Abstract
The fields of precision imaging and drug delivery have revealed a number of tools to improve target specificity and increase efficacy in diagnosing and treating disease. Biological molecules, such as antibodies, continue to be the primary means of assuring targeting of various payloads. However, molecular-scale recognition motifs have emerged in recent decades to achieve specificity through the design of interacting chemical motifs. In this regard, an assortment of bioorthogonal covalent conjugations offer one possibility for in situ complexation under physiological conditions. Herein, a related concept is discussed that leverages interactions from non-covalent or supramolecular motifs to facilitate in situ recognition and complex formation in the body. Classic supramolecular motifs based on host–guest complexation offer one such means of facilitating recognition. In addition, synthetic bioinspired motifs based on oligonucleotide hybridization and coiled-coil peptide bundles afford other routes to form complexes in situ. The architectures to include recognition of these various motifs for targeting enable both monovalent and multivalent presentation, seeking high affinity or engineered avidity to facilitate conjugation even under dilute conditions of the body. Accordingly, supramolecular “click chemistry” offers a complementary tool in the growing arsenal targeting improved healthcare efficacy.
Keywords: Drug delivery, Diagnostics, Therapeutics, Supramolecular Chemistry, Biomaterials
Graphical Abstract
1. INTRODUCTION
A primary goal in drug delivery is the pursuit of technologies to increase the fraction of drug delivered to a site of need.1 One key characteristic of an active pharmaceutical agent is its therapeutic index, a ratio of its toxic dose (TD50) to its effective dose (ED50). Accordingly, drug delivery technologies seek to increase the therapeutic index through two parallel mechanisms: i) attenuating the systemic activity of a drug through encapsulation and/or prodrug methodologies to enable higher dosing without toxicity, and ii) ensuring a larger fraction of the delivered agent reaches the physiological site of need to increase effectiveness of the therapeutic agent.2 Drug delivery can be achieved through passive accumulation of drug carriers, sometimes taking advantage of leaky vasculature that is a pathophysiological hallmark of certain diseases.3,4 The first FDA-approved nanoscale drug delivery technology, Doxil®, was a PEGylated liposomal formulation of doxorubicin that functioned through such a mechanism.5 Accordingly, early efforts in the field of drug delivery often sought to increase the therapeutic index through a combination of sequestering toxic agents within nanoscale carriers and leveraging physiologic features of diseased tissue to promote preferential accumulation.
Another strategy broadly explored in the field of drug delivery to increase the therapeutic index is active targeting. These routes commonly leverage affinity from biological molecules such as antibodies to localize a therapeutic to a site of need, targeting on the basis of a disease-relevant biomarker.6–8 Several antibody-drug conjugates have been recently FDA-approved,9 consisting of a therapeutic agent attached via a labile linker to a monoclonal antibody with affinity for specific biomarkers.10 Antibodies or aptamers can likewise be used for active targeting of nanoscale drug carriers.11–13 These and other methods of active targeting are often limited by the availability of targeting antibodies specific to the disease of interest; the use of larger constructs or drug carriers also limits tissue perfusion, carries risk of off-site accumulation, and may lead to prolonged circulation while shedding active drug systemically.14,15 For example, only 0.001–0.01% of an injected monoclonal antibody, and by logical extension an antibody-drug conjugate, localizes to a tumor site in humans.16,17 Meanwhile, nanoparticles targeted with a clinically validated antibody have demonstrated local accumulation of <1% in vivo.18 As such, there remains a need to explore new technologies in order to more effectively deliver therapeutics to sites of need.
In the field of bioconjugate chemistry, a molecular scale pre-targeting approach has been demonstrated using different bioorthogonal ligations to capture circulating agents at specific sites in the body through spontaneous formation of covalent bonds.19–22 Spatial localization can be achieved within the body by covalent bond formation in situ using two-step application of a pre-targeted entity bearing one component of a bioorthogonal motif followed by application of the second motif attached to a drug or imaging agent (Fig 1).23–27 The attachment of a drug to a motif for click chemistry offers certain prodrug benefits of attenuated systemic activity; such agents also incorporate labile linkages for subsequent release of the active therapeutic via linker hydrolysis following local accumulation. Others have demonstrated so-called “click-to-release” and “catch and release” chemistries wherein an active agent releases from its bioorthogonal motif-bearing prodrug precursor by spontaneous ring isomerization simultaneous to in situ formation of a covalent bond.28,29
Figure 1:
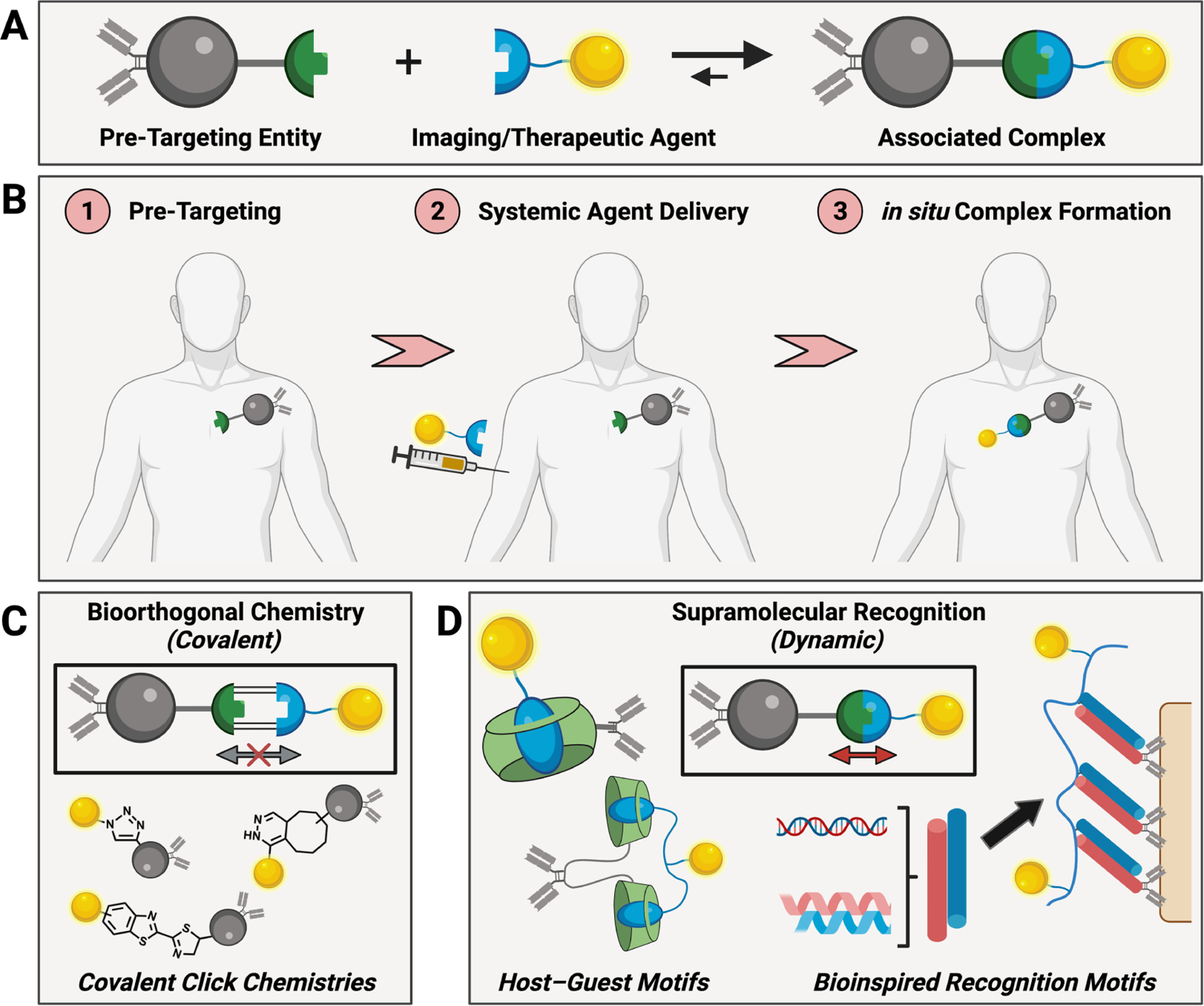
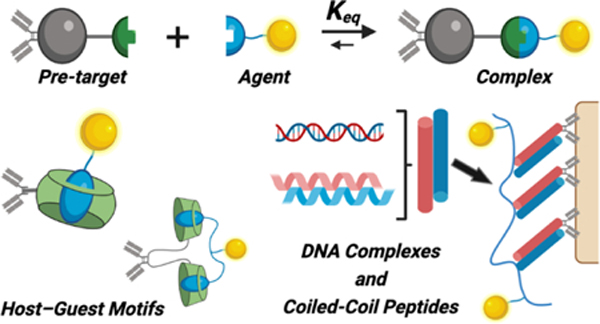
(A) Schematic of dynamic supramolecular interactions used for in situ association, leveraging complementary motifs with one attached to an entity for pre-targeting and another attached to the desired agent to be delivered. (B) Generalized overview of this two-component approach to targeting via in situ complex formation. (C) Examples of common covalent bioorthogonal reactions used in pre-targeting applications for in situ covalent bond formation. (D) Overview of the dynamic supramolecular motifs described herein, which form association complexes through non-covalent molecular recognition.
The present review focuses on related molecular-scale approaches akin to bioorthogonal click chemistry, instead using non-covalent supramolecular interactions for in situ recognition in the body. These synthetic motifs are attractive in the development of drug delivery platforms due to their scalable, tunable, and molecularly well-defined characteristics.30–34 The various motifs used for non-covalent recognition in the body include host–guest macrocycle complexes, complementary oligonucleotide segments, and coiled-coil peptide assemblies (Fig 1D). The mechanisms that underlie recognition incorporate pseudo-specificity through unnaturally high affinity and/or high effective affinity through engineering multivalent motifs to enable avidity. While certain of these motifs (e.g., host–guest) are subject to competition from naturally occurring compounds and thus not fully bioorthogonal, outcomes resembling orthogonality can be realized through motif selection and design to tune affinity well in excess of naturally present competitors, or by engineering avidity to gain advantage.33
Molecular-scale approaches to drug targeting through both in situ covalent bioconjugation and non-covalent recognition offer certain distinctions relative to drug delivery methods using active biological targeting. In one manifestation of this two-step molecular-scale approach, pre-targeting a site of interest with an antibody or related biomolecule maintains the benefits of biological recognition of disease biomarkers. However, separating the drug from the antibody reduces the risk of undesired release during prolonged circulation. In other uses, pre-targeting and capture of a circulating therapeutic using a localized material suffers from a requirement for a priori knowledge in applying the pre-targeting material to guide subsequent administration of the agent, and as such may be more limited in its practical application. Small molecule prodrugs, prepared by modifying a therapeutic with a molecular-scale targeting motif, offer the benefits of attenuated activity in systemic circulation, more extensive tissue perfusion, and rapid clearance owing to small size relative to antibodies or even larger nanoscale carriers. As such, the general concept introduced here for non-covalent molecular recognition of synthetic motifs in the context of in situ targeting of therapeutics and imaging agents should be framed with these benefits and drawbacks in mind. With the aim of specifically focusing on uses of synthetic non-covalent molecular recognition motifs, this review will also (by necessity) not cover voluminous work in the areas of biomolecular-based recognition using antibodies, aptamers, peptides, or other common biomolecular affinity agents. Instances where such affinity agents are used in the context of pre-targeting a synthetic motif for subsequent non-covalent recognition-mediated targeting will be discussed.
