Abstract
Timelapse TIRF microscopy of pH-sensitive GFP (pHluorin) attached to vesicle SNARE proteins is an effective method to visualize single vesicle exocytic events in cell culture. To perform unbiased, efficient identification and analysis of such events, a computer-vision based approach was developed and implemented in Matlab. The analysis pipeline consists of a cell-segmentation and exocytic-event identification algorithm. The computer-vision approach includes tools for investigating multiple parameters of single events, including the half-life of fluorescence decay and peak ΔF/F, as well as whole-cell analysis of frequency of exocytosis. These and other parameters of fusion are used in a classification approach to distinguish distinct fusion modes. Here a newly built GUI performs the analysis pipeline from start to finish. Further adaptation of Ripley’s K function in R Studio is used to distinguish between clustered, dispersed, or random occurrence of fusion events in both space and time.
SUMMARY:
Urbina and Gupton developed automated computer vision software to detect exocytic events marked by pH-sensitive fluorescent probes. Here they demonstrate use of a graphical user interface and RStudio to detect fusion events, analyze and display spatiotemporal parameters of fusion, and classify events into distinct fusion modes.
INTRODUCTION:
VAMP-pHluorin constructs or transferrin receptor (TfR)-pHuji constructs are excellent markers of exocytic events, as these pH-sensitive fluorophores are quenched within the acid vesicle lumen and fluoresce immediately upon fusion pore opening between the vesicle and plasma membrane (Miesenböck et al., 1998). Following fusion pore opening, fluorescence decays exponentially, with some heterogeneity that reveals information about the fusion event. Here, a graphical user interface (GUI) application is described that automatically detects and analyzes exocytic events. This application allows the user to automatically detect exocytic events revealed by pH sensitive markers (Urbina et al. 2018) and generate features from each event that can be used for classification purposes (Urbina et al. 2021) (Figure 1A). In addition, analysis of exocytic event clustering using Ripley’s K function is described.
