ABSTRACT
Many antibiotic resistant uropathogenic Escherichia coli (UPEC) strains belong to clones defined by their multilocus sequence type (ST), with ST131 being the most dominant. Although we have a good understanding of resistance development to fluoroquinolones and third-generation cephalosporins by ST131, our understanding of the virulence repertoire that has contributed to its global dissemination is limited. Here we show that the genes encoding Afa/Dr fimbriae, a group of adhesins strongly associated with UPEC that cause gestational pyelonephritis and recurrent cystitis, are found in approximately one third of all ST131 strains. Sequence comparison of the AfaE adhesin protein revealed a unique allelic variant carried by 82.9% of afa-positive ST131 strains. We identify the afa regulatory region as a hotspot for the integration of insertion sequence (IS) elements, all but one of which alter afa transcription. Close investigation demonstrated that the integration of an IS1 element in the afa regulatory region leads to increased expression of Afa/Dr fimbriae, promoting enhanced adhesion to kidney epithelial cells and suggesting a mechanism for altered virulence. Finally, we provide evidence for a more widespread impact of IS1 on ST131 genome evolution, suggesting that IS dynamics contribute to strain level microevolution that impacts ST131 fitness.
KEYWORDS: adhesins, antibiotic resistance, fimbriae, uropathogenic Escherichia coli, virulence regulation
INTRODUCTION
Uropathogenic Escherichia coli (UPEC) is the most common cause of urinary tract infections (UTIs) and a frequent cause of life-threatening sepsis (1). The majority of UPEC strains belong to global clones that can be differentiated based on their multilocus sequence type (ST), including ST69, ST73, ST95, and ST131 (2–4). ST131 is the most globally dominant high-risk UPEC clone and a major contributor to increasing antibiotic resistance (2, 5). ST131 was first identified in 2008 and is associated with resistance to multiple antibiotic classes, including fluoroquinolones, third-generation cephalosporins and more recently last-line carbapenems and polymyxins (6–9).
While genomic studies have accurately traced the epidemiology and emergence of ST131 (7, 10–12), the precise molecular mechanisms that have led to the dominance of ST131 over other UPEC clones remain unclear. ST131 shares many common virulence factors with non-ST131 UPEC, including adhesins, toxins, capsule polysaccharides, and iron acquisition systems. The Dr family or Afa/Dr chaperone-usher fimbriae represent one important UPEC virulence determinant that have been suggested to display a higher prevalence in ST131 compared with non-ST131 UPEC (4, 13). Afa/Dr fimbriae are frequently associated with UPEC that cause cystitis in children, as well as pyelonephritis and recurrent UTIs in young and pregnant women (14–18). The receptors for Afa/Dr fimbriae are the Dr blood group antigen on the human decay-accelerating factor (DAF), some members of the carcinoembryonic antigen (CEA) family and type IV collagen (18–21). Interactions between Afa/Dr fimbriae and its host receptor stimulate invasion, leading to the avoidance of host immunosurveillance and antibiotic treatment (22–24), and possibly contributing to recurrent infection (14, 18).
Members of the Dr family were initially reported to form both fimbrial and afimbrial structures on the bacterial surface (14, 25–28). However, more recent studies have revealed the afimbrial pattern results from the collapse of flexible fimbrial structures onto the bacterial surface, demonstrating they are indeed assembled as fimbrial organelles (29, 30). Afa/Dr fimbriae are closely related at the DNA level, share a similar genetic organization, and have been described using a range of gene designations including dra, drb, nfa, agg, hda, and afa (used here) (31, 32). Afa/Dr fimbriae are encoded by a cluster of genes organized in two divergent transcriptional units - a primary transcriptional unit comprising genes encoding a regulator (e.g., afaA), chaperone (e.g., afaB), usher (e.g., afaC), repeating major adhesin subunit (e.g., afaE), and tip-associated capping subunit protein (e.g., afaD), as well as a minor transcriptional unit comprising a single regulatory gene (e.g., afaF) (14, 33, 34). The AfaE major adhesin subunit exhibits extensive amino acid sequence diversity and is under positive selection pressure that leads to altered binding phenotypes, including increased binding to the DAF receptor (35). Afa/Dr expression is phase variable due to the concerted action of deoxyadenosine methylase (Dam) and the leucine-responsive regulatory protein (Lrp) that control differential methylation patterns in the afa/dr promoter region (36, 37), as well as activation by integration host factor (IHF) and repression by the histone-like nucleoid-structuring protein H-NS (37, 38). The afa/dr genes have been shown to be located on plasmids or within chromosomal genomic islands (GIs) (39, 40), supporting their capacity to be transferred via mobile genetic elements. Furthermore, the afa-3 allelic variant present in the UPEC cystitis isolate A30 is flanked by IS1 elements that facilitate plasmid to chromosome transfer via IS1-mediated recombination (33, 41).
Previous in vivo studies have demonstrated the role of Afa/Dr adhesins in UPEC virulence. The draE and afaE-III alleles are among the most widely studied and have been associated with chronic tubulointerstitial nephritis (42) and mortality of pregnant rats (43). In this study, we examined the distribution of afa/dr genes among the most common E. coli STs, confirming their high prevalence in ST131. Close examination of the afa/dr gene complexity in ST131 showed the regulatory region is a hotspot for the integration of IS elements, and we demonstrate that IS1 insertion leads to increased Afa fimbrial expression, highlighting the capacity of IS elements to drive altered virulence at the strain level. Finally, we expand these findings by mapping the integration of IS1 within the ST131 pangenome, thus revealing how the widespread occurrence of this mobile genetic element can impact ST131 genome evolution.
RESULTS
Afa/Dr fimbrial genes are highly prevalent in E. coli ST131.
The prevalence of Afa/Dr fimbrial genes in E. coli was initially examined by investigating their distribution in genomes representing the most common STs from EnteroBase, a large publicly available Enterobacteriaceae genome sequence database (44). Analysis of 100 randomly selected genome assemblies from each of the 83 highest-represented STs revealed the afaC usher gene, which is conserved in all afa/dr loci, is present in 18 different STs. The highest incidence was found in ST131 (35%; 35/100) and ST38 (37%; 37/100); followed by ST59 (17%; 17/100), ST405 (14%; 14/100), ST448 (12%; 12/100), and ST648 (12%; 12/100) (Fig. S1A in the supplemental material). Overall, the Afa/Dr fimbrial genes were found in STs from all phylogroups, but at different prevalence rates.
Distribution of Dr family fimbriae in (A) strains belonging to the 83 most common E. coli STs and (B) strains belonging to the five most dominant UPEC clones. E. coli STs are grouped into seven main phylogenetic groups: A (ST10, ST34, ST48, ST93, ST167, ST301, and ST746), B1 (ST448), B2 (ST12 and ST131), C (ST23 and ST410), D (ST38, ST405 and ST69), E (ST335), and F (ST59 and ST648). The percentage of strains containing Afa/Dr fimbriae was determined by tBLASTn against afaCEC958, with a cut-off value of > 90% nucleotide identity and > 80% gene coverage. Download FIG S1, TIF file, 0.3 MB (308.6KB, tif) .
Copyright © 2022 Alvarez-Fraga et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
We were particularly interested in the high prevalence of the Afa/Dr fimbrial genes in ST131, and therefore expanded our screen to perform a detailed comparison of ST131 (phylogroup B2) and four other UPEC dominant STs: ST10 (phylogroup A), ST69 (phylogroup D), ST73 (phylogroup B2), and ST95 (phylogroup B2). This large-scale analysis revealed that the prevalence of Afa/Dr fimbrial genes largely mirrored our results from the 83 ST screen, with 29% (1,157/3,993) of ST131 strains containing the afaC usher gene, compared with ST10 (65/4,731; 1.4%), ST69 (16/805; 2%), ST73 (2/923; 0.2%), and ST95 (1/831; 0.1%) (Fig. S1B).
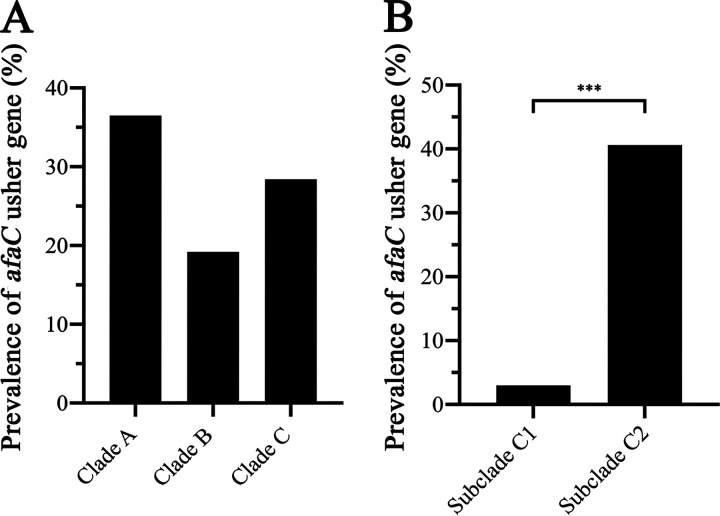
Despite the enormous diversity of UPEC at the genome level, the ST131 lineage represents a monophyletic clone with a well-defined genealogy (7, 10, 12, 45, 46). ST131 is comprised of three major sublineages, clades A, B, and the fluoroquinolone-resistant clade C. A total of 3,857 ST131 strains from the Enterobase data set were categorized into their specific sublineage using clade-defining single nucleotide polymorphisms (SNPs) that we have characterized previously (10). One-hundred and 36 strains could not be classified into any clade, and were excluded from this analysis. Overall, 36.5% (138/378) of clade A strains, 19.2% (66/344) of clade B strains, and 28.4% (890/3,135) of clade C strains possessed the afaC usher gene (Fig. 1A). Within clade C, the prevalence of the Afa/Dr fimbrial genes was significantly more common in strains from the multidrug-resistant subclade C2 (40.6%; 840/2,069) compared with subclade C1 (3%; 30/993) (P < 0.0001; Chi-square test) (Fig. 1B).
FIG 1.
Distribution of Afa/Dr fimbrial genes in strains belonging to the ST131 lineage; (A) clade A, B, and C; and (B) subclade C1 and C2. The percentage of strains containing Afa/Dr fimbriae was determined by tBLASTn against afaCEC958, with a cut-off value of > 90% nucleotide identity and > 80% gene coverage. ST131 strains were classified in clades and subclades based on clade-specific SNPs. One-hundred and 36 ST131 strains could not be classified into any clade, and thus were excluded from the analysis. Chi-squared test was used for statistical analysis (***, P < 0.0001 for pair of C2–C1).
Variation of the AfaE major subunit protein in ST131 is dominated by a unique variant.
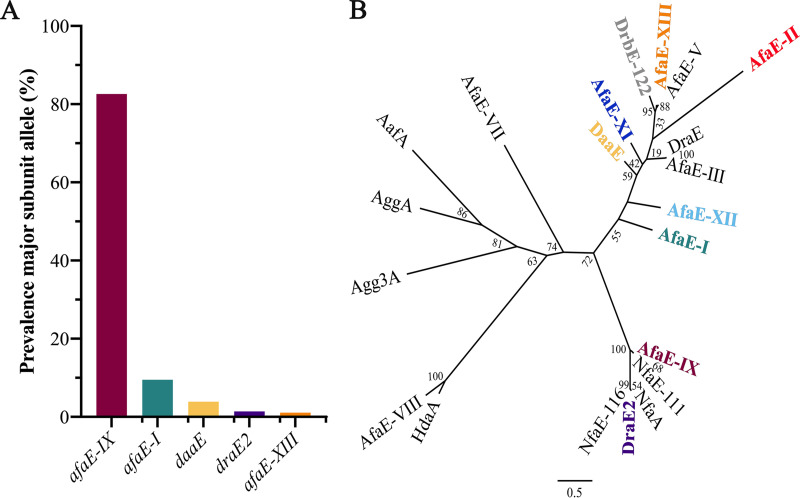
Analysis of the nucleotide sequence of the Afa/Dr major subunit gene in our ST131 data set identified nine allelic variants (Fig. 2A). Among these, 82.9% (907/1,094) of the strains possessed an identical afaE allele, which was the same as the one found in the reference ST131 EC958 genome (referred to as afaE-IX). Other afaE alleles were present at lower levels: afaE-I (9.6%; 105/1,094), daaE (3.7%; 41/1,094), draE2 (1.4%; 15/1,094), afaE-II (0.3%; 3/1,094), drbE-122 (0.1%; 1/1,094), and three previously unreported afaE alleles (referred to as afaE-XIII; 1.1%; 12/1,094), afaE-XI (0.7%; 8/1,094), and afaE-XII (0.2%; 2/1094) (Fig. 2A). The afaE-IX allele was predominant in all ST131 clades, and found at a significantly higher rate in subclade C2 (37.5%) compared with C1 (1.6%) (P < 0.0001; Chi-square test) (Fig. S2A, B).
