ABSTRACT
ERG11 sequencing of 28 Candida auris clade III isolates revealed the presence of concomitant V125A and F126L substitutions. Heterologous expression of Erg11-V125A/F126L in Saccharomyces cerevisiae led to reduced fluconazole and voriconazole susceptibilities. Generation of single substitution gene variants through site-directed mutagenesis uncovered that F126L primarily contributes to the elevated triazole MICs. A similar yet diminished pattern of reduced susceptibility was observed with the long-tailed triazoles posaconazole and itraconazole for the V125A/F126L, F126L, Y132F, and K143R alleles.
KEYWORDS: Candida auris, ERG11, azole resistance, fluconazole resistance, clade III, African clade, heterologous expression, mutagenesis, short- and long-tailed triazoles
INTRODUCTION
Candida auris is an emerging fungal pathogen that has spread across the globe and caused multiple health care center outbreaks. Strains of C. auris are divided into five genetically distinct, geographic clades: South Asian (I), East Asian (II), African (III), South American (IV), and Iranian (V) (1). Initial spread of C. auris to the U.S. and other parts of the world is predicted to have occurred through multiple travel-related introductions (2). Recently, several reports have shown high rates of C. auris candidemia in hospitalized patients with severe COVID-19 (SARS-CoV-2 infection), particularly in severely ill patients in the intensive care unit (ICU) setting (3–5). Interestingly, the pathogenicity of C. auris differs from other species in that it can colonize the skin, persist on hospital surfaces and on medical equipment, and transfer from person to person (6, 7). In addition, C. auris exhibits elevated rates of antifungal resistance. Clinical isolates that demonstrate reduced susceptibility to one or more classes of antifungals, including triazoles, polyenes (amphotericin B), and echinocandins, have been reported with triazole resistance being the most prevalent (8–10).
Triazole antifungals, such as fluconazole, voriconazole, itraconazole, and posaconazole, target the biosynthesis of fungal ergosterol specifically through inhibition of lanosterol 14-alpha-demethylase (Erg11p) that is encoded by the ERG11 gene in yeast. Early reports identified single Erg11 substitutions (F126L, Y132F, or K143R) in strains from multiple clades (9, 11–13). These substitutions were highlighted due to their connection to triazole resistance within other species of Candida, specifically C. albicans (14). In our previous study (15), we identified and analyzed C. auris ERG11 mutations in clinical isolates of clades I and IV. Using a heterologous expression system, we directly linked the Y132F and K143R Erg11 substitutions to fluconazole and voriconazole resistance, whereas other alterations, including I466M, Y501H, and clade-specific polymorphisms, were not associated with elevated MICs.
Here, we investigated triazole resistance in 28 clinical isolates of C. auris clade III obtained from South Africa (n = 21), Australia (n = 5), and the CDC and FDA Antimicrobial Resistance (AR) Isolate Bank (n = 2). ERG11 was amplified and sequenced as described before (15). In agreement with recent reports (8, 16, 17), we identified two ERG11 mutations, T374C and T376C, that lead to two amino acid substitutions, V125A and F126L, respectively, in all 28 isolates (Table 1). Antifungal susceptibility testing was performed in triplicate for each clinical isolate according to CLSI methodology (18, 19) with C. parapsilosis (ATCC 22019) and C. krusei (ATCC 6258) used as quality control strains. MICs were interpreted using tentative breakpoints as suggested by the CDC (https://www.cdc.gov/fungal/candida-auris/c-auris-antifungal.html). These isolates demonstrated reduced triazole susceptibilities, specifically to fluconazole and voriconazole (Table 1). Of note, clinical isolates SA13, SA15, and SA16 demonstrated fluconazole MICs in the susceptible range of less than 32 μg/ml, despite containing the same ERG11 mutations as the other strains (Table 1). This may point to other mechanisms of differential triazole resistance and/or additional alterations in these isolates that specifically influence the fluconazole-Erg11p interaction. Further analyses on these strains are under way.
