ABSTRACT
The failure of antibiotic therapy in respiratory tract infections in cystic fibrosis is partly due to the high tolerance observed in Pseudomonas aeruginosa biofilms. This tolerance is mediated by changes in bacterial metabolism linked to growth in biofilms, opening up potential avenues for novel treatment approaches based on modulating metabolism. The goal of the present study was to identify carbon sources that increase the inhibiting and/or eradicating activity of tobramycin, ciprofloxacin, and ceftazidime against P. aeruginosa PAO1 biofilms grown in a synthetic cystic fibrosis sputum medium (SCFM2) and to elucidate their mode of action. After screening 69 carbon sources, several combinations of antibiotics + carbon sources that showed markedly higher anti-biofilm activity than antibiotics alone were identified. d,l-malic acid and sodium acetate could potentiate both biofilm inhibiting and eradicating activity of ciprofloxacin and ceftazidime, respectively, while citric acid could only potentiate biofilm inhibitory activity of tobramycin. The mechanisms underlying the increased biofilm eradicating activity of combinations ciprofloxacin/d,l-malic acid and ceftazidime/sodium acetate are similar but not identical. Potentiation of ceftazidime activity by sodium acetate was linked to increased metabolic activity, a functional TCA cycle, increased ROS production, and high intracellular pH, whereas the latter was not required for d,l-malic acid potentiation of ciprofloxacin. Finally, our results indicate that the potentiation of antibiotic activity by carbon sources is strain dependent.
KEYWORDS: Pseudomonas aeruginosa, biofilms, cystic fibrosis
INTRODUCTION
Treatment of bacterial infections has become challenging as microbial susceptibility to antibiotics has been eroded by increasing levels of resistance as well as by tolerance associated with biofilm formation (1–3). This is particularly relevant for people with cystic fibrosis (CF) who are at an increased risk for biofilm-associated respiratory tract infections (4–6). Pseudomonas aeruginosa is the most important pulmonary pathogen in CF and contributes significantly to morbidity and mortality (7, 8). Approximately 53% of children with CF younger than 5 years old are already infected with P. aeruginosa (9, 10), and this increases to 75% in adult CF patients (11, 12).
There is increasing evidence that metabolic activity plays an important role in the antimicrobial susceptibility of bacteria (13), and while searching for innovative treatments for P. aeruginosa infections in CF patients, several studies have investigated the use of particular carbon sources to potentiate antibiotic activity. For example, mannitol increased tobramycin sensitivity of P. aeruginosa persister cells up to 1,000-fold (14), while glucose, mannitol, acetate, fumarate, succinate, α-ketoglutarate, pyruvate, and gluconate increased the efficacy of tobramycin against stationary-phase P. aeruginosa cells (15). Fumarate potentiated the activity of tobramycin, allowing complete eradication of P. aeruginosa biofilms (16).
The role of metabolism in antibiotic activity has also been investigated in other organisms. Fructose was reported to sensitize Edwardsiella tarda persisters and biofilms through activating the tricarboxylic acid (TCA) cycle, which increased NADPH production, the proton motive force (PMF), and kanamycin uptake (17). In biofilms formed by Burkholderia cepacia complex (Bcc) bacteria, persister cells that survived treatment with very high tobramycin concentrations downregulated the TCA cycle to avoid the production of reactive oxygen species (ROS) and at the same time activated an alternative pathway, the glyoxylate shunt (18).
In the present study we performed a large-scale screening for carbon sources that could potentiate the activity of antibiotics belonging to different classes against P. aeruginosa biofilms formed in synthetic cystic fibrosis sputum medium 2 (SCFM2), a defined medium mimicking several essential aspects of CF sputum (19–21). For the most promising combinations, we investigated the mechanism behind the potentiating activity.
RESULTS AND DISCUSSION
Screening of carbon sources for potentiating biofilm inhibition by antibiotics.
Based on previous studies about potentiating antibiotic activity by adding carbon sources (16, 22–24), we compiled a list of 69 carbon sources from different classes and determined their potentiating activity against P. aeruginosa PAO1 biofilms. First, we confirmed that P. aeruginosa PAO1 formed biofilm aggregates in SCFM2 in the conditions used in the present study; after 24 h densities had reached approximately 3.2 × 109 CFU/ml and aggregates could be observed (Fig. S1). The MBIC, defined as the lowest concentration of antibiotic inhibiting biofilm growth by at least 90%, of tobramycin, ciprofloxacin, and ceftazidime for P. aeruginosa PAO1 in SCFM2 was 5.0, 0.6, and 3.13 μg/mL, respectively.
We subsequently evaluated the effect of 69 carbon sources on the antimicrobial activity of these three antibiotics (used in a concentration of 0.5 × MBIC) against P. aeruginosa PAO1 biofilms (Fig. S2 to S7). Overall, there were considerable differences between carbon sources (e.g., most of the amino acids stimulated biofilm formation of P. aeruginosa PAO1 in the presence of the three antibiotics while l-serine and l-glutamine inhibited the biofilm formation) and between antibiotics (e.g., activity of ceftazidime but not tobramycin or ciprofloxacin is potentiated by d,l-malic acid). Several combinations caused an additional reduction of at least 30% in the log value of the number of CFU compared to treatment with the antibiotic alone. These carbon sources include 4-methoxycarbonylphenylboronic acid, citric acid, D-(+)-glucosamine hydrochloride, and sodium acetate for tobramycin; d,l-malic acid and sodium succinate dibasic hexahydrate for ciprofloxacin; and D-(-)-iso ascorbic acid, sodium acetate and d-serine for ceftazidime.
