Abstract
The occurrence of food-borne disease outbreaks in Taiwan increased dramatically in 1996, and the incidence has since remained elevated. This increase in outbreaks is correlated with a high rate of isolation of Vibrio parahaemolyticus, which caused between 61 and 71% of the total outbreaks for the period 1996 to 1999. By serotyping, 40 serovars were identified from 3743 V. parahaemolyticus isolates, of which O3:K6 was the most frequently detected. The O3:K6 serovar could have emerged in Taiwan as early as October 1995 and at that time accounted for only 0.6% of the V. parahaemolyticus infections. This level increased suddenly to 50.1% in 1996 and reached a peak (83.8%) in 1997. Comparison of the outbreak profiles for the etiology groups indicates that the high incidence of food-borne disease outbreaks during 1996 to 1999 can be attributed to the extraordinarily high O3:K6 infections. In 1999, the O3:K6 serovar was still prevalent, and accounted for 61.3% of all V. parahaemolyticus infections. Due to its extraordinarily high infection frequency and its capability to spread globally, this organism needs to be intensively monitored internationally.
Vibrio parahaemolyticus is an important etiologic agent of seafood-borne gastroenteritis and has become a leading cause of food-borne disease outbreaks in Taiwan and Japan (9, 12). In Taiwan, food-borne disease data have been systematically collected by the Department of Health since 1986, with an average of 85 outbreaks per annum having been recorded during the period from 1986 to 1995 (12). From this surveillance, we noticed that the incidence of food-borne disease outbreaks increased suddenly in 1996 and has remained high since this time. In addition, we have observed a correlation between this increase in outbreaks and a high isolation rate for V. parahaemolyticus. In this study, we investigated the causes of this increase by analyzing the etiologic agents responsible for food-borne disease outbreaks occurring in Taiwan from 1995 to 1999.
Specimens such as stool, rectal swab, and nose swab were collected by local health authorities from patients and food handlers involved in the food-borne disease outbreaks and sent to the laboratories of the Center for Disease Control, where bacterial pathogen examinations were conducted. Briefly, the specimens were streaked in selective differential media described for Vibrio cholerae and V. parahaemolyticus (7), Salmonella spp. (16), Staphylococcus aureus (5), Bacillus cereus (13), Shigella spp. (14), Aeromonas spp. (15), and Escherichia coli O157:H7 (6). Suspected colonies were initially identified by biochemical tests with triple sugar iron agar (Eiken Chemical Co., Tokyo, Japan), SIM medium (Eiken), and lysine iron agar (Difco Laboratories, Detroit, Mich.). For screening E. coli O157:H7, suspected E. coli colonies were tested with Clig agar slants (Kyokuto, Tokyo, Japan) instead of triple sugar iron agar. After the screening tests, suspected isolates were further identified with API biochemical test kits (bioMérieux, l'Etoile, France), i.e., API 20E for V. cholerae, V. parahaemolyticus, Salmonella spp., Shigella spp., and Aeromonas spp., API ID32 Staph for S. aureus, and API 50CH for B. cereus. Serologic tests for identification and serotyping were performed routinely with the antisera of O antigens and H antigens for E. coli O157:H7 (Difco) and Salmonella spp. (Denka, Seiken Corp., Tokyo, Japan), the antisera of O1 and O139 antigens for V. cholerae (Denka), the antisera of O antigens for Shigella spp. (Denka), the antisera of K antigens for V. parahaemolyticus (Denka), and the antisera of enterotoxins (SEA to SEE) for S. aureus (Denka). Some isolates of serovars K6, K8, K10, K12, K63, and K68 of V. parahaemolyticus were also serotyped with antisera of O antigens (Denka). Laboratory procedures of the Centers for Disease Control and Prevention, Atlanta, Ga., were followed for examination of Clostridium botulinum (2).
Epidemiological data relating to the food-borne disease outbreaks investigated were obtained from standardized case report forms filled in by the county public health authorities. These reports included the dates, times, and locations of suspected food ingestion, the suspected foods, the persons who consumed the food and were poisoned, and basic information about each patient's sex, age, date and time of onset, residency, clinical symptoms, and medical treatment.
