ABSTRACT
Resistance to β-lactams, the most used antibiotics worldwide, constitutes the major problem for the treatment of bacterial infections. In the nosocomial pathogen Acinetobacter baumannii, β-lactamase-mediated resistance to the carbapenem family of β-lactam antibiotics has resulted in the selection and dissemination of multidrug-resistant isolates, which often cause infections characterized by high mortality rates. There is thus an urgent demand for new β-lactamase-resistant antibiotics that also inhibit their targets, penicillin-binding proteins (PBPs). As some PBPs are indispensable for the biosynthesis of the bacterial cell wall and survival, we evaluated their importance for the growth of A. baumannii by performing gene inactivation studies of d,d-transpeptidase domains of high-molecular-mass (HMM) PBPs individually and in combination with one another. We show that PBP3 is essential for A. baumannii survival, as deletion mutants of this d,d-transpeptidase were not viable. The inactivation of PBP1a resulted in partial cell lysis and retardation of bacterial growth, and these effects were further enhanced by the additional inactivation of PBP2 but not PBP1b. Susceptibility to β-lactam antibiotics increased 4- to 8-fold for the A. baumannii PBP1a/PBP1b/PBP2 triple mutant and 2- to 4-fold for all remaining mutants. Analysis of the peptidoglycan structure revealed a significant change in the muropeptide composition of the triple mutant and demonstrated that the lack of d,d-transpeptidase activity of PBP1a, PBP1b, and PBP2 is compensated for by an increase in the l,d-transpeptidase-mediated cross-linking activity of LdtJ. Overall, our data showed that in addition to essential PBP3, the simultaneous inhibition of PBP1a and PBP2 or PBPs in combination with LdtJ could represent potential strategies for the design of novel drugs against A. baumannii.
KEYWORDS: Acinetobacter, β-lactams, inactivation, penicillin-binding proteins
INTRODUCTION
The worldwide spread of antibiotic-resistant bacteria is recognized as a serious public health threat (1–5). Infections caused by such pathogens are notorious for high mortality rates and exert a heavy financial burden on health care institutions. Several multidrug-resistant bacterial species are responsible for the majority of life-threatening infections. Among them, Acinetobacter baumannii plays a prominent role, as it has become increasingly resistant to various antimicrobial agents, including β-lactams, the most used antibiotics worldwide (6). Resistance to the carbapenem family of β-lactams (7) is of special concern, as they were used as the drugs of choice for the treatment of A. baumannii infections (8, 9). The major mechanism of resistance to carbapenems and other β-lactams is their inactivation by β-lactamases (10), enzymes that are widespread among clinical isolates of A. baumannii. Carbapenem-resistant A. baumannii isolates are often multidrug resistant, and the occurrence of infections caused by such strains reaches up to 90% in some clinics, causing staggering mortality rates that can exceed 50% (11–14). Due to its clinical importance, the CDC lists multidrug-resistant A. baumannii as a bacterium that poses an urgent resistance threat in the United States, and it is included in the list of six bacterial pathogens responsible for the majority of health care-associated infections (5, 15).
Carbapenems and other β-lactam antibiotics kill bacteria by targeting the d,d-transpeptidase (TP) domains of their penicillin-binding proteins (PBPs) (16), enzymes involved in the biosynthesis of peptidoglycan, a major component of the cell wall. Peptidoglycan is composed of alternating N-acetylglucosamine (NAG) and N-acetylmuramic acid (NAM) residues linked into linear chains (17). These chains are cross-linked, to various extents, by short peptides that are attached to the NAM residues to form a three-dimensional mesh that entirely surrounds the bacterial cell. The cell wall of Gram-positive bacteria is composed of multiple peptidoglycan layers, while only one layer is typically found in Gram-negative microorganisms (18). Peptidoglycan acts as an exoskeleton, allowing bacteria to withstand high intracellular osmotic pressure (19). Apart from its mechanical role in maintaining cell integrity, peptidoglycan also defines the shape of a bacterial cell and plays a critical role in cell division. To perform these functions, peptidoglycan constantly undergoes modifications to meet the requirements of its bacterial host during various stages of the cellular life cycle (20).
The biogenesis of peptidoglycan is an extremely complex and not yet fully understood process. It is carried out by two multiprotein complexes, the elongasome, which conducts cell elongation, and the divisome, which orchestrates cell division (21). PBPs are essential components of these complexes, and based on their molecular masses, they are referred to as high-molecular-mass (HMM) and low-molecular-mass PBPs (22). HMM PBPs are further subdivided into two classes: class A PBPs are bifunctional enzymes that possess glycosyltransferase and TP domains, while class B PBPs are monofunctional TPs. Glycosyltransferase domains of class A PBPs elongate peptidoglycan chains by covalently linking NAG-NAM units. TP domains of class A and B PBPs cross-link glycan strands by forming a covalent bond between the fourth residue of one pentapeptide chain and the third residue of another (the 4-3 cross-link) (23). There is a growing number of cases demonstrating that in some bacterial species, cross-linking of glycan strands is also performed by l,d-transpeptidases (Ldts), enzymes that are structurally unrelated to PBPs (24). Unlike TPs, which produce 4-3 cross-links, Ldts generate 3-3-cross-linked peptidoglycan. It was recently shown that the cell wall of A. baumannii contains 3-3-cross-linked peptidoglycan, which is generated by the l,d-transpeptidase LdtJ (25).
Bacterial TPs are of special importance as they are the principal targets of β-lactam antibiotics. These antibiotics bind to TP domains with different affinities, which can also vary by pathogen, and this, in turn, determines whether they are effective as drugs. It was demonstrated that TPs of PBP1a and PBP3 from A. baumannii have similar relative affinities for several penicillins and cephalosporins and the carbapenem doripenem (26). In other experiments, it was shown that the β-lactams ceftazidime and aztreonam preferentially inhibit TPs of PBP1a and PBP3 over PBP2 from both A. baumannii and another Gram-negative pathogen, Pseudomonas aeruginosa, whereas amdinocillin showed the opposite trend, preferentially inhibiting PBP2 (27). The carbapenem meropenem efficiently inhibited all three PBPs from both bacteria. However, several species-dependent differences were observed. Of note, according to this study, ceftazidime, aztreonam, and meropenem more potently inhibited PBP3 from P. aeruginosa than its ortholog from A. baumannii, and meropenem was less reactive with PBP1a and PBP2 from P. aeruginosa than with those of A. baumannii.
To better understand the mechanisms of the bactericidal activity of β-lactam antibiotics and to establish a basis for the development of novel antibiotics that efficiently inhibit TPs but resist inactivation by β-lactamases, it is important to understand which enzymes, either individually or in combination with each other, are essential for bacterial survival. Attempts to inactivate TPs have been described for just a few bacterial species. It was first shown in the Gram-negative bacterium Escherichia coli that three individual HMM PBPs, PBP1a, PBP1b, and PBP2, could be successfully deleted, which demonstrated their nonessentiality for bacterial survival (28). The construction of TP deletion mutants of P. aeruginosa revealed that PBP3 is the only essential TP (29). Later, in A. baumannii, the entire genes for PBP1a and PBP1b were inactivated individually, thus abolishing their TP and glycosyltransferase activities (30). It was subsequently demonstrated that the simultaneous inactivation of these two PBPs is lethal (25). In this paper, we evaluated the impact of the inactivation of individual TPs of A. baumannii CIP 70.10 and their combinations on bacterial growth and resistance to β-lactam antibiotics. We also assessed the muropeptide composition of the triple mutant of this strain, where PBP3 remained the only functional TP.
RESULTS AND DISCUSSION
Inactivation of A. baumannii PBPs.
Four TPs (those of PBP1a, PBP1b, PBP2, and PBP3) have been identified in A. baumannii (31). The first two HMM PBPs are class A bifunctional enzymes, while the other two, PBP2 and PBP3, are class B monofunctional TPs. To establish whether any of these TPs are essential for survival, we aimed to inactivate the genes for each of these four individual enzymes (gene names are listed in Table S1 in the supplemental material) from the genome of A. baumannii CIP 70.10, an important β-lactam-susceptible fully sequenced reference strain that is used to study antibiotic resistance in this species (32). For the inactivation of genes encoding PBPs, we utilized a two-step homologous recombination protocol (integration followed by excision) (33) with derivatives of the suicide vector pMo130 (Table S2), which were constructed using primers listed in Table S3. This vector contains the sacB gene, whose presence is toxic for the cell in the presence of sucrose. It allows, following integration, the elimination of cells that still harbor the vector and the selection of cells containing the desired gene deletions or mutations.