2. THERMODYNAMICS OF RECOGNITION
The propensity for a non-covalent complex to form in the dilute environment of the body is governed by the thermodynamics of the particular interaction. In the simplest case of a monovalent interaction, the dynamic process of recognition proceeds as follows:
where [A] and [B] are the concentrations of the free binding pairs and [A•B] is the concentration of the formed complex. From the law of mass action, an equilibrium constant, Keq (sometimes denoted KA), can be derived as follows:
This quantity has standard units of [M−1] for a 1:1 monovalent interaction. Keq is commonly referred to as the affinity of an interaction. It is conventional in some systems to express the reciprocal of this value, 1/Keq=KD, yielding units of [M] and serving to define the dilution concentration for spontaneous dissociation of a complex. A number of synthetic host molecules have been reported to bind with an array of complementary guest motifs, enabling Keq to be tuned by molecular design or in response to a biologically relevant stimulus.35 A higher value of Keq thus signifies a more stable complex that exhibits preferential formation even under dilute conditions. The value of Keq is also related to the rates of dynamic formation, kon, and dissociation, koff, of the complex, as follows:
For a 1:1 monovalent interaction, kon has units of [M−1s−1] and koff has units of [s−1]. The rate of complex formation for some supramolecular motifs has been found to occur near the diffusion limit (~108 M−1s−1);36 when compared to common covalent bioorthogonal conjugations such as azide–DBCO (2.3 M−1s−1)37 and tetrazene–trans-cyclooctene (3100 M−1s−1)38 this suggests a possible benefit of fast association for supramolecular motifs. Typically, higher affinity interactions will have concomitantly slower koff and thus have a longer lifetime of complexation once formed.
The effective doses of different therapeutics vary, but an assumption for serum concentrations on the order of ~[nM] for most drugs defines (roughly) the target Keq needed for complex formation when considering the use of a particular motif in targeting therapeutics; this implies Keq may need to be greater than ~108 M−1 to drive complex formation in vivo. Given this extent of dilution expected for uses in the body, as well as a variety of possible competitors for certain classes of interactions, monovalent affinity may thus not be sufficient for some recognition motifs to facilitate efficient supramolecular complexation. Accordingly, other design approaches may couple multiple lower affinity interactions on a defined scaffold to achieve a higher effective affinity, a phenomenon referred to as avidity. The complexes formed between antibody and antigen, with multivalent display of a specific binding epitope on the antibody, illustrates the use of avidity in nature.39 Binding events in multivalent systems do not necessarily occur simultaneously, but they are likewise not completely independent. The physical tethering of multiple binding motifs creates an elevated local concentration through the close proximity of binding sites to drive complex formation.40 In other instances, both motifs may be presented on multivalent scaffolds, leading to an overall reduction in the effective koff given the asynchronous timescale of dynamic complex exchange for individual binding sites as multiple dynamic interactions drive greater complexation between the two scaffolds.41 In this way, the use of multivalent systems may compensate for the low affinity of an individual motif to facilitate recognition even under the conditions of dilution expected for applications in the body.
3. HOST–GUEST RECOGNITION
Host–guest chemistry, characterized as the non-covalent association of a small molecule guest within the portal of a host macrocycle, is among the most recognizable of supramolecular motifs. The affinity of different interactions can vary substantially, though complexes have been demonstrated that form at high affinity (e.g., Keq >1010 M−1) and are therefore resistant to dilution and native competition, in pursuit of various bioconjugation-based applications.33,42,43 Many synthetic macrocyclic host molecules are known, including crown ethers, cryptands, cyclodextrins (CD), cucurbit[n]urils (CB[n]), calix[n]arenes, and pillar[n]arenes.30 High-affinity designer molecules have also been revealed from host–guest complexes that form stable and highly fluorescent dyes.44,45 Of the motifs used in the context of drug delivery, CD macrocycles prepared enzymatically from starch constitute the most broadly explored and readily available macrocycles.46,47 CDs have rigid conical geometry and are comprised of different numbers of glucopyranoside subunits, to include α-CD (6), β-CD (7), and γ-CD (8), enabling size-mediated selectivity in their binding to different guests. Their hydrophobic interiors and hydrophilic exteriors allow guest encapsulation within the cavity, taking advantage of both hydrophobic and Van der Waals interactions.48 Binding between CDs and their guests occur with Keq values not typically exceeding ~105 M−1, the order expected for binding between β-CD and an adamantane guest.49,50 The CB[n] family of macrocycles, composed of [n] repeating glycoluril subunits, constitutes another useful macrocycle for guest binding in water.51–54 Glycoluril subunits afford a symmetric macrocycle with a rigid hydrophobic cavity and two identical carbonyl-fringed portals. CB[7] macrocycles bind to adamantane-class guests with Keq up to ~1017 M−1, well in excess of what is achievable by other macrocycles or even natural motifs such as biotin-avidin.55–57 High-affinity binding is possible through a combination of the hydrophobic association and volume-filling of the macrocycle cavity coupled along with electrostatic interactions between aliphatic-adjacent protonating groups and the electronegative carbonyl-fringed portal.58 Accordingly, the differing spectrum of affinity offered by CD and CB[n] macrocycles affords distinct opportunities for host–guest recognition and complex formation in the conditions of the body, as described herein.
3.1. Monovalent Host–Guest Recognition
In the context of in situ recognition, protein-based motifs have been extensively explored, yet can exhibit slow biodistribution and clearance.59–61 Long circulation times to reach a target may limit their use to deliver short-lived isotopes for radioimaging and increase the possibility for enzymatic degradation in circulation.62,63 Host–guest motifs, with small molecule guests on the order of ~200 g/mol and macrocycles on the order of 1200 g/mol, may thus offer a variety of possible benefits. CB[n] macrocycles, and in particular the water-soluble and high-affinity CB[7] variant, have been most explored in the context of monovalent host–guest recognition in the body. The types of guest molecules useful for this purpose are amine-containing ferrocene and adamantane derivatives, exhibiting Keq values in the range of 1010−15 M−1 in binding CB[7].64–66 Recognition using these motifs has thus been explored for a variety of imaging and therapeutic applications.
The use of supramolecular host–guest motifs for in situ targeting typically comprises a pre-targeting step followed by subsequent administration of an agent for imaging or therapy. In this context, an antibody may be used for the initial pretargeting to deposit either a host or guest at the site of interest, followed by subsequent addition of the desired agent attached to the complementary binder (Fig 2A).67 Using pre-targeting principles, in situ formation of host–guest complexes have been explored in live nematodes (C. elegans) and mice.68 The studies in nematodes coupled complementary FRET pairs to CB[7] and guest, verifying sequential administration of the motif resulted in complex formation in situ. This system was then explored in mice for in vivo cancer imaging. CB[7] was covalently attached to cetuximab, an antibody recognizing epidermal growth factor receptor that is used clinically in treatment of colorectal, neck, and lung cancers. Following pre-targeting with the CB[7]-antibody conjugate, adamantane linked to a near-infrared cyanine dye was found to accumulate at the tumor site for selective tumor imaging.68
Figure 2:
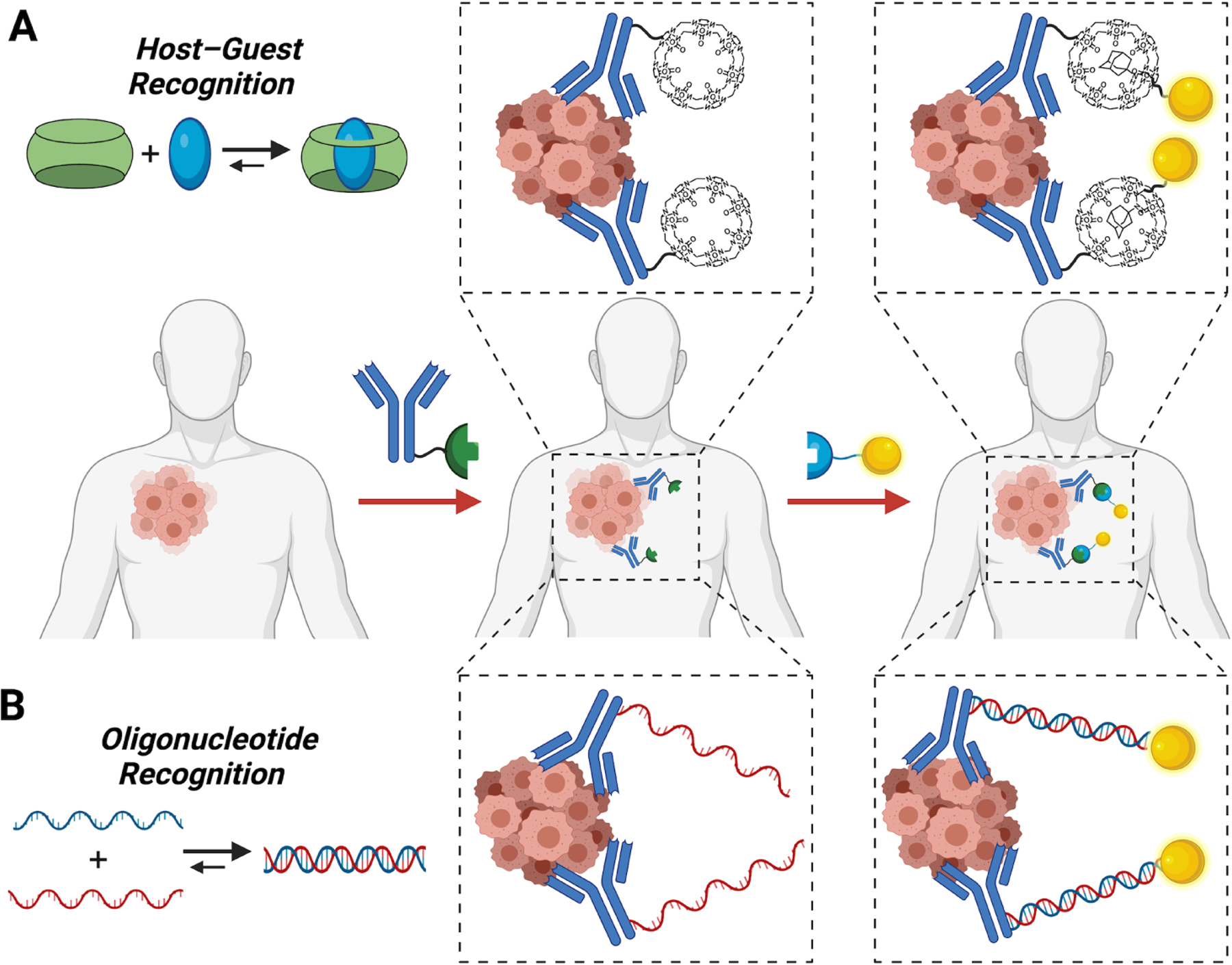
Antibody-based pre-targeting for delivery to sites of disease bearing a specific biomarker, coupled with a secondary delivery approach and in situ targeting driven by (A) monovalent host–guest motifs or (B) complementary oligonucleotide sequences designed for hybridization.
