Abstract
Targeted protein degradation (TPD) is an emerging therapeutic modality with the potential to tackle disease-causing proteins that have historically been highly challenging to target with conventional small molecules. In the 20 years since the concept of a proteolysis-targeting chimera (PROTAC) molecule harnessing the ubiquitin–proteasome system to degrade a target protein was reported, TPD has moved from academia to industry, where numerous companies have disclosed programmes in preclinical and early clinical development. With clinical proof-of-concept for PROTAC molecules against two well-established cancer targets provided in 2020, the field is poised to pursue targets that were previously considered ‘undruggable’. In this Review, we summarize the first two decades of PROTAC discovery and assess the current landscape, with a focus on industry activity. We then discuss key areas for the future of TPD, including establishing the target classes for which TPD is most suitable, expanding the use of ubiquitin ligases to enable precision medicine and extending the modality beyond oncology.
Subject terms: Drug discovery, Chemical biology, Biotechnology
Targeted protein degradation with proteolysis-targeting chimeras (PROTACs) has the potential to tackle disease-causing proteins that have historically been highly challenging to target with conventional small molecules. This article summarizes the first two decades of PROTAC discovery and discusses key areas for the future of this therapeutic modality, including establishing the target classes for which it is most suitable and extending its application beyond oncology.
Introduction
Targeted protein degradation (TPD) is attracting substantial interest owing to its potential to therapeutically modulate proteins that have proved difficult to target with conventional small molecules. Some have been intractable because their active sites are broad, shallow pockets that are difficult to bridge with small molecules; others have ‘smooth’ surfaces that offer few sites for a small molecule to bind. Many of these targets have key roles in cancer and other diseases, and so have remained of great therapeutic interest, despite their recalcitrance to small-molecule inhibitors.
A major class of molecules that may enable such proteins to be modulated through TPD are known as proteolysis-targeting chimera (PROTAC) protein degraders. These are heterobifunctional small molecules consisting of two ligands joined by a linker: one ligand recruits and binds a protein of interest (POI) while the other recruits and binds an E3 ubiquitin ligase. Simultaneous binding of the POI and ligase by the PROTAC induces ubiquitylation of the POI and its subsequent degradation by the ubiquitin–proteasome system (UPS), after which the PROTAC is recycled to target another copy of the POI (Fig. 1). It is this catalytic-type mechanism of action (MoA) and event-driven pharmacology that distinguishes PROTACs from classical inhibitors, which have a one-to-one relationship with the POI and whose pharmacology is driven by stoichiometry and, usually, by interactions with a catalytic site.
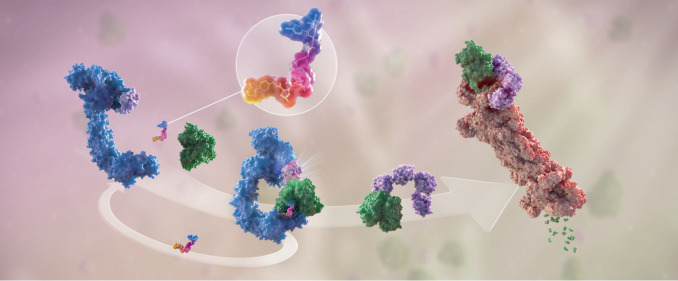
Fig. 1. The mechanism of PROTAC-mediated targeted protein degradation.
Schematic representation of the mechanism of action of proteolysis-targeted chimera (PROTAC) molecules. The PROTAC molecule (enlarged in the circle) is a heterobifunctional molecule bridging a ubiquitin ligase (in blue) and a target protein (in green). As a first step, PROTACs induce the proximity of the ligase and the substrate, such that ubiquitin (in pink) will be conjugated to the recruited substrate by the activity of the ligase. This is a catalytic step that a single PROTAC molecule can perform iteratively, enabling multiple turnover of ubiquitylation reactions, resulting in formation of ubiquitin chains on a substrate. Ubiquitin chains are then recognized by the proteasome (in red), shuttling the ubiquitylated substrate through its proteolytic chamber and degrading the target protein into small peptides (in green). Figure reproduced with permission from Arvinas, Inc.
There are also several other types of targeted protein degrader. Molecular glues, based on the serendipitous discovery that thalidomide and its analogues act as degrader molecules, constitute another important therapeutic class1–8. Although not heterobifunctional in the manner of PROTAC molecules, molecular glues promote ubiquitylation of a POI by enhancing a protein–protein interaction (PPI) between a ligase and a potential substrate (Fig. 2; Box 1).
Fig. 2. Modalities in targeted protein degradation.
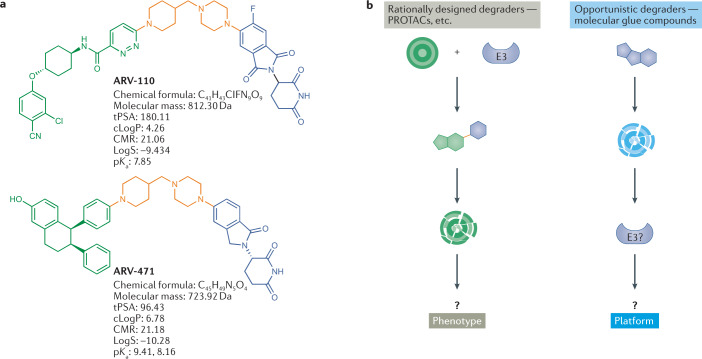
a | Structure and properties of two proteolysis-targeting chimeras (PROTACs) that have entered clinical trials, ARV-110 and ARV-471. PROTACs are composed of a target-binding moiety (green), a linker (orange) and an E3 ligase-binding moiety (blue). ARV-110 and ARV-471 target the androgen receptor and the oestrogen receptor, respectively, while the E3-binding ligand interacts with the cereblon (CRBN) E3 ligase. b | Schematic representation of the two foundational modalities for targeted protein degradation. Left-hand side: discovery of PROTACs composed of a target-binding moiety (green), a linker (orange) and a ligase-binding moiety (blue), enabling the rational discovery of heterobifunctional molecules to degrade a desired target. Right-hand side: opportunistic discovery of degrader molecules, whereby a known molecule is shown to have a degrader effect, making it possible to identify the E3 ligase mediating that degradation and determine whether that degrader mechanism could be expanded to target additional proteins of interest. The pros and cons of these modalities are discussed in the text and in Box 1. CMR, calculated molecular refractivity; tPSA, total polar surface area.
