Abstract
BACKGROUND
Clinicians and machine classifiers reliably diagnose pilocytic astrocytoma (PA) on magnetic resonance imaging (MRI) but less accurately distinguish medulloblastoma (MB) from ependymoma (EP). One strategy is to first rule out the most identifiable diagnosis.
OBJECTIVE
To hypothesize a sequential machine-learning classifier could improve diagnostic performance by mimicking a clinician's strategy of excluding PA before distinguishing MB from EP.
METHODS
We extracted 1800 total Image Biomarker Standardization Initiative (IBSI)-based features from T2- and gadolinium-enhanced T1-weighted images in a multinational cohort of 274 MB, 156 PA, and 97 EP. We designed a 2-step sequential classifier – first ruling out PA, and next distinguishing MB from EP. For each step, we selected the best performing model from 6-candidate classifier using a reduced feature set, and measured performance on a holdout test set with the microaveraged F1 score.
RESULTS
Optimal diagnostic performance was achieved using 2 decision steps, each with its own distinct imaging features and classifier method. A 3-way logistic regression classifier first distinguished PA from non-PA, with T2 uniformity and T1 contrast as the most relevant IBSI features (F1 score 0.8809). A 2-way neural net classifier next distinguished MB from EP, with T2 sphericity and T1 flatness as most relevant (F1 score 0.9189). The combined, sequential classifier was with F1 score 0.9179.
CONCLUSION
An MRI-based sequential machine-learning classifiers offer high-performance prediction of pediatric posterior fossa tumors across a large, multinational cohort. Optimization of this model with demographic, clinical, imaging, and molecular predictors could provide significant advantages for family counseling and surgical planning.
Keywords: Artificial intelligence, Ependymoma, Machine learning, Medulloblastoma, Pilocytic astrocytoma, Posterior fossa tumors, Radiomics
ABBREVIATIONS
- EP
ependymoma
- GLCM
gray level co-occurrence matrix
- GLRLM
gray level run length matrix
- GLSZM
gray level size zone matrix
- IBSI
Imaging Biomarker Standardization Initiative
- LASSO
least absolute shrinkage and selection operator
- LR
logistic regression
- MB
medulloblastoma
- NN
neural net
- NPV
negative predictive value
- PA
pilocytic astrocytoma
- PF
posterior fossa
Brain tumors are the most common solid pediatric tumors, and 45% to 60% occupy the posterior fossa (PF).1 Medulloblastoma (MB), pilocytic astrocytoma (PA), and ependymoma (EA) make up the majority of PF lesions with prevalence of 30% to 40%, 25% to 35%, and 10% to 15%, respectively.2 Although all warrant surgery for primary management and tissue diagnosis, the surgical approach, optimal extent of resection, and potential complications can vary by pathology.3 Without a preoperative diagnosis, the neurosurgeon must be flexible and occasionally adjust the surgical strategy based on intraoperative information. Thus, accurate, early diagnosis could greatly facilitate preoperative planning, treatment decisions, and family discussions.
For the most part, experienced neurosurgeons and neuroradiologists would not require the assistance of machine learning to accurately diagnosis cerebellar PA on magnetic resonance imaging (MRI). However, when faced with an atypical presentation, clinicians may mentally bin imaging features into PA and non-PA categories, and then for the latter try to refine the diagnosis based on classic features, eg, foci of calcification or “candle wax” appearance at 4 ventricular outlets for EP, or highly restricted diffusion in MB. This is not always straightforward, as many PF tumors lack classical features and have significantly overlapping phenotypes. Very heterogeneous cases of calcification, hemorrhage, and cyst formation occur with MB and EP, and a range of features can occur across the spectrum of MB subgroups. These nuances leave room for improvement in diagnostic accuracy and create a potential role for machine learning in assisting clinicians.2,4,5
Radiomics-based machine learning has shown clinical utility for management of neurosurgical problems.6-9 However, previous applications for pediatric PF tumors had limited accuracy and reproducibility, not only because of small cohorts and obscure feature extraction methods10-12 but also because of the failure to adapt machine-learning strategies to the unique situation, eg, applying a single classifier method to a wide range of diagnoses. We initially sought to develop such a single-step, multiclass classifier for pediatric PF tumor and found it to perform asymmetrically across subgroups, thereby needing optimization.
We hypothesized that a sequential radiomic classifier could improve diagnostic performance by mimicking a clinician's strategy of excluding PA before distinguishing MB from EP. We maximize performance by linking separate classifiers that first focus on one set of specific features to rule out PA, and then use a separate set of features to distinguish MB from EP.
