ABSTRACT
Small base-pairing RNAs (sRNAs) and small proteins comprise two classes of regulators that allow bacterial cells to adapt to a wide variety of growth conditions. A limited number of transcripts encoding both of these activities, regulation of mRNA expression by base pairing and synthesis of a small regulatory protein, have been identified. Given that few have been characterized, little is known about the interplay between the two regulatory functions. To investigate the competition between the two activities, we constructed synthetic dual-function RNAs, hereafter referred to as MgtSR or MgtRS, comprised of the Escherichia coli sRNA MgrR and the open reading frame encoding the small protein MgtS. MgrR is a 98-nucleotide (nt) base-pairing sRNA that negatively regulates eptB encoding phosphoethanolamine transferase. MgtS is a 31-amino-acid (aa) small inner membrane protein that is required for the accumulation of MgtA, a magnesium (Mg2+) importer. Expression of the separate genes encoding MgrR and MgtS is normally induced in response to low Mg2+ by the PhoQP two-component system. By generating various versions of this synthetic dual-function RNA, we probed how the organization of components and the distance between the coding and base-pairing sequences contribute to the proper function of both activities of a dual-function RNA. By understanding the features of natural and synthetic dual-function RNAs, future synthetic molecules can be designed to maximize their regulatory impact.
IMPORTANCE Dual-function RNAs in bacteria encode a small protein and also base pair with mRNAs to act as small, regulatory RNAs. Given that only a limited number of dual-function RNAs have been characterized, further study of these regulators is needed to increase understanding of their features. This study demonstrates that a functional synthetic dual-regulator can be constructed from separate components and used to study the functional organization of dual-function RNAs, with the goal of exploiting these regulators.
KEYWORDS: MgrR, MgtS, Hfq, sRNA, small protein, synthetic biology
INTRODUCTION
Small RNAs (sRNAs) play an important role in shaping and regulating cellular responses to changing environmental conditions (reviewed in reference 1). Hundreds of sRNAs have been identified through various deep sequencing methodologies (reviewed in references 2 and 3). Most of these 50- to 300-nucleotide (nt) molecules regulate target mRNAs through base pairing (reviewed in reference 4). In many bacteria, sRNAs depend on the RNA binding protein Hfq (host factor for bacteriophage Qβ) for stability and efficient base pairing with their target mRNAs (reviewed in references 5 and 6). The functional outcomes of sRNA base pairing with the target mRNA include the inhibition of translation, an increase in translation, a change in mRNA degradation, as well as postinitiation decreases in transcription (reviewed in references 7 and 8).
While generally assumed to be noncoding, some sRNAs have been found to encode small open reading frames (sORFs), and for a small subset of the RNAs, this ORF has been shown to be translated. An sRNA capable of base-pairing regulation while also encoding a protein is denoted as a dual-function RNA (reviewed in reference 9). Until now, only a limited number of dual-function RNAs have been identified and characterized, as they tend to be overlooked due to the difficulty in distinguishing true translated ORFs from sORFs that occur randomly. Furthermore, traditional biochemical methods for identifying and purifying proteins commonly miss proteins that are less than 50 amino acids (aa) (reviewed in reference 10).
Despite the difficulties inherent in the study of dual-function RNAs, two Escherichia coli dual-function RNAs, SgrST and AzuCR, have been examined in sufficient depth to describe both the base-pairing and protein-coding activities. Both of these RNAs are involved in regulating carbon metabolism. The 227-nt SgrS sRNA was originally identified in a computational screen for sRNAs in E. coli (11). Subsequently, SgrS was shown to play a role in the cellular response to glucose-phosphate stress and was found to base pair with the ptsG and manXYZ mRNAs, which encode important sugar transporters (12, 13). In the course of these studies, it was discovered that SgrS is translated to produce the 43-amino acid small protein SgrT, which inhibits the transport activity of the glucose permease PtsG (14, 15). Together, SgrS and SgrT counteract the accumulation of sugar phosphates in the cell by reducing the translation and activity of sugar transporters and increasing dephosphorylation of sugar phosphates. Another recently characterized dual-function RNA is AzuCR (16). This 164-nt RNA was initially identified in a bioinformatic search for novel sRNA genes but was reclassified as an mRNA when it was found to encode a 28-aa protein, AzuC (17, 18). AzuC interacts with aerobic glycerol-3-phosphate dehydrogenase (GlpD) to increase its activity (16). However, overexpression of the RNA produces growth defects in galactose and glycerol medium that persist even when a derivative carrying a stop codon mutation in the sORF is overexpressed, suggesting that AzuCR also functions as an sRNA denoted AzuR. Consistent with this observation, the AzuCR transcript is capable of direct base-pairing regulation of cadA, encoding lysine decarboxylase, and galE, encoding UDP-glucose 4-epimerase (16).
One interesting question regarding dual-function RNAs is whether the two functions can be carried out simultaneously by the same molecule or whether the activities interfere with each other. The base-pairing region of SgrS is 15 nt downstream of the SgrT ORF. Mutations to inhibit sgrT translation do not impede SgrS regulation of mRNA targets, but mutations that restrict base pairing increase SgrT translation, indicating that the base-pairing function interferes with translation. Furthermore, translation of the small protein lags behind transcription of the SgrST RNA by approximately 30 min (19). Based on these observations, it was suggested that the base-pairing function predominates, and SgrST molecules that base pair with target mRNAs are unavailable to be translated, as they are codegraded. Once the pool of targeted mRNAs has been depleted, SgrST accumulates and is translated.
In contrast, there is direct overlap between the base-pairing region and sORF of AzuCR such that one function necessarily interferes with the other (16). The introduction of a stop codon at the third codon, which presumably reduces ribosomal occupancy of the transcript, increases base-pairing activity. For AzuCR, translation of the small protein is repressed by the anaerobic growth-induced sRNA FnrS, suggesting one mechanism by which translation of the small protein might be inhibited, allowing base-pairing activity to predominate, is through sRNA base pairing near the ribosome binding site of the sORF (16).
To further explore the features of dual-function RNAs that impact what activity predominates, we chose to construct synthetic dual-function RNAs. We decided to use parts of the transcript corresponding to the sRNA MgrR and the mRNA encoding the small protein MgtS. The sRNA component, MgrR, was identified through Hfq coimmunoprecipitation followed by genome-wide RNA detection on microarrays (20). This sRNA negatively regulates eptB, a lipopolysaccharide (LPS)-modifying enzyme, and ygdQ, a protein of unknown function (21). MgtS is a 31-amino acid small protein that interacts with MgtA, a P-type ATPase Mg2+ importer (22). This small protein was first predicted as a conserved sORF, and further studies showed that it is translated in E. coli (11, 23). The interaction of MgtS with MgtA serves to increase levels of the importer and increase the intracellular concentration of Mg2+. Induction of both components, MgrR and MgtS, is dependent on the two-component system PhoQ/PhoP. In low-Mg2+ conditions, the sensor kinase PhoQ autophosphorylates and donates a phosphate group to PhoP. Phosphorylated PhoP in turn activates a number of different genes, including mgrR and mgtS, which are needed for survival in low Mg2+ (24).
