Significance
Most microbial knowledge comes from experiments in laboratory models, despite the assumption that these artificial systems alter microbial physiology relative the native environment. We tested this assumption for an oral pathogen, Porphyromonas gingivalis, using 93 metatranscriptomes from periodontally healthy and diseased patients and 122 transcriptomes from experimental models. We discovered that a simple in vitro model, midlogarithmic growth in rich media, highly recapitulates P. gingivalis gene expression in the human oral cavity, outperforming other models, including a murine infection model. These results support the biological relevance of decades of laboratory experiments with this pathogen and validate an accessible experimental model for studying P. gingivalis biology. Furthermore, these data provide a conceptual framework for understanding in situ gene expression across microbes.
Keywords: Porphyromonas gingivalis, metatranscriptomics, periodontitis, laboratory models
Abstract
Bacterial behavior and virulence during human infection is difficult to study and largely unknown, as our vast knowledge of infection microbiology is primarily derived from studies using in vitro and animal models. Here, we characterize the physiology of Porphyromonas gingivalis, a periodontal pathogen, in its native environment using 93 published metatranscriptomic datasets from periodontally healthy and diseased individuals. P. gingivalis transcripts were more abundant in samples from periodontally diseased patients but only above 0.1% relative abundance in one-third of diseased samples. During human infection, P. gingivalis highly expressed genes encoding virulence factors such as fimbriae and gingipains (proteases) and genes involved in growth and metabolism, indicating that P. gingivalis is actively growing during disease. A quantitative framework for assessing the accuracy of model systems showed that 96% of P. gingivalis genes were expressed similarly in periodontitis and in vitro midlogarithmic growth, while significantly fewer genes were expressed similarly in periodontitis and in vitro stationary phase cultures (72%) or in a murine abscess infection model (85%). This high conservation in gene expression between periodontitis and logarithmic laboratory growth is driven by overall low variance in P. gingivalis gene expression, relative to other pathogens including Pseudomonas aeruginosa and Staphylococcus aureus. Together, this study presents strong evidence for the use of simple test tube growth as the gold standard model for studying P. gingivalis biology, providing biological relevance for the thousands of laboratory experiments performed with logarithmic phase P. gingivalis. Furthermore, this work highlights the need to quantitatively assess the accuracy of model systems.
The ultimate goal of the vast majority of microbiology research is to understand the processes that shape microbial behavior, ecology, and evolution, ranging from studies of the microbial role in pathogenesis to the microbial contribution to nutrient cycling in the oceans. Experimental laboratory models such as in vitro culture, microcosms, and animal models are the workhorses of these studies and have greatly advanced our understanding of microbial physiology. However, the relationship between an experimental model system and a microbe’s native environment is often not well understood, and recent work has shown that the in situ gene expression signature is distinct from that in experimental model systems for some opportunistic pathogens (1–3).
Microbes in the oral cavity involved in periodontitis, or gum disease, live in the subgingival pocket between the tooth and gum surfaces, where they are in contact with a diverse microbial community, the human immune system, and a unique nutritional environment (4). The ability to directly sample communities from the human subgingival environment has provided a real-time snapshot of microbial gene expression during periodontitis, including establishing the importance of virulence factor expression and butyrate synthesis in periodontitis at the community level (5, 6). However, the majority of these studies have not had the depth and breadth to examine the gene expression of individual taxa within the human oral cavity (7).
Here, we performed a meta-analysis of 93 human oral metatranscriptomes, focusing on the gene expression patterns of the oral pathogen Porphyromonas gingivalis (8). P. gingivalis is an obligate anaerobe and is asaccharolytic, using amino acids as its primary carbon source. This microbe is often associated with chronic periodontitis, and it has been characterized as a keystone pathogen because of its ability to alter the oral immune environment, leading to dysbiosis of the microbial community as a whole (9). We discovered that P. gingivalis highly expressed a number of virulence factors, including the Arg- and Lys-gingipains, and genes related to growth and metabolism during periodontal disease. However, a comparison to 122 transcriptomes from experimental model systems revealed that the P. gingivalis periodontitis transcriptome was not distinct from growth in some common laboratory conditions, specifically logarithmic growth in rich media. Finally, we showed that the global conservation of P. gingivalis gene expression between periodontitis samples and certain laboratory environments is related to low variance in gene expression across environments for P. gingivalis in contrast to other pathogens.
Results
Validation of Mapping Approach and P. gingivalis Pangenome.
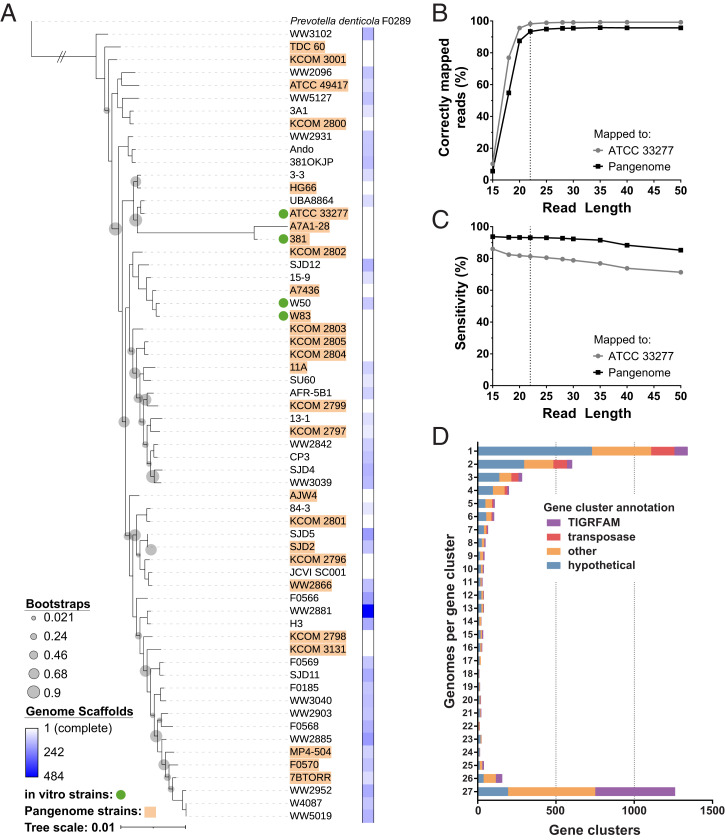
The goal of this work was to analyze the gene expression of P. gingivalis in the human oral cavity. A challenge in this and similar analyses is that metatranscriptomes include short sequencing reads from diverse microbes. Thus, it is important to ensure sequencing reads are assigned to the correct microbe. In our case, human oral subgingival metatranscriptomes often contain reads from over 100 species and may contain multiple strains of P. gingivalis. Our objective was to identify P. gingivalis–derived reads from across the strain diversity of this species while minimizing the inclusion of non–P. gingivalis–derived reads. Correct mapping depends on a number of factors, including sequence read length and the identity of the reference sequence. To test these two factors, we built 10 mock oral metatranscriptomes, each containing 658 bacterial genomes from the Human Oral Microbiome Database and each with a different read length ranging from 15 to 50 base pairs (bp). Then, we mapped the mock metatranscriptomes to the P. gingivalis American Type Culture Collection (ATCC) 33277 genome and to a pangenome of P. gingivalis strains. This pangenome was constructed to represent the taxonomic diversity of the species using 27 high-quality genomes (Fig. 1A and Dataset S1A). For the mock metatranscriptome with 15-bp reads, 25% of all the reads mapped to P. gingivalis ATCC 33277, but only 10% of mapped reads originated from P. gingivalis, indicating high levels of mapping from other members of the in silico community (Fig. 1B). However, as the read length in the metatranscriptome increased, the percentage of mapped reads originating from P. gingivalis increased, plateauing at 99% with 25-bp reads and longer. When mapped to the pangenome, the pattern was similar, with at least 95% of mapped reads originating from P. gingivalis at 25-bp read length and longer. Based on this analysis, we chose 22 bp as the minimum read length for our biological samples to balance the specificity of mapping with the short reads in many of the biological samples (Fig. 1B, Dataset S2A, and SI Appendix, Table S1). Although this approach may detect some reads from other microbes in the community, it is highly selective for P. gingivalis reads. Furthermore, the average read length of the biological samples analyzed below was 61 bp, and the 50-bp mock metatranscriptome analysis shows that these longer reads provide additional stringency (Fig. 1B and SI Appendix, Table S1). Additionally, we asked how many of the total P. gingivalis reads in the mock metatranscriptome were identified via mapping. When we aligned the 22-bp mock metatranscriptome to the single P. gingivalis strain, 81% of the total P. gingivalis reads mapped, but when we aligned to the pangenome, 94% of the total P. gingivalis reads mapped (Fig. 1C and SI Appendix, Table S1). Therefore, we identified more of the P. gingivalis reads when mapping to the pangenome, likely because of the additional accessory genes in the pangenome, and we chose to map our biological samples to the P. gingivalis pangenome.
Fig. 1.
