ABSTRACT
Enterococcus faecium is a major cause of clinical infections, often due to multidrug-resistant (MDR) strains. Whole-genome sequencing (WGS) is a powerful tool to study MDR bacteria and their antimicrobial resistance (AMR) mechanisms. In this study, we used WGS to characterize E. faecium clinical isolates and test the feasibility of rules-based genotypic prediction of AMR. Clinical isolates were divided into derivation and validation sets. Phenotypic susceptibility testing for ampicillin, vancomycin, high-level gentamicin, ciprofloxacin, levofloxacin, doxycycline, tetracycline, and linezolid was performed using the Vitek 2 automated system, with confirmation and discrepancy resolution by broth microdilution, disk diffusion, or gradient diffusion when needed. WGS was performed to identify isolate lineage and AMR genotype. AMR prediction rules were derived by analyzing the genotypic-phenotypic relationship in the derivation set. Phylogenetic analysis demonstrated that 88% of isolates in the collection belonged to hospital-associated clonal complex 17. Additionally, 12% of isolates had novel sequence types. When applied to the validation set, the derived prediction rules demonstrated an overall positive predictive value of 98% and negative predictive value of 99% compared to standard phenotypic methods. Most errors were falsely resistant predictions for tetracycline and doxycycline. Further analysis of genotypic-phenotypic discrepancies revealed potentially novel pbp5 and tet(M) alleles that provide insight into ampicillin and tetracycline class resistance mechanisms. The prediction rules demonstrated generalizability when tested on an external data set. In conclusion, known AMR genes and mutations can predict E. faecium phenotypic susceptibility with high accuracy for most routinely tested antibiotics, providing opportunities for advancing molecular diagnostics.
KEYWORDS: antibiotic resistance, antimicrobial resistance, Enterococcus, whole-genome sequencing, genomics, prediction, VRE, microbiology, antimicrobial agents, genome analysis, vancomycin resistance
INTRODUCTION
Over the last 4 decades, Enterococcus faecium has transformed from a harmless gastrointestinal colonizer into a serious pathogen causing antibiotic-resistant clinical disease and hospital-acquired infections (1–4). The multidrug resistant, hospital-associated strains of E. faecium mostly belong to the clonal complex 17 (CC17) lineage and are a leading cause of nosocomial urinary tract infections, bacteremia, endocarditis, and wound infections (5). The optimal antibiotic treatment for E. faecium infections is a cell wall-active agent, such as ampicillin or vancomycin, which in serious infections may be used in combination with an aminoglycoside like gentamicin for bactericidal synergism (6). Unfortunately, rising rates of resistance have rendered β-lactams, vancomycin, and aminoglycosides increasingly inactive against E. faecium. Consequently, standard empirical antimicrobial therapy regimens containing vancomycin for Gram-positive coverage fail to treat more than 75% of patients with E. faecium bloodstream infections in the United States, resulting in higher mortality rates and health care costs and highlighting the need to improve our diagnostic and therapeutic capabilities for E. faecium (7, 8).
Mechanisms of E. faecium resistance to the most commonly used antimicrobial agents are well described (2). Briefly, vancomycin resistance is conferred by the vancomycin resistance (Van) operon, which disrupts the d-Ala–d-Ala vancomycin binding site on the peptidoglycan cell wall (9–11). In contrast, ampicillin resistance primarily occurs by a more complicated set of alterations in the low-affinity penicillin-binding protein 5 (PBP5) (12). For serious ampicillin- and vancomycin-resistant E. faecium infections, the treatment options are very limited, with only one FDA-approved drug—linezolid—and off-label use of other drugs, such as daptomycin, tigecycline, quinupristin-dalfopristin, and oritavancin. The Clinical and Laboratory Standards Institute (CLSI) also provides interpretative criteria for fluoroquinolones (ciprofloxacin and levofloxacin), tetracyclines (tetracycline and doxycycline), and nitrofurantoin, which may be used in less serious infections.
Whole-genome sequencing (WGS) provides a powerful tool to discover more about E. faecium antimicrobial resistance (AMR) mechanisms and transmission patterns (13, 14). WGS data have primarily been used to predict AMR by applying a rules-based approach, where the presence of one or more known AMR genes or single nucleotide polymorphisms (SNPs) is used to determine resistance to an antibiotic. Alternatively, model-based approaches, which use statistical methods to train a classifier that often determines predictive genetic loci without prior knowledge, have also been used successfully to predict AMR (15, 16). For well-studied organisms and antibiotics for which there is ample understanding of the genotypic-phenotypic relationship, a rules-based approach has the advantage of being easily interpretable and has successfully been used for Staphylococcus aureus, Escherichia coli, and Klebsiella pneumoniae (17–19), with limited results for E. faecium (19–21). Therefore, there remains a need to systematically test the efficacy of a rules-based approach for a broad range of clinically relevant antibiotics in E. faecium.
In this study, we developed and tested AMR prediction rules for E. faecium based on a collection of clinical isolates from a large U.S. academic health center clinical microbiology laboratory. The isolate set was temporally divided into derivation and validation sets. As expected for this organism, 92% of isolates were resistant to more than one drug class, highlighting the high degree of resistance in circulating clinical isolates and the importance of determining antimicrobial susceptibility to guide clinical management. After deriving the genotypic prediction rules using the derivation set, the rules were applied to the validation set and discrepancies were analyzed. The final set of AMR prediction rules was applied to an external data set of previously characterized E. faecium clinical isolates collected from Germany to confirm the accuracy of the prediction rules from a geographically distinct source.
RESULTS
Characterization of isolate sources, sequence type diversity, and AMR profiles.
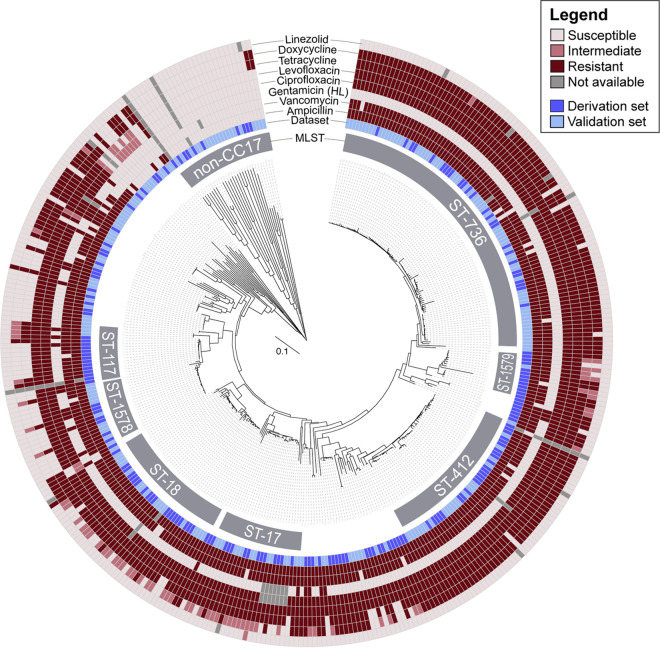
We first examined the population structure of our isolate collection. Using multilocus sequence typing (MLST), the universal typing method that designates sequence types (STs) based on allelic assignments of seven species-specific housekeeping genes (22), we found that over 88% of isolates belonged to the high-risk, hospital-associated clonal complex 17. The most common STs were ST-736 (23%), ST-18 (13%), ST-412 (13%), ST-17 (7%), and ST-117 (5%) (Fig. 1). We identified numerous novel sequence types, including two locally common STs belonging to CC17, now designated ST-1578 and ST-1579 (Fig. 1; see also Table S1 in the supplemental material). Compared to all 1,138 clinical E. faecium isolates in the international PubMLST database, our single hospital data set captured most subgroup founders in CC17 and CC94 (Fig. S1), indicating that the diversity of this collection provides a comprehensive set of isolates to test a generalizable genotypic AMR prediction approach (23).