FIG 2.
(A) Prevalence of the major subunit allelic variants found in the ST131 lineage. The percentage of each afaE allele was determined by tBLASTn with a cut-off value of > 97% nucleotide identity and > 80% gene coverage. The afaE-XI, afaE-II, afaE-XII, and drbE-122 alleles were prevalent at < 1% (not shown). (B) Unrooted ML phylogenetic tree showing the relationship between the AfaE variants. The names in color represent the AfaE variants found in the ST131 lineage. The scale indicates the number of amino acid substitutions; numbers refer to the bootstrap values.
(A) Prevalence of the major subunits allelic variants in ST131 clades. AfaE alleles afaE-IX, afaE-I, daaE and draE2 are shown; five alleles were prevalent at < 1% (not shown): afaE-XIII (0.4%, clade C), afaE-XI (0.2%, clade B and 0.2%, clade C), afaE-II (0.1%, clade C), afaE-XII (0.1%, clade C), and drbE-122 (0.03%, clade C). (B) Prevalence of the major subunits allelic variants in ST131 subclades C1 and C2. AfaE alleles afaE-IX and afaE-I are shown; four alleles were prevalent at < 1% (not shown): afaE-XIII (0.5%, subclade C1 and 0.3%, subclade C2), drbE-122 (0.3%, subclade C1), afaE-XI (0.3%, subclade C2), afaE-II (0.1%, subclade C2), afaE-XII (0.2%, subclade C2). The daaE and afaE-XIV alleles were absent in both subclades. The percentage of each afaE allele was determined by tBLASTn with a cut-off value of > 97% nucleotide identity and > 80% gene coverage. ST131 strains were classified in clades and subclades based on clade-specific SNPs. One-hundred and 36 ST131 strains could not be classified into any clade and thus were excluded from the analysis. Chi-squared test was used for statistical analysis (*** P < 0.0001 for pair of C2–C1). (C) Prevalence of the major subunit allelic variants within E. coli ST38 clone. The daaE, afaE-XII, and afaE-VIII alleles were prevalent at < 1% (not shown). The percentage of each afaE allele was determined by tBLASTn with a cut-off value of > 97% nucleotide identity and > 80% gene coverage. Download FIG S2, TIF file, 0.5 MB (546.3KB, tif) .
Copyright © 2022 Alvarez-Fraga et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Due to the high prevalence of Afa/Dr fimbrial genes in the subset of 100 ST38 genomes, we also downloaded all 1,926 ST38 genomes from EnteroBase and analyzed their overall prevalence and afaE allelic variation. The afaC usher gene was present in 38.6% (743/1,926) of the ST38 strains. The most prevalent major subunit alleles were afaE-XI (33%; 245/743), drbE-122 (21.4%; 159/743), afaE-II (15.7%; 117/743), and draE2 (14.8%; 110/743). Other afaE alleles were present at lower levels: afaE-I (5.8%; 43/743), afaE-IX (6.1%; 45/743), afaE-XIII (1.3%; 10/743), daaE (0.9%; 7/743), afaE-XII (0.8%; 3/743), and afaE-VIII (0.1%, 1/743) (Fig. S2C). We were unable to identify the major subunit allele of three strains. Thus, although both ST131 and ST38 possess a high prevalence of Afa/Dr fimbrial genes, the distribution of afaE allelic variants differs markedly within each clone.
The phylogenetic relationship between all identified AfaE protein variants was examined by generating a multiple-sequence alignment of the mature proteins (Fig. S3). The ST131 AfaE variants could be assigned to previously described distinct phylogenetic groups (35). The dominant AfaE-IX variant forms a discrete group together with the NfaE-111 variant (Fig. 2B). The AfaE-IX and NfaE-111 proteins share 96.2% identity, with all differences centered around one of the predicted DAF binding sides (19, 30, 47) (Fig. S3). The novel AfaE-XIII variant also forms a distinct group, together with AfaE-V (Fig. 2B). Finally, the AfaE-XI and AfaE-XII variants form two distantly related groups (Fig. 2B).
Multiple-sequence alignment of the major subunit variants. The sequences correspond to the mature protein. Each rectangle represents a distinct group, and their phylogenetic relationship is shown in the cladogram. The rectangles in color represent the groups found in the ST131 lineage. The predicted CEA and DAF binding sites are boxed in the protein sequence. The AfaE-XI variant shares a 99.6% identity with the AfaE variant found in the strain VR50. Download FIG S3, TIF file, 2.9 MB (3MB, tif) .
Copyright © 2022 Alvarez-Fraga et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The afa/dr regulatory region is a hotspot for IS integration.
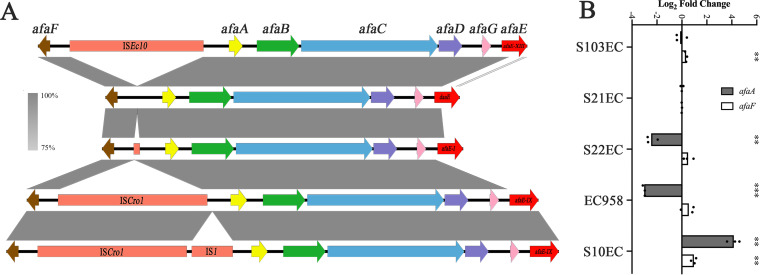
Closer examination of the afa/dr gene cluster in our ST131 genome data set revealed the presence of IS elements in the regulatory region located between the divergently transcribed afaF and afaA genes (Fig. 3A). All strains containing the afaE-IX allele possessed a large 2,824-bp ISCro1 element (IS66 family) 440 bp upstream of the afaA start codon. Five of these strains also possessed an IS1 element 87 bp downstream of the ISCro1 element. The genomic insertion site of ISCro1 appears to be a hotspot for IS integration, as strains possessing other afaE adhesin alleles contain different insertions at this site; strains containing the afaE-I allele possess a 118-bp insertion of unknown origin, while strains containing the afaE-XIII allele possess a 2,530-bp ISEc10 (IS21 family) insertion at this site. Finally, strains containing the daaE, draE2, and afaE-XI alleles did not have any insertions in the afa/dr promoter region (Fig. 3A). We were unable to assemble the afa/dr promoter region associated with the afaE-XIV, afaE-XII, and afaE-II alleles, and thus could not accurately examine the presence of IS elements in strains containing these variants.
FIG 3.
(A) Schematic representation of the Afa/Dr fimbrial gene clusters found in ST131, showing the IS elements located in the promoter region. Brown (afaF) and yellow (afaA) arrows represent genes encoding transcriptional regulators; orange rectangles represent different IS elements; other genes are color-coded as green (chaperone), blue (usher), purple (invasin), pink (genes involved in RNA processing), and red (adhesin major subunit). (B) Transcript levels of afaA and afaF genes in representative ST131 strains. The results are represented as log2 fold change compared with S21EC (which has no IS element in the promoter region). The gapA gene was used as an endogenous control. Experiments were performed in three biological replicates. Unpaired Student's t test was used for statistical analysis (**, P value < 0.01; ***, P value < 0.0001).
To determine the precise genetic location of afa/dr genes representing each afaE allelic variant, we generated complete genome sequences of representative strains: S21EC (daaE; control strain with no insertion), S22EC (afaE-I/118-bp insertion), S10EC (afaE-IX/ISCro1-IS1), and S103EC (afaE-XIII/ISEc10). Summary features of these genomes, including chromosome size and plasmids are presented in Data set S1A; antibiotic resistance genes and major virulence factors were described previously based on analyses of draft genomes (7). The complete genome of EC958 (afaE-IX/ISCro1) has been published previously (48). Analysis of the genome data revealed the afa/dr gene cluster is located within a genomic island (GI) integrated at the pheU-tRNA in S103EC (clade C2) and S22EC (clade B) (GI-S103EC-pheU and GI-S22EC-pheU) and selC-tRNA in EC958 (clade C2) and S10EC (clade C2) (GI-EC958-selC and GI-S10EC-selC) (Fig. S4A). In contrast, the afa/dr gene cluster in S21EC (clade B) is located within an insertion fragment integrated within the ulaE gene (GI-S21EC-ulaE) (Fig. S4A). The sequence variation and different genomic location of the afa/dr genes suggest they have been acquired independently of their clade designation (Fig. S4B).
(A) Comparative genomics between the ST131 representative strains. Prophages (Phi) are indicated by blue boxes; GI are indicated by purple boxes and the GI containing the afa/dr gene cluster are indicated by green boxes. The ulaE gene corresponds to locus tag S21EC_48440. (B) Comparative genomics between the genomic islands carrying the afa/dr operon in the ST131 representative strains. Brown (afaF) and yellow (afaA) arrows represent genes encoding transcriptional regulators; orange arrows represent different IS elements; other genes are color-coded as green (chaperone), blue (usher), purple (invasin), pink (genes involved in RNA processing), and red (adhesin major subunit). Download FIG S4, TIF file, 0.9 MB (982.9KB, tif) .
Copyright © 2022 Alvarez-Fraga et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
(A) Summary features of ST131 representative strains. (B) Chromosomal IS1 insertion sites found in ST131 clone relative to six complete reference genomes. (C) Number of IS1 insertion sites relative to six ST131 complete reference genomes. (D) Chromosomal IS1 insertion sites found in ST131 clone relative to EC958 reference genome. (E) Primers used in this study. Download Data Set S1, XLSX file, 0.2 MB (208.1KB, xlsx) .
Copyright © 2022 Alvarez-Fraga et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
IS integration alters the transcription of afa/dr genes.
