Abstract
Background/Aim: Poorly differentiated thyroid carcinoma (PDTC), anaplastic thyroid carcinoma (ATC), and advanced DTC have poor outcomes. Materials and Methods: We performed next-generation sequencing in nine selected aggressive thyroid cancers. Results: Among the nine patients, the driver gene mutations BRAF V600E (3/9) and NRAS Q61K (1/9) were detected. Other oncogenic mutations included ERBB2 (1/9) and CDK4 (1/9). Telomerase reverse transcriptase (TERT) promoter mutation was found in five cases. Among tumor suppressor genes, mutations in TP53 (3/9), ARID1A (1/9), APC (1/9), MEN1 (1/9), DICER1 (1/9), and MED12 (1/9) were identified. RET fusions were found in two cases, one with PTDC and the other with ATC. The ATC with RET fusion also harbored TP53 and TERT promoter mutations. None of the PDTC cases had BRAF or RAS gene alterations. Conclusion: Since genetic alterations with therapeutic and prognostic implications were detected using next-generation sequencing, this technique is recommended to be performed for patients with aggressive thyroid cancer.
Keywords: Whole exome sequencing, next-generation sequencing, aggressive thyroid cancer, anaplastic thyroid carcinoma, poorly differentiated thyroid carcinoma, advanced differentiated thyroid cancer
Thyroid cancer is one of the most common endocrine malignancies (1,2). Most thyroid follicular cell-derived cancers fall into the category of differentiated thyroid cancer (DTC), which include papillary thyroid carcinoma (PTC) and follicular carcinoma (3,4). DTCs generally demonstrate indolent biological behavior with excellent prognosis, usually treated by surgery with or without radioactive iodine (5). Its prognosis is affected by diverse clinicopathological parameters, including tumor stage, patient age at diagnosis, and tumor size (6). Poorly differentiated thyroid carcinoma (PDTC) and anaplastic thyroid carcinoma (ATC) constitute rare but aggressive subtypes of thyroid carcinoma (7-9). ATC is characterized by undifferentiated follicular thyroid cells with diverse histological patterns with any combination of epithelial, giant cell, or sarcomatoid features (10,11). The prognosis is dismal, with a mortality rate of >90% (12,13). On the other hand, PDTC is known to have intermediate clinicopathological characteristics (14). Although most cases of PDTC and ATC are known to develop from DTC with stepwise accumulation of molecular alteration (15,16), they can develop de novo as well (7,17).
In the past decade, substantial studies have been performed to investigate genetic alterations occurring in thyroid papillary carcinoma (15,18-21). In DTC, most genetic alterations involve mitogen-activated protein kinase (MAPK) and PI3K-AKT pathways (15). Telomerase reverse transcriptase (TERT) promoter mutations in thyroid cancer have been associated with tumor aggressiveness, recurrence, and poor prognosis (22). Tumor suppressor gene mutations, such as TP53 and PTEN, also contribute to tumor aggressiveness (23,24). Chromosomal rearrangements involving RET, NTRK1, NTRK3, ALK, and other genes have been reported in 15% of thyroid cancer cases (25-27). Copy number alterations have also been observed in thyroid cancer (28).
Although the genetic analysis of DTC has been compre-hensively investigated, the biological behavior of aggressive thyroid cancer is not completely understood. Understanding the key molecular alterations in aggressive thyroid cancer is essential for the establishment of targeted therapy.
In this study, we investigated the genomics of clinically aggressive thyroid cancer using whole exome sequencing (WES) and targeted next-generation sequencing (NGS) in association with their clinicopathological characteristics.
Materials and Methods
Study population. We performed next-generation sequencing for nine patients with aggressive thyroid cancers. From 2011 to 2021, we retrospectively collected seven surgically resected thyroid cancer cases with an aggressive course at our institution, which consisted of three advanced DTC, two PDTC, and two ATC. Advanced DTC consisted of thyroid cancer cases with distant site metastasis, recurrent and persistent locally relapsing disease, and stage IV cases. WES was performed on representative tissue sections from each case. We also included two recent cases (one ATC and one PDTC), which underwent targeted NGS as requested by the clinician. In total, the genomic profiling of three cases each of ATC, PDTC, and advanced DTC were studied. The institutional review board approved this retrospective study (No. KNUCH 2017-08-017). Clinicopathological data, including patient age, sex, tumor size, extrathyroidal extension, and lymph node metastasis were retrieved from the electronic medical records of the hospital. Written informed consent from the patients was waived due to the retrospective nature of the study.
Pathological evaluation. The surgical specimens were fixed in 10% neutral buffered formalin and embedded in paraffin blocks. The entire tumor was submitted for microscopic examination for small (<3 cm) tumors. At least three representative sections (one section per 1 cm of tumor) were submitted for each case. For each formalin-fixed, paraffin-embedded (FFPE) tissue block, 4-μm sections were cut and stained with hematoxylin and eosin. Two pathologists with experience in endocrine tumor pathology (MK and JYP) reviewed all available stained slides. Tumors were diagnosed and classified according to the fourth edition of the World Health Organization classification of endocrine organ tumors (29).
Whole exome sequencing.
Library construction and sequencing. DNA was extracted using QIAamp DNA FFPE Tissue Kit (Qiagen, Valencia CA, USA) and RNA was extracted using RNeasy FFPE Kit (Qiagen), according to manufacturer’s recommendations. For exome sequencing, DNA library preparation, capture, and sequencing were conducted by Macrogen (Seoul, Republic of Korea). The captured libraries were prepared from the SureSelect Human All Exon V6 kit (Agilent Technologies, Santa Clara, CA, USA) with an input of 200 ng of sheared DNA according to the manufacturer’s instruction. One hundred bp paired-end sequencing was performed on a NovaSeq 6000 sequencer (Illumina, San Diego, CA, USA) to obtain a mean depth of 200×, according to the manufacturer’s instruction (Illumina). For transcriptome sequencing, the ribosomal RNA was depleted from whole RNA using NEBNext® rRNA depletion Kit (New England Biolabs, Inc., MA, USA). The RNA libraries were prepared from NEBNext® Ultra II RNA Library Prep Kit for Illumina (New England Biolabs, Inc.) according to the manufacturer’s instruction. Then, the final RNA libraries’ quality was evaluated with the Agilent 4200 TapeStation System (Agilent). Following cluster amplification of denatured templates, sequencing was progressed as a paired-end (2×150 bp) on Illumina NextSeq 550 (or 550Dx) according to the manufacturer’s instruction (Illumina).