METHODS
Study Population
For this multicenter, retrospective study, institutional review board approval (No. 33821) was obtained at all participating institutions (Supplementary Table 1, Supplemental Digital Content 1), with waiver of consent. Stanford Children's Hospital served as the host institution, and data-use agreements were obtained at all sites.
We reviewed consecutive imaging spanning July 1997 to May 2020 for MB, PA, and EP of patients under 19 yr old, including both gadolinium-enhanced T1-weighted (T1-MRI) and T2-weighted (T2-MRI) MRI sequences, and surgical specimen for pathologic confirmation. Patients were excluded if the MRI was nondiagnostic because of motion or artifacts.
Feature Extraction and Reduction
The volumetric whole tumor boundary, solid and cystic components inclusive, was performed independently on T1-MRI and T2-MRI using Osirix (Switzerland), with consensus review among experts (K.Y., R.L., A.J.). From each segmentation, we extracted 1800 (900 each from T2-MRI and T1-MRI) Image Biomarker Standardization Initiative (IBSI)-based features13,14 using PyRadiomics (2.2.0.post7 + gac7458e) in the Quantitative Image Feature Pipeline with configurations in Supplementary Appendix 1, Supplemental Digital Content 2.15 Pre-processing and extracted features are described in Supplementary Appendix 2, Supplemental Digital Content 3. A subset of the data was submitted for sparse regression analysis by a least absolute shrinkage and selection operator (LASSO) on RStudio 1.2.5033 (Boston, Massachusetts). LASSO parameters are described in Supplementary Appendix 2, Supplemental Digital Content 3.
Classifier Model Building
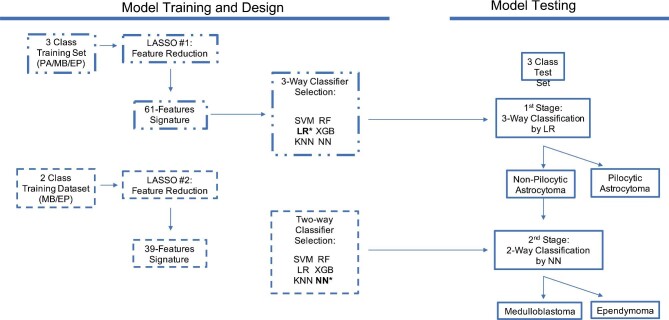
We sought to improve upon a baseline single-stage, 3-way classifier by constructing a 2-stage model (Figure 1). In the first stage, the best performing 3-way multiclass algorithm was selected from 6-candidate classifiers using the initial reduced feature set. In the second stage, feature reduction by LASSO was repeated for the 2 non-PA pathologies (MB and EP) given their lower performances from the first stage. The best performing binary classifier was identified using the new feature set for the 2 pathologies. In the combined final model, the events classified as non-PA were submitted for second-stage classification. Classification performance was assessed for each individual stage as well as for the combined stages.
FIGURE 1.
Workflow for training and testing of a 2-stage classifier, initially with a 3-way classifier and subsequently with a binary classifier, each optimizing over their respective reduced feature sets obtained by sparse regression analyses. Stage 1 passes the negative class to stage 2 for a more specifically trained binary classifier. EP, ependymoma; KNN, k-nearest neighbor; LASSO, least absolute shrinkage and selection operator; LR, logistic regression; MB, medulloblastoma; NN, neural net; PA, pilocytic astrocytoma; RF, random forest; SVM, support vector machine; XGB, eXtreme gradient boost.
Six-candidate classifier models were evaluated for each stage, including support vector machine, logistic regression (LR), k-nearest neighbor, random forest, extreme gradient boosting, and neural net (NN). The cohort underwent resampling to correct for sample imbalance. Training and test sets were randomly allocated from the total cohort in a 75:25 ratio. Optimal classifier parameters were estimated by grid search (Supplementary Appendix 3, Supplemental Digital Content 4). Relative influences of imaging features were calculated with LR based on coefficients used in the weighted sum. Classifier development was performed using Python 3.8.5.
The final radiomics, multiclass classifier was guided by maximizing the F1 score, measured as the weighted average between the precision score (positive predictive value [PPV]) and recall score (sensitivity). Although the F1 score applied to binary classification focuses on the positive data, in multi-class classification, the micro-averaged F1 score provides a holistic view prediction quality for all classes without restricting analysis to the positive class. This stems from classification events necessarily rotating between negative and positive labels as the microaveraging calculation cycles through binarized subgroups. Additionally, because the F1 score is the harmonic mean of precision and recall, in any multiclass microaveraged setting, the accuracy, precision, recall, and F1 score are all equivalent.16
Statistical Analysis
A P-value < .05 was considered statistically significant for all analyses. We calculated sensitivity, specificity, PPV, negative predictive value (NPV), F1 score, and area under the receiver operating characteristic curve (AUC) for each classifier, using the microaverage to pool performance over all samples. Confidence intervals were obtained by bootstrapping of the test sets for 2000 random samples.