The combinations tobramycin/citric acid, ciprofloxacin/d,l-malic acid, and ceftazidime/sodium acetate were selected for more in-depth studies. These carbon sources led to an additional biofilm inhibition of 3.5 log units (tobramycin/citric acid), 1.8 log units (ciprofloxacin/d,l-malic acid), and 2.5 log units (ceftazidime/sodium acetate) (Fig. 1). In order to confirm that the increased biofilm inhibition was due to potentiation of the antibiotic activity and not due to an antimicrobial effect of the carbon source itself, P. aeruginosa PAO1 biofilm formation was also determined in SCFM2 supplemented with the carbon source alone; no effect could be observed (Fig. S8). In addition, as the pH of the supplemented media was adjusted to 6.8 prior to all experiments, pH-related effects on biofilm formation can be ruled out.
FIG 1.
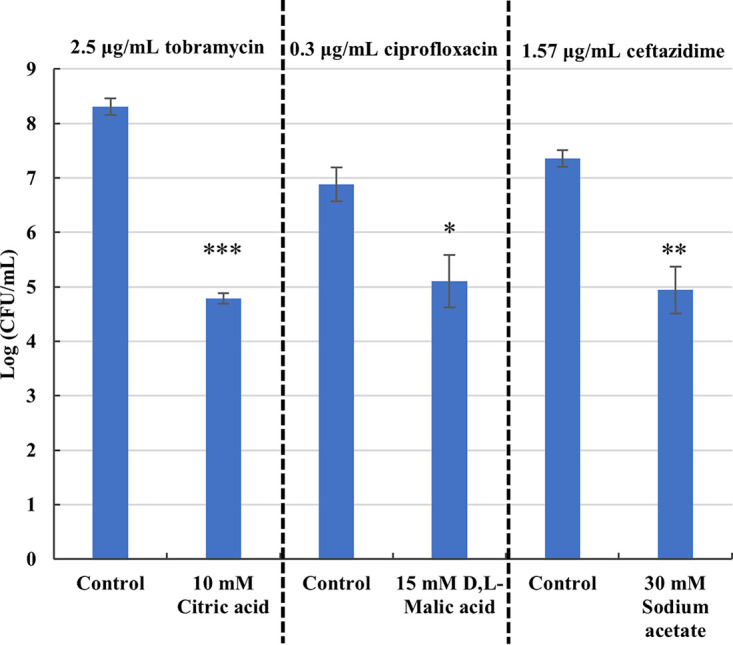
Effect of the selected carbon sources on P. aeruginosa PAO1 biofilm inhibition in SCFM2 by 2.5 μg/mL tobramycin, 0.3 μg/mL ciprofloxacin and 1.56 μg/mL ceftazidime. Values shown are the mean (n ≥ 3). Error bars indicate standard error. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Checkerboard assays with the most effective combinations to assess biofilm eradication.
We conducted a checkerboard assay on 24 h old P. aeruginosa PAO1 biofilms to assess whether effective combinations of carbon source and antibiotic can be found that result in more efficient biofilm eradication. The combination 60 mM d,l-malic acid and 1.2 μg/ml ciprofloxacin significantly reduced the number of CFU from 6.86 to 5.28 log CFU (P < 0.0001) (Fig. 2A). Meanwhile, 60 mM sodium acetate significantly reduced the log CFU of biofilms treated with 12.48 μg/ml ceftazidime from 8.40 to 7.70 (P < 0.05). Similarly, the combination of 120 mM sodium acetate with 6.24 or 12.48 μg/ml ceftazidime significantly reduced the log CFU from 8.36 and 8.40 to 7.26 (P < 0.01) and 6.76 (P < 0.001), respectively (Fig. 2B). For the combination of tobramycin/citric acid no effective concentrations were found that eradicate 24 h old P. aeruginosa PAO1 biofilms in SCFM2 (Fig. S9). For subsequent experiments, the combinations of 12.48 μg/ml ceftazidime/120 mM sodium acetate and 1.2 μg/ml ciprofloxacin/60 mM d,l-malic acid were used.
FIG 2.
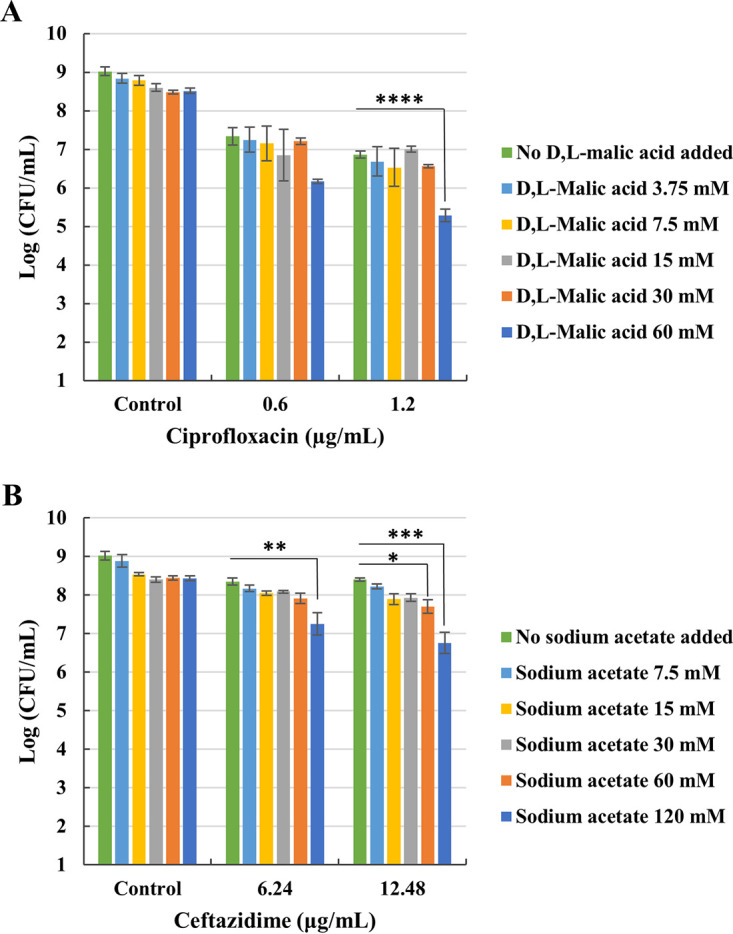
Effect of different combinations of (A) ciprofloxacin/d,l-malic acid and (B) ceftazidime/sodium acetate on eradication of P. aeruginosa PAO1 biofilms. Values shown are the mean (n ≥ 3). Error bars indicate standard error. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
Addition of carbon sources increase metabolic activity of P. aeruginosa PAO1 biofilms.