In previous experiments with P. aeruginosa (a relative of A. baumannii; both are members of the Pseudomonadales order of the Gammaproteobacteria class), the authors generated single-deletion mutants of the TP domains of PBP1a and PBP1b but were unable to obtain their double-deletion mutant (29). The authors subsequently demonstrated that both TP domains can be inactivated simultaneously without affecting bacterial growth. These data strongly indicate that the partial deletion of the genes for PBP1a and PBP1b that encode their TP domains also severely compromised their glycosyltransferase function and that the activity of at least one glycosyltransferase is required for bacterial survival. As the aim of this paper was to evaluate the effects of the inactivation of the TP function of A. baumannii PBPs, we replaced the catalytic serine of PBP1a and PBP1b with alanine, as was reported previously for PBP1a of A. baumannii (30), thereby disabling only their TP function. We also attempted to delete the entire mrcA and mrcB genes individually, which encode both the TP and glycosyltransferase domains of PBP1a and PBP1b, respectively. For monofunctional PBP2 and PBP3, we attempted to delete in frame the entire mrdA and ftsI genes, respectively. We were able to readily generate both types of PBP1a and PBP1b single mutants [ΔPBP1a and ΔPBP1b deletion mutants and PBP1a(S/A) and PBP1b(S/A) inactivation mutants] as well as the deletion mutant of PBP2 (ΔPBP2) (Table S2). However, we were unable to obtain the PBP3 deletion mutant in three separate experiments by testing several hundred individual colonies each time.
To assess the possibility that our failure to delete the gene for PBP3 resulted from the inability of the suicide vector to properly integrate into the chromosome of A. baumannii or to excise from it, we performed PCR analysis to verify each of these events. Analysis of a few colonies obtained after selection for integration confirmed that the suicide vector readily integrates into the upstream chromosomal region of the gene (Fig. S1A), thus creating two regions of homology. Next, we analyzed whether the integrated vector subsequently excises from the chromosome, which can proceed through either of the two homologous regions. In some bacterial cells, the vector will excise in the same way that it integrated, thus restoring the original wild-type chromosomal arrangement. In other cells, excision will result from recombination at the second homologous region, which would create the desired mutation or gene deletion. To analyze the products of excision, we PCR amplified DNA from the liquid bacterial suspension obtained following the first round of sucrose selection. We observed similar amounts of PCR products for both the ftsI gene deletion and the wild-type ftsI gene (Fig. S1B), which indicated that both types of excision events occurred. Subsequent sequencing of the PCR DNA products confirmed the presence of the in-frame deletion of the ftsI gene precisely where it was expected. These data demonstrated that the generation of the ftsI deletion from the chromosome of A. baumannii readily occurs in our experiments. However, the subsequent round of selection on sucrose and analysis of several hundred colonies revealed the presence in all of them (except for a few that are described below) of only the wild-type ftsI gene (Fig. S1C). These results indicated that while the deletion of the gene encoding PBP3 occurs in some colony-forming cells, these cells ultimately lose viability. Combined, our data strongly suggest that PBP3 is essential for A. baumannii survival. These results are in agreement with data generated for P. aeruginosa, where it was also demonstrated that PBP3 is the only PBP that is essential for bacterial growth (29).
Unexpectedly, in a few colonies, PCR identified products characteristic for both the presence of the intact ftsI gene and its deletion (Fig. S1D). Further analysis also detected a third product corresponding to the presence of the integrated suicide vector. This indicated that the vector had become insensitive to sucrose, which allowed cells containing the integrated vector to survive under selective pressure, as was previously described (34). In such an unusual event, a single cell containing the integrated vector would multiply to form a colony despite the presence of sucrose. Subsequently, excision from the chromosome would generate the ftsI gene deletion in some cells and the wild-type genotype in other cells, while the majority of the cells will still harbor the integrated vector. This scenario is in agreement with our PCR analysis that confirmed the presence of all three of these chromosomal arrangements in the colonies. However, when these colonies were subjected to a second round of sucrose selection, we no longer detected the ftsI gene deletion in any of the resulting daughter colonies tested. These results indicated that while the deletion of the ftsI gene occurred upon vector excision from the chromosome of some cells forming the original colonies, these cells are not viable.
It was recently demonstrated, using transposon mutagenesis, that the inactivation of both the mrcA and mrcB genes encoding PBP1a and PBP1b, respectively, is synthetically lethal in A. baumannii (25), and similar outcomes were also reported previously for E. coli (35, 36) and P. aeruginosa (29). To confirm the reliability of the two-step homologous recombination protocol used in our study, we also attempted to create the ΔPBP1a+ΔPBP1b double-deletion mutant in A. baumannii CIP 70.10. As we observed in our experiments with the ftsI gene deletion, we detected the formation of this double mutant by PCR following the first round of sucrose selection in liquid medium. However, as with the ftsI gene deletion, such cells were not viable, as no colonies containing the double mutant were subsequently recovered. These data further confirm that the two-step homologous recombination protocol can reliably be used to demonstrate whether a gene is essential for bacterial survival.
We then proceeded with the generation of mutant variants harboring various combinations of inactivated nonvital TPs of PBP1a, PBP1b, and PBP2. We easily obtained a double mutant where only the function of the TP domains of PBP1a and PBP1b was inactivated by mutation of the catalytic serine residue to alanine [PBP1a(S/A) + PBP1b(S/A) inactivated mutant]. We also succeeded in generating two other double mutants of A. baumannii CIP 70.10, PBP1a(S/A)+ΔPBP2 and PBP1b(S/A)+ΔPBP2 (Table S2). Unlike in P. aeruginosa (29), we were also able to generate a triple mutant where the TPs of PBP1a, PBP1b, and PBP2 were inactivated [PBP1a(S/A)+PBP1b(S/A)+ΔPBP2 mutant]. The viability of this triple mutant is somewhat puzzling given that it lacks all three TPs that are involved in cell elongation; such a mutant also has not been previously reported in any bacterial species.
The presence of all mutations and/or deletions was confirmed by PCR amplifying the corresponding chromosomal regions (using primers listed in Table S3) and subsequently sequencing these products. To verify whether any additional unintended mutations occurred during the construction of the A. baumannii mutants, whole-genome sequencing of the parental strain and mutants was performed. This sequencing further confirmed the presence of all constructed mutations and/or deletions (Table S4). Only one nonspecific change was identified in a gluconate permease gene, where a T→G mutation resulted in the I17S substitution. This mutation first occurred upon the construction of the ΔPBP2 single mutant and subsequently was propagated to all other mutants containing the ΔPBP2 deletion [PBP1a(S/A)+ΔPBP2, PBP1b(S/A)+ΔPBP2, and PBP1a(S/A)+PBP1b(S/A)+ΔPBP2]. As shown below, this mutation had no effect on the growth of the ΔPBP2 single mutant and the PBP1b(S/A)+ΔPBP2 double mutant compared to the parental strain.
Effect of PBP deletion and/or inactivation on bacterial growth.
