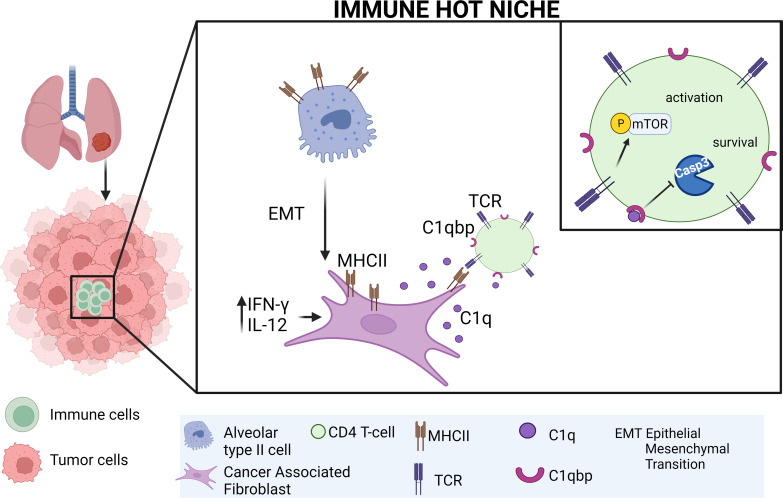
Kerdidani et al. show that immunological hot spots in lung tumors are sustained by fibroblasts. Lung fibroblasts present MHCII cancer antigens locally and at the same time prevent CD4 T cells from apoptosis by activating the complement receptor C1qbp.
Abstract
A key unknown of the functional space in tumor immunity is whether CD4 T cells depend on intratumoral MHCII cancer antigen recognition. MHCII-expressing, antigen-presenting cancer-associated fibroblasts (apCAFs) have been found in breast and pancreatic tumors and are considered to be immunosuppressive. This analysis shows that antigen-presenting fibroblasts are frequent in human lung non-small cell carcinomas, where they seem to actively promote rather than suppress MHCII immunity. Lung apCAFs directly activated the TCRs of effector CD4 T cells and at the same time produced C1q, which acted on T cell C1qbp to rescue them from apoptosis. Fibroblast-specific MHCII or C1q deletion impaired CD4 T cell immunity and accelerated tumor growth, while inducing C1qbp in adoptively transferred CD4 T cells expanded their numbers and reduced tumors. Collectively, we have characterized in the lungs a subset of antigen-presenting fibroblasts with tumor-suppressive properties and propose that cancer immunotherapies might be strongly dependent on in situ MHCII antigen presentation.
Graphical Abstract
Introduction
The series of immunological events that takes place between tumors and tumor-draining LNs forms a cyclic trajectory that is being referred to as the cancer-immunity cycle (Chen and Mellman, 2013). In the first step of these events, tumor antigens are carried to the tumor-draining LNs and are partly transferred to resident dendritic cells (DCs; Ruhland et al., 2020). In LNs, migratory and resident DCs present their antigenic cargo to antigen-inexperienced (naive) T cells, which become differentiated effector cells that egress from LNs and enter tumors. In tumors, CD8 cells exhibit direct killing activity against cancer cells, but they are seriously dependent on CD4 T cells for function and transition to memory cells (Ahrends et al., 2019; Binnewies et al., 2019; Śledzińska et al., 2020; Zander et al., 2019; Bos and Sherman, 2010; Schietinger et al., 2010). Although our current understanding of the functional space in the cancer-immunity cycle is that MHCII cancer antigen presentation primarily occurs in LNs, the contribution of in situ cancer antigen presentation has not been ruled out (Dammeijer et al., 2020; Oh et al., 2020).
A few studies have directly addressed the role of peripheral antigen presentation in CD4 T cell responses (Dammeijer et al., 2020; Doebis et al., 2011; Low et al., 2020; McLachlan and Jenkins, 2007; Schøller et al., 2019). In cancer, three lines of evidence support that the TCRs are stimulated in situ within solid tumors. First, the CD4 TCR repertoire is regionally shaped by the local neoantigen load (Joshi et al., 2019). Second, stemlike CD8 T cells reside in dense MHCII-expressing cell niches within tumors (Jansen et al., 2019). Third, right flank tumors that differ only in one MHCII neoantigen with left flank tumors are infiltrated by higher numbers of neoantigen-specific CD4+ T cells (Alspach et al., 2019). DCs are scarce and immature within solid tumors and are generally considered to exert their primary effects in LNs (Dammeijer et al., 2020; Ferris et al., 2020; Maier et al., 2020; Ruhland et al., 2020; Oh et al., 2020). Because structural tissue cells greatly outnumber professional antigen-presenting cells, express immune genes (Krausgruber et al., 2020), and can be induced to present antigens (Koyama et al., 2019; Low et al., 2020), we hypothesized that they are required for local antigen presentation and anti-tumor immunity.
Fibroblasts are largely considered to be immunosuppressive cells (Biffi and Tuveson, 2021). Accordingly, the recently identified MHCII+ antigen-presenting cancer-associated fibroblasts (apCAFs) in pancreatic adenocarcinoma (PDAC) and breast carcinoma (BC) are presumed to induce immune tolerance (Dominguez et al., 2020; Kieffer et al., 2020; Elyada et al., 2019; Friedman et al., 2020; Sebastian et al., 2020). Here, we show that MHCII-expressing fibroblasts are frequently found in lung non-small cell carcinomas and that they anatomically define areas of high rather than low CD4 T cell density. In orthotopic murine models of lung cancer, fibroblast-specific targeted ablation of MHCII impaired local immunity and accelerated tumor growth. Human and murine apCAFs directly activated the TCRs of effector CD4 T cells and, in parallel, protected them from apoptosis via the C1q receptor C1qbp, while C1qbp overexpression expanded the pool of adoptively transferred CD4 T cells in tumors and reduced tumor burden. Our studies identify and characterize lung apCAFs as tumor-suppressive cells and propose a new conceptual framework under which tumor MHCII immunity can be seen: CD4 T cells require in situ antigen presentation within tumors for effective MHCII immunity to occur.
Results
apCAFs activate adjacent CD4 T cells within human lung tumors
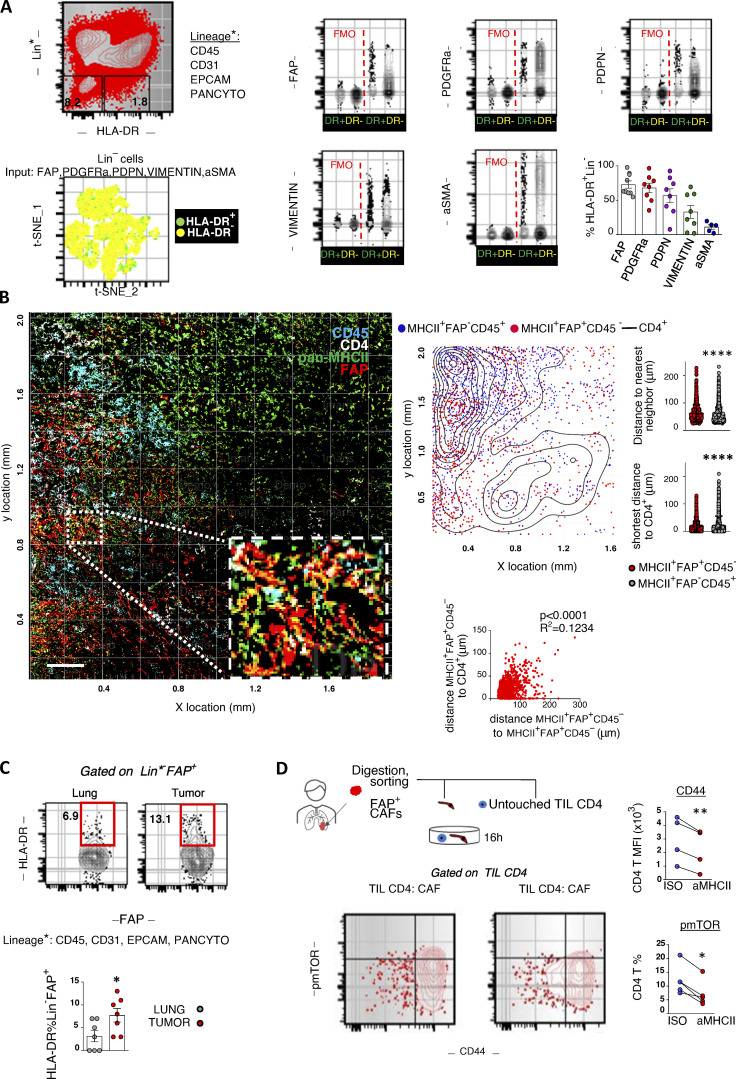
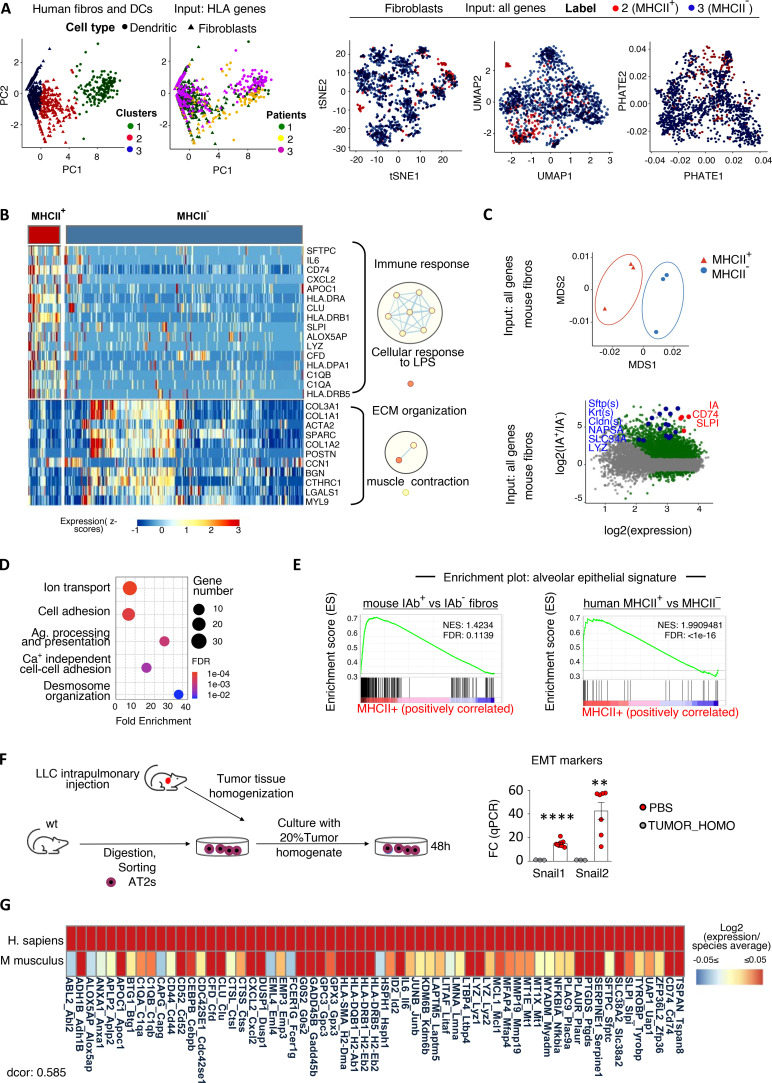
To elaborate on the fibroblastic identity of MHCII+ non-hematopoietic cells, we enzymatically dispersed primary human tumors and analyzed known mesenchymal markers by FACS (Fig. 1 A). MHCII+Lin−CAFs (apCAFs) were phenotypically similar to their MHCII− counterparts and largely coexpressed FAP, PDGFRa, and Podoplanin. Vimentin was expressed at variable levels, while aSMA was lowly expressed (Fig. 1 A). Accordingly, two-dimensional (2D) visualization of lineage− cells in t-distributed stochastic neighbor embedding (t-SNE) plots (input all mesenchymal markers) confirmed that apCAFs do not cluster separately from MHCII− cancer-associated fibroblasts (CAFs). To determine the spatial relationship between apCAFs and CD4 T cells, we analyzed whole-tumor tissue sections by immunofluorescence staining. apCAFs were identified as CD45− cells that coexpressed MHCII and FAP. We noticed that FACS had impressively underestimated the frequencies of fibroblasts compared with imaging. This discrepancy likely reflected differences in detachment efficiency of different cell types following tissue digestion, with fibroblasts being more adherent to the extracellular matrix (ECM) than immune cells and thus more difficult to disconnect. Quantitative analysis showed similar densities of intratumoral MHCII+FAP+CD45− relative to MHCII+FAP−CD45+ hematopoietic antigen-presenting cells, indicating a physiologically relevant abundance of apCAFs. CD4+ T cells accumulated in dense apCAF regions and were identified closer to apCAFs rather than MHCII+FAP−CD45+ cells (Fig. 1 B). We interrogated whether we could identify MHCII fibroblasts in healthy lungs. To rule out the possibility of contamination by non-fibroblastic cell populations, we gated fibroblasts as FSC-AhighLin−FAP+ cells. MHCII was expressed by a subset of normal lung fibroblasts, but it was more frequently detected in CAFs (Fig. 1 C). Collectively, the above data suggest that the tumor microenvironment drives antigen presentation in fibroblasts and that apCAFs create functional immunological spots within lung tumors that might sustain CD4 T cell populations.
Figure 1.
MHCII fibroblasts form CD4 T cell priming spots within human lung tumors. (A) Representative FACS plots of a digested human lung tumor. t-SNE plot illustrates all lineage− cells and was run using expression of all mesenchymal-specific markers of a representative patient as input. Each dot represents a single cell. Cumulative data (n = 5–9 patients). (B) Representative whole-slide imaging for pan-MHCII, FAP, CD45, and CD4 and cellular spatial relationship map from a lung cancer patient. Samples were analyzed by confocal microscopy with a 20× objective. Bars, 200 µm. After acquiring XY coordinates, XY location of MHCII+FAP+CD45− and MHCII+FAP−CD45+ cells was overlaid with CD4 cell density contour. Cell density of MHCII+FAP+CD45− and MHCII+FAP−CD45+ cells calculated as average distance to nearest neighbor. Distance between MHCII+FAP+CD45− or MHCII+FAP−CD45+ and the closest CD4 cell. Co-localization of MHCII+FAP+CD45− and CD4+ cells, depicted as positive correlation between shortest distance of MHCII+FAP+CD45− to CD4+ with average distance of MHCII+FAP+CD45− to nearest neighbor MHCII+FAP+CD45− (representative of n = 3 patients). (C) FACS plots showing MHCII in FSC-AHighLin−FAP+PDGFRa+ fibroblasts of paired digested human lung tumors and tumor-free lungs (n = 7 patients). (D) CD4 T cells and fibroblasts were sorted from the same tumor fragment and co-cultured with pan-MHCII blocking antibody or isotype (ISO) control. Representative FACS plots and cumulative data on phospho-mTOR (pmTOR) and CD44 (n = 4 patients). (B–D) *, P < 0.05; **, P < 0.01; ****, P < 0.0001. aMHCII, pan-MHCII blocking antibody; FMO, fluorescence minus one; MFI, mean fluorescence intensity. Error bars, mean ± SEM; two-tailed unpaired or paired t test.
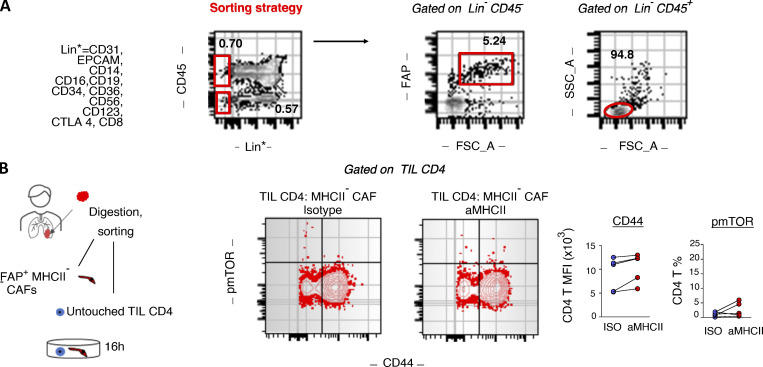
Spatial heterogeneity of T cell clones suggests regional activation by neighboring antigen-presenting cells that present their cognate antigens (Joshi et al., 2019). We reasoned that apCAFs directly presented cancer MHCII peptides to adjacent CD4 T cells. To test this, we co-cultured primary human CD4 T cells and FAP+ CAFs, purified from the same lung tumor fragment, and assessed phosphorylation of the key TCR/CD28–signaling node, mTOR (Fig. 1 D). Primary cell phenotypes are dramatically altered once they are isolated and cultured. To avoid this bias, we used freshly FACS-sorted primary tumor fibroblasts and freshly sorted autologous primary tumor–infiltrating T cells for all our co-cultures. We used the bulk of CAFs rather than MHCII+ CAFs to avoid blocking TCR recognition and did not supplement the culture medium with exogenous cytokines. Under these stringent conditions, ∼1 in 10 intratumoral T cells responded to direct CAF contact by mTOR phosphorylation and CD44 up-regulation, and these effects were abolished with MHCII blockade (Fig. 1 D). Control co-cultures of apCAF-depleted CAFs and CD4 T cells confirmed the dependence of CD4 T cell activation on the apCAF fraction of bulk CAFs (Fig. S1). Thus, human apCAFs acquire exogenous peptides within tumors in vivo and can directly prime adjacent CD4 T cells ex vivo.