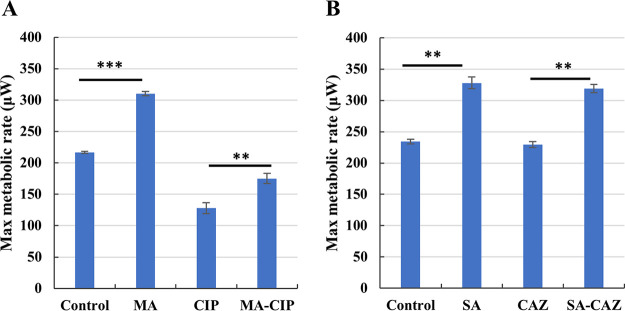
To assess the effect of adding carbon sources on metabolism during biofilm eradication, we used isothermal microcalorimetry. Both 120 mM sodium acetate and 60 mM d,l-malic acid significantly increased the maximum metabolic rate of P. aeruginosa PAO1 biofilms (Fig. 3). Maximum metabolic rates of P. aeruginosa PAO1 biofilms treated with combination of ceftazidime/sodium acetate or ciprofloxacin/d,l-malic acid were significantly higher than biofilms treated with antibiotic alone (Fig. 3). Downregulation of metabolism has previously been observed as a strategy to increase antimicrobial tolerance in bacteria like Mycobacterium tuberculosis (25) and Bcc bacteria (18), and we propose that the increased metabolic activity observed contributes to reduced bacterial tolerance, and more killing in P. aeruginosa PAO1 biofilms. A systematic investigation of the influence of carbon sources with varying degrees of potentiating activity on these metabolic parameters would be required to confirm this hypothesis.
FIG 3.
Maximum metabolic rate of treated and untreated (control) P. aeruginosa PAO1 biofilms. (A) MA: 60 mM d,l-malic acid; CIP: 1.2 μg/ml ciprofloxacin; MA-CIP: 60 mM d,l-malic acid combined with 1.2 μg/ml ciprofloxacin. (B) SA: 120 mM sodium acetate; CAZ: 12.48 μg/ml ceftazidime; SA-CAZ: combination of 120 mM sodium acetate and 12.48 μg/ml ceftazidime. Values shown are the mean (n ≥ 3). Error bars indicate standard error, **, P < 0.01; ***, P < 0.001.
Contribution of SDH, PMF and intracellular pH to the potentiation of biofilm eradicating activity.
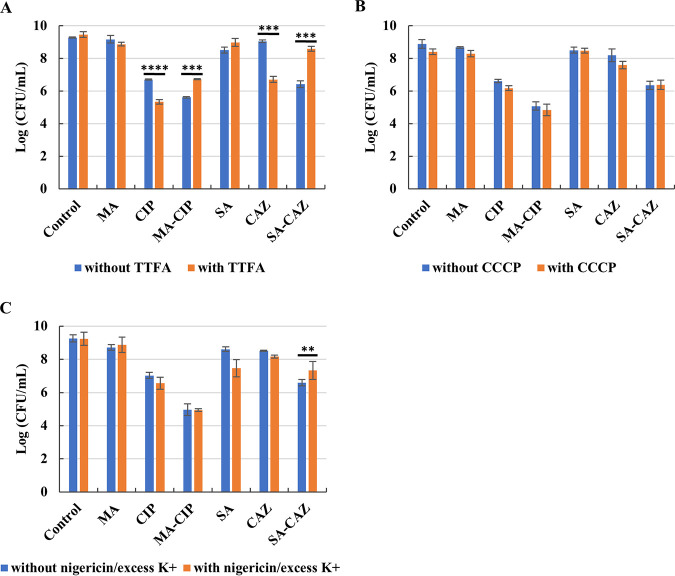
Recent studies have shown that potentiation of antimicrobial activity by adding carbon source is mainly due to increased respiration, and especially increased TCA cycle activity has been linked to increased antibiotic activity (26, 27). To investigate the mechanism underlying the carbon source potentiation of antibiotics in biofilm eradication, the anti-biofilm effect of the antibiotic alone and the combination was investigated in the presence of a SDH inhibitor, a PMF uncoupler, and the K+ ionophore nigericin combined with excess K+.
SDH is responsible for the oxidation of succinate to fumarate in the TCA cycle thereby generating FADH2 (28). Our results show that when ciprofloxacin or ceftazidime were used alone, SDH inhibition increased their antimicrobial activity (P < 0.0001 for ciprofloxacin, P < 0.001 for ceftazidime, Fig. 4A). However, SDH inhibition decreased the antimicrobial activity of the combination of ciprofloxacin and d,l-malic acid (P < 0.001). Similarly, SDH inhibition decreased the antimicrobial activity of the combination of ceftazidime and sodium acetate (P = 0.001) as well. These data suggest that SDH contributes to the potentiating effect of d,l-malic acid on ciprofloxacin and that of sodium acetate on ceftazidime. These data further suggest that a functional TCA cycle was necessary for the potentiation effect of both d,l-malic acid and sodium acetate.
FIG 4.