Pangenome analysis for metatranscripome analyses. (A) Maxiumum Likelihood phylogeny for 62 P. gigivalis strains and the outgroup Prevotella denticola F0289 built using an alignment of 49 core genes in FastTree2. The 27 strains included in the pangenome are highlighted in orange. Bootstraps less than 0.9 are shown with gray bubbles. The completeness of the genome is indicated by the number of genome scaffolds ranging from white (complete genome) to blue (484 genome scaffolds). (B) The percentage of mapped reads that originated from P. gingivalis at each read length in a mock metatranscriptome analysis. A total of 10 mock metatranscriptomes were built with read lengths varying from 15 to 50 bp using 658 genomes in the Human Oral Microbiome Database. These mock metatranscriptomes were each mapped to a single P. gingivalis strain (ATCC 33277) and to the pangenome of 27 strains. (C) The percentage of P. gingivalis reads in the mock metatranscriptome that aligned to the genome or pangenome at each read length. Based on the analysis here and in B, 22 bp was chosen as the minimum read length for an analysis of the biological samples as indicated by the dashed lines. (D) Distribution and annotation of the 4,643 ortholog clusters in our pangenome of 27 P. gingivalis strains. Colors indicate if the ortholog cluster had a TIGRFAM annotation and, if not, if it was annotated as a transposase, another function (other), or hypothetical.
After deciding on the minimum read length of 22 bp and to map to the pangenome, we next defined orthologous genes across all 27 genomes in the pangenome. This step was important to allow for gene-based analyses of the biological samples. We identified 4,643 clusters of genes in our P. gingivalis pangenome (Fig. 1D and Dataset S1). Note that the 27 P. gingivalis genomes each have 1,929 protein coding genes on average. A total of 1,261 clusters included genes from all 27 genomes, and 1,342 clusters only included a single gene (Fig. 1D). Only 16 clusters contained multiple genes from the same genome. A total of 85% of the clusters with genes from all 27 genomes have a functional annotation, while 54% of the clusters containing only one gene are annotated as hypothetical, indicating that many of the known functions are represented by the shared orthologs. Collectively, these orthologs and the mock metatranscriptome benchmarking provide a rigorous and validated approach to analyze the P. gingivalis transcriptional profile in complex metatranscriptomes.
P. gingivalis Transcripts Are Enriched in Metatranscriptomes from One-Third of Periodontally Diseased Patients.
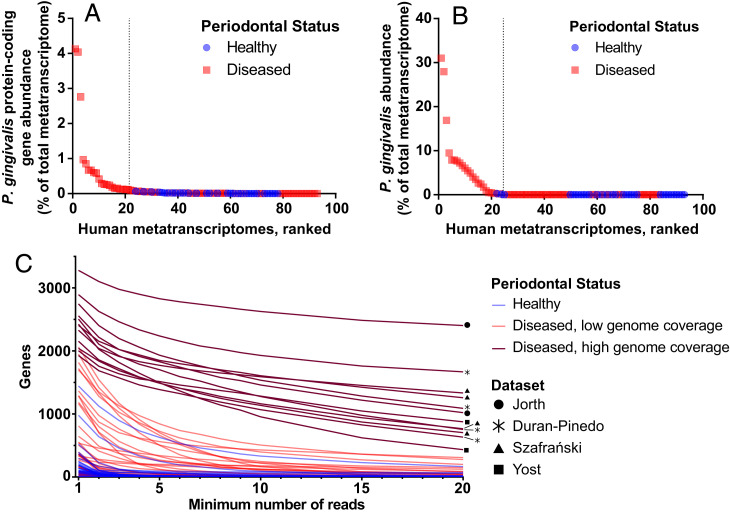
We analyzed 93 previously published metatranscriptomes from six studies for P. gingivalis transcripts (Table 1 and Dataset S2A) (10–15) using the framework determined using the mock metatranscriptomes above and shown in SI Appendix, Fig. S1. These metatranscriptomes included 61 periodontally diseased samples from 38 individuals, including patients with gingivitis, aggressive periodontitis, and chronic periodontitis. In addition, the metatranscriptomes included 32 periodontally healthy samples, each from a separate patient. Most of the samples were collected directly from the human subgingival pocket, but 10 diseased and 10 healthy samples were from saliva. For each metatranscriptome, we mapped all reads longer than 22 bp to the pangenome of 27 P. gingivalis strains. Reads mapping to P. gingivalis protein-coding genes constituted at least 0.1% of the total metatranscriptome in 21 of 61 diseased samples (34%) but not in any of the 32 healthy samples (Fig. 2A and Dataset S2A). Reads mapping to P. gingivalis tended to be more abundant in diseased samples (Mann–Whitney U test, P = 0.07). Similarly, MetaPhlAn identified that P. gingivalis constituted at least 0.1% of the community in 22 of 61 periodontally diseased samples but only 1 of 22 periodontally healthy samples and that P. gingivalis was significantly enriched in the diseased samples (Mann–Whitney U test, P = 0.002; Fig. 2B and Dataset S2). In addition, P. gingivalis was the dominant Porphyromonas species across diseased samples (Dataset S2B). It is important to note that these findings represent the transcriptional abundance of P. gingivalis, and transcript levels do not always correlate with DNA levels in the oral cavity (12). By analyzing the number of genes with mapped reads in each sample, we narrowed our study to 12 human diseased samples with high coverage of the P. gingivalis pangenome (Fig. 2C). These 12 samples are from four different publications and from both chronic and aggressive periodontitis patients. In addition, these samples are each dominated by a single strain of P. gingivalis, but across these samples, there is diversity in the dominant strain (SI Appendix, Fig. S2).
Table 1.
Human oral metatranscriptomic datasets
| Reference | Diseased samples | Healthy samples |
| Belstrøm et al., NPJ Biofilms and Microbiomes, 2017. (saliva) (10) | 10 (chronic periodontitis) | 10 |
| Duran-Pinedo et al., ISME J, 2014 (11). | 7 (chronic periodontitis) | 6 |
| Jorth et al., mBio, 2014 (12). | 3 (aggressive periodontitis) | 3 |
| Nowicki et al., mBio, 2018 (13). | 3 (gingivitis) | 3 |
| Szafrański et al., NPJ Biofilms and Microbiomes, 2015 (14). | 6 (chronic periodontitis) | 10 |
| Yost et al., Genome Medicine, 2015 (15). | 16 (chronic periodontitis, stable site); 16 (chronic periodontitis, progressing site) |
0 |
Fig. 2.
P. gingivalis relative transcriptional abundance across 32 periodontally healthy and 61 periodontally diseased patient samples. (A) P. gingivalis relative abundance based on the proportion of metatranscriptome reads that mapped to the pangenome of P. gingivalis protein-coding genes. Samples are ranked based on abundance, and the dashed line separates samples with P. gingivalis relative abundance less than 0.1%. (B) P. gingivalis relative abundance as determined by MetaPhlAn 3. Samples are ranked based on P. gingivalis abundance, and the dashed line separates samples with P. gingivalis relative abundance less than 0.1%. (C) The read coverage of the P. gingivalis pangenome genes for each metatranscriptome. Each line represents the relationship between gene coverage and read depth for a single metatranscriptome. The 12 periodontitis samples with high genome coverage depth and breadth that were chosen for detailed analyses are colored in dark red and labeled with the dataset name (Table 1).
We also compared the clinical parameter, pocket depth, to P. gingivalis relative abundance (Dataset S2A). Pocket depth measures the separation between the gum and tooth and is one assessment of disease severity; a pocket depth of 1 to 3 mm often indicates periodontal health or gingivitis, while a pocket depth ≥ 4 mm often indicates periodontitis (16). All samples with high coverage of the P. gingivalis genome that were chosen for downstream analysis had a pocket depth of at least 5 mm (SI Appendix, Fig. S3). Together, this meta-analysis of 93 human metatranscriptomes shows that P. gingivalis transcripts are detected rarely in healthy samples and are more abundant in periodontally diseased samples.
Housekeeping and Virulence Functions Are Highly Expressed in Periodontitis.
