ABSTRACT
Echinocandins are frontline antifungal agents in the management of invasive infections due to multidrug resistant Candida auris. The study aimed to evaluate echinocandin resistance in C. auris isolates of multicentric origin, identify the resistance mechanism, and analyze the pharmacodynamic response to caspofungin in a neutropenic mouse model of infection. A total of 199 C. auris isolates originating from 30 centers across India were tested for susceptibility to echinocandins. Isolates with reduced susceptibility were evaluated for FKS1 mutations and in vivo response to caspofungin in a murine model of disseminated candidiasis. In addition, the response to echinocandins was assessed in light of in vitro growth kinetics, chitin content; and transcript levels of chitin synthase and FKS1 genes. We report 10 resistant C. auris isolates with four FKS1 mutations: F635Y (n = 2), F635L (n = 4), S639F (n = 3), and R1354S (n = 1). Of these, F635Y and R1354S exhibited the most profound resistance in mouse model of disseminated infection. S639F and F635L mutations conferred a moderate in vivo resistance, whereas wild-type isolates exhibiting borderline MIC were susceptible in vivo. FKS1 genotype was more accurate predictor of in vivo response than the MIC of the isolates. Isolates with high basal or inducible chitin content exhibited higher in vitro MIC in FKS1 mutant compared to wild type. FKS1 mutations play a major role in clinically relevant echinocandin resistance in C. auris with differential in vivo outcomes. This study could have implications for clinical practice and, therefore, warrants further studies.
KEYWORDS: Candida auris, echinocandins, FKS1, murine model, FKS mutation, chitin, pharmacodynamics, resistance
INTRODUCTION
In the last decade, a paradigm shift in the epidemiology of invasive Candida infections has been reported with frequent isolation of non-albicans Candida species (1, 2). Among these, multidrug-resistant C. auris has become a significant worry due to its rising incidence, its outbreak potential, and association with high mortality in critically ill patients (3–5). Since the first report in 2009 from Japan, C. auris has been reported in 44 countries spanning all the five inhabited continents with multiple outbreaks (5–8).
Although resistance is clade-specific, C. auris overwhelmingly exhibits resistance to fluconazole with variable susceptibility to other triazoles along with substantial resistance to amphotericin B in Clade I (South-Asian clade) (9, 10). This leaves clinicians only with echinocandins as primary therapy for C. auris infections (9, 11). The Joint ESCMID-ECMM and the IDSA clinical practice guidelines recommend echinocandins as first-line treatment options for invasive Candida infections (12, 13). Echinocandins inhibit 1,3-β-d-glucan synthase activity, catalyzing the synthesis of primary fungal cell wall polymer, 1,3-β-d-glucan (14). Although many investigational compounds have demonstrated efficacy against C. auris, none of those has been approved yet as a therapeutic modality (15, 16). However, reports of echinocandin resistance in C. auris have augmented our concerns (9, 17). Echinocandin resistance in Candida species is associated with mutations in the hot spot regions of FKS genes, which encode the catalytic subunit of 1,3-β-d-glucan synthase (18). Previously, Kordalewska et al. (17) demonstrated a poor therapeutic response of S639F FKS1 mutation in C. auris in terms of the reduction in organ fungal burden in a murine model of infection. Further, in C. albicans, chitin upregulation has been found an important factor in promoting echinocandin tolerance and isolates with increased chitin content have demonstrated low in vitro susceptibility (19).
In the present study, we evaluated echinocandin resistance in a collection of C. auris clinical isolates originating from different Indian centers, including six sequential isolates from a transplant patient. All the isolates exhibiting reduced susceptibility to echinocandins were analyzed for FKS1 mutations and evaluated for differential expression of chitin synthase and FKS1 gene. We then analyzed the pharmacodynamic response of these isolates in a mouse model of disseminated candidiasis.
(Part of this work was accepted to be presented at the 30th Meeting of the European Congress on Clinical Microbiology and Infectious Diseases, abstract 9313.)
RESULTS
Antifungal susceptibility testing.
The susceptibility distribution of 199 C. auris to different antifungal agents is summarized in Table 1. The susceptibility of individual isolates is provided in Table S2. MIC90 of all three echinocandins was 0.5 mg/liter. Anidulafungin exhibited higher in vitro activity against C. auris isolates with a geometric mean (GM) MIC 0.18 mg/liter compared to caspofungin (0.38 mg/liter) and micafungin (0.22 mg/liter). Sixteen (8%) and 10 (5%) isolates displayed MICs higher than the ECOFF values proposed for anidulafungin (>0.5 mg/liter) and micafungin (>0.5 mg/liter), respectively, whereas 15 (7.5%) isolates exhibited caspofungin MIC higher than MIC90 (0.5 mg/liter). For amphotericin B, MIC50 and MIC90 were 4-fold and 2-fold higher in our series, respectively, compared to those reported previously (20). For itraconazole and posaconazole, both 50th and 90th percentile MICs were 2-fold higher whereas for voriconazole, the corresponding values were 2-fold and 4-fold lower than the earlier report. For fluconazole, MIC50 and MIC90 were 2-fold lower. Out of 30 centers included in the study, echinocandin-resistant C. auris were isolated from four centers (Fig. S1).
TABLE 1.
Antifungal susceptibility distribution of C. auris isolates to amphotericin B, triazoles, and echinocandins
| MIC (mg/L) |
|||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Antifungal tested | Range | 0.03 | 0.06 | 0.12 | 0.25 | 0.5 | 1 | 2 | 4 | 8 | 16 | 32 | 64 | >64 | MIC50 | MIC90 | GM | ECOFF method and value | % of isolates above ECOFF |
| Caspofungin | 0.125-16 | 0 | 0 | 26 | 62 | 96 | 8 | 2 | 3 | 0 | 2 | 0 | 0 | 0 | 0.5 | 0.5 | 0.38 | ND (0.5)a | 15 (7.5%)a |
| Micafungin | 0.12-16 | 0 | 0 | 92 | 69 | 28 | 7 | 2 | 0 | 0 | 1 | 0 | 0 | 0 | 0.25 | 0.5 | 0.22 | Visual eyeball method (0.5) | 10 (5%) |
| Anidulafungin | 0.03-8 | 12 | 38 | 64 | 35 | 34 | 5 | 6 | 4 | 1 | 0 | 0 | 0 | 0 | 0.25 | 0.5 | 0.18 | Visual eyeball method (0.5) | 16 (8%) |
| Fluconazole | 0.25-64 | 0 | 0 | 0 | 2 | 1 | 6 | 5 | 9 | 16 | 22 | 39 | 36 | 63 | 32 | 64 | 26.70 | Derivatization method (128) | 63 (31.6%) |
| Voriconazole | 0.03-8 | 35 | 8 | 14 | 51 | 44 | 29 | 14 | 3 | 1 | 0 | 0 | 0 | 0 | 0.25 | 1 | 0.27 | Derivatization method (1) | 18 (9.04%) |
| Itraconazole | 0.03-1 | 39 | 23 | 29 | 59 | 46 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.25 | 0.5 | 0.15 | Visual eyeball method (0.5) | 3 (1.50%) |
| Posaconazole | 0.03 to 0.5 | 48 | 41 | 53 | 50 | 7 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.125 | 0.25 | 0.10 | Derivatization method (0.125) | 7 (3.52%) |
| Amphotericin B | 0.03-8 | 3 | 4 | 4 | 2 | 20 | 32 | 88 | 45 | 1 | 0 | 0 | 0 | 0 | 2 | 4 | 1.48 | Visual eyeball method (2) | 46 (23.11%) |
Since ECOFF values were not available for caspofungin, 0.5 was chosen the cutoff value as this value segregated non-wild-type subset from wild-type isolates in our series.