In this study, we demonstrate that it is possible to create a synthetic dual-function RNA from existing individual components, in this case from the transcripts encoding MgrR and MgtS. Additionally, we show that the length and composition of the sequence between the functional elements impact their activities.
RESULTS
Construction of a synthetic dual-function RNA.
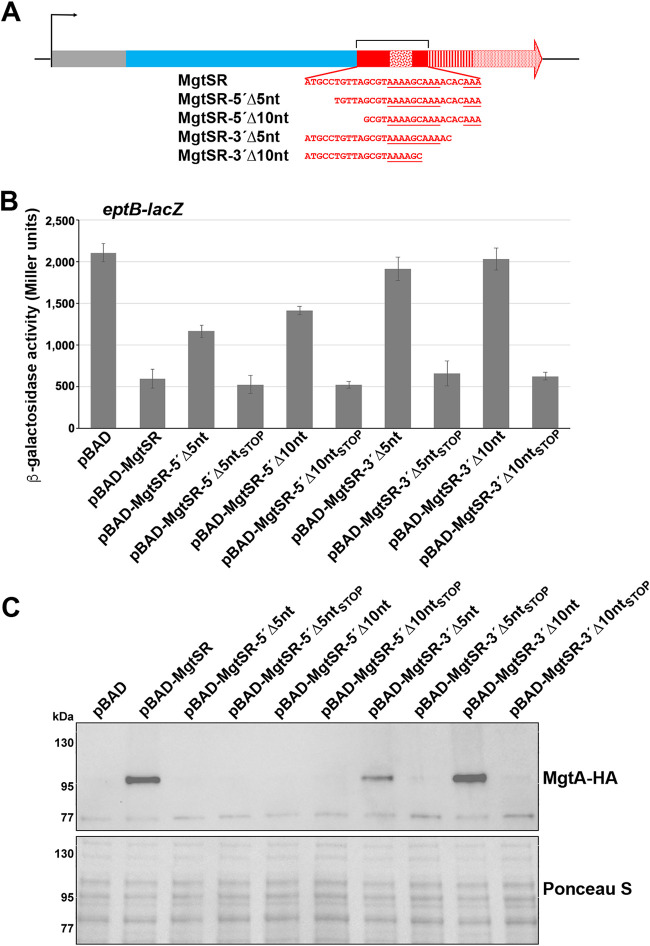
We set out to build a synthetic dual-function RNA from two components that could be easily tested for their regulatory activities. We chose MgrR as the sRNA component and MgtS as the small protein component. The corresponding two genes are transcribed convergently in a PhoPQ-dependent manner in response to low Mg2+ conditions (Fig. 1A). The design of the first synthetic RNA, denoted MgtSR, was modeled after the characterized dual-function RNA, SgrST, whose sORF precedes the base-pairing region by 15 nt (25). Our design included the MgtS ORF, from the start codon to the stop codon (96 nt), immediately followed by 77 nt of native mgrR sequence (see Fig. S1 in the supplemental material). The pBAD24 (pBAD) plasmid carrying the construct provided the ribosome binding site (RBS) for MgtS. The sequence from mgrR includes a 9-nt A-R-N (where A is adenine, R is adenine or guanine, and N is any nucleotide) Hfq binding motif, the seed sequence for base-pairing interactions with MgrR targets, and an intrinsic terminator with a polyU stretch also bound by Hfq (5, 26). The MgtS ORF and the seed sequence of MgrR are separated by 30 nt of native sequence from MgrR (denoted a spacer here) (Fig. 1B). The hybrid gene was cloned behind an arabinose-inducible promoter to control expression.
FIG 1.
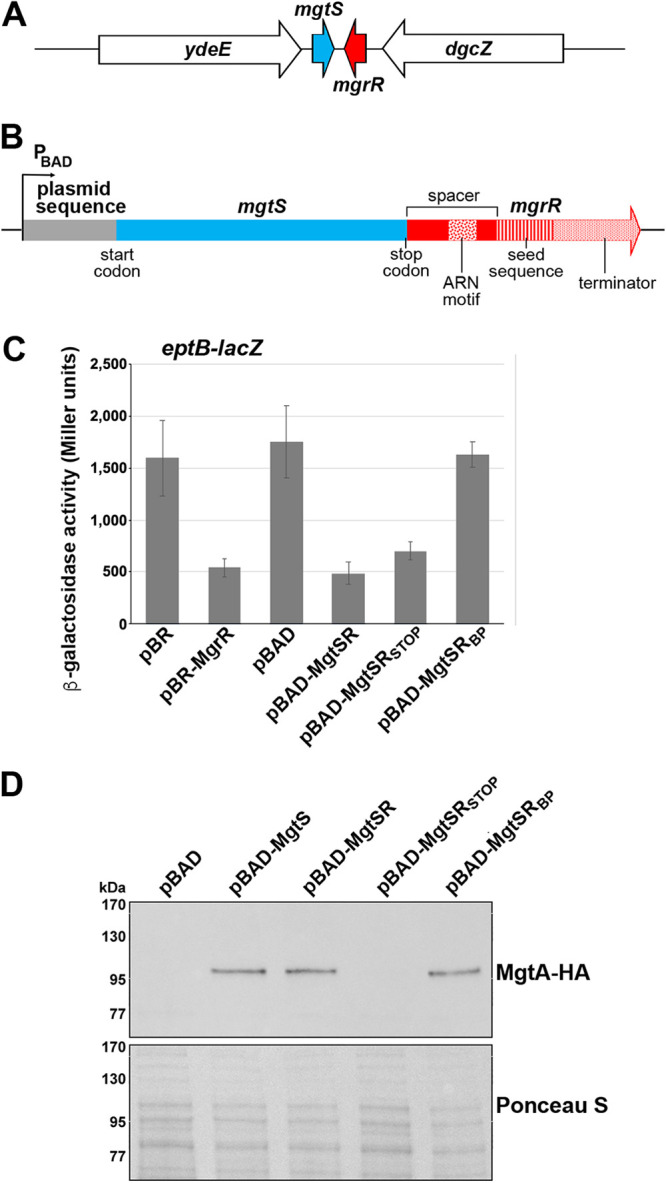
Regulatory activity of a synthetic dual-function RNA. (A) Diagram of the endogenous location of mgtS and mgrR. (B) Schematic of the hybrid dual-function RNA MgtSR. Plasmid sequences are in gray, mgtS sequences are in blue, and mgrR sequences are in red. The ARN, seed, and terminator sequences are indicated by distinct patterns as labeled. (C) Effect of pBR, pBR-MgrR, pBAD, pBAD-MgtSR, pBAD-MgtSRSTOP, or pBAD-MgtSRBP overexpression on an eptB-lacZ fusion in a ΔmgrR strain background (KM238). The graph shows averages from of three biological replicates with error bars representing standard deviation. (D) Immunoblot analysis of MgtA-HA cells in a ΔmgtS strain background (GSO786) transformed with pBAD, pBAD-MgtS, pBAD-MgtSR, pBAD-MgtSRSTOP, or pBAD-MgtSRBP grown to an OD600 of 0.4 in N medium supplemented with 500 μM Mg2+, washed, and then resuspended and grown in N medium supplemented with arabinose but without Mg2+. Samples were collected after 20 min, and MgtA-HA was visualized with anti-HA antibodies to detect MgtA-HA. The Ponceau S stain provides the loading control.
