Abstract
Purpose
Pseudomonas aeruginosa is an important pathogen that causes urinary tract infections. Carbapenems are the drugs of choice for the treatment of P. aeruginosa infections. However, the emergence and spread of carbapenem-resistant P. aeruginosa (CRPA) is a serious global health threat. In this study, we investigated the epidemiology, molecular characteristics, drug resistance, and virulence factors of CRPA isolated from urine samples.
Methods
A total of 124 P. aeruginosa isolates were obtained from urine samples collected between March 2020 and February 2021. Clonal relatedness was evaluated using multilocus sequence typing (MLST) and pulsed-field gel electrophoresis (PFGE). We performed antimicrobial susceptibility tests and investigated the presence of carbapenemase genes and virulence factors in CRPA isolates.
Results
The carbapenem resistance rate of the isolated P. aeruginosa was 46.7% (59/124). A total of 54 (91.5%) out of the 59 CRPA isolates were identified as multidrug-resistant. The majority of the CRPA isolates (81.4%, 48/59) harbored carbapenemase genes, such as blaIMP-6 or blaNDM-1. In an epidemiological analysis using MLST, 88.1% of CRPA isolates were confirmed to be ST773 (50.8%, 30/59) or ST235 (37.3%, 22/59). The CRPA isolates harboring blaIMP-6 and blaNDM-1 belonged to ST235 (PFGE pulsotypes E1-E18, F) and ST773 (PFGE pulsotypes H1-H2, I1-I16) subtypes, respectively. The studied CRPA isolates simultaneously harbored 10 to 14 virulence factors of the 16 virulence factors examined. Nine virulence factor genes (toxA, exoT, plcH, plcN, phzM, phzS, lasB, aprA, and algD) were identified in all CRPA isolates.
Conclusion
Our study shows that P. aeruginosa ST235 harboring blaIMP-6 and ST773 harboring blaNDM-1—known internationally as high-risk clones with multiple virulence factors—are widely spread in the study area. These findings suggest that continuous monitoring is necessary to prevent the further spread of carbapenemase-producing CRPA.
Keywords: carbapenem resistance, metallo-β-lactamase, virulence factor, urinary tract infection
Introduction
Urinary tract infections (UTIs) are one of the most common types of infectious diseases, leading to substantial medical costs each year and showing a high rate of recurrence.1 Pseudomonas aeruginosa is an important pathogen, responsible for about 7–11% of urinary tract infections.2,3
Carbapenems, such as imipenem and meropenem, are effective antibiotics for the treatment of P. aeruginosa infections.4 However, as the rate of carbapenem resistance in P. aeruginosa has rapidly increased in recent years, carbapenem-resistant P. aeruginosa (CRPA) has become a public health concern worldwide.5 The mechanisms of carbapenem resistance in P. aeruginosa are multifactorial and a combination of processes, including the loss of the outer membrane proteins, overexpression of efflux pumps, β-lactamase derepression, and acquisition of carbapenemase genes, could confer resistance to carbapenems in P. aeruginosa.6,7 Particularly, the production of carbapenemases in P. aeruginosa is important since CRPA has been rapidly spreading as a consequence of the acquisition of carbapenemase genes through mobile genetic elements.8 So far, class A, B, and D carbapenemases have been identified in P. aeruginosa, and class B metallo-β-lactamase (MBL) enzymes, including Verona Integron-encoded Metallo-β-lactamase (VIM), imipenemases (IMP), and New Delhi metallo-β-lactamase (NDM), are the most prevalent carbapenemases produced by P. aeruginosa.9
P. aeruginosa produces numerous virulence determinants associated with pathogenicity, including exotoxins, proteases, other enzymes, pigments, secretion systems, alginate capsules, flagella, and pili.10 Among these, Exotoxin A (ETA), encoded by the toxA gene, is an extracellular enzyme that inhibits protein synthesis of host cells by deactivating of elongation factor 2 and stimulates pro-inflammatory cytokine synthesis.11 ExoS, ExoT, ExoU, and ExoY, the effector exotoxins of type III secretion system (T3SS), are considered as an important virulence determinant of P. aeruginosa.12 ExoS and ExoT (bi-functional type III cytotoxins) and ExoY (type-III toxins with adenylate cyclase activity) are responsible for the disruption of the actin cytoskeleton, whereas ExoU is a type III cytotoxic phospholipase that causes the disruption of the host cytoplasmic membrane, resulting in cell lysis.13,14 Meanwhile, hemolytic phospholipase C (PlcH) and nonhemolytic phospholipase C (PlcN) hydrolyze various phospholipids of the host cell membrane: PlcH hydrolyzes host-associated phosphatidylcholine and the sphingomyelin and PlcN hydrolyzes phosphatidylserine and phosphatidylcholine.15 In addition, LasB elastase degrades the structural proteins of cells such as collagen, non-collagen proteins, and elastin, and LasA protease enhances the elastolytic activity of LasB.13,15 Alkaline protease (AprA) is a secreted zinc metalloprotease that degrades components of the host immune system, such as the complement system proteins (C1q, C2, C3), cytokines (IFN-γ, TNF-α), and components of the extracellular matrix (laminins, basal lamina) for tissue invasion and dissemination.13,15 Furthermore, pyoverdin (fluorescein), encoded by the pvdA gene, is a fluorescent yellow-green siderophore that facilitates the uptake of iron for the survival and growth of P. aeruginosa under conditions of iron limitation.16 On the other hand, pyocyanin, a blue-green phenazone-derivative pigment, is involved in the inflammatory response and tissue damage during pathogenesis by inducing cell apoptosis via increasing the levels of reactive oxygen species.17 The biosynthesis of pyocyanin is mediated by phenazine-modifying enzymes, N-methyltransferase PhzM and flavin-dependent hydroxylase PhzS.18 Meanwhile, the pilus proteins, PilA (responsible for the type-IV pilus protein) and PilB (essential for the synthesis of type-IV pili), are involved in bacterial adhesion and biofilm formation.19 Alginate is one of the essential components of pseudomonal biofilms and GDP-mannose dehydrogenase, encoded by the algD gene, is the key enzyme in the alginate biosynthesis pathway.19
An understanding of the epidemiology with molecular characterization of carbapenemase genes is critical to characterize the genetic diversity for confirming clonal diffusion of CRPA and to provide strategies and infection control measures.20 P. aeruginosa clones such as sequence types (STs) ST235, ST111, ST233, ST244, ST357, ST308, ST175, ST277, ST654, and ST298, so-called “international” or “high risk” clones, have been spread worldwide and are frequently associated with multidrug resistance.21 However, the molecular epidemiology and distribution of carbapenemases in CRPA may vary between diverse geographical and clinical origins, and studies on the coexistence of multidrug resistance (MDR) with multiple virulence factors of CRPA isolates causing UTIs are limited.