FKS1 mutation analysis.
Seventeen isolates, including six sequential isolates with reduced echinocandin susceptibility, were subjected to FKS1 sequence analysis. Of these, 10 (5%) isolates exhibiting MIC ≥2 mg/liter for any of the three echinocandins carried a mutation in FKS1; barring one isolate (Table 2). Among them, three harbored previously described S639F mutation. However, a novel mutation, F635Y, was found in two isolates with caspofungin MICs of 4 and 16 mg/liter. Of six sequential isolates from a single patient, four had F635L substitution resulting in elevated MIC to anidulafungin. Two other isolates with caspofungin MIC of 2 mg/liter and 16 mg/liter showed no alteration in the HS1 region of FKS1, however, isolate with 16 mg/liter harbored R1354S mutation in HS2 of FKS1. Two isolates harboring F635Y FKS1 mutation were isolated from two different centers, while as three isolates with S639F mutation were isolated from two centers (Fig. S1). Six isolates exhibiting MIC of 1 mg/liter for caspofungin or anidulafungin carried a wild-type FKS1 genotype. Sequencing of 25 randomly selected caspofungin-susceptible isolates demonstrated wild-type FKS1 in all these isolates (Table S2). The FKS1 nucleotide sequences of all the non-wild-type isolates were submitted to the GenBank with the accession numbers provided in Table 2.
TABLE 2.
Echinocandin susceptibility profile and FKS1 genotypes of 17 C. auris isolates with reduced susceptibility to echinocandin MICs and 4 wild-type comparators analyzed by in vitro and in vivo experimentsa
| MIC (mg/L) |
||||||
|---|---|---|---|---|---|---|
| Isolate ID | Sample | FKS1 mutation | GenBank accession | CAS | AFG | MFG |
| NCCPF470197 | Blood | T1904A(F635Y) | MT199096.1 | 16(R) | 1(S) | 1(S) |
| NCCPF470198 | Urine | C1916T(S639F) | MT199098.1 | 4(R) | 4(R) | 1(S) |
| NCCPF470199 | Urine | C1916T(S639F) | MT199097.1 | 4(R) | 4(R) | 1(S) |
| NCCPF470203 | Blood | C1916T(S639F) | MT199099.1 | 2(R) | 8(R) | 16(R) |
| NCCPF470209 | Blood | T1904A(F635Y) | MT199100.1 | 4(R) | 2(S) | 1(S) |
| NCCPF470237 | Urine | T1903C(F635L) | MT199101.1 | 1(S) | 2(S) | 1(S) |
| NCCPF470238 | Urine | T1903C(F635L) | MT199102.1 | 1(S) | 2(S) | 1(S) |
| NCCPF470240 | Blood | T1903C(F635L) | MT199104.1 | 1(S) | 2(S) | 0.5(S) |
| NCCPF470241 | ETS | T1903C(F635L) | MT199105.1 | 1(S) | 2(S) | 1(S) |
| NCCPF470200 | Urine | C4060A(R1354S) | MW655801.1 | 16(R) | 2(S) | 2(S) |
| NCCPF470201 | Blood | Wild-type | 2(R) | 1(S) | 2(S) | |
| NCCPF470239 | BALF | Wild-type | 0.5(S) | 1(S) | 0.25(S) | |
| NCCPF470242 | ETS | Wild-type | 0.5(S) | 1(S) | 0.25(S) | |
| NCCPF470202 | Blood | Wild-type | 1(S) | 0.5(S) | 0.5(S) | |
| NCCPF470204 | Blood | Wild-type | 1(S) | 0.5(S) | 0.5(S) | |
| NCCPF470205 | Blood | Wild-type | 1(S) | 0.5(S) | 0.5(S) | |
| NCCPF470206 | Blood | Wild-type | 1(S) | 1(S) | 0.25(S) | |
| NCCPF470196 | Blood | Wild-type | 0.12(S) | 0.06(S) | 0.12(S) | |
| NCCPF470208 | Blood | Wild-type | 0.12(S) | 0.25(S) | 0.25(S) | |
| NCCPF470210 | Blood | Wild-type | 0.12(S) | 0.25(S) | 0.25(S) | |
| CBS12372 | Blood | Wild-type | 0.12(S) | 0.12(S) | 0.25(S) | |
BALF, bronchoalveolar lavage fluid; ETS, endotracheal tube secretion. The letters within parenthesis represent interpretation of the susceptibility as per the CDC recommended cutoff values (R, resistant; S, susceptible).
Genotypic analysis of six serial isolates.
Six C. auris were isolated from urine (n = 2), blood (n = 1), bronchoalveolar lavage (n = 1), and endotracheal tube secretions (n = 2) of a 37-year-old male patient with liver transplant (Fig. S2). Detailed case history and therapeutic interventions done in the patient are provided in the supplementary file. In AFLP analysis, three isolates recovered from lower respiratory tract specimens formed one cluster with 98.5% fingerprint similarity (Fig. S3). Two isolates recovered from urine samples formed a second cluster with 98% fingerprint similarity, while the blood isolate was different from the above two clusters.
Pharmacodynamic response to caspofungin.
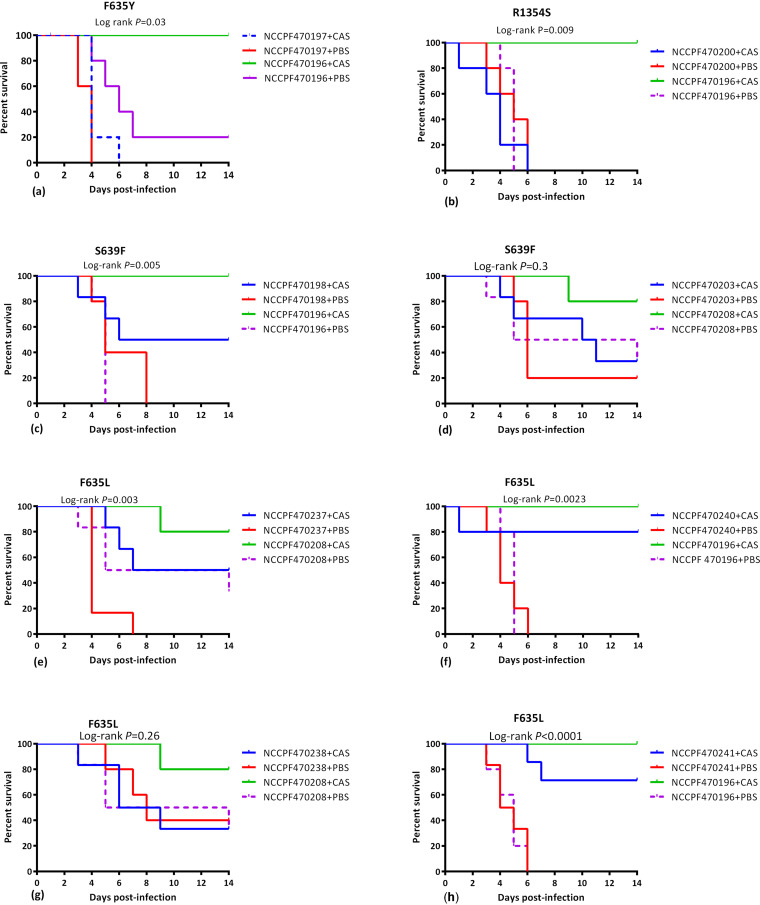
(i) Kaplan-Meier survival kinetics. FKS1 mutants, F635Y and R1354S were unresponsive to caspofungin compared to wild-type isolate NCCPF470196 that showed a complete therapeutic response with 100% survival (Log-rank test P value <0.01) (Fig. 1a and 1b). The median survival of mice infected with these two FKS1 mutant isolates was 4 days on caspofungin therapy (Fig. 2d). FKS1 mutant, S639F, demonstrated a partial response to caspofungin therapy with a 50% survival benefit and median survival of 10 days (Fig. 1c and 2d). However, mice infected with another isolate with the same FKS1 mutation showed statistically insignificant kill kinetics compared to wild-type isolate (Fig. 1d).