FIG 1.
Core genome MLST-based neighbor joining tree demonstrating the relationship between phylogeny and antimicrobial resistance in the derivation and validation sets. Phenotypic susceptibility results are displayed for ampicillin, vancomycin, high-level (HL) gentamicin, ciprofloxacin, levofloxacin, tetracycline, doxycycline, and linezolid.
Using the higher-resolution core genome MLST (cgMLST) system, we examined the relationship between phylogeny and AMR (Fig. 1). Seven percent of isolates were resistant to all three of the key antimicrobials used in the treatment of E. faecium infections—vancomycin, ampicillin, and gentamicin—and belonged to a range of STs within CC17, though most commonly ST-80 and ST-1578. The most common STs, ST-736 and ST-412, had uniform susceptibility to gentamicin and linezolid. Isolates that were not in CC17 tended to be susceptible to all tested antibiotics. While there is clearly a correlation between population structure and antimicrobial resistance, these findings indicate that the population structure can only partially inform the AMR predictions and that further characterization is needed.
Developing a rules-based approach to predicting AMR.
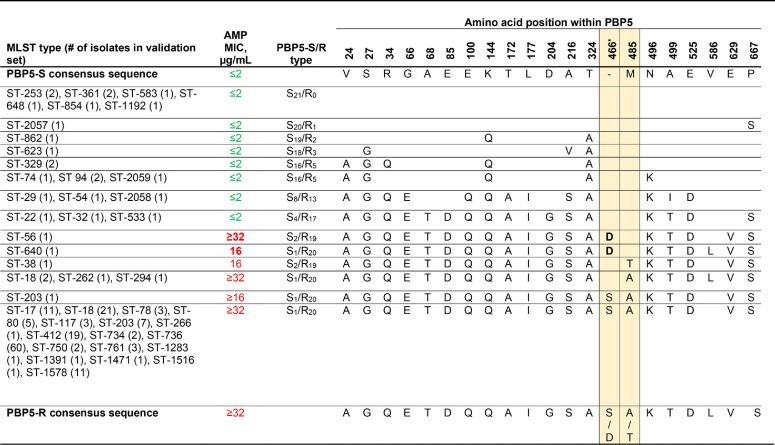
We analyzed the AMR gene content of the derivation set to develop a simple rules-based approach based upon published genes to predict E. faecium resistance. Ampicillin resistance in E. faecium is associated with mutations in the penicillin-binding protein 5 (pbp5) gene, where dominant susceptibility (pbp5-S) and resistance (pbp5-R) alleles have been described (24). PBP5-R and PBP5-S differ by 21 amino acids, which decrease the protein’s ampicillin affinity and result in an increase in the MIC from ≤2 to ≥16 μg/mL, with hybrid alleles exhibiting a range of MICs in between (24). Despite the complex variability in pbp5, most resistance alleles have a mutation in codon 485, which results in a methionine-to-alanine or -threonine substitution and independently increases the ampicillin MIC at least 4-fold (25). In the derivation set, the Met485 substitution was found in every ampicillin-resistant isolate and appeared to be a simple predictor of the presence of an ampicillin resistance pbp5 allele and phenotypic ampicillin resistance.
To predict vancomycin resistance, we used the same approach as FDA-cleared molecular vancomycin-resistant enterococcus (VRE) assays by detecting the vanA or vanB gene as a surrogate for the presence of the vancomycin resistance operon (9, 26). We found vanA in 97% of resistant isolates and vanB in the remaining 3%. We did not detect other vancomycin resistance genes (vanC, vanD, vanE, vanF, vanG, vanL, vanM, or vanN) in any isolates (Table S2).
High-level gentamicin (HLG) resistance was associated with the presence of an intact aac(6′)-Ie-aph(2″)-Ia gene, which encodes a bifunctional aminoglycoside-modifying enzyme that decreases the binding affinity of gentamicin for the ribosome and is the most common described mechanism of HLG resistance in E. faecium (2, 27). Other aminoglycoside-modifying genes, such as aph(3′)-Ia, aph(3′)-IIIa, aac(6′)-Ii, and ant(6′)-Ia, were commonly found but were not associated with high-level gentamicin resistance (Table S3) (28).
Ciprofloxacin and levofloxacin resistances are associated with classic mutations in gyrA (Ser84) and parC (Ser82). Both of these mutations were found exclusively in all fluoroquinolone-resistant isolates and not found in any fluoroquinolone-susceptible isolates of the derivation set. However, it was not possible to distinguish derivation set isolates that were fully susceptible to fluoroquinolones from strains that had intermediate resistance according to the CLSI breakpoints, even when additional genes such as gyrB and parB were incorporated. This is consistent with the ciprofloxacin epidemiologic cutoff value (ECV) of 8 μg/mL being above the CLSI breakpoint for resistance (4 μg/mL) and the levofloxacin ECV of 4 μg/mL coinciding with the intermediate CLSI interpretative category (29). Given that the wild-type fluoroquinolone MIC distributions include both the susceptible and intermediate CLSI interpretive categories, intermediate isolates were categorized with susceptible organisms for the purpose of genotypic prediction.
Numerous tetracycline resistance genes have been reported for enterococci, but resistance to tetracycline among the derivation set isolates appeared to be associated with the presence of either the tet(L) (drug efflux), tet(M) (ribosomal protection), or tet(S) (ribosomal protection) gene, with a sensitivity of 99% and specificity of 87%. Doxycycline resistance had the strongest correlation with the presence of tet(M) alone, but there were 14 isolates that nominally had tet(M) yet were phenotypically susceptible. Alignment to “tet(U)” was found frequently, within 84% of ST-17 isolates and 32% of ST-18 isolates, and in both tetracycline-susceptible and -resistant isolates. Some isolates demonstrated a very high depth of coverage of the tet(U) gene, up to 10 times higher than the rest of the genome. The lack of correlation with phenotypic resistance and the disproportionately high alignment rates are supportive of a previous report that tet(U) is a misannotation of a portion of the rolling-circle replication initiator (rep) gene found in plasmids rather than a tetracycline resistance determinant (30).
Finally, we were not able to initially develop a rules-based method for linezolid based on our derivation set due to a paucity of linezolid-resistant isolates. However, others have shown that linezolid resistance in E. faecium is most commonly due to a G2576T mutation in at least three of six copies of the 23S rRNA gene (2, 31), which is the rule that we incorporated. Other linezolid resistance genes, such as poxtA, optrA, and the cfr-like genes, have been described but were absent or were present only in linezolid-susceptible isolates in our data set (Table S4).
Determining the accuracy of genotypic prediction rules.