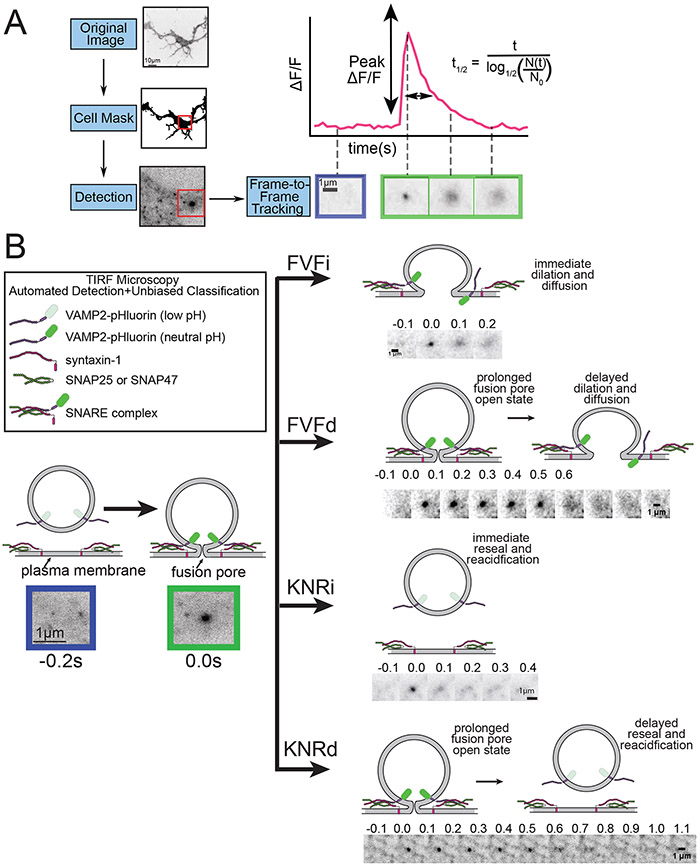
Figure 1 –
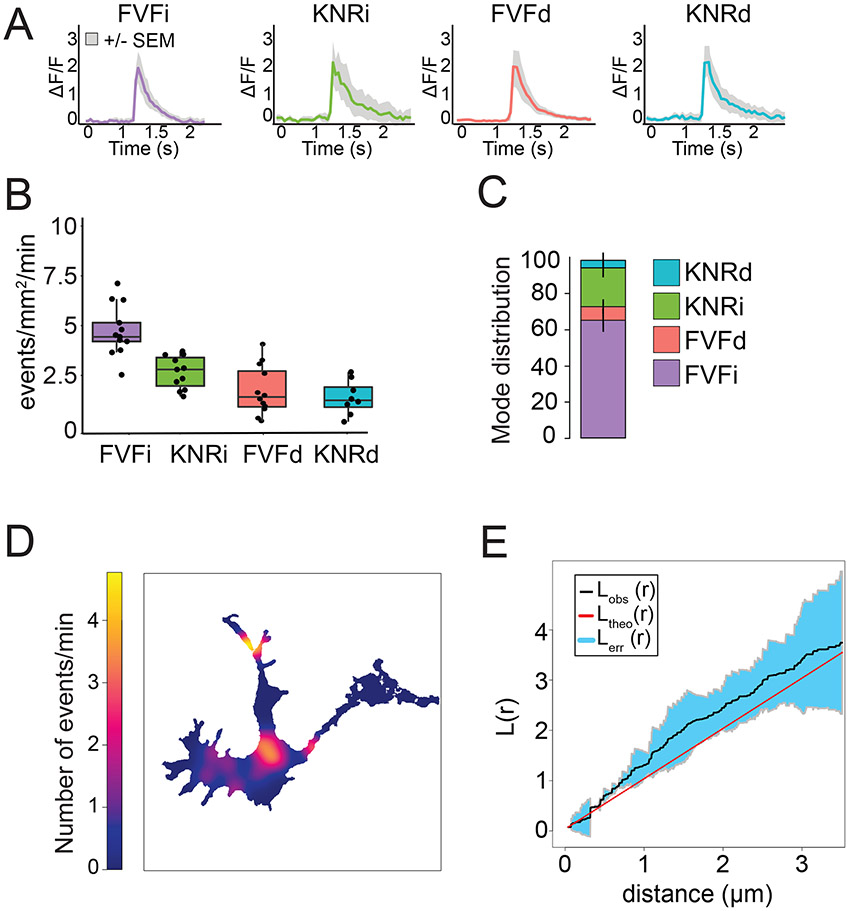
Representation of exocytic analysis and classification. A) Outline of the analysis pipeline for the GUI. Cells are segmented from the background before exocytic events are identified and tracked. Parameters such as the peak ΔF/F and t1/2 are calculated from fluorescent traces of exocytosis in a ROI around the event pre and post fusion. B) illustration of the four modes of exocytosis and example image montages. After fusion, events may proceed instantaneous to FVF or KNR (FVFi and KNRi), or a delay may be present before onset of fusion fate to FVF or KNR (FVFd and KNRd).
The automated classification of exocytic events into different exocytic modes was recently reported (Urbina et al., 2021). Two modes of exocytosis, full-vesicle fusion (FVF) and kiss-and-run fusion (KNR) exocytosis have previously been described (Alabi and Tsien, 2013; Albillos et al., 1997; He and Wu, 2007; Elhamdani et al., 2006). During FVF, the fusion pore dilates and the vesicle is incorporated into the plasma membrane. During KNR, the fusion pore transiently opens and then reseals (Alabi and Tsien, 2013; Albillos et al., 1997; Bowser and Khakh, 2007; Holroyd et al., 2002; Wang et al., 2003). Four modes of exocytosis were identified in developing neurons, two related to FVF and two related to KNR. This work demonstrates that both FVF and KNR can be further subdivided into fusion events that proceed immediate to fluorescence decay (FVFi and KNRi) after fusion pore opening, or exocytic events that exhibit a delay after fusion pore opening before fluorescence decay begins (FVFd and KNRd)(Figure 1B). The classifier identifies the mode of exocytosis for each fusion event. Here this analysis has been incorporated into a GUI that can be installed in MATLAB in Windows and Mac based operating systems. All analysis files can be found at https://github.com/GuptonLab.
PROTOCOL
Choose datasets and directory
1. To select datasets for analysis, navigate to that folder using the “Find Datasets” button (Figure 2A, red box 1). Datafiles will automatically populate the Data Files as a list. There can be more than one dataset in the folder.