FIG 1.
Kaplan-Meier survival curves illustrating the differential survival in mice infected with FKS1 mutants, postadministration of human therapeutic dose of caspofungin. The survival kinetics of FKS1 mutants were compared with two FKS1 wild-type isolates, NCCPF470196 and NCCPF470208 in independent experiments. The data were analyzed by Log-rank test and P values <0.05 were considered significant. CAS, Caspofungin (4 mg/kg); PBS, phosphate-buffered saline.
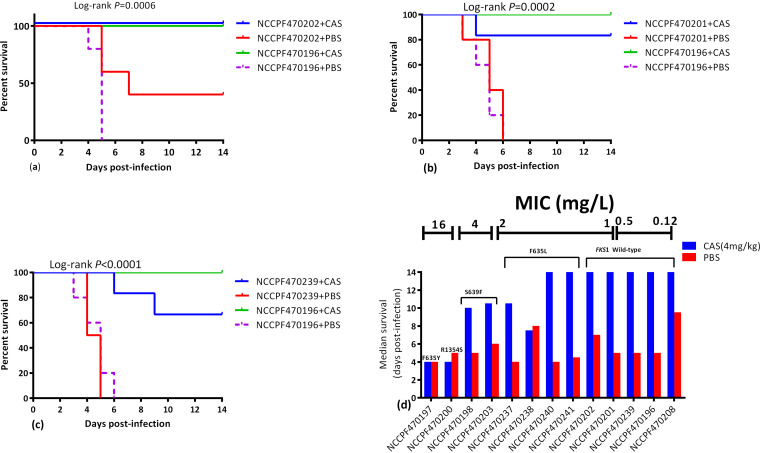
FIG 2.
(a-c) Kaplan-Meier survival curves illustrating the differential survival post administration of caspofungin mice infected with three FKS1 wild-type isolates with borderline susceptibility to echinocandins relative to wild-type susceptible comparator isolate. Kaplan-Meier survival curves were compared by Log-rank test. (d) Median survival days of mice infected with 13 C. auris isolates plotted as a function of FKS1 genotype. The MIC of each isolate/genotype is indicated at the top of the graph. CAS, Caspofungin (4 mg/kg); PBS, phosphate-buffered saline.
Similarly, three isolates with F635L mutation exhibited a partial response to caspofungin therapy with 50–80% survival and median survival days ranging from 7.5–14 (Log-rank P value <0.01) (Fig. 1e, f, and h and 2d). In one F635L FKS1 mutant, the survival rates were not significantly different in caspofungin and vehicle-treated groups (Log-rank P value >0.05) (Fig. 1g).
Among three isolates exhibiting borderline echinocandin MIC (≤2 mg/liter) with wild-type FKS1, two isolates (NCCPF470201 and NCCPF470239) demonstrated a marginally lower response to caspofungin compared to the comparator isolate NCCPF470196 whereas the third isolate (NCCPF470202) showed a complete in vivo response to caspofungin therapy (Fig. 2).
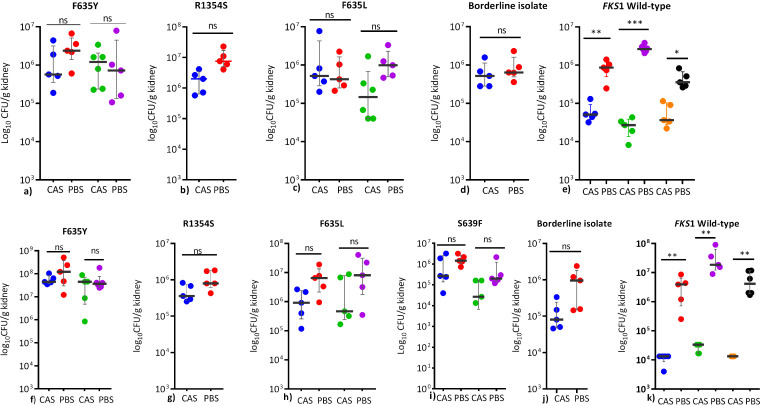
(ii) Fungal kidney burden reduction. A total of 11 isolates, including those with F635Y (NCCPF470196 and NCCPF470209), F635L (NCCPF470238 and NCCPF470240), S639F (NCCPF470199 and NCCPF470203), R1354S (NCCPF470200), and wild type (NCCPF470201, NCCPF470208, NCCPF470196, and CBS12372) were evaluated for fungal burden reduction. In mice receiving either a single shot of caspofungin 1-h postinfection or two shots 24-h apart, no significant reduction in the median log10 CFU/g kidney tissue was observed among all non-WT isolates (Fig. 3a to c and f to i). Also, isolate exhibiting 2 mg/liter MIC to caspofungin but carrying wild-type FKS1 (NCCPF470201) did not show a statistically significant reduction in the kidney burden (Fig. 3d and 3j). In contrast, three susceptible wild-type isolates showed ≥1 Log10 reduction in median CFU counts on caspofungin treatment (P < 0.05) (Fig. 3e). Similarly, on the administration of two shots of caspofungin, only mice infected with three susceptible isolates with wild-type FKS1 demonstrated ≥ 2 Log10 reductions in CFU (P < 0.05) equivalent to 99% clearance of yeast cells compared to untreated controls (Fig. 3k).
FIG 3.
Comparison of in vivo response to a human therapeutic dose of caspofungin among different FKS1 mutants determined as relative log reduction in the C. auris CFU burden in the kidneys after treatment with 4 mg/kg of caspofungin at 1 h postinfection (upper panel, a-e) or 4 mg/kg at 1 h and a repeat dose at 24 h postinfection (lower panel, f-k). Following representative isolates were evaluated: F635Y (NCCPF470197, NCCPF470209), R1354S (NCCPF470200), F635L (NCCPF470238, NCCPF470240), S639F (NCCPF470470199, NCCPF470470203), borderline (NCCPF470201), wild type (NCCPF470196, NCCPF470208, CBS12372. Organ burden assessment for S639F mutant was evaluated at only two doses of caspofungin (i). The organ fungal burden data are shown as median (q1-q3) and were compared using Mann-Whitney U test for two groups or Kruskal-Wallis test for multiple groups with Dunn’s correction factor to minimize type 1 error. ns, nonsignificant, **, P < 0.01, ***, P < 0.001. CAS, Caspofungin (4 mg/kg); PBS, phosphate-buffered saline.
Chitin content; chitin synthase, and FKS1 gene expression.