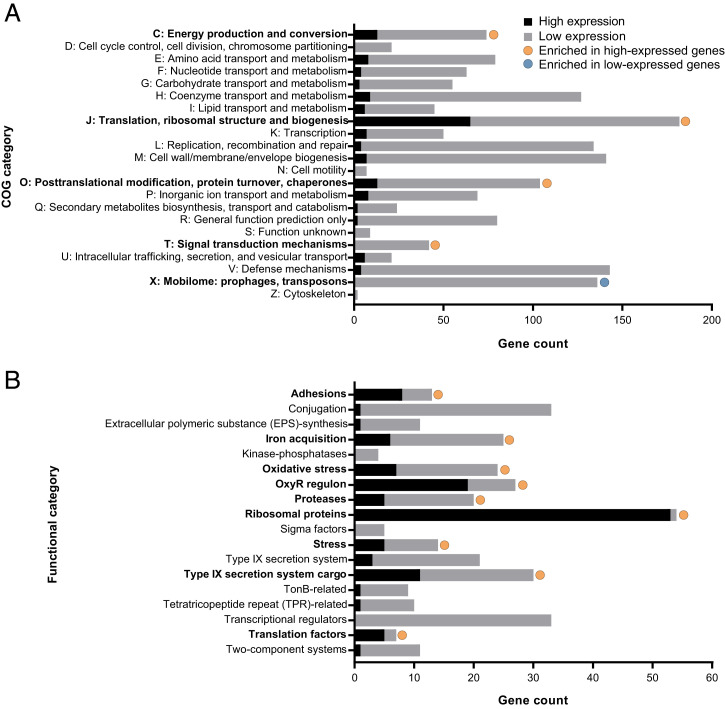
To probe the physiology of P. gingivalis during periodontitis, we first analyzed the function of the 258 most highly expressed P. gingivalis genes on average across the 12 periodontitis samples. This gene set was chosen using an inflection point analysis of the ranked average transcripts per kilobase million (TPM) gene expression counts (SI Appendix, Fig. S4 and Dataset S3). These genes were enriched for clusters of orthologous groups of protein (COG) categories C (energy production and conversion), J (translation, ribosomal structure, and biogenesis), O (posttranslational modification, protein turnover, and chaperones), and U (intracellular trafficking, secretion, and vesicular transport) (Fig. 3A). In addition, COG category X (Mobilome: prophages and transposons) was underrepresented in the highly expressed genes, although one putative IS982 family transposase not classified by COG was among the highly expressed genes. Thus, much of the highly expressed genes were involved in key cellular functions such as translation, protein secretion, and ATPase activities.
Fig. 3.
Functional enrichment of highly expressed P. gingivalis genes during periodontitis. Enrichment of COG categories (A) and hand-curated functional categories (B) was determined using a two-sided Fisher’s exact test, and significance is shown for Padj < 0.05. High expression genes are the 258 genes with the highest mean expression across the 12 periodontitis samples (SI Appendix, Fig. S4), while the low expression genes are the remaining genes.
This finding was further supported using hand-annotated functional categories of genes known to be important for growth, pathogenicity, and colonization (Fig. 3B and Dataset S3) with the caveat that, in this classification scheme, many genes are assigned to multiple functional categories. A total of 53 of 54 ribosomal proteins (98%) were among the top 258 most highly expressed genes, as were 19 of 27 genes (70%) annotated as part of the oxidative stress regulon, including the oxidative stress-responsive regulator OxyR. Functions related to pathogenesis were also significantly enriched. A total of 11 genes annotated as fimbrial proteins were among the 258 most highly expressed genes, including both putative major and minor fimbrial subunits, and genes encoding proteins involved in iron acquisition were significantly enriched in the highly expressed gene set. In addition, the putative Arg- and Lys-gingipains were among the highly expressed genes (rgpA, rgpB, and kgp). Finally, these proteases and the citrullinating peptidylarginine deiminase (ppad) were among the 11 highly expressed genes classified as encoding Type IX Secretion System (T9SS) cargo proteins (11 of 30, 37%), and a gene annotated as a Type VI Secretion system needle protein Hcp was also highly expressed. This analysis shows that P. gingivalis is actively growing and dividing in the human oral cavity, and during periodontitis, P. gingivalis expresses genes that contribute to pathogenesis such as those encoding adhesins, iron acquisition proteins, proteases, and secretion system–related proteins.
The P. gingivalis In Situ Transcriptional Profile Clusters with Transcriptomes from Logarithmic Growth.
An alternative approach to understanding the physiology of P. gingivalis in the human oral cavity is through comparison to in vitro growth conditions that have known physiologies. Thus, we compared P. gingivalis gene expression in the 12 periodontitis samples to 122 transcriptomes of wild-type P. gingivalis collected during growth in experimental model systems (Table 2 and Dataset S2C). A total of 117 of these transcriptomes were from in vitro culture from 12 publications, mostly from growth in rich media under laboratory conditions (17–29). Of these transcriptomes, 85 were from monoculture and 32 from pairwise coculture with either Streptococcus gordonii, Acinetobacter baumannii, or Candida albicans. These in vitro datasets include data from four P. gingivalis strains (Table 2 and Fig. 1). The remaining five transcriptomes are from a murine abscess model of infection using P. gingivalis ATCC 33277 and were generated for this study. To ensure that any differences in gene expression were not due to differences in gene content between the strains in the experimental model systems or the human specimens, we identified a core gene set for all comparative analyses. This core set consisted of 1,500 genes that met at least one of two criteria: 1) present in at least 26 of the 27 genomes in the pangenome and/or 2) had aligned sequencing reads in all 12 human metatranscriptomes with high P. gingivalis coverage and all 122 experimental model system transcriptomes (Dataset S1).
Table 2.
P. gingivalis transcriptomic datasets from experimental model systems
| Reference | P. gingivalis strain(s) | Growth conditions* | Transcriptomes |
| Belvin et al., Infect Immun, 2019 (17). | W83 | Midlogarithmic phase, mycoplasma media ± nitrite | 4 per condition (8 total) |
| Cheng et al., Metallomics, 2019 (18). | ATCC 33277 | Logarithmic phase, THK ± ranitidine bismuth citrate added at midlogarthmic phase and then sampled at 30, 60, 180, and 360 min | 4 per condition, 1 per time point (8 total) |
| Coats et al., Infect Immun, 2019 (19). | ATCC 33277 and 381 | Stationary phase, TYHK | 3 per strain (6 total) |
| Dou et al., Mol Oral Microbiol, 2018 (20). | W83 | Late-logarithmic phase, BYHK with hydrogen peroxide | 3 total |
| Hendrickson et al., Front Microbiol, 2017 (21). | ATCC 33277 | Pelleted cells, held anaerobically in phosphate-buffered saline for 0, 5, 30, 60, 120, 240, and 360 min ± S. gordonii DL1, ± DMSO, or ± 4-aminobenzoate | 2 to 3 per condition and time point (36 total) |
| Hovik et al., J Bacteriol, 2012 (22). | W83 | Hard agar, blood agar plates + HK; midlogarithmic phase, THK; midlogarithmic phase, chemically defined minimal liquid media | 1 per condition (3 total) |
| Jain et al., J Bacteriol, 2019 (23). | ATCC 33277 | Stationary phase, TYHK | 3 total |
| Kin et al., J Oral Microbiol, 2020 (24). | W50 | Logarithmic phase, Oral Bacterial Growth Medium ± Treponema denticola spent media | 3 per condition (6 total) |
| Miller et al., Mol Oral Microbiol, 2018 (25). | ATCC 33277 | Pelleted cells, held aerobically in phosphate-buffered saline for 180 min ± A. baumannii AB0057 | 4 monoculture, 8 coculture (12 total) |
| Moradali et al., ISME J, 2019 (26). | W83 and 381 | Hard and soft agar, 0.3 and 1.5% blood agar plates + THK | 3 per condition per strain (12 total) |
| Moye et al., Appl Environ Microbiol, 2019 (27). | W83 | Midlogarithmic phase, THK ± 0.1% galactose | 3 per condition (6 total) |
| Shen et al., Mol Oral Microbiol, 2020 (28). | ATCC 33277 | Logarithmic phase, TYHK | 5 total |
| Sztukowska et al., mBio, 2018 (29). | ATCC 33277 | Logarithmic phase, CaGHK ± C. albicans hyphae or C. albicans spent media | 3 per condition (9 total) |
| This study. | ATCC 33277 | Murine inner thigh abscess model | 5 total |
*Abbreviations: T = tryptic soy broth; B = brain–heart infusion; H = hemin; K = vitamin K; Y = yeast extract; DMSO = dimethyl sulfoxide; CaG = yeast nitrogen base, 10 mM NaH2PO4 buffer (pH 7.0), 0.05% Bacto tryptone, and 0.4% glucose.
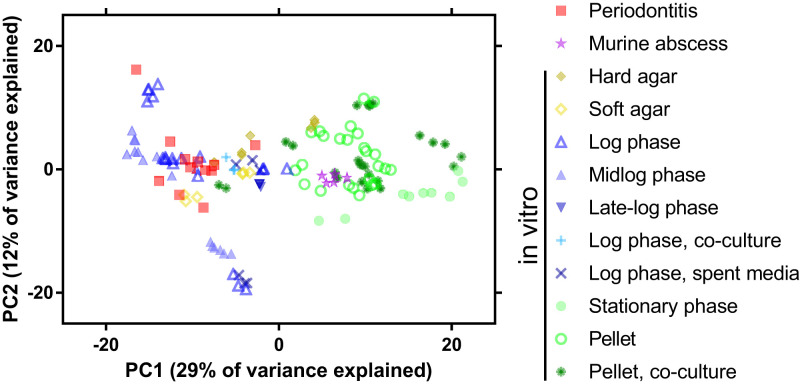
A principal component analysis (PCA) of the regularized log (rlog)-normalized gene expression across these samples showed that the P. gingivalis periodontitis transcriptomes do not cluster separately from the in vitro transcriptomes (Fig. 4 and SI Appendix, Fig. S5). This finding suggests that at a global level, P. gingivalis messenger RNA levels are not distinct between in vitro culture and the human oral cavity. However, the samples did separate across the first principal component (PC1). Specifically, the periodontitis transcriptomes clustered with in vitro transcriptomes collected during logarithmic phase growth or on agar plates and distinct from transcriptomes collected during stationary phase growth, pelleted cells, or the murine abscess (Fig. 4). Of note, both the soft and hard agar samples were from actively growing cultures collected 24 to 30 h after inoculation, before significant colony biofilm formation (26). These findings are supported by a heatmap of Euclidian distances between the read counts of the samples in which the periodontitis samples cluster with midlogarithmic phase and soft agar transcriptomes and separately from cultures grown to late-logarithmic or stationary phase, collected from cell pellets, or harvested from murine abscesses (SI Appendix, Fig. S6). In the Euclidian distance and PCA, the experimental model systems also cluster strongly by the individual RNA sequencing (RNA-seq) studies from which the data were obtained (SI Appendix, Figs. S5A and S6). However, the model systems do not cluster by the P. gingivalis strain used in the experiments (SI Appendix, Figs. S5B and S6).