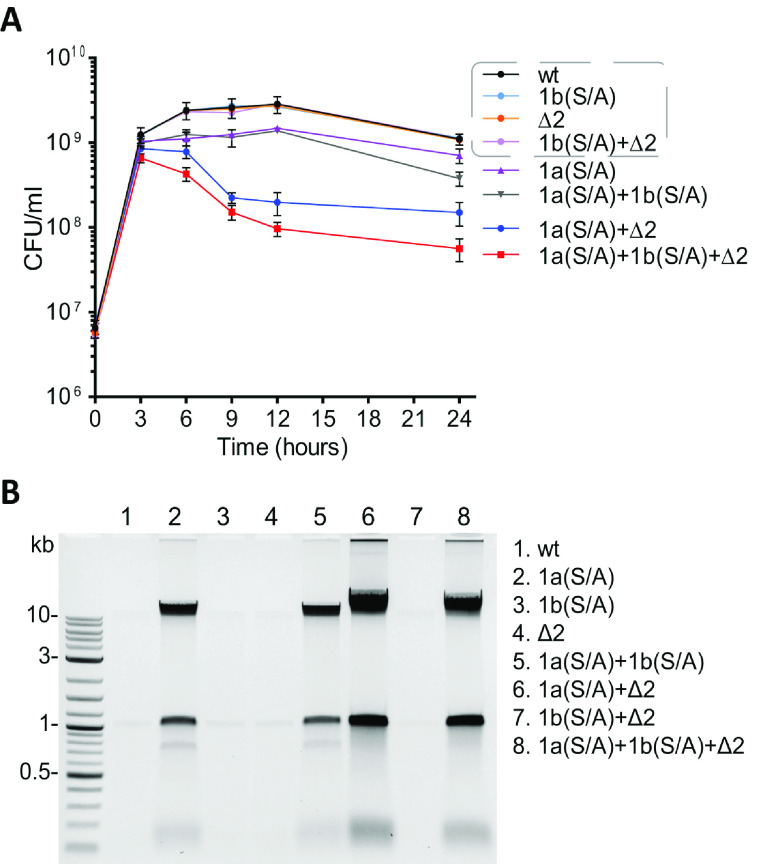
Next, we assessed the effect of the deletional and/or mutational inactivation of TPs of A. baumannii on bacterial growth. Growth curves for the PBP1b(S/A) inactivation mutant and the ΔPBP2 deletion mutant were almost identical to that of the parental strain (Fig. 1A), an indication that their activities can, to a large extent, be compensated for by the remaining TPs. However, mutational inactivation of the TP activity of PBP1a alone [PBP1a(S/A) mutant] resulted in slower growth. This mutant strain did not reach the CFU per milliliter of the parental strain or the PBP1b(S/A) and ΔPBP2 mutants, and a 2-fold decrease in the number of viable cells (compared to the parental strain) was measured after 12 h of growth. These results show that the TP activity of PBP1a is not fully compensated for by the three remaining PBPs, which indicates that the PBP1a TP is more essential for supporting normal growth of A. baumannii than PBP1b or PBP2 TPs. A similar result was recently described for the PBP1a TP mutant of another A. baumannii strain, where a slight retardation of bacterial growth was observed (25). While growth curves were also reported for P. aeruginosa, we cannot directly compare them to our data as our results were generated using CFU per milliliter, and those of P. aeruginosa were based on optical density (OD) measurements (29). The growth curve of the PBP1a(S/A)+PBP1b(S/A) double mutant of A. baumannii was nearly identical to that of the PBP1a(S/A) single mutant up to 12 h of incubation (Fig. 1A). However, a steeper decline in the number of viable cells was observed in the stationary phase (a 2-fold difference in CFU per milliliter was observed at 24 h of incubation). An even more dramatic retardation of growth was observed for the PBP1a(S/A)+ΔPBP2 double mutant. This mutant reached its maximum CFU per milliliter (which was only half that of the parental strain) in 3 h, versus 6 to 9 h for the parental strain, and the greatest difference in CFU per milliliter (∼15-fold) between these two strains was observed after 12 h of growth. Finally, the growth curve for the triple mutant showed a trend similar to that of the PBP1a(S/A)+ΔPBP2 double mutant, but we observed ∼2-fold-lower CFU per milliliter for the triple mutant after 12 h of incubation (Fig. 1A). Compared to the parental strain, a 30-fold decrease in the number of viable cells was measured for the triple mutant at the same time point. Combined, our data indicate that PBP1a of A. baumannii can support a growth rate at the level of the parental strain in the lack of the TP activity of PBP1b and/or PBP2. PBP2 is also capable of supporting this level of growth (but to a lesser extent) in the absence of TP activity of PBP1a and PBP1b. However, PBP1b is significantly less efficient in compensating for the lack of activity of PBP1a and PBP2 when both TPs are inactivated. Finally, while the growth of the triple mutant was significantly impaired compared to the parental strain, it was still viable despite lacking all TPs except for PBP3. We also evaluated the doubling times of the parental strain and the triple mutant in the early-to-mid-exponential phase and found them to be 22 ± 0.4 min and 24 ± 0.3 min, respectively. These data show that in addition to the inhibition of essential PBP3, the simultaneous inhibition of both PBP1a and PBP2 should also be explored as an alternative strategy for the design of new drugs.
FIG 1.
Growth curves of A. baumannii CIP 70.10 and its mutant derivatives (A) and detection of nucleic acids in culture media (B). Strains listed in the gray dashed box of panel A had very similar growth rates. The agarose gel was stained with ethidium bromide and imaged using a ChemiDoc Touch imaging system (Bio-Rad). Detection of nucleic acids was performed independently from the experiment described above for panel A (see Materials and Methods for details). Abbreviations: wt, wild type; Δ2, deletion of PBP2; 1a(S/A), Ser-to-Ala substitution in the TP domain of PBP1a; 1b(S/A), Ser-to-Ala substitution in the TP domain of PBP1b.
As we observed an increase in the viscosity of some mutant cultures, we evaluated whether cell lysis occurred by checking for the presence of nucleic acids in the supernatants of bacterial cultures after 12 h of incubation (see Materials and Methods). Almost no nucleic acids were present in samples of the wild type, the PBP1b(S/A) and ΔPBP2 single mutants, as well as the PBP1b(S/A)+ΔPBP2 double mutant (Fig. 1B). For the PBP1a(S/A) single mutant, there was a significant amount of nucleic acids detected in the growth medium; no further increase was observed with the PBP1a(S/A)+PBP1b(S/A) double mutant. However, the simultaneous inactivation of PBP1a and PBP2 in the PBP1a(S/A)+ΔPBP2 double mutant resulted in an additional substantial increase in the amount of nucleic acids. This increase was similar to that observed for the PBP1a(S/A)+PBP1b(S/A)+ΔPBP2 triple mutant. These data indicate that the inactivation of the PBP1a TP domain is the major contributor to the observed cell lysis.
Susceptibility of PBP mutants of A. baumannii to β-lactam antibiotics.
Next, we assessed the susceptibilities of A. baumannii CIP 70.10 TP mutants to 10 β-lactams of various classes: penicillins (ampicillin and piperacillin), cephalosporins (ceftazidime, ceftriaxone, and cefoxitin), the monobactam aztreonam, carbapenems (imipenem, meropenem, and doripenem), and the β-lactamase inhibitor sulbactam, which has inherent antibacterial activity against A. baumannii and is used for the treatment of infections caused by this multidrug-resistant pathogen. The MICs of ampicillin, piperacillin, and aztreonam against the PBP1a(S/A) mutant decreased 2-fold, while those for all other antibiotics remained unchanged (Table 1). Similarly, a maximum of a 2-fold reduction of MICs (for piperacillin, ceftazidime, cefoxitin, and aztreonam) was observed for the PBP1b(S/A) mutant. Our antibiotic susceptibility data for the PBP1a(S/A) and PBP1b(S/A) mutants are similar to the results for P. aeruginosa PA14 PBP1a and PBP1b single mutants, in which the entire TP domains were deleted, where no or only minor differences in the MICs were noticed (29). Recently, MIC data were reported for A. baumannii ATCC 17978, where the entire mrcA or mrcB gene encoding both the TP and glycosyltransferase domains of PBP1a or PBP1b, respectively, was inactivated (25). Similar to what we observed with our PBP1a(S/A) and PBP1b(S/A) mutants, there was no change observed in the MICs for the carbapenem antibiotics imipenem and meropenem compared to the parental strain. However, 8- to 32-fold differences (increases or decreases) in the MICs of ampicillin, cefoxitin, and aztreonam were reported for the A. baumannii ATCC 17978 ΔmrcA and ΔmrcB mutants, except for the latter with ampicillin, where no change in the MIC was observed. As described above, we observed no change or just a 2-fold decrease in the MICs for our PBP1a(S/A) and PBP1b(S/A) mutants. To evaluate the impact of the simultaneous inactivation of TP and glycosyltransferase activities of PBP1a and PBP1b in A. baumannii CIP 70.10 on antibiotic susceptibility, we determined the MICs for our ΔmrcA and ΔmrcB mutants (ΔPBP1a and ΔPBP1b) and found that they differ from the MICs for the same mutants of A. baumannii ATCC 17978 (Table 1). Significant differences in MICs were also observed between the two parental strains. Combined, these results indicate that discrepancies in the MICs of our PBP1a(S/A), PBP1b(S/A), ΔmrcA, and ΔmrcB mutants and the ΔmrcA and ΔmrcB mutants of A. baumannii ATCC 17978 are likely caused, to a large degree, by differences in parental strains. Finally, the inactivation of PBP2 (ΔPBP2 mutant) affected the MICs of all drugs tested with the exception of cefoxitin; the MICs of piperacillin and aztreonam decreased 4-fold, while the MICs of all remaining antibiotics decreased 2-fold. There were some very insignificant differences in MICs between A. baumannii and P. aeruginosa mutants lacking PBP2, where no changes in the MICs of piperacillin, ceftazidime, doripenem, and aztreonam were observed in the latter (29).
TABLE 1.