Exome sequencing data analysis. Quality control of the sequencing files was performed using FastQC, followed by low quality read trimming using BBDuk (v37.23). The WES reads were alighted to the human reference genome hg19 (GRCh37) using BWA (0.7.10). In order to further process the data, Picard (v0.2.5b9) was employed to mark duplicate reads and GATK (v4.0.6.0) was used to perform local realignment and recalibration. After post-alignment processing of the data, single nucleotide variants (SNVs) were called with MuTect2 (include GATK v4.0.6.0). Variant filtration functions included in the GATK such as CollectSequencingArtifactMetrics, FilterMutectCalls, and FilterByOrientationBias, were applied for the confident somatic mutation calling. Using SnpEFF & SnpSift (v4.4) functions, somatic variants were annotated. The following variants were excluded to reduce false-positives: 1) variants with a minor allele frequency of more than 1% in the genome aggregation, Exome Aggregation Consortium, 1000 genome database, and Korean population databases, 2) variants with oxidized guanine to 8-oxoguanin (OxoG) artifacts, 3) variants with mutated read counts less than 3, 4) variants with total read depth less than 20, and 5) variants with a variant allele frequency less than 3%.
Copy number aberrations were quantified and reported for each gene. It was the segmented normalized copy number exon coverage copy number between each sample. The Circular binary segmentation (CBS) algorithm was used to segment copy number data and identify genomic regions with abnormal copy number.
Transcriptome sequencing data analysis. Quality control of the sequencing files was performed using FastQC, followed by adapter trimming and low quality read trimming using cutadapt (v1.18) and BBDuk (v37.23). STAR (v2.7.2a) (25) was used to map RNA-Seq reads to the human reference genome hg19 (GRCh37). STAR uses an uncompressed suffix array to align reads to a reference genome. A read attempts to sort sequentially, and if there is an unsorted section, it is marked. The results of the previous steps are collected and scored, and the final alignment result is obtained through this. Through this, splice junction, multiple mismatches, and indels can be detected, and it has the advantage of short execution time.
Fusion transcripts were originally identified using STAR-Fusion (v1.7.0). The STAR-Fusion analysis used best practice results optimized for STAR to detect fusion genes. Fusion candidates were manually reviewed and evaluated for clinical significance using Personal Cancer Genome Reporter (PCGR).
Targeted next-generation sequencing. Targeted NGS was performed using a customized cancer panel (ONCOaccuPanel, NGeneBio, Seoul, Republic of Korea) as per the manufacturer’s instruction. The customized cancer panel was designed to detect 323 cancer-related genes with potential SNVs, indels, potential copy number alterations, and six genes with potential fusion variants. For targeted sequencing, DNA and RNA were extracted from FFPE tissue block, using QIAGEN AllPrep DNA/RNA FFPE Kit (Qiagen, Hilden, Germany). After hybridization capture-based target enrichment of 100 ng DNA and 500 ng of RNA, paired-end sequencing (2×150 bp) was performed using a MiSeq sequencer (Illumina) following the manufacturer’s instructions. SNVs, indels, and copy number variations were analyzed using the Burrows–Wheeler Aligner and Genome Analysis Tool Kit. Variants with a total depth of at least 100× and variant allele frequency of at least 3% were included for analysis. Fusion variants were determined using FusionCatcher and STAR-Fusion. Variant interpretation was based on recommendations from the Association for Molecular Pathology, American Society of Clinical Oncology, and College of American Pathologists (30).
PCR analysis for TERT promoter mutation. PCR analysis for TERT promoter mutation was performed for each case. DNA was extracted from 10-μm FFPE tissue blocks. Tumor sections were selected and dissected manually. TERT promoter mutations were analyzed using the real-time PCR clamping method of PNAClamp™ TERT kit (Panagene, Daejeon, Republic of Korea), as per the manufacturer’s instructions. For every run, a positive control of mutation-holding human genomic DNA and a negative control of distilled water were provided.
Results
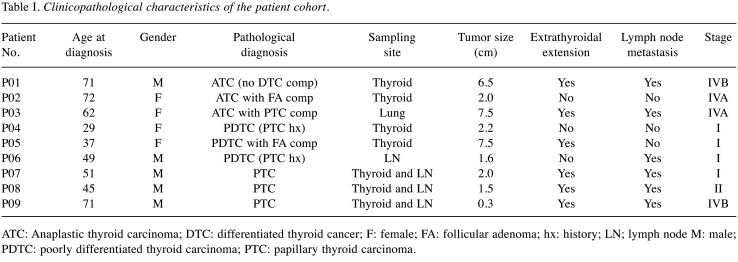
Patient cohort. This study enrolled nine patients, who were designated as P01-P09. There were three cases each of ATC (P01-P03), PDTC (P04-P06), and advanced DTC (P07-P09). Their characteristics are summarized in Table I. The median age was 51 years, ranging from 29 to 72 years. There were five males and four females. Extrathyroidal extension was observed in six cases (6/9), whereas lymph node metastasis was found in six cases (6/9). Four patients had stage I (4/9), one patient had stage II (1/9), and four patients had stage IV (4/9) cancer.
Table I. Clinicopathological characteristics of the patient cohort.