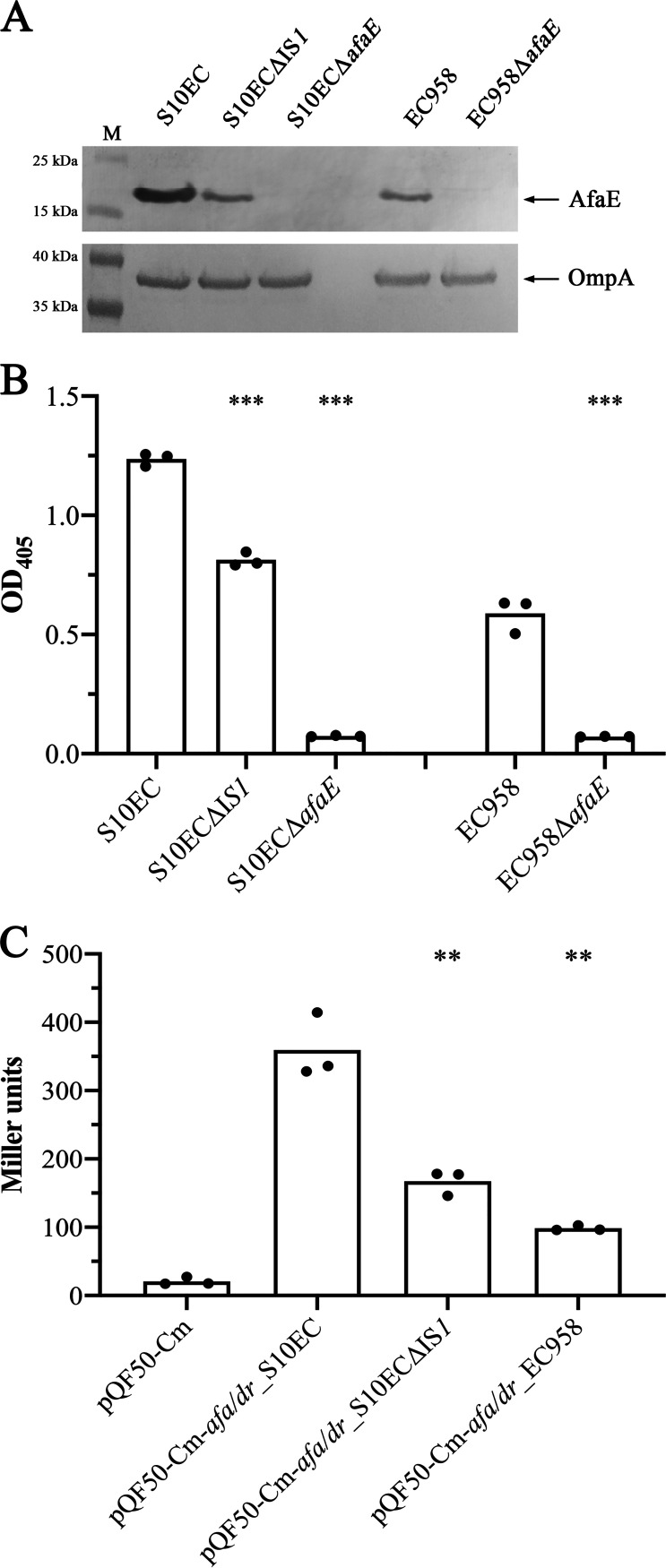
The transposition of IS into non-coding sequences in a bacterial genome can generate several outcomes, including increased transcription of adjacent genes due to the introduction/creation of a strong promoter (49, 50). To investigate the impact of the IS on the transcription of afa genes in ST131, we examined the transcript levels of the afaA and afaF genes in our representative strain set. Compared with S21EC, the transcript level of afaA was reduced in the strains possessing the ISCro1 (2.5- log2 fold change, P = 0.0008) and the 118-bp segment (3-log2 fold change, P < 0.0001), while the presence of ISEc10 had no effect on afaA transcription (P = 0.6286) (Fig. 3B). In contrast, the insertion of IS1 downstream of the ISCro1 in S10EC led to a significant increase in afaA transcript level compared to S21EC (4.2-log2 fold change, P = 0.0002) (Fig. 3B). Analysis of the afaF transcript level revealed a small increase in the strains possessing the ISEc10 (1.3-fold increase, P = 0.0034) and IS1 (2-fold increase, P = 0.0037) compared with the level of afaF transcript in S21EC (Fig. 3B).
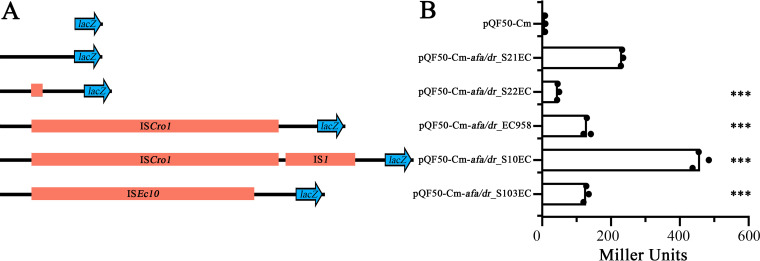
Next, we investigated how the different IS elements impact afa/dr gene transcription by generating a series of promoter-lacZ fusion constructs containing the promoter region from each representative strain cloned into the promoterless lacZ vector pQF50-Cm (Table 1). These plasmids were transformed into EC958Δlac and β-galactosidase levels were measured to assess promoter activity (Fig. 4A). The results were largely congruent with the transcript analyses, with the integration of IS1 resulting in a 2-fold increase in β-galactosidase activity compared to the native promoter (pQF50-Cm-afa/dr_S21EC) and a 3.5-fold increase compared to the ISCro1-promoter (pQF50-Cm-afa/dr_EC958) (Fig. 4B). In contrast, integration of the 118-bp insertion (pQF50-Cm-afa/dr_S22EC), ISEc10 (pQF50-Cm-afa/dr_S103EC), and ISCro1 (pQF50-Cm-afa/dr_EC958) significantly reduced β-galactosidase activity compared to the native promoter (pQF50-Cm-afa/dr_S21EC) (Fig. 4B). No significant β-galactosidase activity was measured in the strain carrying the empty control plasmid pQF50-Cm.
TABLE 1.
Bacterial strains and plasmids used in this work
| Strain or plasmid | Relevant characteristics | Sources or references |
|---|---|---|
| STRAINS | ||
| E. coli TOP10 | F–mcrA Δ(mrr-hsdRMS-mcrBC) Φ80lacZΔM15 ΔlacX74 recA1 araD139 Δ(ara leu) 7697 galU galK rpsL (StrR) endA1 nupG | Invitrogen |
| EC958 | ST131 reference strain (Clade C2). Presence of an ISCro1insertion sequence in the afa promoter region and the afaE-IX allelic variant. | (7, 48, 84) |
| EC958Δlac | EC958lacI-Z::gfp | (79) |
| EC958Δhns | EC958hns::cm; Cmr | (78) |
| EC958ΔihfA | EC958ihfA::cm; Cmr | This study |
| EC958ΔafaE | EC958afaE::cm; Cmr | This study |
| EC958ΔafaC | EC958afaC::cm; Cmr | This study |
| S10EC | ST131 UPEC isolate (Clade C2). Presence of an ISCro1 and IS1 insertion sequences in the afa promoter region and the afaE-IX allelic variant. Presence of an IS1 upstream of the osmB gene. | (7) |
| S10ECΔlac | S10EC lacI-Z::gfp. Cmr cassette removed. | This study |
| S10ECΔIS1 | S10EC IS1. Cmr cassette removed | This study |
| S10ECΔIS1Δlac | S10EC IS1 lacI-Z::gfp. Cmr cassette removed | This study |
| S10ECΔhns | S10EChns::cm; Cmr | This study |
| S10ECΔihfA | S10ECihfA::cm; Cmr | This study |
| S10ECΔafaE | S10ECafaE::cm; Cmr | This study |
| S10ECΔafaC | S10ECafaC::cm; Cmr | This study |
| S103EC | ST131 UPEC isolate (Clade C2). Presence of an ISEc10 insertion sequence in the afa promoter region and the afaE-XIII allelic variant. | (7) |
| S21EC | ST131 UPEC isolate (Clade B). Absence of insertion sequences in the afa promoter region and presence of the daaE allelic variant. | (7) |
| S22EC | ST131 UPEC isolate (Clade B). Presence of a 118 bp unknown sequence in the afa promoter region and the afaE-I allelic variant. | (7) |
| HVM52 | ST131 UPEC isolate (Clade B). Presence of an IS1 upstream of the ugd gene. | (7) |
| HVM277 | ST131 UPEC isolate (Clade B). Presence of an IS1 upstream of the ugd gene. | (7) |
| HVM2044 | ST131 UPEC isolate (Clade B). Presence of an IS1 upstream of the ugd gene. | (7) |
| S12EC | ST131 UPEC isolate (Clade C2). Presence of an IS1 upstream of the afaA and osmB genes. | (7) |
| PLASMIDS | ||
| pKD3 | Template plasmid for cm gene amplification; Cmr | (85) |
| pKOBEG-Gm | λ-red recombinase expressing plasmid; Gmr | (84, 86) |
| pCP20-Gm | FLP expressing plasmid; Gmr | (80) |
| pQF50-Cm | Promoterless lacZ reporter plasmid; Cmr | (87) |
| pQF50-Cm-afa/dr_EC958 | afa promoter region of EC958 cloned in pQF50-Cm | This study |
| pQF50-Cm-afa/dr_S10EC | afa promoter region of S10EC cloned in pQF50-Cm | This study |
| pQF50-Cm-afa/dr_S10ECΔIS1 | pQF50-Cm-afa/dr_S10EC without the IS1 element | This study |
| pQF50-Cm-afa/dr_S103EC | afa promoter region of S103EC cloned in pQF50-Cm | This study |
| pQF50-Cm-afa/dr_S21EC | afa promoter region of S21EC cloned in pQF50-Cm | This study |
| pQF50-Cm-afa/dr_S22EC | afa promoter region of S22EC cloned in pQF50-Cm | This study |
FIG 4.
(A) Schematic representation of the afa/dr promoter-lacZ fusion constructs used in this study. Shown are the different promoter regions cloned immediately upstream of the promoterless lacZ gene in pQF50-Cm. The black lines represent the promoter sequence, and the orange rectangles represent the insertion elements. (B) β-galactosidase assay showing the activity of the different afa/dr promoters. β-galactosidase activity is expressed as Miller units. Experiments were performed in three biological replicates. Unpaired Student's t test was used for statistical analysis (***, P value < 0.0001).
IS1 is responsible for the high level of AfaE expression in the S10EC strain.
To further examine the impact of IS1 integration on Afa expression, the IS1 element was deleted from the genome of the S10EC strain and the expression of the AfaE major subunit was analyzed by Western blotting using an AfaE-IX specific antibody. Analysis of whole cell lysates prepared from S10EC and EC958 revealed increased expression of AfaE in S10EC compared with EC958. When the IS1 element was deleted in S10EC, the expression of AfaE was reduced to a level similar to that observed for EC958 (Fig. 5A). No expression of AfaE was detected in the S10ECΔafaE and EC958ΔafaE mutant strains used as negative controls. We also examined cell-surface expression of AfaE using a whole-cell ELISA in combination with our AfaE-IX antibody. These analyses revealed a 2.1-fold higher level of AfaE expression in the S10EC strain compared with EC958 (P < 0.0001). The amount of AfaE protein on the surface of the S10ECΔIS1 mutant strain was 1.52-fold lower compared with wild-type S10EC (P < 0.0001). No expression was detected in the S10ECΔafaE and EC958ΔafaE mutant strains used as negative controls (Fig. 5B). As a final validation the IS1 element was deleted in the promoter-lacZ fusion construct pQF50-Cm-afa/dr_S10EC and the promoter activity was measured by β-galactosidase assays. Deletion of IS1 led to a 2-fold reduction (P = 0.0028) in the promoter activity compared with the S10EC wild-type promoter; no β-galactosidase activity was identified in the strain carrying the empty plasmid pQF50-Cm (Fig. 5C). Taken together, these data provide direct evidence for the role of IS1 in the increased expression of Afa adhesin in S10EC.
FIG 5.
(A) Western blot analysis of AfaE, performed using whole cell lysates prepared from S10EC, S10ECΔIS1, S10ECΔafaE, EC958, and EC958ΔafaE. Bands corresponding to AfaE (AfaE-IX antibody) and OmpA (loading control, OmpA antibody) are indicated. PageRuler Prestained Protein Ladder (Life Technologies) was used as molecular mass marker (lane M). (B) Graphical presentation of ELISA data showing the level of AfaE protein on the surface of S10EC, S10ECΔIS1, S10ECΔafaE, EC958, and EC958ΔafaE. The primary polyclonal rabbit anti-AfaE-IX and the secondary alkaline phosphatase-conjugated anti-rabbit antibodies were used. (C) β-galactosidase assay showing the activity of the S10EC, S10ECΔIS1, and EC958 afa/dr promoters. β-galactosidase activity is expressed as Miller units. Experiments were performed in three biological replicates. Unpaired Student's t test was used for statistical analysis (**, P value < 0.01; ***, P < 0.0001).
The insertion of IS1 modifies afa/dr regulation.
Afa/Dr fimbrial regulation is complex, involving H-NS (repression), IHF (activation), and Dam methylation (phase variation) (36–38). To test how integration of IS1 affects the regulation of Afa, the hns and ihfA genes were deleted in EC958 and S10EC and the level of AfaE expression was analyzed by whole cell ELISA. EC958 displayed an expected regulatory phenotype, with deletion of hns causing increased AfaE expression (1.67-fold increase compared with wild-type EC958, P = 0.014) and deletion of ihfA leading to decreased AfaE expression (2.4-fold decrease compared with wild-type EC958, P = 0.0014) (Fig. S5A). In contrast, deletion of hns and ihfA in S10EC had no effect of AfaE expression (Fig. S5A). Phase variable expression of AfaE by EC958 and S10EC was measured by flow cytometry and immunofluorescence microscopy. EC958 existed as a heterogeneous population, with both phase-off and phase-on cells identified, and phase-off cells representing the majority of the population (Fig. S5B). In contrast, the population of S10EC was uniform and phase-on (Fig. S5C).