We next evaluated the accuracy of the rules-based genotypic predictions on the Boston validation set. After resolving genotyping or phenotyping errors through repeat testing, the genotypic-phenotypic categorical agreement was generally excellent, with an overall positive predictive value of 98% and negative predictive value of 99%. All drugs achieved a percent categorical agreement above 89.9%, which is the threshold that the FDA considers to be acceptable performance for antimicrobial susceptibility testing (AST) devices compared to a CLSI reference method (Table 1) (32). The very major error (VME) rate, also known as the false-negative rate, was 1.4% or lower for all drugs (Table 1). The major error (ME) rate, or the false-positive rate, was below 3% for ampicillin, vancomycin, HL gentamicin, ciprofloxacin, and levofloxacin (Table 1). The tetracycline and doxycycline ME rates were above the FDA-accepted threshold of 3% (14% and 27%, respectively) due to the presence of tet genes in phenotypically susceptible isolates.
TABLE 1.
Accuracy of genotypic prediction rules V.1 when applied to the Boston validation seta
| Antimicrobial | V.1 rules applied to the Boston validation set |
|||||||
|---|---|---|---|---|---|---|---|---|
| Total no. of isolates | Phenotypic susceptibility rate, % | Genotype used for resistance prediction (V.1) | Categorical agreement, % | Very major error rate (FN), % (95% CI) | Major error rate (FP), % (95% CI) | PPV, % | NPV, % | |
| Ampicillin | 203 | 12 | Mutation of pbp5 485M | 99 | 1.1 (0.13, 4.0)b | 0 (0, 13) | 100 | 92.6 |
| Vancomycin | 204 | 21 | Presence of vanA or vanB | 99 | 0 (0, 2.3) | 2.3 (0.06, 12)c | 99.4 | 100 |
| Gentamicin, high level | 198 | 94 | Presence of aac(6′)-le-aph(2”)-la | 100 | 0 (0, 28) | 0 (0, 1.9) | 100 | 100 |
| Ciprofloxacind | 198 | 15 | Mutation of gyrA (84S) or parC (82S) | 100 | 0 (0, 2.2) | 0 (0, 11) | 100 | 100 |
| Levofloxacind | 205 | 15 | Mutation of gyrA (84S) or parC (82S) | 100 | 0 (0, 2.1) | 0 (0, 11) | 100 | 100 |
| Tetracycline | 205 | 24 | Presence of tet(L), tet(M), or tet(S) | 97 | 0 (0, 2.4) | 14 (5.8, 27) | 95.7 | 100 |
| Doxycycline | 205 | 29 | Presence of tet(M) | 91 | 1.4 (0.16, 4.9) | 27 (16, 40) | 90.0 | 95.7 |
| Linezolidd | 200 | 99 | Mutation of 23S rRNA G2576T in at least 3 alleles | 100 | 0 (0, 84) | 0 (0, 1.8) | 100 | 100 |
FN, false negative; FP, false positive; PPV, positive predictive value; NPV, negative predictive value.
Both isolates that were phenotypically resistant to ampicillin harbored wild-type pbp5 Met485 but had an aspartic acid inserted after position 466 (Asp466′).
A single vancomycin-variable enterococcus, which appeared phenotypically susceptible but could gain vancomycin resistance during treatment, was conservatively classified as a major error.
Antibiotic for which intermediate isolates were considered with susceptible isolates.
We found G2576T mutations in three validation set isolates, which included the only two linezolid-resistant isolates. The two linezolid-resistant isolates each had a linezolid MIC of 8 μg/mL and G2576T mutations in 46% of reads, corresponding to three out of six gene copies. The third isolate had the G2576T mutation in only 16% of reads, or one out of six gene copies, and was phenotypically susceptible to linezolid, with an MIC of 2 μg/mL. Thus, the presence of the G2576T mutation in multiple copies of the 23S rRNA gene appeared to correlate well with phenotypic resistance to linezolid in our data set. Overall, the first iteration of the genotypic prediction rules achieved excellent performance for ampicillin, vancomycin, HL gentamicin, ciprofloxacin, levofloxacin, and linezolid.
Investigating the cause of genotypic-phenotypic discrepancies in the validation set.
To further improve the accuracy of the genotypic prediction rules, we examined the isolates for which the predicted and observed AMR phenotypes did not match. The only isolate with a vancomycin discrepancy was phenotypically susceptible to vancomycin (MIC of 1 μg/mL) yet contained the vanA gene. Further analysis of the vancomycin resistance (Van) operon revealed that the isolate also contained the vanX, vanY, and vanH components of the Van operon with very high coverage but lacked the vanS (sensor) or vanR (regulator) genes that form a two-component regulatory system that is necessary for the expression of the vanHAX gene cluster (33) (Table S5). While this was the only isolate out of 302 vanA-containing isolates in our collection that lacked vanRS, other E. faecium isolates with a “silent” vanA gene via the same mechanism have been described in the literature as vancomycin-variable enterococci (VVE) because they can gain vancomycin resistance during treatment and thereby lead to treatment failure (34–36). To prospectively identify these VVE isolates, the vancomycin genotypic prediction rule can be modified to identify not only vanA/B but also the remaining genes of the Van operon (vanX, vanY, vanH, vanR, and vanS) and flag the presence of vanHAX without vanRS. This example highlights the potential for whole-genome-sequencing-based approaches to identify rich and potentially clinically valuable information beyond what is routinely available today.
Two isolates, belonging to ST-56 and ST-640, were falsely predicted to have ampicillin susceptibility based on a wild-type methionine at position 485 yet were confirmed by multiple phenotypic testing methods to be resistant. Subsequent analysis demonstrated that these isolates contained potentially novel pbp5 alleles encoding an aspartic acid insertion after position 466 (Asp466′ [Table 2]). Resistance occurring in these Met485, Asp466′ isolates support an independent contribution of 466′ insertions to the reduced binding of ampicillin to the active site of PBP5, as suggested by crystal structure studies (25). Updating the rules-based prediction of ampicillin resistance to include either a mutation at 485M or the presence of 466′S/D would correctly classify these unusual organisms and flag them for confirmatory laboratory testing.
TABLE 2.
PBP5 alleles found in the validation set isolatesa
PBP5-S/R types were assigned using the nomenclature of Pietta et al. (24). Blank amino acid positions indicate an identical amino acid as the PBP5-S consensus sequence. AMP, ampicillin. Data for the specific positions discussed in the text are highlighted in yellow. MICs of ≤8 μg/mL, corresponding to the susceptible category, are in green, and MICs of ≥16 μg/mL, corresponding to the resistant category, are in red.
The tetracyclines had strikingly high false-positive rates of 14% for tetracycline and 27% for doxycycline. We hypothesized that distinct allelic forms of tet(L) and tet(M) were responsible for the phenotypic variability. We found that the presence of either the wild-type tet(L) allele or the tet(L) allele with a 34-bp deletion at the N terminus and an SNP at position 61 was highly correlated with both tetracycline and doxycycline resistance. There was no apparent correlation between the tet(L) allelic forms and tetracycline phenotypic susceptibility (Fig. 2), but the tet(L) allele with a 34-bp deletion, which likely requires translation initiation from an alternative upstream start codon (37), was found uniformly in ST-736, ST-412, and ST-203 isolates. Conversely, tet(M) had far more sequence variability, with up to 85 SNPs and 187-bp deletions (Fig. 2). A cluster of isolates, mostly ST-17, ST-18, and ST-117, lacked tet(L) but harbored tet(M) alleles with SNPs or deletions predominantly in domain V of TetM, and they could be distinguished from other tet(M)-containing isolates based on multiple SNPs, such as L528F. Most isolates with tet(M) L528F were resistant to tetracycline but intermediate to doxycycline, but 5 of the 39 isolates were highly susceptible to both tetracycline and doxycycline despite some having identical tet(M) protein sequences as isolates that tested not susceptible to both drugs. Further work is needed to understand the contribution of this L528F-containing tet(M) allele to doxycycline susceptibility, but detection of tet(M) L528F could flag isolates with indeterminant doxycycline susceptibilities to avoid misclassifications.