In the 20 years since the first small-molecule PROTAC was reported in the literature9, the technology has moved from academia to industry, where several biotech and pharmaceutical companies have disclosed programmes in preclinical and early clinical development (Fig. 3). In 2019, the first PROTAC molecules entered clinical testing; in 2020, these trials provided the first clinical proof-of-concept for the modality against two well-established cancer targets: the oestrogen receptor (ER) and the androgen receptor (AR). With this success in hand, the TPD field is now poised to tackle ‘undrugged’ targets and other classes of difficult protein target.
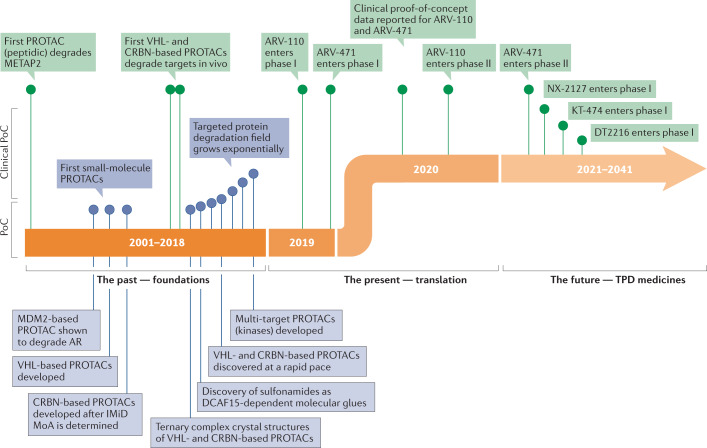
Fig. 3. Timeline of PROTAC discoveries.
The first era of targeted protein degradation (TPD) began with publication of the pivotal proteolysis-targeting chimera (PROTAC) paper by Sakamoto et al.9 in 2001, which was the first demonstration of the concept that protein targets could be intentionally dragged to a ubiquitin ligase to induce their degradation using chemical tools. Between then and today, the field has grown exponentially and has moved from peptide-based tool degraders to multiple classes of fully synthetic small molecules that can induce proximity between a ligase and a protein of interest, leading to its degradation. This foundational era of TPD was capped by the first rational heterobifunctional PROTAC degrader entering clinical trials in 2019, ARV-110, which targets the androgen receptor (AR) by recruiting it to the Cullin–RING ligase 4–cereblon (CRL4–CRBN) ligase complex. The current era of TPD can be considered its initial translational phase, in which multiple molecules designed to degrade disease-causing proteins are entering the clinic with the hope of providing meaningful benefits to patients. DCAF15, DDB1- and CUL4-associated factor 15; IMiD, immunomodulatory imide drug; MoA, mechanism of action; METAP2, methionyl aminopeptidase 2; PoC, proof of concept; VHL, von Hippel–Lindau.
In this Review, we briefly summarize the first two decades of PROTAC development, as it has been extensively reviewed elsewhere (for example10–13) and the current status of clinical translation of TPD. We then focus on discussing key questions relevant to what TPD could achieve therapeutically and what is needed to move the field forwards over the next 20 years.
Box 1 Strategies for discovery of molecular glues.
Rational degrader discovery using the modular proteolysis-targeting chimera (PROTAC) strategy (Fig. 2) can be complemented by the purposeful discovery of degrader compounds known as molecular glues. Molecular glues can enhance complex formation between an E3 ligase and a target by wedging between protein–protein interfaces. Although the best-known examples of molecular glues today are compounds that induce protein degradation, there are many examples of additional intramolecular and intermolecular glues that function outside the ubiquitin–proteasome system (UPS), such as the allosteric protein tyrosine phosphatase non-receptor type 11 (PTPN11; also known as SHP2) inhibitor SHP099 (ref.230), which stabilizes a closed conformation of SHP2 (intramolecular); and cyclosporin231, which induces the proximity of calcineurin and cyclophilin (intermolecular). Two excellent recent reviews describe a plethora of molecular glue approaches, ranging from small molecules to multi-specific antibodies, and paint a promising picture for proximity-induced pharmacology in modern medicine232,233.
Although historically, molecular glue degraders were discovered retrospectively — that is, the mechanism of action of cytotoxic compounds such as thalidomide were determined after their FDA approval — the recent rise in interest in targeted protein degradation as a therapeutic modality has led to a focus on identifying compounds with this mode of action from the beginning of the drug discovery process. The first rational discovery of molecular glues between a ligase and a substrate involved a series of compounds that restored binding affinity between the β-TRCP ligase and its mutated phospho-degron, discovered in a biochemical screen234. More recently, targeted discovery of degrader compounds against oncoproteins in a cell-based system has enabled the discovery of novel cereblon (CRBN)-dependent and CRBN-independent IKAROS family zinc finger 1 (IKZF1) degraders235, suggesting that screening for degraders against a target of choice is a strategy that can yield a bounty of phenotypically relevant degrader molecules. Lastly, multiple recent papers have described agnostic screening approaches for degrader molecules84–86 (as described in more detail in the main text) that were also highlighted by a comprehensive review, which offers guidelines for discovering E3 ligase modulators by phenotypic screening strategies76.
Foundations of TPD
Ubiquitin-dependent proteolysis is a major pathway that degrades intracellular proteins as part of normal cellular maintenance processes. In this pathway, proteins are targeted for degradation by the proteasome in a three-step process involving ubiquitin-activating enzymes (E1), ubiquitin-conjugating enzymes (E2) and, finally, ubiquitin–protein ligases (E3), which coordinate the transfer of ubiquitin molecules to the target protein (substrate)14. The human genome is estimated to encode more than 600 E3 ligases15, each with specificity for a different subset of proteins.
The concept of harnessing this natural degradation system for therapeutic purposes was inspired by early studies of viruses and plants (Box 2), which highlighted the possibility of deliberately designing small molecules that co-opt E3 ligases and recruit them for degradation of a POI. The initial work to co-opt E3 ligases focused on two areas of research. One involved using ligase-recruiting ligands to probe the biology and phenotype of a POI as an alternative to targeted inhibition, gene knockout or gene knockdown. Another area explored engineered E3 ligases as a potential novel therapeutic modality for targeting proteins that had proved difficult to drug with conventional small-molecule inhibitors, such as the small GTPase KRAS16 and the transcription factor MYC17.