The etiology of a food-borne disease outbreak was confirmed based on comparisons of the clinical symptoms, disease incubation period, implicated foods, and laboratory findings from the specimens from the implicated persons.
Table 1 presents the etiologies of food-borne disease outbreaks occurring in Taiwan during the period from 1995 to 1999. A total of 850 outbreaks were reported, and specimens from these outbreaks were examined by the Center for Disease Control. Bacterial agents were responsible for 610 outbreaks (71.8%), with the remainder (28.2%) due to chemical or unknown agents. Among the bacterial agents, V. parahaemolyticus, Salmonella spp., and S. aureus accounted for 63.8, 5.2, and 2.5% of the total outbreaks, respectively. B. cereus, V. cholerae (non-O1, non-O139 strains), S. sonnei, and Aeromonas sp. combined caused only 1.5% (13) of all bacterial outbreaks. The outbreak caused by S. sonnei was imported. No E. coli O157:H7 and C. botulinum outbreaks were identified during this period.
TABLE 1.
Etiologies of food-borne disease outbreaks occurring in Taiwan from 1995 to 1999
| Etiology | No. of outbreaks (%) in (yr):
|
Total no. of outbreaks (%) | ||||
|---|---|---|---|---|---|---|
| 1995 | 1996 | 1997 | 1998 | 1999 | ||
| Bacterial agentsa | 72 (59.5) | 130 (73.4) | 185 (76.4) | 122 (73.9) | 101 (69.7) | 610 (71.8) |
| Vibrio parahaemolyticus | 54 (44.6) | 119 (67.2) | 171 (70.7) | 110 (66.7) | 88 (60.7) | 542 (63.8) |
| Salmonella spp. | 8 (6.6) | 10 (5.6) | 7 (2.9) | 8 (4.8) | 11 (7.6) | 44 (5.2) |
| Staphylococcus aureus | 7 (5.8) | 2 (1.1) | 6 (2.5) | 4 (2.4) | 2 (1.4) | 21 (2.5) |
| Bacillus cereus | 1 (0.8) | 0 (0.0) | 1 (0.4) | 3 (1.8) | 1 (0.7) | 6 (0.7) |
| Vibrio choleraeb | 4 (3.3) | 0 (0.0) | 1 (0.4) | 0 (0.0) | 0 (0.0) | 5 (0.6) |
| Shigella sonneic | 0 (0.0) | 0 (0.0) | 1 (0.4) | 0 (0.0) | 0 (0.0) | 1 (0.1) |
| Aeromonas sp. | 0 (0.0) | 1 (0.6) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 1 (0.1) |
| Chemical and unknown agents | 49 (40.5) | 47 (26.6) | 57 (23.6) | 43 (26.1) | 44 (30.3) | 240 (28.2) |
| Total outbreaks | 121 | 177 | 242 | 165 | 145 | 850 |
All specimens were examined for the bacterial pathogens Vibrio spp., Salmonella spp., S. aureus, B. aureus, Shigella spp., E. coli O157:H7, and Aeromonas spp.
Non-O1, non-O139 strains.
Imported case.
Compared to 1995, outbreaks increased abruptly from 121 to 177 cases in 1996 and have remained high since this time. The highest incidence of disease outbreaks was in 1997, with a total of 242 outbreaks occurring. The number of outbreaks due to chemical and unknown agents continued to remain steady, ranging from 43 to 57, but outbreaks caused by bacterial agents fluctuated from 72 to 185. This fluctuation in bacterial outbreaks is related to the number of V. parahaemolyticus outbreaks. Outbreaks caused by this agent range from 54 (44.6%) to 171 (70.7%) over the period from 1995 to 1999. In contrast, six other bacterial agents, including Salmonella spp. and S. aureus, caused 13 to 20 outbreaks. These data indicate that the high incidence of food-borne disease outbreaks during 1996 through 1999 can be attributed to V. parahaemolyticus.