FIG 2.
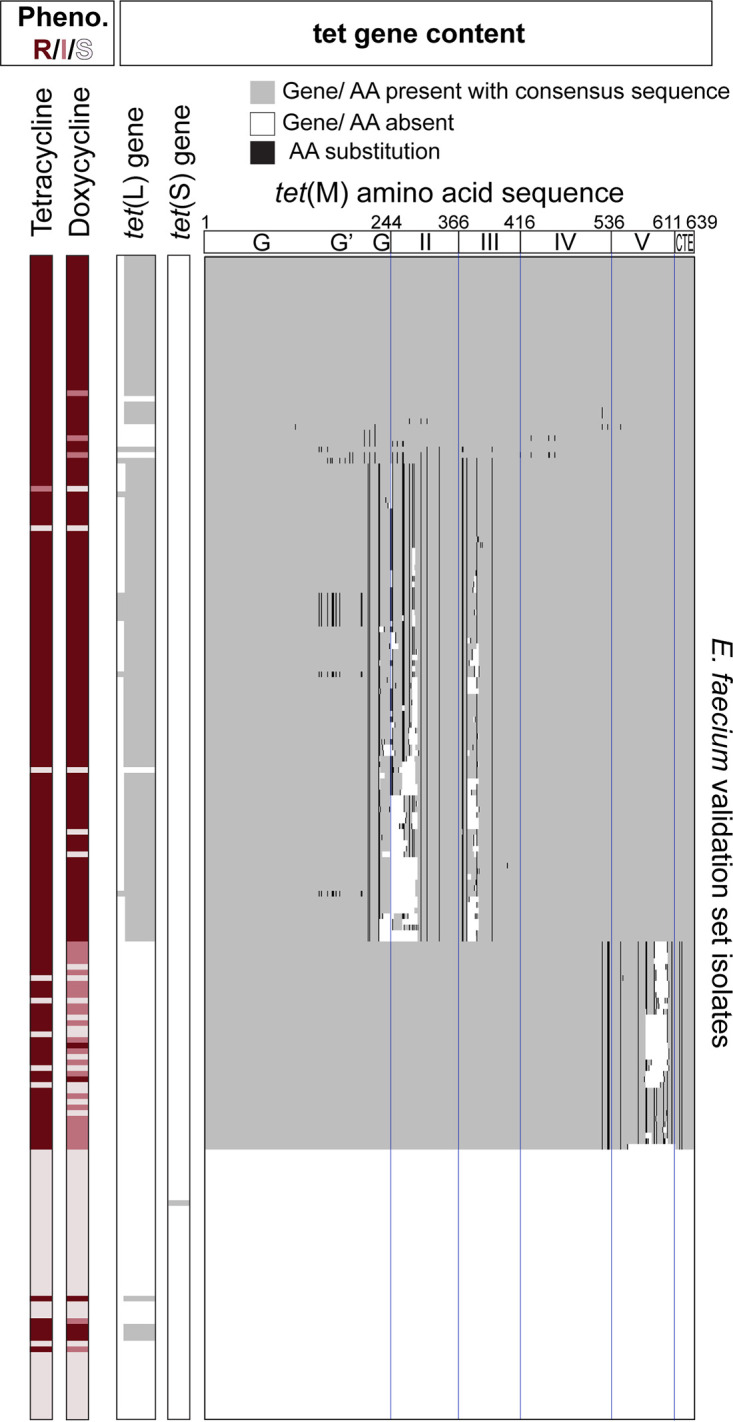
Phenotypic-genotypic correlations between tetracycline and doxycycline susceptibility testing results and tet gene content. Rows represent individual E. faecium validation set isolates. Columns represent phenotypic susceptibility testing results or tet gene content. The allelic form of tet(M) containing deletions and SNPs in the fifth TetM domain comprised the majority of isolates with phenotypic-genotypic discrepancies, and this allelic form could be distinguished from others based on tet(M) L528F. R, resistant; I, intermediate; S, susceptible; AA, amino acid.
Applying updated genotypic prediction rules to an external test data set.
To assess the generalizability of the AMR prediction rules, we assessed the revised rules’ classification accuracy on an external test data set of 50 clinical E. faecium isolates collected from Germany that had been previously published and characterized (19). All isolates belonged to CC17, with 38% ST-117, 14% ST-80, 12% ST-262, and 8% ST-203. The genotypic predictions and phenotypes had 100% categorical agreement for ampicillin, vancomycin, and ciprofloxacin (Table 3). Gentamicin susceptibility testing results were reported for only 13 isolates, for all of which the gentamicin MIC was at or above 128 μg/mL. The genotypic prediction rule only had 69% categorical agreement due to one false negative from an isolate that reportedly contained the aph(2″)-Ih gene and three false positives from isolates that all contained the aac(6′)-Ie-aph(2″)-Ia gene yet had gentamicin MICs of 128 μg/mL, which is 1 dilution below the EUCAST breakpoint (19). These discrepancies may be due to methodological differences in incubation times, broth composition, and antibiotic concentrations used between the two data sets or associated with the phylogenetic background of the isolates, which were all ST-262. Tetracycline had 94% categorical agreement, with two minor errors and one major error in a tet(M)-containing isolate with a tetracycline MIC within 1 dilution of the breakpoint. Phenotypic data for doxycycline and levofloxacin were not available. Finally, linezolid had 100% categorical agreement, where both linezolid-resistant (MIC 32 μg/mL) isolates had at least three alleles with the G2576T mutation. Six isolates tested as linezolid intermediate, but only one of those isolates had the G2576T mutation, and it was found in two alleles. Overall, the prediction rules appeared to generalize very well when applied to an isolate set from a different continent.
TABLE 3.
Accuracy of genotypic prediction rules V.2 when applied to the German test set
| Antimicrobial | Total no. of isolates | Phenotypic susceptibility rate, % | Genotype used for prediction (V.2) | Categorical agreement, % | Very major error rate (FN), % (95% CI) | Major error rate (FP), % (95% CI) | PPV, % | NPV, % |
|---|---|---|---|---|---|---|---|---|
| Ampicillin | 50 | 0 | Mutation of pbp5 485M or presence of 466′S/D | 100 | 0 (0, 7) | NAa | 100 | NA |
| Vancomycin | 50 | 20 | Presence of vanA or vanB Flag isolates with vanHAX but without vanRS as potential VVE | 100 | 0 (0, 8.8) | 0 (0, 31) | 100 | 100 |
| Gentamicin, high levelb | 13 | 30 | Presence of aac(6′)-le-aph(2”)-la | 69 | 10 (0.25, 44) | 100 (29, 100) | 75 | 0 |
| Ciprofloxacinc | 50 | 0 | Mutation of gyrA (84S) or parC (82S) | 100 | 0 (0, 7) | NA | 100 | NA |
| Levofloxacinc | NA | Mutation of gyrA (84S) or parC (82S) | ||||||
| Tetracycline | 50 | 56 | Presence of tet(L), tet(M), or tet(S) | 94 | 0 (0, 17) | 3.6 (0, 18) | 95 | 90 |
| Doxycycline | NA | Presence of tet(M) flag isolates with tet(M) L528F as indeterminant | ||||||
| Linezolidc | 50 | 96 | Mutation of 23S rRNA G2576T in at least 3 alleles | 100 | 0 (0, 84) | 0 (0, 7.4) | 100 | 100 |
NA, not available.