In this study, we aimed to evaluate the prevalence of antibiotic resistance and presence of carbapenemase genes and the various virulence factors and to carry out an epidemiological analysis using both multilocus sequence typing (MLST) and pulsed-field gel electrophoresis (PFGE) to reveal the current spread and cluster analysis of UTI-causative CRPA.
Materials and Methods
Bacterial Isolates
Urine samples were collected into a sterile container and transported immediately to the microbiology Laboratory. If the urine samples were not immediately treated, they were refrigerated for up to 24 hours. Unprocessed urine samples were inoculated onto MacConkey and blood agar plates using a 0.001-mL sterile calibrated loop for semi-quantitative urine culture. The plates were incubated aerobically at 37°C for 18–24 h. Isolates were identified using matrix-assisted laser desorption/ionization time-of-flight mass spectrometry with a MALDI Biotyper (Bruker Daltonics, Bremen, Germany). The common limit of detection that must be placed at the target spot of the MALDI Biotyper could be defined as approximately 5×105 cells, and bacterial identification was reported with high confidence if the log (score) was ≥ 2.00. Stock cultures were stored at −80°C in a 1:1 mixture of 60% glycerol and Nutrient Broth medium (Difco, Detroit, MI, USA). Urine samples with concentrations greater than 105 colony forming units (CFU)/mL of P. aeruginosa were included in this study.
Antimicrobial Susceptibility Testing
Antimicrobial susceptibility tests were performed using the broth microdilution method using Neg MIC Panel Type 44 and MicroScan WalkAway-96 Plus system (Beckman Coulter, Brea, CA, USA). Additionally, the minimum inhibitory concentrations (MICs) of imipenem and meropenem were determined using the E-test (Biomerieux, Marcy l’Etoile, France) on Mueller-Hinton agar (Difco Laboratories, Detroit, MI, USA) according to the Clinical and Laboratory Standards Institute guideline.22 P. aeruginosa ATCC 27853 was used as a quality control strain for antimicrobial susceptibility testing. P. aeruginosa resistant to at least one carbapenem [imipenem (MIC ≥ 8.0 µg/mL) and/or meropenem (MIC ≥ 8.0 µg/mL)] was defined as CRPA. MDR was defined as resistance to at least one agent in three or more antimicrobial classes.23
Detection of Carbapenemase Genes and Virulence Factors
Genomic DNA from P. aeruginosa was extracted using the AccuPrep® Genomic DNA Extraction Kit (Bioneer, Daejeon, Korea) according to the manufacturer’s protocol. The presence of carbapenemase genes (blaKPC, blaIMP, blaVIM, blaNDM, blaGES, and blaOXA-48-like) was detected with polymerase chain reaction (PCR) using specific primers (Table S1). PCR reactions were run using a BioRad C1000 Thermal Cycler (Bio-Rad, California, USA), and the products were subjected to 1.5% agarose gel electrophoresis (Bioneer, Daejeon, Korea). The PCR products were sequenced using an ABI 3730xL DNA analyzer (Applied Biosystems, Foster City, CA, USA). The obtained nucleotide sequences were compared with the sequences in the National Center for Biotechnology Information (NCBI) database using Basic Local Alignment Search Tool (BLAST). We performed PCR to investigate the presence or absence of 16 virulence factor genes, namely toxA, exoS, exoT, exoU, exoY, plcH, plcN, phzM, phzS, lasA, lasB, pilA, pilB, aprA, pvdA, and algD (Table S2).
Multilocus Sequence Typing and Pulsed-Field Gel Electrophoresis
The epidemiological relatedness among the CRPA isolates was determined using MLST and PFGE. MLST typing of P. aeruginosa isolates was carried out using PCR and the DNA sequencing of seven housekeeping (acsA, aroE, guaA, mutL, nuoD, ppsA, and trpE).24 To determine the allelic numbers and STs, the nucleotide sequences were compared with the sequences in the MLST database (http://pubmlst.org/paeruginosa). PFGE analysis was performed as previously described.25 DNA fragments digested with XbaI (Promega, Southampton, UK) were separated using a CHEF-DR® III variable angle system (Bio-Rad Laboratories, Hercules, CA, USA) according to the following conditions: initial switch time, 0.5 s; final switch time, 30 s; voltage, 6 V/cm; included angle, 120°; and run time, 17 hours. The DNA of Salmonella enterica serovar Braenderup H9812 (ATCC BAA-664) digested with XbaI (Promega) was used as a molecular size marker. BioNumerics software (Applied Maths, Kortrijk, Belgium) was used to analyze the PFGE patterns. If the Dice similarity was >80% using Dice correlation for band matching at a 1.5% position tolerance and the unweighted pair group method with arithmetic average, CRPA isolates were classified into the same PFGE cluster and considered to be closely related.
Statistical Analysis
Statistical analyses were performed using SPSS version 21 (IBM SPSS Statistics for Windows 21.0, IBM Corp., Armonk, NY, USA). Fisher’s exact test was performed to compare the antibiotic resistance and virulence factors of P. aeruginosa ST235 and ST773. Statistical significance was set at P < 0.05.
Results
Clinical CRPA Isolates
From March 2020 to February 2021, a total of 124 non-duplicate P. aeruginosa strains were isolated at a concentration of 105 CFU/mL or higher from urine samples of inpatients and outpatients at Konyang University Hospital. CRPA accounted for 47.6% (59/124) of all isolates.