Figure S1.
Human MHCII− CAF-tumor infiltrating CD4 T cell co-cultures. (A) Gating strategy for sorting untouched CD4 T cells and CAFs from the same tumor fragment. CAFs are sorted as live FSC-AhighLin−CD45−FAP+ and untouched CD4 T cells as FSC-AlowLin−CD45+. (B) Purified CD4 T cells and FSC-AhighLin−CD45−FAP+MHCII− CAFs were co-cultured with panMHCII-blocking antibody or isotype (ISO) control. Representative FACS plots and cumulative data on phospho-mTOR (pmTOR) and CD44 (n = 5 patients). *, P < 0.05; **, P < 0.01. Error bars, mean ± SEM; two-tailed paired t test. aMHCII, pan-MHCII blocking antibody; FSC-A, forward scatter-A; MFI, mean fluorescence intensity; SSC-A, side scatter-A.
apCAFs are detected in tumors of IFN-γ–sufficient mice
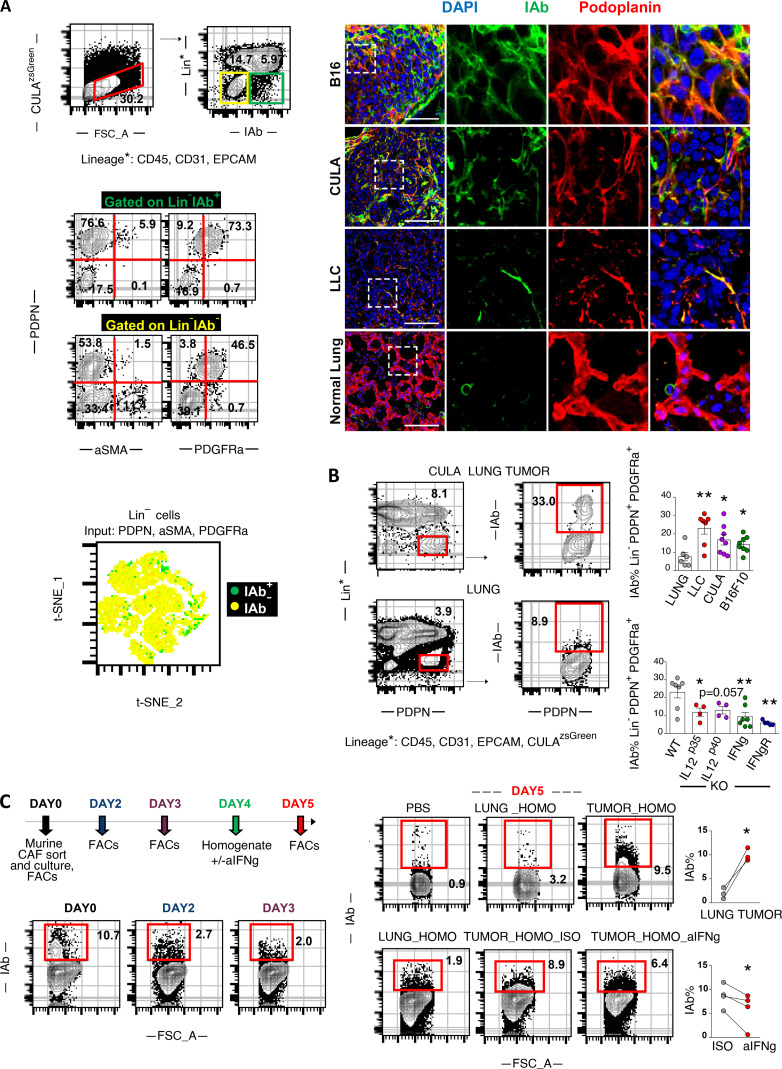
To assess whether apCAFs were present in murine tissues, we analyzed two models of orthotopic lung cancer and one metastatic. We inoculated one commercially available Lewis Lung Carcinoma (LLC) cell line (LLCmCherry) or an autochthonous cancer cell line (CULAzsGreen) in the left lung lobe of syngeneic mice or injected melanoma cells (B16F10mCherry) in the tail vein of mice. Tumors were digested and subjected to FACS analysis. Intratumoral FSC-AhighCD45−EpCAM−CD31−IAb(MHCII)+ non-cancer cells coexpressed Podoplanin and PDGFRa, but not aSMA (Fig. 2 A). Murine FAP antibodies showed nonspecific and unreliable staining and were thus excluded from the analysis. Similar to human apCAFs, 2D visualization of all non-cancer lineage− cells in t-SNE plots indicated that apCAFs are admixed with MHCII− CAFs. Immunofluorescence staining detected MHCII+Podoplanin+ cells within the tumor bed (Fig. 2 A). Similar to what we had observed in humans, MHCII was more frequently expressed by CAFs relative to healthy lung fibroblasts (Fig. 2 B). To determine whether fibroblasts depended on IFN-γ for MHCII expression, we inoculated LLC cells in the lungs of IL-12 p35−/−, IL12p40−/−, IFN-γR−/−, and IFN-γ−/− mice. MHCII expression was decreased in CAFs of IL-12–deficient mice and severely impaired in those of IFN-γ/IFN-γR–deficient versus wild-type mice (Fig. 2 B).
Figure 2.
MHCII expression in fibroblasts is IFN-γ dependent. (A–D) LLC/CULA lung adenocarcinoma cells or B16F10 melanoma cells were injected in the left lung lobe of syngeneic mice or through the tail vein, respectively. (A) Representative FACS plots of a digested mouse CULA lung tumor and expression of mesenchymal-specific genes. Immunofluorescence for IAb (MHCII) and Podoplanin identifies MHCII fibroblasts. Samples were analyzed by confocal microscopy with a 40× objective. Bars, 50 µm. (B) Top: Representative FACS plots of IAb in FSC-AHighLin−Podoplanin+PDGFRa+ fibroblasts of a CULA lung tumor and a healthy lung of wild-type C57BL/6 mice. Bottom left: Cumulative data on IAb expression of LLC, CULA, and B16F10 lung tumors and healthy lungs of wild-type C57BL/6 mice (n = 6–8 per group). Bottom right: Cumulative data on IAb expression of LLC lung tumors of cytokine-deficient mice (n = 4–7 per group). (C) FSC-AHighLin−Podoplanin+PDGFRa+ fibroblasts were isolated from pooled LLC lung tumors and cultured in 3D, as indicated (n = 4 experiments). FSC-A, forward scatter-A; ISO, isotype control; Lung_homo, lung homogenate; Tumor_homo, tumor homogenate. (B and C) *, P < 0.05; **, P < 0.01. Error bars, mean ± SEM; two-tailed unpaired or paired t test.
To probe the contribution of the tumor microenvironment in sustaining MHCII in fibroblasts, we developed an in vitro model system where we sorted PDGFRa+Podoplanin+ fibroblasts from LLCmCherry tumors and cultured them in vitro. Although by day 3 all cultured CAFs had lost MHCII, exposure to fresh lung tumor, but not to healthy lung homogenates, restored MHCII expression (Fig. 2 C). It should be noted that MHCII induction was observed upon three-dimensional (3D) but not 2D culture conditions, consistent with previous observations that 2D cultures induce MHCII loss in PDAC fibroblasts and differentiation to myofibroblasts (Elyada et al., 2019). Notably, neutralizing IFN-γ in tumor homogenates restrained MHCII induction in primary CAFs (Fig. 2 C).
Human and murine apCAF transcriptomes reveal conserved immune phenotypes and epithelial signature
To characterize human antigen-presenting fibroblasts, we leveraged a public single-cell RNA sequencing (scRNAseq) dataset of primary human lung tumor, adjacent unaffected lung, and computational analyses (Lambrechts et al., 2018). There is no standardized computational method for characterizing gene expression levels in a binary form of “high” and “low.” Furthermore, gene expression levels correlate variably with corresponding protein levels. We hypothesized that we would be able to identify antigen-presenting fibroblasts in single fibroblast RNA sequencing (RNAseq) datasets if we used classical DCs (cDCs) as a metric for physiologically relevant MHCII gene expression. Re-clustering of 1,284 single cells annotated as either fibroblasts (798) or cross-presenting cDCs (486; E-MTAB-6149 and E-MTAB-6653; n = 3 patients) via k-means clustering, using as input only input MHCII genes (k = 3 clusters supported by Silhouette scoring), separated fibroblasts into two clusters that shared (MHCII+) or did not share (MHCII−) MHCII expression features with cDCs. Different dimensionality reduction and visualization approaches using as input all 1,785 expressed genes indicated that MHCII+ fibroblasts do not form a distinct cluster among total fibroblasts (Fig. 3 A). Differential expression analysis (DEA) revealed 115 deregulated genes in MHCII+ versus MHCII− fibroblasts, 67 of which were up-regulated (Fig. 3 B). Among the top up-regulated were signature genes of PDAC apCAFs (CD74, SLPI; Elyada et al., 2019), the IL6 gene (known as a prototype inflammatory CAF gene; Biffi and Tuveson, 2021), and components of the complement pathway (CFD, C1QA, C1QB). Among the top down-regulated genes was ACTA2 (encoding the myofibroblastic CAF protein aSMA; Biffi and Tuveson, 2021), a number of collagen genes (COL3A1, COL1A1, COL1A2), and other secreted ECM proteins (SPARC, POSTN, BGN). Comparing pathway activities between MHCII+ and MHCII− fibroblasts revealed enrichment in up-regulated genes that are involved in immunological processes and down-regulated genes involved in ECM organization (Fig. 3 B). Although MHCII+ fibroblasts were interspersed among MHCII− fibroblasts in 2D plots, it was striking that SFTC (encoding for surfactant protein C) was on the top of the list of up-regulated genes, raising the possibility that a significant fraction of MHCII+ fibroblasts may arise from alveolar type II (ATII) cells, which constitutively express SFTC and MHCII (Hasegawa et al., 2017). Three other genes known to be highly expressed by epithelial cells (albeit not considered lineage markers for epithelial cells), CLU (clusterin) and the antimicrobial proteins lysozyme (LYZ) and SLPI, were among the 11 top up-regulated genes (Habiel et al., 2017; Leiva-Juárez et al., 2018). Altogether, our computational analysis of the transcriptome of MHCII+ fibroblasts depicts (i) prominent immune functions, (ii) a profile closer to inflammatory rather than myofibroblastic CAFs (IL6 high, a-SMA low, ECM proteins low), and (iii) strong cues of an alveolar epithelial origin.
Figure 3.
MHCII fibroblasts closely relate to alveolar epithelial cells across mice and humans. (A) Fibroblast and DC scRNAseq datasets from lung tumors and adjacent lungs (E-MTAB-6149, -6653) were extracted and reclustered based only on MHCII genes to identify MHCII fibroblasts (fibros; n = 3 patients). (B) Heatmap of differentially expressed genes (adj. P values < 0.05) between MHCII+ and MHCII− fibroblasts. GO enrichment analysis was performed using DAVID. Network visualization was conducted by Cytoscape's EnrichmentMap. Nodes represent GO biological processes, and edges connect functional terms with overlapping genes. Clusters of related nodes are circled and given labels reflecting broad biological processes. GO terms with an FDR < 0.1 are shown. (C) Multidimensional scaling representation of bulk RNAseq data of sorted MHCII+ and MHCII− fibroblasts from pooled mouse LLC lung tumors (n = 3 experiments). Differential expression between MHCII+ and MHCII− fibroblasts. Deregulated genes with adj. P values < 0.05 are indicated. (D) GO enrichment analysis using up-regulated genes with |Log2FC| > 3 as input. (E) GSEA enrichment plots for alveolar epithelial gene sets. (F) Murine ATII cells were FACS sorted from healthy lungs and exposed ex vivo to tumor homogenate (TUMOR_HOMO). Expression of EMT genes was quantified by qPCR (n = 2 or 3, pooled from two experiments). (G) Homology analysis of gene expression across mouse and human MHCII fibroblasts. Tumor-homo, Tumor homogenate. (F) **, P < 0.01; ****, P < 0.0001. Error bars, mean ± SEM; two-tailed unpaired t test. LPS, lipopolysaccharides; Ag, antigen; NES, negative normalized enrichment score.
To investigate whether the human lung MHCII fibroblast signature was conserved across mice, we sorted MHCII+ and MHCII− CAFs and performed bulk RNAseq (Fig. 3 C). DEA (inputting all expressed 15,675 genes, adjusted [adj.] P < 0.05, |Log2 fold change [FC]| > 3) revealed a large number of de-regulated genes (486), almost all of which (483) were up-regulated in MHCII+ CAFs (Fig. 3 C). As in humans, among the top up-regulated genes were the two MHCII CAF signature genes CD74 and SLPI. Pathway analysis was in concordance with the immune-related pathways that were identified in our human dataset (Fig. 3 D). Intriguingly, the ATII-specific genes SFTPs and many alveolar epithelial cell genes were again found to be increased (CLAUDINs, KRTs, SLC34A, NAPSA, LYZ). ATII cells are long-lived cells that self-renew locally (Barkauskas et al., 2013). To further support the hypothesis that MHCII fibroblasts originate from alveolar epithelial cells, we used alveolar cell type signature gene sets (curated from cluster markers, Materials and methods) and used them as inputs for gene set enrichment analysis (GSEA). GSEA comparing MHCII+ versus MHCII− fibroblasts of mice and humans demonstrated that MHCII+ fibroblasts were significantly enriched in genes that characterized alveolar epithelial cells in both species (Fig. 3 E). Primary ATII cells from healthy mice were induced to express known epithelial-to-mesenchymal transition (EMT) markers in response to tumor homogenates, adding more evidence to our hypothesis that apCAFs may evolve from ATII cells (Fig. 3 F). To align the gene signatures expressed across MHCII fibroblasts in mice and humans, we co-clustered genes based on their transcriptional profile in each species, using genes that were conserved and variable across MHCII fibroblasts in the human dataset (Fig. 3 G). Briefly, 71 out of the 79 genes that were found up-regulated were successfully translated into their mouse homologies using biomaRt. Out of the 71 genes, 67 were found expressed in our mouse dataset. Thus, our analysis denoted conservation of the MHCII fibroblast profile across the two species. Altogether, these results indicate that the process of EMT regulates the in situ differentiation of ATII cells to a subset of fibroblasts with antigen-presenting functions.