In vitro proof of concept for the first fully synthetic PROTAC, dubbed Protac-1, was reported in 2001 (ref.9). Protac-1 was designed to target methionyl aminopeptidase 2 (METAP2), the putative target of the potent angiogenesis inhibitors ovalicin and fumagillin, and consisted of two domains: ovalicin, and a 10-amino acid phosphopeptide from nuclear factor-κB inhibitor-α (NF-κBIα; also known as IκBα) that is recognized by the E3 ligase β-transducin repeat-containing E3 ubiquitin–protein ligase (β-TRCP). Protac-1 acted as a tether between METAP2 and β-TRCP, enabling the ligase to ubiquitylate METAP2 in extracts from unfertilized Xenopus laevis eggs.
The subsequent discovery of a peptide from hypoxia-inducible factor 1 subunit-α (HIF1α) that bound the E3 ligase von Hippel–Lindau tumour suppressor (VHL)18,19 led to the design of cell-penetrating PROTACs that degraded a range of POIs. Technically, these early PROTACs are now considered ‘bioPROTACs’ because they are not fully small-molecule structures but instead contain peptide ligands for the E3 ligase. The discovery of small-molecule mimetics of the HIF1α peptide20,21 opened the door to the rational design of PROTACs based wholly on small molecular structures — for example, the first PROTACs based on the bromodomain protein inhibitor JQ1 that recruited VHL to degrade bromodomain-containing protein 4 (BRD4)22,23.
In parallel with the development of these early PROTAC molecules, the E3 ligase cereblon (CRBN) was identified as the target of thalidomide and its analogues lenalidomide and pomalidomide6, which are known as immunomodulatory imide drugs (IMiDs) in the context of cancer therapy. These drugs co-opt CRBN to target IKAROS family zinc finger 1 (IKZF1) and IKZF3 for degradation3,4,24, and are now considered as pioneering examples of molecular glues.
Additionally, the sulfonamide indisulam, a small-molecule cell-cycle inhibitor used to treat some solid tumours and leukaemias, was shown to function in a similar manner by facilitating the interaction between the E3 ligase damage-specific DNA damage-binding protein 1 (DDB1) and CUL4-associated factor 15 (DCAF15) and the pre-mRNA splicing factor RNA-binding motif protein 39 (RBM39)25–27 and the related splicing factor RBM23 (refs26,28,29).
Over the past two decades, evidence for the broad therapeutic potential of PROTACs and other TPD molecules has moved from studies in cell lysates and cell culture to studies in animals and animal models of disease. In the same time frame, TPD molecules have progressed from being based on peptides to fully synthetic, rationally designed small molecules. Although the exploration of TPD as a therapeutic modality is still in its early days, the realization that TPD as a therapeutic small-molecule paradigm may offer advantages over target inhibition and genomic targeted approaches (for example, small interfering RNAs (siRNAs), antisense oligonucleotides (ASOs), short hairpin RNAs (shRNAs) and CRISPR-based agents) has spurred widespread interest in academia and industry to explore its full potential30.
Box 2 Targeted protein degradation examples from viruses and plants.
At least two dozen viruses are known to hijack the human ubiquitin–proteasome system (UPS) to promote their own survival and replication10. For example, the E6 protein of human papillomavirus type 16 (HPV-16) and type 18 (HPV-18) recruits the human E3 ligase, ubiquitin–protein ligase E3A (UBE3A; also known as E6AP) to ubiquitylate p53, resulting in its degradation236. Human immunodeficiency virus 1 (HIV-1) deploys its viral proteins Vpr and Vpx, to recruit DDB1 and CUL4-associated factor 1 (DCAF1) to target several different human proteins, including DNA repair proteins, for ubiquitylation237,238.
Plant studies revealed that, in addition to proteins, small molecules are capable of inducing UPS-mediated protein degradation. In many species of plants, auxin (indole-3-acetic acid; IAA) functions as a hormone that promotes degradation of the Aux/IAA family of transcriptional repressors, which are involved in regulating plant development. Auxin acts by stabilizing the interaction between Aux/IAA proteins and the plant E3 ligase transport inhibitor response 1 (Tir1)239,240. Another plant hormone, jasmonate, employs the same mechanism241 and many other classes of small molecule in plants promote protein degradation242.
These early examples from viruses and plants highlighted the possibility of deliberately designing small molecules that co-opt E3 ligases and recruit them for degradation of a protein of interest.
More recently, it has been appreciated that viruses provide several examples of the fusion protein and antibody mimic subgroups of bioPROTACs (defined as proteolysis-targeting chimeras (PROTACs) composed of peptide ligands; see main text). For instance, the Vif protein of HIV-1 can be viewed as a fusion protein between an F-box domain and target recognition domain. The F-box domains from HIV-1 Vif, von Hippel–Lindau (VHL) and suppressor of cytokine signalling 2 (SOCS2) have high structural homology and all bind with the RING E3 ligase CUL5–elongin B (ELOB)–ELOC complex. The substrate-binding domain of Vif recruits two human proteins — apolipoprotein B mRNA editing enzyme catalytic subunit 3G (APOBEC3G), which functions to inhibit retroviral replication, and core-binding factor subunit-β (CBF-β)243 — to the E3 ligase for ubiquitylation and degradation by the proteasome. The Vpr protein of HIV-1 (ref.244) and the E6 protein of hepatitis B virus (HBV)245 act as bispecific antibody mimics, whereby one face of the viral protein interacts with the E3 ligase target association proteins DCAF1 and E6AP, while the opposite face binds uracil DNA glycosylase (UNG2) and p53, respectively, to hijack the UPS for target degradation.
Furthermore, while most of the attention on PROTAC technology stems from its potential to treat human disease, applications of the modality in agriculture are also now being explored — in effect, bringing it full circle from the plant-derived small molecules that originally inspired it. As in mammalian and yeast cells, PROTAC molecules can unlock and direct the UPS within crop plants, with the potential to tailor applications to the needs of each crop in the face of climate change and increasing resistance of pests to ageing crop protectants.
