ABSTRACT
Enterococcus faecium is a significant multidrug-resistant pathogen. Bacteriophage cocktails are being proposed to complement antibiotic therapy. After a screen of 8 E. faecium strains against 4 phages, 2 phages (113 and 9184) with the broadest host ranges were chosen for further experiments. Transmission electron microscopy, whole-genome sequencing, comparative genome analyses, and time-kill analyses were performed. Daptomycin (DAP) plus the phage cocktail (113 [myophage] and 9184 [siphopage]) showed bactericidal activity in most regimens, while DAP addition prevented phage 9184 resistance against daptomycin-nonsusceptible E. faecium.
KEYWORDS: Enterococcus faecium, bacteriophage cocktails, bacteriophage-antibiotic combinations, bacteriophages, daptomycin
INTRODUCTION
The World Health Organization has warned of a “postantibiotic era,” in which 10 million persons could die annually from antimicrobial-resistant infections (1, 2). During the past 5 decades, enterococci have surfaced as significant health care-associated pathogens. Among the enterococcal species that are most clinically relevant in the United States, Enterococcus faecium has the highest prevalence of resistance to first-line antibiotics, is more difficult to treat, and has been classified as one of the “ESKAPE” pathogens due to its intrinsic and/or acquired resistance to antimicrobials and its ability to “escape” the action of antibiotics (3, 4). Obligately lytic bacteriophages (phages), which are viruses that specifically target, infect, and kill bacterial cells, have been proposed as an alternative to or in combination with antibiotics as a rescue approach in cases of recalcitrant infections caused by multidrug-resistant (MDR) organisms (5). Phage-antibiotic combinations have shown promise in vitro against MDR pathogens. However, limited evaluations have been conducted against drug-resistant E. faecium (6). Therefore, further preclinical assessments of phage-antibiotic combinations against drug-resistant E. faecium could help assess the potential of this strategy and identify biological factors that might drive optimal phage and antibiotic combinations.
Both monophage therapy (single phage) and “phage cocktails” (multiple phages) have been proposed, with cocktails potentially offering a broader spectrum of activity that might better enable empirical treatment. Both approaches may have the ability to reduce the emergence of resistant mutants, by either combining phages with complementary infection capabilities or sequentially using phages that force evolutionary trade-offs in which the emergence of phage resistance leads to reduced pathogen fitness or reacquired antibiotic susceptibility (7–10). Our group has previously demonstrated in vitro synergistic effects and alterations of resistance development of monophage in combination with several antibiotics against E. faecium strains harboring liaFSR (encoding a three-component regulatory system involved in the cellular membrane stress response) mutations with various susceptibilities to antibiotics and phage (11). However, given the potential advantages of phage cocktails, the objective of this study was to evaluate and compare the abilities of daptomycin (DAP) in addition to monophage and phage cocktails to improve bacterial eradication and prevent resistance development in DAP-nonsusceptible (DNS) E. faecium.
Enterococcus faecium phage 113 (ATCC 19950-B1) and propagating organism E. faecium (ATCC 19950) were purchased commercially from the American Type Culture Collection (ATCC) (Manassas, VA). The recently characterized and previously sequenced E. faecium phages 9181, 9183, and 9184, isolated from a wastewater treatment facility located near Denver, CO, and the propagating organisms E. faecium Com12 (for phage 9181) (isolated from feces of a healthy human volunteer) and 1,141,733 (for phages 9183 and 9184) (clinical isolate) were provided by the Duerkop laboratory (10, 12). DAP was purchased commercially from Sigma Chemical Company (St. Louis, MO). Mueller-Hinton broth II (MHB) (Difco, Detroit, MI) with 50 mg/L calcium and 12.5 mg/L magnesium was used for susceptibility testing and time-kill analyses (TKAs). All MIC determinations were performed according to Clinical and Laboratory Standards Institute (CLSI) guidelines (13). The susceptibility of the above-mentioned four phages against eight randomly selected E. faecium strains was evaluated as previously described (14, 15). Following phage quantification, high, medium, and low phage susceptibilities were defined as phage counts of >107, between 103 and 107, and <103 PFU/mL, respectively. Phage nonsusceptibility was defined as no visual detection of individual plaques and/or no bacterial lawn clearance (11). The two phages with the broadest host ranges (phages 113 and 9184) were chosen for further experiments. Overall, phages 113 and 9184 exhibited cumulative susceptibility against 56.2% of the evaluated strains. Furthermore, a complementary host range was observed as these two phages demonstrated susceptibility against 87.5% of evaluated isolates, promoting further evaluation of their use as a cocktail.