TABLE 1.
Triazole drug susceptibility and Erg11 profiles of 28 C. auris clade III clinical isolates
| Origin | Isolatea | Specimen | Erg11 | MICb (μg/ml) |
|||
|---|---|---|---|---|---|---|---|
| FLC | VRC | POS | ITC | ||||
| AR bank | AR-0381 | N/A | WT | 4 | 0.03 | <0.03 | 0.03 |
| AR-0382 | N/A | WT | 4 | 0.03 | 0.06 | 0.06 | |
| AR-0383 | N/A | V125A/F126L | >128 | 1 | 0.03 | 0.125 | |
| AR-0384 | N/A | V125A/F126L | >128 | 1 | 0.06 | 0.25 | |
| South Africa | SA1 | CVC tip | V125A/F126L | >128 | 1 | 0.125 | 0.25 |
| SA2 | Blood | V125A/F126L | >128 | 0.5 | 0.125 | 0.5 | |
| SA3 | CVC tip | V125A/F126L | >128 | 1 | 0.03 | 0.5 | |
| SA4 | Blood | V125A/F126L | >128 | 1 | 0.03 | 0.5 | |
| SA5 | Tracheal aspirate | V125A/F126L | >128 | 2 | 0.03 | 0.5 | |
| SA6 | Blood | V125A/F126L | >128 | 1 | 0.03 | 0.5 | |
| SA7 | Urine | V125A/F126L | >128 | 1 | 0.125 | 0.5 | |
| SA8 | Urine | V125A/F126L | >128 | 1 | 0.03 | 0.25 | |
| SA9 | Urine | V125A/F126L | >128 | 1 | 0.03 | 0.5 | |
| SA10 | Blood | V125A/F126L | >128 | 1 | 0.125 | 0.5 | |
| SA11 | Blood | V125A/F126L | >128 | 1 | 0.03 | 1 | |
| SA12 | Urine | V125A/F126L | >128 | 1 | 0.03 | 0.25 | |
| SA13 | Urine | V125A/F126L | 8 | 1 | 0.03 | 0.25 | |
| SA14 | Urine | V125A/F126L | >128 | 2 | 0.03 | 0.5 | |
| SA15 | Tracheal aspirate | V125A/F126L | 8 | 1 | 0.03 | 0.5 | |
| SA16 | Tracheal aspirate | V125A/F126L | 8 | 1 | 0.03 | 0.5 | |
| SA17 | Urine | V125A/F126L | >128 | 1 | 0.03 | 0.25 | |
| SA18 | Urine | V125A/F126L | 128 | 0.5 | 0.03 | 0.5 | |
| SA19 | Urine | V125A/F126L | 128 | 2 | 0.03 | 0.25 | |
| SA22 | Urine | V125A/F126L | 128 | 0.5 | 0.25 | 0.5 | |
| SA23 | Urine | V125A/F126L | >128 | 8 | 0.25 | 0.5 | |
| Australia | A3 | Sternum | V125A/F126L | >128 | 1 | 0.25 | 1 |
| A4 | Sternum | V125A/F126L | >128 | 1 | 0.25 | 1 | |
| A6 | Axilla & groin | V125A/F126L | >128 | 0.25 | 0.125 | 1 | |
| A7 | Axilla & groin | V125A/F126L | >128 | 0.25 | 0.25 | 1 | |
| A8 | Catheter specimen of urine (CSU) | V125A/F126L | >128 | 0.5 | 0.25 | 1 | |
aFor reference, susceptibility results are presented for two Erg11 wild-type (WT) strains: AR-0381 (clade II) and AR-0382 (clade I).
bFLC, fluconazole; VRC, voriconazole; POS, posaconazole; ITC, itraconazole.