ATC: Anaplastic thyroid carcinoma; DTC: differentiated thyroid cancer; F: female; FA: follicular adenoma; hx: history; LN; lymph node M: male; PDTC: poorly differentiated thyroid carcinoma; PTC: papillary thyroid carcinoma.
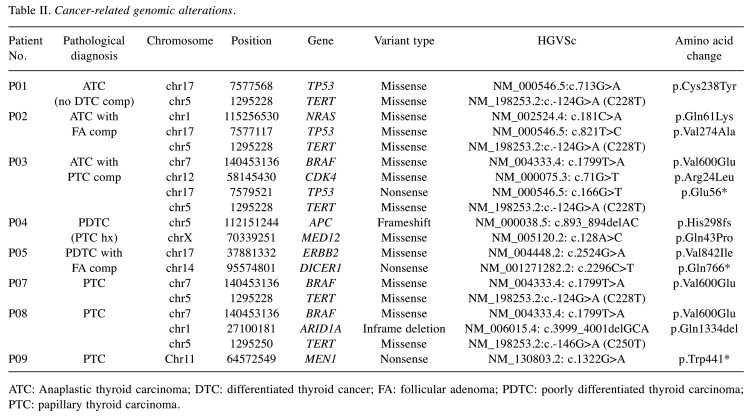
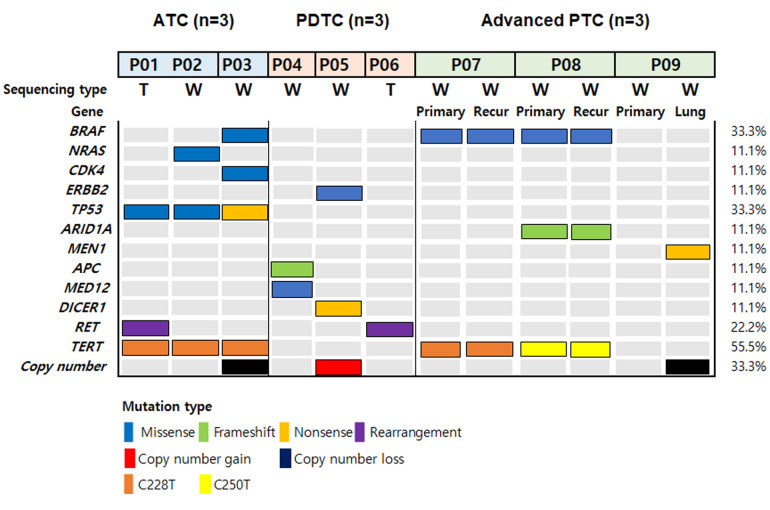
Overall genomic landscape of clinically aggressive thyroid cancer. WES was performed at a mean depth of 123× (range=99×-135×) and targeted NGS was performed at a mean depth of 307× (range=243×-371×). The mean number of pathogenic mutations observed was 3.3 (range=3-4) in ATC, 1.7 (range=1-2) in PDTC, and 1.8 (range=1-3) in advanced DTC. Among cancer-related pathogenic mutations (Table II), BRAF V600E mutation was detected in three patients (3/9). Other oncogenic alterations included NRAS (1/9), ERBB2 (1/9), and CDK4 (1/9). Among tumor suppresser genes, TP53 (3/9), ARID1A (1/9), APC (1/9), MEN1 (1/9), DICER1 (1/9), and MED12 (1/9) were identified. Mutations in TERT promoter were detected in five cases. RET fusions were found in two cases: one PDTC and one ATC. Copy number losses were observed in two cases, whereas copy number gain was noted in one case. The cancer-related mutation heatmap of the patient cohort is shown in Figure 1.
Table II. Cancer-related genomic alterations.
ATC: Anaplastic thyroid carcinoma; DTC: differentiated thyroid cancer; FA: follicular adenoma; PDTC: poorly differentiated thyroid carcinoma; PTC: papillary thyroid carcinoma.
Figure 1. The mutational landscape of anaplastic, poorly differentiated, and advanced thyroid cancers. T: Targeted sequencing; W: whole exome sequencing.
Mutational analysis by cancer subtypes.
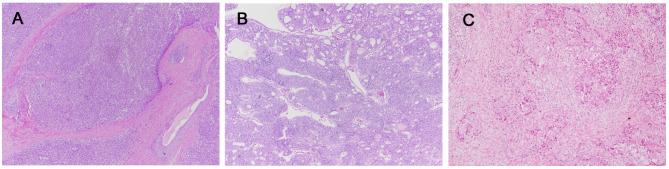
Anaplastic thyroid carcinoma (P01-P03). The entire tumor of P01 showed a sarcomatoid pattern, and a DTC component was not found despite thorough tumor sampling. Interestingly, oncogenic NCOA4-RET fusion was detected. Mutations of TP53 C238Y and TERT C228T were also found.
P02 had ATC on the background of follicular adenoma. NRAS Q61K mutation was found. Additionally, TP53 V274A and TERT C228T mutations were detected.
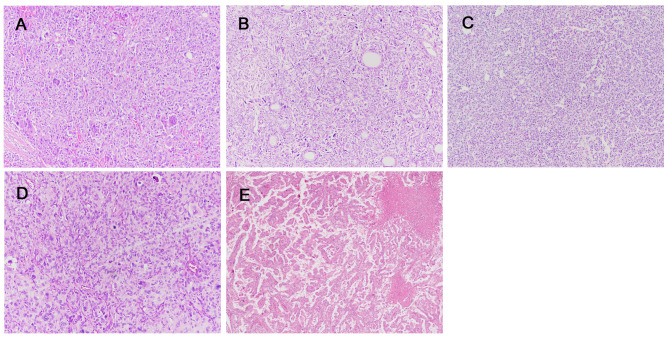
P03 had metastatic ATC in the lung with a PTC component. P03 underwent total thyroidectomy about 30 years ago due to PTC. The patient harbored a BRAF V600E mutation, which was consistent with the histological findings. The oncogenic mutation CDK4 R24L was also found. Additionally, the mutations TP53 E56* and TERT C228T were identified. Other than these genetic alterations, copy number losses of chromosome 8, 10, 13, 16q, 17p, and 18 were observed. Representative photos of each case are presented in Figure 2.