Fig. 4.
PCA of the P. gingivalis transcriptome in human periodontitis samples and experimental model systems. PCA was constructed using rlog-normalized read counts. Sample shapes and colors indicate the growth condition. Logarithmic growth is indicated when the methods were unclear on whether the culture was early-, mid-, or late-logarithmic phase. Pellet indicates cells that were pelleted then held statically. See SI Appendix, Fig. S5 for sample annotation by publication and strain and for the scree plot.
We further identified the genes that had strong correlations with PC1 in the PCA, as this axis separated the samples by growth condition. Putative stress-related genes, including those annotated as RpoE, universal stress protein, and DNA starvation/stationary phase protection protein were positively correlated with PC1, as were transposases and a number of genes encoding proteins with T9SS sorting domains, indicating that these functions had higher expression in stationary phase grown cells, pelleted cells, and within murine abscesses than in the periodontitis metatranscriptomes or during logarithmic growth. In contrast, genes encoding for the heme import system (HmuRY), tetrahydrofolate metabolism, histidine degradation, and transcription- and translation-related proteins were negatively correlated with PC1, indicating that these functions had higher expression in periodontitis and logarithmic growth datasets than in the stationary phase, pelleted, and murine abscess datasets. Thus, while P. gingivalis does not have a distinct periodontitis transcriptome, cells in situ are more transcriptionally similar to those in actively growing cultures than those in nutrient-limited, nongrowing cultures or in the murine abscess.
Growth in Midlogarithmic Phase and on Agar Plates Are Highly Transcriptionally Accurate Infection Models.
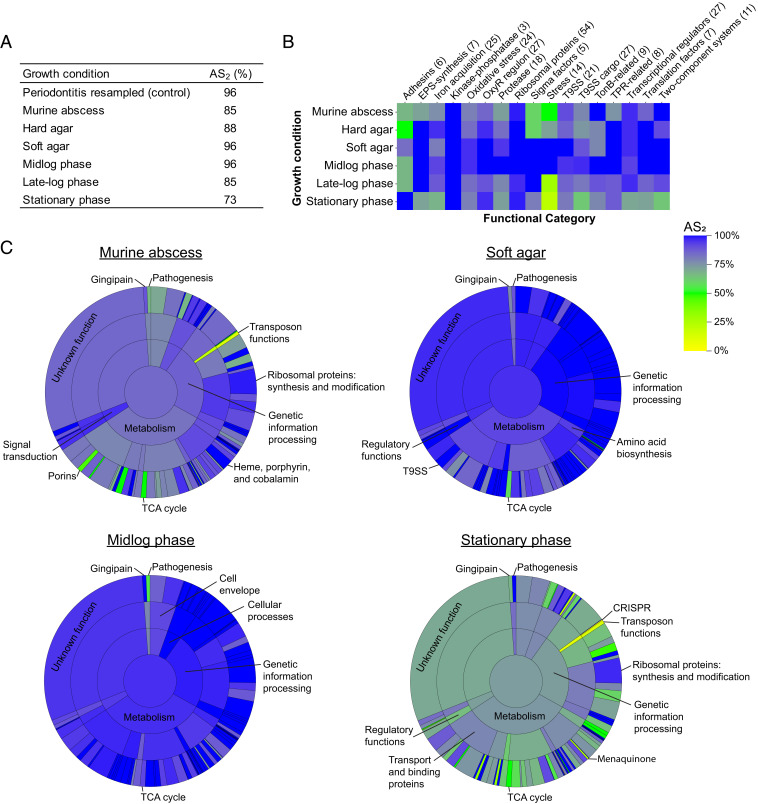
To quantify the extent to which P. gingivalis gene expression in each model mimics that in the human oral cavity, we used an accuracy score (AS) framework recently developed in our laboratory (30). This quantitative framework determines the fraction of genes in an experimental model whose normalized expression falls within a specified number of SDs of the mean of the expression in situ (periodontitis in this case), providing an easily interpretable gauge of model performance. As in our previous studies, we have chosen to focus on genes whose expression falls within two SDs (termed AS2) of the mean in the in situ metatranscriptomes, as two SDs encompass the expression range of ∼95% of the in situ samples for each gene. For example, if a model has an AS2 of 90%, then expression of 90% of a microbe’s genes fall within two SDs of the means of the genes in the in situ metatranscriptomes. While the AS score can be calculated using other SD ranges, such as an AS1 or AS1.5, the false-negative rate increases with more stringent criteria; for example, an AS1.5 has a false-negative rate of ∼13%, while the false-negative rate for AS2 is ∼5% (30). This AS metric is valuable in that it uses P. gingivalis periodontitis gene expression as a benchmark, and it determines accuracy of model systems using all of the core genes.
Before assessing the accuracy of model systems, we performed a control experiment to assess the accuracy of subsampled P. gingivalis periodontitis transcriptomes with the rationale that these analyses will provide an upper AS2 benchmark for the experimental models. To accomplish this, we calculated the AS2 for P. gingivalis gene expression in the periodontitis metatranscriptomes by randomly choosing two P. gingivalis periodontitis transcriptomes as the “model” and comparing their gene expression to the remaining 10 periodontitis transcriptomes and then repeating this analysis 500 times. This analysis resulted in a mean AS2 value of 96%, indicating that across the periodontitis transcriptomes, 96% of the core genes fell within two SDs of the mean expression (Fig. 5A, periodontitis resampled). These results set the upper benchmark for model systems at 96%, as this is the accuracy of the periodontitis samples themselves.
Fig. 5.
Accuracy of P. gingivalis gene expression in experimental model systems relative to periodontally diseased patient samples. (A) Accuracy of relevant growth conditions compared to gene expression during periodontitis based on the percentage of genes that fall within two SDs of the mean gene expression during periodontitis (AS2). Periodontitis resampled is a control to measure the upper AS2 benchmark, in which the AS2 was calculated for 500 randomly resampled pairs of periodontitis transcriptomes. (B) AS2 for hand-curated functional categories across growth conditions. Heatmap depicts the AS2 for within each functional category. The number of genes in each functional category is indicated in parentheses. Abbreviations: EPS = extracellular polymeric substance; T9SS = type IX secretion system; TPR = tetratricopeptide repeat. (C) AS2 for hierarchical TIGRFAM functional categories across growth conditions. “Sunburst” plots depict the AS2 at each hierarchical TIGRFAM level for the indicated experimental model systems. The center circle represents the AS2 for all core genes, the innermost ring represents the AS2 for TIGRFAM meta roles, the middle ring represents the AS2 for TIGRFAM main roles, and the outermost ring represents the AS2 for sub roles. The area of each section corresponds to the number of genes in that functional category. See SI Appendix, Fig. S7 for plots for hard agar and late-logarithmic phase growth.
Next, we calculated the AS2 for common growth models (Fig. 5A, and Dataset S3D). The AS2 values for liquid midlogarithmic growth and soft agar were 96%, equal to the benchmark AS2 of the periodontitis samples. Thus, these P. gingivalis in vitro growth conditions are indistinguishable from periodontitis transcriptomes using this metric. In contrast, in the murine abscess, 85% of the P. gingivalis genes fell within two SDs of the mean periodontitis expression, and stationary phase growth had the lowest AS2 of 73%. When the AS2 was calculated for individual replicates, the P. gingivalis transcriptional profile during growth in soft agar and in liquid midlogarithmic phase were significantly more accurate than that in stationary phase (Dunn’s multiple comparison test, Padj < 0.001), and growth on soft agar was significantly more accurate than in murine abscesses (Dunn’s multiple comparison test, Padj = 0.04; SI Appendix, Fig. S7A).
Accuracy of Experimental Growth Conditions across Functional Categories.