To investigate whether a particular V. parahaemolyticus serovar was involved in the sudden increase in food-borne disease outbreaks, the K serotypes of the recovered V. parahaemolyticus isolates were analyzed. Furthermore, some isolates of several major K serovars were also serotyped with antisera of O antigens. Table 2 presents the prevalence of V. parahaemolyticus K serovars in food-borne disease outbreaks in Taiwan from 1995 to 1999 (inclusive). A total of 3,743 V. parahaemolyticus isolates were recovered and serotyped. Of these, 3,729 isolates were grouped into 40 K serovars and 14 isolates were nontypeable. K6 (O3:K6), K12 (O4:K12), K8 (O4:K8), K10 (O4:K10), and K63 (O3:K63) were ranked as the leading serovars, accounting for 82.6% of the total V. parahaemolyticus isolates. The O3:K6 serovar was the most prevalent over this time period. There were 2,234 isolates of O3:K6 identified in 351 of the 542 V. parahaemolyticus outbreaks over the period investigated. In 1995, O3:K6 was identified in a single outbreak with three isolates recovered, accounting for 0.6% of the V. parahaemolyticus identified in that year. However, in 1996, 372 O3:K6 isolates were identified in 66 outbreaks, accounting for 50.1% of the V. parahaemolyticus. The highest incidence of O3:K6 occurred in 1997 and accounted for 83.8% of the total V. parahaemlyticus isolates detected. The incidence of O3:K6 infections decreased in 1998, but this serovar still accounted for 71.5% of the V. parahaemolyticus and 61.3% in 1999. These data indicate that the prevalence of O3:K6 is correlated with the increase in total V. parahaemolyticus outbreaks in Tawian.
TABLE 2.
Prevalence of the V. parahaemolyticus K serovars in food-borne disease outbreaks in Taiwan from 1995 to 1999
| Serovar | No. of isolates (no. of outbreaks) in (yr):
|
Total no. of isolates (no. of outbreaks) | ||||
|---|---|---|---|---|---|---|
| 1995 | 1996 | 1997 | 1998 | 1999 | ||
| K3 | 4 (1) | 1 (1) | 8 (3) | 6 (2) | 7 (2) | 26 (9) |
| K4 | 9 (4) | 19 (5) | 8 (4) | 2 (2) | 38 (15) | |
| K5 | 1 (1) | 6 (2) | 1 (1) | 8 (4) | ||
| K6 | 3 (1) | 372 (66) | 990 (146) | 523 (75) | 346 (63) | 2234 (351) |
| K7 | 1 (1) | 13 (6) | 1 (1) | 1 (1) | 16 (9) | |
| K8 | 62 (12) | 59 (20) | 57 (11) | 16 (8) | 55 (11) | 249 (62) |
| K9 | 4 (2) | 13 (4) | 6 (3) | 6 (3) | 6 (2) | 35 (14) |
| K10 | 11 (4) | 23 (2) | 7 (5) | 92 (20) | 80 (14) | 213 (45) |
| K11 | 3 (2) | 3 (1) | 8 (2) | 14 (5) | ||
| K12 | 264 (11) | 7 (4) | 8 (2) | 5 (2) | 284 (19) | |
| K13 | 2 (2) | 2 (1) | 4 (3) | |||
| K15 | 22 (5) | 20 (6) | 2 (2) | 29 (2) | 2 (2) | 75 (17) |
| K17 | 1 (1) | 1 (1) | 2 (2) | |||
| K18 | 31 (3) | 1 (1) | 1 (1) | 33 (5) | ||
| K19 | 20 (7) | 20 (7) | ||||
| K22 | 12 (3) | 18 (1) | 1 (1) | 31 (5) | ||
| K25 | 5 (2) | 4 (2) | 9 (4) | |||
| K28 | 1 (1) | 1 (1) | ||||
| K29 | 13 (4) | 12 (5) | 3 (1) | 2 (1) | 4 (1) | 34 (12) |