Antibiotic susceptibilities of A. baumannii CIP 70.10 and its mutant derivativesa
| Strain | Genotype | MIC of antibiotic (μg/mL) |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AMP | PIP | CAZ | CRO | FOX | IPM | MEM | DOR | ATM | SUL | KAN | CIP | ||
| Control | Wild type | 32 | 64 | 8 | 16 | 128 | 0.25 | 0.5 | 0.25 | 64 | 2 | 2 | 0.25 |
| PBP1a(S/A) | mrcA(S/A) | 16 | 32 | 8 | 16 | 128 | 0.25 | 0.5 | 0.25 | 32 | 2 | 2 | 0.125 |
| PBP1b(S/A) | mrcB(S/A) | 32 | 32 | 4 | 16 | 64 | 0.25 | 0.5 | 0.25 | 32 | 2 | 2 | 0.25 |
| ΔPBP2 | ΔmrdA | 16 | 16 | 4 | 8 | 128 | 0.125 | 0.25 | 0.125 | 16 | 1 | 1–2 | 0.25 |
| PBP1a(S/A)+PBP1b(S/A) | mrcA(S/A) mrcB(S/A) | 16 | 16 | 4 | 16 | 64 | 0.25 | 0.25 | 0.125 | 32 | 2 | 2 | 0.125 |
| PBP1a(S/A)+ΔPBP2 | mrcA(S/A) ΔmrdA | 16 | 16 | 8 | 8 | 64 | 0.25 | 0.25 | 0.125 | 16 | 1 | 1–2 | 0.125 |
| PBP1b(S/A)+ΔPBP2 | mrcB(S/A) ΔmrdA | 16 | 16 | 4 | 8 | 64 | 0.25 | 0.25 | 0.25 | 16 | 1 | 1 | 0.25 |
| PBP1a(S/A)+PBP1b(S/A)+ΔPBP2 | mrcA(S/A) mrcB(S/A) ΔmrdA | 8 | 8 | 4 | 4 | 4 | 0.03 | 0.06 | 0.06 | 16 | 0.5 | 1 | 0.125 |
| ΔPBP1a | ΔmrcA | 64 | 32 | 8 | 16 | 256 | 0.25 | 0.5 | 0.25 | 64 | 2 | 2 | 0.125 |
| ΔPBP1b | ΔmrcB | 32 | 32 | 2 | 8 | 64 | 0.25 | 0.5 | 0.25 | 32 | 2 | 2 | 0.25 |
The control was A. baumannii CIP 70.10. ΔPBP2, deletion of PBP2; PBP1a(S/A), Ser-to-Ala substitution in the TP domain of PBP1a; PBP1b(S/A), Ser-to-Ala substitution in the TP domain of PBP1b; ΔPBP1a, deletion of PBP1a; ΔPBP1b, deletion of PBP1b; AMP, ampicillin; PIP, piperacillin; CAZ, ceftazidime; CRO, ceftriaxone; FOX, cefoxitin; IPM, imipenem; MEM, meropenem; DOR, doripenem; ATM, aztreonam; SUL, sulbactam; KAN, kanamycin; CIP, ciprofloxacin.
For the PBP1a(S/A)+PBP1b(S/A) double mutant strain, the MICs of ceftriaxone, imipenem, and sulbactam remained unchanged; the MIC of piperacillin decreased 4-fold; and the MICs of the other antibiotics decreased 2-fold. The inactivation/deletion of TPs of PBP1a(S/A)+ΔPBP2 did not affect the MICs of ceftazidime and imipenem, decreased the MICs of piperacillin and aztreonam 4-fold, and decreased the MICs of the other drugs 2-fold. For the PBP1b(S/A)+ΔPBP2 double mutant strain, the MICs of imipenem and doripenem were unchanged, its MICs for piperacillin and aztreonam decreased 4-fold, and the MICs of the remaining antibiotics decreased 2-fold. Altogether, the MICs of β-lactams against the three double mutants of A. baumannii CIP 70.10 were very similar: they were either identical or differed by 2-fold, with the exception of aztreonam, where a 4-fold decrease of the MICs was observed. Of note, this 4-fold change was observed in all strains where PBP2 was inactivated, which suggests that PBP2 plays a role in resistance to this antibiotic. Overall smaller changes in the MICs of ceftazidime, aztreonam, and doripenem were reported for the three double mutants of P. aeruginosa PA14: they were either identical to the parental strain or differed by only 2-fold (29). However, the authors of that study observed larger differences for imipenem and piperacillin. We emphasize that our PBP single and double mutants and those of P. aeruginosa were constructed differently. For our PBP1a(S/A) and PBP1b(S/A) single mutants, we inactivated the TP domain by mutating the catalytic Ser residue to Ala, while the whole mrdA gene for PBP2 was deleted. The same approach was used for the construction of our double and triple mutants. For the P. aeruginosa PBP1a, PBP1b, and PBP2 single mutants, the TP domains of these enzymes were deleted. For the PBP1a+PBP2 and PBP1b+PBP2 double mutants, the TP domains of PBP1a and PBP1b were deleted, while the gene encoding PBP2 was replaced by a gentamicin resistance cassette. Finally, the authors were not able to construct a strain where both the PBP1a and PBP1b TP domains were deleted. Thus, to construct this double mutant, they inserted a functional copy of the gene encoding PBP1a into the bacterial chromosome under the control of an inducible PBAD promoter. These differences between mutants of A. baumannii and P. aeruginosa could differentially impact their susceptibility to antibiotics.
Finally, the triple mutant strain was most sensitive to all antibiotics tested. The MIC of ceftazidime against this mutant decreased 2-fold; the MICs of ampicillin, ceftriaxone, doripenem, aztreonam, and sulbactam decreased 4-fold; the MICs of piperacillin, imipenem, and meropenem decreased 8-fold; and the MIC of cefoxitin decreased 32-fold. In summary, our data showed that, with the exception of the triple mutant, the inactivation of individual TPs and their combinations in A. baumannii CIP 70.10 results in just a 2- to 4-fold decrease in the MICs of β-lactams against mutant strains.
To evaluate whether the inactivation of A. baumannii TPs resulted in significant alterations of the permeability of the bacterial cell wall, we also tested the MICs of two non-β-lactam antibiotics, kanamycin (an aminoglycoside) and ciprofloxacin (a fluoroquinolone). We observed a maximum of a 2-fold reduction in the MICs of these drugs for some of the mutant strains, including the triple mutant (Table 1), an indication that their cell wall integrity was not significantly compromised.
Muropeptide composition of the A. baumannii CIP 70.10 triple mutant.
We were intrigued by the fact that the triple mutant, in which PBP3 remained the only active TP, was still viable. PBP3 is essential for bacterial cell division but has not been implicated in elongation. We wondered whether the recently discovered A. baumannii l,d-transpeptidase LdtJ (25) compensates for the lack of TP activities of PBP1a, PBP1b, and PBP2 in the triple mutant by producing more cross-linked peptidoglycan. To assess this assumption, we determined the muropeptide composition of A. baumannii CIP 70.10 and its triple mutant using liquid chromatography-mass spectrometry (LC-MS). This methodology allowed us to distinguish between the TP activity that produces 4-3 cross-links and the Ldt activity that generates 3-3-cross-linked peptidoglycan (Fig. S2).
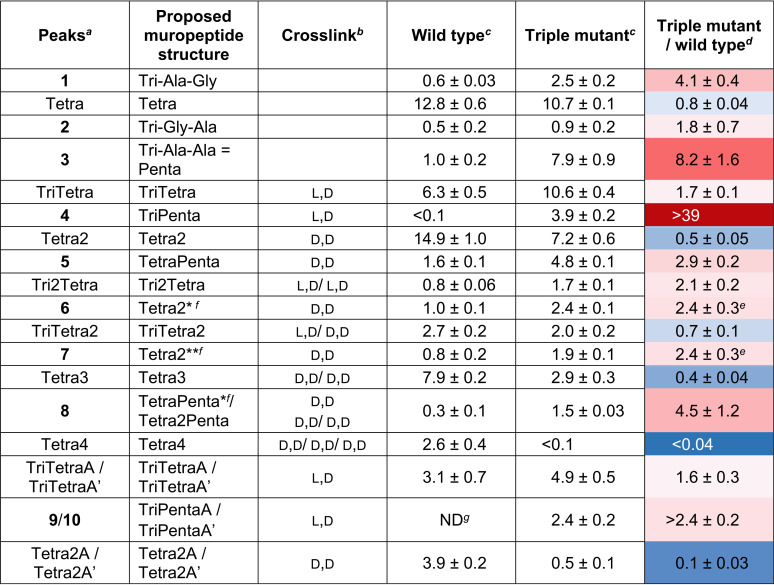
Our experiments revealed dramatic changes in the muropeptide composition of the triple mutant in comparison to the parental strain (Fig. 2A to C, red peaks, and Table 2; Fig. S3). Overall, the lack of activity of the three major TPs resulted in a significant (>8-fold) accumulation of their substrate, the pentapeptide (Fig. 2A, peak 3), which was almost fully consumed in the parental strain. We also note 4- and 2-fold increases of other pentapeptides (peaks 1 and 2) in which –d-Ala-d-Ala in the pentapeptide stem is replaced by –d-Ala-Gly and –Gly-d-Ala, respectively. PBP3, as the only source of TP activity in the mutant, was capable of producing tetrapeptide-containing muropeptides (Tetra2 and Tetra3) at levels that were reduced 2- and 2.5-fold, respectively, compared to those observed in wild-type A. baumannii CIP 70.10 (Table 2). The levels of the two other tetrapeptide-containing muropeptides (Tetra2A and Tetra2A′) were reduced 10-fold, while that of Tetra4 was almost undetectable. We also found that the level of the TriTetra2 muropeptide, which is a product of both Ldts and TPs, was slightly decreased (1.4-fold). Combined, these results showed that PBP3, by itself, is not capable of cross-linking a large number of peptidoglycan strands to fully compensate for the lack of activity of other TPs. In contrast, large quantities (in comparison to the parental strain) of the nonproductive tetrapeptide-containing cyclic species (Fig. 2A, peaks 6 and 7), where two adjacent peptides on the same strand of glycan are cross-linked, were detected in the triple mutant. There is literature precedent for the detection of a cyclic muropeptide in E. coli (37). In addition to the tetrapeptide-containing species, d,d-cross-linked muropeptides containing pentapeptides (peak 5 [TetraPenta] and peak 8 [Tetra2Penta]) were detected in A. baumannii CIP 70.10. These species were found at elevated levels in the triple mutant (Table 2), possibly resulting from the excessive accumulation of the unreacted pentapeptide substrate caused by the lack of the three other major TPs.