Antibiotic Resistance Profile and Distribution of Carbapenemase Genes in the CRPA Isolates
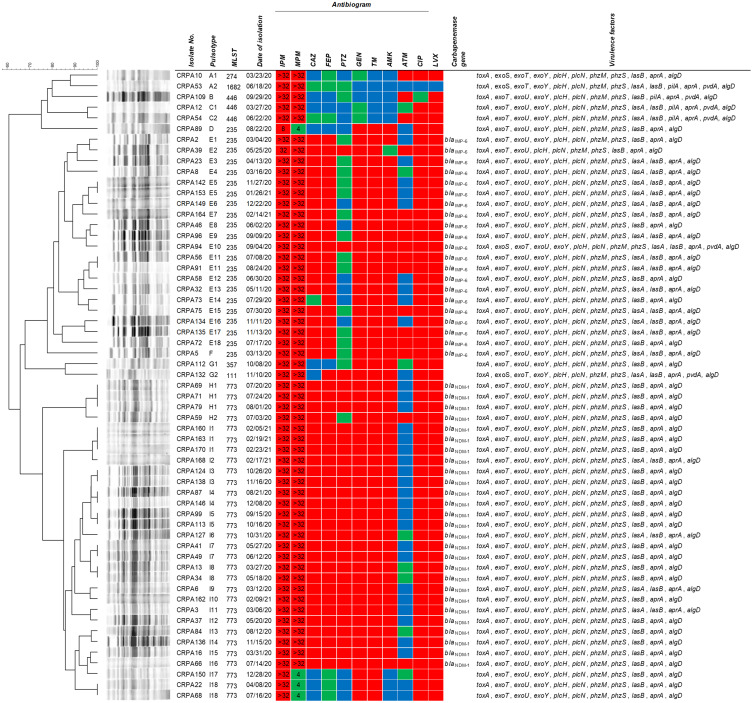
The resistance rates for imipenem and meropenem in 59 CRPA isolates reached 100% (59/59) and 93.2% (55/59), respectively (Figure 1). Imipenem showed an MIC value greater than 32 µg/mL for most of the CRPA isolates (96.6%, 57/59), whereas for one isolate (1.7%), the MIC of imipenem was 32 µg/mL, and for another isolate (1.7%), it was 8 µg/mL. Likewise, meropenem showed an MIC value greater than 32 µg/mL for most of the CRPA isolates (93.2%, 55/59), while for 4 isolates (6.8%), it showed an MIC of 4 µg/mL; the susceptibility of these 4 isolates was deemed intermediate. Regarding other antibiotics, the resistance rates were the highest for levofloxacin (98.3%, 58/59), followed by ciprofloxacin (96.6%, 57/59), gentamicin (91.5%, 54/59), ceftazidime (79.7%, 47/59), piperacillin-tazobactam (50.8%, 30/59), and aztreonam (27.1%, 16/59). Of all the CRPA isolates, 91.5% were MDR. Forty-eight of the fifty-nine CRPA isolates (81.4%) harbored the carbapenemase gene blaNDM-1 (45.8%, 27/59) or blaIMP-6 (35.6%, 21/59). Other carbapenemase genes that were evaluated, such as blaKPC, blaVIM, blaGES, and blaOXA-48-like, were not detected in the CRPA isolates.
Figure 1.
Dendrogram based on pulsed-field gel electrophoresis patterns, multilocus sequence typing, carbapenemase gene, and antibiotic susceptibility profiles of 59 carbapenem-resistant P. aeruginosa. The red, green, and blue squares indicate resistant, intermediate, and susceptible to each antibiotic, respectively. The numbers in the imipenem and meropenem boxes indicate the MICs (µg/mL) for each antibiotic.
Abbreviations: IPM, imipenem; MPM, meropenem; CAZ, ceftazidime; FEP, cefepime; PTZ, piperacillin-tazobactam; GEN, gentamicin; TM, tobramycin; AMK, amikacin; ATM, aztreonam; CIP, ciprofloxacin; LVX, levofloxacin.
Molecular Epidemiology Based on MLST and PFGE
The CRPA isolates comprised seven different STs: ST773 (50.8%, 30/59), ST235 (37.3%, 22/59), ST446 (5.1%, 3/59), ST111 (1.7%, 1/59), ST274 (1.7%, 1/59), ST357 (1.7%, 1/59), and ST1682 (1.7%, 1/59) (Figure 1). For PFGE analysis, the CRPA isolates were classified into 9 clusters and 47 pulsotypes. The results of the PFGE analysis according to ST type were as follows: ST773 comprised PFGE pulsotypes H1-H2 and I1-I18; ST235 showed PFGE pulsotypes D, E1-E18, and F; ST446 showed PFGE pulsotypes B and C1-C2; ST111 showed PFGE pulsotype G2; ST274 showed PFGE pulsotype A1; ST357 showed PFGE pulsotype G1; ST1682 showed PFGE pulsotype A2. All CRPA isolates harboring blaIMP-6 belonged to ST235 and PFGE pulsotypes E1-E18 and F with >80.6% similarity for the E1-E18 pulsotype. All CRPA isolates harboring blaNDM-1 belonged to ST773 and PFGE pulsotypes H1-H2 and I1-I16 with >95.0% similarity for the H1-H2 pulsotype and >85.8% similarity for the I1-I16 pulsotype.
Virulence Factor Genes
All CRPA strains carried at least 10 virulence factors from among the 16 virulence factor genes (Table 1). Nine virulence factor genes (toxA, exoT, plcN, plcH, phzM, phzS, lasB, aprA, and algD) were identified in all CRPA isolates. The exoY and exoU genes showed a high prevalence [98.3% (58/59) and 94.9% (56/59)], respectively. The prevalence of the other virulence factors was as follows: lasA, 39.0% (23/59); pvdA, 10.2% (6/59); exoS, 6.8% (4/59); and pilA, 6.8% (4/59). pilB was not detected in any of the CRPA strains tested. Except for one IMP-6-producing CRPA ST235 isolate with exoS+/exoU+ genotype, the other 58 CRPA isolates had either exoS-/exoU+ (94.8%, 55/58) or exoS+/exoU- (5.2%, 3/58) genotypes (Figure 1). The most frequent combination of virulence factors detected among all CRPA isolates was toxA, exoT, exoU, exoY, plcH, plcN, phzM, phzS, lasB, aprA, and algD (55.9%, 33/59). CRPA isolates harboring 14 virulence factor genes accounted for 6.8% (4/59) of the isolates. The most frequent combination for ST773 comprised 11 virulence factors (toxA, exoT, exoU, exoY, plcH, plcN, phzM, phzS, lasB, aprA, and algD) (86.7%, 26/30). In the case of ST235, a combination of 12 virulence factors (toxA, exoT, exoU, exoY, plcH, plcN, phzM, phzS, lasA, lasB, aprA, and algD) was observed with the highest frequency (63.6%, 14/22). The exoU gene was identified in all CRPA ST235, 357, 446, and 773 isolates, while the exoS gene was identified in the ST111, 274, and 1682 CRPA isolates (Figure 1). The pilA gene was identified only in the CRPA ST446 and 1682 isolates, which did not show carbapenemase production.