Pre-targeting has also been achieved by local injection of a polymer hydrogel presenting CB[7], with subsequent systemic administration of a guest-linked agent.69 A series of guests ranging in Keq from ~109 to 1012 M−1 fused to a near-infrared fluorescent dye were explored to assess the role of affinity for in situ complex formation at the site of the CB[7]-rich depot (Fig 3). These studies identified complexes between CB[7] and an amino-ferrocene guest with Keq of ~1012 M−1 that achieved substantial localization, whereas the dye bound to a different ferrocene guest with Keq of ~109 M−1 showed no accumulation. For the high-affinity case, ~4% of the administered agent homed within a few hours; the remainder was rapidly cleared over this same time. This figure is impressive in context of the typical targeting efficiency achieved by antibodies, referenced previously here. The depot site could be serially reloaded, with site retention of the bound agent for multiple weeks following administration. This same high-affinity guest motif was then conjugated to the chemotherapeutic doxorubicin to create a prodrug for integration with supramolecular targeting. By injecting the CB[7]-rich hydrogel near a tumor, the therapeutic efficacy of supramolecular homing was evaluated in comparison to a prodrug variant with no affinity for CB[7]. In this case, the targeted prodrug demonstrated a significant reduction in tumor growth rate, with the effect extending for weeks following initial dosing.
Figure 3:
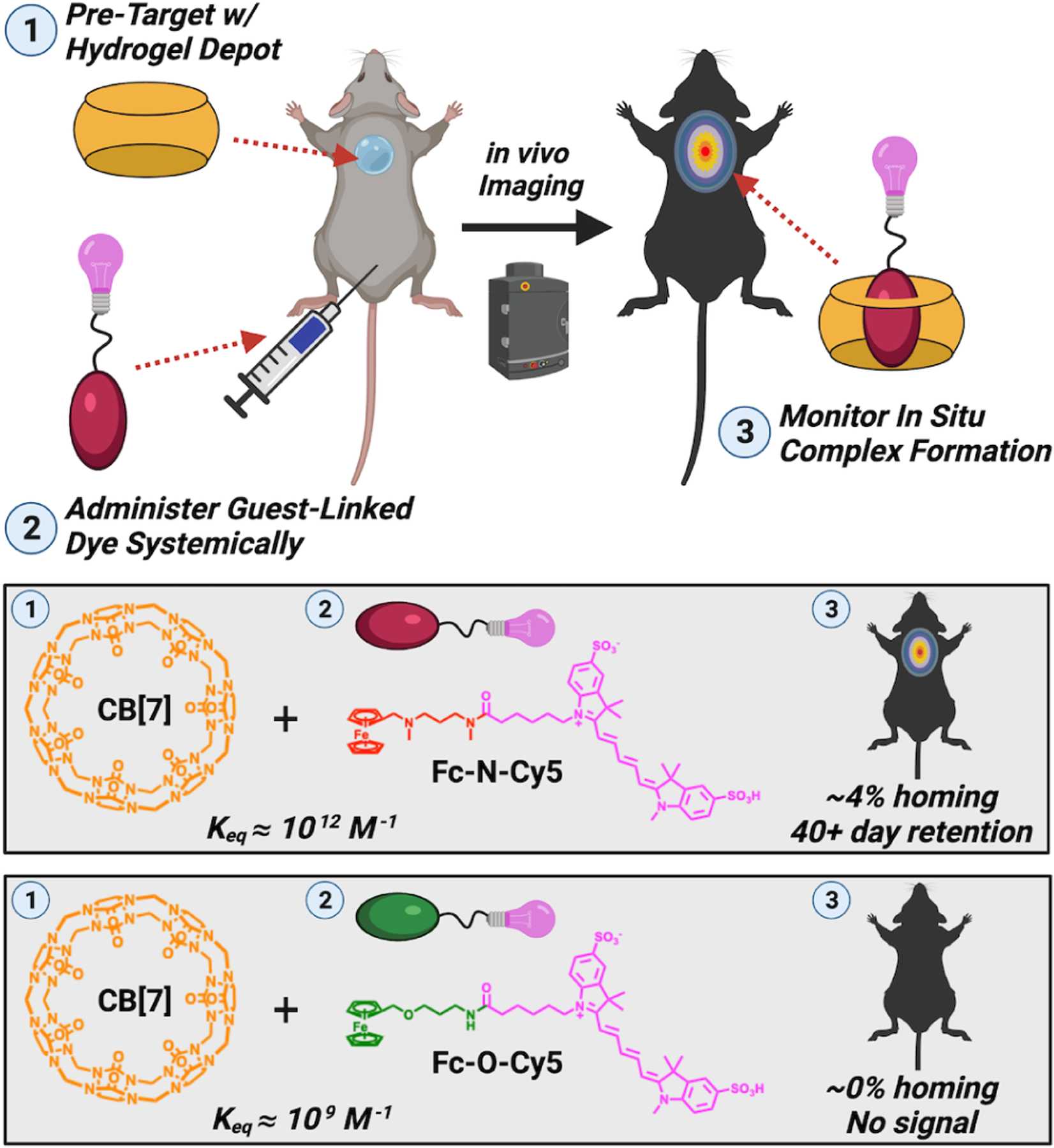
Methodology to assess the affinity needed for in situ recognition and targeting. A hydrogel presenting pendant CB[7] macrocycles was implanted locally at a site. Subsequently, a near-infrared fluorescent probe (Cy5) modified with two ferrocene guests having different affinities for CB[7] (Fc-N:1012 M−1 vs. Fc-O:109 M−1) was administered systemically. Through in vivo imaging, the amount of dye localized and retained at the site presenting CB[7] was then quantified. Subtle differences in the guest structure, altering their resulting affinity for CB[7], led to dramatic changes in the effectiveness of in situ complexation.
3.2. Multivalent Host–Guest Recognition
The uses of CD for in situ complex formation have primarily leveraged multivalent constructs to introduce avidity, thereby compensating for the relatively low Keq of a monovalent CD host–guest complex compared to those observed for CB[7]. In one such design, adamantane-functionalized albumin aggregates were used to pre-target sites for subsequent delivery of β-CD-modified polymers carrying agents for either fluorescence or SPECT imaging modalities (Fig 4).70 Pre-targeting with the multivalent albumin aggregates followed by multivalent agent delivery offered a ~16-fold increase in the accumulation of the agent in the liver and 4.5-fold in the lungs when compared using SPECT imaging to pre-targeting with unmodified albumin aggregates. Further studies using albumin aggregates to pre-target a β-CD-modified polymer leveraged dual-isotope imaging (99mTc on the albumin particles and 111In on the polymer) to validate co-localization of the two components in vivo.71 As such, multivalent scaffolds presenting both host and guest enable the use of CD macrocycles in spite of its modest monovalent affinity.
Figure 4:
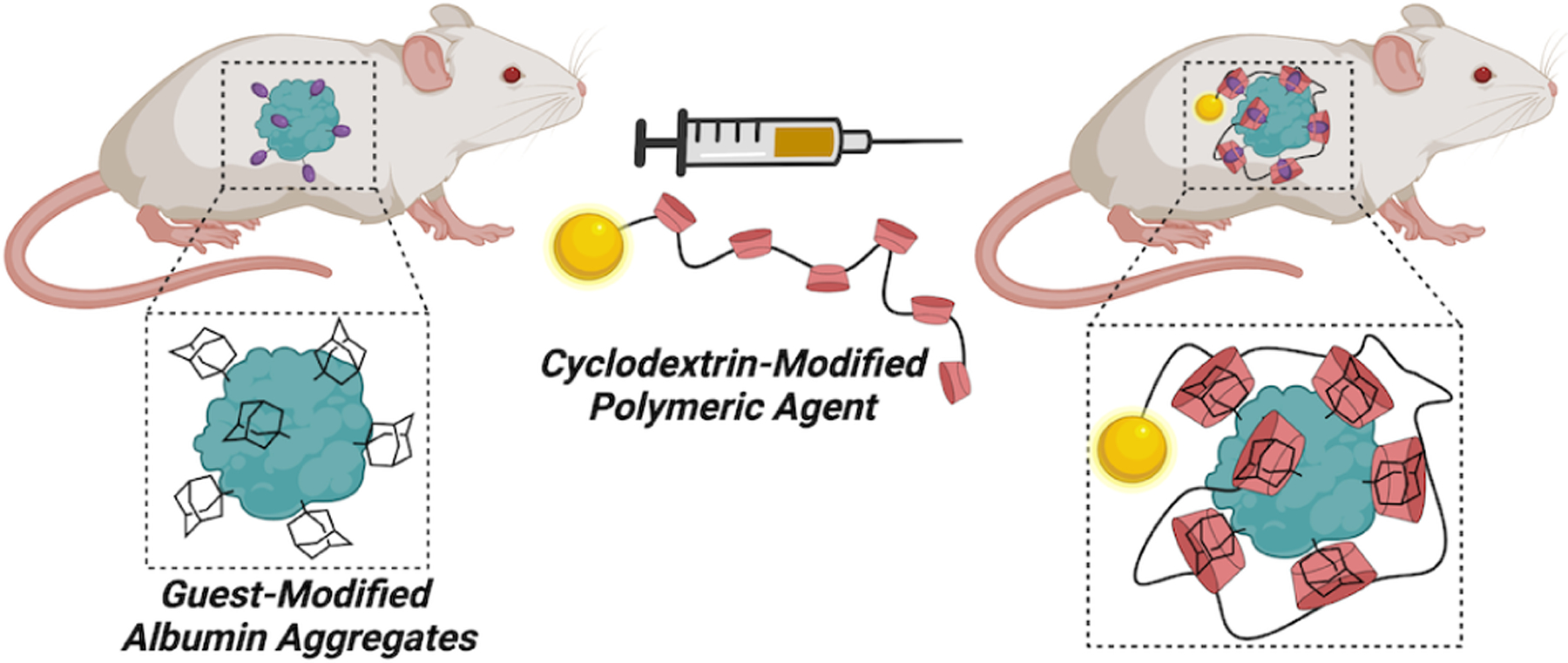
Schematic overview of the use of guest-modified serum albumin aggregates, which following administration preferentially accumulate at sites of disease. Subsequently, a cyclodextrin-modified polymer was administered to enable multivalent in situ complexation and agent delivery to the site bearing these guest-modified albumin aggregates.