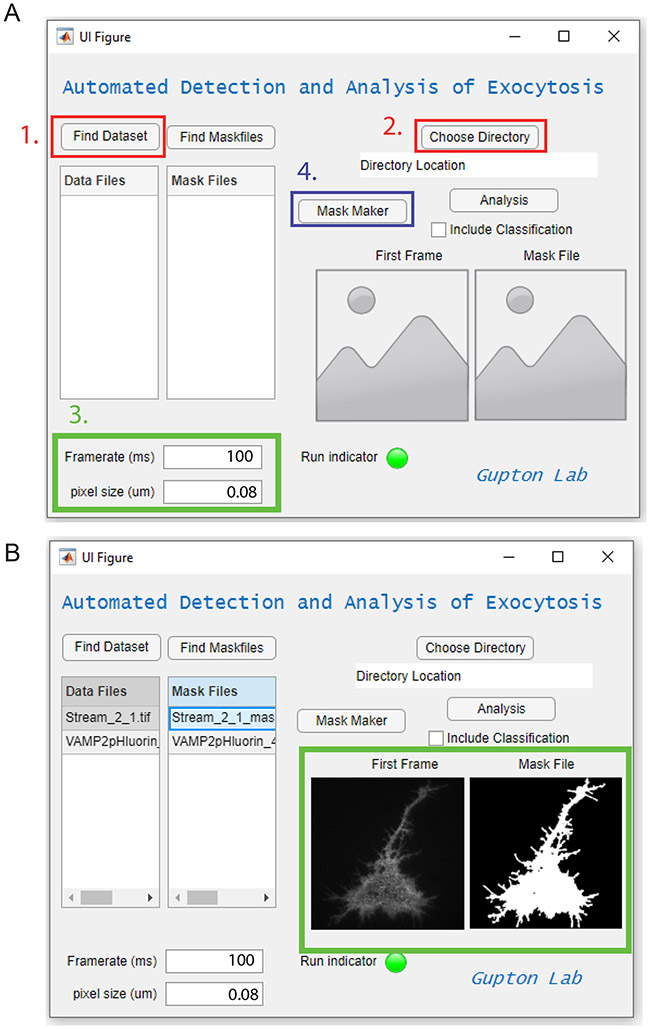
Figure 2 –
Step by step example analysis. A) First, datasets are chosen (1., red box) and a directory is chosen to place analysis files (2., red box). Next, the framerate and pixel size are specified (3., green box). Here, 0.08μm pixel size and 100ms framerate were used. The MaskMaker function button is then pushed (4., blue box). A folder titled “MaskFiles” is automatically created in the chosen directory containing a mask file for each image file in the dataset. B) When datasets are loaded, selecting a data file and/or mask file will display the first frame of the images for ease of comparison (Green box). Mask files may not be completely correct; this mask file can be corrected for errors, or a new mask file may be made manually
1.1 Click the “Choose Directory” button and select the directory where analyzed files will be deposited (Figure 2A, red box 2). A set of folders and finished analysis files as well as intermediate temporary images will be created in this directory when the Analysis button is pushed. Errors will be produced if a directory is not chosen.
Set the pixel size and framerate
2. Input the appropriate frame rate and pixel size of the images the appropriate “Framerate” or “pixel size” box (Figure 2A, green box). If no values are provided (they are set to the default “0”), then the program will search the metadata associated with the file for the framerate and pixel size. If these values cannot be found, the program will default to per-pixel for measurement and perframe for time points.
Choose or make masks
3. Use the “Mask Maker” button to automatically create cell masks for the data in the file data setlist (Figure 2A, blue box). Upon using the “Mask Maker” button, a new folder in the chosen directory will be created called MaskFiles. A mask for each file in the Data Files list will be created from the first 10 frames of the image file (or from all of the frames if less than 10 are in the image file) and deposited in the MaskFiles folder using the proper naming scheme (described below). The mask files will automatically populate the Mask Files list and the user may proceed directly to the analysis.
NOTE: Always visually check mask files and confirm they capture the entire region of interest. The first frame of the data file and the mask file are displayed on the UI when selected. (Figure 2B). The Mask Maker may produce errors in the case of low signal-to-noise, so validating that mask files are appropriate is critical for quality control.
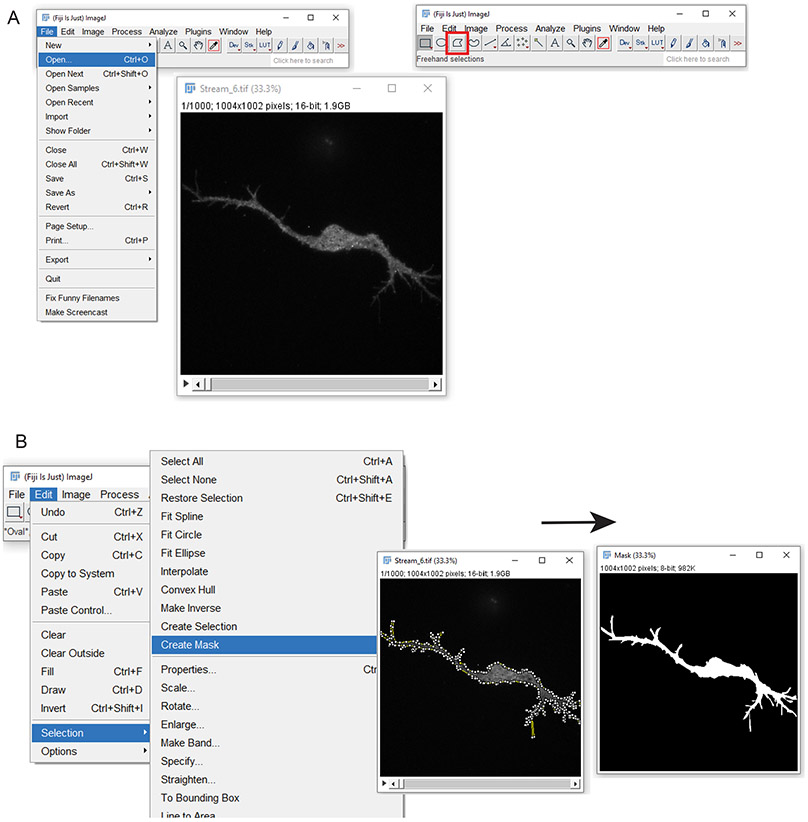
3.1 As an alternative to using Mask Maker if the signal to noise of images is insufficient, create masks manually in ImageJ. First, open the raw data file in ImageJ (Figure 3A). Next, click the “polygon selection” button and click to draw a mask around the cell. Once finished, double-click on the last point to complete the polygon. Once finished, navigate to Edit -> Selection -> Create Mask (Figure 3B). A new mask will be created based on the polygon drawing. Save masks in a designated MaskFiles folder in the chosen directory. The mask file naming scheme must match the corresponding individual data files followed by “_mask_file”. For example, if a data file is named “VAMP2_488_WT_1.tif”, the corresponding mask file must be named “VAMP2_488_WT_1_mask_file.tif”. Use the “Find Maskfiles” button to navigate to the chosen folder of deposited mask files. The masks will populate the Mask Files as a list.