Table 1.
Distribution of Virulence Factors According to Sequence Type (ST) of Carbapenem-Resistant P. aeruginosa Isolates
| No. of Virulence Factors | Virulence Factor Profiles | ST (%) | |||||||
|---|---|---|---|---|---|---|---|---|---|
| ST773 (n=30) | ST235 (n=22) | ST446 (n=3) | ST111 (n=1) | ST274 (n=1) | ST1682 (n=1) | ST357 (n=1) | Total (n=59) | ||
| 10 | toxA, exoT, exoU, plcH, plcN, phzM, phzS, lasB, aprA, algD | 0 | 1 (4.5%) | 0 | 0 | 0 | 0 | 0 | 1 (1.7%) |
| 11 | toxA, exoT, exoU, exoY, plcH, plcN, phzM, phzS, lasB, aprA, algD | 26 (86.7%) | 6 (27.3%) | 0 | 0 | 0 | 0 | 1 (100%) | 33 (55.9%) |
| 11 | toxA, exoS, exoT, exoY, plcH, plcN, phzM, phzS, lasB, aprA, algD | 0 | 0 | 0 | 0 | 1 (100%) | 0 | 0 | 1 (1.7%) |
| 12 | toxA, exoT, exoU, exoY, plcH, plcN, phzM, phzS, lasA, lasB, aprA, algD | 4 (13.3%) | 14 (63.6%) | 0 | 0 | 0 | 0 | 0 | 18 (30.5%) |
| 13 | toxA, exoT, exoU, exoY, plcH, plcN, phzM, phzS, lasB, pilA, aprA, pvdA, algD | 0 | 0 | 1 (33.3%) | 0 | 0 | 0 | 0 | 1 (1.7%) |
| 13 | toxA, exoS, exoT, exoY, plcH, plcN, phzM, phzS, lasA, lasB, aprA, pvdA, algD | 0 | 0 | 0 | 1 (100%) | 0 | 0 | 0 | 1 (1.7%) |
| 14 | toxA, exoT, exoU, exoY, plcH, plcN, phzM, phzS, lasA, lasB, pilA, aprA, pvdA, algD | 0 | 0 | 2 (66.7%) | 0 | 0 | 0 | 0 | 2 (3.4%) |
| 14 | toxA, exoS, exoT, exoY, plcH, plcN, phzM, phzS, lasA, lasB, pilA, aprA, pvdA, algD | 0 | 0 | 0 | 0 | 0 | 1 (100%) | 0 | 1 (1.7%) |
| 14 | toxA, exoS, exoT, exoU, exoY, plcH, plcN, phzM, phzS, lasA, lasB, aprA, pvdA, algD | 0 | 1 (4.5%) | 0 | 0 | 0 | 0 | 0 | 1 (1.7%) |
Comparison of Antibiotic Resistance and Virulence Factors Between ST235 and ST773
In the comparative analysis of ST235 and ST773—the major clones of CRPA isolates in this study—no significant difference between the number of antibiotics to which the strains were resistant was observed (P = 0.655). The prevalence of piperacillin-tazobactam resistance in ST235 and ST773 was 9.1% (2/22) and 86.7% (26/30), respectively. Most of the ST773 isolates showed resistance to piperacillin-tazobactam (P < 0.001) (Table 2). On the other hand, when comparing the prevalence of aztreonam resistance, ST235 and ST773 showed a prevalence of 50.0% (11/22) and 6.7% (2/30), respectively. Most of the ST773 strains were susceptible or intermediate to aztreonam (P = 0.001). There was no significant difference between ST235 and ST773 with respect to the prevalence of resistance for the other antibiotics tested. In the distribution of virulence factors, the lasA gene was identified at a higher rate in ST235 (68.2%, 15/22) than in ST773 (13.3%, 4/30), and this observation was statistically significant (P < 0.001). There was no statistically significant difference between ST235 and ST773 with respect to the distribution of other virulence factors, except for lasA.
Table 2.