Using the same approach as in reference (15), we cloned the Erg11 allele (V125A/F126L) from isolate AR-0384 onto pRS416, a low-copy-number plasmid that contains the S. cerevisiae URA3 marker (ATCC 87521). This construct was then expressed in a haploid strain of S. cerevisiae (BY4741; ATCC 201388) that is auxotrophic for uracil biosynthesis. This heterologous system allowed us to focus solely on the effects of ERG11 mutations on triazole susceptibilities. Multiple clones were passaged on selective medium (synthetic defined medium lacking uracil; SD-Ura), screened by PCR, and the resulting plasmid sequences verified (for primers, see reference [15]). S. cerevisiae that expressed C. auris Erg11-V125A/F126L demonstrated elevated MICs to fluconazole (64 μg/ml) and voriconazole (1 μg/ml). In comparison, expression of an empty vector or Erg11 wild-type alleles from other clades yielded MICs 4- to 8-fold more susceptible (≤16 μg/ml to fluconazole; ≤0.25 μg/ml to voriconazole) (Table 2).
TABLE 2.
Triazole susceptibilities of S. cerevisiae strains that express C. auris Erg11 plasmid constructs
| MICa (μg/ml) |
||||
|---|---|---|---|---|
| Erg11 allele | FLC | VRC | POS | ITC |
| Empty vector | 16 | 0.12 | 0.25 | 0.5 |
| Wild type (clade I) | 16 | 0.25 | 0.25 | 1 |
| Wild type (clade IV) | 16 | 0.12 | 0.25 | 1 |
| I466M | 16 | 0.25 | 0.5 | 2 |
| Y132F | 128 | 2 | 0.5 | 4 |
| K143R | 64 | 1 | 0.5 | 2 |
| V125A/F126L | 64 | 1 | 0.5 | 2 |
| Wild type (clade III) | 16 | 0.25 | 0.25 | 1 |
| V125A | 16 | 0.25 | 0.25 | 1 |
| F126L | 64 | 1 | 0.5 | 4 |
MICs were obtained in both nutrient-rich YPD (yeast extract, peptone, dextrose) and nutrient-limited SD-Ura broth media. We observed a 2-fold or less difference between these media. Fluconazole and voriconazole MICs of the first six strains were previously reported (15); however, these MICs were repeated in tandem with the newly engineered strains and are presented here for comparison. FLC, fluconazole; VRC, voriconazole; POS, posaconazole; ITC, itraconazole.
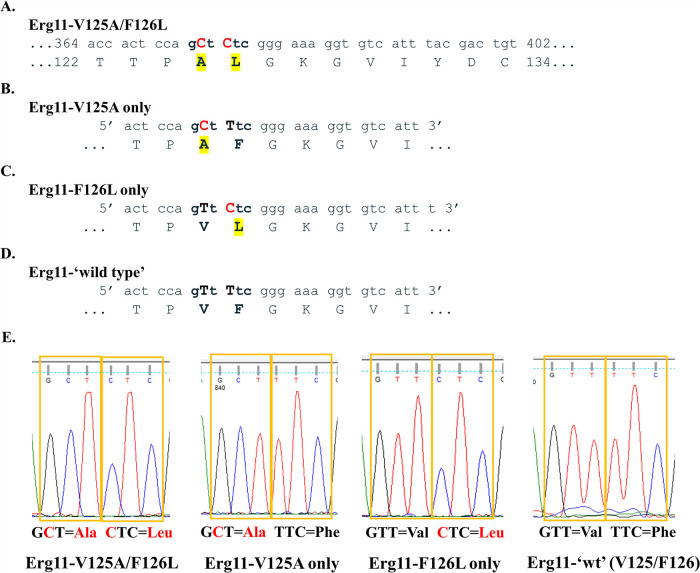
To further dissect the specific role of V125A and F126L substitutions in triazole resistance, we designed mutagenic primers to individually revert each amino acid substitution (Fig. 1). A Phusion site-directed mutagenesis kit (Thermo Scientific; cat. no. F541) was used to introduce the desired wild-type mutations. The resulting C. auris Erg11-V125A and Erg11-F126L plasmid constructs were expressed in S. cerevisiae. In addition, we performed two consecutive rounds of site-directed mutagenesis to produce a strain that carried neither substitution (Erg11-V125/F126) (Fig. 1D and E). This strain represented a de facto clade III wild-type allele. Plasmid sequences of all alleles were confirmed. Subsequent triazole susceptibility assays revealed that cells expressing F126L alone exhibited elevated MICs, similar to V125A/F126L, while V125A alone led to MICs similar to that of the wild-type alleles (Table 2). Our engineered, clade III wild-type allele yielded susceptible MICs, allowing us to conclude that the ERG11 mutations, as opposed to expression levels, were mainly contributing to the observed decreased in susceptibility.