RESULTS
Patient Cohort
A total of 535 patients met the inclusion criteria: 278 (52.0%) MB, 160 (29.9%) PA, and 97 (18.1%) EP (Supplementary Table 2, Supplemental Digital Content 5). Average age at diagnosis was 88.0, 111.6, and 95.4 mo, respectively.
First-Stage Classifier Model: PA vs Non-PA
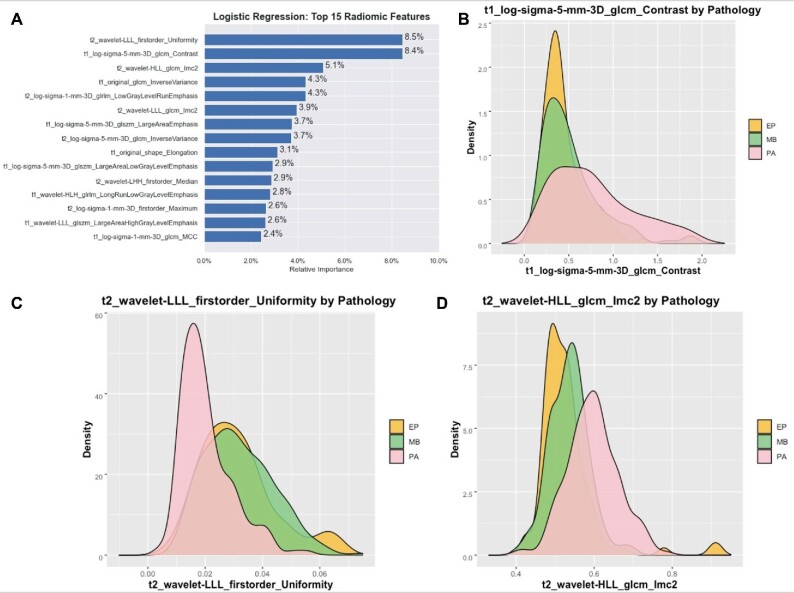
LASSO regression identified 61 relevant IBSI features, with 27 from T1-MRI and 34 from T2-MRI (Supplementary Table 3, Supplemental Digital Content 6), including 2 shapes, 16 first order, 19 gray level co-occurrence matrix (GLCM), 7 gray level run length matrix (GLRLM), and 17 gray level size zone matrix (GLSZM). Among the 6 classifier models, LR had the best performance (F1 score 0.7388, AUC = 0.9013) (Supplementary Table 4, Supplemental Digital Content 7). The binary performance comparison for each pathology was PA vs non-PA (F1 score 0.8809), MB vs non-MB (F1 score 0.7768), and EP vs non-EP (F1 score 0.4761; Table 1). For discerning PA from non-PA, the sensitivity, specificity, PPV, NPV, and accuracy were 0.8222, 0.9775, 0.9487, 0.9157, and 0.9477, respectively. The top 3 relevant features for LR training were T2 uniformity (first-order intensity), T1 contrast (GLCM texture), and T2-information measure of correlation-2 (GLCM texture; Figure 2; Supplementary Table 5, Supplemental Digital Content 8).
TABLE 1.
Binarized and Overall Performance Metrics of the Individual LR on the Holdout Test Set for the 3-Way Classifier Assessing MB, PA, and EP
| Sensitivity | Specificity | PPV | NPV | Accuracy | F1 score | AUC | |
|---|---|---|---|---|---|---|---|
| EP vs non-EP | 0.5172 | 0.8190 | 0.4411 | 0.8600 | 0.7313 | 0.4761 | 0.7671 |
| PA vs non-PA | 0.8222 | 0.9775 | 0.9487 | 0.9157 | 0.9477 | 0.8809 | 0.9890 |
| MB vs non-MB | 0.7833 | 0.8108 | 0.7704 | 0.8219 | 0.7761 | 0.7768 | 0.8909 |
| Microaverage | 0.7388 | 0.8694 | 0.7388 | 0.8694 | 0.8258 | 0.7388 | 0.9013 |
AUC, area under the receiver operating characteristic curve; EP, ependymoma; MB, medulloblastoma; NPV, negative predictive value; PA, pilocytic astrocytoma; PPV, positive predictive value.
FIGURE 2.
A, Bar plot of the top 15 features of the first stage, reduced feature set and their relative influence as calculated by LR, trained to distinguish MB, PA, and EP. Density plots of the top 3 features, including B, T2 uniformity, C, T2 contrast, and D, T2 informational measure of correlation.