(A) ELISA showing the level of AfaE protein expressed on the surface of EC958 and S10EC wild-type and mutant derivatives. The primary polyclonal rabbit anti-AfaE-IX and the secondary alkaline phosphatase-conjugated anti-rabbit antibodies were used. Experiments were performed in three independent replicates. Unpaired student’s t-test was used for statistical analysis (* P < 0.05; ** P < 0.01; *** P < 0.0001). (B) EC958 and (C) S10EC analysis of AfaE expression based on (i) flow cytometry and (ii) immunofluorescence microscopy. For flow cytometry analyses, the fluorescence intensity of EC958 and S10EC cells was measured using anti-AfaE-IX and secondary anti-rabbit AlexaFluor 488 antibodies. The blue lines represent the negative control (EC958ΔafaE and S10ECΔafaE, respectively) used for setting the correct threshold value between noise and positive signal. The red lines represent the samples. The populations of EC958 phase-off, EC958 phase-on, and S10EC phase-on are indicated by arrows. For microscopy, phase contrast (left panels) and corresponding immunofluorescence (right panels) images are presented. Expression of the AfaE protein by EC958 and S10EC was detected primary polyclonal rabbit AfaE-IX antibody in combination with the secondary rabbit IgG FITC antibody. Download FIG S5, TIF file, 1.1 MB (1.1MB, tif) .
Copyright © 2022 Alvarez-Fraga et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Enhanced Afa/Dr fimbrial expression by S10EC leads to increased adherence to kidney epithelial cells.
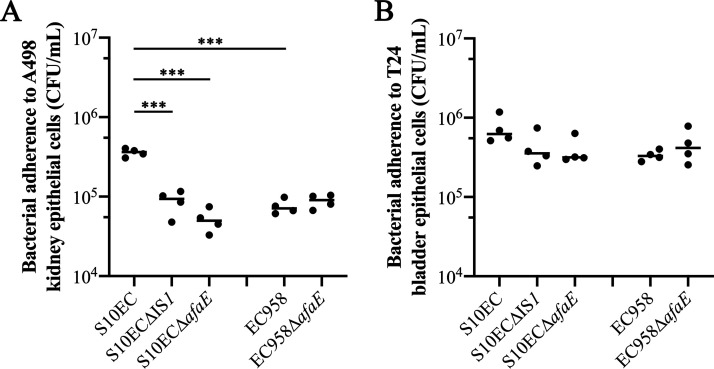
To determine the impact of IS1-mediated enhanced Afa/Dr fimbrial expression on virulence, we examined the capacity of our strains to adhere to and invade human kidney (A498) and bladder (T24) epithelial cells. This analysis revealed higher numbers of S10EC cells adhered to A498 kidney cells compared to EC958 (P < 0.0001). When the IS1 element or the afaE gene were deleted in S10EC, the adherence capacity was reduced to a level similar to EC958 (P < 0.0001) (Fig. 6A). A similar trend was observed for adherence to T24 bladder epithelial cells, but the differences were not statistically significant (Fig. 6B). No significant difference was observed in the capacity of our strains to invade A498 kidney and T24 bladder epithelial cells (Fig. S6A, B). We also extended these analyses to test if increased Afa/Dr fimbrial expression enhanced colonization of the mouse bladder; however, no difference was observed in mixed competitive experiments using wild-type S10EC versus mutants deleted for IS1 or Afa expression (Fig. S6C). Thus, IS1-mediated enhanced Afa/Dr expression leads to increased adherence to kidney cells but does not increase bladder cell adherence, epithelial cell invasion or bladder colonization under the conditions used in this study.
FIG 6.
Adherence of S10EC, S10ECΔIS1, S10ECΔafaE, EC958, and EC958ΔafaE strains to (A) A498 kidney and (B) T24 bladder human epithelial cells. Each data point is the mean of six technical replicates. Experiments were performed in four independent replicates. ANOVA and Šídák's multiple comparisons tests were used for statistical analysis (***, P value < 0.0001).
Invasion of S10EC, S10ECΔIS1, S10ECΔafaE, EC958 and EC958ΔafaE strains to (A) A498 kidney and (B) T24 bladder human epithelial cells. Each data point is the mean of six technical replicates. Experiments were performed in three independent replicates. ANOVA and Šídák's multiple comparisons tests were used. (C) Mouse urinary tract colonization model. Competitive mixed-infection experiment employing a 1:1 mixture of E. coli S10EC/S10ECΔlac, E. coli S10ECΔIS1/S10ECΔlac, E. coli S10ECΔafaC/S10ECΔlac, and E. coli S10ECΔafaC/S10ECΔIS1Δlac. Each symbol represents the log10 competitive index calculated for each mouse at 24-h postinfection. Horizontal bars represent median values. Statistical significance was determined by the two-tailed Wilcoxon’s signed rank test. Download FIG S6, TIF file, 0.5 MB (483.3KB, tif) .
Copyright © 2022 Alvarez-Fraga et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
IS1 contributes to genome evolution in ST131.
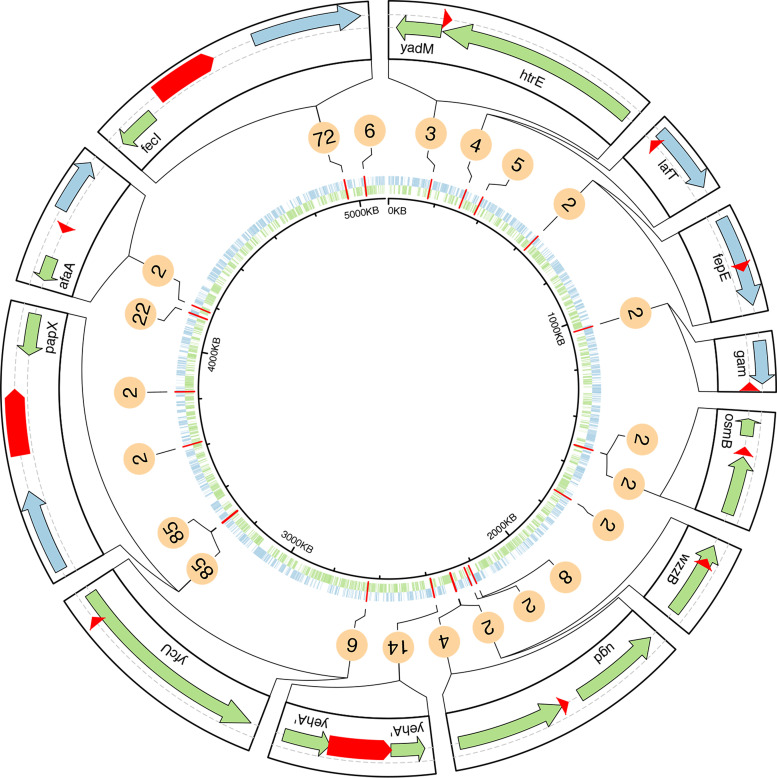
IS1 is found abundantly in E. coli at both plasmid and chromosomal locations (51). The element comprises two adjacent transposase genes, insA and insB, flanked by short, inverted repeat sequences. To evaluate the impact of IS1 on the evolution of ST131 at a chromosomal level, we examined its prevalence and location in our previously characterized set of 95 ST131 genomes, mapped against six completely sequenced reference genomes from this collection. Depending on the strain, we observed 113 to 125 IS1 insertion sites relative to each corresponding reference genome, with a cumulative number of ∼160 unique insertion sites (Fig. S7A, Data set S1B and Data set S1C). We identified 124 insertion sites of IS1 in our in-house ST131 strain collection relative to the EC958 reference genome (Data set S1B and Data set S1C). Among them, 22 were present in two or more strains at a prevalence of 2% to 90%; five insertions were found in EC958 (Fig. 7 and Data set S1D). To determine whether there was an association between IS1 and large mobile genetic elements, we analyzed the location of each of the IS1 identified in our set of 95 ST131 genomes using EC958 as the reference. The analysis revealed a correlation between IS1 location and GIs, with the number of IS1 insertion sites in GIs was significantly higher than the number of sites in the core genome (P = 0.0013) (Fig. S7B). Overall, three different types of insertions were identified: (i) IS1 inserted upstream of a coding sequence (CDS) in the same orientation, such as the afaA, osmB, or ugd genes; (ii) IS1 inserted upstream of a CDS in the opposite orientation, such as yadM; and (iii) IS1 inserted within a CDS, such as yehA, wzzB, or fepE (Fig. 7). To further investigate the impact of IS1 when orientated in the same direction as a downstream gene, we examined the transcript levels of afaA, osmB, and ugd genes in EC958 (no insertion) compared with representative strains containing the IS1 insertion. Consistent with our analyses described above, the transcript level of afaA was increased in strains that possess IS1 compared to EC958 (∼ 7-log2 fold change, P < 0.0001) (Fig. S8A). In contrast, the presence of IS1 upstream of the osmB gene resulted in a reduction in the transcript level of this gene (∼ 2-log2 fold change, P < 0.0001), while no difference was observed in the transcript level of the ugd gene (Fig. S8A). IS1 contains an outward oriented −35 promoter box in its inverted repeat, and its insertion at the correct distance from a potential −10 box can generate a hybrid promoter (52). A close analysis of the IS1 integration sites in the afaA, osmB. and ugd genes revealed the presence of a strong hybrid promoter generated by the insertion of IS1 upstream of the afaA gene, but not the osmB and ugd genes (Fig. S8B).
FIG 7.
Circular genomic map showing the distribution of IS1 elements within our 95 in-house ST131 strain collection (7). The black inner ring represents the E. coli EC958 reference genome. The distribution of minus strand ORFs (green) and plus strand ORFs (blue) are represented. The IS1 elements are indicated in red and the number of strains possessing each IS1 is represented in the circles. The outer ring represents 12 selected IS1 elements with their respective flanking genes. Large red arrows represent IS1 present in the EC958 reference strain. Small red arrows represent IS1 not present in the EC958 strain. Arrows indicate orientation.
(A) Plot showing the cumulative number of IS1 insertion sites within the 95 in-house ST131 strain collection. Paired-end Illumina reads were mapped against six completely sequenced ST131 reference genomes (EC958, S10EC, S21EC, S22EC, S65EC, and S103EC). Each data point represents the number of insertion sites when different reference genomes were combined. (B) Plot showing the location of each of the IS1 insertion sites within the 95 in-house ST131 strain collection. Paired-end Illumina reads were mapped against the EC958 reference genome. Each data point represents the number of IS1 insertion sites found in each strain. Download FIG S7, TIF file, 0.4 MB (453.8KB, tif) .
Copyright © 2022 Alvarez-Fraga et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
(A) Transcript levels of the afaA, osmB, and ugd genes in the ST131 strains carrying an IS1 element upstream of the gene. The results are represented as log2 fold change compared to EC958 (which has not insertion sequences). The gapA gene was used as an endogenous control. Experiments were performed in three biological replicates. Statistically significant differences are represented with asterisks (** P value < 0.01; *** P value < 0.0001). (B) Nucleotide sequence of the IS1-promoter region junctions. The right end of the IS1 is underlined. The −35 region located in the IS1 and the predicted −10 region are boxed. The asterisks show the nucleotides identical to the corresponding E. coli consensus sequences (−10, TATAAT; −35, TTGACA). Download FIG S8, TIF file, 0.9 MB (939.8KB, tif) .
Copyright © 2022 Alvarez-Fraga et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
DISCUSSION
The Afa/Dr family form a group of fimbrial adhesins strongly associated with UPEC that cause gestational pyelonephritis and recurrent cystitis (16, 17, 53–57). Here, we have shown that the afa/dr genes occur frequently in strains from the globally disseminated multidrug resistant ST131 clone, and that the afa/dr promoter is a hotspot for IS integration, in some cases leading to increased expression associated with altered capacity for virulence.