We also designed controls to individually eliminate the activity of each component (see Fig. S1 in the supplemental material). To this end, we constructed a stop codon control, pBAD-MgtSRSTOP, with a stop codon replacing the second codon to prevent translation of the small protein component while maintaining the integrity of the base-pairing region, and pBAD-MgtSRBP, a construct with mutations in the base-pairing region of MgrR to prevent sRNA activity while maintaining translation of the small protein. The expected ∼200-nt RNA was expressed from all constructs, though the transcripts from pBAD-MgtSR, pBAD-MgtSRBP appeared as a doublet, while the transcript for pBAD-MgtSRSTOP is a single band that is present at somewhat higher levels (see Fig. S2A in the supplemental material).
MgtSR is functional.
To determine whether MgtSR is functional, we first assayed the effect of MgtSR overexpression on the activity of an eptB-lacZ fusion, a reporter fusion to a known target of MgrR. β-Galactosidase activity was measured in ΔmgrR cells carrying the eptB-lacZ translational fusion and transformed with pBR322 (pBR), pBR-MgrR, pBAD, pBAD-MgtSR, pBAD-MgtSRSTOP, and pBAD-MgtSRBP. Repression of eptB-lacZ was observed upon overexpression of MgtSR to a similar extent as overexpression of natural MgrR (Fig. 1C). Furthermore, pBAD-MgtSRSTOP derivative was still able to repress the fusion, but the pBAD-MgtSRBP no longer affected eptB-lacZ expression. This result demonstrates that the MgtSR RNA has the ability to act as a base-pairing regulatory RNA.
We similarly tested for functional expression of the MgtS component by carrying out immunoblot analysis to examine the levels of MgtA, a membrane protein that is stabilized by MgtS (22). ΔmgtS cells expressing chromosomally encoded hemagglutinin (HA)-tagged MgtA were transformed with pBAD, pBAD-MgtS, pBAD-MgtSR, pBAD-MgtSRSTOP, and pBAD-MgtSRBP. Overexpression of MgtSR increases MgtA-HA levels under conditions of Mg2+ limitation similar to natural MgtS (Fig. 1D). As expected, pBAD-MgtSRSTOP did not affect MgtA-HA levels while pBAD-MgtSRBP did, demonstrating that a functional small protein is translated from mgtSRBP despite the inability of the transcript to repress eptB-lacZ (Fig. 1D). The β-galactosidase data taken together with the immunoblot analysis indicate that we have successfully constructed a synthetic dual-function RNA that can act as a base-pairing sRNA and also express a functional small protein.
Translation of mgtS is dominant in the absence of a spacer.
To examine how the mgrR sequence that provides a spacer between the mgtS coding sequence and the mgrR base-pairing region impacts the activities of the two MgtSR components, we removed the 30-nt sequence to generate the pBAD-MgtSR-no spacer construct (Fig. 2A; see also Fig. S1). We hypothesized that in the absence of the spacer, the two functions may interfere with each other, resulting in the loss of one or both of the activities. To test this hypothesis, we assayed the eptB-lacZ fusion and MgtA-HA levels for cells transformed with pBR, pBR-MgrR, pBAD, pBAD-MgtSR, and pBAD-MgtSR-no spacer. We discovered that removing the spacer sequence between the components of MgtSR eliminates regulation of the eptB-lacZ fusion (Fig. 2B) but not stabilization of MgtA (Fig. 2C). These results suggest that translation of MgtS predominates and may sterically block MgtSR base-pairing activity (27). To test this prediction, we introduced a stop codon into the pBAD-MgtSR-no spacer construct. This construct, pBAD-MgtSR-no spacerSTOP, rescued eptB-lacZ regulation, consistent with translation of mgtS disrupting base-pairing regulation by MgtSR (Fig. 2B). It is interesting to note that the MgtSR-no spacerSTOP RNA is still functional despite the upstream Hfq binding site being removed, though some ARN sequences dispersed throughout the mgtS coding sequence could conceivably provide an alternative binding site. We noted that the levels of this sRNA are significantly higher than MgtSR, though the levels decrease in a Δhfq mutant, consistent with the MgtSR-no spacerSTOP transcript binding Hfq (see Fig. S2A and B). As expected, the MgtSR-no spacerSTOP RNA, which should not be translated, did not lead to increased accumulation of MgtA-HA (Fig. 2C). Thus, the presence of a 30-nt spacer sequence helps to maintain the function of each component.
FIG 2.