Comparative Analysis of the Antibiotic Resistance Patterns and Virulence Factors According to ST235 Clone (n=22) and ST773 Clone (n=30)
| Variable | ST235 (n=22) (%) | ST773 (n=30) (%) | P-value | |
|---|---|---|---|---|
| Resistance of antibiotics | Impenem | 22 (100.0) | 30 (100.0) | 1.000 |
| Meropenem | 21 (95.5) | 27 (90.0) | 0.629 | |
| Ceftazidime | 20 (90.9) | 27 (90.0) | 1.000 | |
| Cefepime | 21 (95.5) | 27 (90.0) | 0.629 | |
| Piperacillin-tazobactam | 2 (9.1) | 26 (86.7) | <0.001 | |
| Gentamicin | 22 (100.0) | 30 (100.0) | 1.000 | |
| Tobramycin | 22 (100.0) | 30 (100.0) | 1.000 | |
| Amikacin | 21 (95.5) | 27 (90.0) | 0.629 | |
| Aztreonam | 11 (50.0) | 2 (6.7) | 0.001 | |
| Ciprofloxacin | 22 (100.0) | 30 (100.0) | 1.000 | |
| Levofloxacin | 22 (100.0) | 30 (100.0) | 1.000 | |
| Virulence factors | toxA | 22 (100.0) | 30 (100.0) | 1.000 |
| exoS | 1 (4.5) | 0 (0.0) | 0.423 | |
| exoT | 22 (100.0) | 30 (100.0) | 1.000 | |
| exoU | 22 (100.0) | 30 (100.0) | 1.000 | |
| exoY | 21 (95.5) | 30 (100.0) | 0.423 | |
| plcH | 22 (100.0) | 30 (100.0) | 1.000 | |
| plcN | 22 (100.0) | 30 (100.0) | 1.000 | |
| phzM | 22 (100.0) | 30 (100.0) | 1.000 | |
| phzS | 22 (100.0) | 30 (100.0) | 1.000 | |
| lasA | 15 (68.2) | 4 (13.3) | <0.001 | |
| lasB | 22 (100.0) | 30 (100.0) | 1.000 | |
| pilA | 0 (0.0) | 0 (0.0) | 1.000 | |
| pilB | 0 (0.0) | 0 (0.0) | 1.000 | |
| aprA | 22 (100.0) | 30 (100.0) | 1.000 | |
| pvdA | 1 (4.5) | 0 (0.0) | 0.423 | |
| algD | 22 (100.0) | 30 (100.0) | 1.000 | |
Discussion
CRPA causes serious problems, such as limiting the therapeutic options available for the treatment and prolonging the patients’ hospital stays.26,27 The phenotypic and genotypic characterization of CRPA is crucial for providing insights into its epidemiological features.28 In this study, the carbapenem-resistance rate of P. aeruginosa isolated from urine samples was 47.6%. Moreover, 91.5% of the CRPA isolates were multidrug-resistant P. aeruginosa. The rate of carbapenem resistance of P. aeruginosa isolated from various clinical specimens has been reported to vary from 1% to 75.3% in different previous studies.27,29,30 Meanwhile, the carbapenem-resistance rate of P. aeruginosa in clinical urine samples has been reported to be approximately 56%.31
The production of carbapenemases is associated with clinical problems such as treatment failure due to reduced activity against last-resort antibiotics and epidemiological issues due to transmission to various bacteria.32 In this study, blaNDM-1 or blaIMP-6 were detected in 81.4% of the CRPA isolates. This confirmed that carbapenem resistance in the isolated P. aeruginosa strains was mainly mediated by the production of carbapenemases. IMP- and VIM-type MBLs are the most common carbapenemases produced by P. aeruginosa worldwide, followed by the GES variants of Ambler class A.33 IMP-6 has become the most prevalent MBL type among CRPA isolates in South Korea.30,34 Since carbapenemase genes are transmitted by mobile genetic elements, there could be a high potential for rapid dissemination.31,35
P. aeruginosa ST773 and ST235 were the predominant clones among the CRPA isolates in this study. Moreover, 90.0% of ST773 harbored blaNDM-1 and 95.5% of ST235 harbored blaIMP-6. P. aeruginosa ST235, a founder strain of the clonal complex CC235, is known as a representative high-risk clone associated with the multidrug-resistant phenotype worldwide and produces over 60 different variants of β-lactamases, including carbapenemases from classes A and B, along with the expression of multiple virulence factors.21,36 In addition, P. aeruginosa ST111 and ST357 identified in this study have been considered as high-risk clones with a global distribution.21,37 Previous research has confirmed that IMP-6-producing P. aeruginosa ST235 is prevalent nationwide in South Korea and that its IMP-6 gene is located on a class 1 integron.25,30,38 On the other hand, NDM-1-producing P. aeruginosa ST773 was first reported in South Korea in urine and wound samples in 2020, and the NDM-1 gene was found to be located within the integrative and conjugative element on the bacterial chromosome.33 Similar to the findings of previous studies, NDM-1-producing P. aeruginosa ST773 in our study exhibited multidrug resistance to most antibiotics, such as β-lactam-β-lactamase inhibitor combinations (piperacillin-tazobactam), cephalosporins (ceftazidime, cefepime), carbapenems (imipenem, meropenem), aminoglycosides (gentamicin, tobramycin, amikacin), and fluoroquinolones (ciprofloxacin, levofloxacin).33,39 The results of the PFGE analysis showed that P. aeruginosa ST773 and ST235, which were each divided into two clonal groups, were closely related, indicating the possibility of clonal spread.
CRPA isolates showing a co-existence of virulence factors with multidrug resistance determinants have become an emerging threat.40 Data on the interaction between antibiotic resistance and virulence factors is essential to address bacterial infections and to provide management strategies.41 In this study, we investigated the distribution and prevalence of virulence factor genes in CRPA isolated from UTI samples. Nine virulence factor genes (toxA, exoT, plcN, plcH, phzM, phzS, lasB, aprA, and algD) were detected in all CRPA isolates. These results suggest that these factors may play an important role in the pathogenesis of CRPA-mediated UTIs. ExoS, ExoT, ExoU, and ExoY, four effector proteins of the T3SS, have been identified in P. aeruginosa.42,43 In this study, the prevalence of the exoT gene in all CRPA isolates was 100%, followed by exoY (98.3%), exoU (94.9%), and exoS (6.8%). Although P. aeruginosa carries genes encoding T3SS, most strains do not have all four genes as a complete set.43 In particular, almost all P. aeruginosa harbor either exoS or exoU genes, but rarely exhibit both genes.43,44 Similarly, in our study, except for one CRPA isolate harboring both exoU and exoS, the other 58 CRPA isolates harbored either exoU or exoS. The prevalence of the exoS- /exoU+ genotype (94.8%, 55/58) was much higher than that of the exoS+ /exoU- genotype (5.2%, 3/58). Globally, exoS is found in 58%–72%, and exoU in 28–42% of P. aeruginosa isolated from clinical samples.40,43,45 In a recent study investigating the prevalence of exoS according to the presence of carbapenemase genes, exoS prevalence was found to be significantly lower in the CRPA group harboring carbapenemase genes (18.2%) than the CRPA group not harboring carbapenemase genes (77.0%) (P < 0.001).19 Similarly, in our study, the exoS gene was identified in carbapenemase-nonproducing CRPA isolates, except for one carbapenemase-producing CRPA isolate harboring exoS+/exoU+. Further studies are needed to determine the correlation between exoS gene incidence and carbapenemase production. In the distribution of the exoU and exoS genes according to the STs, exoU was identified in all ST235, 357, 446 and 773 isolates and exoS was identified in all ST111, 274, and 1682 isolates, corroborating the findings of previous research.44 Among the elastase-related lasA and lasB genes, lasA was identified in 39.0% of all CRPA isolates, and the prevalence of lasA between the strains ST235 (68.2%) and ST773 (13.3%) differed significantly (P < 0.001). This result is different from a previous finding in which all ST235 CRPA strains carried the lasA gene.44 In contrast, the lasB gene was detected in all the tested CRPA strains isolated from urine, confirming the necessity of this virulence gene for the pathogenesis of UTIs, as observed in previous studies.46 The pili-encoding genes, pilA and pilB, were observed at relatively low percentages (6.8% and 0%, respectively) among the studied CRPA strains. These results were similar to those of a previous study, which showed that the prevalence of pilA and pilB genes was 19.2% and 0%, respectively, in P. aeruginosa derived from children with UTIs.47 In addition, the pilA gene was identified only in carbapenemase-nonproducing ST446 and 1682 CRPA strains; however, additional studies are needed to confirm and further analyze this result. Pyoverdine, a siderophore that aids bacterial iron uptake, plays an essential role in lung infections, such as cystic fibrosis.13,48,49 In our study, 10.2% of the CRPA isolates exhibited the pvdA gene. This result differed from the results of the research carried out by Tielen et al, in which pyoverdine could be detected in all P. aeruginosa isolates from UTIs and catheter-associated UTIs, suggesting that pyoverdine plays an important role in iron-limited conditions such as those of the human urinary tract.50 Therefore, the role of pyoverdine in UTIs remains unclear.