Figure 2. Histopathological findings of anaplastic thyroid carcinoma. A: P01. The entire tumor shows sarcomatoid features, and no differentiated thyroid cancer component was found. B-C: P02. The tumor exhibited findings of an anaplastic thyroid carcinoma (B) with a follicular adenoma component (C). D-E: P03. Metastatic anaplastic thyroid carcinoma (D) in the lung, with a papillary thyroid carcinoma component (E). (Original magnifications: A-D: ×100; E: ×40).
Poorly differentiated thyroid carcinoma (P04-P06). P04 had a previous history of PTC in 2006 and underwent total remnant thyroidectomy in 2013 due to cancer recurrence. Five years later, he was diagnosed with familial adenomatous polyposis (FAP) and associated colon adenocarcinoma. P04 harbored APC H298fs mutation, consistent with FAP. A MED12 Q43P mutation was also observed. Histological features showing cribriform-morular pattern, which is frequently associated with FAP, were not identified.
P05 exhibited a focal follicular adenoma component. Oncogenic ERBB2 V842I and tumor suppressor DICER1 Q766* mutations were identified. Accompanying these genetic alterations, copy number gains of chromosomes 4, 5, 14, and 18 were detected. No other DICER1 syndrome-associated tumors were found in this patient.
P06 had a history of PTC with multiple lymph node metastases for 2 years. Oncogenic CCDC6-RET fusion was detected in PDTC. Representative photos of each case are presented in Figure 3.
Figure 3. Histopathological findings of poorly differentiated thyroid carcinoma. A: P04. B: P05. The tumor shows findings of a poorly differentiated thyroid carcinoma with a follicular adenoma component. C: P06. (Original magnifications: A-B: ×40; C: ×100).
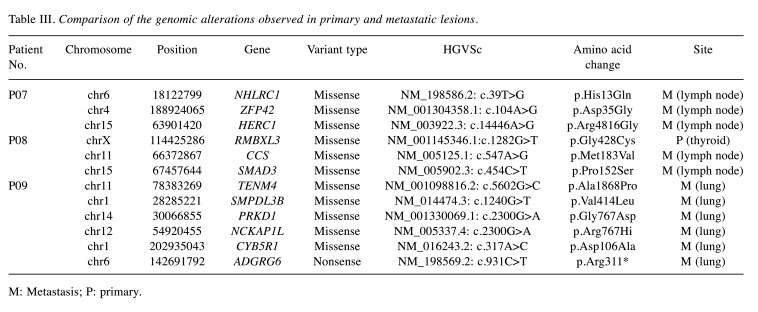
Advanced differentiated thyroid cancer (P07-P09). P07, P08, and P09 had histologically differentiated PTC with distant metastasis and/or regional lymph node recurrence. WES was performed in each patient, and the samples were collected from both the primary tumor and the metastatic foci. Genetic alterations identified in either primary or metastatic lesions are shown in Table III.
Table III. Comparison of the genomic alterations observed in primary and metastatic lesions.
M: Metastasis; P: primary.
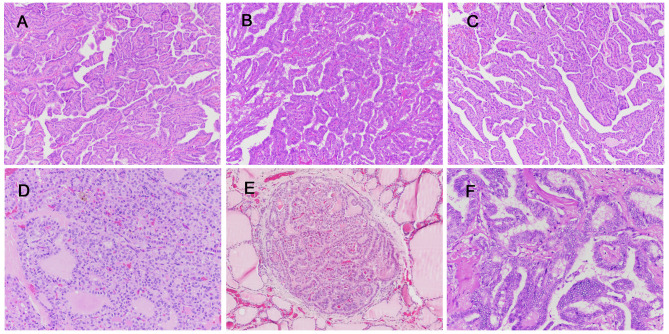
P07 had PTC with a history of neck node recurrences. BRAF V600E and TERT C228T mutations occurred in both the primary site and metastatic lymph nodes. Alterations in ACACB D436G, NHLRC1 H13Q, ZFP42 D35G, and HERC1 R3816G were found only in the metastatic lymph nodes.
P08 had PTC with metastasis to both the neck lymph nodes and soft tissue. The primary tumor and metastatic lymph nodes also shared BRAF V600E, ARID1A Q543*, and TERT C250T mutations. SMAD3 P152S and CCS M183V were detected only in the metastatic lymph nodes.
P09 had lung metastasis at the time of diagnosis. No pathogenic genetic alterations were detected, but MEN1 W441* mutation was found in the metastatic thyroid carcinoma in the lung. Additionally, mutations of TENM4 A1868P, SMPDL3B V414L, PRKD1 G767D, NCKAP1L R767H, CYB5R1 D106A, and ADGRG6 R311* were found only in the metastatic lesions. Copy number losses, involving chromosomes 11q and 22, were also seen in the metastatic lung lesion. Representative photos of each case are presented in Figure 4.
Figure 4. Histopathological findings of advanced differentiated thyroid carcinoma. A-B: P07, primary (A) and metastatic (B). C-D: P08, primary (C) and metastatic (D). E-F: P09, primary (E) and metastatic (F). (Original magnifications: A-C: ×100; D: ×200, E: ×40, F: ×200).
Discussion
In this study, we investigated the genetic alterations that occur in aggressive thyroid cancer. Previous genetic analyses of thyroid cancers have demonstrated the characteristic high frequencies of somatic mutations in the MAPK pathway. However, we found four cases with BRAF and RAS gene alterations. Furthermore, rare but targetable genetic alterations and tumor suppressor gene mutations were detected through sequencing. This could be because our study involved aggressive thyroid cancers, in contrast to previous genetic studies such as The Cancer Genome Atlas study, which primarily focused on low-to-intermediate risk thyroid cancers (31).