Second-Stage Classifier Model: MB vs EP
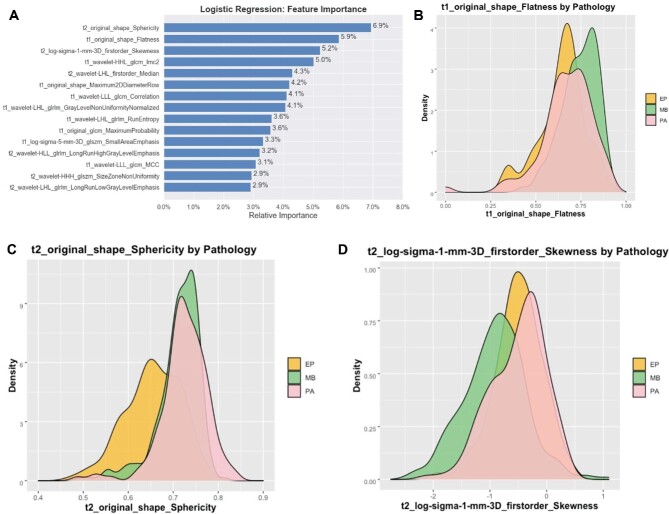
The second LASSO feature reduction over only MB and EP images identified 39 features, with 21 from T1-MRI and 18 from T2-MRI (Supplementary Table 3, Supplemental Digital Content 6), including 4 shapes, 9 first orders, 13 GLCM, 8 GLRLM, and 5 GLSZM. Among the 6 classifier models, NN had the best performance (F1 score 0.9189) (Table 2). Sensitivity, specificity, PPV, NPV, accuracy, and AUC were 0.9189, 0.7000, 0.9189, 0.7000, 0.8723, and 0.9243, respectively. The top 3 relevant features included T2 sphericity (shape), T1 flatness (shape), and T2 skewness (first-order intensity) (Figure 3; Supplementary Table 5, Supplemental Digital Content 8).
TABLE 2.
Binarized and Overall Performance Metrics of the Individual NN Classifier on the Holdout Test Set for the 2-Way Classifier Assessing MB and EP
| Metric | Score |
|---|---|
| Sensitivity | 0.9189 |
| Specificity | 0.7000 |
| PPV | 0.9189 |
| NPV | 0.7000 |
| Accuracy | 0.8723 |
| F1 score | 0.9189 |
| AUC | 0.9243 |
AUC, area under the receiver operating characteristic curve; NPV, negative predictive value; PPV, positive predictive.
FIGURE 3.
A, Bar plot of the top 15 features of the second stage, reduced feature set and their relative influence as calculated by LR, trained to distinguish MB, PA, and EP. Density plots of the top 3 features, including B, sphericity, C, T2 flatness, and D, T2 skewness.
Sequential Model
Finally, the 2 classifiers were performed sequentially such that the output of the initial LR classifier containing non-PA was fed to the NN classifier. The metrics for the combined model were sensitivity 0.9179, specificity 0.9589, PPV 0.9179, NPV 0.9589, accuracy 0.9452, and F1 score 0.9179 (Table 3). Accuracy for the final model was compared to the no information rate and found to be better than random guessing for all sub-groupings (P < .0001).
TABLE 3.
Binarized and Overall Performance Metrics of the Final Sequential Classifier, First Using a LR 3-Way Classifier Followed by a Neural Network Binary Classifier, on a Hold-Out Test Set Assessing MB, PA and EP
| Sensitivity | Specificity | PPV | NPV | F1 score | Accuracy (95% CI) | NIR | |
|---|---|---|---|---|---|---|---|
| EP vs Non-EP | 0.8965 | 0.9714 | 0.8965 | 0.9714 | 0.8965 | 0.9552 (0.9179-0.9851)a | 0.2164 |
| PA vs Non-PA | 0.8666 | 0.9775 | 0.9512 | 0.9354 | 0.9069 | 0.9402 (0.8955-0.9776)a | 0.3358 |
| MB vs Non-MB | 0.9666 | 0.9189 | 0.9062 | 0.9714 | 0.9354 | 0.9402 (0.8955-0.9776)a | 0.4478 |
| Micro-Average | 0.9179 | 0.9589 | 0.9179 | 0.9589 | 0.9179 | 0.9452 (0.9030-9801) | – |
Accuracy for binarized metrics is compared to the no information rate.
CI, confidence interval; EP, ependymoma; MB, medulloblastoma; NIR, no information rate, NPV, negative predictive value; PA, pilocytic astrocytoma; PPV, positive predictive value. aP < .0001 compared to NIR.