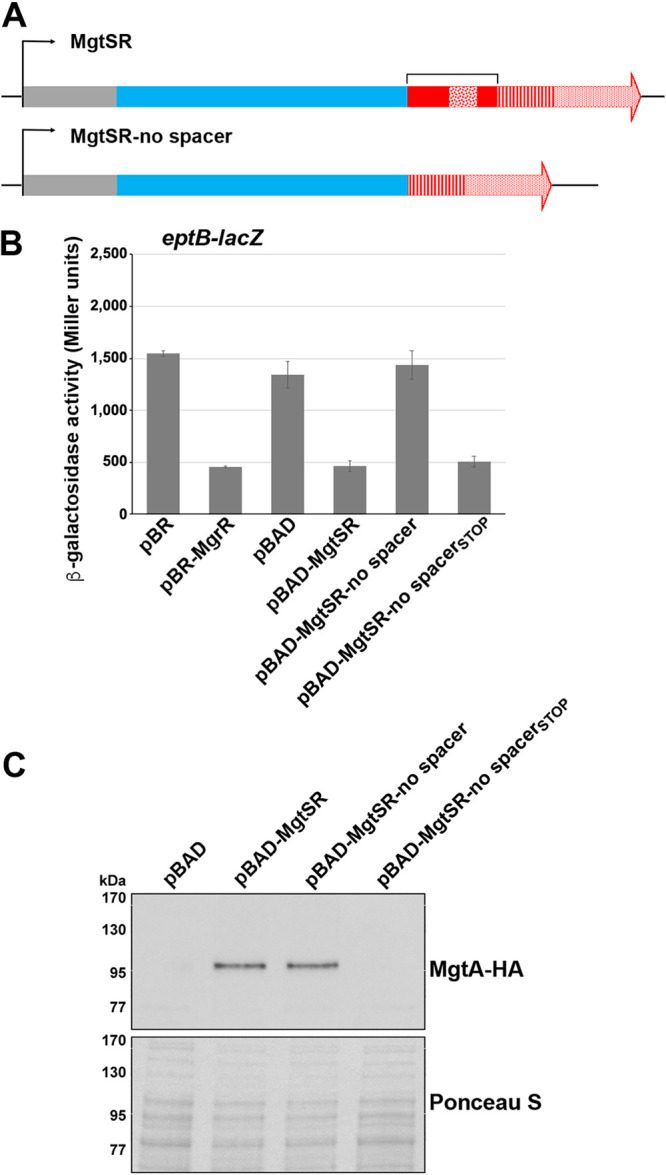
MgtS translation is the dominant activity absent the spacer sequence. (A) Schematic of the hybrid dual-function RNA with the spacer sequence (bracket) removed. Colors and patterns for plasmid (gray), mgtS (blue), mgrR (red), ARN (confetti), seed (striped), and terminator (stippled) sequences are as in Fig. 1B. (B) Effect of pBR, pBR-MgrR, pBAD, pBAD-MgtSR, or pBAD-MgtSR-no spacer overexpression on an eptB-lacZ fusion in a ΔmgrR strain background. The graph shows averages from three biological replicates with error bars representing standard deviation. (C) Immunoblot analysis of MgtA-HA cells in a ΔmgtS strain transformed with pBAD, pBAD-MgtSR, pBAD-MgtSR-no spacer, or pBAD-MgtSR-no spacerSTOP grown to an OD600 of 0.4 in N medium supplemented with 500 μM Mg2+, washed, and then resuspended and grown in N medium supplemented with arabinose but without Mg2+. Samples were collected after 20 min, and MgtA-HA was visualized with anti-HA antibodies to detect MgtA-HA. The Ponceau S stain provides the loading control.
Truncation of the spacer sequence differentially disrupts regulation by each component.
To better understand how the composition of the 30-nt spacer sequence contributes to regulation by each component, we next examined the effects of truncations from either end of the sequence to determine if the 5′ or 3′ end played a more prominent role in maintaining the functions of the synthetic dual-function RNA. We constructed derivatives with 5- or 10-nt truncations from either the 5′ or 3′ ends of the spacer sequence to give pBAD-MgtSR-5′Δ5nt, pBAD-MgtSR-5′Δ10nt, pBAD-MgtSR-3′Δ5nt, and pBAD-MgtSR-3′Δ10nt (Fig. 3A; see also Fig. S1). The levels of these MgtSR deletion derivatives are similar to the full-length MgtSR RNA, except for MgtSR-3′Δ10nt, which is somewhat lower (see also Fig. S2B). We carried out β-galactosidase assays for eptB-lacZ strains carrying each of these plasmids or the control pBAD and pBAD-MgtSR plasmids. Interestingly, the derivatives with truncations near the 3′ end of the 30-nt sequence completely lost the ability to regulate eptB-lacZ, while the constructs with truncations near the 5′ end retained some ability to repress eptB-lacZ (Fig. 3B). When stop codons were introduced into each of the truncation constructs, the RNAs all repressed eptB-lacZ, demonstrating that the truncated constructs retain the potential to base pair and that translation of the sORF is responsible for disrupting regulatory activity.
FIG 3.
Analysis of the regulatory contributions of the spacer sequence. (A) Schematic of the 5-nt and 10-nt truncation mutants of MgtSR. Colors and patterns for plasmid (gray), mgtS (blue), mgrR (red), ARN (confetti), seed (striped), and terminator (stippled) sequences are as in Fig. 1B. (B) Effects of overexpression from pBAD, pBAD-MgtSR, pBAD-MgtSR truncations, or pBAD-MgtSR truncation stop constructs on an eptB-lacZ fusion in a ΔmgrR strain background. The graph shows averages from three biological replicates with error bars representing standard deviation. (C) Immunoblot analysis of MgtA-HA cells in a ΔmgtS strain transformed with pBAD, pBAD-MgtSR, pBAD-MgtSR truncations, or pBAD-MgtSR truncation stop constructs grown to an OD600 of 0.4 in N medium supplemented with 500 μM Mg2+, washed, and then resuspended and grown in N medium supplemented with arabinose but without Mg2+. Samples were collected after 20 min, and MgtA-HA was visualized with anti-HA antibodies to detect MgtA-HA. The Ponceau S stain provides the loading control.
In contrast, the truncations toward the 5′ end of the spacer sequence interfered with the MgtSR effect on MgtA-HA levels, while the 3′ truncation derivatives were less defective (Fig. 3C). Compared to the MgtSR control, no MgtA-HA was observed for pBAD-MgtSR-5′Δ5nt and pBAD-MgtSR-5′Δ10nt, somewhat lower MgtA-HA levels were observed for MgtSR-3′Δ5nt, and equivalent MgtA-HA levels were observed for MgtSR-3′Δ10nt. Thus, while removal of portions of the spacer sequence from the 5′ end had less of an effect on base-pairing activity, these deletions prevented MgtS-mediated stabilization of MgtA-HA. In contrast, the 3′ end truncation derivatives, which lacked base-pairing activity, still stabilized MgtA-HA, indicating MgtS function. Possibly, truncations at the 5′ or 3′ end produce changes in RNA structure that disrupt translation or base pairing, respectively. These results show that the sequence between the ORF and seed region has an impact on the functions of the chimeric RNA.
The spacer sequence of the reverse MgtRS also impacts activity.