Although ATC comprises only 1%-2% of all thyroid cancers, it accounts for more than 50% of thyroid cancer-related deaths. Out of the three ATC cases, two harbored a driver gene mutation of either BRAF or RAS, consistent with previous studies (32). Additionally, all ATC cases had co-altered TP53 and TERT promoters, which are frequently co-mutated in ATC and contribute to aggressive biological behavior (33). Notably, P01 harbored oncogenic NCOA4-RET fusion. Few previous reports have discussed ATC with RET fusion (34). Histologically, the entire tumor had a sarcomatoid appearance without a DTC component despite extensive tumor sampling. Although most previous studies support the notion that ATC develops from well-differentiated thyroid cancer via a stepwise dedifferentiation process involving either the BRAF- or RAS-driven pathway, this rare case may suggest that de novo ATC can develop from oncogenic fusion with additional gain of TP53 and TERT promoter mutation. In P03, oncogenic CDK4 mutation was also identified in addition to BRAF V600E mutation. Alterations in cell cycle genes have been reported in ATC (26,32) that can thus be a future therapeutic target (35-37).
ATC showed more pathogenic alterations compared to PDTC and advanced DTC in this study. This is in line with previous studies, wherein ATC was associated with more pathogenic mutations and a high tumor mutational burden compared with the other subtypes of thyroid cancer (26,32,33,38).
Although PDTC accounts for a small proportion of thyroid cancers, it carries a poor prognosis, with an overall 5-year survival rate of only 60%-70% (14,39). P04 had a history of FAP, characterized by germline APC mutation. Thyroid cancer frequently occurs in individuals with FAP (40), but most FAP cases are associated with the cribriform-morular variant or conventional PTC (41). In this case, the concomitant MED12 mutation might have contributed to the development of PDTC (42). In P05, an oncogenic ERBB2 mutation was detected, and this could be a possible therapeutic target (43). In this patient, the accompanying tumor suppressor gene DICER1 mutation and chromosomal copy number gain, harboring the TERT genomic locus (5p15.33) (44), might be associated with the development of PDTC. In P06, oncogenic RET fusion was also identified. PDTC with a PTC component is closely associated with RET fusion (38), in line with the histological appearance of P06.
BRAF or RAS mutations were not detected in PDTC included in this study. Many previous studies have considered PDTC as an intermediate state between DTC and ATC. However, the proportion of BRAF or RAS mutations in PDTC is consistently low compared with DTC or ATC (7,26,38). Thus, PDTC may involve a different, non-BRAF and non-RAS tumorigenic pathway altogether.
DTC usually presents as a localized cancer with good prognosis; however, DTC with distant metastasis and/or multiple regional lymph node recurrence is associated with significantly high morbidity and mortality. In this study, we performed WES in cases matched for primary thyroid carcinoma and metastases, i.e., in patients P07-P09 who presented with an aggressive clinical course. Previously, WES has been used to evaluate the clonal relationship of matched primary tumors/metastases in several cancers, including breast and colorectal carcinomas (45,46), but only few studies have simultaneously investigated genetic changes in primary PTC and their metastases (47).
In addition to the BRAF V600E mutation, P07 and P08 had the TERT promoter mutation, consistent with their aggressive clinical course. The ARID1A mutation, a component of the SWItch/sucrose non-fermentable (SWI/SNF) nucleosome remodeling complexes (48), might have also contributed to the aggressiveness of the tumor in P08. However, despite WES, no pathogenic mutation was detected in the primary tumor of P09. Thus, the mechanism of tumorigenesis in this sample might involve epigenetic alterations. Alternatively, it could be due to somatic mutations of unknown clinical significance whose biological behaviors still need to be established. On the other hand, metastatic thyroid carcinoma in the lung harbored the pathogenic MEN1 mutation, which might be associated with distant metastasis. Since the primary tumor and metastasis shared most pathogenic mutations except for MEN1, aggressive clinical behavior in advanced DTC might be determined early on during tumorigenesis. However, some somatic alterations, such as SMAD3, NHLRC1, RMBXL3, and ADGRG6, occurred only either in the primary or metastatic tumor in P07-P09, and thus their biological effects remain uncertain.
The histological appearance of advanced DTC harboring TERT promoter mutation or tumor suppressor genes in this study were not distinguishable from localized conventional DTC. Routine molecular studies, including the TERT promoter mutation test, are thus needed for better prognostic stratification of thyroid cancer patients.
In addition to somatic alterations, the copy number alterations involving multiple chromosomes, was identified in P03, P05, and P09. Chromosomal instability has previously been observed in aggressive thyroid cancers (44,49), but their association with tumor aggressiveness is currently unknown. The copy number gain or loss of hotspot regions in thyroid cancer might have affected tumor progression (26,32).
Previous studies have indicated that aberrant DNA repair gene expression may be associated with thyroid cancer dedifferentiation (44). However, genetic alterations in DNA mismatch repair genes (e.g., MSH1 and MSH2) were not detected in this study. Other genetic alterations that are known to be associated with tumor progression in thyroid cancer, such as EIF1AX, PIK3CA, NFE2L2, SPOP, TET2, and ATRX, were also not detected (26). This might be due to the limited sample size.
This study has some limitations. Since tumor-only sequencing was performed in this study, tumor mutational burden and mutational signature were not calculated, but these might have clinical significance. Transcriptome expression analysis was not performed due to the lack of non-neoplastic tissue samples. Moreover, potential subclonal mutations with clinical implications might have been missed in WES due to the limitations in the read depth. Additionally, due to the small sample size in this cohort, a larger cohort analysis should be conducted to validate the major findings in the present study.
Although we analyzed a relatively small number of cases, we were able to recapitulate major pathogenic alterations known to occur in aggressive thyroid cancers. Additionally, rare but targetable genetic alterations and mutations with prognostic implications were found. This can be of benefit to establish a precise therapeutic approach individualized to each patient. Subsequent studies with a large number of aggressive thyroid cancer cases are needed to confirm the results of this study.