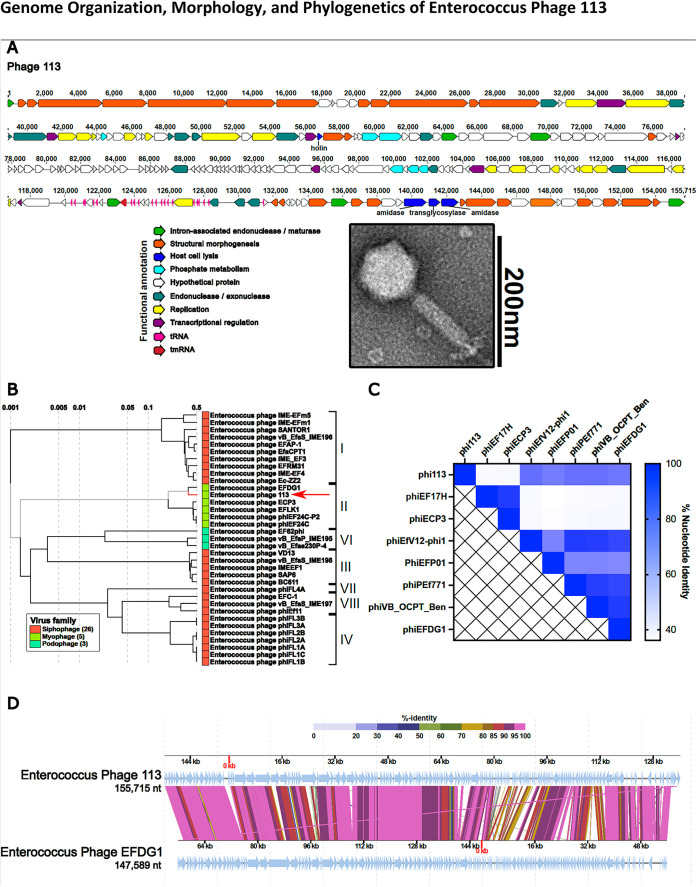
The transmission electron microscopy (TEM) methodology can be found in Text S1A in the supplemental material. TEM on phage 113 demonstrated an icosahedral head and a contractile tail consistent with a myophage morphotype (Fig. 1A), while phage 9184 has an icosahedral head and a noncontractile tail consistent with a siphophage morphotype (previously reported [10, 16]). Whole-genome sequence analysis was performed as previously described (10, 17). DNA sequence analysis demonstrated that phage 113 is 155,715 bp long and is predicted to harbor 192 open reading frames (ORFs). The genome was defined as partial on the basis of partial ORFs on the 5′ and 3′ termini of the genome. The genome is modular in organization except for host cell lysis genes (Fig. 1A). Functional classifications, consisting of intron-associated genes, replication, transcriptional regulation, structural morphogenesis, host cell lysis, phosphate metabolism, and endonuclease or exonuclease, could be predicted for approximately 45% of the phage 113 ORFs. The genome is predicted to encode 21 tRNAs or transfer-messenger RNAs (tmRNAs). Initial genome analysis using the functional annotation workflow, v2021.01, on Phage Galaxy did not detect genes annotated as encoding toxins, antibiotic resistance determinants, or integrases (17). It is generally agreed that phages should lack such genes if they might be considered for therapeutic use (18). DNA sequence analysis of phage 9184 has been previously reported (10).
FIG 1.