DISCUSSION
Preoperative diagnosis of pediatric PF tumors may pose challenges because of similar clinical presentations and atypical or overlapping image features. Although gross total resection remains the intent of surgery when morbidity is acceptable, intraoperative findings may alter the risk-benefit profile and require important decisions under pressure. Moreover, the pathology-specific morbidity of the procedure becomes apparent only after the surgery is underway. Given the important ramifications a preliminary pathological diagnosis, some pediatric neurosurgeons stop the procedure to discuss the findings and goals of surgery with the family.3
Although diagnosis will continue to rely on tissue specimens for the near future, increasing confidence in a relevant machine-learning classifier is desired. Its value will depend on a well-rounded performance across various metrics and for all components in the differential. Here we demonstrated how multiple radiomic models can be sequentially incorporated in a rational design to greatly improve precision.
Model Design and Performance
Radiomics-based, multiclass classifiers have traditionally focused on a single-stage model encompassing a single set of inputs and outputs.10,11 Moreover, many studies are often reduced to the representation of a single summary metric such as the AUC. Although our first-stage 3-way classifier produced a robust microaveraged AUC, the microaveraged F1 score was lower. When the full cohort was binarized, the performance metrics of EP vs non-EP were appreciably poorer than those for PA vs non-PA.
The unbalanced performance prompted our interest to develop a superior and more practical classifier that improves upon the weaker and more challenging MB and EP differentiation. Thus, the second-stage NN classifier was designed to re-evaluate the complete, original PyRadiomics feature list for a new set of LASSO-reduced features and address 2 limitations of a single-stage classifier. First, variables that may have been important for classifying PA were no longer necessary, and second, previously discarded variables from the first stage could become relevant again. In this second stage, we see that F1 score performance is substantially higher than those from binarized subgroupings in the first stage. The AUC for this binary performance was consistent with pilot work by Dong et al,12 who classified 51 patients for MB and EP but without a test set to rule out overfitting.
Regarding metrics reporting, traditionally the AUC is reserved for binary classifiers, as it provides a visual representation of the tradeoffs between sensitivity and specificity when predicting a positive class based on a probability threshold. Thus, although a generalized, microaveraged AUC has been described, the metric is a derived value. It becomes removed from the underlying binary tradeoff it is intended to portray and can differ from other classical metrics.17 Therefore, we elected to optimize over the microaveraged F1 score, which factors the precision and recall values across subgroups. Nevertheless, the F1 score remains an important tool for assessing weakness as evidenced by values in the first-stage binarization performances. For instance, the less populous EP decision-making exhibited a weaker F1 score that was penalized by its underlying precision and recall. The higher micro-averaged F1 score was likely compensated for by the stronger PA and MB performances. Collectively, these helped to motivate a need for our stronger, 2-stage model.
In summary, we see that the sequential model involving a first-stage LR classifier followed by a second-stage NN classifier that mimics human decision strategy, greatly improves the overall performance (Figure 4). Zhou et al10 previously described a smaller cohort with a microaveraged AUC of 0.92 and accuracy of 0.74. Our first-stage AUC performance already matched that reported by the author; however, our micro-averaged F1 score and accuracy score were substantially higher. When the overall model was binarized, EP vs non-EP and MB vs non-MB had a 42.0% and 15.8% improvement in their respective F1 scores. Meanwhile, the microaveraged F1 score also improved 17.9%. For the clinician, these ultimately translate into a higher precision (PPV) and recall (sensitivity). Therefore, the final classifier avoids erroneous labeling by limiting false positives while still detecting most of the positive samples.
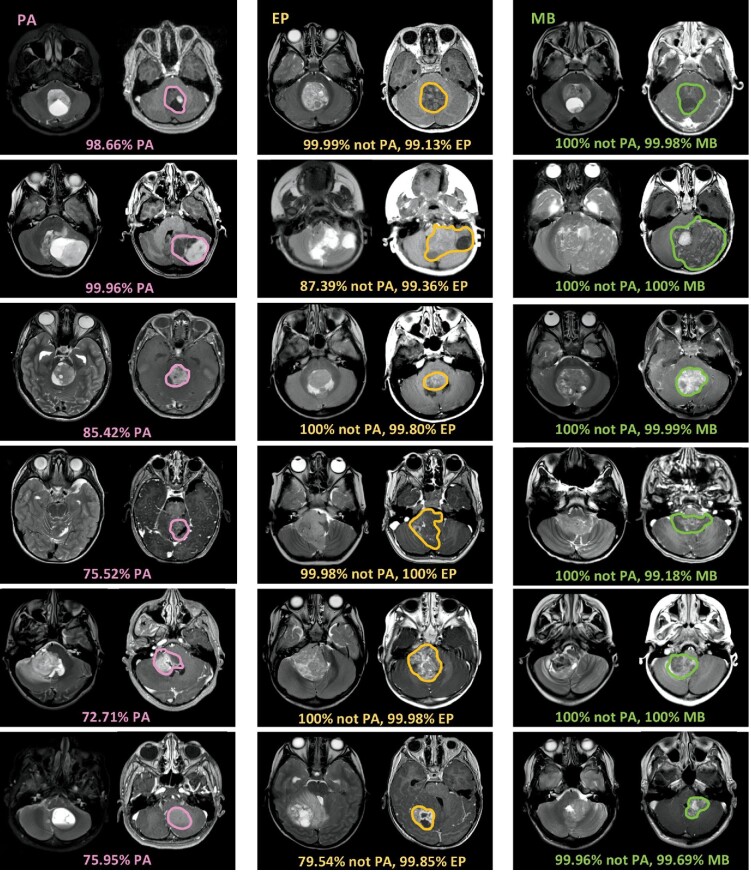
FIGURE 4.