To understand the transcriptional changes driving these differences in accuracy score, we determined the AS2 for individual functional categories (Fig. 5 B and C, SI Appendix, Fig. S7B, and Dataset S3D). These categories include the hand-curated functional classifications used in Fig. 3B and hierarchical TIGRFAM categories, which were hand curated to assign putative sub roles, main roles, and meta roles for all core P. gingivalis genes (Dataset S1) (30–32). Transcript levels of metabolic functions were highly similar between midlogarithmic phase growth and periodontitis, but metabolism is largely dysregulated in stationary phase relative to periodontitis (Fig. 5C). An especially inaccurate metabolic functional category was the tricarboxylic acid (TCA) cycle, which only had high accuracy during midlogarithmic growth (AS2 = 92%) and was the least accurate functional category during growth on soft agar (AS2 = 58%). Low accuracy in the TCA cycle in most growth conditions was due to lower expression in periodontitis of an oxaloacetate decarboxylase, three subunits of the fumarate reductase/succinate dehydrogenase, three subunits of the 2-oxoglutarate oxidoreductase, and an adjacent ferredoxin. In addition, the TIGRFAM meta role “genetic information processing” had decreased accuracy in stationary phase relative to other conditions, including low AS2 values for transposon functions (13%, overexpressed relative to periodontitis), protein stabilization–related genes (47%, predominately overexpressed relative to periodontitis), and transfer RNA aminoacylation genes (60%, both over- and underexpressed relative to periodontitis; Fig. 5C). This is also evident in the low accuracy of the “stress” functional category (AS2 = 21%), which includes overexpressed genes in stationary phase relative to periodontitis encoding for chaperones, Clp protease, universal stress protein, and DNA starvation/stationary phase protection protein (Fig. 5B). However, ribosomal proteins and oxidative stress genes within the OxyR regulon had high accuracy across growth conditions (Fig. 5 B and C).
Virulence-related functions also varied in their accuracy scores. The T9SS AS2 values ranged from 77% in stationary phase transcriptomes to 100% for soft agar transcriptomes (Fig. 5 B and C). In addition, the accuracy scores varied for genes encoding the Arg- and Lys-gingipains and gingipain-associated proteins; the gingipain functional category had an AS2 of 100% in midlogarithmic phase transcriptomes but 67% in stationary phase. Finally, adhesins (Fig. 5B) and pathogenesis (Fig. 5C) had low accuracy in mid- and late-logarithmic phase and a higher accuracy in stationary phase. It is likely that strain differences in fimbriae expression influence these differences, as stationary phase transcriptomes included in the accuracy score analysis were derived using the closely related P. gingivalis strains ATCC 33277 and 381, while the transcriptomes from mid- and late-logarithmic growth were derived using strain W83 (Fig. 1 and Table 2), and W83 is known to have lower fimbriae expression (33). However, strain differences did not fully explain low accuracy in the adhesin and pathogenesis categories, as the murine abscess, infected with P. gingivalis ATCC 33277, had an AS2 of 67% in both adhesins and pathogenesis functional categories. Also, hard agar and soft agar each have replicates from both strain types, but differ in their AS2 for these functions.
We also identified four genes that were not accurately captured by any of the experimental models within two SDs. These genes encode three hypothetical proteins, including one conserved hypothetical protein thought to be involved in iron acquisition, and rubrerythrin, which is important for oxidative stress. Together, this analysis shows that across the majority of functional categories, midlogarithmic growth in liquid culture and growth on soft agar largely recapitulate the P. gingivalis gene expression patterns in the human oral cavity.
P. gingivalis Gene Expression Has Low Variance.
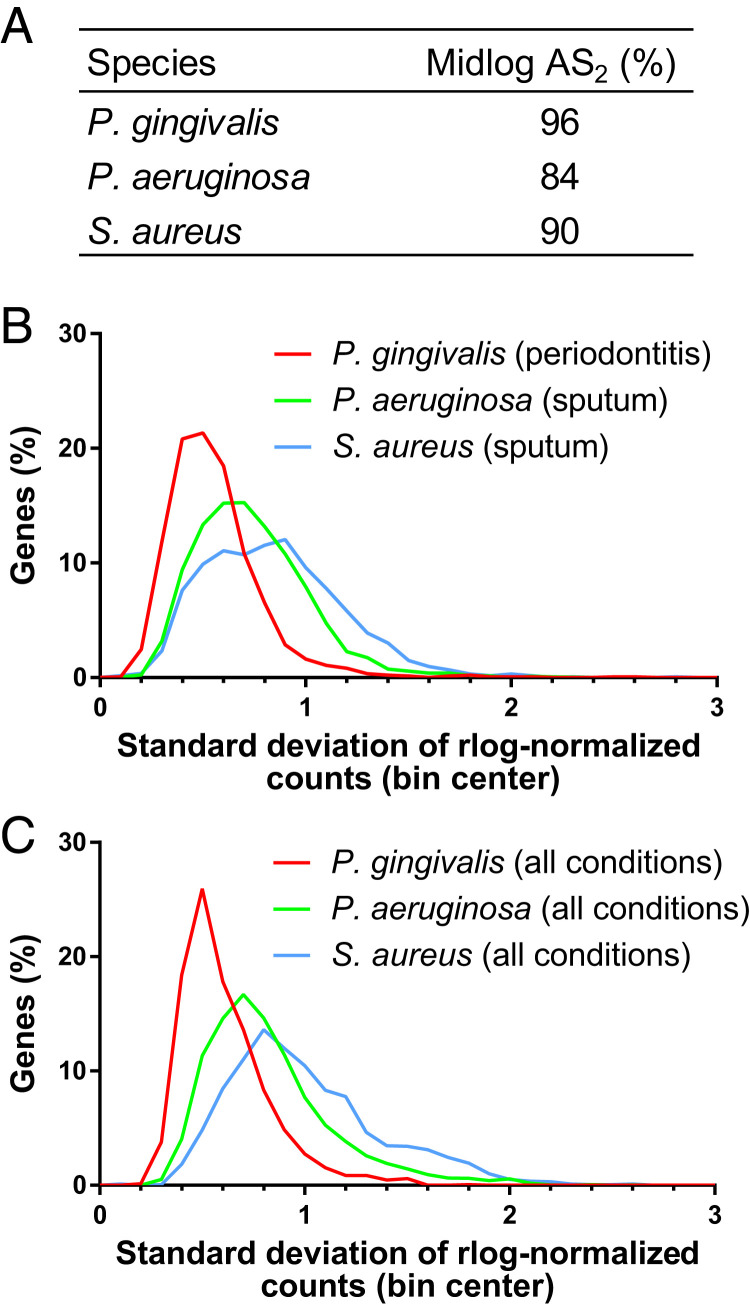
Our analyses showed that for almost all functional categories, the P. gingivalis transcriptome was highly similar between midlogarithmic phase growth, growth on soft agar, and during periodontal disease (Figs. 4 and 5 and SI Appendix, Fig. S6). In contrast, similar analyses for other microbes have found distinct gene expression profiles between human infection and growth in laboratory or animal models using PCAs (1–3). These distinct profiles result in an AS2 of 84 and 90% for P. aeruginosa and S. aureus in midlogarithmic growth, respectively (Fig. 6A). In addition, an analysis of P. aeruginosa across diverse experimental model systems found AS2 values ranging from 81 to 86% for all genes (30).
Fig. 6.
Gene expression variance of P. gingivalis, P. aeruginosa, and S. aureus in human patient samples and in laboratory models. (A) The percentage of genes that fall within two SDs of the mean in situ gene expression (AS2) during midlogarithmic growth. For P. gingivalis, 16 in vitro transcriptomes collected during midlogarithmic growth were compared to 12 periodontitis metatranscriptomes; for P. aeruginosa, 11 in vitro transcriptomes collected during midlogarithmic growth were compared to 20 sputum metatranscriptomes from people with cystic fibrosis; and for S. aureus, 14 in vitro transcriptomes collected during midlogarithmic growth were compared to 10 sputum metatranscriptomes from people with cystic fibrosis. (B) Histogram of the variation in gene expression across human samples. Histogram was constructed with a bin size of 0.1 using the variation in rlog-normalized bacterial gene expression levels across metatranscriptomes collected from periodontitis (P. gingivalis; n = 12) and from cystic fibrosis sputum (P. aeruginosa and S. aureus; n = 20 and n = 10, respectively). (C) Histogram of the variation in gene expression across diverse growth conditions. Histogram was constructed with a bin size of 0.1 using rlog-normalized bacterial gene expression levels across diverse sample types, including human metatranscriptomes, animal models, and in vitro experimental models. A total of 134 samples were analyzed for P. gingivalis, 92 for P. aeruginosa, and 34 for S. aureus.
Therefore, we wanted to better understand the high accuracy scores for P. gingivalis in certain experimental model systems. First, as the AS2 metric is based on the variance of the in situ gene expression, high accuracy of an experimental model system could result from high in situ gene expression variance. To ask if the high AS2 scores were due to high variance in P. gingivalis gene expression during periodontitis, we recalculated the AS2 scores, excluding 54 genes with a high SD during periodontitis. The resulting accuracy scores of the experimental models remained high (SI Appendix, Fig. S8). In addition, we found that the range of the SD of rlog-normalized read counts was smaller for P. gingivalis transcriptomes from periodontitis samples than for P. aeruginosa or S. aureus transcriptomes from sputum samples from people with cystic fibrosis (Fig. 6B). Thus, the high P. gingivalis accuracy scores are not due to high variance in periodontitis, and in fact, P. gingivalis has a smaller in situ gene expression variance than these other microbes.