(A) Whole-genome sequencing reveals a modular organization of phage 113 intermixed with group I introns (bright green). Open reading frames for each phage were determined by the Texas A&M Center for Phage Therapy structural analysis workflow, version 2021.02 (17). Colored open reading frames correspond to functional predictions. The bottom right portion demonstrates the myophage morphotype of phage 113 by TEM. (B) Comparative genome analysis demonstrates that Enterococcus phage 113 clusters with Enterococcus phages of the orthocluster II myophages and most closely resembles phage EFDG1 (19, 20). A proteomic tree was constructed by submitting the Enterococcus phage 113 nucleotide sequence to ViPTree (27). (C) Heat map demonstrating phage 113 nucleotide identity compared to other Enterococcus myophages. The nucleotide identity matrix was determined with progressiveMauve using the Texas A&M Center for Phage Therapy Phage Comparative Genomics workflow, version 2021.01 (17, 28). (D) Enterococcus phages 113 and EFDG1 have high protein sequence homology and similar genome organizations. Colored lines connecting genomes indicate percent protein identity along the length of each genome. The protein-coding sequence alignment was performed using ViPTree (27). nt, nucleotides.
The methodology for the comparative genome analysis can be found in Text S1B. Proteome analysis of the E. faecium phage 113 genome using ViPTree revealed clustering with other enterococcal myophages (Fig. 1B). Whole-genome alignments revealed approximately 40 to 80% nucleotide identity between phage 113 and other enterococcal myophages (Fig. 1C). These results suggest that phage 113 is a myophage that belongs to enterococcal phage orthocluster II, the only enterococcal phage orthocluster containing myophages (19). Genomic alignment of phage 113 with phage EFDG1, the nearest neighbor from the proteomic tree (Fig. 1B), demonstrates high protein homology and a similar arrangement of genes (Fig. 1D) (20). Despite this altered genome arrangement, the gene order remains consistent between phages 113 and EFDG1. Accession numbers can be found in Text S1C.
Time-kill analyses were performed as previously described, with duplicate samples obtained at each time point and DAP tested at subinhibitory concentrations (11). Subinhibitory DAP concentrations (0.25× MIC) were used to be able to measure potential synergistic effects with phages. Given that phage-induced bacterial killing is strain specific, phage cocktail dose optimization was performed to evaluate the theoretical multiplicity of infection (tMOI) that produced optimal observations of synergistic effects (21). Any phage-containing samples were centrifuged at a relative centrifugal force (RCF) of 15,322 for 2 min, with the supernatant removed and replaced with normal saline to reduce the concentration of unadsorbed phages that might alter CFU counts during plating. Synergy was defined as ≥2-log10(CFU/mL) killing compared to the most effective agent (or double-combination regimen) alone at 24 h. Bactericidal activity was defined as a ≥3-log10(CFU/mL) reduction from the baseline. The emergence of DAP and phage resistance (against both phages) was evaluated as previously described by using the 24-h TKA liquid sample (9, 11, 14, 15). Phage counts were assessed as previously described (to determine if the presence or absence of antibiotics had an impact on phage growth) (11, 14, 15).
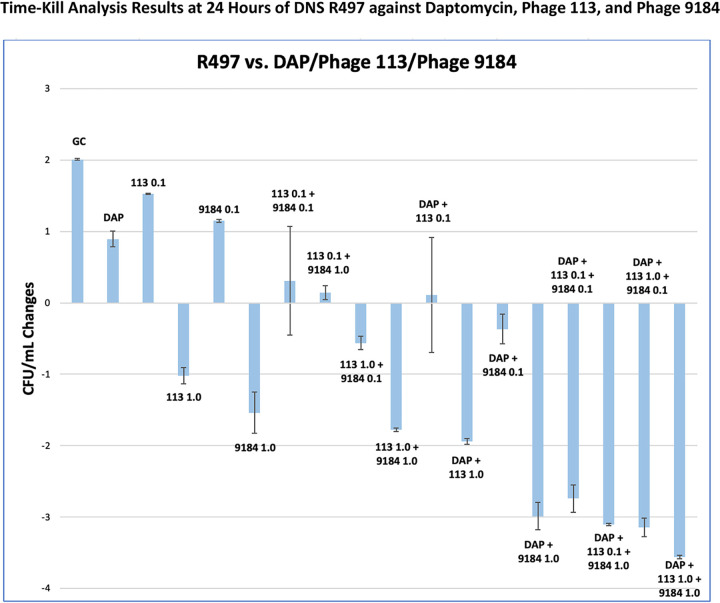
R497 is a DNS isolate harboring mutations in liaFSR (DAP MIC of 16 mg/L) that exhibited high susceptibility to both phage 113 and phage 9184 (22). DAP alone or the single phages did not lead to meaningful bacterial eradication. The phage cocktail caused an ∼2-log10(CFU/mL) reduction at a tMOI of 1.0. However, ∼3.0- to 3.5-log10(CFU/mL) reductions (deemed bactericidal) were noted in nearly all combination regimens with DAP plus phage cocktails, while synergistic effects were observed with the phage cocktail and DAP combination at a tMOI of 0.1 (Fig. 2). Resistance to both phages was observed in isolates collected and rescreened after TKA for all monophage and phage cocktail regimens without DAP. In contrast, phage 9184 resistance was not observed after the addition of DAP (DAP plus phage 9184 and DAP plus phage cocktails). No meaningful differences were observed in phage counts among treatment regimens following the end of TKA.