FIG 2.
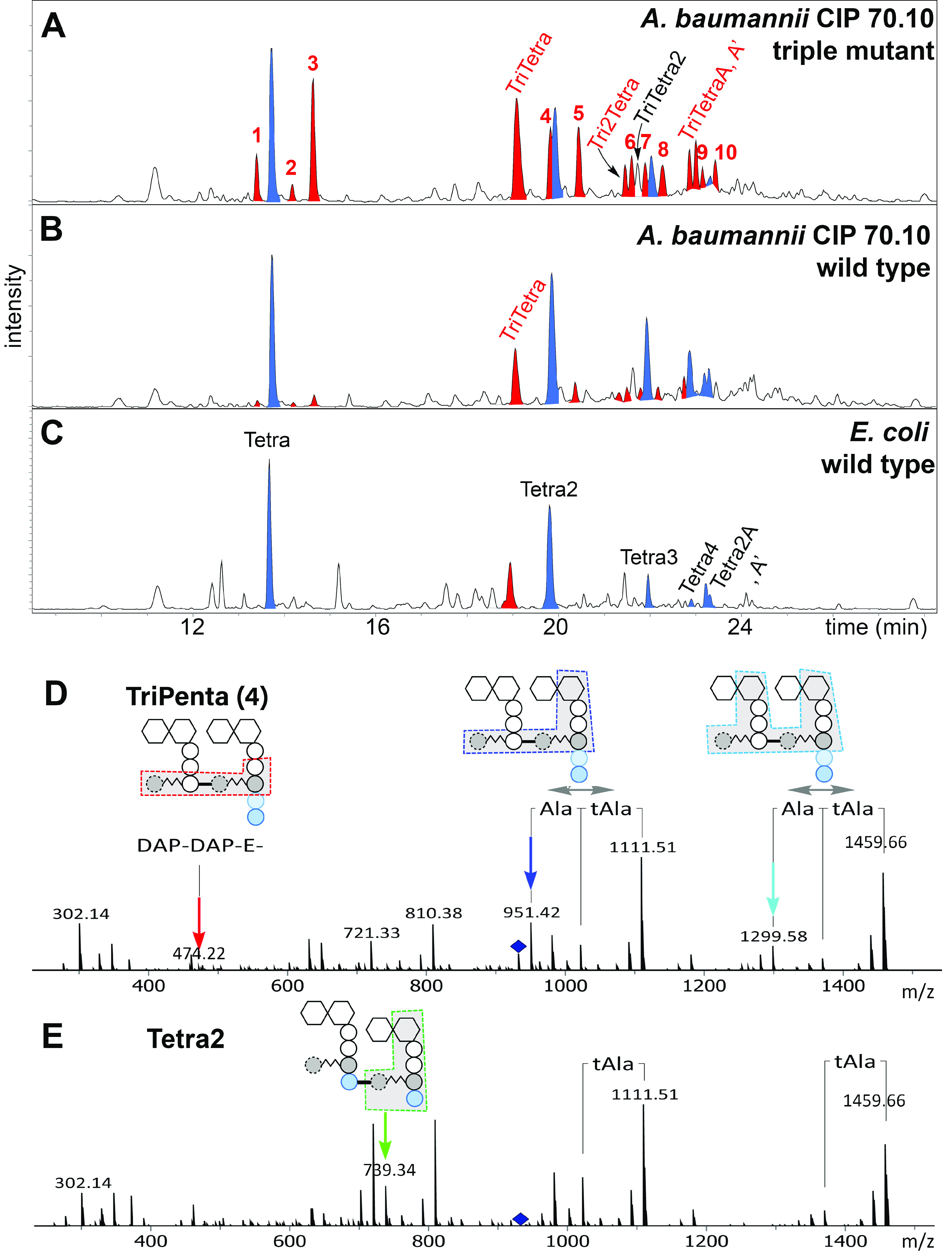
Peptidoglycan analysis of A. baumannii CIP 70.10, its triple mutant, and E. coli DH10B. (A to C) LC-MS chromatograms of muropeptides of the triple mutant (A), the wild type (B), and E. coli DH10B (C). The chromatographic peaks in blue represent muropeptides containing Tetra peptides (as non-cross-linked and cross-linked peptides) of the E. coli sacculus whose presence we also confirmed in A. baumannii. Muropeptides indicated in red were increased the most between wild-type A. baumannii and its triple mutant. (D and E) Collision-induced dissociation mass spectra of peak 4 (TriPenta) (D) and Tetra2 (E). The nature of a cross-link was determined using the presence of signature ions existing in only one form of cross-link. Peaks with green arrows exist only in structures with d,d-cross-links, while peaks with blue, cyan, and red arrows and gray double arrows exist only in structures with l,d-cross-links. The presence of –A-A was confirmed by the presence of fragment ions by the concomitant loss of Ala and tAla, indicated with a gray double arrow. DAP, diaminopimelic acid.
TABLE 2.
Relative abundances of muropeptides of A. baumannii CIP 70.10 and its triple mutant
Peaks labeled by number or structure as shown in Fig. 2.
The identity of the cross-link (d,d versus l,d) was confirmed by LC-MS/MS.
Percentages of extracted ion chromatograms of the peak area given as averages from triplicates and errors.
Numbers are given with a color gradient (the deeper the red, the larger the increase; the deeper the blue, the larger the decrease).
The sum of two related muropeptides, 6 and 7, was used.
Structures are presented in Fig. S3 in the supplemental material.
ND, not detected.
Our analysis also revealed a number of 3-3-cross-linked muropeptides, which are likely produced by the recently identified A. baumannii l,d-transpeptidase LdtJ, whose inactivation in two A. baumannii strains abolished the production of 3-3-cross-linked muropeptides (25). The levels of the Ldt cross-linked TriTetra muropeptide species TriTetra, TriTetraA, and TriTetraA′ increased 1.6- to 1.7-fold and that of the Tri2Tetra species increased 2.1-fold in the triple mutant compared to the parental strain (Table 2). Among muropeptides that were increased in the mutant strain, peak 4 drew our attention. While only a trace amount was found in wild-type A. baumannii, its intensity in the mutant was as high as that of Tetra2 (Fig. 2A) and was elevated more than 40-fold compared to the parental strain (Table 2). It eluted slightly earlier than the Tetra2 peak (Fig. 2A) by LC, and its m/z value was the same as that of a doubly charged Tetra2 species (m/z = 933.4097). LC-tandem MS (MS/MS) of peak 4 and comparison to that of Tetra2 revealed that peak 4 is a new pentapeptide-containing l,d-cross-linked muropeptide, TriPenta (Fig. 2D and E). Two other l,d-cross-linked muropeptides, TriPentaA and TriPentaA′ (peaks 9 and 10 in Fig. 2A), members of the same TriPenta series, were found exclusively in the mutant. The observed increase in the amount of l,d-cross-linked peptidoglycan in the triple mutant may result from the compensatory response to the significant decrease in d,d-cross-linking. It was recently demonstrated that the LdtJ l,d-transpeptidase plays an important role in the biosynthesis of the bacterial cell wall, as its activity becomes essential for the cell viability of A. baumannii in the absence of the activity of only the PBP1a TP (25). These data indicated that the lack of TP activity of PBP1a has to be compensated for by the l,d-transpeptidase activity of LdtJ for bacterial survival. It is logical to assume that the lack of TP activity of PBP1a, PBP1b, and PBP2 in the triple mutant would require an even higher level of compensation by LdtJ, as the remaining TP, PBP3, known to be a major component of the septal wall biosynthesis complex, has not been implicated in peripheral peptidoglycan synthesis (21).