Evaluation of the role of SDH, PMF and intracellular pH on the potentiating effect of additional carbon source on antibiotic during eradication of P. aeruginosa PAO1 biofilms. 24-h-old-biofilm growth treated by antibiotic (or not) in SCFM2 supplemented with or without additional carbon source in the presence or absence of (A) the SDH inhibitor TTFA, (B) the proton motive force disruptor CCCP, (C) the K+ ionophore nigericin and excess levels of K+. MA: 60 mM d,l-malic acid; CIP: 1.2 μg/ml ciprofloxacin; MA-CIP: 60 mM d,l-malic acid combined with 1.2 μg/ml ciprofloxacin; SA: 120 mM sodium acetate; CAZ: 12.48 μg/ml ceftazidime; SA-CAZ: combination of 120 mM sodium acetate and 12.48 μg/ml ceftazidime. Values shown are the mean (n ≥ 3). Error bars indicate standard error, **, P < 0.01; ***, P ≤ 0.001; ****, P < 0.0001.
The PMF uncoupler CCCP had no effect on the biofilm eradicating activity of antibiotics alone or combinations of antibiotic + carbon source, suggesting that the PMF is not involved in the potentiating activity (Fig. 4B).
Finally, equalizing the intra- and extracellular pH levels by adding nigericin and excess K+ led to a loss of potentiating activity for sodium acetate in the combination ceftazidime/sodium acetate, without affecting the potentiating effect of d,l-malic acid in the combination ciprofloxacin/d,l-malic acid (Fig. 4C). Nigericin transports K+ across biological membranes (in exchange for H+), leading to a decrease in intracellular K+ concentration and an increased intracellular H+ concentration (29, 30), and the decreased intracellular pH caused by nigericin/excess K+ was (at least partially) responsible for the loss of potentiating activity for sodium acetate in the combination ceftazidime/sodium acetate.
Collectively, these data indicate that sodium acetate potentiation of ceftazidime requires a fully functional TCA cycle and high intracellular pH, while the latter is not required for d,l-malic acid potentiation of ciprofloxacin.
Role of ROS production in the potentiation of biofilm eradicating activity.
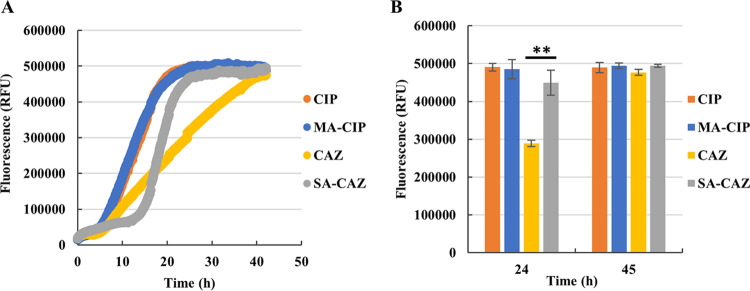
Production of ROS is an important mediator in killing bacteria by bactericidal antibiotics (31–34). ROS production was quantified by measuring fluorescence that resulted from H2DCFDA (Fig. 5). While fluorescence reached similar maximum levels for all treatments, the shape of the curves was markedly different between ceftazidime and ceftazidime + sodium acetate, with the latter curve reaching maximum fluorescence much earlier. The curves for ciprofloxacin and ciprofloxacin + d,l-malic acid were virtually identical. These data suggest that increased ROS production at earlier stages during ceftazidime + sodium acetate (but not ciprofloxacin + d,l-malic acid) treatment could help explain the observed potentiation.
FIG 5.
ROS-induced fluorescence in P. aeruginosa PAO1 biofilms. (A) Fluorescence generated over time. (B) Fluorescence at 24 h and 45 h. CIP: 1.2 μg/ml ciprofloxacin; MA-CIP: 60 mM d,l-malic acid combine with 1.2 μg/ml ciprofloxacin; CAZ: 12.48 μg/ml ceftazidime; SA-CAZ: combination of 120 mM sodium acetate and 12.48 μg/ml ceftazidime. Values shown are the mean (n = 3). Error bars indicate standard error, **, P < 0.01.
Biofilm eradication of selected combinations on other P. aeruginosa strains.
Finally, we assessed potentiation of the biofilm eradicating activity of ciprofloxacin and ceftazidime by d,l-malic acid and sodium acetate in biofilms formed by other P. aeruginosa strains, i.e., LES B58, AA2, AA44, and DK2 (Fig. 6). However, no significant differences were observed between LES B58, AA2, or AA44 biofilms treated with antibiotic alone or the combination. For P. aeruginosa DK2 biofilms, addition of 60 mM malic acid increased biofilm eradication by 1.2 μg/ml ciprofloxacin (1.1 log units), but this difference was not significant (P = 0.258). In previous work it had already been observed that the potentiating effect of mannitol on tobramycin activity against biofilms could be observed in P. aeruginosa strains PAO1 and FRD1, but not in strain 18 A, another strain isolated from a CF patient (14). These data suggest that potentiation of the biofilm eradicating activity of antibiotics is strain dependent, and it remains to be seen whether more active combinations can be identified for these strains. The mechanisms behind the observed strain-dependent effects are currently unclear. Our observations again highlight the need of using relevant strains when evaluating novel antimicrobial treatments.
FIG 6.