The afa genes were found in 18/83 (21.7%) of the most common E. coli STs, spanning all phylogroups. The highest prevalence was observed in strains from ST131 and ST38, both of which represent important multidrug-resistant high-risk clones associated with UTI (3, 58–62). We also identified a different distribution of AfaE allelic variants within these two STs, consistent with their acquisition via independent horizontal gene transfer events. Our results are in line with previous studies using smaller strain collections that identified a significantly higher prevalence of afa/dr genes in ST131 compared with non-ST131 UPEC (4, 13). In the case of ST131, although we observed the predominance of a single previously undefined AfaE-IX allelic variant most closely related to NfaE-111, we speculate that it is unlikely the gain of afa fimbriae genes imparted a central fitness feature that drove its global dissemination. Rather, we suggest that Afa fimbriae modulate ST131 fitness, possibly by influencing colonization of the urinary tract. In this way, we predict that the expression of Afa fimbriae comprises one element of an intricate genetic landscape that contributes to the selective advantage and fitness of ST131. We note that Afa fimbriae also contribute to colonization of the gut by diffusely adhering E. coli (31), and therefore it is possible that the expression of Afa fimbriae by ST131 and ST38 strains increases intestinal colonization, thereby enabling a sustained reservoir for dissemination to extra-intestinal sites during infection as reported in a recent patient investigation (63).
Close analysis of the afa/dr cluster in ST131 revealed a hotspot for IS integration at a position 440-bp upstream of the afaA start codon. This corresponded to the insertion of ISCro1 or ISEc10 elements, as well as a 118-bp sequence of unknown origin, resulting in decreased afaA transcription and/or afa promoter activity. A further five ST131 strains, represented by S10EC, possessed an additional IS1 element immediately downstream of ISCro1, resulting in increased AfaE expression. The activation of gene transcription following the integration of an IS can occur in several ways, including the integration of a new promoter contained within the IS element (64–67) or the generation of a hybrid promoter formed by an outward oriented −35 promoter box at one end of the IS element proximal to a −10 box near the integration site (52, 68, 69). Multiple examples have been described in the literature in relation to antibiotic resistance; for example IS1 integration upstream of the genes encoding the AcrEF efflux pump is associated with increased resistance in Salmonella enterica (70) and E. coli (71), and an IS1-like element is responsible for increased expression of the extended spectrum beta-lactamase TEM-6 in Enterobacteriaceae (72). Here, we describe a mechanism whereby IS1 integration alters the regulation of a well-characterized virulence determinant, Afa/Dr fimbriae, leading to increased constitutive expression in some ST131 strains, and conferring an enhanced adherence phenotype. No significant difference was observed in the capacity of wild-type S10EC versus the mutant strains S10ECΔIS1 and S10ECΔafaE to invade uroepithelial cells or to colonize the mouse bladder in mixed competitive experiments. Although the mouse model of acute UTI has previously been used to demonstrate a role for Afa/Dr fimbriae in bladder colonization (40), we speculate that lack of a colonization defect for the S10EC mutants could be due to a difference in ligand specificity between the S10EC AfaE variant (AfaE-IX) compared to the VR50 AfaE variant (one amino acid difference compared to the AfaE-XI variant), both of which share only 17.3% amino acid identity (Fig. S3).
The contribution of IS to genome evolution and virulence is frequently overlooked. Previous studies of ST131 evolution have examined genome variation caused by single nucleotide polymorphisms without detailed investigation of the impact of IS elements (7, 10–12). Here we used a curated set of 95 ST131 genomes to map the spread of IS1 locations, revealing ∼160 different IS1 insertion sites in our genome data set. The integration of IS elements on the chromosome can impart a range of effects, including insertional inactivation of the target gene, the introduction of new cargo genes to the recipient strain, or altered transcription of adjacent genes (50, 73). Thus, in addition to altered regulation and expression of afa/dr genes caused by IS1, there are likely to be other changes associated with IS1 integration that remain to be characterized. Indeed, comparison of IS1 integration upstream of the afaA, osmB, and ugd genes revealed different outcomes; afaA transcription increased, osmB transcription decreased, and ugd transcription was largely unchanged. This reinforces our finding that although the integration of IS1 can lead to the formation of a hybrid promoter that drives increased transcription of a downstream gene, this only occurs when the insertion is optimally located with respect to an adjacent −10 promoter element.
In summary, we have described the diversity and prevalence of afa/dr fimbrial genes in ST131, revealing the dominance of a new afaE-IX allelic variant. We additionally showed how different IS elements can alter the regulation of afa transcription and provide evidence for a more widespread impact of IS1 on ST131 genome evolution. IS have been shown to play an important role in genome evolution of multiple pathogens, elegantly demonstrated by a recent study of Shigella species (74). Given the high carriage of IS1 and other IS elements on antibiotic resistance plasmids found in ST131, our data support the contention that IS dynamics contribute to strain level microevolution that can affect multiple phenotypes, including resistance, virulence and metabolism.
MATERIALS AND METHODS
Key experimental procedures used in the study are listed below. Extended experimental methods, including (i) whole genome sequencing and analysis, (ii) β-galactosidase assays, (iii) ELISA, (iv) flow cytometry, (v) immunofluorescence, (vi) epithelial cell adhesion and invasion assays, and (vii) mouse UTI model, are provided in Text S1 in the supplementary material.
Extended experimental methods for (i) whole genome sequencing and analysis, (ii) β-galactosidase assays, (iii) ELISA, (iv) flow cytometry, (v) immunofluorescence, (vi) epithelial cell adhesion and invasion assays, and (vii) the mouse UTI model. Download Text S1, DOCX file, 0.03 MB (28.9KB, docx) .
Copyright © 2022 Alvarez-Fraga et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Bacterial strains and culture conditions.
All ST131 strains used in this study were previously described (7) and are listed in Table 1. Bacteria were grown routinely at 37°C in solid and liquid Luria-Bertani (LB) medium. Where appropriate, media was supplemented with 30 μg/mL of chloramphenicol (Cm) and/or 20 μg/mL of gentamicin (Gm).
Bioinformatic analysis.
Assemblies of E. coli strains belonging to ST10, ST69, ST73, ST95, and ST131 were downloaded from EnteroBase in July 2018, resulting in a collection of 11,283 strains (https://enterobase.warwick.ac.uk) (44). Assemblies of E. coli strains belonging to ST38 were downloaded from EnteroBase in November 2020, resulting in a collection of 1,926 strains. In addition, approximately 100 sequence assemblies were randomly chosen from each of the top 83 E. coli STs in the E. coli collection from EnteroBase (8,247 strains). ST131 strains were classified in clades and subclades based on clade-specific SNPs (10). The prevalence of Afa/Dr fimbriae was determined using the BLAST software package and the afaC gene from EC958 (75). The cut-off values used for filtering were > 90% nucleotide identity and > 80% gene coverage. The distribution of each afaE allele in ST38 and ST131 and the afa/dr promoter region in ST131 was analyzed using the BLAST software package. The cut-off values used for filtering were > 97% nucleotide identity and > 80% gene coverage. Analysis of BLAST output files analysis was carried out in R (version 3.6.2) through the R Studio environment (version 1.1.442) using the tidyverse package (version 1.2.1). DNA sequences were visualized using Artemis version 18.1.0 and CLC Main Workbench v8.1.3. The genomic context of genes was analyzed and drawn with Easyfig (76). Protein sequence alignments were performed with ClustalO and phylogenetic trees were generated using IQ-TREE v.1.6.8 and visualized and edited using FigTree v1.4.4. Paired-end Illumina reads from 95 ST131 strains were examined using ISmapper (77) to identify IS1 insertion sites relative to each of the six completely sequenced ST131 reference genomes (EC958, S10EC, S21EC, S22EC, S65EC, and S103EC). To estimate the number of IS1 insertion sites in multiple reference genomes, each insertion site was considered the same when sharing the same flanking genes with the same distances to the left and right genes. The R package circlize (version 0.4.11) was used to visualize the insertion sites of IS1 relative to the EC958 reference genome (HG941718).
RNA extraction and quantitative reverse transcription-PCR (qRT-PCR).
Bacteria were grown to exponential phase (optical density [OD] at 600 nm = 0.6) and stabilized with 2 volumes of RNAprotect bacterial reagent (Qiagen). Total RNA was isolated using RNeasy minikit (Qiagen) following the manufacturer's instructions. RNA samples were treated with RNase-free DNase I to remove contaminating DNA and purified using Qiagen RNeasy columns. cDNA was synthesized using the SuperScript®III First Strand Synthesis System (Life Technologies) according to the manufacturer's instructions. Real-time PCR (qPCR) reactions were performed using SYBR Green Master Mix (Applied Biosystems) on a QuantStudio 6 instrument (Applied Biosystems). The transcript level of each tested gene was normalized relative to the transcript level of the housekeeping gene gapA. All experiments were performed as three independent replicates. Statistical analysis was performed using an unpaired, two-tailed Student's t test. Primers used are listed in Data set S1E.
Plasmid construction.
Molecular methods were performed according to standard protocols as previously described (78). The afa/dr promoter-lacZ fusion constructs were created by PCR amplification of the different afa/dr promoter regions and cloned in front of the promoterless lacZ gene in the plasmid pQF50-Cm via NcoI-BamHI or SalI-BamHI digestion (see Data set S1E for primer list). The resulting plasmids were transformed into the EC958Δlac mutant strain. Transformants were selected following growth on LB agar containing chloramphenicol and checked by PCR followed by sequencing.
Mutant construction.
All mutants were generated using the λ-Red recombinase gene replacement system (79). Briefly, a three-way PCR procedure was performed to amplify the chloramphenicol cassette from plasmid pKD3 with ∼500 bp homologous arms flanking the region targeted for deletion. The fused PCR products were electroporated into the wild-type strains harboring the gentamicin resistant plasmid pKOBEG-G carrying the λ-Red recombinase gene. When necessary, the chloramphenicol resistance cassette was removed using plasmid pCP20-G (80). Mutants were confirmed by PCR followed by sequencing using the primers listed in Data set S1E.
Western blotting.
Whole cell lysates were prepared by pelleting 1 mL of OD600 = 1.0 standardized cell suspensions and resuspending the cells in 50 μL water and 50 μL of 2X loading buffer (2X NuPAGE LDS Sample Buffer, 200 mM DTT) and heating the samples at 100°C for 5 min. SDS-PAGE and Western blotting was performed as previously described (81). Rabbit polyclonal antisera specific for AfaE-IX (1:200) and OmpA (1:50,000) were used as primary antibodies and alkaline phosphatase-conjugated anti-rabbit antisera (Sigma-Aldrich) (1:15,000) was used as the secondary antibody. Blots were developed using BCIP/NBT stock solution.
Epithelial cell assays.
Afa/Dr fimbriae-mediated adherence and invasion was assessed using human T24 bladder (ATCC HTB-4) and A498 kidney (ATCC HTB-44) epithelial cells. T24 cells express the Afa fimbriae target DAF receptor (82). While DAF expression has not been directly demonstrated for A498 cells, it is well-established that the DAF receptor is expressed by human kidney cells (83). Full methods are provided in Text S1 in the supplementary material.
ACKNOWLEDGMENTS
This work was supported by grants APP1181958 and APP2001431 from the National Health and Medical Research Council of Australia (NHMRC). L.A.F. is supported by the Galician Ministry of Culture, Education, and University Planning, and Galician Ministry of Economy, Employment, and Industry (Regional Government of Galicia-Spain) (IN606B-2018/011). The funders had no role in study design, data collection and interpretation, or the decision to submit this work for publication.
Footnotes
This article is a direct contribution from Mark A. Schembri, a Fellow of the American Academy of Microbiology, who arranged for and secured reviews by Mathew Upton, Plymouth University, and Elizabeth Hartland, Hudson Institute of Medical Research.
Contributor Information
Laura Alvarez-Fraga, Email: laura.alvarez.fraga@sergas.es.
Mark A. Schembri, Email: m.schembri@uq.edu.au.
Sheryl Justice, The Ohio State University School of Medicine.