Conflicts of Interest
The Authors have no conflicts of interest to declare in relation to this study.
Authors’ Contributions
JYP provided the conception and design of the study. JHK and KMS drafted the manuscript. JHK analyzed previous articles regarding aggressive thyroid cancers. MSK, Nora JYP and JYP carefully reviewed and revised the manuscript. All Authors read and approved the final manuscript.
Acknowledgements
The Authors are thankful for the support provided by Kyungpook National University Chilgok Hospital molecular pathology laboratory.
References
- 1.Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.La Vecchia C, Malvezzi M, Bosetti C, Garavello W, Bertuccio P, Levi F, Negri E. Thyroid cancer mortality and incidence: a global overview. Int J Cancer. 2015;136(9):2187–2195. doi: 10.1002/ijc.29251. [DOI] [PubMed] [Google Scholar]
- 3.Chen AY, Jemal A, Ward EM. Increasing incidence of differentiated thyroid cancer in the United States, 1988-2005. Cancer. 2009;115(16):3801–3807. doi: 10.1002/cncr.24416. [DOI] [PubMed] [Google Scholar]
- 4.Liska J, Altanerova V, Galbavy S, Stvrtina S, Brtko J. Thyroid tumors: histological classification and genetic factors involved in the development of thyroid cancer. Endocr Regul. 2005;39(3):73–83. [PubMed] [Google Scholar]
- 5.Mazzaferri EL. Long-term outcome of patients with differentiated thyroid carcinoma: effect of therapy. Endocr Pract. 2000;6(6):469–476. doi: 10.4158/EP.6.6.469. [DOI] [PubMed] [Google Scholar]
- 6.Duntas L, Grab-Duntas BM. Risk and prognostic factors for differentiated thyroid cancer. Hell J Nucl Med. 2006;9(3):156–162. [PubMed] [Google Scholar]
- 7.Ibrahimpasic T, Ghossein R, Shah JP, Ganly I. Poorly differentiated carcinoma of the thyroid gland: current status and future prospects. Thyroid. 2019;29(3):311–321. doi: 10.1089/thy.2018.0509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Patel KN, Shaha AR. Poorly differentiated and anaplastic thyroid cancer. Cancer Control. 2006;13(2):119–128. doi: 10.1177/107327480601300206. [DOI] [PubMed] [Google Scholar]
- 9.Wiseman SM, Loree TR, Rigual NR, Hicks WL Jr, Douglas WG, Anderson GR, Stoler DL. Anaplastic transformation of thyroid cancer: review of clinical, pathologic, and molecular evidence provides new insights into disease biology and future therapy. Head Neck. 2003;25(8):662–670. doi: 10.1002/hed.10277. [DOI] [PubMed] [Google Scholar]
- 10.Deeken-Draisey A, Yang GY, Gao J, Alexiev BA. Anaplastic thyroid carcinoma: an epidemiologic, histologic, immunohistochemical, and molecular single-institution study. Hum Pathol. 2018;82:140–148. doi: 10.1016/j.humpath.2018.07.027. [DOI] [PubMed] [Google Scholar]
- 11.Giuffrida D, Gharib H. Anaplastic thyroid carcinoma: current diagnosis and treatment. Ann Oncol. 2000;11(9):1083–1089. doi: 10.1023/a:1008322002520. [DOI] [PubMed] [Google Scholar]
- 12.Sugitani I, Miyauchi A, Sugino K, Okamoto T, Yoshida A, Suzuki S. Prognostic factors and treatment outcomes for anaplastic thyroid carcinoma: ATC Research Consortium of Japan cohort study of 677 patients. World J Surg. 2012;36(6):1247–1254. doi: 10.1007/s00268-012-1437-z. [DOI] [PubMed] [Google Scholar]
- 13.Kebebew E, Greenspan FS, Clark OH, Woeber KA, McMillan A. Anaplastic thyroid carcinoma. Treatment outcome and prognostic factors. Cancer. 2005;103(7):1330–1335. doi: 10.1002/cncr.20936. [DOI] [PubMed] [Google Scholar]
- 14.Asioli S, Erickson LA, Righi A, Jin L, Volante M, Jenkins S, Papotti M, Bussolati G, Lloyd RV. Poorly differentiated carcinoma of the thyroid: validation of the Turin proposal and analysis of IMP3 expression. Mod Pathol. 2010;23(9):1269–1278. doi: 10.1038/modpathol.2010.117. [DOI] [PubMed] [Google Scholar]
- 15.Nikiforov YE, Nikiforova MN. Molecular genetics and diagnosis of thyroid cancer. Nat Rev Endocrinol. 2011;7(10):569–580. doi: 10.1038/nrendo.2011.142. [DOI] [PubMed] [Google Scholar]
- 16.Dobashi Y, Sugimura H, Sakamoto A, Mernyei M, Mori M, Oyama T, Machinami R. Stepwise participation of p53 gene mutation during dedifferentiation of human thyroid carcinomas. Diagn Mol Pathol. 1994;3(1):9–14. doi: 10.1097/00019606-199403010-00003. [DOI] [PubMed] [Google Scholar]
- 17.Parenti R, Salvatorelli L, Magro G. Anaplastic thyroid carcinoma: current treatments and potential new therapeutic options with emphasis on TfR1/CD71. Int J Endocrinol. 2014;2014:685396. doi: 10.1155/2014/685396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Virk RK, Van Dyke AL, Finkelstein A, Prasad A, Gibson J, Hui P, Theoharis CG, Carling T, Roman SA, Sosa JA, Udelsman R, Prasad ML. BRAFV600E mutation in papillary thyroid microcarcinoma: a genotype-phenotype correlation. Mod Pathol. 2013;26(1):62–70. doi: 10.1038/modpathol.2012.152. [DOI] [PubMed] [Google Scholar]