| K32 | 2 (1) | 2 (1) | ||||
| K33 | 1 (1) | 1 (1) | ||||
| K37 | 4 (2) | 1 (1) | 5 (3) | |||
| K38 | 2 (1) | 26 (9) | 2 (2) | 9 (2) | 39 (14) | |
| K41 | 23 (7) | 24 (7) | 3 (2) | 9 (2) | 59 (18) | |
| K44 | 1 (1) | 1 (1) | ||||
| K45 | 1 (1) | 1 (1) | ||||
| K46 | 1 (1) | 1 (1) | ||||
| K48 | 1 (1) | 4 (4) | 1 (1) | 6 (6) | ||
| K53 | 3 (3) | 1 (1) | 1 (1) | 5 (5) | ||
| K54 | 1 (1) | 7 (2) | 8 (3) | |||
| K55 | 1 (1) | 15 (5) | 2 (2) | 18 (8) | ||
| K56 | 32 (4) | 26 (12) | 14 (7) | 2 (2) | 74 (25) | |
| K57 | 2 (1) | 1 (1) | 3 (2) | |||
| K58 | 12 (1) | 12 (1) | ||||
| K59 | 1 (1) | 1 (1) | ||||
| K60 | 3 (1) | 3 (1) | ||||
| K63 | 18 (6) | 72 (18) | 8 (2) | 1 (1) | 13 (2) | 112 (29) |
| K66 | 1 (1) | 1 (1) | ||||
| K68 | 11 (3) | 17 (10) | 18 (9) | 46 (22) | ||
| K69 | 3 (2) | 2 (1) | 5 (3) | |||
| Nontypeable | 2 (1) | 8 (1) | 4 (2) | 14 (4) | ||
| Total | 526 (54) | 742 (119) | 1,181 (171) | 731 (110) | 564 (88) | 3,743 (542) |
| %K6 | 0.6 (1.9) | 50.1 (55.5) | 83.8 (85.4) | 71.5 (68.2) | 61.3 (71.6) | 59.7 (64.8) |
Serovars O4:K8 and O4:K10 caused 107 (19%) outbreaks over the study period. The number of these outbreaks caused by these two serovars showed no significant change during these 5 years. An O4:K68 serovar appeared for the first time in 1997 and had caused 22 outbreaks by 1999. This serovar is rarely implicated in food poisoning, but it had caused six outbreaks in Japan in 1998 (9). The future trend of V. parahaemolyticus O4:K68 infection must be carefully watched.
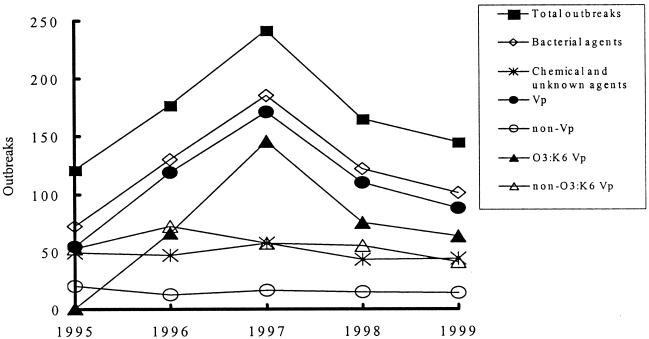
To determine the cause of the high incidence of food-borne disease outbreaks, we compared the outbreak profiles of the classified groups. That is, we compared the yearly total outbreaks with the agent responsible for that outbreak. The agents were classified into etiological groups, either chemical and unknown agents, or bacterial agents (V. parahaemolyticus, non-V. parahaemolyticus, O3:K6 , and non-O3:K6). Figure 1 depicts the profiles of the total outbreaks during the study period and also of the six etiology groups. The outbreak profiles of bacterial agents, V. parahaemolyticus, and O3:K6 coincide with the total outbreaks. In contrast, the outbreak profiles for all other groups showed no correlation with the total outbreak profiles. This profile analysis indicates that the O3:K6 V. parahaemolyticus was the cause of the unusually high incidence of food-borne disease outbreaks over the period from 1996 to 1999.
FIG. 1.
Outbreak profiles of the total outbreaks and the six etiology groups from 1995 to 1999. The etiology groups include bacterial agents, chemical and unknown agents, V. parahaemolyticus (Vp), non-V. parahaemolyticus (non-Vp) (consisting of Salmonella spp., S. aureus, B. cereus, V. cholerae, S. sonnei, and Aeromonas sp.), O3:K6 V. parahaemolyticus (O3:K6 Vp), and non-O3:K6 V. parahaemolyticus (non-O3:K6 Vp).