Status of clinical translation of TPD
The era of rationally designed targeted protein degraders as potential human therapeutics began in 2019 with the entry of two heterobifunctional degraders into first-in-human trials (Fig. 3): the PROTACs ARV-110 (NCT03888612) and ARV-471 (NCT04072952), targeting the AR and the ER, respectively. Both of these degraders have now moved on to phase II trials (Table 1), and have been followed into the clinic by degraders from Bristol Myers Squibb (BMS), Nurix Therapeutics, Kymera Therapeutics, Dialectic Therapeutics, Foghorn Therapeutics and others.
Table 1.
Heterobifunctional PROTAC targeted protein degraders in clinical development
| Company | Degrader | Target | Indications | E3 ligase | ROA | Highest phase | Clinical trial no. (if applicable) |
|---|---|---|---|---|---|---|---|
| Arvinas | ARV-110 | AR | Prostate cancer | CRBN | Oral | Phase II | NCT03888612 |
| Arvinas/Pfizer | ARV-471 | ER | Breast cancer | CRBN | Oral | Phase II | NCT04072952 |
| Accutar Biotech | AC682 | ER | Breast cancer | CRBN | Oral | Phase I | NCT05080842 |
| Arvinas | ARV-766 | AR | Prostate cancer | Undisclosed | Oral | Phase I | NCT05067140 |
| Bristol Myers Squibb | CC-94676 | AR | Prostate cancer | CRBN | Oral | Phase I | NCT04428788 |
| Dialectic Therapeutics | DT2216 | BCL-xL | Liquid and solid tumours | VHL | I.v. | Phase I | NCT04886622 |
| Foghorn Therapeutics | FHD-609 | BRD9 | Synovial sarcoma | Undisclosed | I.v. | Phase I | NCT04965753 |
| Kymera/Sanofi | KT-474 | IRAK4 | Autoimmune diseases (e.g., AD, HS, RA) | Undisclosed | Oral | Phase I | NCT04772885 |
| Kymera | KT-413 | IRAK4 | Diffuse large B cell lymphoma (MYD88-mutant) | CRBN | I.v. | Phase I | |
| Kymera | KT-333 | STAT3 | Liquid and solid tumours | Undisclosed | Undisclosed | Phase I | |
| Nurix Therapeutics | NX-2127 | BTK | B cell malignancies | CRBN | Oral | Phase I | NCT04830137 |
| Nurix Therapeutics | NX-5948 | BTK | B cell malignancies and autoimmune diseases | CRBN | Oral | Phase I | NCT05131022 |
| C4 Therapeutics | CFT8634 | BRD9 | Synovial sarcoma | CRBN | Oral | IND-e | |
| C4 Therapeutics | CFT8919 | EGFR-L858R | Non-small-cell lung cancer | CRBN | Oral | IND-e | |
| Cullgen | CG001419 | TRK | Cancer and other indications | CRBN | Oral | IND-e |
AD, atopic dermatitis; AR, androgen receptor; BCL-xL, B cell lymphoma-extra large; BRD9, bromodomain-containing protein 9; BTK, Bruton’s tyrosine kinase; CRBN, cereblon; EGFR, epidermal growth factor receptor; ER, oestrogen receptor; HS, hidradenitis suppurativa; IND-e, in IND-enabling preclinical studies; IRAK4, interleukin-1 receptor-associated kinase 4; i.v., intravenous; PROTAC, proteolysis-targeting chimera; RA, rheumatoid arthritis; ROA, route of administration; STAT3, signal transducer and activator of transcription 3; TRK, tropomyosin receptor kinase; VHL, von Hippel–Lindau.
Additionally, the clinical development of molecular glue compounds such as CC-90009 and CC-92480, built around the IMiD platform at Celgene (now BMS), as well as molecular glue compounds from C4 Therapeutics and Novartis (Table 2), underscores the advances made in understanding and exploiting protein degradation as a therapeutic modality. Although not heterobifunctional, these new-generation CRBN modulators have been engineered with therapeutic targets in mind (IKZF1/IKZF3 and G1 to S phase transition 1 (GSPT1)), which are recruited by the compounds in a molecular glue mechanism to target them for degradation (Box 1).
Table 2.
Molecular glue targeted protein degraders in clinical development
| Company | Degrader | Target | Indications | E3 ligase | Highest phase | Clinical trial no. (if applicable) |
|---|---|---|---|---|---|---|
| Bristol Myers Squibb | CC-220 | IKZF1/3 | MM | CRBN | Phase II | NCT02773030 |
| Bristol Myers Squibb | CC-92480 | IKZF1/3 | MM | CRBN | Phase II | NCT03989414 |
| Bristol Myers Squibb | CC-90009 | GSPT1 | Acute myeloid leukaemia | CRBN | Phase II | NCT02848001/NCT04336982 |
| Bristol Myers Squibb | CC-99282 | IKZF1/3 | Chronic myeloid leukaemia, non-Hodgkin lymphoma | CRBN | Phase I | NCT04434196/NCT03930953 |
| C4 Therapeutics | CFT7455 | IKZF1/3 | MM | CRBN | Phase I | NCT04756726 |
| Novartis | DKY709 | Helios | Solid tumours (NSCLC) | CRBN | Phase I | NCT03891953 |
CRBN, cereblon; GSPT1, G1 to S phase transition 1; IKZF1, IKAROS family zinc finger 1; MM, multiple myeloma; NSCLC, non-small-cell lung carcinoma.
The two foundational TPD modalities (Fig. 2) — one based on the discovery and rational design of IMiDs pioneered at Celgene/BMS (reviewed extensively elsewhere8,31,32), and the other based on the conceptualization and development of heterobifunctional PROTACs based on rational pairings of an E3 ligase recruiter with a POI-targeting warhead pioneered at Arvinas33–35 — have been catalysts in taking TPD from the bench to the clinic.
Target selection for degraders
Novel CRBN modulators are built on the basic understanding of the mechanism of action of thalidomide analogues, which primarily target the IKZF family of transcription factors, and potentially other C2H2 zinc finger transcription factors, as well as proteins that contain the Gly-β-hairpin loop structural motifs36,37. By contrast, the establishment of rationally designed, heterobifunctional PROTAC degraders started with specific POI targets in mind, which has driven the rapid expansion of this TPD approach across diverse therapeutic targets and disease indications.
For any new therapeutic modality, the selection of well-known targets is important for establishing proof of concept because it introduces just one variable (the modality), not two (the modality and a novel target) into the equation. So, the initial wave of heterobifunctional degraders in the clinic (Table 1) has focused on proteins with a combination of well-characterized biological and biochemical properties, clinically validated roles in diseases with clear unmet medical need, and previously established clinical efficacy for their inhibition. The ER in breast cancer38,39 and the AR in prostate cancer33 are the leading examples of such targets.