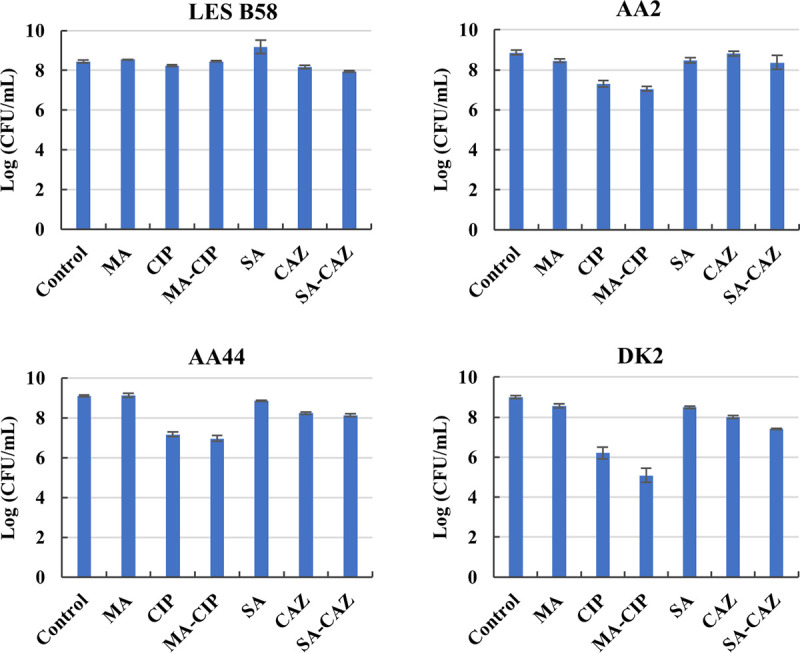
Eradication of biofilms formed by P. aeruginosa strains LES B58, AA2, AA44, and DK2. Control: no carbon source, no antibiotic; MA: 60 mM d,l-malic acid; CIP: 1.2 μg/ml ciprofloxacin; MA-CIP: 60 mM d,l-malic acid + 1.2 μg/ml ciprofloxacin; SA: 120 mM sodium acetate; CAZ: 12.48 μg/ml ceftazidime; SA-CAZ: 120 mM sodium acetate + 12.48 μg/ml ceftazidime. Values shown are the mean (n = 3). Error bars indicate standard error.
Our results show that several carbon sources, mainly organic acids and organic acid salts, can potentiate the biofilm inhibiting and/or eradicating activity of several antibiotics against P. aeruginosa PAO1 biofilms grown in SCFM2. d,l-malic acid and sodium acetate potentiated both biofilm inhibiting and eradicating activity of ciprofloxacin and ceftazidime, respectively, while citric acid only potentiated biofilm inhibitory activity of tobramycin. We found that the mechanisms behind the increased activity behind the combinations of ceftazidime/sodium acetate and ciprofloxacin/d,l-malic acid are distinct. A functional TCA cycle, high intracellular pH, and increased ROS production played a role in the combination ceftazidime/sodium acetate, whereas d,l-malic acid potentiation of ciprofloxacin mainly relied on increased metabolic activity and TCA cycle activity. Our data indicate that the potentiation of anti-biofilm activity of antibiotics by carbon sources is strain dependent; whether a better coverage could be obtained by simultaneously combining antibiotics with different carbon sources remains to be investigated.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Overnight cultures P. aeruginosa PAO1, LES B58, AA2, AA44. and DK2 (35) were routinely prepared by inoculating from −80°C frozen stock into Luria Bertani broth (LB; Lab M, Moss Hall, UK). Pure cultures and serially 10-fold diluted P. aeruginosa solutions were cultured on tryptic soy agar (TSA; Lab M, Moss Hall, UK).
Chemicals.
Carbon sources used and their suppliers are listed in Table S1. Ciprofloxacin, 4,4,4-trifluoro-1-(2-thienyl)-1,3-butanedione (TTFA), carbonyl cyanide 3-chlorophenylhydrazone (CCCP), 2′,7′-dichlorodihydrofluorescein diacetate (H2DCFDA), and hydrogen peroxide solution were obtained from Sigma-Aldrich (Bornem, Belgium). Tobramycin and ceftazidime were obtained from TCI Europe (Zwijndrecht, Belgium). Nigericin and KCl were obtained from Thermo Fischer Scientific (Waltham, USA) and VWR Chemicals (Leuven, Belgium), respectively.
Biofilm formation in SCFM2.
SCFM2 was prepared as described previously with minor modifications (20). Briefly, mucin was autoclaved at 121°C for 20 min prior to adding buffered base instead of being sterilized by UV. The final pH value of SCFM2 was adjusted to 6.8 using NaOH (1 M solution) before use. The optical density at 590 nm (OD590) of a P. aeruginosa overnight culture was measured in LB broth and diluted in SCFM2 to a density of approximately 5 × 105 CFU/mL, i.e., CFU/mL. Two hundred μl of the diluted cultures in SCFM2 was transferred to a flat-bottomed 96-well plate. After 0 h, 1 h, 2 h, 4 h, 6 h, 12 h, 24 h, 48 h, and 72 h of incubation at 37°C, the entire content of the well was removed and the number of CFU in the biofilms were quantified by making a 10-fold serial dilution series and plating (36). All experiments were performed in biological triplicates (with three technical replicates in each biological replicate, i.e., n = 3 × 3). Formation of biofilm aggregates was confirmed by light microscopy after transferring 10 μl to a microscope slide (EVOS FL Auto Cell Imaging System, Thermo Fischer Scientific, Waltham, USA).
Determination of minimum biofilm inhibitory concentration (MBIC).
The MBICs of tobramycin, ciprofloxacin, and ceftazidime in SCFM2 were determined by plate counts. Twofold dilution series of antibiotics were prepared in SCFM2. Concentrations tested ranged from 0.125 to 32 μg/mL (tobramycin), from 0.125 to 128 μg/mL (ciprofloxacin) and from 0.125 to 256 μg/mL (ceftazidime). An P. aeruginosa overnight culture was diluted in SCFM2 to prepare an inoculum suspension containing approximately 5 × 105 CFU/mL. This suspension was added to flat bottom 96-well plates (100 μl/well) together with serial antibiotic dilutions (100 μl/well) and was incubated at 37°C for 24 h. The MBIC was defined as the lowest concentration of antibiotic that inhibited biofilm growth by at least 90% compared to the untreated control (37). All experiments were performed in biological triplicates (with three technical replicates in each biological replicate, i.e., n = 3 × 3).