- 19.Nikiforova MN, Wald AI, Roy S, Durso MB, Nikiforov YE. Targeted next-generation sequencing panel (ThyroSeq) for detection of mutations in thyroid cancer. J Clin Endocrinol Metab. 2013;98(11):E1852–E1860. doi: 10.1210/jc.2013-2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nikiforov YE. Molecular analysis of thyroid tumors. Mod Pathol. 2011;24(Suppl 2):S34–S43. doi: 10.1038/modpathol.2010.167. [DOI] [PubMed] [Google Scholar]
- 21.Xing M. BRAF mutation in papillary thyroid cancer: pathogenic role, molecular bases, and clinical implications. Endocr Rev. 2007;28(7):742–762. doi: 10.1210/er.2007-0007. [DOI] [PubMed] [Google Scholar]
- 22.Liu X, Bishop J, Shan Y, Pai S, Liu D, Murugan AK, Sun H, El-Naggar AK, Xing M. Highly prevalent TERT promoter mutations in aggressive thyroid cancers. Endocr Relat Cancer. 2013;20(4):603–610. doi: 10.1530/ERC-13-0210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schultz KAP, Rednam SP, Kamihara J, Doros L, Achatz MI, Wasserman JD, Diller LR, Brugières L, Druker H, Schneider KA, McGee RB, Foulkes WD. PTEN, DICER1, FH, and their associated tumor susceptibility syndromes: Clinical features, genetics, and surveillance recommendations in childhood. Clin Cancer Res. 2017;23(12):e76–e82. doi: 10.1158/1078-0432.CCR-17-0629. [DOI] [PubMed] [Google Scholar]
- 24.Latteyer S, Tiedje V, König K, Ting S, Heukamp LC, Meder L, Schmid KW, Führer D, Moeller LC. Targeted next-generation sequencing for TP53, RAS, BRAF, ALK and NF1 mutations in anaplastic thyroid cancer. Endocrine. 2016;54(3):733–741. doi: 10.1007/s12020-016-1080-9. [DOI] [PubMed] [Google Scholar]
- 25.Chu YH, Dias-Santagata D, Farahani AA, Boyraz B, Faquin WC, Nosé V, Sadow PM. Clinicopathologic and molecular characterization of NTRK-rearranged thyroid carcinoma (NRTC) Mod Pathol. 2020;33(11):2186–2197. doi: 10.1038/s41379-020-0574-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yoo SK, Song YS, Lee EK, Hwang J, Kim HH, Jung G, Kim YA, Kim SJ, Cho SW, Won JK, Chung EJ, Shin JY, Lee KE, Kim JI, Park YJ, Seo JS. Integrative analysis of genomic and transcriptomic characteristics associated with progression of aggressive thyroid cancer. Nat Commun. 2019;10(1):2764. doi: 10.1038/s41467-019-10680-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kelly LM, Barila G, Liu P, Evdokimova VN, Trivedi S, Panebianco F, Gandhi M, Carty SE, Hodak SP, Luo J, Dacic S, Yu YP, Nikiforova MN, Ferris RL, Altschuler DL, Nikiforov YE. Identification of the transforming STRN-ALK fusion as a potential therapeutic target in the aggressive forms of thyroid cancer. Proc Natl Acad Sci USA. 2014;111(11):4233–4238. doi: 10.1073/pnas.1321937111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matsuse M, Sasaki K, Nishihara E, Minami S, Hayashida C, Kondo H, Suzuki K, Saenko V, Yoshiura K, Mitsutake N, Yamashita S. Copy number alteration and uniparental disomy analysis categorizes Japanese papillary thyroid carcinomas into distinct groups. PLoS One. 2012;7(4):e36063. doi: 10.1371/journal.pone.0036063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lloyd RV, Osamura RY, Kloppel G, Rosai J, editors. Lyon, IARC Press. 2017. WHO classification of tumours of endocrine organs, 4th edn. [Google Scholar]
- 30.Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, Grody WW, Hegde M, Lyon E, Spector E, Voelkerding K, Rehm HL, ACMG Laboratory Quality Assurance Committee Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17(5):405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cancer Genome Atlas Research Network Integrated genomic characterization of papillary thyroid carcinoma. Cell. 2014;159(3):676–690. doi: 10.1016/j.cell.2014.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pozdeyev N, Gay LM, Sokol ES, Hartmaier R, Deaver KE, Davis S, French JD, Borre PV, LaBarbera DV, Tan AC, Schweppe RE, Fishbein L, Ross JS, Haugen BR, Bowles DW. Genetic analysis of 779 advanced differentiated and anaplastic thyroid cancers. Clin Cancer Res. 2018;24(13):3059–3068. doi: 10.1158/1078-0432.CCR-18-0373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xu B, Ghossein R. Genomic landscape of poorly differentiated and anaplastic thyroid carcinoma. Endocr Pathol. 2016;27(3):205–212. doi: 10.1007/s12022-016-9445-4. [DOI] [PubMed] [Google Scholar]
- 34.Dias-Santagata D, Lennerz JK, Sadow PM, Frazier RP, Govinda Raju S, Henry D, Chung T, Kherani J, Rothenberg SM, Wirth LJ. Response to RET-specific therapy in RET fusion-positive anaplastic thyroid carcinoma. Thyroid. 2020;30(9):1384–1389. doi: 10.1089/thy.2019.0477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sherr CJ, Beach D, Shapiro GI. Targeting CDK4 and CDK6: From discovery to therapy. Cancer Discov. 2016;6(4):353–367. doi: 10.1158/2159-8290.CD-15-0894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dai M, Zhang C, Ali A, Hong X, Tian J, Lo C, Fils-Aimé N, Burgos SA, Ali S, Lebrun JJ. CDK4 regulates cancer stemness and is a novel therapeutic target for triple-negative breast cancer. Sci Rep. 2016;6:35383. doi: 10.1038/srep35383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sheppard KE, McArthur GA. The cell-cycle regulator CDK4: an emerging therapeutic target in melanoma. Clin Cancer Res. 2013;19(19):5320–5328. doi: 10.1158/1078-0432.CCR-13-0259. [DOI] [PubMed] [Google Scholar]