FIG 2.
Monotherapy regimens including DAP, monophage, and phage cocktails at various tMOIs (0.1 and 1.0) did not lead to meaningful bacterial eradication. However, DAP in combination with phage cocktails led to bactericidal activity in nearly all combination regimens.
Although DAP combinations with beta-lactams have shown promise in the clinical realm for E. faecium infections, some strains of E. faecium are nonresponsive, and other patient-specific factors (e.g., true allergies) may preclude their use (23–25). We have shown that a phage cocktail in combination with DAP improved the bacterial eradication of a DNS E. faecium strain and that DAP was able to prevent the emergence of phage resistance against one phage. While these data are currently limited to a single strain of E. faecium, this strain is a prototypical isolate with well-characterized genetic alterations (liaFSR mutations) that are often observed in other difficult-to-treat E. faecium clinical strains (26). Furthermore, phage 113 shared genetic similarities with phage EFDG1, which has been shown to have the ability to clear biofilms off abiotic surfaces (20). Further research should evaluate the mechanisms underlying these effects and examine phages against biofilm-producing E. faecium.
Data availability.
The Illumina DNA sequencing reads have been deposited in the National Center for Biotechnology Information under accession number SAMN19047785 (phage 113) and the European Nucleotide Archive under accession number PRJEB39873 (phage 9184). The assembled bacteriophage genomes were submitted to GenBank and were assigned the following accession numbers: MZ147816 (phage 113), MT939240 (phage 9181), MT939241 (phage 9183), and MT939242 (phage 9184).
ACKNOWLEDGMENTS
This work was partially supported by NIAID R01-AI121400 (M.J.R.). B.A.D. is partially supported by NIAID R01-AI141479. C.A.A. is partially supported by NIH/NIAID grants R01-AI148342, R01-AI134637, P01-AI152999, and K24-AI121296. M.J.R. is partially supported by NIAID R01-AI121400 and R21-AI163726-01. T.M., K.L.L., G.S.C. R.K., J.C.A., K.C.S., S.W., and S.M.L. have no conflicts of interest to disclose.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.World Health Organization. 2019. New report calls for urgent action to avert antimicrobial resistance crisis. World Health Organization, Geneva, Switzerland. https://www.who.int/news/item/29-04-2019-new-report-calls-for-urgent-action-to-avert-antimicrobial-resistance-crisis. Accessed 1 August 2021. [Google Scholar]
- 2.O’Neill J. 2014. Antimicrobial resistance: tackling a crisis for the health and wealth of nations. Review on Antimicrobial Resistance, London, United Kingdom. https://amr-review.org/sites/default/files/AMR%20Review%20Paper%20-%20Tackling%20a%20crisis%20for%20the%20health%20and%20wealth%20of%20nations_1.pdf. Accessed 1 August 2021. [Google Scholar]
- 3.Arias CA, Murray BE. 2012. The rise of Enterococcus: beyond vancomycin resistance. Nat Rev Microbiol 10:266–278. 10.1038/nrmicro2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rice LB. 2008. Federal funding for the study of antimicrobial resistance in nosocomial pathogens: no ESKAPE. J Infect Dis 197:1079–1081. 10.1086/533452. [DOI] [PubMed] [Google Scholar]
- 5.Lin DM, Koskella B, Lin HC. 2017. Phage therapy: an alternative to antibiotics in the age of multi-drug resistance. World J Gastrointest Pharmacol Ther 8:162–173. 10.4292/wjgpt.v8.i3.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morrisette T, Kebriaei R, Lev KL, Morales S, Rybak MJ. 2020. Bacteriophage therapeutics: a primer for clinicians on phage-antibiotic combinations. Pharmacotherapy 40:153–168. 10.1002/phar.2358. [DOI] [PubMed] [Google Scholar]