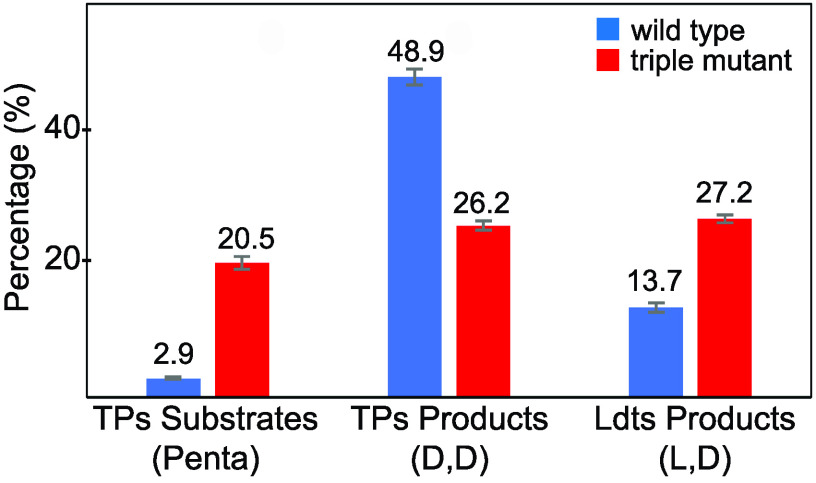
To summarize, our analysis of muropeptides showed that the triple mutant had a 7-fold increase of the combined substrates of d,d-transpeptidases (pentapeptide-containing muropeptides), a 2-fold decrease of the combined products (muropeptides containing the d,d-cross-linkage), and a 2-fold increase of alternative cross-linked muropeptides (containing the l,d-cross-linkage) (Fig. 3). These data also showed that in the absence of all other TPs, the activity of PBP3, in combination with that of the LdtJ l,d-transpeptidase, is sufficient for the viability of the A. baumannii CIP 70.10 triple mutant; however, its growth was dramatically impaired, and it became significantly more sensitive to β-lactams. As LdtJ is the only (or the major) l,d-transpeptidase of A. baumannii that generates 3-3-cross-linked peptidoglycan (25), it can also be explored as a potential target for the development of novel β-lactam antibiotics.
FIG 3.
Effect of the triple mutation on the overall substrates and products of d,d-transpeptidases (TPs) and the products of l,d-transpeptidases (Ldts). Three independently generated peptidoglycan samples were analyzed, and error bars represent the standard deviations.
MATERIALS AND METHODS
Bacterial strains and growth media.
All bacterial strains used in this study (see Table S2 in the supplemental material) were grown at 37°C in Luria-Bertani (LB) or Mueller-Hinton (MH) liquid medium or on LB agar plates unless otherwise specified. Kanamycin (KAN) was added at concentrations of 30 μg/mL to maintain plasmids in Acinetobacter baumannii CIP 70.10 and its derivatives and 50 μg/mL in Escherichia coli DH10B. LB medium was supplemented with 20% sucrose for counterselection against plasmids in the integrants of A. baumannii.
DNA methods.
The suicide vector pMo130 (Addgene plasmid 27388) (38) was used for the mutational inactivation or deletion of genes encoding PBPs (Table S1) in the genome of A. baumannii CIP 70.10 (32). E. coli DH10B was used as a host for pMo130 vector derivatives (Table S2). Plasmid DNA was isolated with the QIAprep spin miniprep kit (Qiagen). Primers for PCR were synthesized by Eurofins Genomics (Table S3). Colony PCR was performed using Q5 Hot Start high-fidelity 2× master mix (New England BioLabs). PCR products were analyzed on a 1% agarose gel and purified with the Wizard SV gel and PCR cleanup system (Promega). DNA sequencing was done by Molecular Cloning Laboratories (MCLAB). Restriction enzymes and T4 DNA ligase (New England BioLabs) were used according to the manufacturer’s recommendations. Electrocompetent cells of A. baumannii CIP 70.10 and its derivatives were prepared as described previously (39). Electroporation was performed with a Bio-Rad MicroPulser using 40 μL cells with 2 μL plasmid DNA in a 0.2-cm cuvette, using the manufacturer’s preset protocol 2 for bacteria. Following electroporation, bacteria were transferred to 1 mL super optimal broth with catabolite repression (SOC) medium and incubated at 30°C for 4 h prior to plating on LB-KAN agar plates. Genomic DNA from the wild type and mutant variants of A. baumannii CIP 70.10 was purified with the DNeasy blood and tissue kit (Qiagen). Whole-genome sequencing, using an Illumina platform, and comparative genomic data analysis were performed by EzBiome (Maryland).
Construction of pMo130 derivatives.
To generate gene deletions, the upstream region (UR) and downstream region (DR) of the targeted genes encoding PBPs were PCR amplified. Amplification primers were designed to generate fragments of approximately 1,000 bp in length that included the first and last 15 bp of the targeted genes (the names of PBPs and matching genes are listed in Table S1, and primers are listed in Table S3). The resulting UR and DR products were purified, ligated, and further amplified using primers that introduced a BamHI site at both ends of the amplified fragment. Following purification on an agarose gel, the PCR products were digested with BamHI, and the fragments were inserted into the pMo130 plasmid at the BamHI site, resulting in the gene knockout vectors pMT308FW, pMT309FW, pMT310FW, and pNS257. To generate the catalytic Ser-to-Ala substitution in the d,d-transpeptidase (TP) domains of PBP1a and PBP1b, primers were designed to amplify the UR and DR of the catalytic Ser residue that included the entire genes for PBP1a and PBP1b and to mutate the codon for Ser (TCT) to that of Ala (GCA). PCR amplification, ligation, and cloning into the pMo130 plasmid were performed as described above, resulting in the pMT321FW and pMT322FW vectors. All vectors were subsequently transformed into E. coli DH10B, plasmids were purified from the transformants, and the sequences were verified by DNA sequencing.
Generation of A. baumannii CIP 70.10 mutants.
To generate deletions of the genes encoding PBP1a, PBP1b, PBP2, and PBP3, each suicide vector (pMT308FW, pMT309FW, pNS257, and pMT310FW) was electroporated into A. baumannii CIP 70.10 cells, and the transformants were selected on LB-KAN agar plates. After incubation at 30°C overnight, individual KAN-resistant (KANr) colonies were analyzed by PCR to identify integrants and to determine (based on the size of the amplified product) whether the crossover had occurred within the homologous chromosomal UR or DR of the targeted gene. In cases where we attempted to inactivate the TP domains of PBP1a and PBP1b by replacing their catalytic Ser with Ala, the pMT321FW and pMT322FW vectors were used for transformation, and KANr colonies were selected for further analysis. The location of the crossover for these two vectors was determined by using primers that specifically anneal to the part of the genes for PBP1a or PBP1b that contains the codon for Ala but not for Ser (Table S3). To select for the rare second recombination event that leads to the excision of the integrated vector from the chromosome of A. baumannii and results in the deletion or mutation of the targeted PBP, sucrose counterselection was performed as described previously (40). Briefly, the integrants were cultured in 5 mL LB broth containing 20% sucrose and passaged daily (1:50). Starting from the second passage, aliquots of the diluted culture were plated onto LB agar supplemented with 10% sucrose. The resulting colonies were replica plated onto LB-KAN agar plates to select the KAN-sensitive colonies, which were individually analyzed by PCR to identify the desired mutants. The deletion of target genes to yield ΔPBP1a, ΔPBP1b, and ΔPBP2 mutants and the Ser-to-Ala mutation of target TP domains to yield PBP1a(S/A) and PBP1b(S/A) mutants were verified by DNA sequencing. A. baumannii single mutants were used to generate all double mutants, ΔPBP1a+ΔPBP2, ΔPBP1b+ΔPBP2, PBP1a(S/A)+ΔPBP2, PBP1b(S/A)+ΔPBP2, and PBP1b(S/A)+PBP1a(S/A), and, subsequently, the triple mutant PBP1a(S/A)+PBP1b(S/A)+ΔPBP2. These mutants were generated and analyzed using the procedures described above. Suicide vectors and A. baumannii CIP 70.10 mutants constructed in this study are listed in Table S2.
Antimicrobial susceptibility testing.
The MICs of selected β-lactam antibiotics were measured according to Clinical and Laboratory Standards Institute (CLSI) (41) recommendations using the broth dilution method in MH medium as described previously (42).
Growth curve.
To monitor bacterial growth, triplicate cultures of the A. baumannii parental strain and its mutant derivatives grown overnight were diluted 1:100 in MH broth and grown for 1.5 to 2 h to mid-exponential phase. The optical density (OD) of the bacterial cultures was adjusted at 600 nm to 0.2, and the cultures were further diluted (5-fold) into MH medium to a final volume of 5 mL. Bacterial growth was monitored for 24 h by plating cells at designated time points onto large (15-cm) agar plates, and cells were counted after incubation overnight. The average values obtained from three independent measurements were plotted as a function of time. Prism 5 (GraphPad Software, Inc.) was used to generate growth curves and calculate standard deviations.