FIG 1.
Molecular dissection of C. auris Erg11 V125A and F126L amino acid substitutions. (A) Region of C. auris clade III ERG11 DNA that displays nucleotide mutations (T374C/T376C) in red and resulting protein alterations (V125A/F126L) in yellow highlight. (B) Forward mutagenic primer used to revert leucine (L) back to phenylalanine (F). After mutagenesis, this construct contained only V125A (pCauErg11-V125A). (C) Forward mutagenic primer used to revert alanine (A) back to wild-type valine (V). After mutagenesis, this construct contained only F126L (pCauErg11-F126L). (D) Forward mutagenic primer used to revert leucine (L) back to phenylalanine (F) using the pCauErg11-F126L plasmid as a template. After mutagenesis, this construct contained both wild-type nucleotides and amino acids (pCauErg11-‘wt’). (E) Plasmid sequencing chromatograms of relevant codons corresponding to the 125th and 126th amino acids following mutagenesis and propagation in Escherichia coli.
Drug binding and cloning studies have demonstrated that certain ERG11 mutations in S. cerevisiae and C. albicans influence susceptibility to all triazoles, while other mutations lead to decreased susceptibility to only short- or long-tailed triazoles (20–22). Therefore, in addition to fluconazole and voriconazole (short-tailed triazoles), we tested each of our strains to determine susceptibility to posaconazole and itraconazole (long-tailed triazoles) (Table 2). Changes in the posaconazole and itraconazole MICs were minimal, although consistent, and with 2- to 4-fold differences between the “resistant” alleles (V125A/F126L, Y132F, or K143R) and the wild-type alleles (Table 2). These results are in alignment with the minimal differences observed in clinical isolates (Table 1) and to those of previous studies that analyzed Y132F and K143R or equivalent changes in C. albicans and S. cerevisiae (21, 23).
Crystallization of C. albicans Erg11 identified residue 126, and the equivalent residue in S. cerevisiae, as being located within the enzyme’s active site and a likely player in substrate binding (24, 25). Furthermore, the authors from that study predicted that alteration of this residue would likely reduce affinity for all triazole drugs but would do so most extensively for short-tailed azoles (24). Because C. auris clade III isolates described in the literature contain both V125A and F126L substitutions, it is likely that these two mutations occurred at nearly the same time in the evolution of this clade. The V125A substitution may simply be a passenger mutation. Alternatively, V125A may increase the stability of the Erg11 enzyme or be advantageous for the yeast in another way and/or in combination with other alterations (e.g., ERG11 copy number variants [8]). Studies have since identified TAC1b transcription factor mutations, linked to increased expression of drug efflux pumps (e.g., CDR1 and/or other unidentified transporters), as an alternate mechanism of triazole resistance in C. auris (26–28). Additionally, a recent report demonstrated an additive effect that concomitant ERG11 (F444L) and TAC1b mutations can have on triazole resistance (29). Of note, the SA23 isolate demonstrated unusually high triazole MICs, which were most noticeable for voriconazole. It is probable that additional mechanisms of triazole resistance are involved and are being investigated in an ongoing study.