To test whether the order of the encoded functions of the synthetic dual-function RNA impacted the activities, we placed the mgtS 5′ untranscribed region (UTR), including the RBS (denoted spacer sequence) and the mgtS coding sequence, between the base-pairing region and terminator sequence of mgrR (Fig. 4A; see also Fig. S1). However, when cells with an eptB-lacZ fusion were transformed with pBAD and pBAD-MgtRS, we did not observe decreased β-galactosidase activity as seen for MgtSR (Fig. 4B). Furthermore, pBAD-MgtRS did not increase MgtA-HA levels as seen for pBAD-MgtSR (Fig. 4C). Northern analysis revealed that MgtRS was not detected, indicating that the transcript might be unstable (see Fig. S2C). We hypothesized that the process of reversing the order of mgtS and mgrR coding elements might have disrupted binding to the Hfq protein if the ARN motif of MgrR is too distant from the terminator to allow for strong Hfq binding. Alternatively, the combinations of elements may have inadvertently introduced an endonuclease cleavage site.
FIG 4.
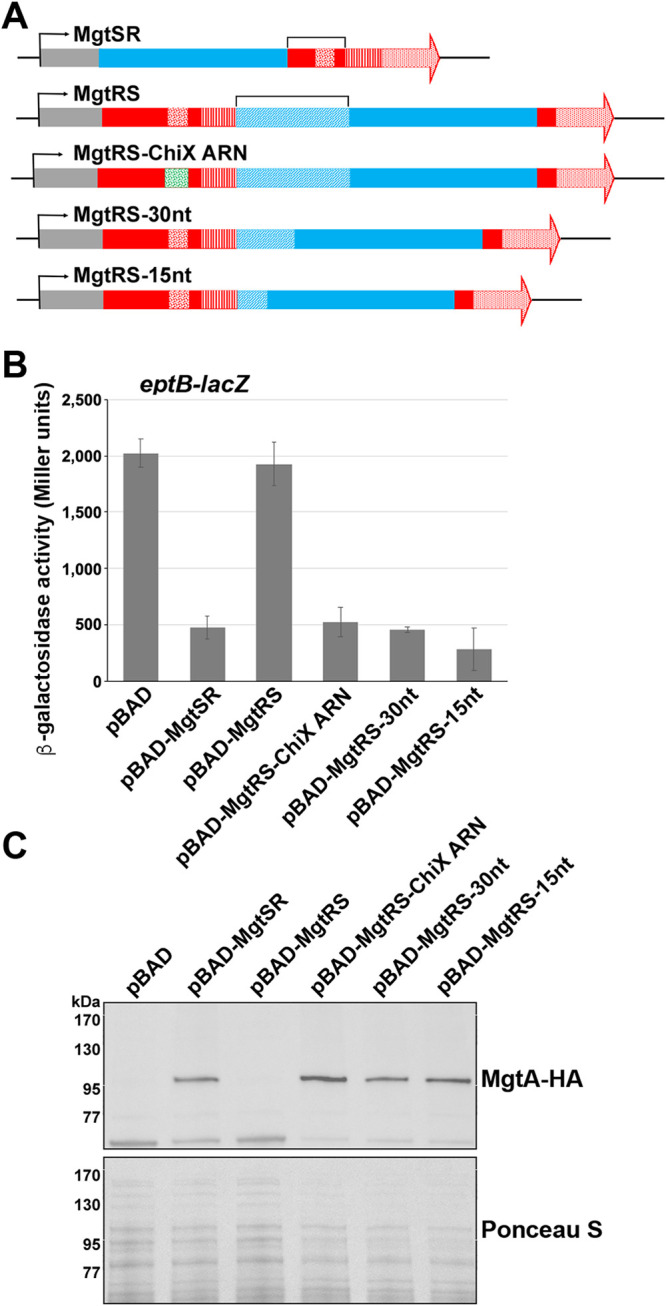
Reversal of components produces a nonviable construct in the absence of an appropriate spacer. (A) Schematic of the inverse hybrid constructs. Colors and patterns for plasmid (gray), mgtS (blue), mgrR (red), ARN (confetti), seed (striped), and terminator (stippled) sequences are as in Fig. 1B. The stippled blue region corresponds to the mgtS sequence outside of the coding region, and the confetti green region denotes the ChiX ARN sequence. (B) Effects of pBAD, pBAD-MgtSR, pBAD-MgtRS, pBAD-MgtRS-ChiX ARN, pBAD-MgtRS-30nt, or pBAD-MgtRS-15nt overexpression on an eptB-lacZ fusion in a ΔmgrR strain background. The graph shows averages from three biological replicates with error bars representing standard deviation. (C) Immunoblot analysis of MgtA-HA cells in a ΔmgtS strain transformed with pBAD, pBAD-MgtSR, pBAD-MgtRS, pBAD-MgtRS-ChiX ARN, pBAD-MgtRS-30nt, or pBAD-MgtRS-15nt grown to an OD600 of 0.4 in N medium supplemented with 500 μM Mg2+, washed, and then resuspended and grown in N medium supplemented with arabinose but without Mg2+. Samples were collected after 20 min, and MgtA-HA was visualized with anti-HA antibodies to detect MgtA-HA. The Ponceau S stain provides the loading control.
To attempt to stabilize the MgtRS derivative and gain activity, we made a number of modifications. First, we replaced the ARN sequence of mgrR in MgtRS with the strong ARN sequence from ChiX (MgtRS-ChiX ARN). Additionally, we made two other variants of MgtRS in which the 58-nt spacer sequence was truncated to 30 nt or 15 nt. We chose these lengths given the 30-nt spacer sequence in the functional MgtSR construct and the 15-nt spacer sequence in SgrS. When eptB-lacZ cells carrying pBAD-MgtRS-ChiX ARN, pBAD-MgtRS-30nt, and pBAD-MgtRS-15nt were assayed for β-galactosidase activity, we observed that the altered versions of MgtRS were all capable of eptB-lacZ repression (Fig. 4B). Additionally, pBAD-MgtRS-ChiX ARN, pBAD-MgtRS-30nt, pBAD-MgtRS-15nt were capable of increasing MgtA-HA levels to a similar degree as MgtSR (Fig. 4C). Northern analysis showed that all of the constructs could be detected, though we also observed some shorter products, and MgtRS-15nt was present at elevated levels (see Fig. S2C). Together, these results show that functional synthetic dual-function RNAs can be generated with the two functional elements, protein coding and base pairing, in either order as long as the appropriate spacer sequence is present.
Synthetic dual-function RNAs complement the ΔmgtS growth defect in low Mg2+.
Finally, we examined the ability of the synthetic dual-function RNAs to complement the low Mg2+ sensitivity of a ΔmgtS strain. While a growth defect was observed in limited Mg2+ media for the ΔmgtS strains carrying the pBAD vector control, the pBAD-MgtSR, pBAD-MgtRS-ChiX ARN, and pBAD-MgtRS-15nt constructs all protected the ΔmgtS strain cells at low intracellular Mg2+ levels similar to pBAD-MgtS (Fig. 5). These data demonstrate that synthetic dual-function RNAs can produce biologically relevant regulatory effects.