Alternatively, we asked if the high accuracy scores of P. gingivalis were due to overall smaller deviations in gene expression for this microbe. We calculated the SD of the rlog-normalized expression of each gene for P. gingivalis, P. aeruginosa, and S. aureus across a wide range of growth conditions, including in situ growth, mouse models, and laboratory growth conditions (Fig. 6C). The SD of P. gingivalis gene expression had a smaller distribution than that for the other two microbes. Specifically, for P. gingivalis, 95% of genes across all conditions had an SD of 1 or less. In contrast, for P. aeruginosa and S. aureus, the 95th percentile was an SD of 1.5 and 1.7, respectively. Together, these findings indicate that the gene expression levels for P. gingivalis are more stable across growth conditions and provide insight into the high similarity of the P. gingivalis transcriptome between rich media in laboratory conditions and the human oral cavity.
Discussion
Next-generation sequencing has provided a portal to understanding the behavior of microbes in their native environments. This study leverages 93 publicly available metatranscriptomes from the human oral cavity to further our understanding of the physiology of P. gingivalis during infection and to identify which experimental model systems best encapsulate this physiology. We discovered that P. gingivalis in the diseased oral cavity has highly similar gene expression to that during midlogarithmic phase in vitro growth. This finding is in contrast to previous work comparing gene expression between infection and laboratory models for diverse bacterial pathogens (1–3, 7, 30). Thus, while great effort is required to develop accurate in vitro models for other opportunistic pathogens such as P. aeruginosa (1, 30, 34, 35), P. gingivalis human gene expression is highly mimicked in a common, easily accessible laboratory model. These data also provide strong support for a need to continuously and quantitatively evaluate current model systems using sceintific data, rather than altering current models or developing new models based on intuition. We anticipate that for many bacteria, particularly niche specialists such as P. gingivalis, simple in vitro growth conditions may be highly useful, biologically relevant model systems.
Our discovery that P. gingivalis gene expression during periodontitis is similar to that in logarithmic in vitro cultures offers important context for understanding the biology of this pathogen. First, these results provide biological relevance for the thousands of in vitro and in vivo experimental results published on this bacterium. The fact that a midlogarithmic test tube model highly recapitulates gene expression in situ significantly advances our understanding of this bacterium during periodontal infection. Logarithmic growth in vitro is generally the most common system used to study P. gingivalis and most other bacteria, and there are decades of data on P. gingivalis physiology during logarithmic phase growth. Furthermore, laboratory growth in rich media offers an accessible, inexpensive experimental model for testing future hypotheses. Second, while one might expect the oral cavity to be a hostile environment because of competition for resources with other microbes and interactions with the host immune system, our findings indicate that in the periodontitis samples analyzed here, P. gingivalis is growing in a nutrient-rich, relatively stress-free environment. Although the transcriptional profile of P. gingivalis was comparably stable during periodontitis in this sample set (Figs. 4 and 6), we cannot rule out that other experimental models best capture P. gingivalis growth under certain conditions, for instance, at very low abundance in the oral cavity or at alternative disease sites such as the human brain in connection to Alzheimer's disease (36). Thus, this study provides a framework for future analyses of how the transcriptome of P. gingivalis changes relative to its abundance, across disease sites, over longitudinal studies of disease and treatment, or in the presence of different coinfecting microbes. Also, this study focused on the transcription of P. gingivalis during disease at the population level, primarily using core genes, and it would be interesting in the future to consider the role of strain-level differences and population-level heterogeneity as well as the accuracy of accessory genes.
In addition, we have specifically identified experimental model systems that best capture the few genes not well mimicked in logarithmic phase in vitro cultures (Fig. 5 and Dataset S3). For example, the murine abscess model was not highly accurate overall, including increased expression of stress-related genes, increased nutrient limitation, and decreased fimbria expression in the abscesses relative to periodontitis (Fig. 5 B and C); however, there are 49 genes captured by this model that were not accurately mimicked by midlogarithmic phase in vitro cultures (Dataset S3). These genes include a putative hemolysin virulence factor as well as several metabolic genes. Thus, if one is interested in this small subset of genes, the mouse would be a preferred model over midlogarithmic phase in vitro culture. Our accuracy score framework also presents future opportunities to characterize the utility of other animal models (37). Moreover, while gene expression is a major determinant of physiology, certain biological questions may additionally require the accurate capture of factors such as gene essentiality, bacterial fitness, or host–microbe interactions. For example, studying disease outcomes requires the use of animal models. Furthermore, a similar accuracy score approach could be used to compare host gene expression between human infection and animal models of periodontitis.
In our framework, the accuracy of experimental models depends both on the variance of gene expression in situ and the differences in gene expression between in situ and laboratory growth. A model could be considered accurate because its gene expression is highly similar to that in situ or because the in situ gene expression is so highly variable that it encapsulates large deviations in model gene expression. We found that P. gingivalis accuracy is likely due to the former, as it has low variability in gene expression across environments, including periodontitis (Fig. 6). Despite this low variability, in vitro stationary-phase cells had a low AS2 (73%), indicating a specific change in gene expression at this growth phase compared to periodontitis. In contrast, the high AS2 for S. aureus during midlogarithmic growth (90%) was due to highly variable gene expression within the in situ sputum samples (Fig. 6). We hypothesize that these differences in S. aureus and P. gingivalis gene expression variability result from the fact that S. aureus is a generalist and P. gingivalis a specialist. Indeed, S. aureus is an opportunistic pathogen that can be found in multiple sites on the body and on multiple hosts (38). In comparison, P. gingivalis is specific to the oral cavities of humans and old-world primates and is predominantly an oral pathogen (Fig. 2) (39). Although P. gingivalis has been proposed to contribute to disease at other body sites, fitness in these alternative sites is likely not a primary driver of this microbe’s evolution. Therefore, it is not unreasonable that a bacterium that is highly niche specific has evolved stable gene expression patterns to be fit in its environment, and these stable expression patterns are manifested in other, nonnative environments. We propose that P. gingivalis has not evolved the transcriptomic plasticity to adapt to diverse habitats, and this smaller regulatory need is also evidenced by the small regulon in P. gingivalis relative to many other bacteria, even relative to genome size (40, 41).
In sum, we further a recently developed framework for studying bacterial in situ gene expression, relative to experimental model systems, including the analyses to determine minimum read length for mapping and the accuracy score metric (30). Through curating hundreds of datasets, this work reveals that P. gingivalis gene expression during periodontitis is highly similar to midlogarithmic in vitro cultures, providing strong evidence for the use of a simple test tube model as the gold standard model for studying P. gingivalis biology. These results have significant implications to bacteriology, as they provide a framework to quantitatively assess the biological relevance of the tens of thousands of in vitro experiments performed in this basic laboratory model system.
Materials and Methods
A summary of the analysis approach is available in SI Appendix, Fig. S1.
Pangenome Construction.
The phylogeny of 62 P. gingivalis strains (all strains in The National Center for Biotechnology Information [NCBI] excluding duplicates as of January 2021) was built in Kbase using the “Build Microbial SpeciesTree” app version 1.7.1 (42). Briefly, this analysis constructs a tree using 49 core genes defined by COG gene families. These core genes were inserted into curated multiple sequence alignments for each COG family, the alignments were concatenated, and the maximum likelihood phylogenetic tree was constructed using FastTree2 version 2.1.10 (43). The tree was annotated using Interactive Tree Of Life (iTOL) v5 (44). Strains chosen for the pangenome included the 20 strains with complete genomes and seven additional genomes to ensure coverage of the species diversity (Fig. 1 and Dataset S1). Gene clusters (orthologs) were constructed for these 27 P. gingivalis genomes using Roary with the flag -i 90 (45). After minimal manual curation, pangenome locus tags were assigned based on the genome fragment order in the Roary output (Dataset S1). TIGRFAM annotations for each gene were assigned using KBase, Kyoto Encyclopedia of Genes and Genomes (KEGG) annotations were assigned using both BlastKOALA and KofamKOALA, and COG terms were assigned using KEGG’s binary relationships to COG in the BRITE database (46, 47).
Mock Metatranscriptome Analysis.
We built our mock metatranscriptome using 658 genomes downloaded from the Human Oral Microbiome Database with habitat labeled as “Oral” or “Nasal, Oral” spanning 82 genera and including 19 P. gingivalis genomes, 10 of which are also in our pangenome (48). Art v.2.5.8 was used to simulate metatranscriptomic reads from the coding sequences of this mock community at 10 lengths varying from 15 to 50 bp with 1× coverage (49). The reads were mapped using Bowtie2 v2.3.5 to the P. gingivalis ATCC 33277 genome and to concatenated genomes of the 27 strains in the pangenome (50). This workflow is available at https://github.com/glew8/Pgingivalis_Metatranscriptome_Analyses.
Murine Abscess RNA-Seq.