Screening of carbon sources for potentiating the biofilm inhibitory activity of antibiotics.
The effect of 69 different carbon sources (Table S1) on the potentiation of antibiotic activity during P. aeruginosa PAO1 biofilm formation was evaluated by exposing the diluted P. aeruginosa PAO1 overnight culture (approximately 5 × 105 CFU/mL) to 0.5 × MBIC of each antibiotic either alone or together with an additional carbon source (delivering 60 mM carbon) in SCFM2 (15). The concentration of 0.5 × MBIC of each antibiotic was used as it has an effect on growth of P. aeruginosa in biofilm aggregates without completely inhibiting it, allowing to identify carbon sources that potentiated the effect of the antibiotic. Following 24 h of incubation at 37°C, plate counting was used to determine the number of CFU. Each antibiotic/carbon source combination was tested in technical triplicates.
Identification of effective combinations for biofilm eradication.
In order to determine whether a selection of carbon sources could increase the biofilm eradicating activity, antibiotics checkerboard assays were conducted. We challenged 24 h old P. aeruginosa PAO1 biofilms (grown in SCFM2 as described above) with different antibiotics and the previously selected carbon sources, including citric acid, d,l- malic acid and sodium acetate, in concentrations normalized to deliver 0, 15, 30, 60, 120, and 240 mM carbon, in SCFM2. Concentrations of tobramycin, ciprofloxacin, and ceftazidime ranged from 0.625 to 40 μg/mL, from 0.075 to 2.4 μg/mL and from 0.195 to 24.96 μg/mL, respectively in 2-fold concentration gradients. After an additional 24 h of incubation at 37°C, cells were harvested after vortexing and sonicating (2 times, 5 min for each), and the number of CFU was determined by plating. Combinations were considered effective if they reduced the number of CFU by more than 90% compared to eradication obtained with the antibiotic alone.
Characterization of metabolic activity with isothermal microcalorimetry.
To measure the heat generated by the metabolic activity in biofilms, the calScreener microcalorimeter (Symcel, Stockholm, Sweden) was used. Biofilms were grown in plastic inserts that fit in the titanium cups of the calScreener and subsequently exposed to various combinations of carbon source and antibiotic. After starting the treatment, the plastic inserts were transferred into the titanium cups, which were placed in a 48-well plate and inserted in the instrument. The heat flow was measured at 37°C for 48 h and the resulting data were analyzed using the CalView software (Symcel).
Evaluation of the role of succinate dehydrogenase (SDH), the proton motive force (PMF), intracellular pH, and ROS production in potentiation of antibiotic activity.
To evaluate the role of SDH in the potentiating activity of selected carbon sources, biofilm eradication experiments were carried out as described above, but in the presence of 250 nM TTFA, an inhibitor of SDH (18). To confirm contribution of the PMF in potentiation of biofilm eradicating activity, 100 μM the proton ionophore uncoupler CCCP was added (29). To explore the importance of intracellular pH in potentiation of biofilm eradicating activity, the potassium ionophore nigericin (10 μM) and excess K+ (150 mM) were used, which are known to equilibrate intracellular and extracellular pH (29). Briefly, 24 h old biofilms were treated by the selected combinations and TTFA, CCCP, or nigericin/excess K+ for 24 h. The number of CFU was determined by plating.
ROS production was measured by staining with fluorescent reporter dye H2DCFDA (38). In every experiment the pH value was adjusted to 6.8 to avoid pH-related effects on the fluorescence of H2DCFDA. After 45 min staining by H2DCFDA (10 μM, 37°C, in the dark), 24 h old biofilms were washed with 0.9% NaCl and treated with 3% H2O2 (control), antibiotic alone, or selected antibiotic/carbon source combinations at 37°C. Fluorescence (λexc= 485 nm, λem= 535 nm) was determined using an Envision multilabel plate reader (Perkin Elmer, Waltham, MA, USA) at 37°C for 45 h. Autofluorescence of cells cultured without staining and uninoculated SCFM2 were taken into consideration when calculating the fluorescence. All experiments described above were performed in biological triplicates (with three technical replicates in each biological replicate, i.e., n = 3 × 3).
Biofilm eradication of selected combinations on P. aeruginosa clinically relevant strains.
The antibiotic potentiating activity of the selected carbon sources on the biofilm eradication of other P. aeruginosa strains was studied, including LES B58, AA2, AA44, and DK2. The same method as described above was used and all experiments were performed in biological triplicates (with three technical replicates in each biological replicate, i.e., n = 3 × 3).
Statistical analysis.
Statistical analysis was performed using SPSS statistical software version 27 (IBM, New York, USA). The normal distribution of the data was verified by the Shapiro-Wilk test. When the data were normally distributed, an independent sample t test or one-way ANOVA with Bonferroni correction was used to analyze the data. Data that were not normally distributed were analyzed by a Mann-Whitney test.