REFERENCES
- 1.Flores-Mireles AL, Walker JN, Caparon M, Hultgren SJ. 2015. Urinary tract infections: epidemiology, mechanisms of infection and treatment options. Nat Rev Microbiol 13:269–284. doi: 10.1038/nrmicro3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Banerjee R, Johnston B, Lohse C, Chattopadhyay S, Tchesnokova V, Sokurenko EV, Johnson JR. 2013. The clonal distribution and diversity of extraintestinal Escherichia coli isolates vary according to patient characteristics. Antimicrob Agents Chemother 57:5912–5917. doi: 10.1128/AAC.01065-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Riley LW. 2014. Pandemic lineages of extraintestinal pathogenic Escherichia coli. Clin Microbiol Infect 20:380–390. doi: 10.1111/1469-0691.12646. [DOI] [PubMed] [Google Scholar]
- 4.Gibreel TM, Dodgson AR, Cheesbrough J, Fox AJ, Bolton FJ, Upton M. 2012. Population structure, virulence potential and antibiotic susceptibility of uropathogenic Escherichia coli from Northwest England. J Antimicrob Chemother 67:346–356. doi: 10.1093/jac/dkr451. [DOI] [PubMed] [Google Scholar]
- 5.Nicolas-Chanoine MH, Blanco J, Leflon-Guibout V, Demarty R, Alonso MP, Canica MM, Park YJ, Lavigne JP, Pitout J, Johnson JR. 2008. Intercontinental emergence of Escherichia coli clone O25:H4-ST131 producing CTX-M-15. J Antimicrob Chemother 61:273–281. doi: 10.1093/jac/dkm464. [DOI] [PubMed] [Google Scholar]
- 6.Rogers BA, Sidjabat HE, Paterson DL. 2011. Escherichia coli O25b-ST131: a pandemic, multiresistant, community-associated strain. J Antimicrob Chemother 66:1–14. doi: 10.1093/jac/dkq415. [DOI] [PubMed] [Google Scholar]
- 7.Petty NK, Ben Zakour NL, Stanton-Cook M, Skippington E, Totsika M, Forde BM, Phan MD, Gomes Moriel D, Peters KM, Davies M, Rogers BA, Dougan G, Rodriguez-Baño J, Pascual A, Pitout JD, Upton M, Paterson DL, Walsh TR, Schembri MA, Beatson SA. 2014. Global dissemination of a multidrug resistant Escherichia coli clone. Proc Natl Acad Sci USA 111:5694–5699. doi: 10.1073/pnas.1322678111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ortiz de la Tabla V, Ortega A, Buñuel F, Pérez-Vázquez M, Marcos B, Oteo J. 2017. Detection of the high-risk clone ST131 of Escherichia coli carrying the colistin resistance gene mcr-1 and causing acute peritonitis. Int J Antimicrob Agents 49:115–116. doi: 10.1016/j.ijantimicag.2016.10.003. [DOI] [PubMed] [Google Scholar]
- 9.Roer L, Hansen F, Stegger M, Sönksen UW, Hasman H, Hammerum AM. 2017. Novel mcr-3 variant, encoding mobile colistin resistance, in an ST131 Escherichia coli isolate from bloodstream infection, Denmark, 2014. Euro Surveill 22. doi: 10.2807/1560-7917.ES.2017.22.31.30584. [DOI] [PubMed] [Google Scholar]
- 10.Ben Zakour NL, Alsheikh-Hussain AS, Ashcroft MM, Khanh Nhu NT, Roberts LW, Stanton-Cook M, Schembri MA, Beatson SA. 2016. Sequential acquisition of virulence and fluoroquinolone resistance has shaped the evolution of Escherichia coli ST131. mBio 7:e00347-16–e00316. doi: 10.1128/mBio.00347-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Price LB, Johnson JR, Aziz M, Clabots C, Johnston B, Tchesnokova V, Nordstrom L, Billig M, Chattopadhyay S, Stegger M, Andersen PS, Pearson T, Riddell K, Rogers P, Scholes D, Kahl B, Keim P, Sokurenko EV. 2013. The epidemic of extended-spectrum-β-lactamase-producing Escherichia coli ST131 is driven by a single highly pathogenic subclone. H30-Rx mBio 4:e00377-13. doi: 10.1128/mBio.00377-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stoesser N, Sheppard AE, Pankhurst L, De Maio N, Moore CE, Sebra R, Turner P, Anson LW, Kasarskis A, Batty EM, Kos V, Wilson DJ, Phetsouvanh R, Wyllie D, Sokurenko E, Manges AR, Johnson TJ, Price LB, Peto TE, Johnson JR, Didelot X, Walker AS, Crook DW, Modernizing Medical Microbiology Informatics Group (MMMIG) . 2016. Evolutionary history of the global emergence of the Escherichia coli epidemic clone ST131. mBio 7:e02162. doi: 10.1128/mBio.02162-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Olesen B, Hansen DS, Nilsson F, Frimodt-Møller J, Leihof RF, Struve C, Scheutz F, Johnston B, Krogfelt KA, Johnson JR. 2013. Prevalence and characteristics of the epidemic multiresistant Escherichia coli ST131 clonal group among extended-spectrum beta-lactamase-producing E. coli isolates in Copenhagen, Denmark. J Clin Microbiol 51:1779–1785. doi: 10.1128/JCM.00346-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Servin AL. 2005. Pathogenesis of Afa/Dr diffusely adhering Escherichia coli. Clin Microbiol Rev 18:264–292. doi: 10.1128/CMR.18.2.264-292.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Arthur M, Johnson CE, Rubin RH, Arbeit RD, Campanelli C, Kim C, Steinbach S, Agarwal M, Wilkinson R, Goldstein R. 1989. Molecular epidemiology of adhesin and hemolysin virulence factors among uropathogenic Escherichia coli. Infect Immun 57:303–313. doi: 10.1128/iai.57.2.303-313.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Foxman B, Zhang L, Tallman P, Palin K, Rode C, Bloch C, Gillespie B, Marrs CF. 1995. Virulence characteristics of Escherichia coli causing first urinary tract infection predict risk of second infection. J Infect Dis 172:1536–1541. doi: 10.1093/infdis/172.6.1536. [DOI] [PubMed] [Google Scholar]
- 17.Germani Y, Begaud E, Duval P, Le Bouguenec C. 1996. Prevalence of enteropathogenic, enteroaggregative, and diffusely adherent Escherichia coli among isolates from children with diarrhea in new Caledonia. J Infect Dis 174:1124–1126. doi: 10.1093/infdis/174.5.1124. [DOI] [PubMed] [Google Scholar]
- 18.Nowicki B, Selvarangan R, Nowicki S. 2001. Family of Escherichia coli Dr adhesins: decay-accelerating factor receptor recognition and invasiveness. J Infect Dis 183 Suppl 1:S24–7. doi: 10.1086/318846. [DOI] [PubMed] [Google Scholar]
- 19.Korotkova N, Cota E, Lebedin Y, Monpouet S, Guignot J, Servin AL, Matthews S, Moseley SL. 2006. A subfamily of Dr adhesins of Escherichia coli bind independently to decay-accelerating factor and the N-domain of carcinoembryonic antigen. J Biol Chem 281:29120–29130. doi: 10.1074/jbc.M605681200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Berger CN, Billker O, Meyer TF, Servin AL, Kansau I. 2004. Differential recognition of members of the carcinoembryonic antigen family by Afa/Dr adhesins of diffusely adhering Escherichia coli (Afa/Dr DAEC). Mol Microbiol 52:963–983. doi: 10.1111/j.1365-2958.2004.04033.x. [DOI] [PubMed] [Google Scholar]
- 21.Carnoy C, Moseley SL. 1997. Mutational analysis of receptor binding mediated by the Dr family of Escherichia coli adhesins. Mol Microbiol 23:365–379. doi: 10.1046/j.1365-2958.1997.2231590.x. [DOI] [PubMed] [Google Scholar]
- 22.Goluszko P, Popov V, Selvarangan R, Nowicki S, Pham T, Nowicki BJ. 1997. Dr fimbriae operon of uropathogenic Escherichia coli mediate microtubule-dependent invasion to the HeLa epithelial cell line. J Infect Dis 176:158–167. doi: 10.1086/514018. [DOI] [PubMed] [Google Scholar]
- 23.Plancon L, Du Merle L, Le Friec S, Gounon P, Jouve M, Guignot J, Servin A, Le Bouguenec C. 2003. Recognition of the cellular beta1-chain integrin by the bacterial AfaD invasin is implicated in the internalization of afa-expressing pathogenic Escherichia coli strains. Cell Microbiol 5:681–693. doi: 10.1046/j.1462-5822.2003.00308.x. [DOI] [PubMed] [Google Scholar]
- 24.Dhakal BK, Kulesus RR, Mulvey MA. 2008. Mechanisms and consequences of bladder cell invasion by uropathogenic Escherichia coli. Eur J Clin Invest 38 Suppl 2:2–11. doi: 10.1111/j.1365-2362.2008.01986.x. [DOI] [PubMed] [Google Scholar]
- 25.Zalewska B, Piątek R, Bury K, Samet A, Nowicki B, Nowicki S, Kur J. 2005. A surface-exposed DraD protein of uropathogenic Escherichia coli bearing Dr fimbriae may be expressed and secreted independently from DraC usher and DraE adhesin. Microbiology (Reading) 151:2477–2486. doi: 10.1099/mic.0.28083-0. [DOI] [PubMed] [Google Scholar]
- 26.Loomis WP, Moseley SL. 1998. Translational control of mRNA processing in the F1845 fimbrial operon of Escherichia coli. Mol Microbiol 30:843–853. doi: 10.1046/j.1365-2958.1998.01117.x. [DOI] [PubMed] [Google Scholar]
- 27.Piatek R, Zalewska B, Kolaj O, Ferens M, Nowicki B, Kur J. 2005. Molecular aspects of biogenesis of Escherichia coli Dr Fimbriae: characterization of DraB-DraE complexes. Infect Immun 73:135–145. doi: 10.1128/IAI.73.1.135-145.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cota E, Jones C, Simpson P, Altroff H, Anderson KL, Du Merle L, Guignot J, Servin A, Le Bouguenec C, Mardon H, Matthews S. 2006. The solution structure of the invasive tip complex from Afa/Dr fibrils. Mol Microbiol 62:356–366. doi: 10.1111/j.1365-2958.2006.05375.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Anderson KL, Cota E, Simpson P, Chen HA, Du Merle L, Bouguénec CL, Matthews S. 2004. Complete resonance assignments of a “donor-strand complemented” AfaE: the afimbrial adhesin from Diffusely Adherent E. coli. J Biomol NMR 29:409–410. doi: 10.1023/B:JNMR.0000032498.94441.08. [DOI] [PubMed] [Google Scholar]
- 30.Anderson KL, Billington J, Pettigrew D, Cota E, Simpson P, Roversi P, Chen HA, Urvil P, Du Merle L, Barlow PN, Medof ME, Smith RA, Nowicki B, Le Bouguénec C, Lea SM, Matthews S. 2004. An atomic resolution model for assembly, architecture, and function of the Dr adhesins. Mol Cell 15:647–657. doi: 10.1016/j.molcel.2004.08.003. [DOI] [PubMed] [Google Scholar]
- 31.Servin AL. 2014. Pathogenesis of human diffusely adhering Escherichia coli expressing Afa/Dr adhesins (Afa/Dr DAEC): current insights and future challenges. Clin Microbiol Rev 27:823–869. doi: 10.1128/CMR.00036-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Le Bouguénec C, Servin AL. 2006. Diffusely adherent Escherichia coli strains expressing Afa/Dr adhesins (Afa/Dr DAEC): hitherto unrecognized pathogens. FEMS Microbiol Lett 256:185–194. doi: 10.1111/j.1574-6968.2006.00144.x. [DOI] [PubMed] [Google Scholar]
- 33.Garcia MI, Labigne A, Le Bouguenec C. 1994. Nucleotide sequence of the afimbrial-adhesin-encoding afa-3 gene cluster and its translocation via flanking IS1 insertion sequences. J Bacteriol 176:7601–7613. doi: 10.1128/jb.176.24.7601-7613.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bilge SS, Clausen CR, Lau W, Moseley SL. 1989. Molecular characterization of a fimbrial adhesin, F1845, mediating diffuse adherence of diarrhea-associated Escherichia coli to HEp-2 cells. J Bacteriol 171:4281–4289. doi: 10.1128/jb.171.8.4281-4289.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Korotkova N, Chattopadhyay S, Tabata TA, Beskhlebnaya V, Vigdorovich V, Kaiser BK, Strong RK, Dykhuizen DE, Sokurenko EV, Moseley SL. 2007. Selection for functional diversity drives accumulation of point mutations in Dr adhesins of Escherichia coli. Mol Microbiol 64:180–194. doi: 10.1111/j.1365-2958.2007.05648.x. [DOI] [PubMed] [Google Scholar]
- 36.van der Woude MW, Low DA. 1994. Leucine-responsive regulatory protein and deoxyadenosine methylase control the phase variation and expression of the sfa and daa pili operons in Escherichia coli. Mol Microbiol 11:605–618. doi: 10.1111/j.1365-2958.1994.tb00340.x. [DOI] [PubMed] [Google Scholar]
- 37.Bilge SS, Apostol JM, Jr, Fullner KJ, Moseley SL. 1993. Transcriptional organization of the F1845 fimbrial adhesin determinant of Escherichia coli. Mol Microbiol 7:993–1006. doi: 10.1111/j.1365-2958.1993.tb01191.x. [DOI] [PubMed] [Google Scholar]