Deletion of MHCII in fibroblasts has a local impact on tumor-infiltrating CD4 T cells
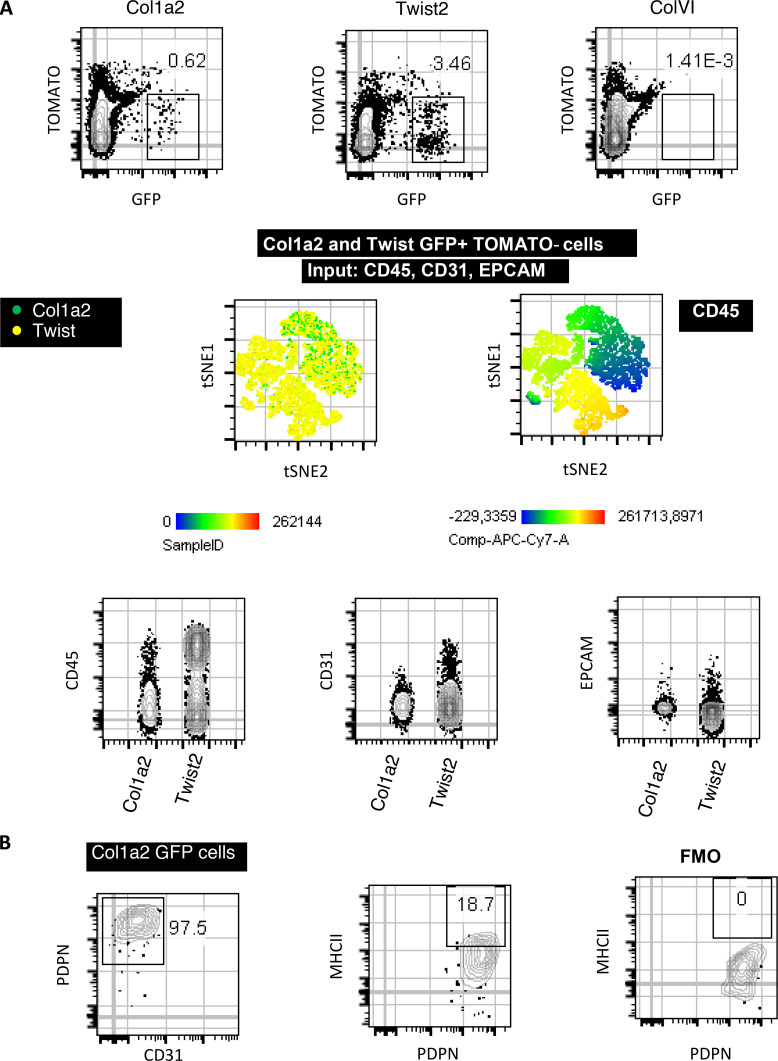
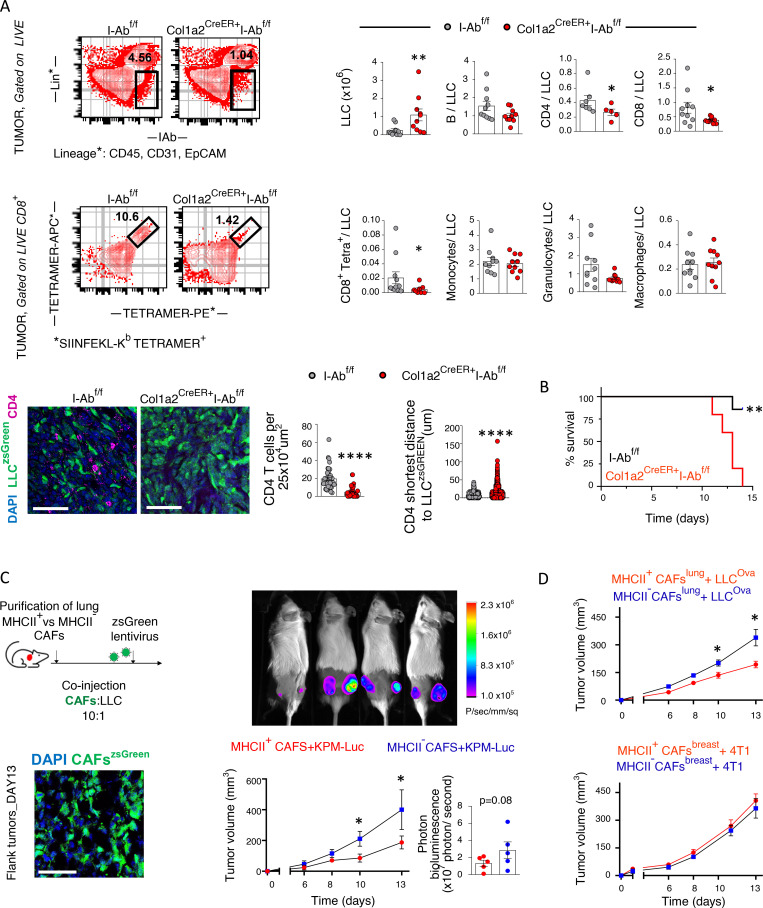
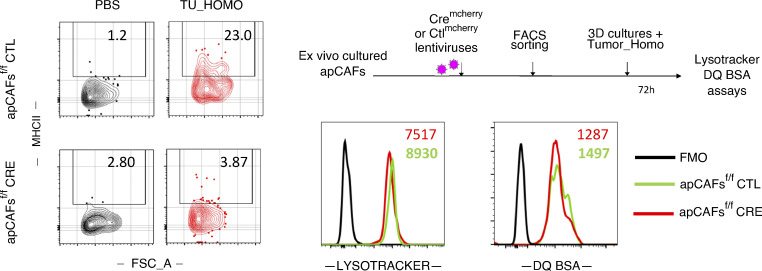
We asked whether cancer antigen presentation by CAFs was a bystander, suppressor, or driver of tumor progression in vivo. We set up pilot experiments starting with three murine lines expressing Cre recombinase (Cre) driven off Twist (Šošić et al., 2003), collagen VI (Armaka et al., 2008), and collagen 1a2 (Col1a2) promoters (Zheng et al., 2002), which are lineage markers for mesenchymal cells. Lineage-restricted expression in LLC lung tumors was determined by Cre-driven GFP expression. An important fraction of intratumoral Twist-driven GFP-expressing cells were CD45+ cells consistent with cells of the hematopoietic lineage, while collagen VI–driven GFP-expressing cells could not be identified in lung tumors (Fig. S2 A). Col1a2-driven GFP-expressing cells in lung tumors of Col1a2-CreER mice were CD45−CD31−EpCAM− and largely Podoplanin+, indicating that they were bona fide fibroblasts (Fig. S2 B). Therefore, we used Col1a2-CreER mice for our subsequent studies. IAb is the MHCII molecule expressed in C57BL/6 mice, and its expression can be deleted from Cre-expressing cell lineages using a floxed IAb gene (I-Ab-fl/fl; Hashimoto et al., 2002). We inoculated LLCmCherry cells that expressed the model antigen ovalbumin (Ova) in the lungs of Col1a2-CreER+I-Abfl/fl and I-Abfl/fl mice. Tumors appeared immunologically colder, with fewer CD4 T cells, CD8 T cells, and Ova-specific CD8 T cells, accompanied by increased tumor burden and decreased survival rates (Fig. 4, A and B). It could be hypothesized that invariant chain overload upon MHCII deletion induced lysosomal stress in apCAFs and impaired their function. To exclude this, we deleted MHCII from apCAFs of I-Abfl/fl mice using Cre lentiviral vectors and studied their lysosomal function ex vivo. Labeling and FACS tracking of acidic organelles by LysoTracker showed no difference between MHCII-deleted and control apCAFs (Fig. S3). Similar results were obtained when proteolysis of DQ-BSA was quantified (Fig. S3). Thus, we consider it unlikely that impaired apCAF function upon MHCII deletion is due to lysosomal stress.
Figure S2.
Lung CAF specificity of the Col1a2-CreER, ColVI-Cre, and Twist2-Cre mouse strains. (A and B) LLC cells were implanted intrapulmonary in the left lung lobe of tamoxiphen-treated Col1a2-CreER/mTmG (Col1a2), ColVI-Cre/mTmG (ColVI), and Twist2-Cre/mTmG mice (Twist) mice. Lung tumors were digested and subjected to FACS analyses. (A) Expression of hematopoietic- (CD45), endothelial- (CD31), and epithelial- (EpCAM) specific markers by lung GFP cells. (B) Twist2 expression of the fibroblast-specific marker Podoplanin and of IAb (MHCII) (n = 2 per group, from two independent experiments). FMO, fluorescence minus one.
Figure 4.
Lung apCAFs restrict tumor growth via MHCII. (A) Top: Col1a2 CreER+I-Abfl/fl and I-Abfl/fl mice were inoculated with LLCmcherryOva or LLCzsGreen. Representative FACS plots of Lin−IAb+ and CD8+tetramer+ cells (gated as tetramer-APC+tetramer-PE+) in digested tumors. Cumulative data of tumor burden and intratumoral immune profiles (n = 3–7 per group, pooled from two experiments). Immune cell number was normalized to cancer cell number. Numbers of cancer cells (tumor burden) and immune cells were assessed by FACS using counting beads. Bottom: Representative immunofluorescence for CD4 and LLCzsGreen cells. Cell density of CD4+ T cells in tumors of Col1a2 CreER+I-Abfl/fl versus I-Abfl/fl mice (cells per 25 × 104 μm2 regions in whole-slide images). Shortest distance between CD4 and LLCzsGreen cells in tumors of Col1a2 CreER+I-Abfl/fl versus I-Abfl/fl mice (n = 2 or 3 mice per group). Samples were analyzed by confocal microscopy with a 20× objective. Bars, 50 µm. (B) Survival curves (representative of two experiments). (C) Left: Experimental scheme and immunofluorescence of subcutaneous tumors after cotransplantation of LLC cells plus zsGreen-expressing MHCII+ versus MHCII− CAFs into syngeneic mice. Samples were analyzed by confocal microscopy with a 20× objective. Bars, 50 µm. Right: Tumor growth after subcutaneous cotransplantation of luciferase-expressing KPM cells plus MHCII+ versus MHCII− CAFs in syngeneic mice (n = 5 mice per group). (D) Top: Tumor growth after subcutaneous cotransplantation of LLCmcherryOva cells plus lung tumor–derived MHCII+ versus MHCII− CAFs in syngeneic mice (C57BL/6; n = 6 mice per group, representative of two experiments). Bottom: Tumor growth after subcutaneous cotransplantation of 4T1 cells plus breast tumor–derived MHCII+ versus MHCII− CAFs in syngeneic mice (BALB/c; n = 8 mice per group, pooled from two experiments). (A–D) *, P < 0.05; **, P < 0.01; ****, P < 0.0001. Error bars, mean ± SEM; two-tailed Mann-Whitney U test.
Figure S3.
Lysosomal processes of MHCII-deleted apCAFs. Purified apCAFs from I-Ab-fl/fl mice were expanded and transduced with CREmcherry or control (CTL) lentiviruses. To induce MHCII, apCAFs were exposed for 72 h in 3D conditions to 30% tumor homogenate (Tumor_Homo). Left: Representative FACS plots of MHCII. Lysosomes were tracked (LysoTracker), and lysosomal proteolysis was assessed (DQ Green BSA) with FACS. Right: Representative FACS plots. FSC-A, forward scatter-A; FMO, fluorescence minus one.
To strengthen our data on the tumor-suppressive function of lung apCAFs in a different in vivo experimental setting, we established a cotransplantation model, where we coinjected cancer cells with primary in vitro expanded low-passage lung apCAFs. We reasoned that, similar to what others have shown, possible confounding effects from resident CAFs would be minimal (Hutton et al., 2021). Compared with control apCAF-depleted lung CAFs (non-apCAFs), lung apCAFs dramatically restricted subcutaneous tumor growth from two different cancer cell lines. Thus, the tumor suppressive effect of lung apCAFs is stable and dominant after in vitro expansion (Fig. 4 C). Intriguingly, these findings could not be reproduced when BC-derived primary apCAFs were used for the cotransplantation studies, raising the possibility that the immunological functions of apCAFs depend on the tissue/cell type of origin (Fig. 4 D).
MHCII antigen presentation is expected to directly affect CD4 T cells. Thus, we sought to explore whether CD4 T cells were required for the in vivo tumor-promoting effect of fibroblast-specific MHCII deletion. CD4 T cell–depleting antibodies abolished differences in tumor burden between Col1a2-CreER+I-Abfl/fl and I-Abfl/fl mice, indicating that MHCII fibroblasts were strongly dependent on CD4 T cells for anti-tumor function (Fig. 5 A). Functionally, purified tumor-infiltrating lymphocytes (TILs) from Col1a2 CreER+I-Abfl/fl versus I-Abfl/fl mice demonstrated inferior cancer cell–specific cytotoxicity (Fig. 5 B). In addition, cancer-specific CD4 T cells were induced to produce high levels of effector cytokines upon ex vivo co-culture with primary lung apCAFs via MHCII (Fig. 5 C). Thus, MHCII antigen presentation contributes to the tumor-suppressive effects of apCAFs by increasing the effector functions of cancer-specific CD4 T cells.
Figure 5.
Lung anti-tumor CD4 T cell responses depend on apCAF MHCII. (A) Col1a2 CreER+I-Abfl/fl and I-Abfl/fl mice were inoculated with LLCOva and treated with CD4 T cell–depleting antibody. Cumulative data of tumor burden (n = 3–6 per group, pooled from two experiments). Absolute numbers of tumor cells were assessed by FACS using counting beads. (B) Col1a2 CreER+I-Abfl/fl versus I-Abfl/fl mice bearing LLCOva lung tumors were adoptively (a.t.) transferred with OTII T cells. TILs were FACS sorted and incubated with CFSE-loaded LLCOva (CSFEhigh = 5 μM CFSE) plus LLC (CSFElow = 0.5 μM CFSE) cells at a 60:1:1 ratio. Cytotoxicity was determined at 16 h by quantifying the ratio of LLCOva to LLC (n = 2–5 per group, pooled from two experiments). Representative and cumulative FACS data are shown. (C) CAFs from LLCOva lung tumor–bearing mice were sorted and co-cultured with OTII T cells from OTII RAG−/− mice at a 1:1 ratio plus MHCII-blocking antibody or isotype (ISO) control. Representative FACS plots of intracellular cytokines (n = 2 or 3 per group, representative of two experiments). (D) Depletion of circulating T cells upon FTY720 treatment. Representative FACS plots. (E) Lung tumor Col1a2 CreER+I-Abfl/fl versus I-Abfl/fl mice were treated with FTY720. Lung tumors were excised and analyzed by FACS (n = 6 or 7 per group, pooled from two experiments). (F and G) Intratumoral lung CD4 T cells were purified from Col1a2 CreER+I-Abfl/fl versus I-Abfl/fl mice (n = 7) and analyzed by bulk RNAseq. Expression heatmaps (mean values) of marker genes (F). MA (M [log ratio] and A [mean average]) plots and GO term enrichment analysis, with nodes in the networks representing biological processes and edges connecting terms with overlapping genes (G). Adj. P values > 0.05, except from: Stat6 0.0042, Stat3:0.0053 (F). Deregulated genes with FDR < 0.01 and GO terms with an FDR < 0.1 are shown (G). (H) Left: Pearson correlation analysis between CD4 effector/effector memory T cell and apCAF signature genes in patients with lung adenocarcinoma and squamous cell carcinoma in The Cancer Genome Atlas [TCGA], GEPIA2). Right: Input gene lists. (A, B, and E) *, P < 0.05; **, P < 0.01. Error bars, mean ± SEM; two-tailed Mann-Whitney U test. aMHCII, pan-MHCII blocking antibody; FSC-A, forward scatter-A; SSC-A, side scatter-A.
RNAseq analysis of FACS-sorted TIL CD4 showed a high number of down-regulated genes in Col1a2-CreER+I-Abfl/fl versus I-Abfl/fl mice (Fig. 5 G). Among top down-regulated were genes involved in gene transcription (CRCT2, KAT7, EP300), cellular energy metabolism (GLUL, LIPA, PRKAG2), and protein ubiquitination and proteosomal degradation (PSMD6, CDC26, TRIM26; Fig. 5 G). Signature genes of T helper type 1 (Th1), Th2, Th17, and Th9 and regulatory T cells (Tregs), cytotoxicity, or exhaustion marker genes were not different between the two groups (Fig. 5 F). Likewise, pathway activities of down-regulated genes showed significant enrichment in macromolecular metabolic processes. This agrees with the known fundamental role of TCR activation in T cell metabolism and effector functions (van der Windt and Pearce, 2012). In addition, when we looked into The Cancer Genome Atlas database, we found a strong positive correlation between the apCAF and CD4 effector/effector memory T cell signature genes in patients with non-small cell lung cancer (Fig. 5 H). Taken together, the results described above suggest that antigen presentation by lung MHCII fibroblasts is a critical driver of anti-tumor MHCII immunity.
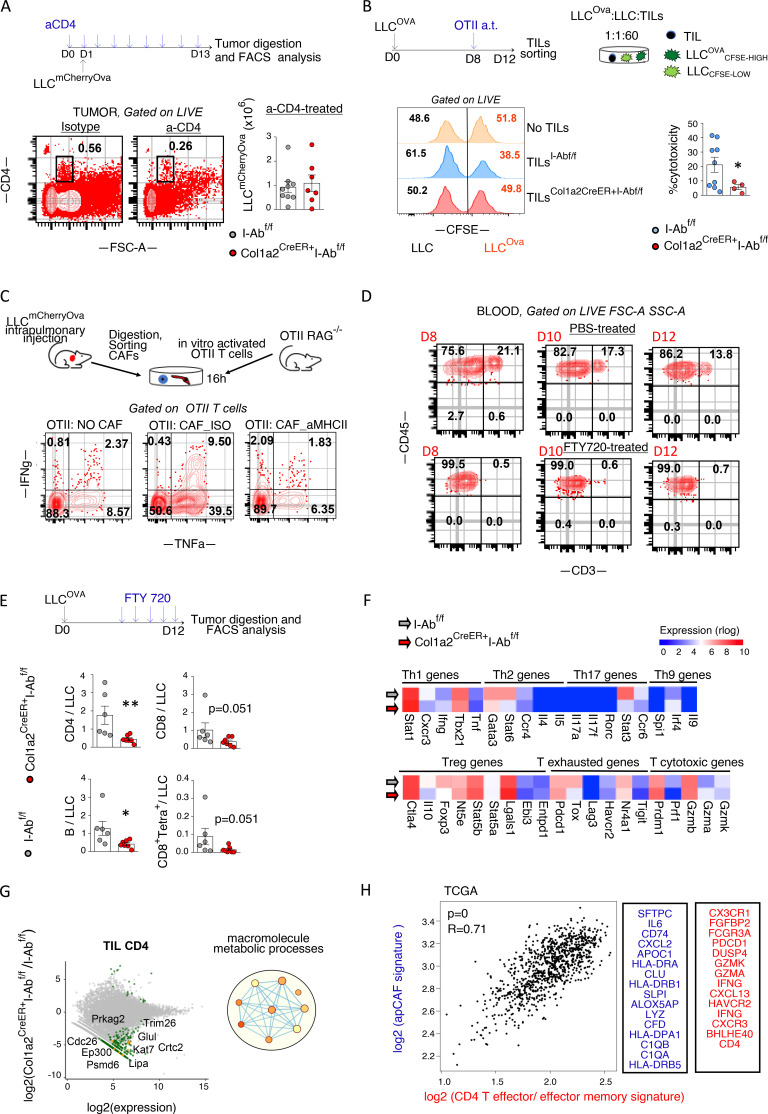
LN fibroblastic reticular cells (FRCs) have contradictory roles in immune responses, such as inhibition of T cell proliferation via nitric oxide (Lukacs-Kornek et al., 2011) and T cell reprogramming toward memory T cells via IL6 (Brown et al., 2019). Albeit FRCs in LNs acquire transcriptional programs typically associated with tumor escape (Riedel et al., 2016), we considered the possibility that immune evasion in Col1a2-Cre+I-Abfl/fl mice resulted from MHCII deletion in FRCs. We assessed MHCII expression in LN FRCs of Col1a2-CreER+I-Abfl/fl versus I-Abfl/fl mice and found a trend toward decreased MHCII+ FRCs (Fig. S4). However, there were no gross differences in nodal CD4+, CD8+ T cells, and B cells (Fig. S4). We questioned whether antigen-presenting CAFs could sustain MHCI and MHCII immunity in the absence of continuous T cell migration from LNs. Therefore, we treated mice with an S1PR antagonist FTY720, which blocks egress of T cells from LNs, for 5 consecutive days, starting on day 7 after LLCmcherryOva transplantation. CD4 and CD8 T cells were again found to be decreased, while Ova peptide-MHCI tetramers stained less of CD8 T cells in lung LLCmcherryOva tumors of FTY720-treated MHCII conditional KO mice (Fig. 5, D and E). These results suggest that targeting MHCII in fibroblasts facilitates immune escape by acting locally and that T cell immunity in Col1a2-CreER+I-Abfl/fl mice is compromised by MHCII loss in CAFs rather than FRCs.