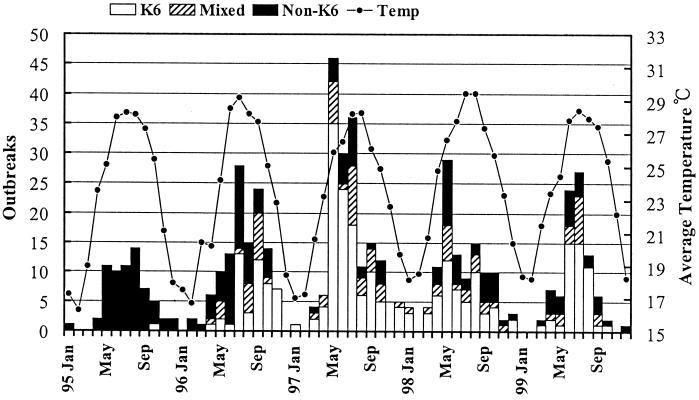
Figure 2 depicts the numbers of monthly food-borne disease outbreaks caused by the V. parahaemolyticus O3:K6 serovar, mixed serovars (consisting of the O3:K6 serovar and other non-O3:K6 serovars), and non-O3:K6 serovars and compares these with the average temperature in Taiwan. V. parahaemolyticus outbreaks were high between May and September for each year investigated. However, frequent outbreaks caused by V. parahaemolyticus were also reported in April and October. The occurrence of V. parahaemolyticus food-borne disease outbreaks can be correlated to temperature. Outbreaks tend to be more prevalent in late spring to early autumn and less prevalent in the cooler months of winter. Figure 2 also shows that the first V. parahaemolyticus O3:K6 serovar was identified in October 1995. The same serovar was identified again in April 1996 and has become more prevalent over time.
FIG. 2.
Monthly food-borne outbreaks caused by V. parahaemolyticus O3:K6 serovar, mixed serovars (consisting of O3:K6 serovar and other non-O3:K6 serovars), and non-O3:K6 serovars versus the average temperature in Taiwan from 1995 to 1999.
This study has investigated the causes behind a high incidence of food-borne disease outbreaks from 1996 to 1999 in Taiwan by analyzing the possible etiologic agents. We present data indicating that a single bacterial agent, V. parahaemolyticus, has caused 61 to 71% of the total food-borne disease outbreaks in Taiwan during a 5-year period. By using the technique of outbreak profile analysis based on etiology groups, we have demonstrated that infections with a single V. parahaemolyticus serovar, O3:K6, were responsible for the unusually high incidence of food-borne disease from 1996 to 1999.
Infections caused by V. parahaemolyticus are usually associated with diverse serovars, but this situation has altered in recent years. Okuda et al. (11) first reported that a new strain of V. parahaemolyticus (O3:K6 serovar) had emerged. This serovar first emerged in February 1996 in Calcutta, India, and was responsible for 50 to 80% of the V. parahaemolyticus infections during the surveillance period of February to August 1996. Soon, it appeared that the emergence of this new serovar was a pandemic. The serovar appeared in several Asian countries, including Taiwan, Bangladesh, Laos, Japan, Korea, and Thailand (8), as well as in North America (3, 4). In Japan, O3:K6 has been identified as the major serovar in V. parahaemolyticus food-borne disease outbreaks in recent years (9). This strain has recently also caused two outbreaks associated with the consumption of raw seafood in the United States (3, 4). In Taiwan, the O3:K6 serovar may have appeared as early as October 1995, and the incidence has increased since April 1996 (Fig. 2). Our data show that this serovar has caused 50.1 to 83.8% of the V. parahaemolyticus infections during the 1996-to-1999 period, in contrast to only 0.6% in 1995. In 1999, the O3:K6 serovar still accounted for 61.3% of the V. parahaemolyticus infections in Taiwan, indicating that the number of infections with this serovar is still high. The O3:K6 serovar is currently dominant in the Asian region and has also appeared on the North American continent (3, 4). The origins of this serovar are unknown, and it may have spread to other regions of the world. This suggests that an intensive monitoring program for this organism is needed globally to monitor its spread and help understand its origins.