In conclusion, the ERG11 allele found in C. auris clade III isolates directly contributes to reduced triazole susceptibility, in particular to fluconazole and voriconazole. Moreover, our mutagenic experiments revealed that the F126L substitution was primarily responsible for the elevated triazole MICs. Results of this study further improve our understanding of triazole resistance mechanisms in C. auris, which can have a direct impact on diagnostic and treatment practices.
ACKNOWLEDGMENTS
This research was supported by the William Paterson University (WPU) Department of Biology and College of Science and Health’s Center for Research to K.R.H. Undergraduate work of B.W. and A.W. was supported by WPU College of Science and Health and of I.S. and G.C.-P. by the NSF Louis Stokes Alliances for Minority Participation (LSAMP) program.
We declare no conflicts of interest.
Contributor Information
Milena Kordalewska, Email: milena.kordalewska@hmh-cdi.org.
Kelley R. Healey, Email: healeyk3@wpunj.edu.
REFERENCES
- 1.Du H, Bing J, Hu T, Ennis CL, Nobile CJ, Huang G. 2020. Candida auris: epidemiology, biology, antifungal resistance, and virulence. PLoS Pathog 16:e1008921. 10.1371/journal.ppat.1008921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chow NA, Gade L, Tsay SV, Forsberg K, Greenko JA, Southwick KL, Barrett PM, Kerins JL, Lockhart SR, Chiller TM, Litvintseva AP, Team USCaI. 2018. Multiple introductions and subsequent transmission of multidrug-resistant Candida auris in the USA: a molecular epidemiological survey. Lancet Infect Dis 18:1377–1384. 10.1016/S1473-3099(18)30597-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chowdhary A, Tarai B, Singh A, Sharma A. 2020. Multidrug-resistant Candida auris infections in critically ill coronavirus disease patients, India, April-July 2020. Emerg Infect Dis 26:2694–2696. 10.3201/eid2611.203504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Villanueva-Lozano H, Trevino-Rangel RJ, Gonzalez GM, Ramirez-Elizondo MT, Lara-Medrano R, Aleman-Bocanegra MC, Guajardo-Lara CE, Gaona-Chavez N, Castilleja-Leal F, Torre-Amione G, Martinez-Resendez MF. 2021. Outbreak of Candida auris infection in a COVID-19 hospital in Mexico. Clin Microbiol Infect 27:813–816. 10.1016/j.cmi.2020.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Prestel C, Anderson E, Forsberg K, Lyman M, de Perio MA, Kuhar D, Edwards K, Rivera M, Shugart A, Walters M, Dotson NQ. 2021. Candida auris outbreak in a COVID-19 specialty care unit - Florida, July-August 2020. MMWR Morb Mortal Wkly Rep 70:56–57. 10.15585/mmwr.mm7002e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Jong AW, Hagen F. 2019. Attack, defend and persist: how the fungal pathogen Candida auris was able to emerge globally in healthcare environments. Mycopathologia 184:353–365. 10.1007/s11046-019-00351-w. [DOI] [PubMed] [Google Scholar]
- 7.Sexton DJ, Bentz ML, Welsh RM, Derado G, Furin W, Rose LJ, Noble-Wang J, Pacilli M, McPherson TD, Black S, Kemble SK, Herzegh O, Ahmad A, Forsberg K, Jackson B, Litvintseva AP. 2021. Positive correlation between Candida auris skin-colonization burden and environmental contamination at a ventilator-capable skilled nursing facility in Chicago. Clin Infect Dis 73:1142–1148. 