Figure 3 -.
Creating a mask file manually in ImageJ. A) First, open the file for making a mask. The “Polygon Selections” button is outlined in red. By clicking around the edge of the cell, a polygon outline is created. B) How to create a mask from the polygon outline. By selecting Edit, Selection, Create Mask, a black and white mask will be created from the polygon (right image).
Analysis and Feature Extraction
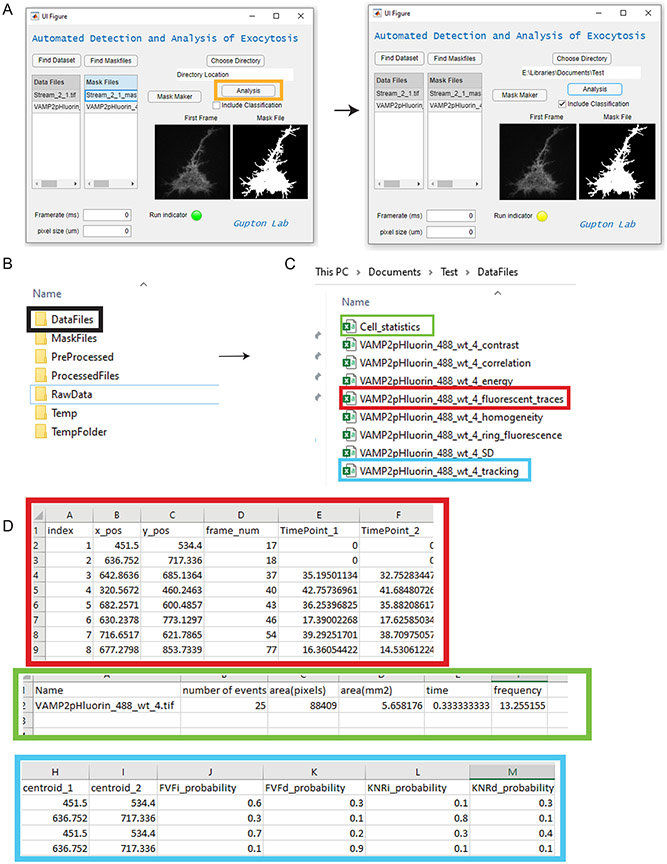
4. After the directory is chosen and the Data Files list and Mask files list are populated, click the “Analysis” button (Figure 4A). The Analysis button will perform a series of automated tasks to analyze the data. It will create individual folders in the chosen directory to deposit analyzed data. While running, the “run indicator” will change from green to yellow (Figure 4A, red box). After the analysis is finished, this will change back to green. Find a DataFiles folder (Figure 4B) with complete set of analysis files (as well as feature extraction files, to be used in classification later) named according to each datafile (Figure 4C):
Figure 4-.
Analysis of exocytic events. A) Demonstration of the analysis button (orange box) and a running analysis. Notice the run indicator turns yellow while an analysis is being performed. B) Example analysis files that are created in the chosen directory when the analysis is complete. DataFiles contain all analysis files of the exocytic event. C) Analysis files generated in the DataFiles Folder. Color boxes represent the open files in subsequent images. D) Three open files, “X_fluorescent_traces.csv” (red) and “X_Cell_statistics”(green), and “X_tracking”(blue). Fluorescent traces contain the x,y position and frame number of each event, as well as fluorescent intensity at each ROI Cell_statistics contains summary information of whole-cell exocytic statistics such as frequency of exocytosis. X_tracking contains position and time information for each exocytic event as well as the probability of each class of exocytosis for each event, represented as a number between 0-1 (>0.5 indicates an event belongs to a particular class).
Classification of exocytic events
5. To perform simultaneous classification of exocytic events with automated detection, check the “Classification” checkbox before clicking on the “Analysis” button. For each exocytic event, a probability score between 0-1 is assigned for each class. An exocytic event is considered classified as one of the four classes if the probability score for that class is > 0.5.