This study has a few limitations. First, no investigation on the mechanism of carbapenem resistance other than the identification of carbapenemases has been conducted in this study. Therefore, further investigation of the other mechanisms of carbapenem resistance, such as lack of OprD porins or overexpression of efflux pumps among the remaining carbapenemase-nonproducing strains, is needed. Second, as this was a single-center study, further studies are required to gain insight into the overall epidemiology of CRPA isolates in South Korea. Nevertheless, this study has important implications for clinicians treating UTIs caused by CRPA strains by providing important information regarding their production of carbapenemases, epidemiological characteristics, and virulence factors.
Conclusion
The present research confirmed that in the study area, most CRPA strains causing UTIs are multidrug-resistant and produce carbapenemases, including the NDM-1-producing ST773 and IMP-6-producing ST235, and have already disseminated in this area. The CRPA isolates also exhibited multiple virulence factors. This finding suggests that it is important to continuously strengthen the monitoring of carbapenemase-producing P. aeruginosa to prevent further spread and that strategies for appropriate antibiotic use need to be established to overcome the treatment failure of CRPA-related infections.
Funding Statement
This work was supported by the Research Resettlement Fund for the new faculty of Konyang University Hospital.
Abbreviations
BLAST, Basic Local Alignment Search Tool; CFU, colony forming units; CRPA, carbapenem-resistant P. aeruginosa; ETA, exotoxin A; IMP, imipenemase; PCR, polymerase chain reaction; MBL, metallo-β-lactamase; MDR, multidrug resistance; MIC, minimum inhibitory concentration; MLST, multilocus sequence typing; NCBI, National Center for Biotechnology Information; NDM, New Delhi metallo-β-lactamase; PFGE, pulsed-field gel electrophoresis; STs, sequence types; T3SS, type III secretion system; UTI, urinary tract infection; VIM, Verona Integron-encoded Metallo-β-lactamase.
Ethics Approval and Informed Consent
The study was reviewed and approved by the Ethical Review Committee of the Konyang University Hospital (No. NON2021-001). The ethical review committee of Konyang University Hospital waived the requirement for informed consent. The retrospective study was considered exempt from the ethical review because only bacterial isolates were taken from patients and patient confidentiality was fully guaranteed. This study was conducted in accordance with the principles of the Declaration of Helsinki.
Consent for Publication
All authors approved the manuscript and gave their consent for submission and publication.
Author Contributions
All authors contributed to data analysis, drafting or revising the article, have agreed on the journal to which the article will be submitted, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
The authors declare that there are no conflicts of interest regarding the publication of this article.
References
- 1.Penaranda C, Chumbler NM, Hung DT. Dual transcriptional analysis reveals adaptation of host and pathogen to intracellular survival of Pseudomonas aeruginosa associated with urinary tract infection. PLoS Pathog. 2021;17(4):e1009534. doi: 10.1371/journal.ppat.1009534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lamas Ferreiro JL, Álvarez Otero J, González González L, et al. Pseudomonas aeruginosa urinary tract infections in hospitalized patients: mortality and prognostic factors. PLoS One. 2017;12(5):e0178178. doi: 10.1371/journal.pone.0178178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hu -Y-Y, Cao J-M, Yang Q, et al. Risk factors for carbapenem-resistant Pseudomonas aeruginosa, Zhejiang Province, China. Emerg Infect Dis. 2019;25(10):1861–1867. doi: 10.3201/eid2510.181699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wi YM, Choi J-Y, Lee J-Y, et al. Antimicrobial effects of β-lactams on imipenem-resistant ceftazidime-susceptible Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2017;61(6):e00054–17. doi: 10.1128/AAC.00054-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hu Y, Liu C, Wang Q, et al. Emergence and expansion of a carbapenem-resistant Pseudomonas aeruginosa clone are associated with plasmid-borne bla (KPC-2) and virulence-related genes. mSystems. 2021;6(3):e00154–121. doi: 10.1128/mSystems.00154-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pang Z, Raudonis R, Glick BR, Lin T-J, Cheng Z. Antibiotic resistance in Pseudomonas aeruginosa: mechanisms and alternative therapeutic strategies. Biotechnol Adv. 2019;37(1):177–192. doi: 10.1016/j.biotechadv.2018.11.013 [DOI] [PubMed] [Google Scholar]
- 7.Yin S, Chen P, You B, et al. Molecular typing and carbapenem resistance mechanisms of Pseudomonas aeruginosa isolated from a Chinese Burn Center from 2011 to 2016. Front Microbiol. 2018;9:1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meletis G, Exindari M, Vavatsi N, Sofianou D, Diza E. Mechanisms responsible for the emergence of carbapenem resistance in Pseudomonas aeruginosa. Hippokratia. 2012;16(4):303–307. [PMC free article] [PubMed] [Google Scholar]