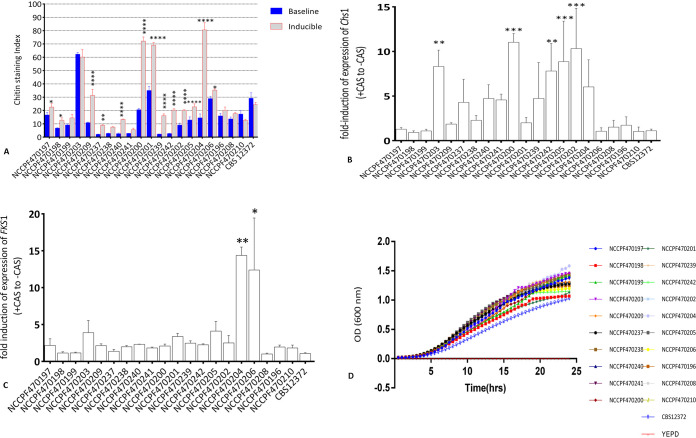
Among three isolates with S639F FKS1 mutation (NCCPF470198, 470199, 470203), NCCPF470203 distinctively exhibited high baseline chitin content (Staining index, 62 ± 1.2) compared to others (NCCPF470198, 6.8 ± 0.3; NCCPF470199, 9.1 ± 0.8) (Fig. 4A). The transcript levels of Chs1 gene in this isolate were also significantly higher (8.2 ± 1.8) compared to other two isolates (NCCPF470198, 1 ± 0.2; NCCPF 470199, 1 ± 0.18, P < 0.01) (Fig. 4B). On exposure with caspofungin, majority of the isolates (13/20, 65%) demonstrated a statistically significant induction (P < 0.05) in the chitin content compared to their respective baseline values. While five of these isolates exhibited staining indices of >30 (baseline staining index of reference isolate, CBS12372) chitin induction in remaining eight isolates was marginal. Five among these 13 isolates also demonstrated a significant upregulation of putative chitin synthase gene, Chs1 (Fig. 4B). However, Chs1 gene expression levels and staining indices correlated well only in S639F and R1354S FKS1 mutant isolates. In contrast, none of the sensu stricto susceptible isolates showed either high chitin content or a significant induction in Chs1 (P > 0.05).
FIG 4.
A. Baseline and caspofungin-induced differential chitin content represented as staining index. Three biological replicates were used for each isolate to determine the chitin levels and data were analyzed by using two-way ANOVA with Sidak’s correction factor for multiple comparison. B&C represent relative fold-induction in the expression of Chs1 and FKS1 on caspofungin treatment, respectively. The data are represented as mean±SD of three biological replicates for each isolate. The data points were compared to reference isolate CBS12372 and analyzed by one-way ANOVA with Bonferroni correction for multiple comparisons. D, Growth curve depicting the in vitro growth rates of C. auris isolates represented as mean OD values at 600 nm measured at 30 min interval through a 24-h growth period. *, P < 0.05, **, P < 0.01, ***, P < 0.001.
Only two isolates with borderline echinocandin MIC demonstrated a significant induction of the FKS1 gene compared to the reference isolate CBS12372 (14.4 ± 1 and 12.4 ± 7.1, P < 0.05) (Fig. 4C).
Growth kinetics.
The growth kinetics experiment showed differential growth rates among isolates. However, these varied growth rate patterns were independent of the underlying FKS1 genotype. While the duration of lag-phase in a secondary broth culture was more or less similar with average 4 h duration among the replicates, the doubling time (g) varied from 1.7 to 2.7 h with CBS12372 of clade IV showing the largest doubling time (2.7 h) consistent with its less steep log phase (Fig. 4D and Table S3). In addition, NCCPF470203, -470208, and -470238 had also relatively higher generation time compared to other isolates.
DISCUSSION
Echinocandins are frontline drugs in the management of invasive Candida infections, including those due to multidrug-resistant C. auris (12, 13). Invasive candidiasis due to C. auris is associated with mortality rates of 30–60% (21). Therefore, the emergence of echinocandin resistance in C. auris is a significant concern. The resistance prevalence and resistance rate were 4.1% (8/194) and 5.5% (11/199), respectively, in this series as per the cutoff values suggested by CDC. MIC distributions of the present series were largely in concordance with those reported by Arendrup et al. (20) with MIC50/MIC90 for anidulafungin and micafungin as 0.25/0.5; however, the MIC90 (0.5 mg/liter) and GM (0.22) were higher for micafungin by 2-fold in our series. Interestingly, the ECOFF values derived by eye-ball method by Arendrup et al. (20) could clearly delineate non-wild-type isolates in our study.
Definitive breakpoints for interpretation of echinocandin susceptibility in C. auris are not available yet. CDC has proposed tentative breakpoints based on the modal distribution of 100 isolates originating from diverse geographic locales, and these cutoff values do not take into account the PK/PD parameters of echinocandins in the C. auris infection model (22). Four of our isolates exhibited anidulafungin MIC 4-fold lower than the proposed cutoff (≥4 mg/liter), but those isolates carried a mutation, F635L in FKS1. These findings, therefore, suggest a review of existing breakpoints of echinocandin susceptibility in C. auris that can capture these newly emerged FKS1 non-wild-type isolates. Considering the presence of a genetic marker of echinocandin resistance at known hot spot positions, 5% (10/199) of our isolates exhibited bonafide resistance to at least one echinocandin. This estimation fits within the ballpark of 2–9.5% resistance rate reported in earlier studies (9, 10, 17, 23).
Owing to its persistence on inanimate surfaces and acquisition from health care workers or fomites, C. auris demonstrates horizontal transmission (4, 24). In the present study, none of the patients with resistant isolates were on prior echinocandin therapy before the isolation of resistant isolate. Therefore, development of echinocandin-resistant C. auris infection would suggest acquisition of already resistant strains probably from the nosocomial environment. Also, the patient that yielded six sequential isolates from different clinical specimens was on amphotericin B therapy at the time of isolation of first resistant isolate (Fig. S2). In our study, 10 resistant isolates were reported from four health care facilities with F635Y and S639F genotypes reported from two centers each (Fig. S1). The lack of any evidence of transfer of these patients harboring resistant strains between the health care facilities suggests minimal chances of acquisition of resistance from a common source.
The isolation of mixed populations of both FKS1 wild-type and non-wild-type isolates from different clinical specimens of an immunosuppressed patient makes an interesting case study. Although, AFLP analysis showed >92% similarity in banding patterns suggesting a clonal origin, however, given the absence of unanimous criteria for determining clonality in AFLP typing, the possibility of heterogeneous subpopulations implying multimodal acquisition of C. auris by this patient could not be ruled out. A recent study also demonstrated heterogeneous populations of echinocandin-resistant and susceptible C. auris isolates in a single patient (25). It also lies within the realm of possibility that this patient had a unimodal acquisition of C. auris followed by in-host microevolutionary genomic changes giving rise to niche-specific heterogeneous populations of C. auris (26).
Both experimental and clinical data have demonstrated therapeutic failure and increased all-cause mortality associated with a mutation in the FKS1 gene (17, 27). In the present study, 10 isolates harbored a mutation in the FKS1 gene with F635Y, F635L, and R1354S mutations conferring resistance in seven isolates, apart from already known S639F and S639P mutations (9, 17, 28, 29). It is pertinent to mention that Carolus et al., (30) recently reported Δ635 in one of their experimentally evolved lineages of caspofungin-resistant C. auris coupled with a compensatory mutation M690I in hot spot 3 of FKS1. F635Y and F635L, occur at the first residue of the HS1 of the FKS1 subunit in C. auris, and its equivalent in C. albicans is mutation at F641 (17, 31). F635L substitution conferred a reduced susceptibility pattern different from that of F635Y with high anidulafungin MIC (2 mg/liter) and a modest increase in the MIC of caspofungin and micafungin (1 mg/liter each). The side chain, R of the substituted amino acid at F635 seems to confer variable levels of resistance. While replacement with aromatic amino acid, tyrosine (Y), seems to confer high-level resistance with a less in vivo response, substitution with aliphatic amino acid, Leucine (L) results in low-level resistance with partial in vivo response. However, this rationale may not hold across all the species as in C. albicans, substitution with aliphatic amino acid, serine at the homologous position 641 also led to high-level resistance (32). Interestingly, one isolate exhibiting high-level resistance demonstrated R1354S in HS2 of FKS1. In C. albicans, mutation at the homologous position, i.e, R1361H has been reported in 6% of the echinocandin-resistant isolates. However, unlike R1354S, this mutation feebly impacts the sensitivity of glucan synthase to the echinocandins (low IC50) with less likelihood of resulting in a breakthrough infection (32).