FIG 5.
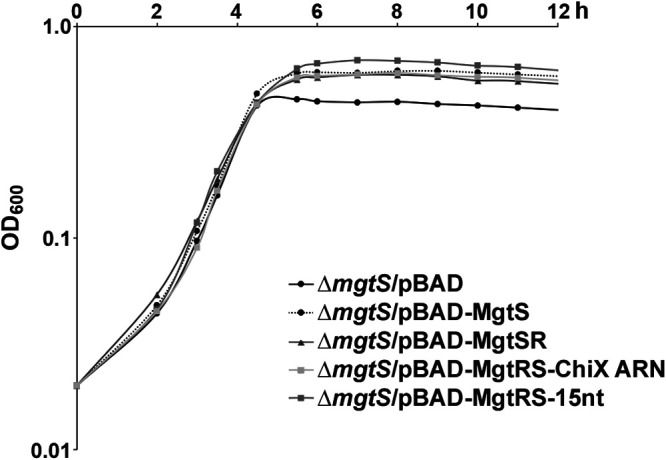
MgtSR and MgtRS constructs allow growth in low magnesium. ΔmgtS::kan strain (GSO229) transformed with pBAD, pBAD-MgtS, pBAD-MgtSR, pBAD-MgtRS-ChiX, or pBAD-MgtRS-15nt grown overnight in LB was diluted to an OD600 of ∼0.02 (time zero) in 25 ml of the N medium with no Mg2+ and grown at 37°C. Growth was tracked by OD600 over 12 h.
DISCUSSION
Dual-function RNAs are an intriguing yet largely unexplored class of regulatory molecules. Given the underidentification of small proteins, these RNA transcripts are likely more prevalent than has currently been reported. To better understand the organization and competition between the components of a dual-function RNA, we designed and constructed synthetic dual-function RNAs from the individual genes encoding the MgrR sRNA and MgtS small protein (Fig. 1A). These hybrid RNAs are capable of regulating by base pairing and are translated to give functional MgtS protein. We used derivatives of the synthetic constructs to examine how the organization of the components affects the regulatory activities of these RNA molecules.
Elements necessary for dual function.
The MgtSR and MgtRS chimeras and their derivatives underscore what elements are necessary to construct a dual-function RNA. First, translation of the sORF likely requires an unobstructed ribosome binding site. Proper sRNA function requires an unobstructed seed sequence for base pairing along with Hfq binding sites, including a Rho-independent terminator. How these elements are spaced relative to one another has an impact on the two activities. We suspect that when the ARN sequence for Hfq binding was moved far from the Hfq binding site at the Rho-dependent terminator in the MgtRS construct, Hfq was no longer able to stabilize the RNA. This was overcome by replacing the mgrR ARN sequence with the longer chiX ARN sequence (Fig. 4). The length of the sequence that genetically separates the components of dual-function RNA also is important for maintaining the independent regulatory activity of each component. Most deletions that reduced the sequence separating the sORF and the seed sequence in MgtSR eliminated base-pairing activity (Fig. 2 and 3). The observation that all of these derivatives regained base-pairing activity upon the introduction of a stop codon suggests that the main obstacle to base pairing is occlusion by ribosome binding, although some of the deletions might also affect Hfq binding. For most of these deletion constructs, the MgtS activity predominated, possibly because this activity is encoded first. Interestingly, however, with a limited distance between the two genetic elements, base pairing can also interfere with translation as illustrated for the 5′Δ5 nt and 5′Δ10 nt derivatives.
In natural dual-function RNAs, overlap or close proximity of the ORF and base-pairing regions can lead to competition between the activities. For AzuCR, mutations that prevent translation of the ORF enhance regulation by the sRNA component (16). For SgrS, where the components are separated by 15 nt, the introduction of mutations to disrupt translation of the encoded small protein, SgrT, do not change base-pairing regulation of mRNA targets by SgrS (19). Nevertheless, mutations that disrupted base pairing actually led to an increase in SgrT levels (19). This is consistent with the model in which the stability of SgrS RNA dictates that translation will occur after base-pairing regulation is completed and suggests that a single transcript only acts as an mRNA or an sRNA, not both simultaneously. Overall, we expect that separation of the parts of a dual-function RNA by a spacer sequence gives each component more autonomy despite existing within the same RNA molecule. However, based on what has been observed for AzuCR and SgrST, we suggest that an individual MgtSR or MgtRS molecule is either translated or acts as a base-pairing RNA, but this hypothesis needs further testing.
Evolution of dual-function RNAs.
In thinking about the components of dual-function sRNAs, it is interesting to consider how these regulatory molecules evolve (28) and whether there are more barriers to evolution of base pairing or expression of a small protein as the second feature. Phylogenetic analyses of SgrST and the RNAIII dual-function RNA from Staphylococcus aureus suggest that evolution in both directions is possible. For example, all SgrST homologs across enteric species carry the highly conserved 13-nt seed sequence, complementary to the ptsG RNA near the 3′ end of the RNA, while some species with an SgrS homolog, such as Yersinia, lack a functional SgrT ORF, suggesting that the sRNA served as the precursor RNA, which evolved an mRNA function (29). In contrast, the 26-aa δ-hemolysin encoded by RNAIII appears to be more broadly conserved than the base-pairing regions, indicating an mRNA evolved base-pairing capability (30).
Possibility of exploiting dual-function RNAs.
Dual-function RNAs present an interesting opportunity for synthetic biology applications. As we have demonstrated in this work, the creation of an artificial dual-function RNA from existing sRNA and small protein components is possible and can be tuned to favor one or the other activity. Given that these molecules can regulate both the translation and activity of a protein, dual function RNAs can provide an effective means to rapidly alter both mRNA levels and protein activity and thus allow efficient control of a specific process. Furthermore, if the two activities regulate different pathways, a single dual-function RNA can provide a regulatory link between the two pathways. By modulating when each component is active, these molecules could be used to regulate two pathways under mutually exclusive conditions. Dual-function RNAs represent an opportunity to design transcripts that can effectively control gene expression using two different regulatory mechanisms. Ultimately, there is much to be explored about the regulatory potential of natural and synthetic dual-function RNAs.
MATERIALS AND METHODS
Bacterial strains and plasmid construction.
Bacterial strains, plasmids, and oligonucleotides used in this study are listed in Tables S1 to S3 in the supplemental material. E. coli strains are derivatives of wild-type MG1655 (crl+). To generate the ΔmgtS Δhfq strain, Δhfq::cat-sacB from GSO954 was P1 transduced into GSO786. The sequences of all plasmid inserts were confirmed.
Bacterial growth.