Clinical proof of concept for PROTACs
Before 2020, four key questions remained unanswered about heterobifunctional degrader molecules. Would they have drug-like properties? Would they be safe in humans? Would they work against the target protein as expected? Would they have a therapeutic effect?
In 2020, initial positive data reported from the phase I trials of ARV-110 and ARV-471 answered all four foundational questions in the affirmative — not just for the compounds themselves but for the entire TPD field. Given the importance of these findings for the TPD modality, we briefly summarize them here.
The AR degrader ARV-110 was evaluated in heavily pretreated patients with metastatic castration-resistant prostate cancer (mCRPC). The phase I trial (NCT03888612) was an excellent test case for PROTAC degraders because AR is a well-known driver of prostate cancer, and patients with mCRPC, especially this heavily pretreated subset, have limited therapeutic options owing to insensitivity or resistance to anti-androgenic therapies that are the mainstays of prostate cancer treatment. Initial trial data showed that ARV-110 was well tolerated at doses up to 420 mg. The data also demonstrated ARV-110-mediated degradation of the protein target in tumours — the first such evidence for a PROTAC molecule in humans — and revealed early signs of antitumour activity, as measured by reductions in levels of prostate specific antigen (PSA) and/or by RECIST (Response Evaluation Criteria in Solid Tumors). These data support the continued development of ARV-110, and the phase II ARDENT trial began in October 2020 with a dose of 420 mg (ref.40).
ARV-471, an ER degrader, entered clinical trials as a monotherapy for patients with ER+/HER2− locally advanced or metastatic breast cancer. The interim data for ARV-471 revealed a manageable tolerability profile with robust signals of clinical efficacy41, a 42% clinical benefit rate in a heavily pretreated population and evidence of better ER degradation than fulvestrant and other clinical-stage selective ER degraders (SERDs) when compared at the same stage of development. ARV-471 has now progressed to phase II (VERITAC; NCT04072952) in metastatic breast cancer as a single agent, and a phase Ib study evaluating ARV-471 in combination with the cyclin-dependent kinase 4/cyclin-dependent kinase 6 (CDK4/CDK6) inhibitor palbociclib has also commenced. ARV-471 is now being co-developed by Arvinas and Pfizer to treat ER+/HER2− metastatic breast cancer.
The early but strong clinical profiles for these two PROTAC degraders, in two very different populations, in which they demonstrated desirable safety, efficacious exposure and signs of clinical efficacy with meaningful benefits for patients, are solidifying the therapeutic viability of the modality. Twenty years after its theoretical conception, at least 15 targeted protein degraders, among them heterobifunctional PROTACs and molecular glues, had entered the clinic by the end of 2021 (ref.30) (Tables 1,2), and many more are expected to follow.
Outlook for the next 20 years of TPD
Beyond the current slate of compounds in clinical testing and others in the development pipelines of multiple companies, where can — and even should — the TPD field aim to go in the next 20 years? What challenges and possibilities remain for the field, and what more might be accomplished when those are explored and addressed? What additional tools — E3 ligases, ligands and targeted protein degrader classes — might be useful in which diseases?
The basic science behind TPD has grown exponentially and matured substantially in the past few years (for example, as reviewed in refs42–44). In our opinion, the next milestones in this ‘new era’ of TPD will focus on four clinical translation inflexion points, namely, defining and clinically demonstrating the target classes best served by degradation over inhibition; expanding the scope of E3 ubiquitin ligases employed clinically in a targeted fashion to enable precision medicine; extending the clinical reach of the modality beyond oncology; and validating TPD modalities beyond IMiDs and PROTACs (Box 3) in clinical settings. In this section, we discuss each of these points and what excites us for the next 20 years of degrader discovery and development.
Box 3 Alternative degrader approaches to PROTACs.
The success of proteolysis-targeting chimeras (PROTACs) in hijacking the ubiquitin–proteasome system (UPS) for targeted protein degradation (TPD) has motivated research into other classes of heterobifunctional molecules that depend on non-UPS pathways to degrade a protein of interest (POI)246,247. These classes include autophagy-targeting chimeras (AUTACs), autophagosome-tethering compounds (ATTECs), lysosome-targeting chimeras (LYTACs) and antibody-based PROTACs (AbTACs). Collectively, AUTACs and ATTECs have also been categorized as macroautophagy degradation-targeting chimeras (MADTACs)248. Like PROTACs, AUTECs and ATTECs focus on intracellular triggering of TPD, while LYTACs and AbTACs trigger intercellular TPD via an extracellular process.
AUTACs link a warhead for the POI to a guanine derivative that tags the protein for degradation by the autophagy machinery. Takahashi et al.249 have demonstrated an autophagy-dependent mechanism for an AUTAC that S-guanylated enhanced-GRP (EGFP) in mouse embryonic fibroblasts. Their study also explored the generality of directed autophagic degradation by making AUTACs against methionyl aminopeptidase 2 (METAP2), FKBP prolyl isomerase 1A (FKBP1A; also known as FKBP12) and BET family proteins, all of which were found to be effective degraders.
ATTECs link a POI warhead to a ligand that binds to the autophagy protein LC3 (microtubule-associated protein 1 light chain 3α) glue, thereby bypassing the ubiquitin pathways by directly tethering the POI to the autophagosome. Li et al.196 have shown that ATTECs targeting mutant huntingtin (mHTT) directed the protein to the autophagosome for degradation in both cells and animals, and rescued phenotypes relevant to Huntington disease.
LYTACs bind a membrane-bound POI and the extracellular domains of a lysosome-shuttling receptor, which then drags the POI into the lysosome for degradation. The first LYTACs used a monoclonal antibody (mAb) conjugate: the mAb was directed against the POI, while the conjugated glycopolypeptides, which contained multiple serine-O-mannose-6-phosphonate (M6Pn) residues, interacted with the cation-independent M6P receptor (CI-M6PR) for internalization and lysosomal degradation of the POI. LYTACs against the APOE4 allele of apolipoprotein E (APOE), epidermal growth factor receptor (EGFR), CD71 and PDL1 all degraded their respective targets in various cell lines250.