The genome of numerous O3:K6 isolates has been studied by several molecular techniques, including the arbitrarily primed PCR method (8, 11), ribotyping (1), pulsed-field gel electrophoresis analysis (PFGE; 1, 17), and toxRS sequence analysis (8). These studies conclude that the O3:K6 strain which emerged recently in Asia and the United States actually belongs to a single clone. However, Bag et al. (1) reported that a certain degree of genomic reassortment has occurred among the O3:K6 strains. Molecular typing of O3:K6 isolates recovered in Taiwan and several other Asian countries has recently been conducted by Wong and colleagues (17). From this study, eight different but genetically closely related PFGE pulsotypes were identified. In accordance with other studies (1, 8), Wong et al. demonstrated that the newest O3:K6 strains which caused pandemics in many Asian countries, including Taiwan, originated from a common ancestor with minor genomic changes.
The factors which influenced the emergence of the O3:K6 serovar are of great scientific interest. Two recent studies show that, compared to other O3:K6 strains isolated before 1996 and compared to non-O3:K6 reference strains, the recent O3:K6 strains show no significant differences in the levels of thermostable direct hemolysin, antibiotic susceptibility, and survival rate under the same environmental stresses (i.e., extreme temperatures, low pH, and high salinity) (11, 17). Recently, Nasu et al. (10) found that these newly emerged O3:K6 strains have a filamentous phage (f237) in common. The genome of f237 contains 10 open reading frames (ORFs). The ORF8 encodes a protein with a similar combination of motifs to a Drosophila adhesive protein encoded by plx (18). These authors point out that if ORF8 encodes a protein that has an adhesive function, the recent V. parahaemolyticus O3:K6 strains that possess ORF8 could be more adhesive, possibly adhering to host intestine cells or to the surface of marine plankton. This adhesiveness could account for the high potency of V. parahaemolyticus O3:K6 infection. Indeed, this adhesiveness hypothesis could account for the high incidence of O3:K6, but other possibilities, such as alternative virulence factor(s), competitive capability with other V. parahaemolyticus strains, and its population distribution in the natural environment, also need to be investigated.
Acknowledgments
We gratefully acknowledge J. M. Chen, the Central Weather Bureau, Taipei, Taiwan, for providing the temperature data. We also thank S. Y. Li, the Center for Disease Control, Taipei, Taiwan, for her critical review of the manuscript.
REFERENCES
- 1.Bag P K, Nandi S, Bhadra R K, Ramamurthy T, Bhattacharya S K, Nishibuchi M, Hamabata T, Yamasaki S, Takeda Y, Nair G B. Clonal diversity among recently emerged strains of Vibrio parahaemolyticus O3:K6 associated with pandemic spread. J Clin Microbiol. 1999;37:2354–2357. doi: 10.1128/jcm.37.7.2354-2357.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Centers for Disease Control. Botulism in the United States, 1899–1977. In: Gunn R A, editor. Handbook for epidemiologists, clinicians, and laboratory workers. U.S. Atlanta, Ga: Department of Health, Education, and Welfare; 1979. pp. 8–12. [Google Scholar]
- 3.Centers for Disease Control and Prevention. Outbreak of Vibrio parahaemolyticus infections associated with eating raw oysters—Pacific Northwest, 1997. Morb Mortal Wkly Rep. 1998;47:457–462. [PubMed] [Google Scholar]
- 4.Centers for Disease Control and Prevention. Outbreak of Vibrio parahaemolyticus infection associated with eating raw oysters and clams harvested from Long Island Sound-Connecticut, New Jersey, and New York, 1998. Morb Mortal Wkly Rep. 1999;48:48–51. [PubMed] [Google Scholar]