Cells were grown to the indicated optical density at 600 nm (OD600) in Luria-Bertani broth (LB) overnight or N-minimal medium [7.5 mM (NH4)2SO4, 0.5 mM K2SO4, 1 mM KH2PO4, 0.4% glycerol, 5 mM KCl, 100 mM Tris-HCl (pH 7.5), 10 μM vitamin B1, and 0.1% Casamino Acids] with either 500 μM or 0 μM Mg2+ (31). pBAD24 and pBRplac plasmid constructs were induced with 0.4% arabinose or 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG). Where indicated, media contained antibiotics with the following concentrations: ampicillin (100 μg/ml), chloramphenicol (25 μg/ml), and kanamycin (30 μg/ml).
β-Galactosidase assays.
Cultures were grown in LB to an OD600 of ∼1.0 with arabinose (0.2%). Cells (100 μl) were added to 700 μl of Z buffer (60 mM Na2HPO4·7H2O, 40 mM NaH2PO4·H2O, 10 mM KCl, 1 mM MgSO4·7H2O, and 50 mM β-mercaptoethanol). After adding 15 μl of freshly prepared 0.1% SDS and 30 μl of chloroform to each sample, the cells were vortexed for 30 s and then incubated at room temperature for 15 min to lyse the cells. The assay was initiated by the addition of 100 μl of o-nitrophenyl-β-d-galactopyranoside (ONPG) (4 mg/ml) to each sample in 10-s intervals. The samples were incubated at room temperature before the reactions were terminated by the addition of 500 μl of 1 M Na2CO3, after which A420 and A550 values were determined with a spectrophotometer, and the absorbance data were used to calculate Miller units.
Immunoblot analysis.
After growth in the indicated medium, cells corresponding to 0.5 OD600 were collected by centrifugation; resuspended in 1× phosphate-buffered saline (PBS) (KD Medical), 7 μl of 2× Laemmli buffer (Bio-Rad), and 2 μl of β-mercaptoethanol; and 10 μl was loaded on a Mini-Protean TGX 5% to 20% Tris-glycine gel (Bio-Rad) and run in 1× Tris glycine-SDS (KD Medical) buffer. The proteins were electrotransferred to nitrocellulose membranes (Invitrogen) for 1 h at 100 V. Membranes were blocked with 5% nonfat milk (Bio-Rad) in 1× PBS with 0.1% of Tween 20 (PBS-T) for 1 h and probed with a 1:1,000 dilution of anti-HA antiserum (Abcam) in the same PBS-T buffer with 5% milk for 1 h. After the incubation with the anti-HA antiserum, membranes were incubated with a 1:20,000 dilution of horseradish peroxidase (HRP)-labeled goat anti-rabbit antibody (Pierce). All blots were washed 4× with PBS-T and then developed with an Amersham ECL Western blotting detection kit (GE Healthcare).
Total RNA isolation.
Cells corresponding to the equivalent of 10 OD600 were collected by centrifugation and snap-frozen in liquid nitrogen. RNA was extracted according to the standard TRIzol (Thermo Fisher Scientific) protocol. Briefly, 1 ml of room temperature TRIzol was added to cell pellets, which were resuspended thoroughly, and incubated for 5 min at room temperature. After the addition of 200 μl of chloroform and thorough mixing by inversion, samples were incubated for 10 min at room temperature. After samples were centrifuged for 10 min at 4°C at 20,000 × g, the upper phase (∼0.6 ml) was transferred into a new tube and 500 μl of isopropanol was added. Samples again were mixed thoroughly by inversion, incubated for 10 min at room temperature, and centrifuged at 20,000 × g for 15 min at 4°C. RNA pellets were washed twice with 75% ethanol and then dried at room temperature. RNA was resuspended in 20 to 50 μl of diethyl pyrocarbonate (DEPC) water and quantified using a NanoDrop (Thermo Fisher Scientific).
Northern analysis.
Total RNA (5 to 10 μg per lane) was separated on denaturing 8% polyacrylamide gels containing 6 M urea (1:4 mix of UreaGel Complete to UreaGel 8 (National Diagnostics) with 0.08% ammonium persulfate) in 1× Tris-borate-EDTA (TBE) buffer at 300 V for 90 min. The RNA was transferred to a Zeta-Probe GT membrane (Bio-Rad) at 20 V for 16 h in 0.5× TBE, UV cross-linked, and probed with 32P-labeled oligonucleotides (listed in Table S1 in the supplemental material) in ULTRAhyb-oligo buffer (Ambion Inc.) at 45°C. Membranes were rinsed twice with 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)/0.1% SDS at room temperature and once with 0.2× SSC/0.1% SDS at room temperature, washed for 25 min with 0.2× SSC/0.1% SDS at 45°C, and followed by a final rinse with 0.2× SSC/0.1% SDS at room temperature before autoradiography was performed with HyBlot CL film (Denville Scientific Inc.).
Growth curves.
Colonies of the ΔmgtS::kan strain (GSO229) transformed with pBAD, pBAD-MgtS, pBAD-MgtSR, pBAD-MgtRS-ChiX, and pBAD-MgtRS-15nt grown overnight in LB were diluted to an OD600 of ∼0.02 (time zero) in 25 ml of the N medium with no Mg2+ and grown at 37°C. Growth was monitored for 22 h.
ACKNOWLEDGMENTS
We thank members of the Storz group for comments on the manuscript.
This research was supported by the Intramural Research Program of the Eunice Kennedy Shriver National Institute of Child Health and Human Development.
Footnotes
Supplemental material is available online only.
Contributor Information
Gisela Storz, Email: storzg@mail.nih.gov.