AbTACs are bispecific antibodies that recruit membrane-bound E3 ligases to a membrane POI for degradation by the lysosome degradation pathway. Cotton et al.251 recently debuted AbTACs by describing AC-1, consisting of a recombinant antibody (R3) to the extracellular domain of the E3 ligase RING finger protein 43 (RNF43) and the PDL1-binding antibody atezolizumab. In MDA-MB-231 cells, AC-1 induced the degradation of PDL1 via an RNF43- and lysosomal-dependent mechanism.
Targets best suited for degradation
As described above, the first wave of clinical-stage protein degraders is aimed at classically drugged targets that have clinically validated roles in disease and readily available chemical matter. Success against these targets has begun to solidify PROTACs as a therapeutic modality and underscores the potential of these molecules to become best-in-class medicines by way of degrading a target instead of inhibiting it. However, the true promise of the modality is reaching targets that are currently difficult to drug with existing modalities or have not yet been drugged at all.
To date, traditional small-molecule drug discovery research for intracellular targets has often focused on developing high-affinity inhibitors that target either the active site or an allosteric site on an enzyme to shut down the function of the POI (occupancy-driven pharmacology). Although this has been a highly effective approach, it has left potential drug targets undrugged or underdrugged45. PROTACs bring the degradation function to the target (event-driven pharmacology), negating the need for an active site and redefining undruggable targets as simply undrugged.
The optimal targets for PROTAC therapy, which we have dubbed ‘Tenets of PROTAC targets’ (Fig. 4) can have several common characteristics, including: a change away from the natural state, via overexpression, mutation, aggregation, isoform expression or localization, that results in a disease-causing gain of function; a binding surface that is approachable by an E3 ligase; and ideally, an unstructured region to thread into the proteasome46,47. Proteins that have evolved resistance mutations to targeted therapies, proteins with scaffolding functions and proteins that are considered ‘undruggable’ with other modalities can also be highly suitable PROTAC targets.
Fig. 4. The tenets of PROTAC targets.
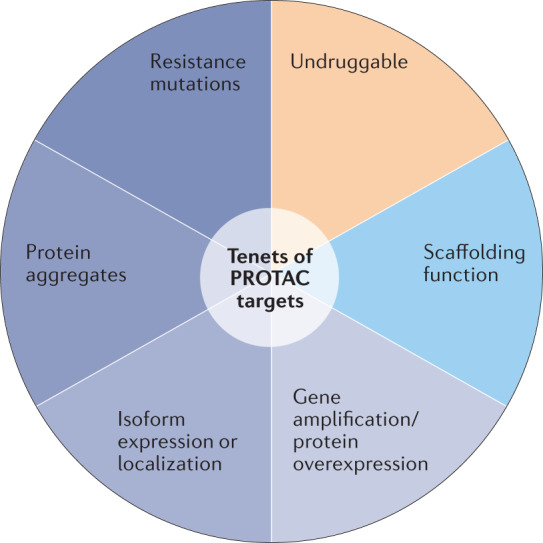
Proteolysis-targeting chimeras (PROTACs) bring the protein degrader function to the target; they do not need to bind within a biologically functional active site. This expands the accessible targets well beyond those that are druggable by traditional stoichiometric inhibition and provides novel ways to achieve selectivity. Proteins that may be best suited to therapeutic intervention by targeted protein degradation instead of stoichiometric inhibition include proteins with disease-causing gain of function owing to mutation, overexpression, aggregation or the differential expression or localization of protein isoforms. From a structural perspective, PROTAC targets need a small-molecule binding surface that is approachable by an E3 ligase, and ideally have an unstructured region that can be threaded into the proteasome.
The early PROTAC targets focused on POIs that had existing ligands, in the form of available inhibitors, but were still associated with clear unmet medical need. There are several reasons that existing ligands failed to fully benefit patients, such as incomplete inhibition, narrow therapeutic window and partial selectivity. By incorporating the ligand into a PROTAC, which often acts in an iterative or ‘pseudo-catalytic’ fashion by binding and facilitating the interaction of its ligase and POI targets, the POI can be degraded without requiring a large excess of drug, thereby resulting in more complete target blockade and a wider therapeutic window22,48. PROTACs may also benefit from cooperative PPIs between the E3 ligase and the POI. This can have the advantages of improved potency49 and selectivity50.
Although ‘PROTACable’ POIs do not need an enzyme active site, they do need a small-molecule binding site that is approachable by an E3 ligase. Using these sites does not require a high-affinity ligand if coupled to the right E3 ligand, but moderate affinity (≥1–500 nM) is typically needed, and access to the POI surface near the binding site by a recruited E3 ligase is essential. Achieving such binding affinities can often be challenging and has promoted research into alternative degraders (Box 3). Selection of the ligand-binding site is particularly important in the case of scaffolding proteins, where the POI may only be partially exposed within a given complex. It may be possible to degrade a POI by targeting a neighbouring protein within a protein complex (the bystander effect). This approach may prove useful in degrading scaffolding proteins in which the surface of the POI is mostly buried within the complex51 or the POI is a membrane-associated protein.
Cell-surface proteins are currently not considered optimal targets for PROTAC therapy as the UPS resides inside the cell. However, Bensimon et al.52 challenged this view by showing that multi-transmembrane proteins from the solute carrier (SLC) transporter family could be degraded using the dTAG technology53. Furthermore, they developed d9A-2, a pomalidomide-based PROTAC that degraded SLC family 9 member A1 (SLC9A1) — and, to a lesser extent, other SLC9 family members — leading to impaired pH homeostasis and cytotoxicity in multiple cancer cell lines, via a mechanism consistent with UPS-dependent TPD52. More recently, Wang et al.54 showed that programmed cell death ligand 1 (PDL1) — an immune checkpoint protein that resides on the cell surface to carry out its natural function but circulates between the surface and the cytoplasm — could be degraded using a CRBN-based PROTAC linked to the BMS-37 PDL1 warhead in MC-38 cells.
Although the ligand affinity for the POI can be low, an ideal E3 ligase ligand would be a potent binder with a slow off-rate. PROTACs that covalently bind the POI lose the ability to turn over and act iteratively55, whereas PROTACs that covalently bind an E3 ligase would reduce the three-body assembly kinetics into a two-body problem and enhance the catalytic efficiency. Currently there are limited methods to selectively covalently bind to a protein56–58, but advancements in bio-orthogonal chemistry could result in longer-lasting PROTACs with higher catalytic efficiencies.