Model prediction probability output from automated 2-stage algorithm. PA, EP, and MB are shown in columns 1, 2, and 3, respectively. In the first step, the model outputs probability for PA. In the second step, the model outputs probability for EP or MB. Some cases are straightforward. For example, the model prediction probability is very high on classic hemispheric PA with cyst and enhancing nodule (pink arrows). Some PA tumors have atypical features (*), including darker T2 signal, or more irregular or hemorrhagic appearance that might mimic MB or EP. Nevertheless, the model correctly identifies PA pathology, albeit at a slightly lower probability. Once deemed not-PA, tumors are automatically routed to second step to distinguish EP vs MB. Some EP and MB tumors demonstrate classic features, such as extension along Luschka and deformation around the brainstem (yellow arrows) or midline/hemispheric location, characteristic of EP and MB, respectively. However, some EP and MB tumors show overlap in features on human visual inspection. Model probability prediction outputs for EP and MB are shown.
Feature Interpretation
An important contribution of radiomics is its ability to preserve feature descriptions throughout the machine-learning pipeline, thereby avoiding the “black-box” classification seen with deep learning.6 Consequently, a radiomics approach allows significant features to be reviewed for model validation as well as informing future human performance. We examined the most important features contributing to our 2 stages and saw how known, qualitative radiographic elements for PA, MB, and EP are also quantitatively captured.
Among the 3 classes studied here, PA is the most distinguishable on routine MRI because of characteristically high T2 signal due to its low cellular density and frequent cystic component.2,4,5 Our first-stage model similarly prioritized this T2 signal by ranking T2 uniformity, a measure of signal homogeneity, as its most important variable (Figure 2; Supplementary Figure 1, Supplemental Digital Content 9). The predictive value of this feature is visibly appreciable on the corresponding density plot by the strong separation between PA and the other 2 tumors. As suspected, MB and EP strongly overlapped for T2 uniformity, thereby explaining why it was not preserved in the second-stage feature set. Additionally, we saw PA occupy a broader and larger distribution for T1 contrast, which can be expected given their avid contrast enhancement.5 Specifically, the bright voxels for PA can juxtapose with the nonenhancing cystic components and lead to higher contrast values relative to MB and EP.
The second-stage binary classifier shows how MB and EP can be further distinguished when in the absence of PA. We confirmed that T2 sphericity and T1 flatness (computationally the inverse of true flatness) are greater for MB than for EP (Figure 3; Supplementary Figure 2, Supplemental Digital Content 10). This suggests that MB embodies a more spherical conformation than EP, perhaps attributable to how each tumor tends to occupy their local anatomic compartments. The histopathological origination of MB and EP has been attributed to the cerebellar vermis and fourth ventricular floor, respectively.18 Because EP adapt to the surrounding ventricle or cistern, classically extending through the fourth ventricle apertures, they may display a greater surface area for a given tumor volume.2,18 Meanwhile, the T2 sphericity and T1 flatness values are less informative about PA. Converse to the first feature set, we see the distribution of values for PA strongly overlap with either of the other tumors. Thus, removal of PA from the second stage enabled us to recover imaging predictors that would have been overlooked in a single-stage classifier.
Limitations
As with other retrospective and radiomic studies, our work is subject to several limitations. Additional imaging sequences such diffusion-weighted imaging (DWI)/apparent diffusion coefficient, although known to have predictive information among PF pediatric tumors, could not be included because of low sample size and uneven distribution across tumor types. Also, many DWI scans were nondiagnostic because of dental artifacts; and a large number of the T1 and T2 MRI scans in our cohort represented preoperative navigation protocols that lacked DWI. As another limitation of radiomics, texture analysis was strictly derived from the isolated tumor volume and did not incorporate many common qualitative elements identifiable to human readers such as degree of anatomic laterality, additional neuro-axis involvement, and perilesional edema.2,18 Awareness of these additional predictors suggests that the accuracy featured in our work can only further improve when incorporated into human workflow.