Gentamicin was only tested on a subset of isolates using an alternative phenotypic method from that used in the derivation and validation sets. For all three isolates that were phenotypically susceptible, the gentamicin MIC was 128 μg/mL, which is 1 dilution below the EUCAST breakpoint, and the isolate contained the aac(6′)-Ie-aph(2″)-Ia gene.
Antibiotic for which intermediate isolates were considered with susceptible isolates.
DISCUSSION
E. faecium is one of the most common and difficult-to-treat multidrug-resistant pathogens, yet systematic, genome-wide surveys of its antibiotic resistance mechanisms in clinical isolates have not been performed. In this study, we collected, sequenced, and analyzed a large and diverse set of clinical E. faecium isolates from Massachusetts General Hospital (MGH) and found a surprisingly small set of AMR genes and SNPs that we incorporated into simple antibiotic resistance prediction rules. When applied to a validation set that was prospectively collected from the same hospital, our rules predicted phenotypic susceptibility testing results with an average categorical agreement of 98% across eight commonly used antibiotics. When applied to a geographically distant test set, the genotypic-phenotypic agreement was 97% across six antibiotics analyzed. In addition to supporting rule generalizability, the diverse sample set enabled the detection of rare alleles, such as the ampicillin-resistant Asp466′ and Met485 pbp5 alleles, enhanced the generalizability of the prediction rules, and may elucidate antibiotic resistance mechanisms. Our data suggest that genomic methods are potentially useful for guiding E. faecium antibiotic selection.
We were able to resolve all genotype-phenotype discrepancies in the validation set except for tetracycline and doxycycline, which had false-positive rates of 14% and 27%, respectively, when the presence of tet(L), tet(M), or tet(S) was used for resistance prediction. The majority of false-positive genotypic predictions occurred with one allelic form of tet(M) that contained deletions and SNPs in the fifth TetM domain, which makes critical contacts with 23S rRNA (38). In the short term, L528F or other SNPs in the fifth TetM domain may be used as a proxy to flag isolates with “indeterminant” genotypic predictions. Future studies may examine the mechanism for the observed variability, such as expression differences due to variation in the tet(M) promoter sequence, and assess how the tet(L) and tet(M) allele variation impacts resistance to other important tetracycline-derived antibiotics like minocycline and tigecycline. Finally, while tet(S) was included in the prediction rule, its contribution to tetracycline resistance is unclear because it was found as the sole tet gene, in its wild-type form, in only three isolates: two tetracycline- and doxycycline-resistant derivation set isolates but also one tetracycline- and doxycycline-susceptible validation set isolate.
Our study also had some notable limitations. The phenotypic susceptibility testing results for the majority of isolates were generated by the Vitek 2 automated instrument, rather than the broth microdilution reference method, though previous studies have demonstrated 99 to 100% essential agreement between the two methods with the drugs included in our prediction rules (39, 40). In addition, our genotypic prediction rules included most, but not all, antibiotics with CLSI breakpoints for E. faecium. Daptomycin has obvious clinical importance in the treatment of vancomycin-resistant E. faecium infections, but resistant isolates remain very rare in most hospitals, the phenotypic gold standard is prone to error (41), the genotypic resistance mechanism is incompletely characterized, and daptomycin susceptibility results are available for only a subset of isolates because of selective testing of E. faecium in our laboratory. Likewise, enterococci are not routinely tested for quinupristin-dalfopristin, rifampin, teicoplanin, and minocycline susceptibility at our institution, leading to a dearth of derivation data. More generally, we specifically determined phenotypic categories using CLSI breakpoints rather than the ECVs for maximal translatability to clinical practice, though ECV-based predictions may have led to higher agreement due to their better correlation with wild-type and non-wild-type categories (42). Like for most other genotype-based predictions, we did not explicitly predict the intermediate category, though doing so may have had the most notable performance improvement on the doxycycline predictions. While these simple rules appeared to generalize well when applied to a test set of German clinical E. faecium isolates, further testing with geographically diverse isolates is warranted, as geographic variations in resistance mechanisms have been described (43). In particular, additional acquired resistance genes that are not included in our rules, such as optrA and poxtA for linezolid, may need to be included in genotypic prediction strategies when applied in settings where those antibiotics are used more commonly and selective pressures are stronger (44). Finally, these genotypic prediction rules may need to be updated in the future to reflect emerging mechanisms of AMR.
While WGS is not yet routinely implemented in most clinical microbiology labs due to several factors, including the lack of FDA-cleared in vitro diagnostics and cost, our findings can be used to improve existing E. faecium diagnostics and inform the development and interpretation of future testing strategies. Rapid molecular VRE detection methods used in many clinical laboratories currently only predict vancomycin resistance but could be modified to predict resistance to other highly relevant antibiotics, like ampicillin. Implementing our genotypic prediction rules that are based on essential sites like pbp5 485M and 466′ rather than the complete set of mutations that may exist in a given gene may simplify assay design and improve generalizability compared to other rules-based prediction methods (19). The development and validation of prediction rules like ours rely heavily on existing literature but can be used to supplement more agnostic, unsupervised approaches and provide interpretability for clinicians (16). Finally, our findings also underscore the value of genomic surveillance in enhancing our understanding of the molecular epidemiology of AMR and improving patient care through the development of more rapid methods to determine AMR.
MATERIALS AND METHODS
Microbiologic species identification and susceptibility testing.
For all isolates, species identification was performed using matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) mass spectrometry (Vitek MS; bioMérieux). Initial susceptibility testing was performed using the AST-GP75 card on the bioMérieux Vitek 2 instrument for ampicillin, high-level gentamicin, ciprofloxacin, levofloxacin, linezolid, vancomycin, tetracycline, and doxycycline (validated within our laboratory). Confirmatory testing was performed by broth microdilution (Sensititre Gram-positive GPALL3F AST plate; Thermo Fisher Scientific), CLSI reference disk diffusion (45), and gradient diffusion (Etest; bioMérieux) when needed. Categorical interpretations were assigned using CLSI document M100 (30th edition) breakpoints (46). Intermediate isolates were considered with susceptible isolates for ciprofloxacin, levofloxacin, and linezolid and with resistant isolates for vancomycin, tetracycline, and doxycycline, based on the epidemiologic cutoff value (ECV).
Clinical isolate collection.
The derivation and validation sets were collected from the Massachusetts General Hospital (MGH) Microbiology Laboratory. The derivation set included a random sampling of E. faecium isolates collected from January 2016 to December 2017. The validation set was collected from January 2018 to September 2019 with an attempt to enrich for organisms with unusual phenotypic resistance to linezolid, high-level gentamicin, or daptomycin, though no daptomycin-resistant isolates were available. Duplicate isolates collected from the same patient (e.g., same isolation site over multiple days or multiple isolation sites on the same day) were removed from the data sets. The derivation set included 177 isolates and the validation set included 205 isolates. Further details regarding isolate collection are available in the Supplemental Methods. All isolates were collected under a research protocol reviewed by the MGH internal review board (IRB; protocol 2017P000376). The test set (phenotypic AST and whole-genome sequencing data) was obtained from a previous study (19), in which a convenience set of E. faecium isolates from Germany was tested via broth microdilution, which we reinterpreted using CLSI clinical breakpoints.
DNA extraction and sequencing.