10.1093/cid/ciab327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chow NA, Munoz JF, Gade L, Berkow EL, Li X, Welsh RM, Forsberg K, Lockhart SR, Adam R, Alanio A, Alastruey-Izquierdo A, Althawadi S, Arauz AB, Ben-Ami R, Bharat A, Calvo B, Desnos-Ollivier M, Escandon P, Gardam D, Gunturu R, Heath CH, Kurzai O, Martin R, Litvintseva AP, Cuomo CA. 2020. Tracing the evolutionary history and global expansion of Candida auris using population genomic analyses. mBio 11:e03364-19 10.1128/mBio.03364-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lockhart SR, Etienne KA, Vallabhaneni S, Farooqi J, Chowdhary A, Govender NP, Colombo AL, Calvo B, Cuomo CA, Desjardins CA, Berkow EL, Castanheira M, Magobo RE, Jabeen K, Asghar RJ, Meis JF, Jackson B, Chiller T, Litvintseva AP. 2017. Simultaneous emergence of multidrug-resistant Candida auris on 3 continents confirmed by whole-genome sequencing and epidemiological analyses. Clin Infect Dis 64:134–140. 10.1093/cid/ciw691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lyman M, Forsberg K, Reuben J, Dang T, Free R, Seagle EE, Sexton DJ, Soda E, Jones H, Hawkins D, Anderson A, Bassett J, Lockhart SR, Merengwa E, Iyengar P, Jackson BR, Chiller T. 2021. Notes from the field: transmission of pan-resistant and echinocandin-resistant Candida auris in health care facilities - Texas and the District of Columbia, January-April 2021. MMWR Morb Mortal Wkly Rep 70:1022–1023. 10.15585/mmwr.mm7029a2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rhodes J, Abdolrasouli A, Farrer RA, Cuomo CA, Aanensen DM, Armstrong-James D, Fisher MC, Schelenz S. 2018. Genomic epidemiology of the UK outbreak of the emerging human fungal pathogen Candida auris. Emerg Microbes Infect 7:43. 10.1038/s41426-018-0045-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Munoz JF, Gade L, Chow NA, Loparev VN, Juieng P, Berkow EL, Farrer RA, Litvintseva AP, Cuomo CA. 2018. Genomic insights into multidrug-resistance, mating and virulence in Candida auris and related emerging species. Nat Commun 9:5346. 10.1038/s41467-018-07779-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chowdhary A, Prakash A, Sharma C, Kordalewska M, Kumar A, Sarma S, Tarai B, Singh A, Upadhyaya G, Upadhyay S, Yadav P, Singh PK, Khillan V, Sachdeva N, Perlin DS, Meis JF. 2018. A multicentre study of antifungal susceptibility patterns among 350 Candida auris isolates (2009–17) in India: role of the ERG11 and FKS1 genes in azole and echinocandin resistance. J Antimicrob Chemother 73:891–899. 10.1093/jac/dkx480. [DOI] [PubMed] [Google Scholar]
- 14.Flowers SA, Colon B, Whaley SG, Schuler MA, Rogers PD. 2015. Contribution of clinically derived mutations in ERG11 to azole resistance in Candida albicans. Antimicrob Agents Chemother 59:450–460. 10.1128/AAC.03470-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Healey KR, Kordalewska M, Jimenez Ortigosa C, Singh A, Berrio I, Chowdhary A, Perlin DS. 2018. Limited ERG11 mutations identified in isolates of Candida auris directly contribute to reduced azole susceptibility. Antimicrob Agents Chemother 62:e01427-18. 10.1128/AAC.01427-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maphanga TG, Naicker SD, Kwenda S, Munoz JF, van Schalkwyk E, Wadula J, Nana T, Ismail A, Coetzee J, Govind C, Mtshali PS, Mpembe RS, Govender NP, For G-S. 2021. In vitro antifungal resistance of Candida auris isolates from bloodstream infections, South Africa. Antimicrob Agents Chemother 65:e0051721. 10.1128/AAC.00517-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Naicker SD, Maphanga TG, Chow NA, Allam M, Kwenda S, Ismail A, Govender N-SA. 2021. Clade distribution of Candida auris in South Africa using whole genome sequencing of clinical and environmental isolates. Emerg Microbes Infect 10:1300–1308. 10.1080/22221751.2021.1944323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.CLSI. 2017. National Committee for Clinical Laboratory Standards. Reference method for broth dilution antifungal susceptibility testing of yeasts. 4th ed Clinical and Laboratory Standards Institute document M27. Wayne, PA. [Google Scholar]