CONCLUSION
This study demonstrates how a staged, radiomics-based machine-learning model can assist a clinician in the preoperative diagnosis of PA, MB, and EP. A micro-averaged F1 score of 0.9179 was achieved; however, the binarized F1 scores for each individual tumor type were also high performing. This was made possible after the set of features most important for distinguishing PAs were identified separately from that most important for MB and EP. Future work can continue to incorporate additional image sequences, semantic features, and clinical variables to improve performance.
Funding
This study did not receive any funding or financial support.
Disclosures
The authors have no personal, financial, or institutional interest in any of the drugs, materials, or devices described in this article. Dr Zhang is funded by the National Institutes of Health (5T32CA009695-27). Dr Yeom is funded by the ABTA (DG1800019).
Supplementary Material
Supplemental Digital Content 1. Supplementary Table 1. Listing of contributing institutions by pathology.
Supplemental Digital Content 2. Supplementary Appendix 1. Configuration files for radiomic feature extraction.
Supplemental Digital Content 3. Supplementary Appendix 2. Parameters for image preprocessing, feature extraction, and feature reduction.
Supplemental Digital Content 4. Supplementary Appendix 3. Final hyperparameters following grid search for 6 classifiers evaluated in the first and second stages.
Supplemental Digital Content 5. Supplementary Table 2. Comparison of clinical features between patients with MB, PA, and EP.
Supplemental Digital Content 6. Supplementary Table 3. A list of the variables identified by feature reduction and submitted for model training in the first and second stage of the final classifier.
Supplemental Digital Content 7. Supplementary Table 4. Performance metrics by 6-candidate, 3-way classifiers of MB, PA, and EP for consideration in the first stage of the overall model.
Supplemental Digital Content 8. Supplementary Table 5. Listing of the top 5 features retained for the first- and second-stage classifiers as calculated by LR and their qualitative interpretations.
Supplemental Digital Content 9. Supplementary Figure 1. Distinctive radiomic features of PA. Overall, quantitative features of brightness of gadolinium-enhanced T1-MRI and uniformity derived from T2-MRI were selected as most contributory features that distinguished from MB or EP. A, Despite variations in the volume of tumor enhancement, higher levels of brightness, or higher tissue contrast calculated from gadolinium-enhanced T1 MRI, were unique to PA tumors. Examples of bright enhancement characteristic of PA are shown, whether well-circumscribed (white arrows) or ill-defined (black arow) along margins of enhancement. B, Similarly, lower uniformity in pixel distribution within the tumor measured on T2-MRI-characterized PA.
Supplemental Digital Content 10. Supplementary Figure 2. Distinctive radiomic features of PF MB and EP. Shape-based features calculated from both T1 MRI and T2 MRI were robust features that distinguished MB and EP. For example, spherical or rounder morphology (green contour) was characteristic of MB compared to EP, which tended to envelope the brain, deform, and insinuate along CSF spaces, resulting in more irregular contours (yellow outline). Texture features extracted from filtered images are difficult to resolve by human eye. For example, based on one such texture feature (eg, t1_wavelet-LLL_GLCM_Correlation), EP tended to show less correlation between tumor voxels, potentially reflecting a more complex phenotype of EP on gadolinium-enhanced T1-MRI. Macroscopic examples of complex tumor patterns of EP are shown (within the tumor volume outlined in yellow), including irregular and rim-like as well as amorphous, nodular, and curvilinear enhancement. In comparison, despite a wide range, from solid to faint or no enhancement, MB tended (tumor contained within the green outline) to display less complex or irregular patterns of tumor enhancement.
COMMENT
This is a well-written and timely article on the application of Radionics for pediatric brain tumor diagnosis. There have been a number of recent of publications using a machine learning approach to aid diagnosis of posterior fossa tumors utilizing various imaging sequences. This article is unique in that it is the first multi-center, international study using machine learning to differentiate the most common pediatric posterior fossa tumors. Furthermore to aid the development of their model they only utilized the most common MR sequences ie. T1 with contrast and T2 weighted imaging. Their approach was to exclude pilocytic astrocytoma before attempting to differentiate ependymoma from medulloblasta which mimics the clinical sieve approach to these tumors. Correctly they integrated multiple machine learning approaches as there is no single algorithm or method that can single-handedly work with great efficiency and accuracy. Using Radiomics-based machine learning with a three-way logistic regression classifier they firstly distinguished pilocytic astrocytoma with T2-Uniformity and T1-Contrast emerging as the most relevant Image Biomarker Standardization Initiative features. Once a tumor was established as a non pilocytic astrocytoma, a two-way neural net classifier was used to distinguish medulloblastoma from ependymoma with T2-Sphericity and T1-Flatness as most relevant ISBI features. Furthermore the authors machine performance for MB versus EP is welcome as a recently published multi institutional study reported significant accuracy problems with the diagnosis of ependymoma (though they only used T2 weighted imaging).1
The Machine learning-assisted model as described will hopefully be (in the future when fully developed to include other imaging data such as DWI) useful for individual patient prognostication and facilitate preoperative strategy planning and discussions with family.