Exposure of C. albicans to low echinocandin levels has been shown to stimulate chitin synthase gene expression resulting in elevated chitin synthesis and increased tolerance to echinocandins (33). Mutation in the FKS1 gene in conjunction with high chitin has been observed in resistant C. albicans isolates (34). In our study, exposure with corresponding 2-fold sub-MIC caspofungin concentration significantly upregulated Chs1 and FKS1 gene in five and two isolates, respectively. This upregulation of chitin synthase and FKS1 gene could partially be responsible for the marginal increase in MIC in pharmacodynamically inconsequential reduced susceptibility in those isolates. Further, one of the three isolates with S639F mutation had high baseline chitin content and exhibited a distinct susceptibility pattern with high-level MIC to anidulafungin and micafungin.
To get a sense of the clinical relevance of FKS1 mutations observed in our isolates, we evaluated the pharmacodynamic impact of these mutations in a disseminated infection model, using a clinically relevant dose of caspofungin (4 mg/kg) (17). Interestingly, F635Y and R1354S demonstrated poorest response to caspofungin in terms of the survival enhancement and reduction in organ fungal burden. On the other hand, S639F and F635L showed a moderate in vivo resistance to caspofungin with substantial improvement in the median survival days. Although, evaluation of fungal burden in kidney were consistent with overall mortality, there was no difference between mutant strains and wild-type intermediate susceptible strains as they showed only a 1 log reduction in burden, which limited the power of the analysis. In contrast, wild-type isolates with borderline MICs and sensu stricto susceptible isolates showed a comparable in vivo response to caspofungin therapy. This finding is in line with the observation that elevated MICs without an underlying FKS1 mutation generally do not result in therapeutic failure (17, 35). Although in vitro MIC of the isolates tested largely correlated with their in vivo outcome, however, in isolates with MIC between 1 and 2 mg/liter, FKS1 genotype rather than the MIC was predictive of in vivo outcome (Fig. 2d).
In C. albicans and C. glabrata, increased chitin content in FKS1 mutants was associated with attenuated growth translating to hypovirulence in animal models of infection (19). In the present study, two isolates, NCCPF470197 and NCCPF470200 with mutations F635Y and R1354S, respectively, that displayed a in vivo resistance had comparable growth kinetics (Table S3). However, one isolate harboring S639F mutation (NCCPF470203) with higher baseline chitin showed an attenuated growth pattern (generation time = 2.7 h) compared to the other isolate (NCCPF470198; generation time = 2.0 h) with the same mutation (Fig. 4D). This attenuated growth phenotype presumably led to hypovirulence in a murine model and statistically insignificant kill kinetics in treated and untreated groups (Fig. 1d). In contrast, the FKS1 mutant, R1354S, showed a similar higher inducible chitin and a corresponding upregulation in chitin synthase, but its growth pattern and kill kinetics without exposure to caspofungin were comparable to that of wild-type isolate, NCCPF470196. Similarly, of four F635L mutants, three isolates had kill kinetics comparable to that of susceptible control isolates with 100% mortality in 6–7 days postinfection in vehicle-treated groups (Fig. 1e and f). This is suggestive that there was no fitness trade-off associated with this mutation. Nevertheless, the fitness of these FKS1 mutants warrants further evaluation in animal models of infection.
One of our study's limitations is that it did not evaluate the pharmacodynamic impact of FKS1 mutations on micafungin and anidulafungin. Evaluating in vitro and in vivo response to SCY-078 that has shown promising efficacy against invasive candidiasis would have been interesting. (36). Inclusion of two different wild-type comparators in the survival experiments may have led to the skewed survival curves. Further, the inclusion of six serial isolates whose clonality was assessed by AFLP could potentially confound the prevalence of echinocandin resistance. The disparity in the pharmacodynamics of isolates carrying the same FKS1 mutation as was observed in S639F and F635L mutants could be due to the confounding effect of genetic differences beyond FKS1 impinging upon the virulence and immune response. As such, the independent influence of these mutations on the pharmacodynamics of caspofungin could be more accurately assessed in a genetic model using the same parental strain for different FKS1 mutations.
Our study demonstrates that echinocandin resistance is emerging among Indian C. auris clinical isolates. We report previously unknown mutations, F635Y, F635L, and R1354S in FKS1, conferring echinocandin resistance to C. auris. We also demonstrate that FKS1 mutations could lead to variable pharmacodynamics, with some mutants showing poor response to echinocandin therapy while others may respond partially. The study suggests that C. auris isolates with echinocandin MIC ≥1 should be evaluated for FKS1 mutation. However, isolates with a marginal rise in MIC but carrying wild-type FKS1 may respond well to the treatment. The study also suggests the adjunctive role of isolate-dependent transcriptional upregulation of chitin synthesis in conferring reduced susceptibility in C. auris; however, a definitive conclusion cannot be made unless more studies are conducted.
Finally, this study has the potential to be a steppingstone for further studies in exploring the nuances of in vivo impact of FKS1 mutation in C. auris.
MATERIALS AND METHODS
Isolates and growth conditions.
A total of 199 C. auris isolates from 194 invasive candidiasis cases originating from 30 centers of the country during September 2017 through March 2020 were evaluated in this study. The isolates evaluated in this study are part of a nation-wide antimicrobial resistance surveillance program conducted under the auspices of Indian Council of Medical Research, New Delhi (https://main.icmr.nic.in/sites/default/files/guidelines/candida_Auris.pdf) and stored at nodal coordinating center, Postgraduate Institute of Medical Education and Research, Chandigarh, as per the advisory of Indian Council of Medical Research, New Delhi (https://main.icmr.nic.in/sites/default/files/guidelines/candida_Auris.pdf).
The identity of each isolate was confirmed by matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometry (Bruker Daltonic GmbH, Bremen, Germany) as per previously described protocol (37).
Antifungal susceptibility testing.
Antifungal drug susceptibility testing was done according to the CLSI broth microdilution method following the M27-protocol (38). The susceptibility data were interpreted as per tentative breakpoints suggested by the Centers for Disease Control and Prevention (CDC), Atlanta, (22) and epidemiological cutoff values proposed by Arendrup et al. (20).
ITS and FKS1 sequencing analysis.
All 17 isolates having higher MIC (≥1mg/L) to any echinocandin, and 29 randomly selected susceptible isolates were used for further analysis. Genomic DNA was extracted using phenol-chloroform-isoamyl alcohol according to an earlier described method (39). The identification of those isolates was confirmed by sequencing the internal spacer (ITS) region of rDNA. The primer pairs Cau_HS1-F/Cau_HS1-R and Cau_HS2-F/Cau_HS2-R were designed to amplify hot spot region-1 (HS1) and hot spot region-2 (HS2) of FKS1 gene (XM_018312389.1) using the NCBI’s primer blast tool (https://www.ncbi.nlm.nih.gov/tools/primer-blast/). Primers and cycling conditions used for amplification and sequencing analysis are provided in the supplementary file. Cycling conditions for FKS1 amplification included initial denaturation at 94°C for 2 min, 45 cycles of 94°C for 15 s, annealing at 60°C (HS1), or 56°C (HS2) for 30 s, and a final extension at 68°C for 1 min. Gene sequencing was performed using the BigDye Terminator ready reaction kit (version 3.1; Applied Biosystems, Foster City, CA, USA) in ABI Genetic Analyzer 3500Dx (Applied Biosystems, Foster City, CA, USA). The raw sequences were analyzed using Seqman software (DNA Star, Laser Gene, ABI) for nucleotide substitutions. Protein sequences were obtained using the online ExPASy tool (http://www.expasy.org/translate), and multiple sequence alignment was performed in BioEdit.