- 5.Chiou C S, Wei H L, Yang L C. Comparison of pulsed-field gel electrophoresis and coagulase gene restriction profile analysis techniques in the molecular typing of Staphyloccoccus aureus. J Clin Microbiol. 2000;38:2186–2190. doi: 10.1128/jcm.38.6.2186-2190.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cubbon M D, Coia J E, Hanson M F, Thomson-Carter F M. A comparison of immunomagnetic separation, direct culture and polymerase chain reaction for the detection of verocytotoxin-producing Escherichia coli O157 in human faeces. J Med Microbiol. 1996;44:219–222. doi: 10.1099/00222615-44-3-219. [DOI] [PubMed] [Google Scholar]
- 7.Elliot E L, Kaysner C A, Tamplin M L. V. cholerae, V. parahaemolyticus, V. vulnificus and other Vibrio spp. In: Jackson G J, editor. Bacteriological analytical manual. 7th ed. Arlington, Va: AOAC International; 1992. pp. 111–140. [Google Scholar]
- 8.Matsumoto C, Okuda J, Ishibashi M, Iwanaga M, Garg P, Rammamurthy T, Wong H C, Depaola A, Kim Y B, Albert M J, Nishibuchi M. Pandemic spread of an O3:K6 clone of Vibrio parahaemolyticus and emergence of related strains evidenced by arbitrarily primed PCR and toxRS sequence analyses. J Clin Microbiol. 2000;38:578–585. doi: 10.1128/jcm.38.2.578-585.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.National Institute of Infectious Disease, Japan. Vibrio parahaemolyticus, Japan, 1996–1998. Infect Agents Surveillance Rep. 1999;20:1–2. [Google Scholar]
- 10.Nasu H, Iida T, Sugahara T, Yamaichi Y, Park K S, Yokoyama K, Makino K, Shinagawa H, Honda T. A filamentous phage associated with recent pandemic Vibrio parahaemolyticus O3:K6 strains. J Clin Microbiol. 2000;38:2156–2161. doi: 10.1128/jcm.38.6.2156-2161.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Okuda J, Ishibashi M, Hayakawa E, Nishino T, Takeda Y, Mukhopadhyay A K, Garg S, Bhattacharya S K, Nair G B, Nishibuchi M. Emergence of a unique O3:K6 clone of Vibrio parahaemolyticus in Calcutta, India, and isolation of strains from the same clonal group from Southeast Asian travelers arriving in Japan. J Clin Microbiol. 1997;35:3150–3155. doi: 10.1128/jcm.35.12.3150-3155.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pan T M, Wang T K, Lee C L, Chien S W, Horng C B. Foodborne disease outbreaks due to bacteria in Taiwan, 1986 to 1995. J Clin Microbiol. 1997;35:1260–1262. doi: 10.1128/jcm.35.5.1260-1262.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shinagawa K. Int. J. Food Microbiol. 10:125–41. 1990. Analytical methods for Bacillus cereus and other Bacillus species. [DOI] [PubMed] [Google Scholar]
- 14.Vargas M, Gascon J, Jimenez De Anta M T, Vila J. Prevalence of Shigella enterotoxins 1 and 2 among Shigella strains isolated patients with traveler's diarrhea. J Clin Microbiol. 1999;37:3608–3611. doi: 10.1128/jcm.37.11.3608-3611.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.von Graevenitz A, Altwegg M. Aeromonas and Plesiomonas. In: Balows A, Hausler W J, Herrmann K L, Isenberg H D, Shadomy H J, editors. Manual of clinical microbiology. 5th ed. Washington, D.C.: American Society for Microbiology; 1991. pp. 396–401. [Google Scholar]
- 16.Wang T K, Tseng T C, Lee J H, Wang W T, Tsai J L, Ho S I, Pan T M. Analysis of Salmonella serovars in Taiwan by the phase induction method. Chin J Microbiol Immunol. 1994;27:13–24. [PubMed] [Google Scholar]
- 17.Wong H C, Liu S H, Wang T K, Lee C L, Chiou C S, Liu D P, Nishibuchi M, Lee B K. Characteristics of Vibrio parahaemolyticus O3:K6 from Asia. Appl Environ Microbiol. 2000;66:3981–3986. doi: 10.1128/aem.66.9.3981-3986.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang S D, Kassis J, Olde B, Mellerick D M, Odenwald W F. Pollux, a novel Drosophila adhesion molecule, belongs to a family of proteins expressed in plants, yeast, nematodes, and man. Genes Dev. 1996;10:1108–1119. doi: 10.1101/gad.10.9.1108. [DOI] [PubMed] [Google Scholar]