Spatiotemporal Analysis of Exocytosis using Ripley’s K values
6. A separate “soma” and “neurite” mask must first be created. First, segment the soma from the neurite. There is no unbiased method for segmenting the soma from the neurites, so the user should be blinded to condition experiment and use best judgement; an ellipsoid with no obvious neurite extensions is suggested.
6.1 Open ImageJ/Fiji.
6.2 Drag-and-drop the mask file or use file->open and then select the mask file.
6.3 Use the color-picker tool and click on any of the black pixels in the background of the mask to set the color to black.
6.4 Use the polygon-selection or freehand to draw around the soma, separating it from the neurites.
6.5 Click edit -> selection -> make mask. A new image will open with the circled-soma segmented from the rest of the image. Save this soma mask file.
6.5 Without moving the drawn region, click on the header of the original mask file.
6.6 Click Edit -> Fill to fill-in the circled soma so that only the neurites remain the mask.
6.7 Save this as the neurites mask file.
6.8 Open “neurite_2D_network” in Matlab.
6.9 Change the “neurite_mask_name.m” to the name of the neurite mask.
6.10 Click “run” to skeletonize the neurite mask file.
6.11 Open “CSV_mask_creator.m” in Matlab.
6.12 Change the “mask_file_name” to the desired neurite or soma mask for Ripley’s K analysis. Click run.
6.13 Repeat 1.12 for every mask file.
RStudio setup
7. Open Rstudio.
7.1 Install the package “spatstat” in RStudio by going to Tools -> Install Packages and typing in “spatstat” followed by clicking Install.
Note: This only needs to be performed once per Rstudio Installation.
7.2 Run the library spatstat at the beginning of each session.
7.3 Read in the .csv of the mask files for each of the neurons to be analyzed.
7.4 Read in a file with the x,y,t positions of the exocytic events, as well as the neurite-specific file of the x,y positions for the 2D network.
7.5 Convert the data into a point-process format:
soma_data = ppp(exocytic_x_positions,exocytic_y_positions, mask = mask.csv).
Neurite_data = lpp(linear_x_positions,linear_y_positions, mask = mask.csv)
time_data = lpp(time_position, mask = mask.csv)
7.6 Use plot(density(soma_data,0.4)) to create a heat map to visualize clustering.
Note: The number “0.4” here represents how smoothed-out the density function should be. It can be changed to fit user data in a meaningful way, but if comparisons are to be performed between different heatmaps, the number must be the same between them.
7.7 Export or save image from Rstudio. If a heatmap requires further editing, choose an appropriate file type (SVG or EPS).
Ripley’s Analysis
8.1 First, run the envelope function for each cell. This function simulated complete spatial randomness (CSR) to test the Ripley’s K value of the exocytic event point pattern against.
Data_envelope_1 = envelope(soma_data_1, Kest, nsim = 19, savefuns = TRUE)
Data_envelope_2 = envelope(soma_data_2, Kest, nsim = 19, savefuns = TRUE)
…
8.2 Next, pool these envelopes together and create one estimate of CSR for the group:
Pool_csr = pool(Data_envelope_1, Data_envelop_2,…)
Next, run the Ripley’s K function for all data points.
Data_ripleys_k_1 = Kest(soma_data_1, ratio = TRUE)
Data_ripleys_k_2 = Kest(soma_data_2, ratio = TRUE)
…
8.3 Once complete, pool the Ripley’s K values together and bootstrap their confidence intervals:
Data_pool = pool(Data_ripleys_k_1, Data_ripleys_k_2,…)
8.4 Bootstrap:
Final_Ripleys_K = varblock(fun = Kest, Data_pool)
8.5 Plot
8.6 If a hard-statistical difference is required, Studentised Permutation Test is included in the spatstat package to test for a difference between groups of point patterns:
Test_difference = studpermu.test(all_points_to_test, exocytic_events ~ group, nperm = np).
REPRESENTATIVE RESULTS:
Here the GUI (Figure 2A) was utilized to analyze exocytic events from three VAMP2-pHluorin expressing neurons at 3 DIV using TIRF (total internal reflection fluorescence) microscopy. E15.5 cortical neurons were isolated, followed by transfection with VAMP2-pHluorin and plating using the protocols as outlined in Winkle et al., 2016 and Viesselmann et al., 2010. The methodology of imaging parameters is as outlined in Urbina et al, 2018. Briefly, TIRF microscopy was used to image the basal plasma membrane of neurons every 100 ms for 2 min. Figure 2-4 shows a step-by-step guide to analysis of exocytic events. The folder where the neuron images are located is selected, and a directory to deposit the final analysis datafiles is chosen (Figure 2A). Using the MaskMaker function, a mask is generated for the neurons, which is inspected in the GUI (Figure 2B). In this instance, the cell mask is of good quality and the analysis can proceed. Should a mask be insufficient, a mask can be created in ImageJ (Figure 3). After using the MaskMaker function or creating a mask in ImageJ and selecting the directory where the mask files are located, the analysis is performed (Figure 4A). Results are generated in the DataFiles folder when the analysis is finished (the yellow indicator changes back to green)(Figure 4B).