4. OLIGONUCLEOTIDE HYBRIDIZATION
The association of complementary strands of DNA, forming its canonical double helix, is one of the most recognizable non-covalent motifs in the living world. Synthetic oligonucleotides thus offer a tunable and biologically relevant affinity motif for non-covalent complex formation, toward many therapeutic uses.72–74 This is highlighted by the decades of work evaluating the therapeutic potential of small interfering RNAs (siRNA), where therapeutic function arises specifically from recognition and binding to target mRNA to transiently inhibit protein expression.75,76 Oligonucleotide strand complexation, a process known as hybridization, is driven by Watson-Crick base pairing with lateral hydrogen bond formation between complementary bases offering an enthalpic driving force.77–79 The vertical stacking of aromatic bases in the formed helical structure also contributes a favorable driving force for hybridization via hydrophobic and π-π interactions.80 The number of base pairs, and by extension the number of hydrogen bonds and π-π interactions, dictates the binding affinity between oligonucleotide strands; this affinity is highly dependent on environmental conditions such as temperature, concentration, and osmolarity.81,82 For example, the complexation of model 10-base strands in 3 mM buffer exhibits a Keq of ~5*107 M−1 at 15°C, reducing to ~3*105 M−1 at 35°C as non-covalent interactions become less favorable.82 Meanwhile, 20-base strands have Keq values (~108 M−1) that are much less temperature-dependent over the same range. For both lengths, affinity also increases by ~1–2 orders of magnitude for interactions in a buffer of higher salt (10 mM). Accordingly, the design of oligonucleotide sequences for recognition in the body must account for specific operating conditions to ensure reliable complex formation. As the focus here is on the use of synthetic non-covalent recognition motifs for targeting applications, the many important uses of aptamers for recognition of biomolecules falls outside the present scope of this review; the reader is encouraged to explore other relevant reviews on this topic.83–85
4.1. Monovalent Oligonucleotide Hybridization
One benefit of oligonucleotide-based recognition arises from its ease of synthetic modification with molecular cargo.86–89 This design tool enables an array of therapeutics or imaging agents to be appended to oligonucleotide strands. One salient example of this approach is found in the field of molecular beacons, wherein binding to a target DNA or RNA strand triggers a hybridization-mediated unfolding of the beacon and (typically) an increase in fluorescence relative to a quenched state in the folded form.90,91 Early work using this technology in vivo relied on aptamer-mediated recognition to facilitate beacon rearrangement for imaging.92,93 Related aptamer-targeted technologies have also been used to deliver drugs bound via intercalation with double-stranded regions of the probe.94 Other technologies evaluated in vitro suggest the possibility that aptamer-based constructs with a pendant oligonucleotide tail can be used for pre-targeting, with subsequent delivery of a probe coupled to the complementary oligonucleotide strand.95 A similar pre-targeting approach was also demonstrated in vitro using copper-free click chemistry to modify the cell surface with oligonucleotides, subsequently delivering a complementary strand linked to a probe for imaging.96 However, the use of oligonucleotide hybridization specifically for targeting molecular beacons in vivo has been less commonly explored.
Targeting via monovalent oligonucleotide hybridization has been demonstrated in the context of antibody-mediated pre-targeting for PET-CT imaging (Fig 2B).97 In this work, the cetuximab antibody was modified with a 17-mer L-DNA segment. Subsequently, a mirror-image 17-mer L-DNA segment connected to a 64Cu radionuclide chelator was administered for localization by in situ hybridization. Biodistribution studies performed in vivo demonstrated significant tumor accumulation and contrast enhancement when using this pre-targeting approach for radionuclide delivery.
In a related context, synthetic oligonucleotide analogues known as peptide nucleic acids (PNAs) may also enable recognition in the context of targeting. The nucleobases of PNAs form stable duplexes with DNA or RNA segments, and may also be designed to recognize other PNAs.98–101 Accordingly, PNA recognition has been used in the context of a two-step pre-targeting.102 In this work, a PNA-modified protein was first administered for passive accumulation at sites of infection or tumors, and subsequently a PNA radiolabelled with 99mTc was administered for localization by in situ hybridization.
4.2. Multivalent Oligonucleotide Hybridization
Efforts to increase the effective binding affinity of complementary oligonucleotide strands have entailed developing multivalent scaffolds to introduce avidity into the process of targeting. In one example, recognition via oligonucleotide hybridization of complementary oligonucleotides has been demonstrated to refill a locally applied hydrogel depot.103 In this design, an alginate hydrogel modified with oligodeoxynucleotide (ODN) strands was applied locally. Subsequent systemic application of alginate modified with the complementary ODN strands enabled local accumulation at the depot through strand recognition. A control of non-complementary ODN sequences exhibited no increased accumulation. The ODN-targeted platform was evaluated for functional use in vivo in the delivery of a chemotherapeutic, doxorubicin, to the site of a tumor. Mice treated weekly by systemic application of ODN-modified alginate strands conjugated to doxorubicin showed a significant reduction in tumor growth compared to controls, attributable to ODN hybridization localizing the drug-modified polymer to the site of the depot.
Certain therapeutic benefits arise when using multivalent scaffolds apart from increasing the effective Keq of recognition. One such example is found in efforts to pre-target using oligonucleotide-modified antibodies followed by subsequent recognition on the cell surface of a multivalent oligonucleotide scaffold (Fig 5).104,105 The therapeutic effect of this approach arises from induction of apoptosis due to receptor multimerization on the cell surface, leading to demonstrations for a new class of drug-free macromolecular therapeutics.106 Efforts to prepare these constructs with oligonucleotides have relied on morpholino oligomers, synthetic analogs of oligonucleotides consisting of DNA bases attached to a backbone of methylenemorpholine rings linked through phosphorodiamidate groups, intended to facilitate enhanced stability in serum.107 The first design leveraged an antibody fragment (Fab’) against a marker for B-cell lymphoma (CD20), fusing this to a morpholino strand for pre-targeting cancer cells. A polymer based on N-(2-hydroxypropyl)methacrylamide (HPMA) with pendant complimentary morpholino strands was then administered to multimerize the CD20 cell surface receptors and induce apoptosis.104 These constructs demonstrated therapeutic function in a disseminated B-cell lymphoma model in mice, demonstrating a key benefit of this approach against metastatic disease. Subsequent work on this concept utilized an intact anti-CD20 antibody (Obinutuzumab) for pre-targeting with morpholino strands and induced multimerization with morpholino-modified human serum albumin.105 By combining the intact antibody with multivalent crosslinking, this approach enabled two synergistic modes to induce apoptosis.
Figure 5:
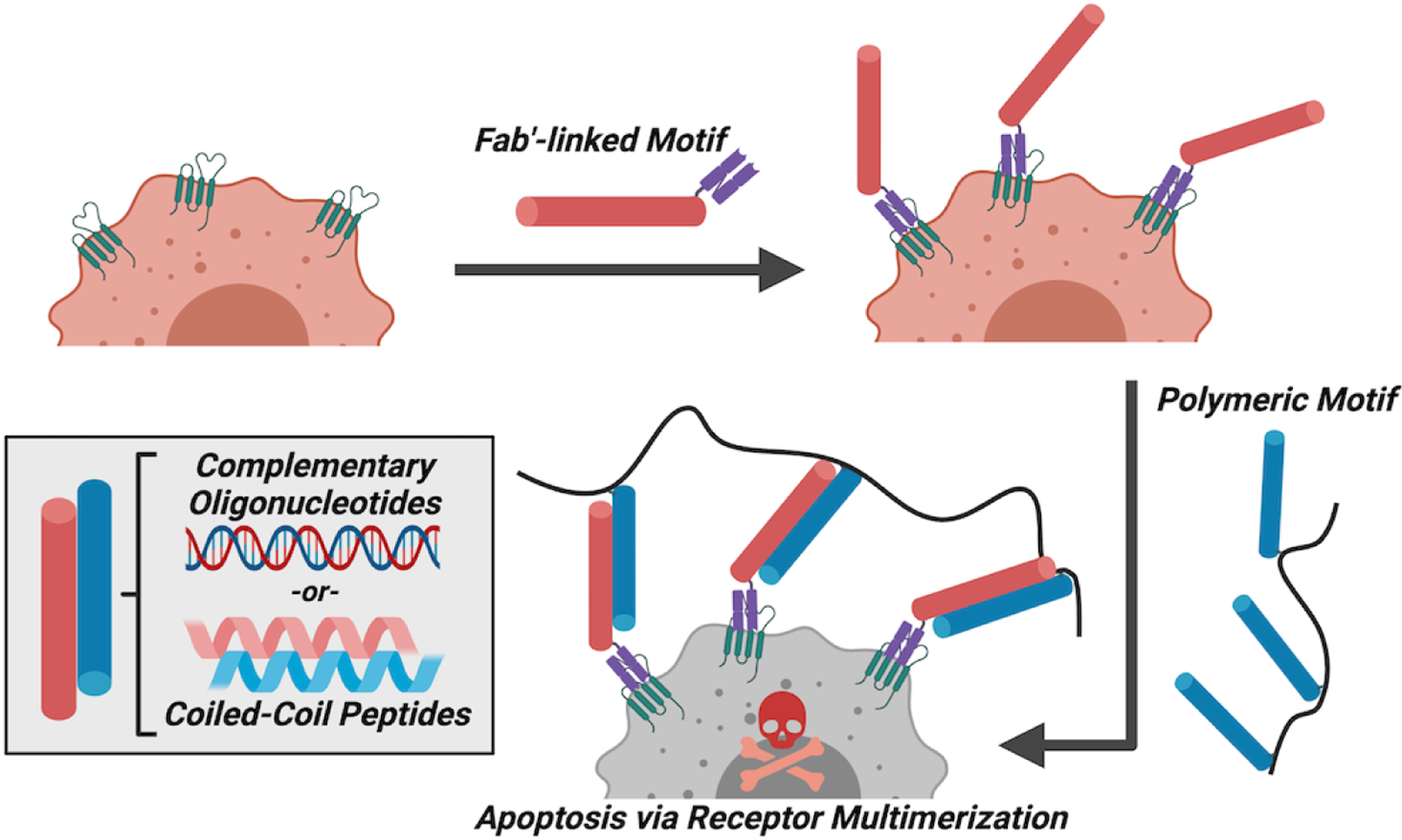
Schematic overview of the concept of drug-free macromolecular therapeutics. This concept has been demonstrated for motifs derived from both oligonucleotide hybridization and coiled-coil peptide association. A cancer cell of interest was first pre-targeted with a reagent for biological recognition, an antibody or antibody fragment, that was modified with one-half of the desired recognition motif. Subsequently, a multivalent polymer bearing the complementary recognition motif was administered. The polymer scaffold, by simultaneously binding multiple surface-presented motifs, serves to non-covalently link the receptors on the cell leading to triggered cell death.
4.3. In Situ Strand Displacement
Oligonucleotide complexes can be designed to engage in strand displacement through binding to unhybridized segments flanking a double-stranded segment, an approach used to facilitate polymer de-gelling, site-specific drug release, and surface regeneration.108–110 This displacement is often initiated through toehold-mediated strand exchange, wherein a single-stranded oligonucleotide binds to an exposed portion of its complementary strand that is otherwise engaged in a double helix, triggering dissociation of the initial complex as the replacement strand hybridizes.111 Recently, this mechanism was utilized to regenerate antithrombotic functionality of a surface (Fig 6).110 To combat degradation of an antithrombotic agent presented on the device, strand displacement was designed to replace the degraded agent and restore antithrombotic functionality of the surface. This approach demonstrated a significant reduction in fibrin formation. Though not used in vivo, recognition-mediated strand displacement offers many possible opportunities to externally control the properties of biomedical device interfaces in situ.