- 7.Lehman SM, Mearns G, Rankin D, Cole RA, Smrekar F, Branston SD, Morales S. 2019. Design and preclinical development of a phage product for the treatment of antibiotic-resistant Staphylococcus aureus infections. Viruses 11:88. 10.3390/v11010088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chan BK, Abedon ST, Loc-Carrillo C. 2013. Phage cocktails and the future of phage therapy. Future Microbiol 8:769–783. 10.2217/fmb.13.47. [DOI] [PubMed] [Google Scholar]
- 9.Kebriaei R, Lev K, Morrisette T, Stamper KC, Abdul-Mutakabbir JC, Lehman SM, Morales S, Rybak MJ. 2020. Bacteriophage-antibiotic combination strategy: an alternative against methicillin-resistant phenotypes of Staphylococcus aureus. Antimicrob Agents Chemother 64:e00461-20. 10.1128/AAC.00461-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Canfield GS, Chatterjee A, Espinosa J, Mangalea MR, Sheriff EK, Keidan M, McBride SW, McCollister BD, Hang HC, Duerkop BA. 2021. Lytic bacteriophages facilitate antibiotic sensitization of Enterococcus faecium. Antimicrob Agents Chemother 65:e00143-21. 10.1128/AAC.00143-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Morrisette T, Lev KL, Kebriaei R, Abdul-Mutakabbir JC, Stamper KC, Morales S, Lehman SM, Canfield GS, Duerkop BA, Arias CA, Rybak MJ. 2020. Bacteriophage-antibiotic combinations for Enterococcus faecium with varying bacteriophage and daptomycin susceptibilities. Antimicrob Agents Chemother 64:e00993-20. 10.1128/AAC.00993-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Palmer KL, Carniol K, Manson JM, Heiman D, Shea T, Young S, Zeng Q, Gevers D, Feldgarden M, Birren B, Gilmore MS. 2010. High-quality draft genome sequences of 28 Enterococcus sp. isolates. J Bacteriol 192:2469–2470. 10.1128/JB.00153-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clinical and Laboratory Standards Institute. 2021. Performance standards for antimicrobial susceptibility testing; 31st informational supplement. CLSI document M100. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 14.Mazzocco A, Waddell TE, Lingohr E, Johnson RP. 2009. Enumeration of bacteriophages using the small drop plaque assay system. Methods Mol Biol 501:81–85. 10.1007/978-1-60327-164-6_9. [DOI] [PubMed] [Google Scholar]
- 15.O’Flynn G, Ross RP, Fitzgerald GF, Coffey A. 2004. Evaluation of a cocktail of three bacteriophages for biocontrol of Escherichia coli O157:H7. Appl Environ Microbiol 70:3417–3424. 10.1128/AEM.70.6.3417-3424.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ackermann H-W. 2007. 5500 phages examined in the electron microscope. Arch Virol 152:227–243. 10.1007/s00705-006-0849-1. [DOI] [PubMed] [Google Scholar]
- 17.Ramsey J, Rasche H, Maughmer C, Criscione A, Mijalis E, Liu M, Hu JC, Young R, Gill JJ. 2020. Galaxy and Apollo as a biologist-friendly interface for high-quality cooperative phage genome annotation. PLoS Comput Biol 16:e1008214. 10.1371/journal.pcbi.1008214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Plaut RD, Stibitz S. 2019. Regulatory considerations for bacteriophage therapy products, p 337–339. In Górski A, Międzybrodzki R, Borysowski J (ed), Phage therapy: a practical approach. Springer Nature, Cham, Switzerland. [Google Scholar]