Evaluation of doubling times of the wild-type strain and its triple mutant.
To measure the generation time, cultures of the parental and triple mutant A. baumannii strains grown overnight were diluted 100-fold in MH broth and grown for 2 h. Next, the cultures were adjusted to an OD at 600 nm (OD600) of 0.2 and subsequently diluted into fresh MH medium to generate a final inoculum of 3 × 102 CFU/mL. Bacteria were plated every hour (from 0 to 8 h) on agar plates in triplicate, and cells were counted after incubation overnight. The CFU per milliliter were plotted versus time, and the generation time was calculated from the exponential phase of the curve using the equation Nt = N0(ekt), where Nt is the CFU per milliliter at time t, N0 is the initial CFU per milliliter, and k describes the growth rate constant. Data were generated from three independent experiments.
Detection of cell lysis.
Parental and mutant A. baumannii cells were grown overnight in 5 mL of MH medium. The next morning, cultures were diluted by adding ∼5 × 102 CFU/mL into fresh medium. After 12 h of incubation, 1-mL aliquots were centrifuged at 10,000 × g for 10 min, and 15-μL samples were analyzed on a 1% agarose gel with ethidium bromide for the presence of nucleic acids in the supernatant.
Peptidoglycan analysis.
Bacterial cultures of the A. baumannii CIP 70.10 parental strain and its PBP1a(S/A)+PBP1b(S/A)+ΔPBP2 triple mutant were grown to OD600 values of 5.7 and 3, respectively, and the cells were collected by centrifugation for 20 min at 4°C. Pellets were resuspended in 20 mL cold water, slowly added to 20 mL boiling 10% sodium dodecyl sulfate (SDS), and boiled for another hour with stirring, and solubilization then continued overnight with stirring at 42°C. Samples were centrifuged at 100,000 × g for 1 h, and the pellets were washed three times with water. Finally, the pellets were resuspended in 3 mL of 10 mM Tris-HCl buffer (pH 7.6), and a 0.25% final concentration of trypsin was added to digest the samples for 1.5 h at 37°C in a shaker incubator. Reactions were stopped with a 1% final concentration of SDS and boiling for 20 min. Peptidoglycan was pelleted by centrifugation and washed 3 times with water as described above. The final pellets were resuspended in 2 mL water, and aliquots were stored at −20°C for further analysis. The sacculus prep of each strain was incubated with mutanolysin in 50 mM TES [N-tris(hydroxymethyl)methyl-2-aminoethanesulfonic acid] overnight at 37°C. Reactions were stopped by boiling for 5 min. The resulting mixture was reduced with sodium borohydride for 1 h, acidified with phosphoric acid to pH 3 to 4, and centrifuged. The supernatant was analyzed using ultraperformance liquid chromatography-mass spectrometry (UPLC-MS). The UPLC-MS instrument consisted of a Waters Acquity UPLC H-Class system coupled with a Bruker impact II ultrahigh-resolution Qq-time of flight mass spectrometer using Hystar 5.0 SR1 software. The Bruker electrospray ionization source was operated in the positive-ion mode with the following parameters: an end plate offset voltage of −500 V, a capillary voltage of 1,800 V, and nitrogen as both a nebulizer (4 × 105 Pa) and dry gas (7 L/min) at 200°C. Mass spectra were accumulated over the mass range of m/z 200 to 3,000. LC separations were performed on an Acquity UPLC HSS T3 column (1.7 μm, 2.1 by 150 mm) with a Vanguard precolumn (1.8 μm, 2.1 by 5 mm) at 40°C. A 35-min gradient consisted of a 5-min hold at 100% solvent A–0% solvent B and a 30-min linear gradient to 82% solvent A–18% solvent B (solvent A is 0.1% formic acid in water, and solvent B is 0.1% formic acid in acetonitrile) at a flow rate of 0.4 mL/min. LC flow during the first 3 min of each run was diverted to the waste. The chemical structures of some muropeptides of A. baumannii were confirmed by comparison of their retention times and high-resolution accurate masses (high-resolution mass spectrometry [HRMS]) to those of the known muropeptides of the E. coli sacculus (Fig. 2; Fig. S3 and S4 and Table S5). The chemical structures of other muropeptides found in A. baumannii strains were elucidated using HRMS and collisionally induced dissociation MS/MS experiments (Fig. S4 to S8). Fragmentation of the protonated molecules was evaluated under single-collision conditions with argon gas at collision energy levels ranging between 36 and 45 eV.
For the nomenclature of muropeptide products, we followed the literature precedent (43), with slight modifications. The names of cross-linked peptides are given as donor-acceptor partners. For example, TriTetra indicates that a tripeptide donor and a tetrapeptide acceptor are cross-linked to each other. Tetra2 means that two tetrapeptides are cross-linked to each other. Muropeptide names ending in “A” indicate that in the NAG-NAM unit, NAM is replaced by anhydroNAM (NAG-anhNAM). The chemical structures and the corresponding cartoon representations of muropeptides identified in this study are given in Fig. S3.
ACKNOWLEDGMENT
This work was supported by grant 1R01AI155723 from the NIH/NIAID (S.B.V.).
Footnotes
Supplemental material is available online only.
Contributor Information
Marta Toth, Email: mtoth1@nd.edu.
Sergei B. Vakulenko, Email: svakulen@nd.edu.
REFERENCES
- 1.O’Neill J. 2016. Review on antimicrobial resistance: tackling drug-resistant infections globally: final report and recommendations. Wellcome Trust, London, United Kingdom. [Google Scholar]
- 2.Laxminarayan R, Duse A, Wattal C, Zaidi AK, Wertheim HF, Sumpradit N, Vlieghe E, Hara GL, Gould IM, Goossens H, Greko C, So AD, Bigdeli M, Tomson G, Woodhouse W, Ombaka E, Peralta AQ, Qamar FN, Mir F, Kariuki S, Bhutta ZA, Coates A, Bergstrom R, Wright GD, Brown ED, Cars O. 2013. Antibiotic resistance—the need for global solutions. Lancet Infect Dis 13:1057–1098. 10.1016/S1473-3099(13)70318-9. [DOI] [PubMed] [Google Scholar]
- 3.United Nations General Assembly. 2016. Political declaration of the high-level meeting of the general assembly on antimicrobial resistance. United Nations General Assembly 2016. Global health and foreign policy, seventy-first session, agenda item 127. United Nations, New York, NY. [Google Scholar]
- 4.Organisation for Economic Co-operation and Development. 2018. Stemming the superbug tide: just a few dollars. Organisation for Economic Co-operation and Development, Paris, France. https://www.oecd.org/health/health-systems/Stemming-the-Superbug-Tide-Policy-Brief-2018.pdf. [Google Scholar]
- 5.Centers for Disease Control and Prevention. 2019. Antibiotic resistance threats in the United States, 2019. Centers for Disease Control and Prevention, US Department of Health and Human Services, Atlanta, GA. https://www.cdc.gov/drugresistance/pdf/threats-report/2019-ar-threats-report-508.pdf. [Google Scholar]
- 6.Lima LM, Silva B, Barbosa G, Barreiro EJ. 2020. β-Lactam antibiotics: an overview from a medicinal chemistry perspective. Eur J Med Chem 208:112829. 10.1016/j.ejmech.2020.112829. [DOI] [PubMed] [Google Scholar]
- 7.Papp-Wallace KM, Endimiani A, Taracila MA, Bonomo RA. 2011. Carbapenems: past, present, and future. Antimicrob Agents Chemother 55:4943–4960. 10.1128/AAC.00296-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Doi Y, Murray GL, Peleg AY. 2015. Acinetobacter baumannii: evolution of antimicrobial resistance—treatment options. Semin Respir Crit Care Med 36:85–98. 10.1055/s-0034-1398388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nguyen M, Joshi SG. 10 May 2021. Carbapenem resistance in Acinetobacter baumannii, and their importance in hospital-acquired infections: a scientific review. J Appl Microbiol 10.1111/jam.15130. [DOI] [PubMed]
- 10.Bush K. 2018. Past and present perspectives on β-lactamases. Antimicrob Agents Chemother 62:e01076-18. 10.1128/AAC.01076-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.World Health Organization. 2017. Central Asian and Eastern European surveillance of antimicrobial resistance. World Health Organization Regional Office for Europe, Copenhagen, Denmark. [Google Scholar]