Figure 6:
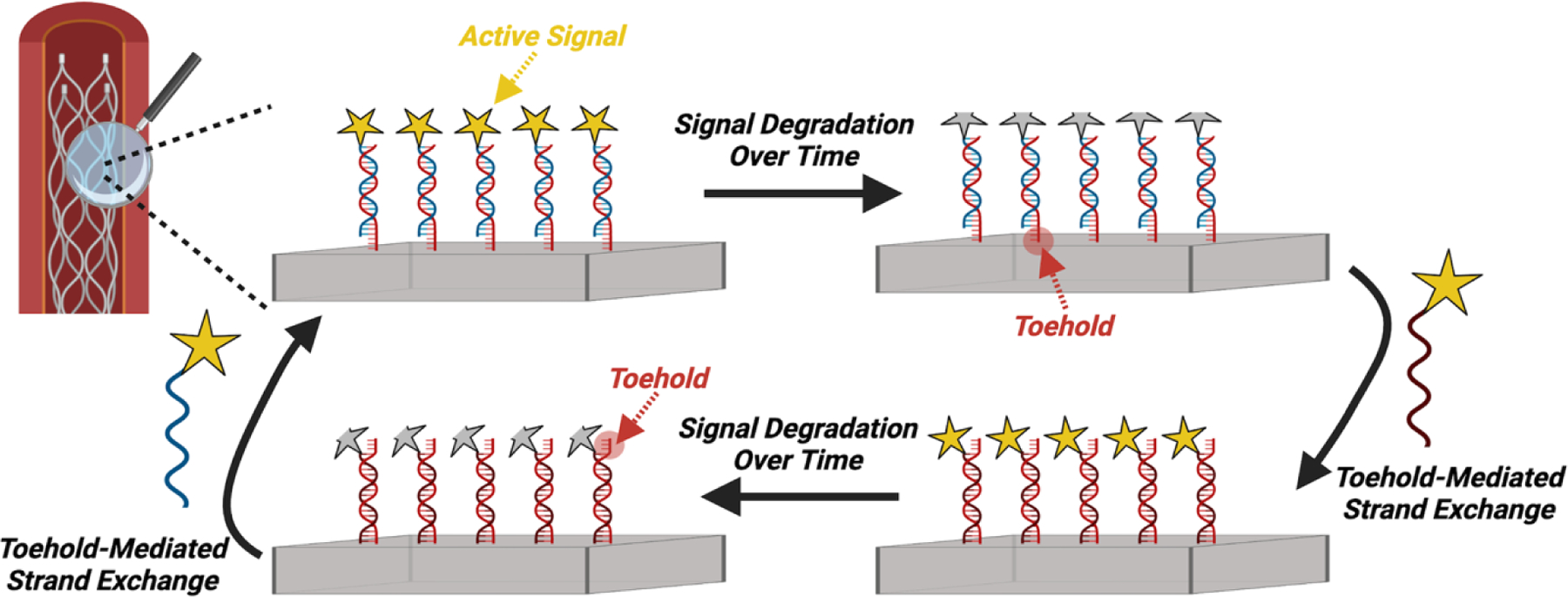
Schematic of in situ surface regeneration using a toehold-mediated strand exchange approach. This general strategy illustrates a route to use designed recognition motifs to regenerate the presentation of active sites on a device surface.
5. PEPTIDE COILED-COIL FORMATION
Engineered coiled-coil peptides, characterized by the arrangement of alpha-helical peptides into a superhelix bundle, afford recognition properties with utility in the design of functional materials and systems.112,113 Their biological relevance as a common structural motif found in nature have inspired significant study into both the mechanisms of formation and strategies for sequence manipulation to realize coiled-coils motifs comprised of a different number (n=2–6) of both homo- or hetero-[n]meric alpha-helical peptides.114–117 Various naturally derived and de novo designs have thus been demonstrated for coiled-coil recognition, with some synthetic heterodimeric variants having Keq values up to 1014 M-1.118–121 Such interactions are thus comparable to (or higher than) high-affinity host–guest or oligonucleotide motifs. The predictable nature of these associations has been used to recreate the complex higher-ordered structures of natural proteins with synthetic variants, for instance in the preparation of discrete cage-like assemblies.122,123 In addition, coiled-coil motifs have been incorporated as a modular associating unit in non-covalent preparation of modular drug carriers.124–127 Accordingly, these interaction offer another class of synthetic non-covalent interactions with promise for in situ recognition in the body.
5.1. Multivalent Coiled-Coil Recognition
As with work in oligonucleotide systems, coiled-coil interactions have been explored in conjunction with routes for pre-targeting as well as scaffolds for multivalent presentation toward the concept of drug-free macromolecular therapeutics.106 In one example, one component of a heterodimeric coiled-coil was attached multivalently to HPMA with the complementary alpha-helical segment attached to a Fab’ with reactivity against CD20 (Fig 5).128 This platform showed in vivo efficacy in a mouse model of B-cell lymphoma, functioning by crosslinking the surface-bound Fab’ on cell surface receptors to induce apoptosis.129 The immunogenicity of this platform was studied in vitro and in vivo, pointing to no specific immunogenicity for the coil-forming peptide motif; this study explored the same motif prepared from D-isomer peptides and found the enantiomeric peptide coiled-coils to behave similarly to the originally used L-amino acids.130 Multi-fluorophore imaging of this system further verified in situ assembly of the two components on B-cell membranes when administered by this two-step pre-targeting approach, noting the importance of the delay time between administration of the first and second component to enable localization.131 This system was also found to function when the multivalent HPMA component was replaced with human serum albumin modified with multiple copies of one of the coil-forming peptide segments.132 Related work demonstrated the ability to target cancer cells presenting one-half of a coiled-coil motif with liposomes presenting the complementary peptide, demonstrating in situ homing in a zebrafish model.133 Accordingly, systems based on pre-targeting and multivalent recognition may also use synthetic coiled-coil motifs to facilitate recognition in the body.
6. CONCLUSIONS
In the continued pursuit of new routes to enhance efficacy in diagnosing and treating disease, strategies for recognition on the molecular scale hold promise. In particular, the use of these synthetic motifs offers new routes to reliably and efficiently perform in situ conjugation under dilute conditions in the body, while in the presence of salts, proteins, lipids, and other “sticky” biological entities. The use of small molecules affords rapid and extensive tissue perfusion. To date, bioorthogonal covalent conjugations have offered one means of achieving this outcome of in situ recognition. Herein, a related concept leveraging the noncovalent association of synthetic supramolecular motifs is described. Through motif selection and design, high-affinity interactions can be realized to enable quasi-specificity and orthogonality in the body. Many of these motifs offer kinetic advantages over traditional bioorthogonal chemistries, such as the ability to associate with diffusion-governed interaction rates. Moreover, the synthetic origins of these motifs enable facile multivalent display on polymers, nanoparticles, or related scaffolds to engage avidity and further enhance recognition specificity. This approach has even revealed a new therapeutic class based on drug-free macromolecular architectures.
There remain challenges that must be navigated to more fully exploit the potential of these supramolecular tools for in situ targeting. The two-step targeting used in many systems, while advantageous in limiting off-site accumulation and systemic drug shedding of often toxic drugs, introduces complexities and variability with respect to the timing of administration of each component. The benefit of broader and biologically specific systemic surveillance when pre-targeting is done using antibodies is not captured in cases where a locally implanted material depot is used as the pre-targeting entity. This requirement for a priori knowledge of the desired site of action also limits uses for the latter case in disseminated diseases such as metastatic cancer, yet may remain relevant for applications in regenerating active signals on implanted biomedical devices. There are also remaining challenges to better integrate supramolecular targeting motifs with relevant methods in prodrug chemistry, such as incorporating analyte- or enzyme-sensitive linkers for site-specific drug activation following homing.
The emerging concept to use non-covalent association of supramolecular motifs offers inspiration to reimagine the diagnosis and treatment of disease. With nature as inspiration for specific non-covalent recognition in physiological conditions, recreating these concepts using synthetic tools is a path primed for many possible applications. Accordingly, the concept of supramolecular “click chemistry” for in situ targeting offers a promising direction ripe for further evaluation.
SYNOPSIS.
Synthetic supramolecular recognition motifs offer new routes for pre-targeting and in situ complexation, yielding a means for site-specific targeting of drugs or imaging agents.
ACKNOWLEDGMENT
MJW gratefully acknowledges funding support for this work from the National Institutes of Health (R35GM137987), the National Science Foundation (BMAT, 1944875), a 3M Non-Tenured Faculty Award (3M Company), and the University of Notre Dame “Advancing our Vision” initiative. All schematics and graphics were created using BioRender.com.
Footnotes
ASSOCIATED CONTENT
Supporting Information
There is no online supporting information for this work.
The authors declare no competing financial interests.