- 9.Yoon E-J, Jeong SH. Mobile carbapenemase genes in Pseudomonas aeruginosa. Front Microbiol. 2021;12:614058. doi: 10.3389/fmicb.2021.614058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sultan M, Arya R, Kim KK. Roles of two-component systems in Pseudomonas aeruginosa virulence. Int J Mol Sci. 2021;22(22):12152. doi: 10.3390/ijms222212152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Horii T, Muramatsu H, Monji A, Miyagishima D. Release of exotoxin A, peptidoglycan and endotoxin after exposure of clinical Pseudomonas aeruginosa isolates to carbapenems in vitro. Chemotherapy. 2005;51(6):324–331. doi: 10.1159/000088955 [DOI] [PubMed] [Google Scholar]
- 12.Galle M, Jin S, Bogaert P, Haegman M, Vandenabeele P, Beyaert R. The Pseudomonas aeruginosa type III secretion system has an exotoxin S/T/Y independent pathogenic role during acute lung infection. PLoS One. 2012;7(7):e41547. doi: 10.1371/journal.pone.0041547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Newman JW, Floyd RV, Fothergill JL. The contribution of Pseudomonas aeruginosa virulence factors and host factors in the establishment of urinary tract infections. FEMS Microbiol Lett. 2017;364:15. doi: 10.1093/femsle/fnx124 [DOI] [PubMed] [Google Scholar]
- 14.Lee VT, Pukatzki S, Sato H, et al. Pseudolipasin A is a specific inhibitor for phospholipase A2 activity of Pseudomonas aeruginosa cytotoxin ExoU. Infect Immun. 2007;75(3):1089–1098. doi: 10.1128/IAI.01184-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chadha J, Harjai K, Chhibber S. Revisiting the virulence hallmarks of Pseudomonas aeruginosa: a chronicle through the perspective of quorum sensing. Environ Microbiol. 2021. doi: 10.1111/1462-2920.15784 [DOI] [PubMed] [Google Scholar]
- 16.Bashir A, Tian T, Yu X, Meng C, Ali M, Li L. Pyoverdine-mediated killing of Caenorhabditis elegans by Pseudomonas syringae MB03 and the role of iron in its pathogenicity. Int J Mol Sci. 2020;21(6):2198. doi: 10.3390/ijms21062198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Behzadi P, Baráth Z, Gajdács M. It’s not easy being green: a narrative review on the microbiology, virulence and therapeutic prospects of multidrug-resistant Pseudomonas aeruginosa. Antibiotics. 2021;10(1):42. doi: 10.3390/antibiotics10010042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gohain N. Studies on the structure and function of phenazine modifying enzymes PhzM and PhzS involved in the biosynthesis of pyocyanin; 2009.
- 19.Bogiel T, Depka D, Rzepka M, Kwiecińska-Piróg J, Gospodarek-Komkowska E. Prevalence of the genes associated with biofilm and toxins synthesis amongst the Pseudomonas aeruginosa clinical strains. Antibiotics. 2021;10(3):241. doi: 10.3390/antibiotics10030241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Osawa K, Shigemura K, Kitagawa K, et al. Molecular characteristics of carbapenem-resistant Pseudomonas aeruginosa isolated from urine in Hyogo, Japan. Int J Urol. 2019;26(1):127–133. doi: 10.1111/iju.13818 [DOI] [PubMed] [Google Scholar]
- 21.Del Barrio-Tofiño E, López-Causapé C, Oliver A. Pseudomonas aeruginosa epidemic high-risk clones and their association with horizontally-acquired β-lactamases: 2020 update. Int J Antimicrob Agents. 2020;56(6):106196. doi: 10.1016/j.ijantimicag.2020.106196 [DOI] [PubMed] [Google Scholar]
- 22.CLSI. Performance Standards for Antimicrobial Disk Susceptibility Testing, 30th M100. PA: Clinical and Laboratory Standards Institute; 2020. [Google Scholar]
- 23.Magiorakos AP, Srinivasan A, Carey RB, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2012;18(3):268–281. doi: 10.1111/j.1469-0691.2011.03570.x [DOI] [PubMed] [Google Scholar]
- 24.Curran B, Jonas D, Grundmann H, Pitt T, Dowson CG. Development of a multilocus sequence typing scheme for the opportunistic pathogen Pseudomonas aeruginosa. J Clin Microbiol. 2004;42(12):5644–5649. doi: 10.1128/JCM.42.12.5644-5649.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hong JS, Yoon E-J, Lee H, Jeong SH, Lee K. Clonal dissemination of Pseudomonas aeruginosa sequence type 235 isolates carrying blaIMP-6 and emergence of blaGES-24 and blaIMP-10 on Novel Genomic Islands PAGI-15 and −16 in South Korea. Antimicrob Agents Chemother. 2016;60(12):7216–7223. doi: 10.1128/AAC.01601-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meradji S, Barguigua A, Zerouali K, et al. Epidemiology of carbapenem non-susceptible Pseudomonas aeruginosa isolates in Eastern Algeria. Antimicrob Resist Infect Control. 2015;4:27. doi: 10.1186/s13756-015-0067-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.El-Mahdy R, El-Kannishy G. Virulence factors of carbapenem-resistant Pseudomonas aeruginosa in hospital-acquired infections in Mansoura, Egypt. Infect Drug Resist. 2019;12:3455–3461. doi: 10.2147/IDR.S222329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gajdács M, Kárpáti K, Stájer A, Zanetti S, Matthew Gavino D. Insights on carbapenem-resistant Pseudomonas aeruginosa: phenotypic characterization of relevant isolates. Acta Biologica Szegediensis. 2021;65(1):105–112. doi: 10.14232/abs.2021.1.105-112 [DOI] [Google Scholar]