In general, PROTAC physicochemical property space falls beyond the ‘rule of 5’ (refs59,60) owing to the bifunctional nature of the molecules. The rule of 5 was developed in the 1990s as a guideline for assembling high-throughput screening (HTS) libraries. However, several orally bioavailable compounds have been developed that fall outside the rule of 5 parameters. These compounds typically have higher molecular weights and a higher number of hydrogen-bond acceptors than the rule dictates, but particular attention must also be paid to hydrogen-bond donors, topological polar surface area and cLogP61, as well as lipophilic efficiency62–64 and other physicochemical parameters65, including those summarized in a recent review66. Short linkers can be used to modulate PROTAC molecular weights67 and properties65, but ‘linker-ology’ does more than link the POI and E3 warheads together and influence properties. The linker can contribute to the PROTAC solution conformation68 and degradation efficiency (DC50/Dmax)69, and influence the E3 ligase–POI interactions and presentation of the POI within the zone of ubiquitylation (Fig. 5).
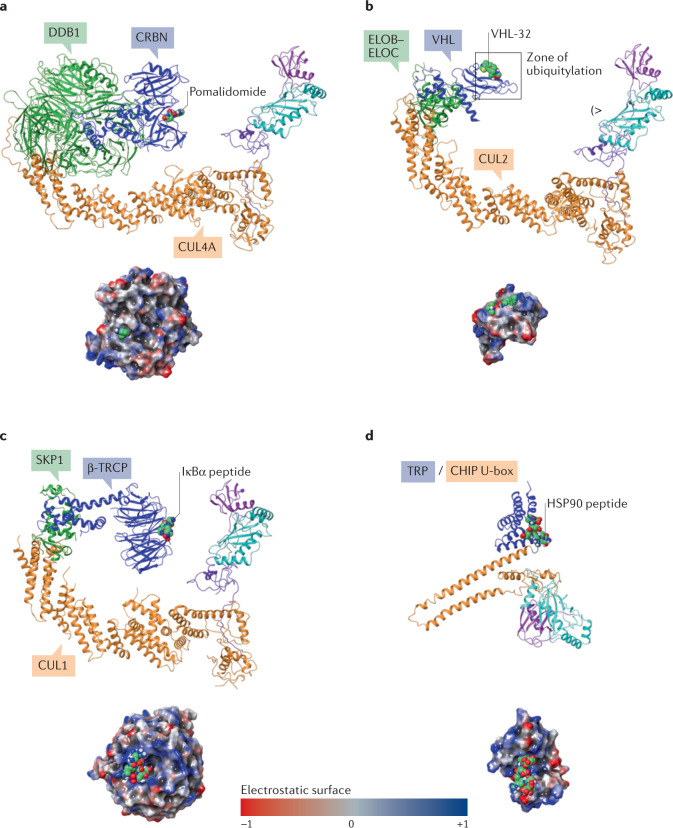
Fig. 5. Example Cullin–RING ligases and their substrate adaptors.
Examples of Cullin–RING ligases (CRLs) and a U-box E3 ligase modelled as complete E3–E2–ubiquitin (Ub) complexes: CRL4–cereblon (CRBN) (panel a); CRL2–von Hippel–Lindau (VHL) (panel b); CRL1–β-transducin repeat-containing E3 ubiquitin–protein ligase (β-TRCP) (panel c); CHIP (STIP1 homology and U-box-containing protein 1) (panel d). The assemblies demonstrate the different size, shape and electrostatic potential surface of each ligase, all potentially enabling degradation of a wide variety of proteins of interest when co-opted as ligases for degradation (Cullin/RING-box protein 1 (RBX1) subunits are coloured orange/purple, respectively; adaptor proteins are green; substrate receptors are blue; ligands are sea green). The surface-rendered substrate receptors in the lower images show the electrostatic potential coloured surface of the binding protein/domain of the substrate as viewed from the E2 (indicated by (>). The U-box CHIP E3 ligase is a single protein and is coloured by domain in panel d (U-box: orange; TRP, tetratricopeptide repeat: blue). The zone of ubiquitylation is defined by the reach of E2–Ub and is shown for CRL2–VHL in panel b. The high-probability ubiquitylation sites on the protein of interest are lysine residues that can be positioned within the zone and within ~4 Å of the reactive thioester between E2 and ubiquitin. The crystal structure protein databank (PDB) codes used to generate the models are: CRBN (2HYE223, 3UGB224, 6BN7 (ref.130)), VHL (6R7F225, 3UGB, 5T35 (ref.129)), β-TRCP (6TTU106, 3UGB) and CHIP (2C2L226, 6S53 (ref.227)). ELOB/ELOC, elongin B–elongin C.
However, short linkers limit the number of ways the E3 ligase and POI come together and, ultimately, how the POI fits into the full E3–E2–ubiquitin complex. To take full advantage of favourable PROTAC-induced E3–POI interactions a wider set of PROTACable E3 ligases will be needed, as discussed in the next section.
Overall, understanding the PROTACability of a target is key, especially as emerging, and already validated targets, are considered for classical small-molecule inhibition or targeted protein degradation as a therapeutic modality. To assess the PROTACable genome, which can be defined as the share of druggable targets that could be targeted by the PROTAC modality, Schneider et al.70 have provided the first systematic assessment of drug targets according to their ability to be degraded by PROTAC molecules. They based their analysis on several aspects of protein degradation mentioned here, such as availability of a small-molecule binder against the target, information on the endogenous ubiquitylation status of the target and target protein half-life. Their perspective offers the first comprehensive assessment of future potential targets with an eye towards protein degradation and can serve as a valuable resource to researchers in the protein degradation field.
Expanding the ligase landscape
Recently, it was disclosed that both ARV-110 and ARV-471 use CRBN as the E3 ligase that is recruited to their respective targets, AR and ER, to catalyse their ubiquitylation and proteasomal degradation41,71. Taken together with established CRBN modulators, such as the IMiD class that targets IKZF1/IKZF3 (refs3,4,72–74) and a new generation of CRBN modulators that target GSPT1 (refs1,75) (reviewed elsewhere13,76), CRBN appears to be emerging as a preferred E3 ligase for the first wave of TPD therapeutics in clinical trials. A notable exception is a BCL-xL degrader known as DT2216 developed by Dialectic Therapeutics that uses VHL as the recruiting ligase77,78, which is in phase I trials (Table 1).