ACKNOWLEDGMENTS
This work was supported by the Chinese Scholarship Council (CSC, NO. 201806150021). We thank Symcel for providing us access to the calScreener and for their continued support.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Aedo SJ, Tang J, Brynildsen MP. 2021. Metabolites potentiate nitrofurans in nongrowing Escherichia coli. Antimicrob Agents Chemother 65:e00858-20. 10.1128/AAC.00858-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Windels EM, Michiels JE, Van den Bergh B, Fauvart M, Michiels J. 2019. Antibiotics: combatting tolerance to stop resistance. mBio 10:e02095-19. 10.1128/mBio.02095-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tuomanen E, Durack DT, Tomasz A. 1986. Antibiotic tolerance among clinical isolates of bacteria. Antimicrob Agents Chemother 30:521–527. 10.1128/AAC.30.4.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Heltshe SL, Cogen J, Ramos KJ, Goss CH. 2017. Cystic fibrosis: the dawn of a new therapeutic era. Am J Respir Crit Care Med 195:979–984. 10.1164/rccm.201606-1250PP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aali M, Caldwell A, House K, Zhou J, Chappe V, Lehmann C. 2017. Iron chelation as novel treatment for lung inflammation in cystic fibrosis. Med Hypotheses 104:86–88. 10.1016/j.mehy.2017.05.029. [DOI] [PubMed] [Google Scholar]
- 6.Harrington NE, Sweeney E, Harrison F. 2020. Building a better biofilm—formation of in vivo-like biofilm structures by Pseudomonas aeruginosa in a porcine model of cystic fibrosis lung infection. Biofilm 2:100024. 10.1016/j.bioflm.2020.100024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heirali A, Thornton C, Acosta N, Somayaji R, Laforest Lapointe I, Storey D, Rabin H, Waddell B, Rossi L, Arrieta MC, Surette M, Parkins MD. 2020. Sputum microbiota in adults with CF associates with response to inhaled tobramycin. Thorax 75:1058–1064. 10.1136/thoraxjnl-2019-214191. [DOI] [PubMed] [Google Scholar]
- 8.Retsch-Bogart GZ, Quittner AL, Gibson RL, Oermann CM, McCoy KS, Montgomery AB, Cooper PJ. 2009. Efficacy and safety of inhaled aztreonam lysine for airway pseudomonas in cystic fibrosis. Chest 135:1223–1232. 10.1378/chest.08-1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smith WD, Bardin E, Cameron L, Edmondson CL, Farrant KV, Martin I, Murphy RA, Soren O, Turnbull AR, Wierre-Gore N, Alton EW, Bundy JG, Bush A, Connett GJ, Faust SN, Filloux A, Freemont PS, Jones AL, Takats Z, Webb JS, Williams HD, Davies JC. 2017. Current and future therapies for Pseudomonas aeruginosa infection in patients with cystic fibrosis. FEMS Microbiol Lett 364. 10.1093/femsle/fnx121. [DOI] [PubMed] [Google Scholar]
- 10.Kidd TJ, Ramsay KA, Vidmar S, Carlin JB, Bell SC, Wainwright CE, Grimwood K, Francis PW, Dakin C, Cheney J, George N, Robertson CF, Moodie M, Carzino R, Carter R, Armstrong DS, Cooper PJ, McKay K, Martin AJ, Whitehead B, Hunter J, Byrnes CA, Tiddens HA, Graniel K, Gerbrands K, Mott L, ACFBAL Study Investigators. 2015. Pseudomonas aeruginosa genotypes acquired by children with cystic fibrosis by age 5-years. J Cyst Fibros 14:361–369. 10.1016/j.jcf.2014.12.007. [DOI] [PubMed] [Google Scholar]
- 11.Françoise A, Héry-Arnaud G. 2020. The microbiome in cystic fibrosis pulmonary disease. Genes (Basel) 11:536. 10.3390/genes11050536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sanders DB, Fink AK. 2016. Background and epidemiology. Pediatr Clin North Am 63:567–584. 10.1016/j.pcl.2016.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Crabbé A, Jensen PØ, Bjarnsholt T, Coenye T. 2019. Antimicrobial tolerance and metabolic adaptations in microbial biofilms. Trends Microbiol 27:850–863. 10.1016/j.tim.2019.05.003. [DOI] [PubMed] [Google Scholar]
- 14.Barraud N, Buson A, Jarolimek W, Rice SA. 2013. Mannitol enhances antibiotic sensitivity of persister bacteria in Pseudomonas aeruginosa biofilms. PLoS One 8:e84220. 10.1371/journal.pone.0084220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meylan S, Porter CBM, Yang JH, Belenky P, Gutierrez A, Lobritz MA, Park J, Kim SH, Moskowitz SM, Collins JJ. 2017. Carbon sources tune antibiotic susceptibility in Pseudomonas aeruginosa via tricarboxylic acid cycle control. Cell Chem Biol 24:195–206. 10.1016/j.chembiol.2016.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koeva M, Gutu AD, Hebert W, Wager JD, Yonker LM, O'Toole GA, Ausubel FM, Moskowitz SM, Joseph-McCarthy D. 2017. An antipersister strategy for treatment of chronic Pseudomonas aeruginosa infections. Antimicrob Agents Chemother 61:e00987-17. 10.1128/AAC.00987-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Su YB, Peng B, Han Y, Li H, Peng XX. 2015. Fructose restores susceptibility of multidrug-resistant Edwardsiella tarda to kanamycin. J Proteome Res 14:1612–1620. 10.1021/pr501285f. [DOI] [PubMed] [Google Scholar]