- 29.Gonçalves IR, Dantas RC, Ferreira ML, Batistão DW, Gontijo-Filho PP, Ribas RM. Carbapenem-resistant Pseudomonas aeruginosa: association with virulence genes and biofilm formation. Braz J Microbiol. 2017;48(2):211–217. doi: 10.1016/j.bjm.2016.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hong DJ, Bae IK, Jang I-H, Jeong SH, Kang H-K, Lee K. Epidemiology and characteristics of metallo-β-lactamase-producing Pseudomonas aeruginosa. Infect Chemother. 2015;47(2):81–97. doi: 10.3947/ic.2015.47.2.81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ohadian Moghadam S, Afshar D, Nowroozi MR, Behnamfar A, Farzin A. Molecular epidemiology of carbapenemase-producing Pseudomonas aeruginosa isolated from an Iranian University Hospital: evidence for spread of high-risk clones. Infect Drug Resist. 2020;13:1583–1592. doi: 10.2147/IDR.S253756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hammoudi Halat D, Ayoub Moubareck C. The current burden of carbapenemases: review of significant properties and dissemination among gram-negative bacteria. Antibiotics. 2020;9(4):186. doi: 10.3390/antibiotics9040186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hong JS, Song W, Park M-J, Jeong S, Lee N, Jeong SH. Molecular characterization of the first emerged ndm-1-producing Pseudomonas aeruginosa isolates in South Korea. Microb Drug Resist. 2021;27(8):1063–1070. doi: 10.1089/mdr.2020.0374 [DOI] [PubMed] [Google Scholar]
- 34.Hong JS, Kim JO, Lee H, Bae IK, Jeong SH, Lee K. Characteristics of metallo-β-lactamase-producing Pseudomonas aeruginosa in Korea. Infect Chemother. 2015;47(1):33–40. doi: 10.3947/ic.2015.47.1.33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Walters MS, Grass JE, Bulens SN, et al. Carbapenem-resistant Pseudomonas aeruginosa at US emerging infections program sites, 2015. Emerg Infect Dis. 2019;25(7):1281–1288. doi: 10.3201/eid2507.181200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Treepong P, Kos VN, Guyeux C, et al. Global emergence of the widespread Pseudomonas aeruginosa ST235 clone. Clin Microbiol Infect. 2018;24(3):258–266. doi: 10.1016/j.cmi.2017.06.018 [DOI] [PubMed] [Google Scholar]
- 37.Kocsis B, Toth A, Gulyas D, et al. Acquired qnrVC1 and blaNDM-1 resistance markers in an international high-risk Pseudomonas aeruginosa ST773 clone. J Med Microbiol. 2019;68(3):336–338. doi: 10.1099/jmm.0.000927 [DOI] [PubMed] [Google Scholar]
- 38.Seok Y, Bae IK, Jeong SH, Kim SH, Lee H, Lee K. Dissemination of IMP-6 metallo-β-lactamase-producing Pseudomonas aeruginosa sequence type 235 in Korea. J Antimicrob Chemother. 2011;66(12):2791–2796. doi: 10.1093/jac/dkr381 [DOI] [PubMed] [Google Scholar]
- 39.Khan A, Shropshire WC, Hanson B, et al. Simultaneous infection with Enterobacteriaceae and Pseudomonas aeruginosa harboring multiple carbapenemases in a returning traveler colonized with candida auris. Antimicrob Agents Chemother. 2020;64(2):e01466–1419. doi: 10.1128/AAC.01466-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ellappan K, Belgode Narasimha H, Kumar S. Coexistence of multidrug resistance mechanisms and virulence genes in carbapenem-resistant Pseudomonas aeruginosa strains from a tertiary care hospital in South India. J Glob Antimicrob Resist. 2018;12:37–43. doi: 10.1016/j.jgar.2017.08.018 [DOI] [PubMed] [Google Scholar]
- 41.Gajdács M, Baráth Z, Kárpáti K, et al. No correlation between biofilm formation, virulence factors, and antibiotic resistance in Pseudomonas aeruginosa: results from a laboratory-based in vitro study. Antibiotics. 2021;10(9):1134. doi: 10.3390/antibiotics10091134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Azimi S, Kafil HS, Baghi HB, et al. Presence of exoY, exoS, exoU and exoT genes, antibiotic resistance and biofilm production among Pseudomonas aeruginosa isolates in Northwest Iran. GMS Hyg Infect Control. 2016;11:Doc04. doi: 10.3205/dgkh000264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hauser AR. The type III secretion system of Pseudomonas aeruginosa: infection by injection. Nat Rev Microbiol. 2009;7(9):654–665. doi: 10.1038/nrmicro2199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee J-Y, Peck K, Ko KS. Selective advantages of two major clones of carbapenem-resistant Pseudomonas aeruginosa isolates (CC235 and CC641) from Korea: antimicrobial resistance, virulence and biofilm-forming activity. J Med Microbiol. 2013;62(7):1015–1024. doi: 10.1099/jmm.0.055426-0 [DOI] [PubMed] [Google Scholar]
- 45.Alonso B, Fernández-Barat L, Di Domenico EG, et al. Characterization of the virulence of Pseudomonas aeruginosa strains causing ventilator-associated pneumonia. BMC Infect Dis. 2020;20(1):909. doi: 10.1186/s12879-020-05534-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bogiel T, Prażyńska M, Kwiecińska-Piróg J, Mikucka A, Gospodarek-Komkowska E. Carbapenem-resistant Pseudomonas aeruginosa strains-distribution of the essential enzymatic virulence factors genes. Antibiotics. 2020;10(1):8. doi: 10.3390/antibiotics10010008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pobiega M, Maciag J, Pomorska-Wesolowska M, et al. Urinary tract infections caused by Pseudomonas aeruginosa among children in Southern Poland: virulence factors and antibiotic resistance. J Pediatr Urol. 2016;12(1):36.e31–36. doi: 10.1016/j.jpurol.2015.05.034 [DOI] [PubMed] [Google Scholar]
- 48.Mittal R, Aggarwal S, Sharma S, Chhibber S, Harjai K. Urinary tract infections caused by Pseudomonas aeruginosa: a minireview. J Infect Public Health. 2009;2(3):101–111. doi: 10.1016/j.jiph.2009.08.003 [DOI] [PubMed] [Google Scholar]
- 49.Visca P, Imperi F, Lamont IL. Pyoverdine siderophores: from biogenesis to biosignificance. Trends Microbiol. 2007;15(1):22–30. doi: 10.1016/j.tim.2006.11.004 [DOI] [PubMed] [Google Scholar]
- 50.Tielen P, Narten M, Rosin N, et al. Genotypic and phenotypic characterization of Pseudomonas aeruginosa isolates from urinary tract infections. Int J Med Microbiol. 2011;301(4):282–292. doi: 10.1016/j.ijmm.2010.10.005 [DOI] [PubMed] [Google Scholar]