- 19.Bolocan AS, Upadrasta A, de Almeida Bettio PH, Clooney AG, Draper LA, Ross RP, Hill C. 2019. Evaluation of phage therapy in the context of Enterococcus faecalis and its associated diseases. Viruses 11:366. 10.3390/v11040366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Khalifa L, Brosh Y, Gelman D, Coppenhagen-Glazer S, Beyth S, Poradosu-Cohen R, Que Y-A, Beyth N, Hazan R. 2015. Targeting Enterococcus faecalis biofilms with phage therapy. Appl Environ Microbiol 81:2696–2705. 10.1128/AEM.00096-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weber-Dabrowska B, Jończyk-Matysiak E, Żaczek M, Łobocka M, Łusiak-Szelachowska M, Górski A. 2016. Bacteriophage procurement for therapeutic purposes. Front Microbiol 12:1177. 10.3389/fmicb.2016.01177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Diaz L, Tran TT, Munita JM, Miller WR, Rincon S, Carvajal LP, Wollam A, Reyes J, Panesso D, Rojas NL, Shamoo Y, Murray BE, Weinstock GM, Arias CA. 2014. Whole-genome analyses of Enterococcus faecium isolates with diverse daptomycin MICs. Antimicrob Agents Chemother 58:4527–4534. 10.1128/AAC.02686-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chuang Y-C, Chen P-Y, Lin C-Y, Chen Y-C, Wang J-T, Chang S-C. 2018. A retrospective clinical comparison of daptomycin vs daptomycin and a beta-lactam antibiotic for treating vancomycin-resistant Enterococcus faecium bloodstream infections. Sci Rep 8:1632. 10.1038/s41598-018-19986-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kebriaei R, Rice SA, Singh KV, Stamper KC, Dinh AQ, Rios R, Diaz L, Murray BE, Munita JM, Tran TT, Arias CA, Rybak MJ. 2018. Influence of inoculum effect on the efficacy of daptomycin monotherapy and in combination with beta-lactams against daptomycin-susceptible Enterococcus faecium harboring LiaSR substitutions. Antimicrob Agents Chemother 62:e00315-18. 10.1128/AAC.00315-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kebriaei R, Stamper KC, Singh KV, Khan A, Rice SA, Dinh AQ, Tran TT, Murray BE, Arias CA, Rybak MJ. 2020. Mechanistic insights into the differential efficacy of daptomycin plus beta-lactam combinations against daptomycin-resistant Enterococcus faecium. J Infect Dis 222:1531–1539. 10.1093/infdis/jiaa319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Davlieva M, Tovar-Yanez A, DeBruler K, Leonard PG, Zianni MR, Arias CA, Shamoo Y. 2016. An adaptive mutation in Enterococcus faecium LiaR associated with antimicrobial peptide resistance mimics phosphorylation and stabilizes LiaR in an activated state. J Mol Biol 428:4503–4519. 10.1016/j.jmb.2016.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nishimura Y, Yoshida T, Kuronishi M, Uehara H, Ogata H, Goto S. 2017. ViPTree: the viral proteomic tree server. Bioinformatics 33:2379–2380. 10.1093/bioinformatics/btx157. [DOI] [PubMed] [Google Scholar]
- 28.Darling AE, Mau B, Perna NT. 2010. progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One 5:e11147. 10.1371/journal.pone.0011147. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental text. Download aac.01623-21-s0001.pdf, PDF file, 0.04 MB (41.5KB, pdf)
Data Availability Statement
The Illumina DNA sequencing reads have been deposited in the National Center for Biotechnology Information under accession number SAMN19047785 (phage 113) and the European Nucleotide Archive under accession number PRJEB39873 (phage 9184). The assembled bacteriophage genomes were submitted to GenBank and were assigned the following accession numbers: MZ147816 (phage 113), MT939240 (phage 9181), MT939241 (phage 9183), and MT939242 (phage 9184).