Purified DNA was extracted from isolates as previously described (47), with minor modifications (see the Supplemental Methods). Library preparation was performed with the Nextera DNA library preparation kit using a protocol adapted from that of Baym et al. (48). Paired-end, 150-bp sequencing was performed on Illumina platforms (MiSeq, HiSeq, and NextSeq). Sequencing reads were quality filtered using Trimmomatic (49), with a threshold quality score of 20 for leading, trimming, and sliding-window trimming and a minimum read length of 36. Eight isolates with under 20× genome coverage were excluded from the analysis.
Species confirmation and MLST.
Species identification was confirmed using Kraken v1.0 (http://ccb.jhu.edu/software/kraken/). Multilocus sequencing typing (MLST) was performed on whole-genome sequences using SRST2 (50) with an E. faecium allele table retrieved from PubMLST containing 1,518 sequence types (STs). GoeBURST analysis was performed at the single-locus variant level with visualization of the first two groups: hospital-associated CC17 and clonal complex 94 (CC94). Members of a clonal complex share at least four alleles with the central sequence type (ST-17 to CC17 and ST-94 to CC94). Core genome MLST (cgMLST) (51) assignment and generation of a neighbor joining tree (ignoring missing values) were performed with Ridom SeqSphere+ v.8.0.0 software on contigs from de novo assembly using SPAdes v.3.15.2 (default parameters with the isolate flag). Phylogenetic tree visualization was performed with ggtree v.2.2.1 (52) in RStudio v.1.3.959.
Resistance gene and SNP identification.
Resistance genes were detected using SRST2 v.0.2.0 (with a minimum coverage cutoff of 90%, maximum divergence cutoff of 10%, minimum depth of 5×, and maximum of 10 mismatches per read with the option to report all consensus sequences and pileups) with the ARG-ANNOT v.3 (53) database supplemented with gyrA (GenBank accession no. NC_017960.1 and NCBI:protein accession no. WP_002288365.1), parC (GenBank accession no. NC_017960.1 and NCBI:protein accession no. WP_002296998.1), pbp5-S (GenBank accession no. GG670325.1 and NCBI:protein accession no. EEV61481), and 23S rRNA (GenBank accession no. CP046077.1) sequences. Additional van genes, aminoglycoside-modifying genes, and linezolid resistance genes were detected using CARD v.3.0.8, as noted in Tables S2, S3, and S4, respectively. Consensus sequences generated by SRST2 for pbp5, gyrA, parC, and tet(M) were imported to AliView (54) for single nucleotide polymorphism (SNP) identification. The 23S rRNA G2576T allele frequency was determined using the base calls mapping to position 2576 within the pileups generated by SRST2. Specifically, the allele fraction was the ratio of the number of reads with 2576T to the total number of aligned reads covering that position. Allele fractions were visually confirmed in Integrated Genomics Viewer. The complete resistance gene and SNP information for each isolate was collated in Table S5. Diagnostic accuracy and confidence intervals were determined with the epiR v.1.0-15 R package.
Implementation of genotypic predictions.
Antimicrobial resistance predictions were made by implementing the rules (Supplemental Methods) with a custom R script using the genotypic information described above as the input.
Genotypic-phenotypic discrepancy resolution.
Fifteen validation set isolates with genotypic-phenotypic discrepancies were revived and underwent repeat phenotyping, repeat genotyping, or both. Repeat testing resolved discrepancies in four isolates and confirmed the original results in the remaining isolates. Two samples were found to contain two or more colony morphotypes of E. faecium, which were individually isolated, phenotyped, and sequenced.
Ethics approval and consent to participate.
This study was approved by the Mass General Brigham Institutional Review Board with a waiver of written informed consent.
Data availability.
The raw sequencing reads have been deposited to the NCBI BioProject database under accession number PRJNA771404.
ACKNOWLEDGMENTS
M.N.A. and D.S.K. are cofounders, consultants, and shareholders of Day Zero Diagnostics. J.T.B., J.X., L.A.D., J.M.P., E.S.R., and V.M.P. declare that they have no competing interests.
Funding was provided by the Ragon Institute.
M.N.A, E.S.R., V.M.P., and D.S.K. contributed to the study design, data acquisition, analysis, and data interpretation. J.T.B., J.X., L.A.D., and J.M.P. contributed to the data acquisition and analysis. All authors discussed the results and contributed to the final manuscript.
We are very grateful to all of the MGH Microbiology Laboratory staff for their unwavering dedication to providing high-quality clinical data every day, Fatima Hussain for her sequencing assistance, Matthew Hayward for sharing his phylogenetic and genomic expertise, Seth Bloom for his scientific and clinical insights, and Sarah Turbett and John Branda for their microbiologic expertise and support.
Footnotes
Supplemental material is available online only.
Contributor Information
Virginia M. Pierce, Email: vmpierce@mgh.harvard.edu.
Douglas S. Kwon, Email: dkwon@mgh.harvard.edu.
REFERENCES
- 1.Weiner-Lastinger LM, Abner S, Edwards JR, Kallen AJ, Karlsson M, Magill SS, Pollock D, See I, Soe MM, Walters MS, Dudeck MA. 2020. Antimicrobial-resistant pathogens associated with adult healthcare-associated infections: summary of data reported to the National Healthcare Safety Network, 2015–2017. Infect Control Hosp Epidemiol 41:1–18. 10.1017/ice.2019.296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arias CA, Murray BE. 2012. The rise of the Enterococcus: beyond vancomycin resistance. Nat Rev Microbiol 10:266–278. 10.1038/nrmicro2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.CDC. 2019. Antibiotic resistance threats in the United States, 2019. US Department of Health and Human Services, CDC, Atlanta, GA. [Google Scholar]
- 4.Tacconelli E, Carrara E, Savoldi A, Harbarth S, Mendelson M, Monnet DL, Pulcini C, Kahlmeter G, Kluytmans J, Carmeli Y, Ouellette M, Outterson K, Patel J, Cavaleri M, Cox EM, Houchens CR, Grayson ML, Hansen P, Singh N, Theuretzbacher U, Magrini N, Group W, WHO Pathogens Priority List Working Group. 2018. Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect Dis 18:318–327. 10.1016/S1473-3099(17)30753-3. [DOI] [PubMed] [Google Scholar]
- 5.Lebreton F, van Schaik W, McGuire AM, Godfrey P, Griggs A, Mazumdar V, Corander J, Cheng L, Saif S, Young S, Zeng Q, Wortman J, Birren B, Willems RJ, Earl AM, Gilmore MS. 2013. Emergence of epidemic multidrug-resistant Enterococcus faecium from animal and commensal strains. mBio 4:e00534-13. 10.1128/mBio.00534-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arias CA, Contreras GA, Murray BE. 2010. Management of multidrug-resistant enterococcal infections. Clin Microbiol Infect 16:555–562. 10.1111/j.1469-0691.2010.03214.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.DiazGranados CA, Zimmer SM, Klein M, Jernigan JA. 2005. Comparison of mortality associated with vancomycin-resistant and vancomycin-susceptible enterococcal bloodstream infections: a meta-analysis. Clin Infect Dis 41:327–333. 10.1086/430909. [DOI] [PubMed] [Google Scholar]