Amplified fragment length polymorphism genotypic analysis.
Amplified fragment analysis of six sequential isolates from a liver transplant patient was performed to assess the clonality of isolates as per our earlier described method with a few modifications (10). The detailed method is described in the supplementary file.
Pharmacodynamic assessment of echinocandin resistance in a murine model.
A neutropenic murine model of invasive candidiasis was developed to evaluate the in vivo response of different FKS1 mutations. The pharmacodynamic response to a human-equivalent dose (4 mg/kg bodyweight) of caspofungin (MSD Pharmaceuticals Pvt. Ltd., India) was then evaluated based on two endpoints: survival kinetics over a 14-day period postinfection and reduction in fungal kidney burden (secondary endpoint). 6-week-old, pathogen-free, female swiss albino mice, weighing 20–25 g were used for all animal experiments. Animal groups were housed in presterilized filter-top cages and allowed access to food and water ad libitum. All animals were kept in the same room, following same light and dark cycle, and same pattern of handling.
(i) Kaplan-Meier estimator. 123 mice were randomized into 22 treatment/infection arms by simple random sampling with 5 or 6 animals per arm and intravenously challenged with cell suspensions of C. auris followed by administration of a clinically relevant dose of caspofungin. Control arms received equal volume of sterile PBS. Eleven isolates were chosen for Kaplan-Meier survival analysis to represent each of the FKS1 mutations [F635Y (n = 1), R1354S (n = 1), S639F (n = 2), F635L (n = 4)], and those with borderline MIC but wild-type FKS1 [n = 3]. The survival kinetics’ of the mice infected with these 11 isolates were compared to those infected with two susceptible wild-type isolates (NCCPF470197, NCCPF470208).
(ii) Fungal kidney burden quantification. The fungal reduction in kidney tissue was assessed after intraperitoneal administration of caspofungin. Following isolates were evaluated: F635Y (n = 2), R1354S (n = 1), S639F (n = 2), F635L (n = 2), isolates with borderline MIC but wild-type FKS1 (n = 1) and susceptible comparator isolates (n = 3). The fungal burden was assessed at a single or two doses of caspofungin (4 mg/kg each). Control arms received equal volume of sterile PBS. To evaluate the efficacy of single dose of caspofungin, 92 mice were randomized into 18 groups with 5 or 6 animals per arm. Following isolates were evaluated: F635Y (n = 2), R1354S (n = 1), F635L (n = 2), isolates with borderline MIC but wild-type FKS1 (n = 1) and susceptible comparator isolates (n = 3). To evaluate the efficacy of two doses of caspofungin administered 24 h apart after infection, 112 mice were randomized into 22 groups and following isolates were evaluated: F635Y (n = 2), R1354S (n = 1), S639F (n = 2) F635L (n = 2), isolates with borderline MIC but wild-type FKS1 (n = 1) and susceptible comparator isolates (n = 3).
Further details of methods are provided in supplementary data. The sample size for each animal experiment was calculated using the resource equation method (40). All animal studies, including the plan of the study were approved by the Institutional Animal Ethics Committee (IAEC) of the Postgraduate Institute of Medical Education and Research, Chandigarh, under registration no. 47/GO/R/SL/Bi/S/99/CPCSEA, and the experiments were performed under biosafety level -2 conditions.
Histopathology.
Histological analysis of representative kidney tissue was performed to confirm the infection (Fig. S4) as per the method described by Balish et al. (41). The kidneys were aseptically removed from euthanized mice and fixed in 10% neutral-buffered formaldehyde. The tissues were then processed in graded alcohol (50%, 70%, 90%, and100%) and Xylene (70% and 100%) solutions and embedded in paraffin. Thin sections (∼5 μ) of tissues were cut, mounted on a slide, stained with Periodic acid-Schiff and counterstained with hematoxylin.
Gene expression analyses.
To quantify the transcript levels, primers were designed for sequences corresponding to FKS1 and putative chitin synthase gene (Chs1). Briefly, 1 × 106 cells were seeded into 10 ml YEPD broth and incubated for 8 h. Each isolate was then exposed to 2-fold lower sub-MIC caspofungin concentration for 4 h. The detailed method for total RNA extraction and RT-qPCR are provided in the supplementary file. Relative expression was calculated as fold change on exposure to caspofungin compared to untreated control, using the 2-ΔΔCt method (42).
Chitin content estimation.
A previously described flow cytometry-based method was used to measure the baseline and caspofungin-inducible cell wall chitin contents (43). Cell suspensions of each isolate in YEPD broth were exposed to respective 2-fold lower sub-MIC caspofungin concentrations, as described above. Cells were washed twice with PBS, and approximately 1 × 106 cells were treated with 2.5 mg/liter calcofluor white (CFW) for 15 min. Cells were washed and subjected to flowcytometry using BD FACS CantoTM II (Becton, Dickinson, San Jose, CA, USA). Mean intensity (MI) of stained and unstained cells were measured, and the staining index was calculated using the following equation:
Growth kinetics.
All the isolates listed in Table 2 with or without an FKS1 mutation were examined for differential growth rates using growth kinetics analysis. Approximately 1 × 106 cells were inoculated in 200 μl YEPD broth in a 96-well flat-bottom microtiter plate and incubated at 30°C for 24 h with continuous double –orbital shaking. Optical densities were recorded at 600 nm every 30 min in Epoch 2 Microplate Reader (Biotek, Agilent Technologies).
Statistical analyses.
Survival curves of mice following infection were plotted using Kaplan-Meier analysis, and kill kinetics were assessed using the log-rank test. The renal fungal burdens were compared using nonparametric Mann-Whitney U test for two-groups or Kruskal-Wallis test for more than two groups. Gene expression and chitin content were analyzed using ANOVA. P values <0.05 were considered significant. The data were analyzed in GraphPad Prism software version 6.0 (GraphPad, Inc., San Diego, CA).
ACKNOWLEDGMENTS
We acknowledge Ujjwayini Ray, Apollo Gleneagles Hospitals, Kolkata; Dr. Anjali Shetty, PD Hinduja National Hospital and Medical Research Centre, Hospital, Mumbai; Dr. Kamini Walia, ICMR, New Delhi; and all those who sent C. auris isolates to the nodal center as per the ICMR notification. We also acknowledge the technical support of Mr. Sourav Agnihotri in sequencing and technical guidance of Imran Ibni Gani Rather in conducting animal experiments.
This work was supported by the grant from Indian Council of Medical Research (ICMR), New Delhi, under project number AMR/149/2018-ECD-II. D.S. was supported by independent junior research fellowship grant 21/12/2014(II)EU-V from University Grants Commission, New Delhi.
The authors have no potential conflicts of interest. The funding agency did not participate or interfere in any stage of the study. All authors had full access to experimental data and take responsibility for its integrity and accuracy of the data analysis.
SMR, RAP, and AC designed the study. DS, RAP, and NK performed the experiments. RAP analyzed the data. RAP, DS, and SMR wrote the manuscript. All authors critically reviewed the article for important intellectual content.
Footnotes
Supplemental material is available online only.
Contributor Information
Shivaprakash M. Rudramurthy, Email: mrshivprakash@gmail.com.
Arunaloke Chakrabarti, Email: arunaloke@hotmail.com.