Figure S4.
FACS analysis of tumor-draining LNs of the Col1a2-CreER strain. LLC cells were implanted intrapulmonary in the left lung lobe of Col1a2-CreER+I-Abfl/fl and I-Abfl/fl mice. After 14 d, mesothoracic/tumor-draining LNs were digested and analyzed by FACS. From left to right: MHCII expression in CD45−CD31−EpCAM−Podoplanin+. Frequencies of CD4+, CD8+ T cells, and B220+ B cells. n = 3–5 per group, pulled from two independent experiments. Error bars, mean ± SEM.
apCAFs rescue intratumoral CD4 cells from apoptosis via the C1q receptor C1qbp
Effector T cells have less stringent and incompletely understood activation requirements compared with naive T cells (Chen and Flies, 2013). Since we could barely detect any of the classical costimulatory molecules (CD80, CD86, CD40) in MHCII fibroblasts, we mined their single-cell transcriptome looking for other immune-related molecules with ligand-receptor functions. Human MHCII+ fibroblasts overexpressed TYROPB and CD47, which ligate to SIRP receptors CD52, and ANXA1, which bind to SIGLEC10 and ALX/FPR2, respectively, and C1q, which binds to cC1qR/calreticulin and gC1qR/C1qbp. None but the receptor of the globular head of C1q (i.e., gC1qr/C1qbp) was found to be expressed on the surface of primary human TIL CD4 cells (Fig. 6 A). C1qbp is located in most cell types in the mitochondria, where it is responsible for maintaining transcription of mitochondrial proteins. In the cytoplasm, it acts as a regulator of RNA stability and plays a critical role in mRNA splicing. The role of C1q as an extracellular signaling molecule and C1qbp as a plasma membrane receptor on T cells has been rarely reported (Kouser et al., 2015; Ling et al., 2018; West et al., 2018) and is largely unexplored. We set out to investigate whether MHCII fibroblast-derived C1q amplified TCR signaling on CD4 T cells via membrane C1qbp. First, we confirmed increased C1q gene expression by apCAFs versus MHCII− CAFs (non-apCAFs) in distinct patients (Fig. 6 A). There was also lower C1q gene expression in TIL CD4 than in apCAFs (Fig. 6 A). Then, we co-cultured human effector CD4 TILs and CAFs in the presence of C1qbp-blocking antibodies. Surprisingly, we did not observe any effect on TCR signaling, but a decrease in the numbers of viable T cells (Fig. 6 B). This led us to hypothesize that C1q is an extrinsic pro-survival signal for CD4 T cells. To test this, we cultured stimulated peripheral blood CD4 T cells for 1 h under serum starvation conditions. Purified C1q acted directly on T cells to almost completely rescue them from apoptosis in a dose-dependent manner (Fig. 6 C). Moreover, aC1qbp substantially inhibited the pro-survival effect of C1q on T cells (Fig. 6 D).
Figure 6.
Exogenous C1q is anti-apoptotic, and human apCAF C1q increases viability of intratumoral CD4 T cells. (A) Immunofluorescence for C1qbp shows surface expression in human intratumoral CD4 T cells. Samples were analyzed by confocal microscopy with a 40× objective. Bars, 40 µm. MHCII+ CAFs, MHCII− CAFs, and intratumoral CD4+ T cells were FACS sorted from human lung tumors and analyzed for C1q by qPCR (n = 4 patients). Pt, patient. (B) CD4 T cells and CAFs were sorted from the same tumor fragment and co-cultured with aC1qbp-blocking antibody or isotype (ISO) control. Absolute (Abs.) numbers of live T cells were assessed by FACS with counting beads (n = 5 patients). (C) Peripheral blood (PB) CD4 T cells from a healthy donor were sorted and activated with aCD3/aCD2/aCD28 beads before undergoing serum starvation with or without purified C1q. Representative FACS plots of two experiments. (D) As in C, plus aC1qbp-blocking antibody or isotype control. Cumulative data of cell viability measured by Annexin V//dead stain by FACS (n = 6 donors). Representative FACS plots of caspase-3 activity. (E) CD4 T cells, total CAFs, and MHCII− CAFs were sorted from the same tumor fragment. T-CAFs were co-cultured with aC1qbp-blocking antibody or isotype control. T cell apoptosis was assessed using Annexin V and PI staining. Representative FACS plots and cumulative data are shown (n = 3 patients). (A–E) *, P < 0.05; ***, P < 0.001; ****, P < 0.0001. Error bars, mean ± SEM; one–tailed unpaired or paired t test. FSC-A, forward scatter-A; PBMC, peripheral blood mononuclear cell.
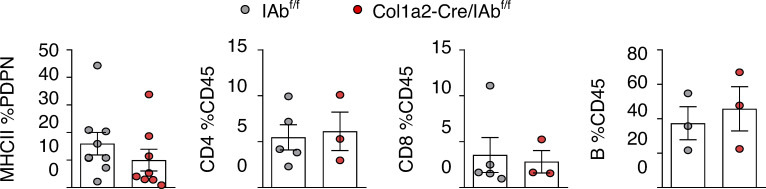
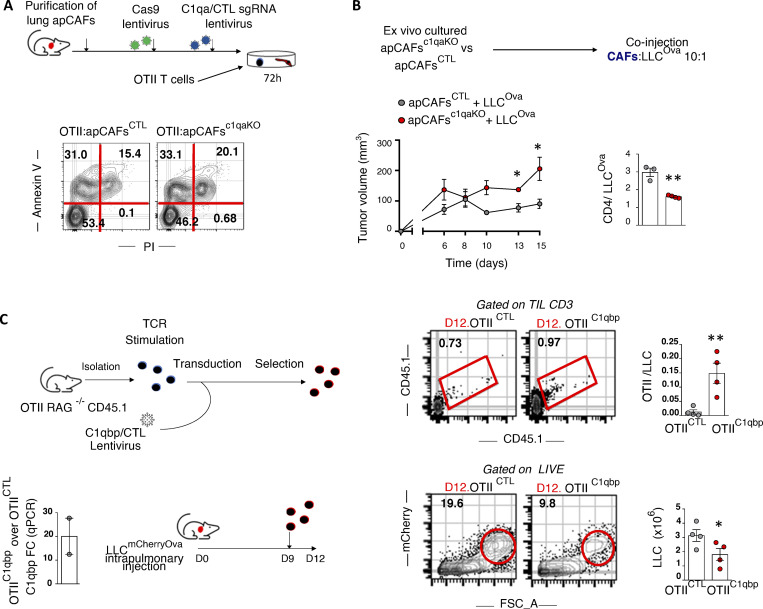
To study the dependence of intratumoral CD4 T cells on apCAF-derived C1q, we depleted apCAFs from the CAF-TIL CD4 co-cultures. apCAF depletion abrogated the apoptotic effect of aC1qbp, indicating that aC1qbp blocks apCAF-derived C1q (Fig. 6 E). We consider it unlikely that these results have been confounded by an autocrine effect of T cell intrinsic C1q because of the low C1q in TIL CD4 (Fig. 6 A). To further substantiate the pro-survival effect of apCAF-derived C1q on T cells, we sorted primary murine lung apCAFs and knocked out C1qa via CRISPR-Cas9. CD4 T cells showed higher apoptosis rates when co-cultured with apCAFc1qaKO versus control apCAFs. (Fig. 7 A). Accordingly, subcutaneous tumors grew bigger when apCAFc1qaKO (versus control apCAFs) and cancer cells were cotransplanted, accompanied by lower CD4 T cell numbers (Fig. 7 B). In addition, when cancer-specific OTII T cells were induced to overexpress C1qbp and were adoptively transferred in lung tumor–bearing mice, an increased number of cancer-specific T cells was identified within lung tumors, accompanied by a decrease in tumor burden (Fig. 7 C). Altogether, these data suggest that CD4 T cells depend on C1qbp for their survival within tumors and point to apCAFs as a potent C1q source.
Figure 7.
In vivo MHCII tumor immunity depends on apCAF C1q, CD4 T cell C1qbp. (A) C1q was knocked out of primary lung apCAFs via CRISPR-Cas9. MHCII+CAFc1qa KO versus control (CTL) MHCII+CAFs were co-cultured with OTII T cells. Apoptosis was measured by Annexin V/dead staining. Representative FACS plots of two experiments. (B) MHCII+CAFc1qa KO versus control MHCII+CAFs were cotransplanted with LLCmcherryOva cells in syngeneic mice. Tumor growth was measured. Cumulative data of CD4 T cell numbers normalized to cancer cell numbers. Absolute numbers were assessed by FACS with counting beads. (n = 3 or 4 mice per group, pooled from two experiments). (C) C1qbp overexpressing OTII T cells were adoptively transferred in LLCmcherryOva lung tumor–bearing mice. Representative FACS plots of LLCmcherryOva cells and OTII cells in digested lung tumors. Cumulative data (n = 4 per group, pooled from two experiments). (A–C) *, P < 0.05; **, P < 0.01. Error bars, mean ± SEM; two-tailed unpaired t test. FSC-A, forward scatter-A; sgRNA, single-guide RNA.
Discussion
A functional dichotomy is currently thought to exist between draining LNs and tumors, whereby T cells are primed in LNs and exert effector functions in tumors. Here, we show that effector CD4 T cells require in situ stimulation by a subset of fibroblasts for effective immunity to occur in lung cancer. Fibroblast-specific–targeted ablation of MHCII induced an LN-independent impairment in local immunity, accelerating tumor growth. Primary human lung fibroblasts directly activated the TCRs of tumor-infiltrating CD4 T cells and, at the same time, protected them from apoptosis via the C1q receptor C1qbp, which unveiled a previously unrecognized function of C1q/C1qbp as an anti-apoptotic ligand/receptor pair for CD4 T cells. Our studies introduce fibroblasts as key peripheral cancer antigen-presenting cells and propose a new concept in tumor immunity: the final outcome of LN CD4 T cell priming depends on intratumoral MHCII antigen-presenting cells.
In tumor slides, MHCII fibroblastic cells were not identified within macroscopically visible tertiary lymphoid structures (TLSs), suggesting that intratumoral fibroblastic antigen-presenting niches are distinct to TLSs. It is likely that TLSs are mainly sites of B cell somatic hypermutation and plasma cell generation (Cabrita et al., 2020; Helmink et al., 2020; Petitprez et al., 2020), while intratumoral antigen-presenting fibroblasts form immunological niches for effector T cells. MHCII fibroblasts did not express classical costimulatory molecules but were still able to activate the TCR/CD28 pathway in effector T cells and enhance T cell metabolism. We consider two possibilities that may explain this contradiction: (i) costimulatory and coinhibitory receptors of effector T cells display great diversity and promiscuity (thus, MHCII fibroblasts mediate effector T cell activation via nonclassical costimulatory molecules; Chen and Flies, 2013), and (ii) antigen recognition suffices to activate effector T cells (Dubey et al., 1996).
C1q is known to regulate basic metabolic processes of T cells (Kouser et al., 2015; Ling et al., 2018; West et al., 2018) but has been disregarded as an extracellular signal. This is also related to the fact that the receptor of its globular head, C1qbp, has a very short intra-cytoplasmic tail and must complex with other membrane molecules to trigger intracellular signaling (Hosszu et al., 2012; Shi et al., 2017). Our results provide the first, to the best of our knowledge, demonstration that a complement component is anti-apoptotic, pointing specifically to exogenous C1q as a critical pro-survival ligand for CD4 T cells via C1qbp. The outcome of T cell therapy is highly dependent on T cell survival within tumors (Charo et al., 2005; Yamamoto et al., 2019). We showed that C1qbp overexpression increases the numbers of adoptively transferred tumor-specific CD4 T cells within tumors. Conceivably, it is likely that the increased efficacy of C1qbp-OTII cells is not only the result of apCAF-derived C1q. Nonetheless, our results showing that (i) apCAFs express high levels of C1q, (ii) they are a key antigen-presenting cell within lung tumors, (iii) apCAF-derived C1q acts directly on T cells to rescue them from apoptosis, and (iv) TIL CD4 cells reside within apCAF spots altogether suggest an important role for apCAF-derived C1q as a pro-survival factor for TIL CD4 cells.
Single-cell/bulk RNAseq approaches revealed a conserved transcriptome signature among murine and human lung apCAFs that was enriched for alveolar epithelial genes, suggesting that lung antigen-presenting fibroblasts may arise from alveolar epithelial cells through the process of EMT. Albeit no safe conclusions can be made without experimental evidence from lineage tracing studies, the following arguments indicate that lung apCAFs originate from ATII rather than cancer or mesothelial cells, the last being considered as likely precursors of PDAC apCAFs (Dominguez et al., 2020; Hutton et al., 2021). (i) In the human dataset, tumor cells clustered away from fibroblasts, while in the mouse dataset, cancer cells were brightly fluorescent and negatively selected. Furthermore, we never detected fluorescence in murine fibroblasts that were cultured up to passage 5. (ii) In both human and mouse datasets, the most up-regulated genes were surfactant proteins, which are highly specific for ATII cells. (iii) Constitutive MHCII expression is a hallmark of ATII cells (Hasegawa et al., 2017). (iv) ATII cells can transit to fibroblasts through the process of EMT (Goldmann et al., 2018; Hill et al., 2019). (v) Murine ATII cells underwent EMT ex vivo when exposed to tumor homogenate. The antigen-presenting functions of ATII cells are well recognized, but their immunological impact has not been extensively investigated (Wosen et al., 2018). Although we did not extensively study the function of other types of apCAFs, purified BC apCAFs did not show tumor-suppressive properties in our cotransplantation studies. Thus, lung antigen-presenting fibroblasts might be unique in their ability to activate effector T cells, and it is tempting to speculate that this could be due to their ATII origin, if such is proven to be. If the apCAF function greatly depends on tissue/cell type of origin, several other questions are raised. Are there specific cell types that are more likely to give rise to stimulatory apCAFs? What are the unique features of stimulatory versus nonstimulatory apCAFs? Are there stimulatory versus nonstimulatory subsets within the same tumor? Given the emergence of tumor stroma as a novel immunotherapeutic target, these are clearly therapeutically relevant and not merely theoretical questions.
One would expect that should lung apCAFs originate from ATII cells, they would create a distinct cluster when plotted in 2D scRNAseq plots with other fibroblasts. There are a few explanations as to why this was not observed. First, apCAFs are rare among total fibroblasts, and most dimensionality reduction approaches are biased in favor of frequent cell states. This is also supported by the fact that in most PDAC and BC scRNAseq studies, apCAFs do not form a separate cluster, but are rather found enriched in a cluster (Dominguez et al., 2020; Elyada et al., 2019; Friedman et al., 2020; Kieffer et al., 2020; Sebastian et al., 2020). Second, the identity of ATII cells is closer to that of fibroblasts after they have undergone EMT transition. Indeed, at the protein level, they expressed equal levels of key fibroblasts markers, such as FAP and PDGFRa and no EpCAM. They also displayed a characteristic fibroblastic morphology, and they grew easily in culture (primary alveolar epithelial cells are notoriously difficult to grow; Sahai et al., 2020).