- 12.Spellberg B, Bonomo RA. 2014. The deadly impact of extreme drug resistance in Acinetobacter baumannii. Crit Care Med 42:1289–1291. 10.1097/CCM.0000000000000181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wong D, Nielsen TB, Bonomo RA, Pantapalangkoor P, Luna B, Spellberg B. 2017. Clinical and pathophysiological overview of Acinetobacter infections: a century of challenges. Clin Microbiol Rev 30:409–447. 10.1128/CMR.00058-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zilberberg MD, Nathanson BH, Sulham K, Fan W, Shorr AF. 2016. Multidrug resistance, inappropriate empiric therapy, and hospital mortality in Acinetobacter baumannii pneumonia and sepsis. Crit Care 20:221. 10.1186/s13054-016-1392-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rice LB. 2008. Federal funding for the study of antimicrobial resistance in nosocomial pathogens: no ESKAPE. J Infect Dis 197:1079–1081. 10.1086/533452. [DOI] [PubMed] [Google Scholar]
- 16.Cochrane SA, Lohans CT. 2020. Breaking down the cell wall: strategies for antibiotic discovery targeting bacterial transpeptidases. Eur J Med Chem 194:112262. 10.1016/j.ejmech.2020.112262. [DOI] [PubMed] [Google Scholar]
- 17.Vollmer W, Blanot D, de Pedro MA. 2008. Peptidoglycan structure and architecture. FEMS Microbiol Rev 32:149–167. 10.1111/j.1574-6976.2007.00094.x. [DOI] [PubMed] [Google Scholar]
- 18.Silhavy TJ, Kahne D, Walker S. 2010. The bacterial cell envelope. Cold Spring Harb Perspect Biol 2:a000414. 10.1101/cshperspect.a000414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koch AL. 2003. Bacterial wall as target for attack: past, present, and future research. Clin Microbiol Rev 16:673–687. 10.1128/CMR.16.4.673-687.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johnson JW, Fisher JF, Mobashery S. 2013. Bacterial cell-wall recycling. Ann N Y Acad Sci 1277:54–75. 10.1111/j.1749-6632.2012.06813.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Egan AJF, Errington J, Vollmer W. 2020. Regulation of peptidoglycan synthesis and remodelling. Nat Rev Microbiol 18:446–460. 10.1038/s41579-020-0366-3. [DOI] [PubMed] [Google Scholar]
- 22.Sauvage E, Kerff F, Terrak M, Ayala JA, Charlier P. 2008. The penicillin-binding proteins: structure and role in peptidoglycan biosynthesis. FEMS Microbiol Rev 32:234–258. 10.1111/j.1574-6976.2008.00105.x. [DOI] [PubMed] [Google Scholar]
- 23.Goffin C, Ghuysen JM. 1998. Multimodular penicillin-binding proteins: an enigmatic family of orthologs and paralogs. Microbiol Mol Biol Rev 62:1079–1093. 10.1128/MMBR.62.4.1079-1093.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aliashkevich A, Cava F. 10 June 2021. l,d-Transpeptidases: the great unknown among the peptidoglycan cross-linkers. FEBS J 10.1111/febs.16066. [DOI] [PubMed]
- 25.Kang KN, Kazi MI, Biboy J, Gray J, Bovermann H, Ausman J, Boutte CC, Vollmer W, Boll JM. 2021. Septal class A penicillin-binding protein activity and l,d-transpeptidases mediate selection of colistin-resistant lipooligosaccharide-deficient Acinetobacter baumannii. mBio 12:e02185-20. 10.1128/mBio.02185-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Papp-Wallace KM, Senkfor B, Gatta J, Chai W, Taracila MA, Shanmugasundaram V, Han S, Zaniewski RP, Lacey BM, Tomaras AP, Skalweit MJ, Harris ME, Rice LB, Buynak JD, Bonomo RA. 2012. Early insights into the interactions of different β-lactam antibiotics and β-lactamase inhibitors against soluble forms of Acinetobacter baumannii PBP1a and Acinetobacter sp. PBP3. Antimicrob Agents Chemother 56:5687–5692. 10.1128/AAC.01027-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Penwell WF, Shapiro AB, Giacobbe RA, Gu RF, Gao N, Thresher J, McLaughlin RE, Huband MD, DeJonge BL, Ehmann DE, Miller AA. 2015. Molecular mechanisms of sulbactam antibacterial activity and resistance determinants in Acinetobacter baumannii. Antimicrob Agents Chemother 59:1680–1689. 10.1128/AAC.04808-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Denome SA, Elf PK, Henderson TA, Nelson DE, Young KD. 1999. Escherichia coli mutants lacking all possible combinations of eight penicillin binding proteins: viability, characteristics, and implications for peptidoglycan synthesis. J Bacteriol 181:3981–3993. 10.1128/JB.181.13.3981-3993.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen W, Zhang YM, Davies C. 2017. Penicillin-binding protein 3 is essential for growth of Pseudomonas aeruginosa. Antimicrob Agents Chemother 61:e01651-16. 10.1128/AAC.01651-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Boll JM, Crofts AA, Peters K, Cattoir V, Vollmer W, Davies BW, Trent MS. 2016. A penicillin-binding protein inhibits selection of colistin-resistant, lipooligosaccharide-deficient Acinetobacter baumannii. Proc Natl Acad Sci USA 113:E6228–E6237. 10.1073/pnas.1611594113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cayo R, Rodriguez MC, Espinal P, Fernandez-Cuenca F, Ocampo-Sosa AA, Pascual A, Ayala JA, Vila J, Martinez-Martinez L. 2011. Analysis of genes encoding penicillin-binding proteins in clinical isolates of Acinetobacter baumannii. Antimicrob Agents Chemother 55:5907–5913. 10.1128/AAC.00459-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Krahn T, Wibberg D, Maus I, Winkler A, Puhler A, Poirel L, Schluter A. 2015. Complete genome sequence of Acinetobacter baumannii CIP 70.10, a susceptible reference strain for comparative genome analyses. Genome Announc 3:e00850-15. 10.1128/genomeA.00850-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Biswas I. 2015. Genetic tools for manipulating Acinetobacter baumannii genome: an overview. J Med Microbiol 64:657–669. 10.1099/jmm.0.000081. [DOI] [PubMed] [Google Scholar]
- 34.Trebosc V, Gartenmann S, Royet K, Manfredi P, Totzl M, Schellhorn B, Pieren M, Tigges M, Lociuro S, Sennhenn PC, Gitzinger M, Bumann D, Kemmer C. 2016. A novel genome-editing platform for drug-resistant Acinetobacter baumannii reveals an AdeR-unrelated tigecycline resistance mechanism. Antimicrob Agents Chemother 60:7263–7271. 10.1128/AAC.01275-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Paradis-Bleau C, Markovski M, Uehara T, Lupoli TJ, Walker S, Kahne DE, Bernhardt TG. 2010. Lipoprotein cofactors located in the outer membrane activate bacterial cell wall polymerases. Cell 143:1110–1120. 10.1016/j.cell.2010.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Suzuki H, Nishimura Y, Hirota Y. 1978. On the process of cellular division in Escherichia coli: a series of mutants of E. coli altered in the penicillin-binding proteins. Proc Natl Acad Sci USA 75:664–668. 10.1073/pnas.75.2.664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Harz H, Burgdorf K, Holtje JV. 1990. Isolation and separation of the glycan strands from murein of Escherichia coli by reversed-phase high-performance liquid chromatography. Anal Biochem 190:120–128. 10.1016/0003-2697(90)90144-X. [DOI] [PubMed] [Google Scholar]
- 38.Hamad MA, Zajdowicz SL, Holmes RK, Voskuil MI. 2009. An allelic exchange system for compliant genetic manipulation of the select agents Burkholderia pseudomallei and Burkholderia mallei. Gene 430:123–131. 10.1016/j.gene.2008.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Aranda J, Poza M, Pardo BG, Rumbo S, Rumbo C, Parreira JR, Rodriguez-Velo P, Bou G. 2010. A rapid and simple method for constructing stable mutants of Acinetobacter baumannii. BMC Microbiol 10:279. 10.1186/1471-2180-10-279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Amin IM, Richmond GE, Sen P, Koh TH, Piddock LJ, Chua KL. 2013. A method for generating marker-less gene deletions in multidrug-resistant Acinetobacter baumannii. BMC Microbiol 13:158. 10.1186/1471-2180-13-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Clinical and Laboratory Standards Institute. 2018. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard, 11th ed. CLSI document M07. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 42.Stewart NK, Smith CA, Antunes NT, Toth M, Vakulenko SB. 2019. Role of the hydrophobic bridge in the carbapenemase activity of class D β-lactamases. Antimicrob Agents Chemother 63:e02191-18. 10.1128/AAC.02191-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Glauner B. 1988. Separation and quantification of muropeptides with high-performance liquid chromatography. Anal Biochem 172:451–464. 10.1016/0003-2697(88)90468-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material. Download AAC.01729-21-s0001.pdf, PDF file, 2.2 MB (2.2MB, pdf)