REFERENCES
- 1.Chakrabarti A, Sood P, Rudramurthy SM, Chen S, Kaur H, Capoor M, Chhina D, Rao R, Eshwara VK, Xess I, Kindo AJ, Umabala P, Savio J, Patel A, Ray U, Mohan S, Iyer R, Chander J, Arora A, Sardana R, Roy I, Appalaraju B, Sharma A, Shetty A, Khanna N, Marak R, Biswas S, Das S, Harish BN, Joshi S, Mendiratta D. 2015. Incidence, characteristics and outcome of ICU-acquired candidemia in India. Intensive Care Med 41:285–295. 10.1007/s00134-014-3603-2. [DOI] [PubMed] [Google Scholar]
- 2.Logan C, Martin-Loeches I, Bicanic T. 2020. Invasive candidiasis in critical care: challenges and future directions. Intensive Care Med 46:2001–2014. 10.1007/s00134-020-06240-x. [DOI] [PubMed] [Google Scholar]
- 3.Schelenz S, Hagen F, Rhodes JL, Abdolrasouli A, Chowdhary A, Hall A, Ryan L, Shackleton J, Trimlett R, Meis JF, Armstrong-James D, Fisher MC. 2016. First hospital outbreak of the globally emerging Candida auris in a European hospital. Antimicrob Resist Infect Control 5:1–7. 10.1186/s13756-016-0132-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Biswal M, Rudramurthy SM, Jain N, Shamanth AS, Sharma D, Jain K, Yaddanapudi LN, Chakrabarti A. 2017. Controlling a possible outbreak of Candida auris infection: lessons learnt from multiple interventions. J Hosp Infect 97:363–370. 10.1016/j.jhin.2017.09.009. [DOI] [PubMed] [Google Scholar]
- 5.Jeffery-Smith A, Taori SK, Schelenz S, Jeffery K, Johnson EM, Borman A, Manuel R, Brown CS, Candida auris Incident Management Team. 2018. Candida auris: a review of the literature. Clin Microbiol Rev 31:1–18. 10.1128/CMR.00029-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kean R, Brown J, Gulmez D, Ware A, Ramage G. 2020. Candida auris: a decade of understanding of an enigmatic pathogenic yeast. JoF 6:30. 10.3390/jof6010030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Satoh K, Makimura K, Hasumi Y, Nishiyama Y, Uchida K, Yamaguchi H. 2009. Candida auris sp. nov., a novel ascomycetous yeast isolated from the external ear canal of an inpatient in a Japanese hospital. Microbiol Immunol 53:41–44. 10.1111/j.1348-0421.2008.00083.x. [DOI] [PubMed] [Google Scholar]
- 8.Hu S, Zhu F, Jiang W, Wang Y, Quan Y, Zhang G, Gu F, Yang Y. 2021. Retrospective analysis of the clinical characteristics of Candida auris Infection worldwide from 2009 to 2020. Front Microbiol 12:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chowdhary A, Prakash A, Sharma C, Kordalewska M, Kumar A, Sarma S, Tarai B, Singh A, Upadhyaya G, Upadhyay S, Yadav P, Singh PK, Khillan V, Sachdeva N, Perlin DS, Meis JF. 2018. A multicentre study of antifungal susceptibility patterns among 350 Candida auris isolates (2009–17) in India: role of the ERG11 and FKS1 genes in azole and echinocandin resistance. J Antimicrob Chemother 73:891–899. 10.1093/jac/dkx480. [DOI] [PubMed] [Google Scholar]
- 10.Rudramurthy SM, Chakrabarti A, Paul RA, Sood P, Kaur H, Capoor MR, Kindo AJ, Marak RSK, Arora A, Sardana R, Das S, Chhina D, Patel A, Xess I, Tarai B, Singh P, Ghosh A. 2017. Candida auris candidaemia in Indian ICUs: analysis of risk factors. J Antimicrob Chemother 72:1794–1801. 10.1093/jac/dkx034. [DOI] [PubMed] [Google Scholar]
- 11.Escandón P, Chow NA, Caceres DH, Gade L, Berkow EL, Armstrong P, Rivera S, Misas E, Duarte C, Moulton-Meissner H, Welsh RM, Parra C, Pescador LA, Villalobos N, Salcedo S, Berrio I, Varón C, Espinosa-Bode A, Lockhart SR, Jackson BR, Litvintseva AP, Beltran M, Chiller TM. 2019. Molecular epidemiology of candida auris in Colombia Reveals a highly related, countrywide colonization with regional patterns in amphotericin B resistance. Clin Infect Dis 68:15–21. 10.1093/cid/ciy411. [DOI] [PubMed] [Google Scholar]
- 12.Arendrup MC, Boekhout T, Akova M, Meis JF, Cornely OA, Lortholary O, Arikan-Akdagli S, Cuenca-Estrella M, Dannaoui E, van Diepeningen AD, Groll AH, Guarro J, Guinea J, Hope W, Lackner M, Lass-Flörl C, Lagrou K, Lanternier F, Meletiadis J, Munoz P, Pagano L, Richardson MD, Roilides E, Tortorano AM, Ullmann AJ. 2014. ESCMID and ECMM joint clinical guidelines for the diagnosis and management of rare invasive yeast infections. Clin Microbiol Infect 20:76–98. 10.1111/1469-0691.12360. [DOI] [PubMed] [Google Scholar]
- 13.Pappas PG, Kauffman CA, Andes DR, Clancy CJ, Marr KA, Ostrosky-Zeichner L, Reboli AC, Schuster MG, Vazquez JA, Walsh TJ, Zaoutis TE, Sobel JD. 2016. Clinical practice guideline for the management of candidiasis: 2016 update by the Infectious Diseases Society of America. Clin Infect Dis 62:409–417. 10.1093/cid/civ1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Katiyar SK, Alastruey-Izquierdo A, Healey KR, Johnson ME, Perlin DS, Edlind TD. 2012. Fks1 and Fks2 are functionally redundant but differentially regulated in candida glabrata: implications for echinocandin resistance. Antimicrob Agents Chemother 56:6304–6309. 10.1128/AAC.00813-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rudramurthy SM, Colley T, Abdolrasouli A, Ashman J, Dhaliwal M, Kaur H, Armstrong-James D, Strong P, Rapeport G, Schelenz S, Ito K, Chakrabarti A. 2019. In vitro antifungal activity of a novel topical triazole PC945 against emerging yeast Candida auris. J Antimicrob Chemother 74:2943–2949. 10.1093/jac/dkz280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ramachandran R, Shrivastava M, Narayanan NN, Thakur RL, Chakrabarti A, Roy U. 2018. Evaluation of antifungal efficacy of three new cyclic lipopeptides of the class bacillomycin from bacillus subtilis RLID 12.1. Antimicrob Agents Chemother 62:e01457-17. 10.1128/AAC.01457-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kordalewska M, Lee A, Park S, Berrio I, Chowdhary A, Zhao Y, Perlin DS. 2018. Understanding echinocandin resistance in the emerging pathogen candida auris. Antimicrob Agents Chemother 62:1–9. 10.1128/AAC.00238-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Park S, Kelly R, Kahn JN, Robles J, Hsu MJ, Register E, Li W, Vyas V, Fan H, Abruzzo G, Flattery A, Gill C, Chrebet G, Parent SA, Kurtz M, Teppler H, Douglas CM, Perlin DS. 2005. Specific substitutions in the echinocandin target Fks1p account for reduced susceptibility of rare laboratory and clinical Candida sp. isolates. Antimicrob Agents Chemother 49:3264–3273. 10.1128/AAC.49.8.3264-3273.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ben-Ami R, Garcia-Effron G, Lewis RE, Gamarra S, Leventakos K, Perlin DS, Kontoyiannis DP. 2011. Fitness and virulence costs of Candida albicans FKS1 hot spot mutations associated with echinocandin resistance. J Infect Dis 204:626–635. 10.1093/infdis/jir351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Arendrup MC, Prakash A, Meletiadis J, Sharma C, Chowdhary A. 2017. Comparison of EUCAST and CLSI reference microdilution MICs of eight antifungal compounds for Candida auris and associated tentative epidemiological cutoff values. Antimicrob Agents Chemother 61:1–10. 10.1128/AAC.00485-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Spivak ES, Hanson KE. 2018. Candida auris: an emerging fungal pathogen. J Clin Microbiol 56:e01588-17. 10.1128/JCM.01588-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Centers for Disease Control and Prevention. 2020. Antifungal susceptibility testing and interpretation: Candida auris. CDC, Atlanta, GA. https://www.cdc.gov/fungal/candida-auris/c-auris-antifungal.html. [Google Scholar]