The apCAFs that were recently identified in PDAC and BC are presumed to induce T cell anergy or differentiation to T regulatory cells (Elyada et al., 2019; Friedman et al., 2020; Sebastian et al., 2020). This presumption opposes the increase in Tregs that has been observed upon fibroblast deletion in PDAC (Özdemir et al., 2014). In our models, MHCII deletion in fibroblasts did not alter CD4 T cell subsets. However, it caused a profound decrease in the metabolic rate of tumor-infiltrating CD4 T cells. This finding fits perfectly well with the pivotal role of T cell metabolism in antigen-specific responses (van der Windt and Pearce, 2012). Further supporting the idea that non-hematopoietic cells can act as stimulating rather than tolerizing antigen-presenting cells, MHCII intestinal epithelial cells activate CD4 T cells in graft-versus-host disease, and diverse types of non-hematopoietic lung antigen-presenting cells prime T resident memory cells (Koyama et al., 2019; Low et al., 2020).
IFN-γ drives MHCII expression in epithelial cells (Koyama et al., 2019). Our results also demonstrate that the tumor microenvironment controls MHCII in fibroblasts via IFN-γ. That said, positive clinical responses to immune checkpoint blockade are associated with increased IFN-γ and MHCII expression (Grasso et al., 2020; Johnson et al., 2016; Rodig et al., 2018). Accordingly, BC responses to aTGFβ are associated with increased MHCII expression in fibroblasts (Grauel et al., 2020). Thus, on the basis of our models, another mechanism behind the success of checkpoint inhibitors in lung cancer could be their ability to sustain IFN-γ–dependent apCAF spots, which in turn activate the unleashed effector T cells within tumors.
Current immunotherapeutic treatments largely focus on immune cells. Our works point to mesenchymal cells as key partners in cancer therapy. We already envisage a few possible scenarios for the immunotherapeutic exploitation of our findings: in vivo delivery of tumor antigens to apCAFs (in vivo vaccination) and amplification of the antigen-presenting fibroblastic spots with IFN-γ–inducing drugs, such as checkpoint inhibitors or aTGFβ. Conceivably, the above strategies are expected to require combinatorial T cell–directed therapies for maximum clinical benefits.
Materials and methods
Human samples
Paired human lung tumor and macroscopically healthy lung resection specimens were obtained from patients with lung adenocarcinoma and lung squamous cell carcinoma at Sotiria Chest Hospital. The study was approved by the Sotiria Chest Hospital ethical committee board, and informed consent was obtained from all patients. lung adenocarcinomas and lung squamous cell carcinomas were clinically scored and staged according to the International Union against Cancer tumor, node, metastasis (TNM) staging system. The clinical and pathological characteristics of all patients included in this study are summarized (Table S1).
Mice
Col1a2-cre/ERT,-ALPP mice (The Jackson Laboratory; ID #029235), Tg (Col6a1-cre)1Gkl (ColVI-Cre) mice (MGI:3775430), B6.129X1-Twist2tm1.1(cre)Dor/J (Twist2-Cre) mice (The Jackson Laboratory; ID #008712), mTmG mice (The Jackson Laboratory; ID #007576), IL-12 p35 KO (The Jackson Laboratory; ID #002691), and IL-12 p40 KO (The Jackson Laboratory; ID #002693) were provided by G. Kollias (Biomedical Sciences Research Center “Alexander Fleming” [BSRC Al. Fleming], Vari, Greece); OTII mice (The Jackson Laboratory; ID #004194) by V. Andreakos (Biomedical Research Foundation Academy of Athens [BRFAA], Athens, Greece); IFN-γR (The Jackson Laboratory; ID #003288) and IFN-γ KO mice (The Jackson Laboratory; ID #002287) from J.P. Gorvel (Centre d’immunologie de Marseille-Luminy [CIML], Marseille, France); B6(Cg)-Tyrc-2J/J (B6-albino) and BALB/c mice by Animal House Facility (BSRC Al. Fleming, Vari, Greece). B6.129X1-H2-Ab1tm1Koni/J (I-AB-flox, ID #013181) and B6.SJL-Ptprca (CD45.1; ID #002014) mice were purchased from The Jackson Laboratory. For MHCII deletion in fibroblasts, the Col1a2-CreER mice were crossed to I-AB-flox mice that possess a loxP-flanked neo cassette upstream of exon 1 and a single loxP site downstream of exon 1 of the H2-Ab1 locus. For T cell responses against Ova-expressing cancer cells, OTII mice that express the mouse α-chain and β-chain T cell receptor that pairs with the CD4 coreceptor and is specific for the chicken Ova 323–339 peptide in the context of I-Ab were crossed to Rag1 KO mice and mice that express the CD45.1 (Ly5.1 PTP) alloantigen. For reporter studies, Col1a2-CreER, ColVI-Cre, and Twist2-Cre were crossed to mTmG mice.
Cell lines
The LLC cell line, obtained from American Type Culture Collection, the C57BL/6-derived urethane-induced lung adenocarcinoma CULA cell line, and Ad-Cre–treated KRASG12D;Trp53f/f C57BL/6-derived cell line KPM, provided by G. Stathopoulos (Helmholtz Zentrum München, Neuherberg, Germany), the murine mammary carcinoma cell line 4T1, provided by Biomedcode Hellas, and the C57BL/6-derivered melanoma cell line B16F10, provided by V. Kostourou (BSRC Al. Fleming, Vari, Greece), were transduced with zsGreen and/or OVAmCherry and/or Luc-ZsGreen lentiviruses and were sorted based on fluorescent protein expression. For lentiviral production, HEK lenti-X 293 T cells were transfected with pHIV-Zsgreen (Addgene; #18121), MIGR1-OVA-IRES-mCherry, pHIV–Luc-ZsGreen (Addgene; #39196), or pLM-CMV-R-Cre (Addgene; #27546) lentiviral plasmids using polyethylenimine (Polysciences). Cells were maintained in DMEM or RPMI 1640, containing 10% heat-inactivated FBS, 1% L-glutamine, and 1% penicillin/streptomycin (Gibco). All the cells tested negative for the presence of mycoplasma contamination using a PCR-based technology. Cell lines were not authenticated.
Sample preparation and staining
Human and murine tissue specimens were perfused with PBS and cut into pieces. Minced samples were immediately processed or cryopreserved using BioCool (SP Scientific) in Recovery Cell Culture Freezing Medium (GIBCO; BRL cat. no. 12648-010) enriched with 10% DMSO. Human and mouse tumor and lung tissue fragments were enzymatically digested in 10% FBS/HBSS (Gibco) using Collagenase IV (Sigma-Aldrich; 1 mg/ml; cat. no. C7657), Dispase II (Roche; 1 mg/ml; cat. no. SCM133), and Dnase I (Sigma-Aldrich; 0.09 mg/ml; cat. no. DN25) for 1 h at 37°C with agitation. Murine LNs were digested in 10% FBS/HBSS using Collagenase P (Sigma-Aldrich; 1 mg/ml; cat. no. 11 249 002 001), Dispase II (Roche; 1 mg/ml), and Dnase I (Sigma-Aldrich; 0.09 mg/ml) for 1 h at 37°C with agitation. Human tumor cells were passed through a 100-µm cell strainer, and murine tumor cells were passed through a 70-µm cell strainer. Murine splenocytes were mechanically dissociated by passing through a 40-µm cell strainer. Cells were washed with FACS buffer (2% FBS/PBS/1.5 mM EDTA), centrifuged, and resuspended in FACS buffer. Nonspecific binding was blocked by incubating cells with human anti-Fc Receptor antibodies (TrueStain; Biolegend; cat. no. 101320) or anti-mouse CD16/32 Fc block (Biolegend; cat. no. 101310).
Staining of human tissue was performed with the following antibodies (all from Biolegend, unless otherwise stated): CD45-PE, (HI30, lot# B255235, cat. no. 304007), CD31-PE, (WM59, lot# B267973, cat. no. 303105), EpCAM-PE, (9C4, lot# B283019, cat. no. 324205), FAP-Alexa700 (lot# A6V10218119; R&D Biosystems; cat. no. MAB3715), Podoplanin APC-Fire 750 (NC80, lot# B266728, cat. no. 337023), CD140a (PDGFRa)-PE-Cy7 (16A1, lot# B250540, cat. no. 323507), CD152 (CTLA-4)-PE (L3D10, lot# B243881, cat. no. 369603), CD40-BV421 (5C3, lot# B251089, cat. no. 334331), CD80-BV510 (2D10, lot# B240242, cat. no. 305233), CD86-APC (lot# 10990; BD; cat. no. 555660), CD8-PERCP-Cy5.5 (SK1, lot# B261859, cat. no. 301031), HLA-DR-BV785 (L243, lot# B251938, cat. no. 307641), HLA-DR-DP-DQ-APC (TU39, lot# B275397, cat. no. 361713), CD44-APC (IM7, cat. no. 361713, B232055, cat. no. 103011), CD14-PE (63D3, lot# B233137, cat. no. 301805), CD16-PE (3G8, lot# B238509, cat. no. 302007), CD19-PE (HIB19, lot# B239940, cat. no. 302207), CD34-PE (581, lot# B266670, cat. no. 343505), CD36-PE (5–271, cat. no. 336205, B286437, cat. no. 336205), CD56-PE (5.1H11, lot# B252299, cat. no. 304605), CD123-PE (6H6, lot# B240986, cat. no. 306005), and CD253-PE (RIK2, lot# B278666, cat. no. 308215) for 30 min at 4°C.
Murine cells were stained with the following antibodies (all from Biolegend, unless otherwise stated): CD45-APC-CY7 (30F11, lot# B290025, cat. no. 103115), CD3-PE-CY7 (1432C11, lot# 4304567; eBioscience; cat. no. 25–0031-82), B220-PERCP (RA36B2, lot# 6032134; eBioscience; cat. no. 45–0452-82), CD8-PERCP (536.7, lot# B210430, cat. no. 126610), CD4-FITC (GK1.5, lot# B288660; BD; Pharmigen, cat. no. 553651), CD45-Alexa700 (30F11, lot# B309099, cat. no. 123128), CD31-Alexa700 (390, lot# B303280, cat. no. 102443), EpCAM-Alexa700 (68.8, lot# B292046, cat. no. 334331), CD140a (PDGFRa)-PE (APA5, lot# B244566, cat. no. 334331) or BV785 (cat. no. 118239), Podoplanin-PE Cy7 (8.1.1, lot# B267028, cat. no. 127412), MHCII-APC-Cy7 (M5/114.15.2, lot# B290142, cat. no. 107627), CD45.1-Alexa700 (A20, lot#B254604, cat. no. 110723), CD11b (M1/70, lot# 3014897; BD Pharmigen; cat. no. 557397), CD11c (HL3, lot# 07852; BD Pharmigen; cat. no. 553802), CD19 (1D3/CD19, lot# B288633, cat. no. 115507), B220 (RA3CB2, lot# 57924; BD Pharmigen; cat. no. 553089), CD49b (DX5, lot# B278344, cat. no. 103506), CD105 (MJ7/18, lot# B300564, cat. no. 110723), MHCII (M5/114.15.2, lot# B221906, cat. no. 107607), and TER119 (TER119, lot# B293614, cat. no. 116207). Tetramers SIINFEKL-PE and SIINFEKL-APC were provided by the National Institutes of Health.
For intracellular staining, cells were fixed and permeabilized using the Intracellular Fixation & Permeabilization Buffer Set (eBioscience; cat. no. 88–8823-88), followed by staining for human samples with PAN-Cytokeratin-PE (C11, lot# 532104; Sigma-Aldrich; cat. no. sAB4700668), Vimentin–Alexa Fluor 674 (EPR3776, lot# GR219 216–9; Abcam; cat. no. ab194719), a-Sma-FITC (114, lot# 087M4798; Sigma-Aldrich; cat. no. F3777), and Alexa Fluor 647 anti-mouse (lot# 1910774; Invitrogen; cat. no. A21235) secondary antibody. For murine samples, also a-SMA-FITC (Sigma-Aldrich; cat. no. F3777) and Vimentin–Alexa Fluor 674 (Abcam; cat. no. ab194719) were used.
For intracellular staining of phosphorylated proteins, cells were fixed and permeabilized using the Intracellular Fixation & Permeabilization Buffer Set (eBioscience; cat. no. 88–8823-88) per the manufacturer’s recommendations for detection of intracellular phosphorylated proteins, followed by staining with pmTor-PE-Cy7 (eBiosciences; cat. no. 25–9718-42).
Sytox-Green viability dye (Thermo Fisher Scientific; cat. no. S7020), Zombie NIR (Biolegend; cat. no. 423105), Zombie Violet (Biolegend; cat. no. 423113), or BD Horizon, Fixable, Viabilility, Stain 700 (BD Pharmigen; cat. no. 564997) were used to exclude dead cells.
For Annexin V/propidium iodide (PI) stain, up to 5 × 104 cells were resuspended in Binding Buffer (0.01 M Hepes, pH 7.4, 0.14 M NaCl, and 2.5 mM CaCl2) and stained with Annexin V FITC (lot# B284572, cat. no. 640906) for 30 min at 4°C. Cells were washed and resuspended in PI–Binding Buffer solution (0.1 mg/ml; Sigma-Aldrich; cat. no. P4170). For Caspase-3 stain, the NucView 488 Caspase-3 Assay Kit for Live Cells (Biotium) was used according to the manufacturer’s instructions.
Functional ex vivo assays
For human immunological assays, CAFs and CD4+ T cells were harvested from the same tumor fragment and dispersed into single-cell suspensions as stated above. CAFs were sorted as FSC-AHighCD45−CD31−EpCAM−FAP+PDGFRa+ cells. To preserve functional TCRs, intratumoral CD4+ T cells were sorted as FSC-ALowSSC-ALowCD45+CD14−CD15−CD16−CD19−CD34−CD36−CD56−CD123−CD8−CTLA-4− cells. CAFs were co-cultured overnight with CD4+ T cells at a 1:1 ratio in the presence of pan-HLA antibody (5 µg/ml; Biolegend; cat. no. 361702) or GC1q R antibody (10 µg/ml; Abcam; cat. no. ab24733) or isotypic controls (mouse IgG2a [Biolegend; cat. no. 361702] and mouse IgG1 [Abcam; cat. no. ab170190], respectively). Cells were co-cultured in complete RPMI (Gibco) medium.
Classical Ficoll-Pague (StemCell; cat. no. 07861) density gradient centrifugation was followed for peripheral blood mononuclear cell isolation from human peripheral blood. Untouched CD4+ T cells were negatively selected from peripheral blood mononuclear cells using anti-phycoerythrin (PE) MicroBeads (Miltenyi Biotech) after incubation with the following PE-conjugated antibodies: CD14, CD15, CD16, CD19, CD34, CD36, CD56, CD123, CD253, and CD8 according to the manufacturer’s instructions. Human T Cell Activation/Expansion Kit (Miltenyi Biotech) was used for activation of CD4+ T cells according to the manufacturer’s protocol. T cells were cultured in Click’s medium supplemented with 10% heat-inactivated human serum (Sigma-Aldrich; H4522), 1% L-glutamine and 1% penicillin/streptomycin, 1% sodium pyruvate, 1% MEM nonessential amino acids, 2 mM Hepes, β-mercaptoethanol, and 50 U/ml huIL-2 (Peprotech; cat. no. 200–02). For selected experiments, T cells were cultured in serum-depleted medium with or without native human C1q protein (6 μg/ml; Abcam; ab96363) or GC1q R antibody (10 µg/ml; Abcam; cat. no. ab24733).
For murine 3D CAF cultures, CAFs sorted as FSC-AHighCancer cellszsGreen−CD45−CD31−EpCAM−PDPN+PDGFRa+ cells were resuspended in liquified Matrigel (Corning; 354254) at 4°C. Approximately 30,000 cells/well were seeded in a 25-μl drop of Matrigel in the center of wells of a 48-well tissue culture plate. The matrix was allowed to set for 15 min at 37°C before adding 400 μl complete RPMI into each well. At day 5 of culture, RPMI medium was replaced with fresh RPMI supplemented with 30% healthy lung or tumor lung homogenate plus/minus anti–IFN-γ (10 μg/ml; Biolegend; cat. no. 505705). To obtain tissue homogenates, tumors or healthy lungs were excised, weighed, and homogenized in PBS (30% weight/volume) on ice. Samples were centrifuged at 13,000 rpm for 10 min at 4°C, and supernatants were added in wells containing Matrigel-encapsulated cells.
For ex vivo cytotoxicity assays, 4 d after adoptive transfer of OTII cells to LLCmcherryOva lung tumor–bearing mice, lung tumors were excised, and untouched total intratumoral T cells were FACS sorted from tumor single-cell suspensions as FSC-AlowLIVECancercellmcherry−CD45+Ter119−CD105−CD11b−CD11c−CD19−B220−CD49b−CD25−. LLCmcherryOva and LLC cells were preconditioned with IFN-γ (20 U/ml) overnight to up-regulate MHC expression and the next day were labeled with 5 μΜ or 0.5 CFSE, respectively. Cells were mixed and plated together with T cells at a 60:1:1 ratio on V-bottom 96-well plates, centrifuged at 1,000 rpm for 10 min, and incubated for 16 h. Cytotoxicity was calculated as cytotoxicity = 100% × {1 − [(CFSElow/CSFEhigh)control/(CFSElow/CSFEhigh)experimental]}.