The murine inner thigh abscess was performed by inoculating Balb/c mice with 1.5 × 109 P. gingivalis ATCC 33277 cells collected in the midlogarithmic growth phase and washed with sterile, prereduced phosphate-buffered saline (PBS). Abscess material was collected 48 h postinoculation by making a small incision adjacent to the abscess and collecting the secreted material using a sterile cotton swab which was then dispersed into sterile, prereduced PBS. Each sample was briefly centrifuged at 150 × g for 3 min at 4 °C to pellet any eukaryotic cells present from the abscess, after which the supernatant was centrifuged at 5,000 × g at 4 °C for 5 min to pellet bacterial cells. After removing the supernatant, the bacterial pellet was used for total RNA extraction using the Invitrogen TRIzol Max Bacterial RNA Isolation Kit (Thermo Fisher Scientific) following the manufacturer’s protocol. Ribosomal RNA depletion, strand-specific library construction, and 150-bp paired-end RNA sequencing were performed by Novogene using the Illumina NovaSeq 6000 platform.
Quality Analyses, Mapping, and Counting of RNA-Seq Datasets.
Using only the forward reads from each dataset if paired end, RNA-seq sequencing read quality was confirmed with FastQC v0.11.8 (51). Reads were trimmed to remove adapters from the 3′ end of the reads using Cutadapt 2.6 (AGA TCG GAA GAG CAC ACG TCT GAA CTC CAG TCA C and AAG TCG GAG GCC AAG CGG TCT TAG GAA GAC AA for Illumina- and BGISEQ-sequenced reads, respectively) (52). Trimmed reads that were at least 22 bp were mapped to the concatenated genomes of 27 P. gingivalis strains (Dataset S1) using Bowtie2 v2.3.5 with default parameters (50). featureCounts (subread-2.0.1) was used to assign reads to protein-coding genes with the flags -s 0 (unstranded) and -O (allowMultiOverlap) in R 4.0.2 (53, 54) so that each read was assigned to a single locus or to neighboring genes. Then, read counts were summed for each ortholog to account for strain-level differences across samples. At each step, MultiQC v1.9 was used to track analysis quality (55).
MetaPhlAn and StrainPhlAn Analyses.
The taxonomy of the human periodontitis datasets was estimated using MetaPhlAn 3.0.6 with marker gene version mpa_v30_CHOCOPhlAn_201901, the minimum read length set to 22 bp, and viruses included (56). The dominant P. gingivalis strain in the metatranscriptomes, relative to the 27 reference genomes in the pangenome, was determined using StrainPhlAn 3.0 with the minimum read length set to 22 bp (56, 57). iTOL v5 was used to visualize the population structure (44).
Gene Expression Analyses.
Gene counts were normalized using TPM by dividing the raw counts by the average length for each ortholog to determine reads per kilobase (RPK), summing the RPK for each sample and dividing by 1,000,000 to determine the scaling factor, and then dividing the RPK by the scaling factor for each gene. Highly expressed genes based on TPM were identified using R package inflection version 1.3.4 (58). Enrichment was determined using a two-sided Fisher’s exact test, and P values were corrected for multiple testing using the Benjamini–Hochberg method in R. rlog normalization of core genes was performed on all metatranscriptomes and transcriptomes together using DESeq2 with blind = TRUE in R (59). The PCA was built using rlog-normalized counts of the 500 core genes with highest variability across samples using the command plotPCA in DESeq2. The Euclidian distance matrix was calculated from the rlog-normalized counts of the 1,500 core genes using the R function dist, and the heatmap was produced in pheatmap version 1.0.12 (60).
AS Analyses.
Accuracy scores were calculated and graphed using rlog-normalized read counts for the 1,500 P. gingivalis core genes (Dataset S1C) in R version 4.0.2 with the following packages: tidyverse version 1.3.0, cowplot version 1.0.0, readr version 1.3.1, dplyr version 1.0.2, tidyr version 1.1.2, tibble version 3.0.3, purrr version 0.3.4, ggsunburst version 0.3.0, zeallot version 0.1.0, ggplot2 version 3.3.2, and reshape version 0.8.8 (61–71). Scripts are modified from Cornforth et al. and are available at https://github.com/glew8/Pgingivalis_Metatranscriptome_Analyses (30). This analysis calculates the mean and SD of normalized read counts for each gene in a target environment, in this case periodontitis. Then, using these values, the analysis determines the number of SDs away from the target mean gene expression for each gene in each replicate in a model (the z-score). The median z-score across replicates is identified for each gene, and this value is outputted as the “penalty.” Finally, the AS2 is calculated by determining the percentage of penalties that fall between −2 and 2. For AS2 calculations across functional categories, the approach is the same, but only the genes within the given functional category are considered. To understand how transcriptionally variable genes during periodontitis impact the AS, the AS2 analysis was also performed excluding 54 genes with a SD of rlog-normalized read counts greater than 1 across the 12 periodontitis samples (Dataset S3C).
P. aeruginosa and S. aureus Analysis.
Raw read counts of P. aeruginosa transcriptomes in experimental model systems, human sputum, and human wounds were obtained from Cornforth et al. (30). P. aeruginosa analyses were limited to 4,945 soft core genes that were 1) present in 277 of 291 (95%) high-quality genomes as analyzed by Roary using the same parameters as the P. gingivalis pangenome construction above and 2) had orthologs in P. aeruginosa PAO1. Raw read counts of S. aureus transcriptomes in experimental model systems, human sputum, and human wounds were obtained from Ibberson and Whiteley (2). S. aureus analyses were limited to 1,960 core genes in a 15-strain pangenome (2). Raw gene counts were rlog normalized with DESeq2, and the AS2 analysis was performed.
Supplementary Material
Acknowledgments
We thank the M.W. laboratory for the discussion of this manuscript, especially Daniel Cornforth for his valuable suggestions. This research was supported in part through research cyberinfrastructure resources and services provided by the Partnership for an Advanced Computing Environment at the Georgia Institute of Technology, Atlanta, GA. This work was supported by NIH Grants R01DE023193 to M.W. and R.J.L., R01DE020100 to M.W., F32DE027281 and K99DE031018 to G.R.L., and R01DE011111 and R01DE012505 to R.J.L.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2116637119/-/DCSupplemental.
Data Availability
RNA-seq reads from P. gingivalis infection for the five murine abscesses are available in the NCBI Sequence Read Archive in BioProject PRJNA762090 at https://www.ncbi.nlm.nih.gov/bioproject/762090. The other 210 datasets used in this publication are previously published as shown in Tables 1 and 2, and Dataset S2. The orthologs and curated annotations for the pangenome are available in Dataset S1. P. aeruginosa and S. aureus datasets are available from refs. 2 and 30.
References
- 1.Cornforth D. M., et al. , Pseudomonas aeruginosa transcriptome during human infection. Proc. Natl. Acad. Sci. U.S.A. 115, E5125–E5134 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ibberson C. B., Whiteley M., The Staphylococcus aureus transcriptome during cystic fibrosis lung infection. MBio 10, e02774-19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Frick-Cheng A. E., et al. , The gene expression profile of uropathogenic Escherichia coli in women with uncomplicated urinary tract infections is recapitulated in the mouse model. MBio 11, e01412-20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lamont R. J., Koo H., Hajishengallis G., The oral microbiota: Dynamic communities and host interactions. Nat. Rev. Microbiol. 16, 745–759 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Solbiati J., Frias-Lopez J., Metatranscriptome of the oral microbiome in health and disease. J. Dent. Res. 97, 492–500 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Duran-Pinedo A. E., Metatranscriptomic analyses of the oral microbiome. Periodontol. 2000 85, 28–45 (2020). [DOI] [PubMed] [Google Scholar]
- 7.Deng Z. L., Sztajer H., Jarek M., Bhuju S., Wagner-Döbler I., Worlds apart - Transcriptome profiles of key oral microbes in the periodontal pocket compared to single laboratory culture reflect synergistic interactions. Front. Microbiol. 9, 124 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bostanci N., Belibasakis G. N., Porphyromonas gingivalis: An invasive and evasive opportunistic oral pathogen. FEMS Microbiol. Lett. 333, 1–9 (2012). [DOI] [PubMed] [Google Scholar]
- 9.Darveau R. P., Hajishengallis G., Curtis M. A., Porphyromonas gingivalis as a potential community activist for disease. J. Dent. Res. 91, 816–820 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Belstrøm D., et al. , Metagenomic and metatranscriptomic analysis of saliva reveals disease-associated microbiota in patients with periodontitis and dental caries. NPJ Biofilms Microbiomes 3, 23 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Duran-Pinedo A. E., et al. , Community-wide transcriptome of the oral microbiome in subjects with and without periodontitis. ISME J. 8, 1659–1672 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jorth P., et al. , Metatranscriptomics of the human oral microbiome during health and disease. MBio 5, e01012-14 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nowicki E. M., et al. , Microbiota and metatranscriptome changes accompanying the onset of gingivitis. MBio 9, e00575-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Szafrański S. P., et al. , Functional biomarkers for chronic periodontitis and insights into the roles of Prevotella nigrescens and Fusobacterium nucleatum; A metatranscriptome analysis. NPJ Biofilms Microbiomes 1, 15017 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yost S., Duran-Pinedo A. E., Teles R., Krishnan K., Frias-Lopez J., Functional signatures of oral dysbiosis during periodontitis progression revealed by microbial metatranscriptome analysis. Genome Med. 7, 27 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Van der Velden U., The Dutch periodontal screening index validation and its application in The Netherlands. J. Clin. Periodontol. 36, 1018–1024 (2009). [DOI] [PubMed] [Google Scholar]