- 8.Kadri SS, Lai YL, Warner S, Strich JR, Babiker A, Ricotta EE, Demirkale CY, Dekker JP, Palmore TN, Rhee C, Klompas M, Hooper DC, Powers JH, III, Srinivasan A, Danner RL, Adjemian J, National Institutes of Health Antimicrobial Resistance Outcomes Research Initiative. 2021. Inappropriate empirical antibiotic therapy for bloodstream infections based on discordant in-vitro susceptibilities: a retrospective cohort analysis of prevalence, predictors, and mortality risk in US hospitals. Lancet Infect Dis 21:241–251. 10.1016/S1473-3099(20)30477-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Faron ML, Ledeboer NA, Buchan BW. 2016. Resistance mechanisms, epidemiology, and approaches to screening for vancomycin-resistant Enterococcus in the health care setting. J Clin Microbiol 54:2436–2447. 10.1128/JCM.00211-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Altun O, Almuhayawi M, Ullberg M, Ozenci V. 2013. Clinical evaluation of the FilmArray blood culture identification panel in identification of bacteria and yeasts from positive blood culture bottles. J Clin Microbiol 51:4130–4136. 10.1128/JCM.01835-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Babady NE, Gilhuley K, Cianciminio-Bordelon D, Tang YW. 2012. Performance characteristics of the Cepheid Xpert vanA assay for rapid identification of patients at high risk for carriage of vancomycin-resistant enterococci. J Clin Microbiol 50:3659–3663. 10.1128/JCM.01776-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rybkine T, Mainardi JL, Sougakoff W, Collatz E, Gutmann L. 1998. Penicillin-binding protein 5 sequence alterations in clinical isolates of Enterococcus faecium with different levels of beta-lactam resistance. J Infect Dis 178:159–163. 10.1086/515605. [DOI] [PubMed] [Google Scholar]
- 13.Diaz L, Tran TT, Munita JM, Miller WR, Rincon S, Carvajal LP, Wollam A, Reyes J, Panesso D, Rojas NL, Shamoo Y, Murray BE, Weinstock GM, Arias CA. 2014. Whole-genome analyses of Enterococcus faecium isolates with diverse daptomycin MICs. Antimicrob Agents Chemother 58:4527–4534. 10.1128/AAC.02686-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gorrie C, Higgs C, Carter G, Stinear TP, Howden B. 2019. Genomics of vancomycin-resistant Enterococcus faecium. Microb Genom 5:e000283. 10.1099/mgen.0.000283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Su M, Satola SW, Read TD. 2019. Genome-based prediction of bacterial antibiotic resistance. J Clin Microbiol 57:e01405-18. 10.1128/JCM.01405-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Anahtar MN, Yang JH, Kanjilal S. 2021. Applications of machine learning to the problem of antimicrobial resistance: an emerging model for translational research. J Clin Microbiol 59:e01260-20. 10.1128/JCM.01260-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stoesser N, Batty EM, Eyre DW, Morgan M, Wyllie DH, Del Ojo Elias C, Johnson JR, Walker AS, Peto TE, Crook DW. 2013. Predicting antimicrobial susceptibilities for Escherichia coli and Klebsiella pneumoniae isolates using whole genomic sequence data. J Antimicrob Chemother 68:2234–2244. 10.1093/jac/dkt180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gordon NC, Price JR, Cole K, Everitt R, Morgan M, Finney J, Kearns AM, Pichon B, Young B, Wilson DJ, Llewelyn MJ, Paul J, Peto TE, Crook DW, Walker AS, Golubchik T. 2014. Prediction of Staphylococcus aureus antimicrobial resistance by whole-genome sequencing. J Clin Microbiol 52:1182–1191. 10.1128/JCM.03117-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bortolaia V, Kaas RS, Ruppe E, Roberts MC, Schwarz S, Cattoir V, Philippon A, Allesoe RL, Rebelo AR, Florensa AF, Fagelhauer L, Chakraborty T, Neumann B, Werner G, Bender JK, Stingl K, Nguyen M, Coppens J, Xavier BB, Malhotra-Kumar S, Westh H, Pinholt M, Anjum MF, Duggett NA, Kempf I, Nykasenoja S, Olkkola S, Wieczorek K, Amaro A, Clemente L, Mossong J, Losch S, Ragimbeau C, Lund O, Aarestrup FM. 2020. ResFinder 4.0 for predictions of phenotypes from genotypes. J Antimicrob Chemother 75:3491–3500. 10.1093/jac/dkaa345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tyson GH, Sabo JL, Rice-Trujillo C, Hernandez J, McDermott PF. 2018. Whole-genome sequencing based characterization of antimicrobial resistance in Enterococcus. Pathog Dis 76:fty018. 10.1093/femspd/fty018. [DOI] [PubMed] [Google Scholar]
- 21.Ekwanzala MD, Dewar JB, Kamika I, Momba MNB. 2020. Comparative genomics of vancomycin-resistant Enterococcus spp. revealed common resistome determinants from hospital wastewater to aquatic environments. Sci Total Environ 719:137275. 10.1016/j.scitotenv.2020.137275. [DOI] [PubMed] [Google Scholar]
- 22.Homan WL, Tribe D, Poznanski S, Li M, Hogg G, Spalburg E, Van Embden JD, Willems RJ. 2002. Multilocus sequence typing scheme for Enterococcus faecium. J Clin Microbiol 40:1963–1971. 10.1128/JCM.40.6.1963-1971.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leavis HL, Bonten MJ, Willems RJ. 2006. Identification of high-risk enterococcal clonal complexes: global dispersion and antibiotic resistance. Curr Opin Microbiol 9:454–460. 10.1016/j.mib.2006.07.001. [DOI] [PubMed] [Google Scholar]
- 24.Pietta E, Montealegre MC, Roh JH, Cocconcelli PS, Murray BE. 2014. Enterococcus faecium PBP5-S/R, the missing link between PBP5-S and PBP5-R. Antimicrob Agents Chemother 58:6978–6981. 10.1128/AAC.03648-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sauvage E, Kerff F, Fonze E, Herman R, Schoot B, Marquette JP, Taburet Y, Prevost D, Dumas J, Leonard G, Stefanic P, Coyette J, Charlier P. 2002. The 2.4-A crystal structure of the penicillin-resistant penicillin-binding protein PBP5fm from Enterococcus faecium in complex with benzylpenicillin. Cell Mol Life Sci 59:1223–1232. 10.1007/s00018-002-8500-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Teixeira LM, Carvalho MGS, Facklam RR, Shewmaker PL. 2015. Enterococcus, p 403–421. In Jorgensen JH, Pfaller MA, Carroll KC (ed), Manual of clinical microbiology, 11th ed. ASM Press, Washington, DC. [Google Scholar]
- 27.Mingeot-Leclercq MP, Glupczynski Y, Tulkens PM. 1999. Aminoglycosides: activity and resistance. Antimicrob Agents Chemother 43:727–737. 10.1128/AAC.43.4.727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Costa Y, Galimand M, Leclercq R, Duval J, Courvalin P. 1993. Characterization of the chromosomal aac(6′)-Ii gene specific for Enterococcus faecium. Antimicrob Agents Chemother 37:1896–1903. 10.1128/AAC.37.9.1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.European Committee for Antimicrobial Susceptibility Testing. 2012. Breakpoint tables for interpretation of MICs and zone diameters. EUCAST, Växjö, Sweden. http://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/Breakpoint_table_v_2.0_120221.pdf. [Google Scholar]