For assessment of lysosomal stress, apCAFs from I-Abfl/fl mice sorted as FSC-AHighCancer cells-zsGreen−CD45−CD31−EpCAM−PDPN+PDGFRa+MHCII+ cells were infected with CREmcherry or control lentiviruses. Low-passaged apCAF I-Abfl/fl and apCAF I-Abfl/fl-Cre were exposed to tumor homogenate for 72 h in 3D cultures supplemented with 30% tumor homogenate, as above, and then incubated with LysoTracker Green DND-26 (Thermo Fisher Scientific; L7526) for 1 h and DQ Green BSA (Thermo Fisher Scientific; D12050) for 2 h, following the manufacturer’s instructions.
For CRISPR-Cas9 gene editing of C1qa in apCAFs, CAFs sorted as FSC-AHighCancerCellmcherry−CD45−CD31−EpCAM−PDPN+PDGFRa+MHCII+ were first transduced with ready-to-use Cas9-GFP lentiviral particles (Sigma-Aldrich), and subsequently with guide RNA (gRNA) lentiviruses. Control and C1qa gRNA-containing lentiviruses were generated from transfection of HEK lenti-X cells with NegativeControl1, MMPD0000004118, and MMPD0000004120 gRNAs purchased from Sigma-Aldrich in gRNA:hPGK-puro-2A-tBFP background vector. C1qaKO and control apCAFs were sorted as GFP and BFP double-positive cells and cultured in 1:1 ratio with in vitro activated OTII T cells for 72 h in RPMI containing 1% heat-inactivated FBS, 1% L-glutamine, and 1% penicillin/streptomycin and 20% tumor homogenate. For in vitro activation of OTII T cells, splenocytes from OT-II/Rag1−/−/CD45.1 mice were cultured in a-CD28–coated plates (Biolegend; cat. no. 102115; 1 μg/ml) with complete RPMI (10% FBS, 1% penicillin/streptomycin, 50 μM β-mercaptoethanol) supplemented with 1 μg/ml OVA Peptide (323–339; Genscript, cat. no. RP10610; DAY 0). Untouched CD45.1 OTII cells were magnetically sorted from cultured splenocytes on day 2 using anti-PE MicroBeads (Miltenyi Biotech) after incubation with the following PE-conjugated antibodies: CD11b, CD11c, CD19, B220, CD49b, CD105, MHCII, and TER119, according to the manufacturer’s instructions. T cells were cultured and expanded in the aforementioned medium supplemented with 1 μg/ml OVA Peptide (323–339) and 50 U/ml mIL-2 (Peprotech; cat. no. 212–12).
For CAF-OTII T cell co-cultures, CAFs sorted as FSC-AHighCancerCellsmcherry−CD45−CD31−EpCAM−PDPN+PDGFRa+ cells were treated with MHCII blocking antibodies (5 μg/m; BioXcell; BE0108) or isotype control (Biolegend; 400643) and cultured at a 1:1 ratio with in vitro activated OTII T cells overnight. The next day, cells were stimulated with 50 ng/ml PMA, 1 µg/ml ionomycin, and Brefeldin A (Biolegend) for 4 h, before harvesting and staining intracellularly for IFN-γ (Biolegend; 505807) and TNFα (Biolegend; 506303).
To study EMT in ATII cells, lungs from healthy mice were harvested and digested. ATII cells were sorted as CD45−CD31−EpCam+MHCII+ and cultured in SAGM medium (Small Airway Epithelial Cell Growth Medium; LONZA; CC-3118) for 5 d. On day 5, medium was supplemented with 20% tumor homogenate for 48 h. For assessment of EMT, total RNA was extracted from ATII cells as described below.
In vivo studies
Gender- and age-matched mice over 8 wk old were used for all studies. All mice were housed under standard special pathogen-free conditions at BSRC Al. Fleming. All animal procedures were approved by the Veterinary Administration Bureau, Prefecture of Athens, Greece, under compliance with the national law and the EU Directives and were performed in accordance with the guidance of the Institutional Animal Care and Use Committee of BSRC Al. Fleming.
For the lung cancer model, mice were anesthetized via i.p. injection with xylazine and ketamine. Cancer cells (2 × 105) resuspended in 50 μl DMEM (Gibco) and enriched with 20% growth-factor–reduced ECM (Matrigel; BD Biosciences) were intrapleurally injected in the lung parenchyma of mice using a 29G needle (BD Biosciences). For the metastatic cancer model, mice were injected i.v. with 4 × 105 B16F10 cells in 100 µl DMEM medium. For the orthotopic breast tumor model, 4T1 (105) cells resuspended in 50 μl of PBS were implanted into the third mammary fat pad. For all experiments with inducible Col1a2-CreER mice, tamoxifen (50 mg/kg; Sigma-Aldrich; T5648) dissolved in corn oil (Sigma-Aldrich; C8267) was given i.p. once per day every day except weekends for the duration of the experiment (5 injections/week, 2 wk), starting on day 0 of cancer cell injection in the lungs.
Monoclonal antibody aCD4/GK1.5 (American Type Culture Collection/TIB-207) was administered i.p. 2 d before tumor implantation and continued three times per week for the duration of the study (150 μg/mouse). FTY720 (Cayman Chemicals) was administered i.p. for 5 consecutive days, starting on day 7 after tumor cell transplantation at 20 mg per mouse.
For the adoptive T cell transfer experiments, T cells from OT-II/Rag1/CD45.1 mice were isolated and in vitro expanded/activated as above. For the cytotoxicity assay on day 7, 1,000,000 T cells per mouse were injected i.v. in LLCmcherryOva tumor-bearing mice. For the C1qbp-OTII experiments on day 4, OTII cells were transduced with C1qbp lentiviral particles (Lenti ORF, C1qbp; Origene; NM_007573) versus Lenti-ORF Control Particles at a multiplicity of infection of 2. The transduction was performed for 24 h in medium supplemented with 8 μg/ml polybrene (Sigma-Aldrich). 2 d after lentiviral transduction, cells were placed in puromycin selection for 2 wk (2 μg/ml; Sigma-Aldrich; P8833). On day 22, 100,000 T cells per mouse were injected i.v. in LLC-OVA tumor-bearing mice.
For cotransplantation studies, CAFs sorted as FSC-AHighCancerCellmcherry−CD45−CD31−EpCAM−PDPN+PDGFRa+ MHCII+ or MHCII− CAFs from lung or breast cancer tumors were cultured in DMEM containing 10% heat-inactivated FBS, 1% L-glutamine, and 1% penicillin/streptomycin (Gibco) in collagen-treated plates (collagen I rat tail; Corning). MHCII+ and MHCII− CAFs were infected with zsGreen lentiviruses and were sorted based on GFP expression. C1qaKO and control MHCII+ CAFs were sorted as GFP/BFP double-positive as stated above. Low-passaged CAFs and cancer cells (LLCmcherryOva [lung], 4T1 [breast], or KPM-Luc-ZsGreen [lung; IVIS Imaging]) were combined in a 10:1 ratio (1,000,000 CAFs/100,000 cancer cells, resuspended in 100 μl DMEM) and implanted bilaterally on both flanks of B6-albino and C57BL/6 mice. Flank tumor volume in B6-albino and C57BL/6 mice (V) was measured by assessing length and width by digital caliper every 2 or 3 d and was calculated as V = (length × width2)/2. Alternatively, tumor volume in B6-albino mice was measured by bioluminescent imaging. Mice were i.p. injected (150 mg/kg) with a 30mg/ml solution of D-luciferin (IVISbrite; PerkinElmer) in filtered PBS. Quantitative imaging was performed using the In-Vivo Xtreme Imaging System (Bruker).
RNA extraction and quantitative PCR (qPCR)
For assessment of human C1q expression, total RNA was extracted from apCAFs, non-apCAFs, blood, and intratumoral CD4+T cells that were isolated as stated above, using the Single Cell RNA Purification Kit (Norgen Biotek) according to the manufacturer’s instructions. Superscript II reverse-transcriptase (Thermo Fisher Scientific) was used for cDNA synthesis and SYBR Green (Thermo Fisher Scientific) for qPCR performed on the CFX96 Touch Real-Time PCR Detection System from Bio-Rad. Transcript levels of C1q were determined relative to U6 reference gene, using the ΔΔCt method. The following primer sets were used: human C1qb forward (Fw): 5′-TAAAAGGAGAGAAAGGGCTTCCAGGG-3′ and reverse (Rv): 5′-TGGCCTTGTAGTCTCCCGATTCACC-3′ and human U6, Fw: 5′-CTCGCTTCGGCAGCACA-3′ and Rv: 5′-AACGCTTCACGAATTTGCGT-3′. Assessment of murine Snail1 and Snail2 (Slug) expression was performed as above using the following primer sets: murine Snail1 Fw: 5′-ACATCCGAAGCCACACG-3′ and Rv: 5′-GTCAGCAAAAGCACGGTTG-3′, murine Snail2 Fw: 5′-ACACATTAGAACTCACACTGGG-3′ and Rv: 5′-TGGAGAAGGTTTTGGAGCAG-3′, and murine B2M Fw: 5′-TTCTGGTGCTTGTCTCACTGA-3′ and Rv: 5′-CAGTATGTTCGGCTTCCCATTC-3′. Assessment of murine C1qa KO was performed as above using the following primer set: Fw: 5′-ATCGAAAAGGACCCCGCAAA-3′ and Rv: 5′-CTCAGGCCGAGGGGAAAATG-3′.
Flow cytometry
FACS analysis or sorting was performed using FACSCANTO II or BD FACSARIA III or BD FACSCelesta (BD Biosciences), and data were analyzed using Flowjo software. For calculation of absolute numbers of tumor cells (burden) or immune cells, counting beads (123Count eBeads; Thermo Fisher Scientific) were used. Ebeads are 7-µm microparticles that have higher side scatter than cell populations and contain fluorescent dyes excited by the blue (488 nm) or violet (405 nm) laser line. They are supplied at a known concentration (∼1,000 beads/µl). Ebeads were thoroughly mixed by vortexing and were carefully suspended and mixed with cell suspensions. The following equation was used for calculation of absolute counts according to the manufacturer’s recommendations: Absolute count (cells/µl) = [(cell count × ebead volume)/(ebead count × cell volume)] × ebead concentration.
Immunohistochemistry/immunofluorescence
For mouse studies, lungs were excised from healthy and tumor-bearing mice and fixed in 4% freshly prepared paraformaldehyde for 14–18 h at 4°C. After fixation and incubation in 30% sucrose, samples were embedded in optimal cutting temperature compound (VWR; cat. no. 361603E), then sectioned (10-µm thick) onto SuperFrost Plus microscope slides (Thermo Fisher Scientific). Slides were washed using PBS followed by 1-h incubation in blocking solution (1% BSA, 0.1% Saponin, and PBS). Sections were incubated with primary antibodies Anti-Mouse MHC-II 1:250 (EBiosciences; 14–5321-81), Anti-Mouse Podoplanin eFluor 660 1:500 (EBiosciences; 50–5381-82), and Recombinant Anti-CD4 antibody (EPR19514; Abcam; ab183685) diluted in BSA 1%/PBS overnight at 4°C. After rinsing with 0.1% Saponin/PBS, sections were incubated with secondary antibody for MHC-II (Goat anti-Rat IgG [H+L] Alexa Fluor 594; Invitrogen) or for CD4 (Goat anti-rabbit IgG [H+L] Alexa Fluor 647; Invitrogen) for 1 h at room temperature. Nuclei were stained with DAPI, and slides were mounted using Fluoroshield mounting media (Sigma-Aldrich; F6182). Images were acquired with a TCS SP8X White Light Laser confocal system (Leica).
For immunofluorescence stainings of human tissues, fresh samples were immersed in cryoprotectant (OCT), frozen, and then cryosectioned in 5-µm-thick sections on charged glass slides. After fixing with methanol for 15 min at room temperature, Fc receptor Blocker (Innovex; NB309-5S) and Background Buster (Innovex; NB306-7) reagents were used for blocking Fc receptors and eliminating background staining, respectively. Then, sections were incubated with anti-FAP (R&D; AF3715) 1:100, anti-HLA DR+DP+DQ (Abcam; ab7856) 1:200, and anti-CD4 (Abcam; ab133616) 1:100 overnight at 4°C. After rinsing, sections were incubated with secondary antibodies (Goat anti-Rabbit IgG [H+L] Cross-Adsorbed ReadyProbes Secondary Antibody; Alexa Fluor 594; cat. no. R37117; Goat anti-Mouse IgG [H+L] Highly Cross-Adsorbed Secondary Antibody; Alexa Fluor 488 cat. no. A-11029; and Donkey anti-Sheep IgG [H+L] Cross-Adsorbed Secondary Antibody; Alexa Fluor 647 cat. no. A-21448). After thorough rinsing, sections were incubated with FITC anti-human CD45 Antibody (Biolegend; 304005) 1:100 overnight at 4°C. Nuclei were stained with DAPI, and slides were mounted using Fluoroshield mounting media (Sigma-Aldrich; F6182). Images were acquired with a TCS SP8X White Light Laser confocal system (Leica).
Immunofluorescence in T cells for the expression and localization of C1qbp were conducted in sorted intratumoral T cells. After sorting, T cells were washed twice and cytospun at 400 g for 4 min. After cytospin, slides were let dry for 5–10 min, and then cells were fixed for 15 min using 4% paraformaldehyde. After fixation, cells were washed twice with PBS 1× and permeabilized for 10 min using 0.1% Triton-X/PBS. Blocking was performed using 2% BSA/2% normal goat serum in 0.1% Triton-X 100/PBS. Cells were stained overnight at 4°C using Recombinant Anti-GC1q R antibody (EPR23238-6; Abcam; ab270033).
Image acquisition and analysis
Images were acquired using a TCS SP8X confocal system (Leica). We selected a fluorophore panel, which allowed for simultaneous visualization of three targets, and a nuclear stain (DAPI). During acquisition, fluorophores were excited with 405 nm (UV Laser), 488 nm (Argon), 555 nm, 594 nm, and 647 nm (White Light Laser). For images shown in Fig. 1, analysis was performed using Leica LASX. Tile scanning was performed in slides stained for pan-MHCII, CD45, CD4, and FAP, and nuclei were stained using DAPI. Autostitching using 10% overlap was performed by Imaris Stitcher. Imaris 9.6 was used for subsequent image manipulations. After creating a colocalization channel between pan-MHCII, CD45 (or CD45 inverted), and FAP (or FAP inverted), all channels were used to define “primary objects” (surfaces—spots) used to analyze the image (distances, cell number, XY positioning, etc.). Shortest distance calculation, average distance to nearest neighbor, and object identification tools were used for data acquisition and image analysis. Cell density analysis was performed by identifying each “object” in fields of 250,000 µm2. Data exported from Imaris 9.6, including XY location of CD4+, MHCII+FAP+CD45−, and MHCII+FAP− CD45+ objects, were used for density plot creation using custom R scripts.
Identification of human MHCII fibroblasts in scRNAseq datasets
Processed data were collected from publicly available, paired human lung tumor and healthy lung scRNAseq datasets (E-MTAB-6149 and E-MTAB-6653). The log2cpm values of the 22,180 ensembl gene IDs were used. The transition from the raw sequence read data to the gene expression table can be found in the original publication (Lambrechts et al., 2018). Processed cells annotated as fibroblasts were included in the analysis. Patient #1 was excluded from the analysis because of the small number of fibroblasts (nine cells). Patient #2 was excluded from the analysis because of the small number of healthy fibroblasts and DCs (5 and 11 cells, respectively). Only fibroblasts of patients #3, #4, and #5 were analyzed (n = 643, 155, and 486, respectively). To identify MHCII+ and MHCII− fibroblasts, cells annotated as cross-presenting DCs in the same dataset were used as positive control for MHCII expression. The nine MHCII genes (HLA-DRA, HLA-DRB5, HLA-DRB1, HLA-DQA1, HLA-DQA2, HLA-DQB1, HLA-DQB2, HLA-DPA1, and HLA-DPB1) were used for reclustering fibroblasts and DCs with k-means function of cluster R package (Mächler et al., 2012) with k = 3 (tuned for k = 3:10), supported by mean Silhouette information scoring, calculated by Silhouette function of cluster R package. This resulted in the splitting of the cells into three groups (low, middle, and high MHCII expression). Fibroblasts of the middle and high groups were considered MHCII+ fibroblasts (206 cells), while those of the low group were considered MHCII− fibroblasts. Gene IDs that had zero expression in all fibroblasts were removed from the analysis (16,877 IDs were kept). 15,487 of them were successfully translated into gene names using the biomaRt R package. After the characterization of MHCII+ and MHCII− fibroblasts, genes with non-zero expression values in more than 25% of each group were kept (1,699 genes for apCAFs and 1,388 genes for non-apCAFs). The union of both lists was considered the list of the genes that are expressed in at least one group. This resulted in 1,785 genes.