- 19.CLSI. 2020. Performance Standards for Antifungal Susceptibility Testing of Yeasts. 2nd ed Clinical and Laboratory Standards Institute document M60. Wayne, PA. [Google Scholar]
- 20.Sagatova AA, Keniya MV, Wilson RK, Sabherwal M, Tyndall JD, Monk BC. 2016. Triazole resistance mediated by mutations of a conserved active site tyrosine in fungal lanosterol 14alpha-demethylase. Sci Rep 6:26213. 10.1038/srep26213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xiang MJ, Liu JY, Ni PH, Wang S, Shi C, Wei B, Ni YX, Ge HL. 2013. Erg11 mutations associated with azole resistance in clinical isolates of Candida albicans. FEMS Yeast Res 13:386–393. 10.1111/1567-1364.12042. [DOI] [PubMed] [Google Scholar]
- 22.Chau AS, Mendrick CA, Sabatelli FJ, Loebenberg D, McNicholas PM. 2004. Application of real-time quantitative PCR to molecular analysis of Candida albicans strains exhibiting reduced susceptibility to azoles. Antimicrob Agents Chemother 48:2124–2131. 10.1128/AAC.48.6.2124-2131.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sanglard D, Ischer F, Koymans L, Bille J. 1998. Amino acid substitutions in the cytochrome P-450 lanosterol 14alpha-demethylase (CYP51A1) from azole-resistant Candida albicans clinical isolates contribute to resistance to azole antifungal agents. Antimicrob Agents Chemother 42:241–253. 10.1128/AAC.42.2.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Keniya MV, Sabherwal M, Wilson RK, Woods MA, Sagatova AA, Tyndall JDA, Monk BC. 2018. Crystal structures of full-length lanosterol 14alpha-demethylases of prominent fungal pathogens Candida albicans and Candida glabrata provide tools for antifungal discovery. Antimicrob Agents Chemother 62:e01134-18. 10.1128/AAC.01134-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sagatova AA, Keniya MV, Wilson RK, Monk BC, Tyndall JD. 2015. Structural insights into binding of the antifungal drug fluconazole to Saccharomyces cerevisiae lanosterol 14alpha-demethylase. Antimicrob Agents Chemother 59:4982–4989. 10.1128/AAC.00925-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rybak JM, Doorley LA, Nishimoto AT, Barker KS, Palmer GE, Rogers PD. 2019. Abrogation of triazole resistance upon deletion of CDR1 in a clinical isolate of Candida auris. Antimicrob Agents Chemother 63:e00057-19. 10.1128/AAC.00057-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rybak JM, Munoz JF, Barker KS, Parker JE, Esquivel BD, Berkow EL, Lockhart SR, Gade L, Palmer GE, White TC, Kelly SL, Cuomo CA, Rogers PD. 2020. Mutations in TAC1B: a novel genetic determinant of clinical fluconazole resistance in Candida auris. mBio 11:e00365-20. 10.1128/mBio.00365-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mayr EM, Ramirez-Zavala B, Kruger I, Morschhauser J. 2020. A zinc cluster transcription factor contributes to the intrinsic fluconazole resistance of Candida auris. mSphere 5:e00279-20. 10.1128/mSphere.00279-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li J, Coste AT, Liechti M, Bachmann D, Sanglard D, Lamoth F. 2021. Novel ERG11 and TAC1b mutations associated with azole resistance in Candida auris. Antimicrob Agents Chemother 65:e02663-20. 10.1128/AAC.02663-20. [DOI] [PMC free article] [PubMed] [Google Scholar]