REFERENCES
- (1).Langer R Drug Delivery and Targeting. Nature 1998, 392 (6679 Suppl), 5–10. [PubMed] [Google Scholar]
- (2).Pattni BS; Chupin VV; Torchilin VP New Developments in Liposomal Drug Delivery. Chemical Reviews 2015, pp 10938–10966. 10.1021/acs.chemrev.5b00046. [DOI] [PubMed]
- (3).Izci M; Maksoudian C; Manshian BB; Soenen SJ The Use of Alternative Strategies for Enhanced Nanoparticle Delivery to Solid Tumors. Chem. Rev 2021, 121 (3), 1746–1803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Golombek SK; May J-N; Theek B; Appold L; Drude N; Kiessling F; Lammers T Tumor Targeting via EPR: Strategies to Enhance Patient Responses. Adv. Drug Deliv. Rev 2018, 130, 17–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Doxil® — The First FDA-Approved Nano-Drug: Lessons Learned. J. Control. Release 2012, 160 (2), 117–134. [DOI] [PubMed] [Google Scholar]
- (6).Alley SC; Okeley NM; Senter PD Antibody-Drug Conjugates: Targeted Drug Delivery for Cancer. Curr. Opin. Chem. Biol 2010, 14 (4), 529–537. [DOI] [PubMed] [Google Scholar]
- (7).Emerich DF; Thanos CG Targeted Nanoparticle-Based Drug Delivery and Diagnosis. J. Drug Target 2007, 15 (3), 163–183. [DOI] [PubMed] [Google Scholar]
- (8).Omstead DT; Sjoerdsma J; Bilgicer B Polyvalent Nanoobjects for Precision Diagnostics. Annu. Rev. Anal. Chem 2019, 12 (1), 69–88. [DOI] [PubMed] [Google Scholar]
- (9).Drago JZ; Modi S; Chandarlapaty S Unlocking the Potential of Antibody–drug Conjugates for Cancer Therapy. Nature Reviews Clinical Oncology 2021, pp 327–344. 10.1038/s41571-021-00470-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Beck A; Goetsch L; Dumontet C; Corvaïa N Strategies and Challenges for the next Generation of Antibody-Drug Conjugates. Nat. Rev. Drug Discov 2017, 16 (5), 315–337. [DOI] [PubMed] [Google Scholar]
- (11).Lv S; Sylvestre M; Prossnitz AN; Yang LF; Pun SH Design of Polymeric Carriers for Intracellular Peptide Delivery in Oncology Applications. Chem. Rev 2021. 10.1021/acs.chemrev.0c00963. [DOI] [PubMed]
- (12).Kunjachan S; Pola R; Gremse F; Theek B; Ehling J; Moeckel D; Hermanns-Sachweh B; Pechar M; Ulbrich K; Hennink WE; Storm G; Lederle W; Kiessling F; Lammers T Passive versus Active Tumor Targeting Using RGD- and NGR-Modified Polymeric Nanomedicines. Nano Lett 2014, 14 (2), 972–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Omstead DT; Mejia F; Sjoerdsma J; Kim B; Shin J; Khan S; Wu J; Kiziltepe T; Littlepage LE; Bilgicer B In Vivo Evaluation of CD38 and CD138 as Targets for Nanoparticle-Based Drug Delivery in Multiple Myeloma. J. Hematol. Oncol 2020, 13 (1), 145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Wang W; Singh S; Zeng DL; King K; Nema S Antibody Structure, Instability, and Formulation. J. Pharm. Sci 2007, 96 (1), 1–26. [DOI] [PubMed] [Google Scholar]
- (15).Coats S; Williams M; Kebble B; Dixit R; Tseng L; Yao N-S; Tice DA; Soria J-C Antibody-Drug Conjugates: Future Directions in Clinical and Translational Strategies to Improve the Therapeutic Index. Clin. Cancer Res 2019, 25 (18), 5441–5448. [DOI] [PubMed] [Google Scholar]
- (16).Epenetos AA; Snook D; Durbin H; Johnson PM; Taylor-Papadimitriou J Limitations of Radiolabeled Monoclonal Antibodies for Localization of Human Neoplasms. Cancer Res 1986, 46 (6), 3183–3191. [PubMed] [Google Scholar]
- (17).Bornstein GG Antibody Drug Conjugates: Preclinical Considerations. AAPS J 2015, 17 (3), 525–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Dai Q; Wilhelm S; Ding D; Syed AM; Sindhwani S; Zhang Y; Chen YY; MacMillan P; Chan WCW. Quantifying the Ligand-Coated Nanoparticle Delivery to Cancer Cells in Solid Tumors. ACS Nano 2018, 12 (8), 8423–8435. [DOI] [PubMed] [Google Scholar]
- (19).Devaraj NK The Future of Bioorthogonal Chemistry. ACS Central Science 2018, pp 952–959. 10.1021/acscentsci.8b00251. [DOI] [PMC free article] [PubMed]
- (20).Chang PV; Prescher JA; Sletten EM; Baskin JM; Miller IA; Agard NJ; Lo A; Bertozzi CR Copper-Free Click Chemistry in Living Animals. Proc. Natl. Acad. Sci. U. S. A 2010, 107 (5), 1821–1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Prescher JA; Dube DH; Bertozzi CR Chemical Remodelling of Cell Surfaces in Living Animals. Nature 2004, 430 (7002), 873–877. [DOI] [PubMed] [Google Scholar]
- (22).Baskin JM; Prescher JA; Laughlin ST; Agard NJ; Chang PV; Miller IA; Lo A; Codelli JA; Bertozzi CR Copper-Free Click Chemistry for Dynamic in Vivo Imaging. Proc. Natl. Acad. Sci. U. S. A 2007, 104 (43), 16793–16797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Brudno Y; Pezone MJ; Snyder TK; Uzun O; Moody CT; Aizenberg M; Mooney DJ Replenishable Drug Depot to Combat Post-Resection Cancer Recurrence. Biomaterials 2018, 178, 373–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Evans HL; Slade RL; Carroll L; Smith G; Nguyen QD; Iddon L; Kamaly N; Stöckmann H; Leeper FJ; Aboagye EO; Spivey AC Copper-Free Click—a Promising Tool for Pre-Targeted PET Imaging. Chem. Commun 2012, pp 991–993. 10.1039/c1cc16220a. [DOI] [PubMed]
- (25).Wang H; Sobral MC; Snyder T; Brudno Y; Gorantla VS; Mooney DJ Clickable, Acid Labile Immunosuppressive Prodrugs for in Vivo Targeting. Biomater Sci 2020, 8 (1), 266–277. [DOI] [PubMed] [Google Scholar]
- (26).Palvai S; Bhangu J; Akgun B; Moody CT; Hall DG; Brudno Y In Vivo Targeting Using Arylboronate/Nopoldiol Click Conjugation. Bioconjugate Chemistry 2020, pp 2288–2292. 10.1021/acs.bioconjchem.0c00453. [DOI] [PMC free article] [PubMed]
- (27).Rossin R; Verkerk PR; van den Bosch SM; Vulders RCM; Verel I; Lub J; Robillard MS In Vivo Chemistry for Pretargeted Tumor Imaging in Live Mice. Angew. Chem. Int. Ed Engl 2010, 49 (19), 3375–3378. [DOI] [PubMed] [Google Scholar]
- (28).Versteegen RM; Rossin R; ten Hoeve W; Janssen HM; Robillard MS Click to Release: Instantaneous Doxorubicin Elimination upon Tetrazine Ligation. Angew. Chem. Int. Ed Engl 2013, 52 (52), 14112–14116. [DOI] [PubMed] [Google Scholar]
- (29).Mejia Oneto JM; Khan I; Seebald L; Royzen M In Vivo Bioorthogonal Chemistry Enables Local Hydrogel and Systemic Pro-Drug To Treat Soft Tissue Sarcoma. ACS Cent Sci 2016, 2 (7), 476–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Liu Z; Nalluri SKM; Stoddart JF Surveying Macrocyclic Chemistry: From Flexible Crown Ethers to Rigid Cyclophanes. Chem. Soc. Rev 2017, 46 (9), 2459–2478. [DOI] [PubMed] [Google Scholar]
- (31).Khvorova A; Watts JK The Chemical Evolution of Oligonucleotide Therapies of Clinical Utility. Nat. Biotechnol 2017, 35 (3), 238–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Webber MJ; Langer R Drug Delivery by Supramolecular Design. Chem. Soc. Rev 2017, 46 (21), 6600–6620. [DOI] [PubMed] [Google Scholar]
- (33).Schreiber CL; Smith BD Molecular Conjugation Using Non-Covalent Click Chemistry. Nat Rev Chem 2019, 3 (6), 393–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Webber MJ; Appel EA; Meijer EW; Langer R Supramolecular Biomaterials. Nat. Mater 2016, 15 (1), 13–26. [DOI] [PubMed] [Google Scholar]
- (35).Braegelman AS; Webber MJ Integrating Stimuli-Responsive Properties in Host-Guest Supramolecular Drug Delivery Systems. Theranostics 2019, 9 (11), 3017–3040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Tang H; Fuentealba D; Ko YH; Selvapalam N; Kim K; Bohne C Guest Binding Dynamics with cucurbit[7]uril in the Presence of Cations. J. Am. Chem. Soc 2011, 133 (50), 20623–20633. [DOI] [PubMed] [Google Scholar]
- (37).Ning X; Guo J; Wolfert MA; Boons G-J Visualizing Metabolically Labeled Glycoconjugates of Living Cells by Copper-Free and Fast Huisgen Cycloadditions. Angew. Chem. Int. Ed Engl 2008, 47 (12), 2253–2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Taylor MT; Blackman ML; Dmitrenko O; Fox JM Design and Synthesis of Highly Reactive Dienophiles for the Tetrazine-Trans-Cyclooctene Ligation. J. Am. Chem. Soc 2011, 133 (25), 9646–9649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Sela-Culang I; Kunik V; Ofran Y The Structural Basis of Antibody-Antigen Recognition. Front. Immunol 2013, 4, 302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Erlendsson S; Teilum K Binding Revisited-Avidity in Cellular Function and Signaling. Front Mol Biosci 2020, 7, 615565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Vauquelin G; Charlton SJ Exploring Avidity: Understanding the Potential Gains in Functional Affinity and Target Residence Time of Bivalent and Heterobivalent Ligands. Br. J. Pharmacol 2013, 168 (8), 1771–1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Liu W; Samanta SK; Smith BD; Isaacs L Synthetic Mimics of Biotin/(strept)avidin. Chem. Soc. Rev 2017, 46 (9), 2391–2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Zou L; VandenBerg MA; Webber MJ Single-Molecule Nanoscale Drug Carriers with Quantitative Supramolecular Loading. Molecular Systems Design & Engineering 2020, pp 197–204. 10.1039/c9me00088g. [DOI]
- (44).Liu W; Johnson A; Smith BD Guest Back-Folding: A Molecular Design Strategy That Produces a Deep-Red Fluorescent Host/Guest Pair with Picomolar Affinity in Water. J. Am. Chem. Soc 2018, 140 (9), 3361–3370. [DOI] [PubMed] [Google Scholar]
- (45).Liu W; McGarraugh H; Smith B Fluorescent Thienothiophene-Containing Squaraine Dyes and Threaded Supramolecular Complexes with Tunable Wavelengths between 600–800 Nm. Molecules 2018, p 2229. 10.3390/molecules23092229. [DOI] [PMC free article] [PubMed]
- (46).Crini G Review: A History of Cyclodextrins. Chemical Reviews 2014, pp 10940–10975. 10.1021/cr500081p. [DOI] [PubMed]
- (47).Jansook P; Ogawa N; Loftsson T Cyclodextrins: Structure, Physicochemical Properties and Pharmaceutical Applications. Int. J. Pharm 2018, 535 (1–2), 272–284. [DOI] [PubMed] [Google Scholar]
- (48).Hu Q-D; Tang G-P; Chu PK Cyclodextrin-Based Host-Guest Supramolecular Nanoparticles for Delivery: From Design to Applications. Acc. Chem. Res 2014, 47 (7), 2017–2025. [DOI] [PubMed] [Google Scholar]