Cormac G. Gavin
London, United Kingdom
REFERENCES
- 1. Quon JL, Bala W, Chen LC et al. Deep learning for pediatric posterior fossa tumor detection and classification: a multi-institutional study. American Journal of Neuroradiology. 2020;41(9):1718-1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
REFERENCES
- 1. Pollack IF. Brain tumors in children. N Engl J Med. 1994;331(22):1500-1507. [DOI] [PubMed] [Google Scholar]
- 2. Brandão LA, Young Poussaint T. Posterior fossa tumors. Neuroimaging Clin N Am. 2017;27(1):1-37. [DOI] [PubMed] [Google Scholar]
- 3. Albright AL, Pollack IF. Surgical treatment. In: Principles and Practice of Pediatric Neurosurgery. 3rd ed. Thieme Medical Publishers, Inc.; 2014. [Google Scholar]
- 4. Mata-Mbemba D, Donnellan J, Krishnan P, Shroff M, Muthusami P. Imaging features of common pediatric intracranial tumours: a primer for the radiology trainee. Can Assoc Radiol J. 2018;69(1):105-117. [DOI] [PubMed] [Google Scholar]
- 5. Kerleroux B, Cottier JP, Janot K, Listrat A, Sirinelli D, Morel B. Posterior fossa tumors in children: radiological tips & tricks in the age of genomic tumor classification and advance MR technology. J Neuroradiol. 2020;47(1):46-53. [DOI] [PubMed] [Google Scholar]
- 6. Quon JL, Bala W, Chen LC et al. Deep learning for pediatric posterior fossa tumor detection and classification: a multi-institutional study. AJNR Am J Neuroradiol. 2020;41(9):1718-1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Park CJ, Han K, Kim H et al. MRI features may predict molecular features of glioblastoma in isocitrate dehydrogenase wild-type lower-grade gliomas. AJNR Am J Neuroradiol. 2021;42(3):448-456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lohmann P, Elahmadawy MA, Gutsche R et al. FET PET radiomics for differentiating pseudoprogression from early tumor progression in glioma patients post-chemoradiation. Cancers (Basel). 2020;12(12):3835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chaddad A, Kucharczyk MJ, Daniel P et al. Radiomics in glioblastoma: current status and challenges facing clinical implementation. Front Oncol. 2019;9:374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhou H, Hu R, Tang O et al. Automatic machine learning to differentiate pediatric posterior fossa tumors on routine MR imaging. AJNR Am J Neuroradiol. 2020;41(7):1279-1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rodriguez Gutierrez D, Awwad A, Meijer L et al. Metrics and textural features of MRI diffusion to improve classification of pediatric posterior fossa tumors. AJNR Am J Neuroradiol. 2014;35(5):1009-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dong J, Li L, Liang S et al. Differentiation between ependymoma and medulloblastoma in children with radiomics approach. Acad Radiol. 2020;28(3):318-327. [DOI] [PubMed] [Google Scholar]
- 13. Zwanenburg A, Vallières M, Abdalah MA et al. The Image Biomarker Standardization Initiative: standardized quantitative radiomics for high-throughput image-based phenotyping. Radiology. 2020;295(2):328-338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. van Griethuysen JJM, Fedorov A, Parmar C et al. Computational radiomics system to decode the radiographic phenotype. Cancer Res. 2017;77(21):e104-e107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mattonen SA, Gude D, Echegaray S, Bakr S, Rubin DL, Napel S. Quantitative imaging feature pipeline: a web-based tool for utilizing, sharing, and building image-processing pipelines. J Med Imaging (Bellingham). 2020;7(4):042803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Grandini M, Bagli E, Visani G. Metrics for multi-class classification: an overview. arXiv preprint arXiv:200805756. 2020. [Google Scholar]
- 17. Hand DJ, Till RJ. A simple generalisation of the area under the ROC curve for multiple class classification problems. Machine Learning. 2001;45(2):171-186. [Google Scholar]
- 18. Raybaud C, Ramaswamy V, Taylor MD, Laughlin S. Posterior fossa tumors in children: developmental anatomy and diagnostic imaging. Childs Nerv Syst. 2015;31(10):1661-1676. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.