- 18.Van Acker H, Sass A, Bazzini S, De Roy K, Udine C, Messiaen T, Riccardi G, Boon N, Nelis HJ, Mahenthiralingam E, Coenye T. 2013. Biofilm-grown Burkholderia cepacia complex cells survive antibiotic treatment by avoiding production of reactive oxygen species. PLoS One 8:e58943. 10.1371/journal.pone.0058943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Palmer KL, Aye LM, Whiteley M. 2007. Nutritional cues control Pseudomonas aeruginosa multicellular behavior in cystic fibrosis sputum. J Bacteriol 189:8079–8087. 10.1128/JB.01138-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Turner KH, Wessel AK, Palmer GC, Murray JL, Whiteley M. 2015. Essential genome of Pseudomonas aeruginosa in cystic fibrosis sputum. Proc Natl Acad Sci USA 112:4110–4115. 10.1073/pnas.1419677112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Willsey GG, Eckstrom K, LaBauve AE, Hinkel LA, Schutz K, Meagher RJ, LiPuma JJ, Wargo MJ. 2019. Stenotrophomonas maltophilia differential gene expression in synthetic cystic fibrosis sputum reveals shared and cystic fibrosis strain-specific responses to the sputum environment. J Bacteriol 201:e00074-19. 10.1128/JB.00074-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Allison KR, Brynildsen MP, Collins JJ. 2011. Metabolite-enabled eradication of bacterial persisters by aminoglycosides. Nature 473:216–220. 10.1038/nature10069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang JH, Bhargava P, McCloskey D, Mao N, Palsson BO, Collins JJ. 2017. Antibiotic-induced changes to the host metabolic environment inhibit drug efficacy and alter immune function. Cell Host Microbe 22:757–765.e3. 10.1016/j.chom.2017.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peng B, Bin Su Y, Li H, Han Y, Guo C, Tian YM, Peng XX. 2015. Exogenous alanine and/or glucose plus kanamycin kills antibiotic-resistant bacteria. Cell Metab 21:249–262. 10.1016/j.cmet.2015.01.008. [DOI] [PubMed] [Google Scholar]
- 25.Baek SH, Li AH, Sassetti CM. 2011. Metabolic regulation of mycobacterial growth and antibiotic sensitivity. PLoS Biol 9:e1001065. 10.1371/journal.pbio.1001065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ye JZ, Lin XM, Cheng ZX, Su YB, Li WX, Ali FM, Zheng J, Peng B. 2018. Identification and efficacy of glycine, serine and threonine metabolism in potentiating kanamycin-mediated killing of Edwardsiella piscicida. J Proteomics 183:34–44. 10.1016/j.jprot.2018.05.006. [DOI] [PubMed] [Google Scholar]
- 27.Ye JZ, Bin Su Y, Lin XM, Lai SS, Li WX, Ali F, Zheng J, Peng B. 2018. Alanine enhances aminoglycosides-induced ROS production as revealed by proteomic analysis. Front Microbiol 9:29. 10.3389/fmicb.2018.00029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Whitehouse DG, May B, Moore AL. 2019. Respiratory chain and ATP synthase, reference module in biomedical sciences. Elsevier. 10.1016/B978-0-12-801238-3.95732-5. [DOI] [Google Scholar]
- 29.Crabbé A, Ostyn L, Staelens S, Rigauts C, Risseeuw M, Dhaenens M, Daled S, Van Acker H, Deforce D, Van Calenbergh S, Coenye T. 2019. Host metabolites stimulate the bacterial proton motive force to enhance the activity of aminoglycoside antibiotics. PLoS Pathog 15:e1007697. 10.1371/journal.ppat.1007697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gao G, Liu F, Xu Z, Wan D, Han Y, Kuang Y, Wang Q, Zhi Q. 2021. Evidence of nigericin as a potential therapeutic candidate for cancers: A review. Biomed Pharmacother 137:111262. 10.1016/j.biopha.2021.111262. [DOI] [PubMed] [Google Scholar]
- 31.Kottur J, Nair DT. 2016. Reactive oxygen species play an important role in the bactericidal activity of quinolone antibiotics. Angew Chem Int Ed Engl 55:2397–2400. 10.1002/anie.201509340. [DOI] [PubMed] [Google Scholar]
- 32.Van Acker H, Coenye T. 2017. The role of reactive oxygen species in antibiotic-mediated killing of bacteria. Trends Microbiol 25:456–466. 10.1016/j.tim.2016.12.008. [DOI] [PubMed] [Google Scholar]
- 33.Belenky P, Ye JD, Porter CB, Cohen NR, Lobritz MA, Ferrante T, Jain S, Korry BJ, Schwarz EG, Walker GC, Collins JJ. 2015. Bactericidal antibiotics induce toxic metabolic perturbations that lead to cellular damage. Cell Rep 13:968–980. 10.1016/j.celrep.2015.09.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kohanski MA, Dwyer DJ, Hayete B, Lawrence CA, Collins JJ. 2007. A common mechanism of cellular death induced by bactericidal antibiotics. Cell 130:797–810. 10.1016/j.cell.2007.06.049. [DOI] [PubMed] [Google Scholar]
- 35.De Soyza A, Hall AJ, Mahenthiralingam E, Drevinek P, Kaca W, Drulis-Kawa Z, Stoitsova SR, Toth V, Coenye T, Zlosnik JEA, Burns JL, Sá-Correia I, De Vos D, Pirnay JP, J Kidd T, Reid D, Manos J, Klockgether J, Wiehlmann L, Tümmler B, Mcclean S, Winstanley C. 2013. Developing an international Pseudomonas aeruginosa reference panel. Microbiologyopen 2:1010–1023. 10.1002/mbo3.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Darch SE, Kragh KN, Abbott EA, Bjarnsholt T, Bull JJ, Whiteley M. 2017. Phage inhibit pathogen dissemination by targeting bacterial migrants in a chronic infection model. mBio 8:e00240-17. 10.1128/mBio.00240-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Macià MD, Rojo-Molinero E, Oliver A. 2014. Antimicrobial susceptibility testing in biofilm-growing bacteria. Clin Microbiol Infect 20:981–990. 10.1111/1469-0691.12651. [DOI] [PubMed] [Google Scholar]
- 38.Van Acker H, Gielis J, Acke M, Cools F, Cos P, Coenye T. 2016. The role of reactive oxygen species in antibiotic-induced cell death in Burkholderia cepacia complex bacteria. PLoS One 11:e0159837. 10.1371/journal.pone.0159837. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental figures. Download AAC.01875-21-s0001.pdf, PDF file, 0.2 MB (178.7KB, pdf)
Supplemental Table S1. Download AAC.01875-21-s0002.xlsx, XLSX file, 0.03 MB (26.6KB, xlsx)