- 38.White-Ziegler CA, Villapakkam A, Ronaszeki K, Young S. 2000. H-NS controls pap and daa fimbrial transcription in Escherichia coli in response to multiple environmental cues. J Bacteriol 182:6391–6400. doi: 10.1128/JB.182.22.6391-6400.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lalioui L, Jouve M, Gounon P, Le Bouguenec C. 1999. Molecular cloning and characterization of the afa-7 and afa-8 gene clusters encoding afimbrial adhesins in Escherichia coli strains associated with diarrhea or septicemia in calves. Infect Immun 67:5048–5059. doi: 10.1128/IAI.67.10.5048-5059.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Beatson SA, Ben Zakour NL, Totsika M, Forde BM, Watts RE, Mabbett AN, Szubert JM, Sarkar S, Phan MD, Peters KM, Petty NK, Alikhan NF, Sullivan MJ, Gawthorne JA, Stanton-Cook M, Nhu NT, Chong TM, Yin WF, Chan KG, Hancock V, Ussery DW, Ulett GC, Schembri MA. 2015. Molecular analysis of asymptomatic bacteriuria Escherichia coli strain VR50 reveals adaptation to the urinary tract by gene acquisition. Infect Immun 83:1749–1764. doi: 10.1128/IAI.02810-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Le Bouguenec C, Garcia MI, Ouin V, Desperrier JM, Gounon P, Labigne A. 1993. Characterization of plasmid-borne afa-3 gene clusters encoding afimbrial adhesins expressed by Escherichia coli strains associated with intestinal or urinary tract infections. Infect Immun 61:5106–5114. doi: 10.1128/iai.61.12.5106-5114.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Goluszko P, Moseley SL, Truong LD, Kaul A, Williford JR, Selvarangan R, Nowicki S, Nowicki B. 1997. Development of experimental model of chronic pyelonephritis with Escherichia coli O75:K5:H-bearing Dr fimbriae: mutation in the dra region prevented tubulointerstitial nephritis. J Clin Invest 99:1662–1672. doi: 10.1172/JCI119329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wroblewska-Seniuk K, Selvarangan R, Hart A, Pladzyk R, Goluszko P, Jafari A, Du Merle L, Nowicki S, Yallampalli C, Le Bouguénec C, Nowicki B. 2005. Dra/AfaE adhesin of uropathogenic Dr/Afa+ Escherichia coli mediates mortality in pregnant rats. Infect Immun 73:7597–7601. doi: 10.1128/IAI.73.11.7597-7601.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhou Z, Alikhan NF, Mohamed K, Fan Y, Achtman M, Agama Study Group . 2020. The EnteroBase user’s guide, with case studies on Salmonella transmissions, Yersinia pestis phylogeny, and Escherichia core genomic diversity. Genome Res 30:138–152. doi: 10.1101/gr.251678.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kallonen T, Brodrick HJ, Harris SR, Corander J, Brown NM, Martin V, Peacock SJ, Parkhill J. 2017. Systematic longitudinal survey of invasive Escherichia coli in England demonstrates a stable population structure only transiently disturbed by the emergence of ST131. Genome Res 27:1437–1449. doi: 10.1101/gr.216606.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schembri MA, Zakour NL, Phan MD, Forde BM, Stanton-Cook M, Beatson SA. 2015. Molecular characterization of the multidrug resistant Escherichia coli ST131 clone. Pathogens 4:422–430. doi: 10.3390/pathogens4030422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Van Loy CP, Sokurenko EV, Samudrala R, Moseley SL. 2002. Identification of amino acids in the Dr adhesin required for binding to decay-accelerating factor. Mol Microbiol 45:439–452. doi: 10.1046/j.1365-2958.2002.03022.x. [DOI] [PubMed] [Google Scholar]
- 48.Forde BM, Ben Zakour NL, Stanton-Cook M, Phan MD, Totsika M, Peters KM, Chan KG, Schembri MA, Upton M, Beatson SA. 2014. The complete genome sequence of Escherichia coli EC958: a high quality reference sequence for the globally disseminated multidrug resistant E. coli O25b:H4-ST131 clone. PLoS One 9:e104400. doi: 10.1371/journal.pone.0104400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vandecraen J, Chandler M, Aertsen A, Van Houdt R. 2017. The impact of insertion sequences on bacterial genome plasticity and adaptability. Crit Rev Microbiol 43:709–730. doi: 10.1080/1040841X.2017.1303661. [DOI] [PubMed] [Google Scholar]
- 50.Siguier P, Gourbeyre E, Chandler M. 2014. Bacterial insertion sequences: their genomic impact and diversity. FEMS Microbiol Rev 38:865–891. doi: 10.1111/1574-6976.12067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wagner A, de la Chaux N. 2008. Distant horizontal gene transfer is rare for multiple families of prokaryotic insertion sequences. Mol Genet Genomics 280:397–408. doi: 10.1007/s00438-008-0373-y. [DOI] [PubMed] [Google Scholar]
- 52.Prentki P, Teter B, Chandler M, Galas DJ. 1986. Functional promoters created by the insertion of transposable element IS1. J Mol Biol 191:383–393. doi: 10.1016/0022-2836(86)90134-8. [DOI] [PubMed] [Google Scholar]
- 53.Poitrineau P, Forestier C, Meyer M, Jallat C, Rich C, Malpuech G, De Champs C. 1995. Retrospective case-control study of diffusely adhering Escherichia coli and clinical features in children with diarrhea. J Clin Microbiol 33:1961–1962. doi: 10.1128/jcm.33.7.1961-1962.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nowicki B, Svanborg-Edén C, Hull R, Hull S. 1989. Molecular analysis and epidemiology of the Dr hemagglutinin of uropathogenic Escherichia coli. Infect Immun 57:446–451. doi: 10.1128/iai.57.2.446-451.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hart A, Nowicki BJ, Reisner B, Pawelczyk E, Goluszko P, Urvil P, Anderson G, Nowicki S. 2001. Ampicillin-resistant Escherichia coli in gestational pyelonephritis: increased occurrence and association with the colonization factor Dr adhesin. J Infect Dis 183:1526–1529. doi: 10.1086/320196. [DOI] [PubMed] [Google Scholar]
- 56.Hart A, Pham T, Nowicki S, Whorton EB, Jr, Martens MG, Anderson GD, Nowicki BJ. 1996. Gestational pyelonephritis–associated Escherichia coli isolates represent a nonrandom, closely related population. Am J Obstet Gynecol 174:983–989. doi: 10.1016/S0002-9378(96)70337-X. [DOI] [PubMed] [Google Scholar]
- 57.Stapleton A, Moseley S, Stamm WE. 1991. Urovirulence determinants in Escherichia coli isolates causing first-episode and recurrent cystitis in women. J Infect Dis 163:773–779. doi: 10.1093/infdis/163.4.773. [DOI] [PubMed] [Google Scholar]
- 58.Mathers AJ, Peirano G, Pitout JD. 2015. The role of epidemic resistance plasmids and international high-risk clones in the spread of multidrug-resistant Enterobacteriaceae. Clin Microbiol Rev 28:565–591. doi: 10.1128/CMR.00116-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hertz FB, Nielsen JB, Schønning K, Littauer P, Knudsen JD, Løbner-Olesen A, Frimodt-Møller N. 2016. Erratum to: Population structure of drug-susceptible,-resistant and ESBL-producing Escherichia coli from community-acquired urinary tract infections. BMC Microbiol 16:114. doi: 10.1186/s12866-016-0725-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gauthier L, Dortet L, Cotellon G, Creton E, Cuzon G, Ponties V, Bonnin RA, Naas T. 2018. Diversity of Carbapenemase-producing Escherichia coli isolates in France in 2012–2013. Antimicrob Agents Chemother 62. doi: 10.1128/AAC.00266-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pietsch M, Eller C, Wendt C, Holfelder M, Falgenhauer L, Fruth A, Grössl T, Leistner R, Valenza G, Werner G, Pfeifer Y, RESET Study Group . 2017. Molecular characterisation of extended-spectrum β-lactamase (ESBL)-producing Escherichia coli isolates from hospital and ambulatory patients in Germany. Vet Microbiol 200:130–137. doi: 10.1016/j.vetmic.2015.11.028. [DOI] [PubMed] [Google Scholar]
- 62.Alghoribi MF, Gibreel TM, Farnham G, Al Johani SM, Balkhy HH, Upton M. 2015. Antibiotic-resistant ST38, ST131 and ST405 strains are the leading uropathogenic Escherichia coli clones in Riyadh, Saudi Arabia. J Antimicrob Chemother 70:2757–2762. doi: 10.1093/jac/dkv188. [DOI] [PubMed] [Google Scholar]
- 63.Forde BM, Roberts LW, Phan MD, Peters KM, Fleming BA, Russell CW, Lenherr SM, Myers JB, Barker AP, Fisher MA, Chong TM, Yin WF, Chan KG, Schembri MA, Mulvey MA, Beatson SA. 2019. Population dynamics of an Escherichia coli ST131 lineage during recurrent urinary tract infection. Nat Commun 10:3643. doi: 10.1038/s41467-019-11571-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Glansdorff N, Charlier D, Zafarullah M. 1981. Activation of gene expression by IS2 and IS3. Cold Spring Harbor Symp Quant Biol 45 Pt 1:153–156. doi: 10.1101/sqb.1981.045.01.024. [DOI] [PubMed] [Google Scholar]
- 65.Charlier D, Piette J, Glansdorff N. 1982. IS3 can function as a mobile promoter in E. coli. Nucleic Acids Res 10:5935–5948. doi: 10.1093/nar/10.19.5935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ciampi MS, Schmid MB, Roth JR. 1982. Transposon Tn10 provides a promoter for transcription of adjacent sequences. Proc Natl Acad Sci USA 79:5016–5020. doi: 10.1073/pnas.79.16.5016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.DeShazer D, Wood GE, Friedman RL. 1994. Molecular characterization of catalase from Bordetella pertussis: identification of the katA promoter in an upstream insertion sequence. Mol Microbiol 14:123–130. doi: 10.1111/j.1365-2958.1994.tb01272.x. [DOI] [PubMed] [Google Scholar]
- 68.Dalrymple B. 1987. Novel rearrangements of IS30 carrying plasmids leading to the reactivation of gene expression. Mol Gen Genet 207:413–420. doi: 10.1007/BF00331609. [DOI] [PubMed] [Google Scholar]
- 69.Leelaporn A, Firth N, Byrne ME, Roper E, Skurray RA. 1994. Possible role of insertion sequence IS257 in dissemination and expression of high- and low-level trimethoprim resistance in staphylococci. Antimicrob Agents Chemother 38:2238–2244. doi: 10.1128/AAC.38.10.2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Olliver A, Vallé M, Chaslus-Dancla E, Cloeckaert A. 2005. Overexpression of the multidrug efflux operon acrEF by insertional activation with IS1 or IS10 elements in Salmonella enterica serovar typhimurium DT204 acrB mutants selected with fluoroquinolones. Antimicrob Agents Chemother 49:289–301. doi: 10.1128/AAC.49.1.289-301.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kobayashi K, Tsukagoshi N, Aono R. 2001. Suppression of hypersensitivity of Escherichia coli acrB mutant to organic solvents by integrational activation of the acrEF operon with the IS1 or IS2 element. J Bacteriol 183:2646–2653. doi: 10.1128/JB.183.8.2646-2653.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Goussard S, Sougakoff W, Mabilat C, Bauernfeind A, Courvalin P. 1991. An IS1-like element is responsible for high-level synthesis of extended-spectrum beta-lactamase TEM-6 in Enterobacteriaceae. J Gen Microbiol 137:2681–2687. doi: 10.1099/00221287-137-12-2681. [DOI] [PubMed] [Google Scholar]