- 30.Caryl JA, Cox G, Trimble S, O’Neill AJ. 2012. “tet(U)” is not a tetracycline resistance determinant. Antimicrob Agents Chemother 56:3378–3379. 10.1128/AAC.05957-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sinclair A, Arnold C, Woodford N. 2003. Rapid detection and estimation by pyrosequencing of 23S rRNA genes with a single nucleotide polymorphism conferring linezolid resistance in enterococci. Antimicrob Agents Chemother 47:3620–3622. 10.1128/AAC.47.11.3620-3622.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.US Food and Drug Administration. 2009. Class II special controls guidance document: antimicrobial susceptibility test (AST) systems. US Food and Drug Administration, Rockville, MD. [Google Scholar]
- 33.Arthur M, Molinas C, Courvalin P. 1992. The VanS-VanR two-component regulatory system controls synthesis of depsipeptide peptidoglycan precursors in Enterococcus faecium BM4147. J Bacteriol 174:2582–2591. 10.1128/jb.174.8.2582-2591.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thaker MN, Kalan L, Waglechner N, Eshaghi A, Patel SN, Poutanen S, Willey B, Coburn B, McGeer A, Low DE, Wright GD. 2015. Vancomycin-variable enterococci can give rise to constitutive resistance during antibiotic therapy. Antimicrob Agents Chemother 59:1405–1410. 10.1128/AAC.04490-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gagnon S, Levesque S, Lefebvre B, Bourgault AM, Labbe AC, Roger M. 2011. vanA-containing Enterococcus faecium susceptible to vancomycin and teicoplanin because of major nucleotide deletions in Tn1546. J Antimicrob Chemother 66:2758–2762. 10.1093/jac/dkr379. [DOI] [PubMed] [Google Scholar]
- 36.Szakacs TA, Kalan L, McConnell MJ, Eshaghi A, Shahinas D, McGeer A, Wright GD, Low DE, Patel SN. 2014. Outbreak of vancomycin-susceptible Enterococcus faecium containing the wild-type vanA gene. J Clin Microbiol 52:1682–1686. 10.1128/JCM.03563-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fiedler S, Bender JK, Klare I, Halbedel S, Grohmann E, Szewzyk U, Werner G. 2016. Tigecycline resistance in clinical isolates of Enterococcus faecium is mediated by an upregulation of plasmid-encoded tetracycline determinants tet(L) and tet(M). J Antimicrob Chemother 71:871–881. 10.1093/jac/dkv420. [DOI] [PubMed] [Google Scholar]
- 38.Arenz S, Nguyen F, Beckmann R, Wilson DN. 2015. Cryo-EM structure of the tetracycline resistance protein TetM in complex with a translating ribosome at 3.9-A resolution. Proc Natl Acad Sci USA 112:5401–5406. 10.1073/pnas.1501775112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bobenchik AM, Hindler JA, Giltner CL, Saeki S, Humphries RM. 2014. Performance of Vitek 2 for antimicrobial susceptibility testing of Staphylococcus spp. and Enterococcus spp. J Clin Microbiol 52:392–397. 10.1128/JCM.02432-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ligozzi M, Bernini C, Bonora MG, De Fatima M, Zuliani J, Fontana R. 2002. Evaluation of the VITEK 2 system for identification and antimicrobial susceptibility testing of medically relevant gram-positive cocci. J Clin Microbiol 40:1681–1686. 10.1128/JCM.40.5.1681-1686.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Satlin MJ, Nicolau DP, Humphries RM, Kuti JL, Campeau SA, Lewis Ii JS, Weinstein MP, Jorgensen JH. 2020. Development of daptomycin susceptibility breakpoints for Enterococcus faecium and revision of the breakpoints for other enterococcal species by the Clinical and Laboratory Standards Institute. Clin Infect Dis 70:1240–1246. 10.1093/cid/ciz845. [DOI] [PubMed] [Google Scholar]
- 42.Ellington MJ, Ekelund O, Aarestrup FM, Canton R, Doumith M, Giske C, Grundman H, Hasman H, Holden MTG, Hopkins KL, Iredell J, Kahlmeter G, Koser CU, MacGowan A, Mevius D, Mulvey M, Naas T, Peto T, Rolain JM, Samuelsen O, Woodford N. 2017. The role of whole genome sequencing in antimicrobial susceptibility testing of bacteria: report from the EUCAST Subcommittee. Clin Microbiol Infect 23:2–22. 10.1016/j.cmi.2016.11.012. [DOI] [PubMed] [Google Scholar]
- 43.Wardenburg KE, Potter RF, D’Souza AW, Hussain T, Wallace MA, Andleeb S, Burnham CD, Dantas G. 2019. Phenotypic and genotypic characterization of linezolid-resistant Enterococcus faecium from the USA and Pakistan. J Antimicrob Chemother 74:3445–3452. 10.1093/jac/dkz367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Egan SA, Shore AC, O’Connell B, Brennan GI, Coleman DC. 2020. Linezolid resistance in Enterococcus faecium and Enterococcus faecalis from hospitalized patients in Ireland: high prevalence of the MDR genes optrA and poxtA in isolates with diverse genetic backgrounds. J Antimicrob Chemother 75:1704–1711. 10.1093/jac/dkaa075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Clinical and Laboratory Standards Institute. 2018. Performance standards for antimicrobial disk susceptibility tests, 13th ed. CLSI standard M02. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 46.Clinical and Laboratory Standards Institute. 2020. Performance standards for antimicrobial susceptibility testing. 30th ed. CLSI supplement M100. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 47.Anahtar MN, Bowman BA, Kwon DS. 2016. Efficient nucleic acid extraction and 16S rRNA gene sequencing for bacterial community characterization. J Vis Exp 2016(110):53939. 10.3791/53939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Baym M, Kryazhimskiy S, Lieberman TD, Chung H, Desai MM, Kishony R. 2015. Inexpensive multiplexed library preparation for megabase-sized genomes. PLoS One 10:e0128036. 10.1371/journal.pone.0128036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Inouye M, Dashnow H, Raven LA, Schultz MB, Pope BJ, Tomita T, Zobel J, Holt KE. 2014. SRST2: rapid genomic surveillance for public health and hospital microbiology labs. Genome Med 6:90. 10.1186/s13073-014-0090-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.de Been M, Pinholt M, Top J, Bletz S, Mellmann A, van Schaik W, Brouwer E, Rogers M, Kraat Y, Bonten M, Corander J, Westh H, Harmsen D, Willems RJ. 2015. Core genome multilocus sequence typing scheme for high-resolution typing of Enterococcus faecium. J Clin Microbiol 53:3788–3797. 10.1128/JCM.01946-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yu G, Smith DK, Zhu H, Guan Y, Lam TT-Y. 2017. ggtree: an r package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods Ecol Evol 8:28–36. 10.1111/2041-210X.12628. [DOI] [Google Scholar]
- 53.Gupta SK, Padmanabhan BR, Diene SM, Lopez-Rojas R, Kempf M, Landraud L, Rolain JM. 2014. ARG-ANNOT, a new bioinformatic tool to discover antibiotic resistance genes in bacterial genomes. Antimicrob Agents Chemother 58:212–220. 10.1128/AAC.01310-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Larsson A. 2014. AliView: a fast and lightweight alignment viewer and editor for large datasets. Bioinformatics 30:3276–3278. 10.1093/bioinformatics/btu531. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material. Download AAC.01196-21-s0001.pdf, PDF file, 0.4 MB (371.7KB, pdf)
Supplemental Table S5. Download AAC.01196-21-s0002.xlsx, XLSX file, 0.1 MB (110.5KB, xlsx)
Data Availability Statement
The raw sequencing reads have been deposited to the NCBI BioProject database under accession number PRJNA771404.