Datafiles are automatically generated and named according to the raw data files provided. Assuming the datafile is named X:
X_tracking: This file includes x,y position and frame number of each event as well as bounding boxes which can be used to draw boxes around each event. Age indicates the number of frames past the initial detection where an event is a distinct gaussian puncta. If classification is checked, the classification results will appear in this file.
X_fluorescent traces: This file includes x,y position and frame number of each event. In addition, it includes fluorescent intensity measures in a region of interest around each event 2 seconds before and 10 seconds following the peak ΔF/F for each event (indicated by the Timepoint columns).
X_cell_statistics: This file includes the cell area,total image time, and automatically calculated frequency of exocytic events for each cell (in events/mm2/minute).
Feature extraction files include:
X_contrast: Contrast. A measure of the intensity contrast between a pixel and its neighbor over the whole image.
X_correlation: Correlation. A measure of how correlated a pixel is to its neighbor over the whole image.
X_energy Total energy. Defined as the squared sum of the pixel intensity.
X_homogeneity measures the closeness of the distribution of elements in the ROI to the ROI diagonal.
X_ring_fluorescence: the average fluorescence of border pixels.
X_SD: Standard Deviation. This is defined as the standard deviation of the ROI.
Examples of average fluorescence traces +/− SEM from each exocytic class were plotted from X_fluorescent traces file (Figure 5A)._Using the Cell_statistics file, the frequency of exocytosis for each class was plotted for each neuron (Figure 5B). With the Classification checkbox clicked, the program assigns each exocytic event to a class, plotted in Figure 5C. Following classification, the Ripley’s K analysis code to determined if exocytic events are random, clustered, or dispersed in space and time. Density heatmaps of the localization of exocytic events (Figure 5D) are generated. This reveals expected clustered “hotspots” in distinct regions of the neuron. Next, Ripley’s K analysis was performed for the soma, neurite, and clustering over time (Figure 5E). The Ripley’s K value and SEM (black line and blue shaded region, respectively) rise about the line of complete spatial randomness (red dotted line), suggesting statistically significant clustering.
Figure 5 –
Representative results. A) Average fluorescence traces +/− SEM from each exocytic class B) Frequency of events plotted for three murine cortical neurons for each exocytic class. These data values were plotted from “X_Cell_statistics”, using the classes assigned in “X_tracking”. C) Distribution of the classes of exocytosis for the same three cells used in A). Here, the ratio of each mode is plotted. D) Density plot of where exocytic events are occurring as generated in the Ripley’s K Analysis portion of the protocol. This can be interpreted as a “heat map” of the spatial likelihood of where events are occurring. E) Ripley’s K analysis of three cells used for A) and B). The red line indicates what value completely spatially random distribution of exocytic events would be. The black line indicates the aggregate Ripley’s K value for the three cells in this example, and the blue shaded region represents the confidence interval. Here, the shaded region notably falls outside of the line of complete spatial randomness between ~0.25-1 μm, suggesting exocytic events are clustered at those distances.
DISCUSSION:
Critical steps within the protocol
Considerations that should be regarded when using the exocytic detection and analysis software are described here. The program only accepts lossless compression .tif files as input. The .tif image files may be 8 bit, 16 bit, or 32 bit grayscale (single channel) images. Other image formats must be converted into one of these types before input. For reference, examples used here are 16 bit grayscale images.
Inherent in the automated detection process, the timelapse image sets are processed for automated background subtraction and photobleaching correction. For background subtraction, the pixels outside of the masked region of the mask file are averaged over the timelapse of the whole image, and the average value is subtracted from the image set. For photobleaching correction, a mono-exponential decay fit is applied to the average fluorescence of pixels in the mask over the course of the video, with the corrected intensity adjusted as follows:
Corrected intensity = (Intensity at time t) ÷ exp-k×t where k = decay constant
Therefore, no pre-processing is necessary prior to input. These processes, however, critically rely on the image mask to effectively separate the cell from the background, and thus a proper cell mask is necessary for good results.
The automated cell mask creator requires a uniform signal with sufficient signal-to-noise ratio (ideally, at least 2x the standard deviation of the average background signal) to perform well. Empirically, the CAAX box tagged with a fluorescent protein, which inserts into the plasma membrane upon prenylation works well. There is no requirement that the signal be maintained throughout the timelapse imaging of exocytosis, as a suitable mask can be created from a high signal in the first 10 frames of the sequence. However, if the cell morphology changes significantly during the imaging paradigm, care should be taken.