Differential expression and pathway analysis of human datasets
To identify the differentially expressed genes between the MHCII+ and MHCII−, we used the FindAllMarkers function of the Seurat R package (Satija et al., 2015), giving as input non-scaled read values of the two clusters. We kept cells that had at least 10 features and removed ribosomal and mitochondrial genes. This resulted in 79 statistically significant up-regulated genes (adj. P value < 0.05, 67 with logFC > 1) and 55 statistically significant down-regulated genes (adj. P value < 0.05, 48 with logFC < −1). The 67 up-regulated and the 55 down-regulated genes were submitted to DAVID (Huang et al., 2009) for gene ontology (GO) term enrichment analysis. DAVID analysis was performed as previously described (Huang et al., 2009) using the οfficial gene names (hg19) as background. GO using DAVID (6.8; Huang et al., 2009) was performed for terms classified as biological processes. An enrichment map was created in Cytoscape (v3.7.2; Shannon et al., 2003) using all significantly enriched GO terms (false discovery rate [FDR] < 0.1), with a gene set size between 10 and 1,000. Overlap was used as a metric, and the cutoff was set to 0.5. Clustering was performed using AutoAnnotate (v1.3.2; Kucera et al., 2016) with overlap used as edge weight values.
Bulk mouse RNAseq data acquisition, processing, and analysis
Intratumoral CD45+CD3+CD4+ T cells were FACS sorted in RL buffer (lysis buffer of cells before RNA isolation). Total RNA was isolated by using NucleoSpin RNA (Macherey-Nagel). RNA integrity was assessed on an Agilent Bioanalyzer RNA 6000 Pico Chip, and the library was constructed using NEB Next Ultra RNA Library Prep Kit. The sequencing platform Ion Torrent PROTON and 3′-untranslated region sequencing strategy was used. Quality of FASTQ files, obtained after Ion Proton sequencing, was assessed using FastQC (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/) following the software recommendations. Alignment of sequencing reads to the reference genome was performed using the software HISAT2 (version 2.1.0; Kim et al., 2019) with the genome reference version mm10. Bam files containing reads that were uniquely aligned were summarized to read counts table using the GenomicRanges package (Lawrence et al., 2013) through MetaseqR2 pipeline (Fanidis and Moulos, 2021) and default settings for 3′-untranslated region data. The resulting gene counts table was subjected to DEA with metaseqr2 function, using DESeq2 algorithm for the normalization and statistical testing. Genes with less than five counts in 75% of the samples were excluded from downstream analysis. DEA thresholds were set for FDR equal to 0.01 and for logFC + −1, returning 365 down-regulated and 19 up-regulated genes. The 365 down-regulated genes were submitted to DAVID for GO term enrichment analysis. DAVID analysis was performed as previously described (Huang et al., 2009) using the official gene names (mm10) as background. GO using DAVID (6.8; Huang et al., 2009) was performed for terms classified as biological processes, as described earlier.
MHCII+ CAFs and MHCII− CAFs from tumor-bearing mice were FACS sorted as FSC-AΗighCD45−CD31−EpCAM−mCherry−PDPL+PDGFRa+MHCII+ and FSC-AΗighCD45−CD31−EpCAM−mCherry−PDPL+PDGFRa+MHCII− cells, respectively. Total RNA was isolated by using Single Cell RNA Purification Kit (Norgen Biotek) according to the manufacturer’s instructions. RNA integrity was assessed on an Agilent Bioanalyzer RNA 6000 Pico Chip, and the library was constructed using NEB Next Ultra RNA Library Prep Kit. The sequencing platform Illumina Novaseq 6000 and Pair-end 150 strategy were used. Quality of FASTQ files was assessed using FastQC, following the software recommendations. Alignment of sequencing reads to the reference genome was performed using a two-way alignment procedure of initial mapping with STAR (version 2.7.3; Dobin et al., 2013) and remapping of the unmapped reads with bowtie2 (version 2.3.5.1; Langmead and Salzberg, 2012) packages with the genome reference version mm10. The raw bam files were summarized to read counts table using the GenomicRanges through MetaseqR2 R package pipeline, and the resulting gene counts table was subjected to DEA using the R package DESeq2 again through MetaseqR2 pipeline. DEA thresholds set for P value equal to 0.05 and for logFC 1.5 returned 2,216 down-regulated and 164 up-regulated genes. The 418 down-regulated genes with logFC > 3 were submitted to DAVID for GO term enrichment analysis, as described earlier.
GSEA of CAFs for alveolar genes
GSEA was performed with (v4.1.0) software. The normalized counts table produced by DESeq2 was used for the GSEA according to software recommendations on the standard GSEA run. For human CAFs, the gene set included marker genes of an alveolar cluster that was identified in the same dataset (Lambrechts et al., 2018). For murine CAFs, we used a gene set that included marker genes of a murine alveolar cluster that was identified in another dataset (Strunz et al., 2020). The rest of the analysis was performed with the default thresholds.
Cross-species homology analysis
Up-regulated genes in MHCII+ versus MHCII− fibroblasts were identified independently for each species, as above. The gene symbols of the genes found up-regulated in the human MHCII+ fibroblast (67 genes) were translated to their mouse homologues through the biomaRt R package (“useMart” and “getLDS” functions). This returned 63 gene homology pairs. For each gene that returned a homologue, the relative gene expression value for each species was indicated by dividing the average gene expression value in the MHCII+ cluster across each species plus a regularization constant (10e−4) by the average expression of all single fibroblasts (human dataset) or all samples (mouse dataset) plus a regularization constant (10e−4). The normalized expression matrices were log2-normalized, and their correlation was calculated by Pearson correlation distance.
Statistics and reproducibility
For the G-squared tests of independence, the g2Test_univariate function of Rfast R package was used. The P value was calculated only for the upper tail through the pchisq function of the same package. The Pearson correlation values were calculated with cor base R function.
Visualization
Principal component analysis was performed using the prcomp base R function, with centering but no scaling. Multidimensional scaling plots were drawn in R with ggplot2. Box plots were generated using the ggplot2 R package and default parameters. MA (M [log ratio] and A [mean average])plots and GO terms dot plot were generated using the geom_point function of ggplot2 R package. Plots were generated using the ggplot2 with dropping. Heatmaps were generated using the pheatmap function of pheatmap R package. Three different dimensionality reduction/visualization methods were applied to the gene expression matrix of all human single-cell fibroblasts (1,294 cells): PHATE (Moon et al., 2019) was applied with the “phateR” R package with default parameters (knn = 5, decay = 40, t = auto), returning a 2D embedding. Uniform Manifold Approximation and Projection (Becht et al., 2019) was applied with the “umap” R package with default configuration parameters (n_neighbors = 15, metric = euclidean, n_epochs = 200, min_dist = 0.1), returning a 2D embedding. t-SNE (van der Maaten and Hinton, 2008) was applied with the “Rtsne” R package with default parameters (perplexity = 30, theta = 0.5, max_iter = 1,000), returning a 2D embedding.
Online supplemental material
Fig. S1 shows the gating strategy for sorting untouched CD4 T cells and CAFs and co-cultures of human MHCII− CAFs with tumor-infiltrating CD4 T cells. Fig. S2 illustrates the lung CAF specificity of the Col1a2-CreER, ColVI-Cre, and Twist2-Cre mouse strains. Fig. S3 shows the lysosomal processes of MHCII-deleted apCAFs. Fig. S4 shows the FACS analysis results of tumor-draining LNs of the Col1a2-CreER strain. Table S1 lists the main data from the patients involved in this study.
Supplementary Material
lists main patient data, including number of patients enrolled in the study, age, gender, histology, smoking history, and TNM stage.
Acknowledgments
We would like to thank G. Kollias for critical insights and discussions and for reviewing the manuscript. We acknowledge G. Kollias, V. Koliaraki, E. Andreakos, J.P. Gorvel, and G. Stathopoulos for sharing mouse models, cell lines, and protocols, and the National Institutes of Health Tetramer Core Facility for SIINFEKL tetramers. We thank BSRC Al. Fleming flow cytometry, animal house, imaging, and genomics facilities and the bioinformatics facility of The Greek Research Infrastructure for Personalised Medicine (MIS 5002802). The graphical abstract was created with BioRender.com.
The study was supported by a Hellenic Foundation for Research and Innovation research grant (HFRI-1289) to M. Tsoumakidou and a Fondation Santé research grant to M. Tsoumakidou. D. Kerdidani was supported by the project Reinforcement of Postdoctoral Researchers—2nd Cycle (MIS-5033021) implemented by the Greek State Scholarships Foundation and co-financed by Greece and the European Union (European Social Fund) through the Operational Program Human Resources Development, Education and Lifelong Learning. I. Angelidis has received funding from the European Respiratory Society and the European Union’s H2020 research and innovation program under the RESPIRE4 Marie Sklodowska-Curie Actions grant agreement No 847462.
Author contributions: Conception and design study, M. Tsoumakidou; Development of methodology, M. Tsoumakidou; Acquisition of data, D. Kerdidani, E. Aerakis, I. Angelidis, K. Douka, M.-A. Maniou, V. Ntafis, K. Goudevenou, P. Stamoulis, and A. Prados; Analysis and interpretation of data, D. Kerdidani, E. Aerakis, K.-M. Verrou, C. Tzaferis, A. Prados, and M. Tsoumakidou; Providing human samples, I. Vamvakaris, E. Kaniaris, K. Vachlas, E. Sepsas, K. Potaris, and A. Koutsopoulos; Writing the manuscript, M. Tsoumakidou; Preparing the figures, D. Kerdidani, E. Aerakis, K.-M. Verrou, K. Vachlas, P. Stamoulis, C. Tzaferis, A. Prados, and M. Tsoumakidou; Study supervision, M. Tsoumakidou.
Data availability
Murine CD4 T cell bulk RNAseq and CAF RNAseq data generated in this study are available through National Center for Biotechnology Information Gene Expression Omnibus (GEO) repository with accession numbers GSE164659 and GSE164653, respectively. Human scRNAseq data used in this study are available through ArrayExpress database (E-MTAB-6149, E-MTAB-6653; Lambrechts et al., 2018). Other relevant data are available from the corresponding author upon reasonable request.
References
- Ahrends, T., Busselaar J., Severson T.M., Bąbała N., de Vries E., Bovens A., Wessels L., van Leeuwen F., and Borst J.. 2019. CD4+ T cell help creates memory CD8+ T cells with innate and help-independent recall capacities. Nat. Commun. 10:5531. 10.1038/s41467-019-13438-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alspach, E., Lussier D.M., Miceli A.P., Kizhvatov I., DuPage M., Luoma A.M., Meng W., Lichti C.F., Esaulova E., Vomund A.N., et al. 2019. MHC-II neoantigens shape tumour immunity and response to immunotherapy. Nature. 574:696–701. 10.1038/s41586-019-1671-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armaka, M., Apostolaki M., Jacques P., Kontoyiannis D.L., Elewaut D., and Kollias G.. 2008. Mesenchymal cell targeting by TNF as a common pathogenic principle in chronic inflammatory joint and intestinal diseases. J. Exp. Med. 205:331–337. 10.1084/jem.20070906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkauskas, C.E., Cronce M.J., Rackley C.R., Bowie E.J., Keene D.R., Stripp B.R., Randell S.H., Noble P.W., and Hogan B.L.. 2013. Type 2 alveolar cells are stem cells in adult lung. J. Clin. Invest. 123:3025–3036. 10.1172/JCI68782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becht, E., McInnes L., Healy J., Dutertre C.-A., Kwok I.W.H., Ng L.G., Ginhoux F., and Newell E.W.. 2019. Dimensionality reduction for visualizing single-cell data using UMAP. Nat. Biotechnol. 37:38–44. 10.1038/nbt.4314 [DOI] [PubMed] [Google Scholar]
- Biffi, G., and Tuveson D.A.. 2021. Diversity and Biology of Cancer-Associated Fibroblasts. Physiol. Rev. 101:147–176. 10.1152/physrev.00048.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binnewies, M., Mujal A.M., Pollack J.L., Combes A.J., Hardison E.A., Barry K.C., Tsui J., Ruhland M.K., Kersten K., Abushawish M.A., et al. 2019. Unleashing Type-2 Dendritic Cells to Drive Protective Antitumor CD4+ T Cell Immunity. Cell. 177:556–571.e16. 10.1016/j.cell.2019.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bos, R., and Sherman L.A.. 2010. CD4+ T-cell help in the tumor milieu is required for recruitment and cytolytic function of CD8+ T lymphocytes. Cancer Res. 70:8368–8377. 10.1158/0008-5472.CAN-10-1322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, F.D., Sen D.R., LaFleur M.W., Godec J., Lukacs-Kornek V., Schildberg F.A., Kim H.J., Yates K.B., Ricoult S.J.H., Bi K., et al. 2019. Fibroblastic reticular cells enhance T cell metabolism and survival via epigenetic remodeling. Nat. Immunol. 20:1668–1680. 10.1038/s41590-019-0515-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabrita, R., Lauss M., Sanna A., Donia M., Skaarup Larsen M., Mitra S., Johansson I., Phung B., Harbst K., Vallon-Christersson J., et al. 2020. Tertiary lymphoid structures improve immunotherapy and survival in melanoma. Nature. 577:561–565. 10.1038/s41586-019-1914-8 [DOI] [PubMed] [Google Scholar]
- Charo, J., Finkelstein S.E., Grewal N., Restifo N.P., Robbins P.F., and Rosenberg S.A.. 2005. Bcl-2 overexpression enhances tumor-specific T-cell survival. Cancer Res. 65:2001–2008. 10.1158/0008-5472.CAN-04-2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, L., and Flies D.B.. 2013. Molecular mechanisms of T cell co-stimulation and co-inhibition. Nat. Rev. Immunol. 13:227–242. 10.1038/nri3405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, D.S., and Mellman I.. 2013. Oncology meets immunology: the cancer-immunity cycle. Immunity. 39:1–10. 10.1016/j.immuni.2013.07.012 [DOI] [PubMed] [Google Scholar]
- Dammeijer, F., van Gulijk M., Mulder E.E., Lukkes M., Klaase L., van den Bosch T., van Nimwegen M., Lau S.P., Latupeirissa K., Schetters S., et al. 2020. The PD-1/PD-L1-Checkpoint Restrains T cell Immunity in Tumor-Draining Lymph Nodes. Cancer Cell. 38:685–700.e8. 10.1016/j.ccell.2020.09.001 [DOI] [PubMed] [Google Scholar]
- Dobin, A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., and Gingeras T.R.. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 29:15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doebis, C., Menning A., Neumann K., Ghani S., Schlawe K., Lauer U., Hamann A., Huehn J., and Syrbe U.. 2011. Accumulation and local proliferation of antigen-specific CD4+ T cells in antigen-bearing tissue. Immunol. Cell Biol. 89:566–572. 10.1038/icb.2010.128 [DOI] [PubMed] [Google Scholar]
- Dominguez, C.X., Müller S., Keerthivasan S., Koeppen H., Hung J., Gierke S., Breart B., Foreman O., Bainbridge T.W., Castiglioni A., et al. 2020. Single-Cell RNA Sequencing Reveals Stromal Evolution into LRRC15+ Myofibroblasts as a Determinant of Patient Response to Cancer Immunotherapy. Cancer Discov. 10:232–253. 10.1158/2159-8290.CD-19-0644 [DOI] [PubMed] [Google Scholar]
- Dubey, C., Croft M., and Swain S.L.. 1996. Naive and effector CD4 T cells differ in their requirements for T cell receptor versus costimulatory signals. J. Immunol. 157:3280–3289. [PubMed] [Google Scholar]
- Elyada, E., Bolisetty M., Laise P., Flynn W.F., Courtois E.T., Burkhart R.A., Teinor J.A., Belleau P., Biffi G., Lucito M.S., et al. 2019. Cross-Species Single-Cell Analysis of Pancreatic Ductal Adenocarcinoma Reveals Antigen-Presenting Cancer-Associated Fibroblasts. Cancer Discov. 9:1102–1123. 10.1158/2159-8290.CD-19-0094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanidis, D., and Moulos P.. 2021. Integrative, normalization-insusceptible statistical analysis of RNA-Seq data, with improved differential expression and unbiased downstream functional analysis. Brief. Bioinform. 22:bbaa156. 10.1093/bib/bbaa156 [DOI] [PubMed] [Google Scholar]
- Ferris, S.T., Durai V., Wu R., Theisen D.J., Ward J.P., Bern M.D., Davidson J.T. IV, Bagadia P., Liu T., Briseño C.G., et al. 2020. cDC1 prime and are licensed by CD4+ T cells to induce anti-tumour immunity. Nature. 584:624–629. 10.1038/s41586-020-2611-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman, G., Levi-Galibov O., David E., Bornstein C., Giladi A., Dadiani M., Mayo A., Halperin C., Meirav Pevsner-Fischer M., Lavon H., et al. 2020. Cancer-associated fibroblast compositions change with breast cancer progression linking the ratio of S100A4+ and PDPN+ CAFs to clinical outcome. Nat. Can. 1:692–708. 10.1038/s43018-020-0082-y [DOI] [PubMed] [Google Scholar]