Tina M. Henkin, Ohio State University
REFERENCES
- 1.Nitzan M, Rehani R, Margalit H. 2017. Integration of bacterial small RNAs in regulatory networks. Annu Rev Biophys 46:131–148. 10.1146/annurev-biophys-070816-034058. [DOI] [PubMed] [Google Scholar]
- 2.Adams PP, Storz G. 2020. Prevalence of small base-pairing RNAs derived from diverse genomic loci. Biochim Biophys Acta Gene Regul Mech 1863:194524. 10.1016/j.bbagrm.2020.194524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hör J, Gorski SA, Vogel J. 2018. Bacterial RNA biology on a genome scale. Mol Cell 70:785–799. 10.1016/j.molcel.2017.12.023. [DOI] [PubMed] [Google Scholar]
- 4.Carrier MC, Lalaouna D, Massé E. 2018. Broadening the definition of bacterial small RNAs: characteristics and mechanisms of action. Annu Rev Microbiol 72:141–161. 10.1146/annurev-micro-090817-062607. [DOI] [PubMed] [Google Scholar]
- 5.Updegrove TB, Zhang A, Storz G. 2016. Hfq: the flexible RNA matchmaker. Curr Opin Microbiol 30:133–138. 10.1016/j.mib.2016.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Woodson SA, Panja S, Santiago-Frangos A. 2018. Proteins that chaperone RNA regulation. Microbiol Spectr 6:6.4.21. 10.1128/microbiolspec.RWR-0026-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wagner EGH, Romby P. 2015. Small RNAs in bacteria and archaea: who they are, what they do, and how they do it. Adv Genet 90:133–208. 10.1016/bs.adgen.2015.05.001. [DOI] [PubMed] [Google Scholar]
- 8.Chen J, Morita T, Gottesman S. 2019. Regulation of transcription termination of small RNAs and by small RNAs: molecular mechanisms and biological functions. Front Cell Infect Microbiol 9:201. 10.3389/fcimb.2019.00201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Raina M, King A, Bianco C, Vanderpool CK. 2018. Dual-function RNAs. Microbiol Spectr 6:6.5.06. 10.1128/microbiolspec.RWR-0032-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Storz G, Wolf YI, Ramamurthi KS. 2014. Small proteins can no longer be ignored. Annu Rev Biochem 83:753–777. 10.1146/annurev-biochem-070611-102400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wassarman KM, Repoila F, Rosenow C, Storz G, Gottesman S. 2001. Identification of novel small RNAs using comparative genomics and microarrays. Genes Dev 15:1637–1651. 10.1101/gad.901001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rice JB, Vanderpool CK. 2011. The small RNA SgrS controls sugar-phosphate accumulation by regulating multiple PTS genes. Nucleic Acids Res 39:3806–3819. 10.1093/nar/gkq1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vanderpool CK, Gottesman S. 2004. Involvement of a novel transcriptional activator and small RNA in post-transcriptional regulation of the glucose phosphoenolpyruvate phosphotransferase system. Mol Microbiol 54:1076–1089. 10.1111/j.1365-2958.2004.04348.x. [DOI] [PubMed] [Google Scholar]
- 14.Wadler CS, Vanderpool CK. 2007. A dual function for a bacterial small RNA: SgrS performs base pairing-dependent regulation and encodes a functional polypeptide. Proc Natl Acad Sci USA 104:20454–20459. 10.1073/pnas.0708102104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lloyd CR, Park S, Fei J, Vanderpool CK. 2017. The small protein SgrT controls transport activity of the glucose-specific phosphotransferase system. J Bacteriol 199:e00869-16. 10.1128/JB.00869-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Raina M, Aoyama JJ, Bhatt S, Paul BJ, Storz G. 2021. AzuCR RNA modulates carbon metabolism as a dual-function RNA in Escherichia coli. bioRxiv 10.1101/2021.04.27.441574. [DOI] [PMC free article] [PubMed]
- 17.Chen S, Lesnik EA, Hall TA, Sampath R, Griffey RH, Ecker DJ, Blyn LB. 2002. A bioinformatics based approach to discover small RNA genes in the Escherichia coli genome. Biosystems 65:157–177. 10.1016/s0303-2647(02)00013-8. [DOI] [PubMed] [Google Scholar]
- 18.Hemm MR, Paul BJ, Miranda-Rios J, Zhang A, Soltanzad N, Storz G. 2010. Small stress response proteins in Escherichia coli: proteins missed by classical proteomic studies. J Bacteriol 192:46–58. 10.1128/JB.00872-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Balasubramanian D, Vanderpool CK. 2013. Deciphering the interplay between two independent functions of the small RNA regulator SgrS in Salmonella. J Bacteriol 195:4620–4630. 10.1128/JB.00586-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang A, Wassarman KM, Rosenow C, Tjaden BC, Storz G, Gottesman S. 2003. Global analysis of small RNA and mRNA targets of Hfq. Mol Microbiol 50:1111–1124. 10.1046/j.1365-2958.2003.03734.x. [DOI] [PubMed] [Google Scholar]
- 21.Moon K, Gottesman S. 2009. A PhoQ/P-regulated small RNA regulates sensitivity of Escherichia coli to antimicrobial peptides. Mol Microbiol 74:1314–1330. 10.1111/j.1365-2958.2009.06944.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang H, Yin X, Wu Orr M, Dambach M, Curtis R, Storz G. 2017. Increasing intracellular magnesium levels with the 31-amino acid MgtS protein. Proc Natl Acad Sci USA 114:5689–5694. 10.1073/pnas.1703415114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hemm MR, Paul BJ, Schneider TD, Storz G, Rudd KE. 2008. Small membrane proteins found by comparative genomics and ribosome binding site models. Mol Microbiol 70:1487–1501. 10.1111/j.1365-2958.2008.06495.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Prost LR, Miller SI. 2008. The Salmonellae PhoQ sensor: mechanisms of detection of phagosome signals. Cell Microbiol 10:576–582. 10.1111/j.1462-5822.2007.01111.x. [DOI] [PubMed] [Google Scholar]
- 25.Raina M, Storz G. 2017. SgrT, a small protein that packs a sweet punch. J Bacteriol 199:e00130-17. 10.1128/JB.00130-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kwiatkowska J, Wroblewska Z, Johnson KA, Olejniczak M. 2018. The binding of class II sRNA MgrR to two different sites on matchmaker protein Hfq enables efficient competition for Hfq and annealing to regulated mRNAs. RNA 24:1761–1784. 10.1261/rna.067777.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Takyar S, Hickerson RP, Noller HF. 2005. mRNA helicase activity of the ribosome. Cell 120:49–58. 10.1016/j.cell.2004.11.042. [DOI] [PubMed] [Google Scholar]
- 28.Dutcher HA, Raghavan R. 2018. Origin, Evolution, and Loss of bacterial small RNAs. Microbiol Spectr 6:6.2.12. 10.1128/microbiolspec.RWR-0004-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Horler RSP, Vanderpool CK. 2009. Homologs of the small RNA SgrS are broadly distributed in enteric bacteria but have diverged in size and sequence. Nucleic Acids Res 37:5465–5476. 10.1093/nar/gkp501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Verdon J, Girardin N, Lacombe C, Berjeaud JM, Hechard Y. 2009. Delta-hemolysin, an update on a membrane-interacting peptide. Peptides 30:817–823. 10.1016/j.peptides.2008.12.017. [DOI] [PubMed] [Google Scholar]
- 31.Nelson DL, Kennedy EP. 1971. Magnesium transport in Escherichia coli. Inhibition by cobaltous ion. J Biol Chem 246:3042–3049. 10.1016/S0021-9258(18)62288-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 and S2, Tables S1 to S3. Download JB.00345-21-s0001.pdf, PDF file, 1.3 MB (1.2MB, pdf)