- (49).Eftink MR; Andy ML; Bystrom K; Perlmutter HD; Kristol DS ChemInform Abstract: Cyclodextrin Inclusion Complexes: Studies of the Variation in the Size of Alicyclic Guests. ChemInform 1989. 10.1002/chin.198946125. [DOI]
- (50).Chen G; Jiang M Cyclodextrin-Based Inclusion Complexation Bridging Supramolecular Chemistry and Macromolecular Self-Assembly. Chemical Society Reviews 2011, p 2254. 10.1039/c0cs00153h. [DOI] [PubMed]
- (51).Lee JW; Samal S; Selvapalam N; Kim H-J; Kim K Cucurbituril Homologues and Derivatives: New Opportunities in Supramolecular Chemistry. Acc. Chem. Res 2003, 36 (8), 621–630. [DOI] [PubMed] [Google Scholar]
- (52).Kim J; Jung I-S; Kim S-Y; Lee E; Kang J-K; Sakamoto S; Yamaguchi K; Kim K New Cucurbituril Homologues: Syntheses, Isolation, Characterization, and X-Ray Crystal Structures of Cucurbit[n]uril (n = 5, 7, and 8). Journal of the American Chemical Society 2000, pp 540–541. 10.1021/ja993376p. [DOI]
- (53).Lagona J; Mukhopadhyay P; Chakrabarti S; Isaacs L The Cucurbit[n]uril Family. Angew. Chem. Int. Ed Engl 2005, 44 (31), 4844–4870. [DOI] [PubMed] [Google Scholar]
- (54).Barrow SJ; Kasera S; Rowland MJ; del Barrio J; Scherman OA Cucurbituril-Based Molecular Recognition. Chem. Rev 2015, 115 (22), 12320–12406. [DOI] [PubMed] [Google Scholar]
- (55).Shetty D; Khedkar JK; Park KM; Kim K Can We Beat the Biotin-Avidin Pair?: cucurbit[7]uril-Based Ultrahigh Affinity Host-Guest Complexes and Their Applications. Chem. Soc. Rev 2015, 44 (23), 8747–8761. [DOI] [PubMed] [Google Scholar]
- (56).Cao L; Śekutor M; Zavalij PY; Mlinarić-Majerski K; Glaser R; Isaacs L Cucurbit[7]uril⋅guest Pair with an Attomolar Dissociation Constant. Angew. Chem. Int. Ed Engl 2014, 53 (4), 988–993. [DOI] [PubMed] [Google Scholar]
- (57).Rekharsky MV; Mori T; Yang C; Ko YH; Selvapalam N; Kim H; Sobransingh D; Kaifer AE; Liu S; Isaacs L; Chen W; Moghaddam S; Gilson MK; Kim K; Inoue Y A Synthetic Host-Guest System Achieves Avidin-Biotin Affinity by Overcoming Enthalpy-Entropy Compensation. Proc. Natl. Acad. Sci. U. S. A 2007, 104 (52), 20737–20742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Sigwalt D; Šekutor M; Cao L; Zavalij PY; Hostaš J; Ajani H; Hobza P; Mlinarić-Majerski K; Glaser R; Isaacs L Unraveling the Structure-Affinity Relationship between Cucurbit[n]urils (n = 7, 8) and Cationic Diamondoids. J. Am. Chem. Soc 2017, 139 (8), 3249–3258. [DOI] [PubMed] [Google Scholar]
- (59).Herce HD; Schumacher D; Schneider AFL; Ludwig K; Mann FA; Fillies M; Kasper M-A; Reinke S; Krause E; Leonhardt H; Cardoso MC; Hackenberger CPR Cell-Permeable Nanobodies for Targeted Immunolabelling and Antigen Manipulation in Living Cells. Nat. Chem 2017, 9 (8), 762–771. [DOI] [PubMed] [Google Scholar]
- (60).Marquez BV; Lapi SE Pretargeted Immuno-PET: Overcoming Limitations of Space and Time. J. Nucl. Med 2016, 57 (3), 332–333. [DOI] [PubMed] [Google Scholar]
- (61).Hnatowich DJ; Virzi F; Rusckowski M Investigations of Avidin and Biotin for Imaging Applications. J. Nucl. Med 1987, 28 (8), 1294–1302. [PubMed] [Google Scholar]
- (62).Beck A; Wurch T; Bailly C; Corvaia N Strategies and Challenges for the next Generation of Therapeutic Antibodies. Nat. Rev. Immunol 2010, 10 (5), 345–352. [DOI] [PubMed] [Google Scholar]
- (63).Rothbauer U; Zolghadr K; Tillib S; Nowak D; Schermelleh L; Gahl A; Backmann N; Conrath K; Muyldermans S; Cardoso MC; Leonhardt H Targeting and Tracing Antigens in Live Cells with Fluorescent Nanobodies. Nat. Methods 2006, 3 (11), 887–889. [DOI] [PubMed] [Google Scholar]
- (64).Moghaddam S; Yang C; Rekharsky M; Ko YH; Kim K; Inoue Y; Gilson MK New Ultrahigh Affinity Host-Guest Complexes of cucurbit[7]uril with bicyclo[2.2.2]octane and Adamantane Guests: Thermodynamic Analysis and Evaluation of M2 Affinity Calculations. J. Am. Chem. Soc 2011, 133 (10), 3570–3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (65).Jeon WS; Moon K; Park SH; Chun H; Ko YH; Lee JY; Lee ES; Samal S; Selvapalam N; Rekharsky MV; Sindelar V; Sobransingh D; Inoue Y; Kaifer AE; Kim K Complexation of Ferrocene Derivatives by the cucurbit[7]uril Host: A Comparative Study of the Cucurbituril and Cyclodextrin Host Families. J. Am. Chem. Soc 2005, 127 (37), 12984–12989. [DOI] [PubMed] [Google Scholar]
- (66).Zou L; Braegelman AS; Webber MJ Dynamic Supramolecular Hydrogels Spanning an Unprecedented Range of Host-Guest Affinity. ACS Appl. Mater. Interfaces 2019, 11 (6), 5695–5700. [DOI] [PubMed] [Google Scholar]
- (67).Strebl MG; Yang J; Isaacs L; Hooker JM Adamantane/Cucurbituril: A Potential Pretargeted Imaging Strategy in Immuno-PET. Mol. Imaging 2018, 17, 1536012118799838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (68).Li M; Kim S; Lee A; Shrinidhi A; Ko YH; Lim HG; Kim HH; Bae KB; Park KM; Kim K Bio-Orthogonal Supramolecular Latching inside Live Animals and Its Application for in Vivo Cancer Imaging. ACS Appl. Mater. Interfaces 2019, 11 (47), 43920–43927. [DOI] [PubMed] [Google Scholar]
- (69).Zou L; Braegelman AS; Webber MJ Spatially Defined Drug Targeting by in Situ Host–Guest Chemistry in a Living Animal. ACS Central Science 2019, pp 1035–1043. 10.1021/acscentsci.9b00195. [DOI] [PMC free article] [PubMed]
- (70).Spa SJ; Welling MM; van Oosterom MN; Rietbergen DDD; Burgmans MC; Verboom W; Huskens J; Buckle T; van Leeuwen FWBA Supramolecular Approach for Liver Radioembolization. Theranostics 2018, 8 (9), 2377–2386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (71).Welling MM; Spa SJ; van Willigen DM; Rietbergen DDD; Roestenberg M; Buckle T; van Leeuwen FWB In Vivo Stability of Supramolecular Host–guest Complexes Monitored by Dual-Isotope Multiplexing in a Pre-Targeting Model of Experimental Liver Radioembolization. Journal of Controlled Release 2019, pp 126–134. 10.1016/j.jconrel.2018.11.020. [DOI] [PubMed]
- (72).Mehta M; Deeksha; Tewari D; Gupta G; Awasthi R; Singh H; Pandey P; Chellappan DK; Wadhwa R; Collet T; Hansbro PM; Kumar SR; Thangavelu L; Negi P; Dua K; Satija S Oligonucleotide Therapy: An Emerging Focus Area for Drug Delivery in Chronic Inflammatory Respiratory Diseases. Chem. Biol. Interact 2019, 308, 206–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (73).Crooke ST Therapeutic Applications of Oligonucleotides. Biotechnology 1992, 10 (8), 882–886. [DOI] [PubMed] [Google Scholar]
- (74).Juliano RL The Delivery of Therapeutic Oligonucleotides. Nucleic Acids Research 2016, pp 6518–6548. 10.1093/nar/gkw236. [DOI] [PMC free article] [PubMed]
- (75).Reynolds A; Leake D; Boese Q; Scaringe S; Marshall WS; Khvorova A Rational siRNA Design for RNA Interference. Nature Biotechnology 2004, pp 326–330. 10.1038/nbt936. [DOI] [PubMed]
- (76).Whitehead KA; Langer R; Anderson DG Knocking down Barriers: Advances in siRNA Delivery. Nat. Rev. Drug Discov 2009, 8 (2), 129–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (77).Carlon E; Heim T Thermodynamics of RNA/DNA Hybridization in High-Density Oligonucleotide Microarrays. Physica A: Statistical Mechanics and its Applications 2006, pp 433–449. 10.1016/j.physa.2005.09.067. [DOI]
- (78).SantaLucia J; Hicks D The Thermodynamics of DNA Structural Motifs. Annual Review of Biophysics and Biomolecular Structure 2004, pp 415–440. 10.1146/annurev.biophys.32.110601.141800. [DOI] [PubMed]
- (79).SantaLucia J A Unified View of Polymer, Dumbbell, and Oligonucleotide DNA Nearest-Neighbor Thermodynamics. Proceedings of the National Academy of Sciences 1998, pp 1460–1465. 10.1073/pnas.95.4.1460. [DOI] [PMC free article] [PubMed]
- (80).Elder RM; Pfaendtner J; Jayaraman A Effect of Hydrophobic and Hydrophilic Surfaces on the Stability of Double-Stranded DNA. Biomacromolecules 2015, pp 1862–1869. 10.1021/acs.biomac.5b00469. [DOI] [PubMed]
- (81).Owczarzy R; Dunietz I; Behlke MA; Klotz IM; Walder JA Thermodynamic Treatment of Oligonucleotide Duplex-Simplex Equilibria. Proceedings of the National Academy of Sciences 2003, pp 14840–14845. 10.1073/pnas.2335948100. [DOI] [PMC free article] [PubMed]
- (82).Bielec K; Sozanski K; Seynen M; Dziekan Z; Ten Wolde PR; Holyst R Kinetics and Equilibrium Constants of Oligonucleotides at Low Concentrations. Hybridization and Melting Study. Phys. Chem. Chem. Phys 2019, 21 (20), 10798–10807. [DOI] [PubMed] [Google Scholar]
- (83).Keefe AD; Pai S; Ellington A Aptamers as Therapeutics. Nat. Rev. Drug Discov 2010, 9 (7), 537–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (84).Hermann T; Patel DJ Adaptive Recognition by Nucleic Acid Aptamers. Science 2000, 287 (5454), 820–825. [DOI] [PubMed] [Google Scholar]
- (85).Ni S; Zhuo Z; Pan Y; Yu Y; Li F; Liu J; Wang L; Wu X; Li D; Wan Y; Zhang L; Yang Z; Zhang B-T; Lu A; Zhang G Recent Progress in Aptamer Discoveries and Modifications for Therapeutic Applications. ACS Appl. Mater. Interfaces 2021, 13 (8), 9500–9519. [DOI] [PubMed] [Google Scholar]
- (86).Roberts TC; Langer R; Wood MJA Advances in Oligonucleotide Drug Delivery. Nature Reviews Drug Discovery 2020, pp 673–694. 10.1038/s41573-020-0075-7. [DOI] [PMC free article] [PubMed]
- (87).Urban E; Noe CR Structural Modifications of Antisense Oligonucleotides. Il Farmaco 2003, pp 243–258. 10.1016/s0014-827x(03)00022-3. [DOI] [PubMed]
- (88).Glazier DA; Liao J; Roberts BL; Li X; Yang K; Stevens CM; Tang W Chemical Synthesis and Biological Application of Modified Oligonucleotides. Bioconjug. Chem 2020, 31 (5), 1213–1233. [DOI] [PubMed] [Google Scholar]
- (89).Hsu N-S; Lee C-C; Kuo W-C; Chang Y-W; Lo S-Y; Wang AH-J Development of a Versatile and Modular Linker for Antibody-Drug Conjugates Based on Oligonucleotide Strand Pairing. Bioconjug. Chem 2020, 31 (7), 1804–1811. [DOI] [PubMed] [Google Scholar]
- (90).Tan W; Wang K; Drake TJ Molecular Beacons. Curr. Opin. Chem. Biol 2004, 8 (5), 547–553. [DOI] [PubMed] [Google Scholar]
- (91).Tyagi S; Kramer FR Molecular Beacons: Probes That Fluoresce upon Hybridization. Nat. Biotechnol 1996, 14 (3), 303–308. [DOI] [PubMed] [Google Scholar]
