV-2 replication in early stage of COVID-19. J Clin Invest 131:e148517. doi: 10.1172/JCI148517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xia H, Cao Z, Xie X, Zhang X, Chen JY, Wang H, Menachery VD, Rajsbaum R, Shi PY. 2020. Evasion of type I interferon by SARS-CoV-2. Cell Rep 33:108234. doi: 10.1016/j.celrep.2020.108234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Konno Y, Kimura I, Uriu K, Fukushi M, Irie T, Koyanagi Y, Nakagawa S, Sato K. 2020. SARS-CoV-2 ORF3b is a potent interferon antagonist whose activity is further increased by a naturally occurring elongation variant. Cell Rep 32:108185. doi: 10.1016/j.celrep.2020.108185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Banerjee AK, Blanco MR, Bruce EA, Honson DD, Chen LM, Chow A, Bhat P, Ollikainen N, Quinodoz SA, Loney C, Thai J, Miller ZD, Lin AE, Schmidt MM, Stewart DG, Goldfarb D, De Lorenzo G, Rihn SJ, Voorhees RM, Botten JW, Majumdar D, Guttman M. 2020. SARS-CoV-2 disrupts splicing, translation, and protein trafficking to suppress host defenses. Cell 183:1325–1339.e1321. doi: 10.1016/j.cell.2020.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thoms M, Buschauer R, Ameismeier M, Koepke L, Denk T, Hirschenberger M, Kratzat H, Hayn M, Mackens-Kiani T, Cheng J, Straub JH, Stürzel CM, Fröhlich T, Berninghausen O, Becker T, Kirchhoff F, Sparrer KMJ, Beckmann R. 2020. Structural basis for translational shutdown and immune evasion by the Nsp1 protein of SARS-CoV-2. Science 369:1249–1255. doi: 10.1126/science.abc8665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rajah MM, Hubert M, Bishop E, Saunders N, Robinot R, Grzelak L, Planas D, Dufloo J, Gellenoncourt S, Bongers A, Zivaljic M, Planchais C, Guivel-Benhassine F, Porrot F, Mouquet H, Chakrabarti L, Buchrieser J, Schwartz O. 2021. SARS-CoV-2 alpha, beta and delta variants display enhanced spike-mediated syncytia formation. bioRxiv doi: 10.1101/2021.06.11.448011. [DOI] [PMC free article] [PubMed]
- 50.Yan Y, Tan KS, Li C, Tran T, Chao SS, Sugrue RJ, Shi L, Chow V, Wang D-Y. 2016. Human nasal epithelial cells derived from multiple subjects exhibit differential responses to H3N2 influenza virus infection in vitro. J Allergy Clin Immunol 138:276–281. doi: 10.1016/j.jaci.2015.11.016. [DOI] [PubMed] [Google Scholar]
- 51.Bost P, Giladi A, Liu Y, Bendjelal Y, Xu G, David E, Blecher-Gonen R, Cohen M, Medaglia C, Li H, Deczkowska A, Zhang S, Schwikowski B, Zhang Z, Amit I. 2020. Host-viral infection maps reveal signatures of severe COVID-19 patients. Cell 181:1475–1488.e1412. doi: 10.1016/j.cell.2020.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Distribution and abundance of airway epithelial cell types in infected versus uninfected sc-seq libraries. (A) Clustering of sc-seq transcriptomes from uninfected (top panel), 3-dpi (middle panel), and 28-dpi (bottom panel) NECs. Each color-coded cluster corresponds to the same cell type annotation displayed for the pooled UMAP in Fig. 3A. (B) Number of cell transcriptomes from each of the airway epithelial cell types in the uninfected, 3-dpi, and 28-dpi libraries. Download FIG S1, TIF file, 2.7 MB (2.7MB, tif) .
Copyright © 2022 Gamage et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Increased KI67+ cells at 28 dpi. (A to C) IF staining for KI67 (red), DAPI nuclear stain (blue), and P63 (green) from (A) uninfected (UI), (B) 3-dpi, and (C) 28-dpi infected NECs. Arrows indicate cells staining positive for KI67. Representative images obtained from at least 3 different donor-derived NEC cross-sections processed for IF staining are shown. (D) Percentage of KI67+cells within NEC structures at each of the indicated time points. Each dot represents data obtained from a different human donor-derived NEC cross-section. Data are represented as the mean ± SEM. The indicated P values are derived from Student's t test. Download FIG S2, TIF file, 2.7 MB (2.7MB, tif) .
Copyright © 2022 Gamage et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
SARS-CoV-2 viral reads detected from 3- and 28-dpi NECs. (A) Viral reads per cell detected in cell transcriptomes derived from uninfected (D0), 3-dpi (D3), and 28-dpi (D28) NEC samples. (B) Coverage plot of pooled SARS-CoV-2 reads from 3-dpi and 28-dpi NEC samples, overlaid with the SARS-CoV-2 genome, demonstrating that the majority of viral reads map to the 3′ genome terminus. Download FIG S3, TIF file, 1.1 MB (793.7KB, tif) .
Copyright © 2022 Gamage et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Data Availability Statement
All sc-seq data generated from uninfected and SARS-CoV-2-infected NECs for this study have been deposited in NCBI’s Gene Expression Omnibus and are accessible through GEO series accession number GSE182475.