- 23.Lockhart SR, Etienne KA, Vallabhaneni S, Farooqi J, Chowdhary A, Govender NP, Colombo AL, Calvo B, Cuomo CA, Desjardins CA, Berkow EL, Castanheira M, Magobo RE, Jabeen K, Asghar RJ, Meis JF, Jackson B, Chiller T, Litvintseva AP. 2017. Simultaneous emergence of multidrug-resistant Candida auris on 3 continents confirmed by whole-genome sequencing and epidemiological analyses. Clin Infect Dis 64:134–140. 10.1093/cid/ciw691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eyre DW, Sheppard AE, Madder H, Moir I, Moroney R, Quan TP, Griffiths D, George S, Butcher L, Morgan M, Newnham R, Sunderland M, Clarke T, Foster D, Hoffman P, Borman AM, Johnson EM, Moore G, Brown CS, Walker AS, Peto TEA, Crook DW, Jeffery KJM. 2018. A Candida auris outbreak and its control in an intensive care setting. N Engl J Med 379:1322–1331. 10.1056/NEJMoa1714373. [DOI] [PubMed] [Google Scholar]
- 25.Zhu Y, O’Brien B, Leach L, Clarke A, Bates M, Adams E, Ostrowsky B, Quinn M, Dufort E, Southwick K, Erazo R, Haley VB, Bucher C, Chaturvedi V, Limberger RJ, Blog D, Lutterloh E, Chaturvedi S. 2020. laboratory analysis of an outbreak of Candida auris in New York from 2016 to 2018: impact and lessons learned. J Clin Microbiol 58:01503–01519. 10.1128/JCM.01503-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Beekman CN, Ene IV. 2020. Short-term evolution strategies for host adaptation and drug escape in human fungal pathogens. PLoS Pathog 16:e1008519. 10.1371/journal.ppat.1008519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Beyda ND, John J, Kilic A, Alam MJ, Lasco TM, Garey KW. 2014. FKS mutant Candida glabrata: risk factors and outcomes in patients with candidemia. Clin Infect Dis 59:819–825. 10.1093/cid/ciu407. [DOI] [PubMed] [Google Scholar]
- 28.Chow NA, Muñoz JF, Gade L, Berkow EL, Li X, Welsh RM, Forsberg K, Lockhart SR, Adam R, Alanio A, Alastruey-Izquierdo A, Althawadi S, Araúz AB, Ben-Ami R, Bharat A, Calvo B, Desnos-Ollivier M, Escandón P, Gardam D, Gunturu R, Heath CH, Kurzai O, Martin R, Litvintseva AP, Cuomo CA. 2020. Tracing the evolutionary history and global expansion of Candida auris using population genomic analyses. mBio 11:e03364-19. 10.1128/mBio.03364-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Berkow EL, Lockhart SR. 2018. Activity of CD101, a long-acting echinocandin, against clinical isolates of Candida auris. Diagn Microbiol Infect Dis 90:196–197. 10.1016/j.diagmicrobio.2017.10.021. [DOI] [PubMed] [Google Scholar]
- 30.Carolus H, Pierson S, Muñoz JF, Subotić A, Cruz RB, Cuomo CA, Van Dijck P. 2021. Genome-wide analysis of experimentally evolved candida auris reveals multiple novel mechanisms of multidrug resistance. mBio 12:1–19. 10.1128/mBio.03333-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Katiyar S, Pfaller M, Edlind T. 2006. Candida albicans and Candida glabrata clinical isolates exhibiting reduced echinocandin susceptibility. Antimicrob Agents Chemother 50:2892–2894. 10.1128/AAC.00349-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Perlin DS. 2011. Current perspectives on echinocandin class drugs. Future Microbiol 6:441–457. 10.2217/fmb.11.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Walker L.a, Munro C.a, de Bruijn I, Lenardon MD, McKinnon A, Gow NAR. 2008. Stimulation of chitin synthesis rescues Candida albicans from echinocandins. PLoS Pathog 4:e1000040. 10.1371/journal.ppat.1000040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Imtiaz T, Lee KK, Munro CA, MacCallum DM, Shankland GS, Johnson EM, MacGregor MS, Bal AM. 2012. Echinocandin resistance due to simultaneous FKS mutation and increased cell wall chitin in a Candida albicans bloodstream isolate following brief exposure to caspofungin. J Med Microbiol 61:1330–1334. 10.1099/jmm.0.045047-0. [DOI] [PubMed] [Google Scholar]
- 35.Perlin DS. 2015. Echinocandin resistance in Candida. Clin Infect Dis 61:S612–S617. 10.1093/cid/civ791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wring S, Borroto-Esoda K, Solon E, Angulo D. 2019. SCY-078, a novel fungicidal agent, demonstrates distribution to tissues associated with fungal infections during mass balance studies with intravenous and oral [14C]SCY-078 in albino and pigmented rats. Antimicrob Agents Chemother 63:e02119-19. 10.1128/AAC.02119-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ghosh AK, Paul S, Sood P, Rudramurthy SM, Rajbanshi A, Jillwin TJ, Chakrabarti A. 2015. Matrix-assisted laser desorption ionization time-of-flight mass spectrometry for the rapid identification of yeasts causing bloodstream infections. Clin Microbiol Infect 21:372–378. 10.1016/j.cmi.2014.11.009. [DOI] [PubMed] [Google Scholar]
- 38.Clinical Laboratory Standards Institute. 2017. Reference Method for Broth Dilution Antifungal susceptibility testing of yeast. Clin Lab Stand Inst 32:1–23. [Google Scholar]
- 39.Turenne CY, Sanche SE, Hoban DJ, Karlowsky JA, Kabani AM. 1999. Rapid Identification of fungi by using the ITS2 genetic region and an automated fluorescent capillary electrophoresis system. J Clin Microbiol 37:1846–1851. 10.1128/JCM.37.6.1846-1851.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Charan J, Kantharia N. 2013. How to calculate sample size in animal studies? J Pharmacol Pharmacother 4:303–306. 10.4103/0976-500X.119726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Balish E, Warner T, Pierson CJ, Bock DM, Wagner RD. 2001. Oroesophageal candidiasis is lethal for transgenic mice with combined natural killer and T-cell defects. Med Mycol 39:261–268. 10.1080/mmy.39.3.261.268. [DOI] [PubMed] [Google Scholar]
- 42.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25:402–408. 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 43.Costa-de-Oliveira S, Silva AP, Miranda IM, Salvador A, Azevedo MM, Munro CA, Rodrigues AG, Pina-Vaz C. 2013. Determination of chitin content in fungal cell wall: an alternative flow cytometric method. Cytometry 83A:324–328. 10.1002/cyto.a.22250. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material. Download AAC.01652-21-s0001.pdf, PDF file, 0.8 MB (860.5KB, pdf)