- Goldmann, T., Zissel G., Watz H., Drömann D., Reck M., Kugler C., Rabe K.F., and Marwitz S.. 2018. Human alveolar epithelial cells type II are capable of TGFβ-dependent epithelial-mesenchymal-transition and collagen-synthesis. Respir. Res. 19:138. 10.1186/s12931-018-0841-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grasso, C.S., Tsoi J., Onyshchenko M., Abril-Rodriguez G., Ross-Macdonald P., Wind-Rotolo M., Champhekar A., Medina E., Torrejon D.Y., Shin D.S., et al. 2020. Conserved Interferon-γ Signaling Drives Clinical Response to Immune Checkpoint Blockade Therapy in Melanoma. Cancer Cell. 38:500–515.e3. 10.1016/j.ccell.2020.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grauel, A.L., Nguyen B., Ruddy D., Laszewski T., Schwartz S., Chang J., Chen J., Piquet M., Pelletier M., Yan Z., et al. 2020. TGFβ-blockade uncovers stromal plasticity in tumors by revealing the existence of a subset of interferon-licensed fibroblasts. Nat. Commun. 11:6315. 10.1038/s41467-020-19920-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habiel, D.M., Camelo A., Espindola M., Burwell T., Hanna R., Miranda E., Carruthers A., Bell M., Coelho A.L., Liu H., et al. 2017. Divergent roles for Clusterin in Lung Injury and Repair. Sci. Rep. 7:15444. 10.1038/s41598-017-15670-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa, K., Sato A., Tanimura K., Uemasu K., Hamakawa Y., Fuseya Y., Sato S., Muro S., and Hirai T.. 2017. Fraction of MHCII and EpCAM expression characterizes distal lung epithelial cells for alveolar type 2 cell isolation. Respir. Res. 18:150. 10.1186/s12931-017-0635-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto, K., Joshi S.K., and Koni P.A.. 2002. A conditional null allele of the major histocompatibility IA-beta chain gene. Genesis. 32:152–153. 10.1002/gene.10056 [DOI] [PubMed] [Google Scholar]
- Helmink, B.A., Reddy S.M., Gao J., Zhang S., Basar R., Thakur R., Yizhak K., Sade-Feldman M., Blando J., Han G., et al. 2020. B cells and tertiary lymphoid structures promote immunotherapy response. Nature. 577:549–555. 10.1038/s41586-019-1922-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill, C., Li J., Liu D., Conforti F., Brereton C.J., Yao L., Zhou Y., Alzetani A., Chee S.J., Marshall B.G., et al. 2019. Autophagy inhibition-mediated epithelial-mesenchymal transition augments local myofibroblast differentiation in pulmonary fibrosis. Cell Death Dis. 10:591. 10.1038/s41419-019-1820-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosszu, K.K., Valentino A., Vinayagasundaram U., Vinayagasundaram R., Joyce M.G., Ji Y., Peerschke E.I., and Ghebrehiwet B.. 2012. DC-SIGN, C1q, and gC1qR form a trimolecular receptor complex on the surface of monocyte-derived immature dendritic cells. Blood. 120:1228–1236. 10.1182/blood-2011-07-369728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, W., Sherman B.T., and Lempicki R.A.. 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4:44–57. 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- Hutton, C., Heider F., Blanco-Gomez A., Banyard A., Kononov A., Zhang X., Karim S., Paulus-Hock V., Watt D., Steele N., et al. 2021. Single-cell analysis defines a pancreatic fibroblast lineage that supports anti-tumor immunity. Cancer Cell. 39:1227–1244.e20. 10.1016/j.ccell.2021.06.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen, C.S., Prokhnevska N., Master V.A., Sanda M.G., Carlisle J.W., Bilen M.A., Cardenas M., Wilkinson S., Lake R., Sowalsky A.G., et al. 2019. An intra-tumoral niche maintains and differentiates stem-like CD8 T cells. Nature. 576:465–470. 10.1038/s41586-019-1836-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, D.B., Estrada M.V., Salgado R., Sanchez V., Doxie D.B., Opalenik S.R., Vilgelm A.E., Feld E., Johnson A.S., Greenplate A.R., et al. 2016. Melanoma-specific MHC-II expression represents a tumour-autonomous phenotype and predicts response to anti-PD-1/PD-L1 therapy. Nat. Commun. 7:10582. 10.1038/ncomms10582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi, K., de Massy M.R., Ismail M., Reading J.L., Uddin I., Woolston A., Hatipoglu E., Oakes T., Rosenthal R., Peacock T., et al. TRACERx consortium . 2019. Spatial heterogeneity of the T cell receptor repertoire reflects the mutational landscape in lung cancer. Nat. Med. 25:1549–1559. 10.1038/s41591-019-0592-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieffer, Y., Hocine H.R., Gentric G., Pelon F., Bernard C., Bourachot B., Lameiras S., Albergante L., Bonneau C., Guyard A., et al. 2020. Single-Cell Analysis Reveals Fibroblast Clusters Linked to Immunotherapy Resistance in Cancer. Cancer Discov. 10:1330–1351. 10.1158/2159-8290.CD-19-1384 [DOI] [PubMed] [Google Scholar]
- Kim, D., Paggi J.M., Park C., Bennett C., and Salzberg S.L.. 2019. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 37:907–915. 10.1038/s41587-019-0201-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouser, L., Madhukaran S.P., Shastri A., Saraon A., Ferluga J., Al-Mozaini M., and Kishore U.. 2015. Emerging and Novel Functions of Complement Protein C1q. Front. Immunol. 6:317. 10.3389/fimmu.2015.00317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyama, M., Mukhopadhyay P., Schuster I.S., Henden A.S., Hülsdünker J., Varelias A., Vetizou M., Kuns R.D., Robb R.J., Zhang P., et al. 2019. MHC Class II Antigen Presentation by the Intestinal Epithelium Initiates Graft-versus-Host Disease and Is Influenced by the Microbiota. Immunity. 51:885–898.e7. 10.1016/j.immuni.2019.08.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krausgruber, T., Fortelny N., Fife-Gernedl V., Senekowitsch M., Schuster L.C., Lercher A., Nemc A., Schmidl C., Rendeiro A.F., Bergthaler A., and Bock C.. 2020. Structural cells are key regulators of organ-specific immune responses. Nature. 583:296–302. 10.1038/s41586-020-2424-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucera, M., Isserlin R., Arkhangorodsky A., and Bader G.D.. 2016. AutoAnnotate: A Cytoscape app for summarizing networks with semantic annotations. F1000 Res. 5:1717. 10.12688/f1000research.9090.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambrechts, D., Wauters E., Boeckx B., Aibar S., Nittner D., Burton O., Bassez A., Decaluwé H., Pircher A., Van den Eynde K., et al. 2018. Phenotype molding of stromal cells in the lung tumor microenvironment. Nat. Med. 24:1277–1289. 10.1038/s41591-018-0096-5 [DOI] [PubMed] [Google Scholar]
- Langmead, B., and Salzberg S.L.. 2012. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 9:357–359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence, M., Huber W., Pagès H., Aboyoun P., Carlson M., Gentleman R., Morgan M.T., and Carey V.J.. 2013. Software for computing and annotating genomic ranges. PLOS Comput. Biol. 9:e1003118. 10.1371/journal.pcbi.1003118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leiva-Juárez, M.M., Kolls J.K., and Evans S.E.. 2018. Lung epithelial cells: therapeutically inducible effectors of antimicrobial defense. Mucosal Immunol. 11:21–34. 10.1038/mi.2017.71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling, G.S., Crawford G., Buang N., Bartok I., Tian K., Thielens N.M., Bally I., Harker J.A., Ashton-Rickardt P.G., Rutschmann S., et al. 2018. C1q restrains autoimmunity and viral infection by regulating CD8+ T cell metabolism. Science. 360:558–563. 10.1126/science.aao4555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low, J.S., Farsakoglu Y., Amezcua Vesely M.C., Sefik E., Kelly J.B., Harman C.C.D., Jackson R., Shyer J.A., Jiang X., Cauley L.S., et al. 2020. Tissue-resident memory T cell reactivation by diverse antigen-presenting cells imparts distinct functional responses. J. Exp. Med. 217:e20192291. 10.1084/jem.20192291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukacs-Kornek, V., Malhotra D., Fletcher A.L., Acton S.E., Elpek K.G., Tayalia P., Collier A.R., and Turley S.J.. 2011. Regulated release of nitric oxide by nonhematopoietic stroma controls expansion of the activated T cell pool in lymph nodes. Nat. Immunol. 12:1096–1104. 10.1038/ni.2112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mächler, M., Rousseeuw P., Struyf A., Hubert M., and Hornik K.. 2012. Cluster: Cluster Analysis Basics and Extensions. R packager version 2.1.2. https://cran.r-project.org/package=cluster
- Maier, B., Leader A.M., Chen S.T., Tung N., Chang C., LeBerichel J., Chudnovskiy A., Maskey S., Walker L., Finnigan J.P., et al. 2020. A conserved dendritic-cell regulatory program limits antitumour immunity. Nature. 580:257–262. 10.1038/s41586-020-2134-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLachlan, J.B., and Jenkins M.K.. 2007. Migration and accumulation of effector CD4+ T cells in nonlymphoid tissues. Proc. Am. Thorac. Soc. 4:439–442. 10.1513/pats.200606-137MS [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon, K.R., van Dijk D., Wang Z., Gigante S., Burkhardt D.B., Chen W.S., Yim K., Elzen A.V.D., Hirn M.J., Coifman R.R., et al. 2019. Visualizing structure and transitions in high-dimensional biological data. Nat. Biotechnol. 37:1482–1492. 10.1038/s41587-019-0336-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh, S.A., Wu D.-C., Cheung J., Navarro A., Xiong H., Cubas R., Totpal K., Chiu H., Wu Y., Comps-Agrar L., et al. 2020. PD-L1 expression by dendritic cells is a key regulator of T-cell immunity in cancer. Nat. Can. 1:681–691. 10.1038/s43018-020-0075-x [DOI] [PubMed] [Google Scholar]
- Özdemir, B.C., Pentcheva-Hoang T., Carstens J.L., Zheng X., Wu C.C., Simpson T.R., Laklai H., Sugimoto H., Kahlert C., Novitskiy S.V., et al. 2014. Depletion of carcinoma-associated fibroblasts and fibrosis induces immunosuppression and accelerates pancreas cancer with reduced survival. Cancer Cell. 25:719–734. 10.1016/j.ccr.2014.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petitprez, F., de Reyniès A., Keung E.Z., Chen T.W., Sun C.M., Calderaro J., Jeng Y.M., Hsiao L.P., Lacroix L., Bougoüin A., et al. 2020. B cells are associated with survival and immunotherapy response in sarcoma. Nature. 577:556–560. 10.1038/s41586-019-1906-8 [DOI] [PubMed] [Google Scholar]
- Riedel, A., Shorthouse D., Haas L., Hall B.A., and Shields J.. 2016. Tumor-induced stromal reprogramming drives lymph node transformation. Nat. Immunol. 17:1118–1127. 10.1038/ni.3492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodig, S.J., Gusenleitner D., Jackson D.G., Gjini E., Giobbie-Hurder A., Jin C., Chang H., Lovitch S.B., Horak C., Weber J.S., et al. 2018. MHC proteins confer differential sensitivity to CTLA-4 and PD-1 blockade in untreated metastatic melanoma. Sci. Transl. Med. 10:eaar3342. 10.1126/scitranslmed.aar3342 [DOI] [PubMed] [Google Scholar]
- Ruhland, M.K., Roberts E.W., Cai E., Mujal A.M., Marchuk K., Beppler C., Nam D., Serwas N.K., Binnewies M., and Krummel M.F.. 2020. Visualizing Synaptic Transfer of Tumor Antigens among Dendritic Cells. Cancer Cell. 37:786–799.e5. 10.1016/j.ccell.2020.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahai, E., Astsaturov I., Cukierman E., DeNardo D.G., Egeblad M., Evans R.M., Fearon D., Greten F.R., Hingorani S.R., Hunter T., et al. 2020. A framework for advancing our understanding of cancer-associated fibroblasts. Nat. Rev. Cancer. 20:174–186. 10.1038/s41568-019-0238-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satija, R., Farrell J.A., Gennert D., Schier A.F., and Regev A.. 2015. Spatial reconstruction of single-cell gene expression data. Nat. Biotechnol. 33:495–502. 10.1038/nbt.3192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schietinger, A., Philip M., Liu R.B., Schreiber K., and Schreiber H.. 2010. Bystander killing of cancer requires the cooperation of CD4(+) and CD8(+) T cells during the effector phase. J. Exp. Med. 207:2469–2477. 10.1084/jem.20092450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schøller, A.S., Fonnes M., Nazerai L., Christensen J.P., and Thomsen A.R.. 2019. Local Antigen Encounter Is Essential for Establishing Persistent CD8+ T-Cell Memory in the CNS. Front. Immunol. 10:351. 10.3389/fimmu.2019.00351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebastian, A., Hum N.R., Martin K.A., Gilmore S.F., Peran I., Byers S.W., Wheeler E.K., Coleman M.A., and Loots G.G.. 2020. Single-Cell Transcriptomic Analysis of Tumor-Derived Fibroblasts and Normal Tissue-Resident Fibroblasts Reveals Fibroblast Heterogeneity in Breast Cancer. Cancers (Basel). 12:1307. 10.3390/cancers12051307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon, P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., and Ideker T.. 2003. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13:2498–2504. 10.1101/gr.1239303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, H., Fang W., Liu M., and Fu D.. 2017. Complement component 1, q subcomponent binding protein (C1QBP) in lipid rafts mediates hepatic metastasis of pancreatic cancer by regulating IGF-1/IGF-1R signaling. Int. J. Cancer. 141:1389–1401. 10.1002/ijc.30831 [DOI] [PubMed] [Google Scholar]
- Śledzińska, A., Vila de Mucha M., Bergerhoff K., Hotblack A., Demane D.F., Ghorani E., Akarca A.U., Marzolini M.A.V., Solomon I., Vargas F.A., et al. 2020. Regulatory T Cells Restrain Interleukin-2- and Blimp-1-Dependent Acquisition of Cytotoxic Function by CD4+ T Cells. Immunity. 52:151–166.e6. 10.1016/j.immuni.2019.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Šošić, D., Richardson J.A., Yu K., Ornitz D.M., and Olson E.N.. 2003. Twist regulates cytokine gene expression through a negative feedback loop that represses NF-kappaB activity. Cell. 112:169–180. 10.1016/S0092-8674(03)00002-3 [DOI] [PubMed] [Google Scholar]
- Strunz, M., Simon L.M., Ansari M., Kathiriya J.J., Angelidis I., Mayr C.H., Tsidiridis G., Lange M., Mattner L.F., Yee M., et al. 2020. Alveolar regeneration through a Krt8+ transitional stem cell state that persists in human lung fibrosis. Nat. Commun. 11:3559. 10.1038/s41467-020-17358-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Maaten, L., and Hinton G.. 2008. Visualizing data using t-SNE. J. Mach. Learn. Res. 9:2579–2605. [Google Scholar]
- van der Windt, G.J., and Pearce E.L.. 2012. Metabolic switching and fuel choice during T-cell differentiation and memory development. Immunol. Rev. 249:27–42. 10.1111/j.1600-065X.2012.01150.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- West, E.E., Kolev M., and Kemper C.. 2018. Complement and the Regulation of T Cell Responses. Annu. Rev. Immunol. 36:309–338. 10.1146/annurev-immunol-042617-053245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wosen, J.E., Mukhopadhyay D., Macaubas C., and Mellins E.D.. 2018. Epithelial MHC Class II Expression and Its Role in Antigen Presentation in the Gastrointestinal and Respiratory Tracts. Front. Immunol. 9:2144. 10.3389/fimmu.2018.02144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto, T.N., Lee P.H., Vodnala S.K., Gurusamy D., Kishton R.J., Yu Z., Eidizadeh A., Eil R., Fioravanti J., Gattinoni L., et al. 2019. T cells genetically engineered to overcome death signaling enhance adoptive cancer immunotherapy. J. Clin. Invest. 129:1551–1565. 10.1172/JCI121491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zander, R., Schauder D., Xin G., Nguyen C., Wu X., Zajac A., and Cui W.. 2019. CD4+ T Cell Help Is Required for the Formation of a Cytolytic CD8+ T Cell Subset that Protects against Chronic Infection and Cancer. Immunity. 51:1028–1042.e4. 10.1016/j.immuni.2019.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, B., Zhang Z., Black C.M., de Crombrugghe B., and Denton C.P.. 2002. Ligand-dependent genetic recombination in fibroblasts : a potentially powerful technique for investigating gene function in fibrosis. Am. J. Pathol. 160:1609–1617. 10.1016/S0002-9440(10)61108-X [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
lists main patient data, including number of patients enrolled in the study, age, gender, histology, smoking history, and TNM stage.
Data Availability Statement
Murine CD4 T cell bulk RNAseq and CAF RNAseq data generated in this study are available through National Center for Biotechnology Information Gene Expression Omnibus (GEO) repository with accession numbers GSE164659 and GSE164653, respectively. Human scRNAseq data used in this study are available through ArrayExpress database (E-MTAB-6149, E-MTAB-6653; Lambrechts et al., 2018). Other relevant data are available from the corresponding author upon reasonable request.