- 73.Mahillon J, Chandler M. 1998. Insertion sequences. Microbiol Mol Biol Rev 62:725–774. doi: 10.1128/MMBR.62.3.725-774.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hawkey J, Monk JM, Billman-Jacobe H, Palsson B, Holt KE. 2020. Impact of insertion sequences on convergent evolution of Shigella species. PLoS Genet 16:e1008931. doi: 10.1371/journal.pgen.1008931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 76.Sullivan MJ, Petty NK, Beatson SA. 2011. Easyfig: a genome comparison visualizer. Bioinformatics 27:1009–1010. doi: 10.1093/bioinformatics/btr039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hawkey J, Hamidian M, Wick RR, Edwards DJ, Billman-Jacobe H, Hall RM, Holt KE. 2015. ISMapper: identifying transposase insertion sites in bacterial genomes from short read sequence data. BMC Genomics 16:667. doi: 10.1186/s12864-015-1860-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tan L, Moriel DG, Totsika M, Beatson SA, Schembri MA. 2016. differential regulation of the surface-exposed and secreted SslE lipoprotein in extraintestinal pathogenic Escherichia coli. PLoS One 11:e0162391. doi: 10.1371/journal.pone.0162391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Phan MD, Peters KM, Sarkar S, Lukowski SW, Allsopp LP, Gomes Moriel D, Achard ME, Totsika M, Marshall VM, Upton M, Beatson SA, Schembri MA. 2013. The serum resistome of a globally disseminated multidrug resistant uropathogenic Escherichia coli clone. PLoS Genet 9:e1003834. doi: 10.1371/journal.pgen.1003834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cherepanov PP, Wackernagel W. 1995. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene 158:9–14. doi: 10.1016/0378-1119(95)00193-a. [DOI] [PubMed] [Google Scholar]
- 81.Ulett GC, Webb RI, Schembri MA. 2006. Antigen-43-mediated autoaggregation impairs motility in Escherichia coli. Microbiology (Reading) 152:2101–2110. doi: 10.1099/mic.0.28607-0. [DOI] [PubMed] [Google Scholar]
- 82.Zalewska-Piątek B, Olszewski M, Lipniacki T, Błoński S, Wieczór M, Bruździak P, Skwarska A, Nowicki B, Nowicki S, Piątek R. 2020. A shear stress micromodel of urinary tract infection by the Escherichia coli producing Dr adhesin. PLoS Pathog 16:e1008247. doi: 10.1371/journal.ppat.1008247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Medof ME, Walter EI, Rutgers JL, Knowles DM, Nussenzweig V. 1987. Identification of the complement decay-accelerating factor (DAF) on epithelium and glandular cells and in body fluids. J Exp Med 165:848–864. doi: 10.1084/jem.165.3.848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Totsika M, Beatson SA, Sarkar S, Phan MD, Petty NK, Bachmann N, Szubert M, Sidjabat HE, Paterson DL, Upton M, Schembri MA. 2011. Insights into a multidrug resistant Escherichia coli pathogen of the globally disseminated ST131 lineage: genome analysis and virulence mechanisms. PLoS One 6:e26578. doi: 10.1371/journal.pone.0026578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA 97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Chaveroche MK, Ghigo JM, d'Enfert C. 2000. A rapid method for efficient gene replacement in the filamentous fungus Aspergillus nidulans. Nucleic Acids Res 28:E97. doi: 10.1093/nar/28.22.e97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Farinha MA, Kropinski AM. 1990. Construction of broad-host-range plasmid vectors for easy visible selection and analysis of promoters. J Bacteriol 172:3496–3499. doi: 10.1128/jb.172.6.3496-3499.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Distribution of Dr family fimbriae in (A) strains belonging to the 83 most common E. coli STs and (B) strains belonging to the five most dominant UPEC clones. E. coli STs are grouped into seven main phylogenetic groups: A (ST10, ST34, ST48, ST93, ST167, ST301, and ST746), B1 (ST448), B2 (ST12 and ST131), C (ST23 and ST410), D (ST38, ST405 and ST69), E (ST335), and F (ST59 and ST648). The percentage of strains containing Afa/Dr fimbriae was determined by tBLASTn against afaCEC958, with a cut-off value of > 90% nucleotide identity and > 80% gene coverage. Download FIG S1, TIF file, 0.3 MB (308.6KB, tif) .
Copyright © 2022 Alvarez-Fraga et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
(A) Prevalence of the major subunits allelic variants in ST131 clades. AfaE alleles afaE-IX, afaE-I, daaE and draE2 are shown; five alleles were prevalent at < 1% (not shown): afaE-XIII (0.4%, clade C), afaE-XI (0.2%, clade B and 0.2%, clade C), afaE-II (0.1%, clade C), afaE-XII (0.1%, clade C), and drbE-122 (0.03%, clade C). (B) Prevalence of the major subunits allelic variants in ST131 subclades C1 and C2. AfaE alleles afaE-IX and afaE-I are shown; four alleles were prevalent at < 1% (not shown): afaE-XIII (0.5%, subclade C1 and 0.3%, subclade C2), drbE-122 (0.3%, subclade C1), afaE-XI (0.3%, subclade C2), afaE-II (0.1%, subclade C2), afaE-XII (0.2%, subclade C2). The daaE and afaE-XIV alleles were absent in both subclades. The percentage of each afaE allele was determined by tBLASTn with a cut-off value of > 97% nucleotide identity and > 80% gene coverage. ST131 strains were classified in clades and subclades based on clade-specific SNPs. One-hundred and 36 ST131 strains could not be classified into any clade and thus were excluded from the analysis. Chi-squared test was used for statistical analysis (*** P < 0.0001 for pair of C2–C1). (C) Prevalence of the major subunit allelic variants within E. coli ST38 clone. The daaE, afaE-XII, and afaE-VIII alleles were prevalent at < 1% (not shown). The percentage of each afaE allele was determined by tBLASTn with a cut-off value of > 97% nucleotide identity and > 80% gene coverage. Download FIG S2, TIF file, 0.5 MB (546.3KB, tif) .
Copyright © 2022 Alvarez-Fraga et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Multiple-sequence alignment of the major subunit variants. The sequences correspond to the mature protein. Each rectangle represents a distinct group, and their phylogenetic relationship is shown in the cladogram. The rectangles in color represent the groups found in the ST131 lineage. The predicted CEA and DAF binding sites are boxed in the protein sequence. The AfaE-XI variant shares a 99.6% identity with the AfaE variant found in the strain VR50. Download FIG S3, TIF file, 2.9 MB (3MB, tif) .
Copyright © 2022 Alvarez-Fraga et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
(A) Comparative genomics between the ST131 representative strains. Prophages (Phi) are indicated by blue boxes; GI are indicated by purple boxes and the GI containing the afa/dr gene cluster are indicated by green boxes. The ulaE gene corresponds to locus tag S21EC_48440. (B) Comparative genomics between the genomic islands carrying the afa/dr operon in the ST131 representative strains. Brown (afaF) and yellow (afaA) arrows represent genes encoding transcriptional regulators; orange arrows represent different IS elements; other genes are color-coded as green (chaperone), blue (usher), purple (invasin), pink (genes involved in RNA processing), and red (adhesin major subunit). Download FIG S4, TIF file, 0.9 MB (982.9KB, tif) .
Copyright © 2022 Alvarez-Fraga et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
(A) Summary features of ST131 representative strains. (B) Chromosomal IS1 insertion sites found in ST131 clone relative to six complete reference genomes. (C) Number of IS1 insertion sites relative to six ST131 complete reference genomes. (D) Chromosomal IS1 insertion sites found in ST131 clone relative to EC958 reference genome. (E) Primers used in this study. Download Data Set S1, XLSX file, 0.2 MB (208.1KB, xlsx) .
Copyright © 2022 Alvarez-Fraga et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
(A) ELISA showing the level of AfaE protein expressed on the surface of EC958 and S10EC wild-type and mutant derivatives. The primary polyclonal rabbit anti-AfaE-IX and the secondary alkaline phosphatase-conjugated anti-rabbit antibodies were used. Experiments were performed in three independent replicates. Unpaired student’s t-test was used for statistical analysis (* P < 0.05; ** P < 0.01; *** P < 0.0001). (B) EC958 and (C) S10EC analysis of AfaE expression based on (i) flow cytometry and (ii) immunofluorescence microscopy. For flow cytometry analyses, the fluorescence intensity of EC958 and S10EC cells was measured using anti-AfaE-IX and secondary anti-rabbit AlexaFluor 488 antibodies. The blue lines represent the negative control (EC958ΔafaE and S10ECΔafaE, respectively) used for setting the correct threshold value between noise and positive signal. The red lines represent the samples. The populations of EC958 phase-off, EC958 phase-on, and S10EC phase-on are indicated by arrows. For microscopy, phase contrast (left panels) and corresponding immunofluorescence (right panels) images are presented. Expression of the AfaE protein by EC958 and S10EC was detected primary polyclonal rabbit AfaE-IX antibody in combination with the secondary rabbit IgG FITC antibody. Download FIG S5, TIF file, 1.1 MB (1.1MB, tif) .
Copyright © 2022 Alvarez-Fraga et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Invasion of S10EC, S10ECΔIS1, S10ECΔafaE, EC958 and EC958ΔafaE strains to (A) A498 kidney and (B) T24 bladder human epithelial cells. Each data point is the mean of six technical replicates. Experiments were performed in three independent replicates. ANOVA and Šídák's multiple comparisons tests were used. (C) Mouse urinary tract colonization model. Competitive mixed-infection experiment employing a 1:1 mixture of E. coli S10EC/S10ECΔlac, E. coli S10ECΔIS1/S10ECΔlac, E. coli S10ECΔafaC/S10ECΔlac, and E. coli S10ECΔafaC/S10ECΔIS1Δlac. Each symbol represents the log10 competitive index calculated for each mouse at 24-h postinfection. Horizontal bars represent median values. Statistical significance was determined by the two-tailed Wilcoxon’s signed rank test. Download FIG S6, TIF file, 0.5 MB (483.3KB, tif) .
Copyright © 2022 Alvarez-Fraga et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
(A) Plot showing the cumulative number of IS1 insertion sites within the 95 in-house ST131 strain collection. Paired-end Illumina reads were mapped against six completely sequenced ST131 reference genomes (EC958, S10EC, S21EC, S22EC, S65EC, and S103EC). Each data point represents the number of insertion sites when different reference genomes were combined. (B) Plot showing the location of each of the IS1 insertion sites within the 95 in-house ST131 strain collection. Paired-end Illumina reads were mapped against the EC958 reference genome. Each data point represents the number of IS1 insertion sites found in each strain. Download FIG S7, TIF file, 0.4 MB (453.8KB, tif) .
Copyright © 2022 Alvarez-Fraga et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
(A) Transcript levels of the afaA, osmB, and ugd genes in the ST131 strains carrying an IS1 element upstream of the gene. The results are represented as log2 fold change compared to EC958 (which has not insertion sequences). The gapA gene was used as an endogenous control. Experiments were performed in three biological replicates. Statistically significant differences are represented with asterisks (** P value < 0.01; *** P value < 0.0001). (B) Nucleotide sequence of the IS1-promoter region junctions. The right end of the IS1 is underlined. The −35 region located in the IS1 and the predicted −10 region are boxed. The asterisks show the nucleotides identical to the corresponding E. coli consensus sequences (−10, TATAAT; −35, TTGACA). Download FIG S8, TIF file, 0.9 MB (939.8KB, tif) .
Copyright © 2022 Alvarez-Fraga et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Extended experimental methods for (i) whole genome sequencing and analysis, (ii) β-galactosidase assays, (iii) ELISA, (iv) flow cytometry, (v) immunofluorescence, (vi) epithelial cell adhesion and invasion assays, and (vii) the mouse UTI model. Download Text S1, DOCX file, 0.03 MB (28.9KB, docx) .
Copyright © 2022 Alvarez-Fraga et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.