- 17.Belvin B. R., Gui Q., Hutcherson J. A., Lewis J. P., The Porphyromonas gingivalis hybrid cluster protein Hcp is required for growth with nitrite and survival with host cells. Infect. Immun. 87, e00572-18 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cheng T., et al. , Bismuth drugs tackle Porphyromonas gingivalis and attune cytokine response in human cells. Metallomics 11, 1207–1218 (2019). [DOI] [PubMed] [Google Scholar]
- 19.Coats S. R., et al. , The distinct immune-stimulatory capacities of Porphyromonas gingivalis strains 381 and ATCC 33277 are determined by the fimB allele and gingipain activity. Infect. Immun. 87, e00319-19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dou Y., et al. , Role of extracytoplasmic function sigma factor PG1660 (RpoE) in the oxidative stress resistance regulatory network of Porphyromonas gingivalis. Mol. Oral Microbiol. 33, 89–104 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hendrickson E. L., et al. , Insights into dynamic polymicrobial synergy revealed by time-coursed RNA-Seq. Front. Microbiol. 8, 261 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Høvik H., Yu W. H., Olsen I., Chen T., Comprehensive transcriptome analysis of the periodontopathogenic bacterium Porphyromonas gingivalis W83. J. Bacteriol. 194, 100–114 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jain S., et al. , Identification of PGN_1123 as the gene encoding lipid A deacylase, an enzyme required for Toll-like receptor 4 evasion, in Porphyromonas gingivalis. J. Bacteriol. 201, e00683-18 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kin L. X., et al. , Metabolic cooperativity between Porphyromonas gingivalis and Treponema denticola. J. Oral Microbiol. 12, 1808750 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miller D. P., Wang Q., Weinberg A., Lamont R. J., Transcriptome analysis of Porphyromonas gingivalis and Acinetobacter baumannii in polymicrobial communities. Mol. Oral Microbiol. 33, 364–377 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moradali M. F., Ghods S., Angelini T. E., Davey M. E., Amino acids as wetting agents: Surface translocation by Porphyromonas gingivalis. ISME J. 13, 1560–1574 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moye Z. D., Gormley C. M., Davey M. E., Galactose impacts the size and intracellular composition of the asaccharolytic oral pathobiont Porphyromonas gingivalis. Appl. Environ. Microbiol. 85, e02268-18 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shen D., et al. , Role of the RprY response regulator in P. gingivalis community development and virulence. Mol. Oral Microbiol. 35, 231–239 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sztukowska M. N., et al. , Community development between Porphyromonas gingivalis and Candida albicans mediated by InlJ and Als3. MBio 9, e00202-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cornforth D. M., Diggle F. L., Melvin J. A., Bomberger J. M., Whiteley M., Quantitative framework for model evaluation in microbiology research using Pseudomonas aeruginosa and cystic fibrosis infection as a test case. MBio 11, e03042-19 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fuchs S., et al. , AureoWiki—The repository of the Staphylococcus aureus research and annotation community. Int. J. Med. Microbiol. 308, 558–568 (2018). [DOI] [PubMed] [Google Scholar]
- 32.Haft D. H., et al. , TIGRFAMs and genome properties in 2013. Nucleic Acids Res. 41, D387–D395 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kuboniwa M., Inaba H., Amano A., Genotyping to distinguish microbial pathogenicity in periodontitis. Periodontol. 2000 54, 136–159 (2010). [DOI] [PubMed] [Google Scholar]
- 34.Turner K. H., Wessel A. K., Palmer G. C., Murray J. L., Whiteley M., Essential genome of Pseudomonas aeruginosa in cystic fibrosis sputum. Proc. Natl. Acad. Sci. U.S.A. 112, 4110–4115 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Palmer K. L., Aye L. M., Whiteley M., Nutritional cues control Pseudomonas aeruginosa multicellular behavior in cystic fibrosis sputum. J. Bacteriol. 189, 8079–8087 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dominy S. S., et al. , Porphyromonas gingivalis in Alzheimer’s disease brains: Evidence for disease causation and treatment with small-molecule inhibitors. Sci. Adv. 5, eaau3333 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Genco C. A., Van Dyke T., Amar S., Animal models for Porphyromonas gingivalis-mediated periodontal disease. Trends Microbiol. 6, 444–449 (1998). [DOI] [PubMed] [Google Scholar]
- 38.Richardson E. J., et al. , Gene exchange drives the ecological success of a multi-host bacterial pathogen. Nat. Ecol. Evol. 2, 1468–1478 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ménard C., Mouton C., Clonal diversity of the taxon Porphyromonas gingivalis assessed by random amplified polymorphic DNA fingerprinting. Infect. Immun. 63, 2522–2531 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lozada-Chávez I., Janga S. C., Collado-Vides J., Bacterial regulatory networks are extremely flexible in evolution. Nucleic Acids Res. 34, 3434–3445 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Madan Babu M., Teichmann S. A., Aravind L., Evolutionary dynamics of prokaryotic transcriptional regulatory networks. J. Mol. Biol. 358, 614–633 (2006). [DOI] [PubMed] [Google Scholar]
- 42.Arkin A. P., et al. , KBase: The United States Department of Energy systems biology knowledgebase. Nat. Biotechnol. 36, 566–569 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Price M. N., Dehal P. S., Arkin A. P., FastTree 2–Approximately maximum-likelihood trees for large alignments. PLoS One 5, e9490 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Letunic I., Bork P., Interactive tree of life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 47, W256–W259 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Page A. J., et al. , Roary: Rapid large-scale prokaryote pan genome analysis. Bioinformatics 31, 3691–3693 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kanehisa M., Sato Y., Morishima K., BlastKOALA and GhostKOALA: KEGG tools for functional characterization of genome and metagenome sequences. J. Mol. Biol. 428, 726–731 (2016). [DOI] [PubMed] [Google Scholar]
- 47.Aramaki T., et al. , KofamKOALA: KEGG Ortholog assignment based on profile HMM and adaptive score threshold. Bioinformatics 36, 2251–2252 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Escapa I. F., et al. , New insights into human nostril microbiome from the expanded Human Oral Microbiome Database (eHOMD): A resource for the microbiome of the human aerodigestive tract. mSystems 3, e00187-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huang W., Li L., Myers J. R., Marth G. T., ART: A next-generation sequencing read simulator. Bioinformatics 28, 593–594 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Langmead B., Salzberg S. L., Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Andrews S., FastQC: A quality control tool for high throughput sequence data (2010). https://www.bioinformatics.babraham.ac.uk/projects/fastqc/
- 52.Martin M., Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17, 3 (2011). [Google Scholar]
- 53.Liao Y., Smyth G. K., Shi W., featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014). [DOI] [PubMed] [Google Scholar]
- 54.R Core Team, R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, Vienna, Austria, 2020). [Google Scholar]
- 55.Ewels P., Magnusson M., Lundin S., Käller M., MultiQC: Summarize analysis results for multiple tools and samples in a single report. Bioinformatics 32, 3047–3048 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Beghini F., et al. , Integrating taxonomic, functional, and strain-level profiling of diverse microbial communities with bioBakery 3. eLife 10, e65088 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Truong D. T., Tett A., Pasolli E., Huttenhower C., Segata N., Microbial strain-level population structure and genetic diversity from metagenomes. Genome Res. 27, 626–638 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Christopoulos D. T., inflection: Finds the inflection point of a curve (2019).
- 59.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kolde R., pheatmap: Pretty heatmaps (2019).
- 61.Wickham H., et al. , Welcome to the tidyverse. J. Open Source Softw. 4, 1686 (2019). [Google Scholar]
- 62.Wilke C. O., cowplot: Streamlined plot theme and plot annotations for 'ggplot2' (2019).
- 63.Wickham H., Hester J., Francois R., readr: Read rectangular text data (2018).
- 64.Wickham H., François R., Henry L., Müller K., dplyr: A grammar of data manipulation (2020).
- 65.Müller K., Wickham H., tibble: Simple data frames (2020).
- 66.Henry L., Wickham H., purrr: Functional programming tools (2020).
- 67.Santesmasses D., ggsunburst: Adjacency diagrams with ggplot2 (2020).
- 68.Teetor N., zeallot: Multiple, unpacking, and destructuring assignment (2018).
- 69.Wickham H., tidyr: Tidy messy data (2020).
- 70.Wickham H.,ggplot2: Elegant Graphics for Data Analysis (Springer-Verlag New York, 2016). [Google Scholar]
- 71.Wickham H., Reshaping data with the reshape package. J. Stat. Softw. 21, 1–20 (2007). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq reads from P. gingivalis infection for the five murine abscesses are available in the NCBI Sequence Read Archive in BioProject PRJNA762090 at https://www.ncbi.nlm.nih.gov/bioproject/762090. The other 210 datasets used in this publication are previously published as shown in Tables 1 and 2, and Dataset S2. The orthologs and curated annotations for the pangenome are available in Dataset S1. P. aeruginosa and S. aureus datasets are available from refs. 2 and 30.