When using the automated detection software, include the framerate and pixel size for accurate temporal and spatial outputs. If no framerate or pixel size are declared, the output will be per-pixel and per-frame. As a rule-of-thumb, vesicle diameters in developing neurons are on the scale of ~100 nm (Plooster et al., 2017), and thus the automated detection of events may work for vesicles similar in size. As it stands, there is no hard limitation on the size of vesicles that can be detected (by pixel area), as the automated detection relies on gaussian-shaped intensity over a large range of gaussian widths. Fusion of vesicles smaller than the width of a pixel can be detected accurately if the intensity of the event meets the signal-to-noise criteria of 2x the standard deviation of the average background signal as the fluorescence diffraction expands over multiple pixels.
This program was developed for detecting exocytic events in developing neurons. However, the software has been exploited to successfully detect exocytic events in other cell lines (Urbina et al., 2018), indicating the detection algorithm is robust. Although we have used the software for detecting exocytic events in non-neuronal cell types, differences in exocytic event mode between cell types (Urbina, FL., Gupton, S. 2020) indicate that classification algorithm may not be suitable for other cell types. Exocytic events in developing neurons were originally classified using three different methods: Hierarchical clustering, Dynamic Time Warping, and PCA, revealing four different classes (Urbina et al, 2021). Here all three classifiers are employed in the GUI to categorize exocytic events into one of these established four classes. For each exocytic event, a probability score between 0-1 is assigned for each of the four possible classes. Any class with a probability score > 0.5 is considered to be of that class. This classification has only been used in developing neurons to date. Whether these classes exist in other cell types or at later developmental time points in neurons is not known. A large number of events with probability scores of <0.5 for any of the classes suggest the classification procedure may not be appropriate for the exocytic events currently being evaluated. If low probability scores are assigned to exocytic events of different cell types, this would suggest that de novo classification is needed as new or alternate modes of exocytosis may exist. The same classification methods used here would need to be applied to the automatically detected exocytic events.
To explore the spatial and temporal clustering of exocytic events, Ripley’s K analysis (Ripley et. Al,. 1976) is exploited. Analyzing clustering for neurons involves three separate analyses: One for the soma, one for the neurites, and one for time. The reason for splitting the soma and neurites is to account for the extreme morphology of neurites, which are often thin enough that they can be treated as a 2D network, and apply a 2D variant of Ripley’s K analysis. For time, a 1D Ripley’s K analysis is implemented for temporal clustering. Ripley’s K analysis is a robust method for detecting clustering of point processes and colocalization. The graph of Ripley’s K can be interpreted as such: the Ripley’s K value+confidence interval has a 5% chance to fall outside of the line of complete spatial randomness at any point, analogous to a p value of 0.05. Where the value falls outside of the line of complete spatial randomness, exocytic events are clustered (above CSR) or are separated at regular intervals (below CSR) at those distances (x-axis).
Limitations and modifications of the technique
The creation of the mask file relies on a high initial signal-to-noise of the cell. pH sensitive markers may not always perform well in illuminating a cell, depending on what protein the probe is attached to. Another option for making mask files if the signal to noise of the exocytic marker insufficiently highlights the cell border is to employ a second fluorescent channel/image for mask creation. If using a different fluorescence marker (tagRFP-CAAX, for example) to make masks, when choosing a dataset, first navigate to the folder with the images to make masks from (for example, a folder containing the tagRFP-CAAX images). Use the Mask Maker button here. Importantly, remember to rename the mask files to match the exocytic data file to be analyzed with the above naming scheme (following the example above, the mask files named “CAAX_1_mask_file.tif” will need to be renamed to “VAMP2_488_WT_1_mask_file.tif” to match the VAMP2 image set to be analyzed). Once mask files are appropriately named, return to the “choose dataset” button again to navigate to exocytic dataset files to analyze.
The detection of exocytosis is remarkably sensitive, and exocytic events were accurately detected at signal-to-noise ratios as low as a ΔF/F of 0.01. The sensitivity of the detection depends partially on the variance of the background fluorescence, and events less than 4 standard deviations above the background signal will not be detected.
The detection of exocytic events relies on their transient nature. As a feature of the analysis of transient events, our detection code explores a 20-second window around the exocytic event. In some cell types, kiss-and-stay exocytosis may “stay” for a much longer temporal window than in developing neurons, and the automated detection may not robustly capture these less transient events. This feature is an easily modifiable variable for those comfortable with MATLAB. Similarly to the limitation of the 20 second ROI, exocytic events that appear but do not disappear before the last time frame of the video may not be counted as true exocytosis, which may influence the frequency, which is calculated based on the entire length of the time series.
The mask maker included is automated by design, and performs well on segmenting complex shapes from the background; however, the fully-automated design limits the range of signal-to-noise and signal type that can be used to generate a mask automatically. If the user cannot include a fluorescent cell marker that is uniform and of a significant signal-to-noise ratio, the cell mask will need to be drawn manually.