- 38.Duan H, Li Y, Hu P, Gao J, Ying J, Xu W, Zhao D, Wang Z, Ye J, Lizaso A, He Y, Wu H, Liang Z. Mutational profiling of poorly differentiated and anaplastic thyroid carcinoma by the use of targeted next-generation sequencing. Histopathology. 2019;75(6):890–899. doi: 10.1111/his.13942. [DOI] [PubMed] [Google Scholar]
- 39.Ibrahimpasic T, Ghossein R, Carlson DL, Nixon I, Palmer FL, Shaha AR, Patel SG, Tuttle RM, Shah JP, Ganly I. Outcomes in patients with poorly differentiated thyroid carcinoma. J Clin Endocrinol Metab. 2014;99(4):1245–1252. doi: 10.1210/jc.2013-3842. [DOI] [PubMed] [Google Scholar]
- 40.Soravia C, Sugg SL, Berk T, Mitri A, Cheng H, Gallinger S, Cohen Z, Asa SL, Bapat BV. Familial adenomatous polyposis-associated thyroid cancer: a clinical, pathological, and molecular genetics study. Am J Pathol. 1999;154(1):127–135. doi: 10.1016/S0002-9440(10)65259-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nieminen TT, Walker CJ, Olkinuora A, Genutis LK, O’Malley M, Wakely PE, LaGuardia L, Koskenvuo L, Arola J, Lepistö AH, Brock P, Yilmaz AS, Eisfeld AK, Church JM, Peltomäki P, de la Chapelle A. Thyroid carcinomas that occur in familial adenomatous polyposis patients recurrently harbor somatic variants in APC, BRAF, and KTM2D. Thyroid. 2020;30(3):380–388. doi: 10.1089/thy.2019.0561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ibrahimpasic T, Xu B, Landa I, Dogan S, Middha S, Seshan V, Deraje S, Carlson DL, Migliacci J, Knauf JA, Untch B, Berger MF, Morris L, Tuttle RM, Chan T, Fagin JA, Ghossein R, Ganly I. Genomic alterations in fatal forms of non-anaplastic thyroid cancer: Identification of MED12 and RBM10 as novel thyroid cancer genes associated with tumor virulence. Clin Cancer Res. 2017;23(19):5970–5980. doi: 10.1158/1078-0432.CCR-17-1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sugishita Y, Kammori M, Yamada O, Poon SS, Kobayashi M, Onoda N, Yamazaki K, Fukumori T, Yoshikawa K, Onose H, Ishii S, Yamada E, Yamada T. Amplification of the human epidermal growth factor receptor 2 gene in differentiated thyroid cancer correlates with telomere shortening. Int J Oncol. 2013;42(5):1589–1596. doi: 10.3892/ijo.2013.1848. [DOI] [PubMed] [Google Scholar]
- 44.Paulsson JO, Backman S, Wang N, Stenman A, Crona J, Thutkawkorapin J, Ghaderi M, Tham E, Stålberg P, Zedenius J, Juhlin CC. Whole-genome sequencing of synchronous thyroid carcinomas identifies aberrant DNA repair in thyroid cancer dedifferentiation. J Pathol. 2020;250(2):183–194. doi: 10.1002/path.5359. [DOI] [PubMed] [Google Scholar]
- 45.Wei Q, Ye Z, Zhong X, Li L, Wang C, Myers RE, Palazzo JP, Fortuna D, Yan A, Waldman SA, Chen X, Posey JA, Basu-Mallick A, Jiang BH, Hou L, Shu J, Sun Y, Xing J, Li B, Yang H. Multiregion whole-exome sequencing of matched primary and metastatic tumors revealed genomic heterogeneity and suggested polyclonal seeding in colorectal cancer metastasis. Ann Oncol. 2017;28(9):2135–2141. doi: 10.1093/annonc/mdx278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ng CKY, Bidard FC, Piscuoglio S, Geyer FC, Lim RS, de Bruijn I, Shen R, Pareja F, Berman SH, Wang L, Pierga JY, Vincent-Salomon A, Viale A, Norton L, Sigal B, Weigelt B, Cottu P, Reis-Filho JS. Genetic heterogeneity in therapy-naïve synchronous primary breast cancers and their metastases. Clin Cancer Res. 2017;23(15):4402–4415. doi: 10.1158/1078-0432.CCR-16-3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Masoodi T, Siraj AK, Siraj S, Azam S, Qadri Z, Albalawy WN, Parvathareddy SK, Al-Sobhi SS, Al-Dayel F, Alkuraya FS, Al-Kuraya KS. Whole-exome sequencing of matched primary and metastatic papillary thyroid cancer. Thyroid. 2020;30(1):42–56. doi: 10.1089/thy.2019.0052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Guan B, Wang TL, Shih IeM. ARID1A, a factor that promotes formation of SWI/SNF-mediated chromatin remodeling, is a tumor suppressor in gynecologic cancers. Cancer Res. 2011;71(21):6718–6727. doi: 10.1158/0008-5472.CAN-11-1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kunstman JW, Juhlin CC, Goh G, Brown TC, Stenman A, Healy JM, Rubinstein JC, Choi M, Kiss N, Nelson-Williams C, Mane S, Rimm DL, Prasad ML, Höög A, Zedenius J, Larsson C, Korah R, Lifton RP, Carling T. Characterization of the mutational landscape of anaplastic thyroid cancer via whole-exome sequencing. Hum Mol Genet. 2015;24(8):2318–2329. doi: 10.1093/hmg/ddu749. [DOI] [PMC free article] [PubMed] [Google Scholar]