- (92).Shi H; He X; Wang K; Wu X; Ye X; Guo Q; Tan W; Qing Z; Yang X; Zhou B Activatable Aptamer Probe for Contrast-Enhanced in Vivo Cancer Imaging Based on Cell Membrane Protein-Triggered Conformation Alteration. Proc. Natl. Acad. Sci. U. S. A 2011, 108 (10), 3900–3905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (93).Yan L. ‘an; Shi H; He X; Wang K; Tang J; Chen M; Ye X; Xu F; Lei Y A Versatile Activatable Fluorescence Probing Platform for Cancer Cells in Vitro and in Vivo Based on Self-Assembled Aptamer/carbon Nanotube Ensembles. Anal. Chem 2014, 86 (18), 9271–9277. [DOI] [PubMed] [Google Scholar]
- (94).Zhu G; Zheng J; Song E; Donovan M; Zhang K; Liu C; Tan W Self-Assembled, Aptamer-Tethered DNA Nanotrains for Targeted Transport of Molecular Drugs in Cancer Theranostics. Proc. Natl. Acad. Sci. U. S. A 2013, 110 (20), 7998–8003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (95).Zhou J; Soontornworajit B; Snipes MP; Wang Y Development of a Novel Pretargeting System with Bifunctional Nucleic Acid Molecules. Biochem. Biophys. Res. Commun 2009, 386 (3), 521–525. [DOI] [PubMed] [Google Scholar]
- (96).Shi P; Zhao N; Lai J; Coyne J; Gaddes ER; Wang Y Polyvalent Display of Biomolecules on Live Cells. Angew. Chem. Int. Ed Engl 2018, 57 (23), 6800–6804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (97).Schubert M; Bergmann R; Förster C; Sihver W; Vonhoff S; Klussmann S; Bethge L; Walther M; Schlesinger J; Pietzsch J; Steinbach J; Pietzsch H-J Novel Tumor Pretargeting System Based on Complementary L-Configured Oligonucleotides. Bioconjug. Chem 2017, 28 (4), 1176–1188. [DOI] [PubMed] [Google Scholar]
- (98).Nielsen PE; Egholm M; Berg RH; Buchardt O Sequence-Selective Recognition of DNA by Strand Displacement with a Thymine-Substituted Polyamide. Science 1991, 254 (5037), 1497–1500. [DOI] [PubMed] [Google Scholar]
- (99).Goldman JM; Zhang LA; Manna A; Armitage BA; Ly DH; Schneider JW High Affinity γPNA Sandwich Hybridization Assay for Rapid Detection of Short Nucleic Acid Targets with Single Mismatch Discrimination. Biomacromolecules 2013, 14 (7), 2253–2261. [DOI] [PubMed] [Google Scholar]
- (100).Gupta A; Mishra A; Puri N Peptide Nucleic Acids: Advanced Tools for Biomedical Applications. J. Biotechnol 2017, 259, 148–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (101).Nielsen PE Peptide Nucleic Acid. A Molecule with Two Identities. Accounts of Chemical Research 1999, pp 624–630. 10.1021/ar980010t. [DOI]
- (102).Rusckowski M; Qu T; Chang F; Hnatowich DJ Pretargeting Using Peptide Nucleic Acid. Cancer 1997, 80 (12 Suppl), 2699–2705. [DOI] [PubMed] [Google Scholar]
- (103).Brudno Y; Silva EA; Kearney CJ; Lewin SA; Miller A; Martinick KD; Aizenberg M; Mooney DJ Refilling Drug Delivery Depots through the Blood. Proc. Natl. Acad. Sci. U. S. A 2014, 111 (35), 12722–12727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (104).Chu T-W; Yang J; Zhang R; Sima M; Kopeček J Cell Surface Self-Assembly of Hybrid Nanoconjugates via Oligonucleotide Hybridization Induces Apoptosis. ACS Nano 2014, 8 (1), 719–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (105).Li L; Wang J; Li Y; Radford DC; Yang J; Kopeček J Broadening and Enhancing Functions of Antibodies by Self-Assembling Multimerization at Cell Surface. ACS Nano 2019, 13 (10), 11422–11432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (106).Yang J; Li L; Kopeček J Biorecognition: A Key to Drug-Free Macromolecular Therapeutics. Biomaterials 2019, 190–191, 11–23. [DOI] [PubMed]
- (107).Summerton J; Weller D Morpholino Antisense Oligomers: Design, Preparation, and Properties. Antisense and Nucleic Acid Drug Development 1997, pp 187–195. 10.1089/oli.1.1997.7.187. [DOI] [PubMed]
- (108).Murakami Y; Maeda M DNA-Responsive Hydrogels That Can Shrink or Swell. Biomacromolecules 2005, 6 (6), 2927–2929. [DOI] [PubMed] [Google Scholar]
- (109).Battig MR; Soontornworajit B; Wang Y Programmable Release of Multiple Protein Drugs from Aptamer-Functionalized Hydrogels via Nucleic Acid Hybridization. J. Am. Chem. Soc 2012, 134 (30), 12410–12413. [DOI] [PubMed] [Google Scholar]
- (110).McNamara SL; Brudno Y; Miller AB; Ham HO; Aizenberg M; Chaikof EL; Mooney DJ Regenerating Antithrombotic Surfaces through Nucleic Acid Displacement. ACS Biomater Sci Eng 2020, 6 (4), 2159–2166. [DOI] [PubMed] [Google Scholar]
- (111).Zhang DY; Winfree E Control of DNA Strand Displacement Kinetics Using Toehold Exchange. Journal of the American Chemical Society 2009, pp 17303–17314. 10.1021/ja906987s. [DOI] [PubMed]
- (112).Utterström J; Naeimipour S; Selegård R; Aili D Coiled Coil-Based Therapeutics and Drug Delivery Systems. Adv. Drug Deliv. Rev 2021, 170, 26–43. [DOI] [PubMed] [Google Scholar]
- (113).Yu YB Coiled-Coils: Stability, Specificity, and Drug Delivery Potential. Adv. Drug Deliv. Rev 2002, 54 (8), 1113–1129. [DOI] [PubMed] [Google Scholar]
- (114).Burkhard P; Stetefeld J; Strelkov SV Coiled Coils: A Highly Versatile Protein Folding Motif. Trends Cell Biol 2001, 11 (2), 82–88. [DOI] [PubMed] [Google Scholar]
- (115).Harbury PB; Zhang T; Kim PS; Alber T A Switch between Two-, Three-, and Four-Stranded Coiled Coils in GCN4 Leucine Zipper Mutants. Science 1993, 262 (5138), 1401–1407. [DOI] [PubMed] [Google Scholar]
- (116).Slovic AM; Lear JD; DeGrado WF De Novo Design of a Pentameric Coiled-Coil: Decoding the Motif for Tetramer versus Pentamer Formation in Water-Soluble Phospholamban. J. Pept. Res 2005, 65 (3), 312–321. [DOI] [PubMed] [Google Scholar]
- (117).Zaccai NR; Chi B; Thomson AR; Boyle AL; Bartlett GJ; Bruning M; Linden N; Sessions RB; Booth PJ; Leo Brady R; Woolfson DN A de Novo Peptide Hexamer with a Mutable Channel. Nature Chemical Biology 2011, pp 935–941. 10.1038/nchembio.692. [DOI] [PMC free article] [PubMed]
- (118).Moll JR; Ruvinov SB; Pastan I; Vinson C Designed Heterodimerizing Leucine Zippers with a Ranger of pIs and Stabilities up to 10(−15) M. Protein Sci 2001, 10 (3), 649–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (119).Reinke AW; Grant RA; Keating AE A Synthetic Coiled-Coil Interactome Provides Heterospecific Modules for Molecular Engineering. J. Am. Chem. Soc 2010, 132 (17), 6025–6031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (120).Woolfson DN The Design of Coiled-Coil Structures and Assemblies. Fibrous Proteins: Coiled-Coils, Collagen and Elastomers 2005, pp 79–112. 10.1016/s0065-3233(05)70004-8. [DOI] [PubMed]
- (121).Fletcher JM; Boyle AL; Bruning M; Bartlett GJ; Vincent TL; Zaccai NR; Armstrong CT; Bromley EHC; Booth PJ; Brady RL; Thomson AR; Woolfson DN A Basis Set of de Novo Coiled-Coil Peptide Oligomers for Rational Protein Design and Synthetic Biology. ACS Synth. Biol 2012, 1 (6), 240–250. [DOI] [PubMed] [Google Scholar]
- (122).Fletcher JM; Harniman RL; Barnes FRH; Boyle AL; Collins A; Mantell J; Sharp TH; Antognozzi M; Booth PJ; Linden N; Miles MJ; Sessions RB; Verkade P; Woolfson DN Self-Assembling Cages from Coiled-Coil Peptide Modules. Science 2013, 340 (6132), 595–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (123).Gradišar H; Božič S; Doles T; Vengust D; Hafner-Bratkovič I; Mertelj A; Webb B; Šali A; Klavžar S; Jerala R Design of a Single-Chain Polypeptide Tetrahedron Assembled from Coiled-Coil Segments. Nat. Chem. Biol 2013, 9 (6), 362–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (124).Pola R; Laga R; Ulbrich K; Sieglová I; Král V; Fábry M; Kabešová M; Kovář M; Pechar M Polymer Therapeutics with a Coiled Coil Motif Targeted against Murine bcl1 Leukemia. Biomacromolecules 2013, 14 (3), 881–889. [DOI] [PubMed] [Google Scholar]
- (125).Pechar M; Pola R; Laga R; Ulbrich K; Bednárová L; Maloň P; Sieglová I; Král V; Fábry M; Vaněk O Coiled Coil Peptides as Universal Linkers for the Attachment of Recombinant Proteins to Polymer Therapeutics. Biomacromolecules 2011, 12 (10), 3645–3655. [DOI] [PubMed] [Google Scholar]
- (126).Park WM; Champion JA Thermally Triggered Self-Assembly of Folded Proteins into Vesicles. J. Am. Chem. Soc 2014, 136 (52), 17906–17909. [DOI] [PubMed] [Google Scholar]
- (127).Dhankher A; Hernandez ME; Howard HC; Champion JA Characterization and Control of Dynamic Rearrangement in a Self-Assembled Antibody Carrier. Biomacromolecules 2020, pp 1407–1416. 10.1021/acs.biomac.9b01712. [DOI] [PMC free article] [PubMed]
- (128).Wu K; Liu J; Johnson RN; Yang J; Kopecek J Drug-Free Macromolecular Therapeutics: Induction of Apoptosis by Coiled-Coil-Mediated Cross-Linking of Antigens on the Cell Surface. Angew. Chem. Int. Ed Engl 2010, 49 (8), 1451–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (129).Wu K; Yang J; Liu J; Kopeček J Coiled-Coil Based Drug-Free Macromolecular Therapeutics: In Vivo Efficacy. J. Control. Release 2012, 157 (1), 126–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (130).Kverka M; Hartley JM; Chu T-W; Yang J; Heidchen R; Kopeček J Immunogenicity of Coiled-Coil Based Drug-Free Macromolecular Therapeutics. Biomaterials 2014, 35 (22), 5886–5896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (131).Zhang R; Yang J; Chu T-W; Hartley JM; Kopeček J Multimodality Imaging of Coiled-Coil Mediated Self-Assembly in a “Drug-Free” Therapeutic System. Advanced Healthcare Materials 2015, pp 1054–1065. 10.1002/adhm.201400679. [DOI] [PMC free article] [PubMed]
- (132).Zhang L; Fang Y; Li L; Yang J; Radford DC; Kopeček J Human Serum Albumin-Based Drug-Free Macromolecular Therapeutics: Apoptosis Induction by Coiled-Coil-Mediated Cross-Linking of CD20 Antigens on Lymphoma B Cell Surface. Macromol. Biosci 2018, 18 (11), e1800224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (133).Yang J; Shimada Y; Olsthoorn RCL; Snaar-Jagalska BE; Spaink HP; Kros A Application of Coiled Coil Peptides in Liposomal Anticancer Drug Delivery Using a Zebrafish Xenograft Model. ACS Nano 2016, 10 (8), 7428–7435. [DOI] [PubMed] [Google Scholar]