The significance with respect to existing methods
User based analysis of exocytosis is a time-consuming process and subject to personal bias, as events are not always clearly separated from other fluorescence. The use of an automated analysis program to correctly identify and analyze exocytic events in an unbiased manner increases analysis efficiency and improves reproducibility and rigor.
Any future applications of the technique
Not only does this exocytic event detection work for accurately capturing pH sensitive fluorescence in developing neurons but other cell types as well (Urbina et al, 2018). Whether classification works for other cell types will require determination. Future applications of this technique may be useful at synapses, in non-neuronal cell types, or with novel markers of exocytic vesicle docking and fusion, such as recently described pHmScarlet (Liu et al. 2021 ).
ACKNOWLEDGMENTS:
We thank Dustin Revell and Reginald Edwards for testing code and the GUI. Funding was provided by the National Institutes of Health supported this research: including R01NS112326 (SLG), R35GM135160 (SLG), and F31NS103586 (FLU).
Footnotes
A complete version of this article that includes the video component is available at http://dx.doi.org/10.3791/62400.
DISCLOSURES:
The authors declare nothing to disclose
REFERENCES:
- Alabi AA, and Tsien RW. 2013. Perspectives on Kiss-and-Run: Role in Exocytosis, Endocytosis, and Neurotransmission. Annu. Rev. Physiol 75:393–422. doi: 10.1146/annurev-physiol-020911-153305. [DOI] [PubMed] [Google Scholar]
- Albillos A, Dernick G, Horstmann H, Almers W, de Toledo GA, and Lindau M. 1997. The exocytotic event in chromaffin cells revealed by patch amperometry. Nature. 389:509–512. doi: 10.1038/39081. [DOI] [PubMed] [Google Scholar]
- Bowser DN, and Khakh BS. 2007. Two forms of single-vesicle astrocyte exocytosis imaged with total internal reflection fluorescence microscopy. Proc. Natl. Acad. Sci 104:4212–4217. doi: 10.1073/pnas.0607625104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elhamdani A, Azizi F, and Artalejo CR. 2006. Double patch clamp reveals that transient fusion (kiss-and-run) is a major mechanism of secretion in calf adrenal chromaffin cells: High calcium shifts the mechanism from kiss-and-run to complete fusion. J. Neurosci 26:3030–3036. doi: 10.1523/JNEUROSCI.5275-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holroyd P, Lang T, Wenzel D, De Camilli P, and Jahn R. 2002. Imaging direct, dynamin-dependent recapture of fusing secretory granules on plasma membrane lawns from PC12 cells. Proc. Natl. Acad. Sci 99:16806–16811. doi: 10.1073/pnas.222677399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu A, Huang X, He W, Xue F, Yang Y, Liu J, Chen L, Yuan L, and Xu P (2021). pHmScarlet is a pH-sensitive red fluorescent protein to monitor exocytosis docking and fusion steps. Nat. Commun 12, 1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miesenböck G, De Angelis DA, and Rothman JE (1998). Visualizing secretion and synaptic transmission with pH-sensitive green fluorescent proteins. Nature 394, 192–195. [DOI] [PubMed] [Google Scholar]
- Plooster M, Menon S, Winkle CC, Urbina FL, Monkiewicz C, Phend KD, Weinberg RJ, and Gupton SL (2017). TRIM9-dependent ubiquitination of DCC constrains kinase signaling, exocytosis, and axon branching. Mol. Biol. Cell 28, 2374–2385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripley BD (1976). The second-order analysis of stationary point processes. Journal of Applied Probability 13, 255–266. [Google Scholar]
- Urbina FL, Gomez SM, and Gupton SL (2018). Spatiotemporal organization of exocytosis emerges during neuronal shape change. J. Cell Biol 217, 1113–1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbina FL, Urbina FL, Gupton SL. (2020). SNARE-Mediated Exocytosis in Neuronal Development. Front Mol Neurosci. 13:133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menon S, Goldfarb D, Edwards R, Ben Major M, Brennwald P, S.L. Gupton Correspondence, and Gupton SL. 2021. TRIM67 regulates exocytic mode and neuronal morphogenesis via SNAP47. Cell Rep. 34. doi: 10.1016/j.celrep.2021.108743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C-T, Lu J-C, Bai J, Chang PY, Martin TFJJ, Chapman ER, and Jackson MB. 2003. Different domains of synaptotagmin control the choice between kiss-and-run and full fusion. Nature. 424:943–947. doi: 10.1038/nature01857. [DOI] [PubMed] [Google Scholar]
- Viesselmann C, Ballweg J, Lumbard D, and Dent EW. 2010. Nucleofection and primary culture of embryonic mouse hippocampal and cortical neurons. J. Vis. Exp doi: 10.3791/2373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkle CC, Hanlin CC, and Gupton SL (2016). Utilizing Combined Methodologies to Define the Role of Plasma Membrane Delivery During Axon Branching and Neuronal Morphogenesis. J. Vis. Exp [DOI] [PMC free article] [PubMed] [Google Scholar]