However, spurred by the therapeutic potential of TPD, the past few years have seen the beginning of a renaissance in the study of the UPS and E3 ligases, in both academia and the pharmaceutical industry, that may well see CRBN and VHL supplanted as the initial ‘workhorses’ of TPD. With more than 600 human ubiquitin E3 ligases79,80 to potentially explore, the question becomes not ‘if’ new E3 ligase-based TPD therapeutics will reach patients, but ‘when’ — and for what diseases?
As noted earlier, degrader discovery has converged upon two main paths for discovering molecules that result in specific target degradation (Fig. 2; Box 1). The first stems from an opportunistic approach, in which a particular compound is shown to cause degradation of a target, leading to retrospective discovery of the E3 ligase responsible for its degradation. Examples here include the approved IMiD class, which relies on CRBN to degrade IKZF1/IKZF3 (refs4,5) via a molecular glue mechanism; sulfonamides previously in clinical trials81,82 that have since been shown to use DCAF15 to degrade RBM39 (refs25,27,29), also via a molecular glue mechanism elucidated by elegant structural biology in recent work by multiple groups26,28,83; and, most recently, a unique class of cyclin-dependent kinase 12 (CDK12) inhibitors, exemplified by CR8, which, unexpectedly, has been shown to directly co-opt DDB1 as an E3 ligase for the degradation of cyclin K (CCNK)84–86. In fact, not only CR8, but four separate classes of compounds, each discovered by a different screening method, were found to have molecular glue-like properties in potentiating the interaction of DDB1 and CDK12, resulting in CCNK degradation84–87. These findings suggest that many more compounds, including those approved or already in the clinic, could have a hitherto overlooked degrader contribution to their mechanism of action.
All these molecular glue compounds had been shown to be efficacious without a priori knowledge that they were, in fact, protein degraders. Their intricate mechanisms of action — in which the ligase and target become glued together — have been reviewed extensively42,88,89, most recently by Chamberlain and colleagues13.
The other path to degrader discovery is rationally driven by the selected therapeutic target, affording the ability to tailor the ligase to the POI by using an inhibitor of choice against the POI and a ligase-recruiting molecule that are connected by a linker. This approach to PROTAC discovery is exemplified by VHL-linked, MDM2-linked and inhibitor of apoptosis (IAP)-linked degraders in preclinical discovery settings34,90,91, and by ARV-110 and ARV-471 and other clinical compounds that recruit CRBN. The modular nature of such PROTACs could allow ‘ligase-hopping’ — the ability to swap ligase ‘handles’ that offer the most potent degradation and the best physicochemical properties — and can potentially enable the employment of E3 ligases with unique characteristics that could offer advantages against a particular target or in a given therapeutic setting.
This wave of rational TPD therapeutics will probably continue to use E3 ligases that have been evaluated and targeted with degraders over the past decade — namely, VHL, CRBN and potentially IAPs. The reach afforded by just these few ligases, coupled with clinically validated inhibitors that could be converted into degrader warheads, already appears large92.
Accordingly, it has become routine to generate and characterize a new degrader in cell culture experiments and animal models of disease, using either VHL or CRBN as a recruiting ligase93. The modular nature of the modality and its ability to achieve near-immediate degradation of a wide range of target proteins allows rapid evaluation of such basic degraders and characterization of their functions in cells through acute protein loss. Although use of small-molecule PROTACs in an endogenous setting is limited to protein targets that have ligands available, Buckley et al.94 have developed a tagging system based on HaloTag fusions that can be targeted by HaloPROTACs, and Nabet et al.53,95 have developed a heterologous tagging platform, dTAG, that allows validation of unliganded or yet-to-be-liganded proteins as degradation targets and is enabled for both VHL and CRBN. HaloPROTACs, dTAGs and other degradation proof-of-concept strategies, such as the AID system, have been reviewed extensively elsewhere96,97 and now serve as crucial target validation tools when approaching a new therapeutic POI.
The need for new E3 ligases
Given how well PROTAC degraders that use VHL, CRBN and other E3 ligases with known ligands work, and given the modular nature of the PROTAC concept, one might well ask whether there is truly a need to find and use new E3 ligases for TPD. We would argue that the opportunity is ripe to tap into unexplored E3 ligases for protein degradation. Although the generation of VHL- and CRBN-based PROTAC molecules as research tools has become routine, their preclinical and clinical development can still pose challenges. Particularly in oncology, resistance mechanisms to therapeutic agents can arise rapidly; this creates concerns that tumour cells could mutate to evade degraders that rely on non-essential ligases, such as CRBN and VHL, whose genomic loss or deletion results in no discernible effect on cellular viability or phenotype as defined by CERES scores in DepMap98, for example. In fact, preclinical studies of degraders that use CRBN or VHL to target multiple protein classes have detected emerging resistance that occurs via mutation and/or downregulation of components of the ubiquitin ligase machinery99–101. Similarly, in the clinical setting in multiple myeloma, resistance to IMiDs such as pomalidomide and lenalidomide has been shown to include genomic alterations in both the target protein (IKZF1/IKZF3) and components of the CRBN ligase machinery102,103. These and other challenges leave the door open to opportunities for identifying and developing alternative E3 ligases.
Recognizing the need for additional E3 ligases naturally leads to the question: what about an E3 ligase must be understood in order to co-opt it effectively for TPD? In the most basic sense, ubiquitin E3 ligases serve as scaffolds between a target protein and an activated ubiquitin E2 conjugating enzyme charged with a ubiquitin molecule (E2–Ub), by binding both molecules, then catalysing the transfer of ubiquitin from the active site cysteine of the E2 onto a lysine side chain of a target protein104–107. Ubiquitin ligases achieve this catalysis via multiple mechanisms; for this reason, the more than 600 E3 ligases in the human genome fall into different classes, depending on which ubiquitin ligation strategy they use (reviewed in refs80,108–110). As a handy online tool, the Structural Genomics Consortium (SGC) has created UbiHub, a database of E3 ligases and other protein classes in the UPS111.
To recap briefly: the largest class of ubiquitin ligases, RING ligases, bind to the target protein and E2–Ub simultaneously and transfer ubiquitin directly onto a lysine residue of the target protein. The second class, HECT ligases, bind the target then use an intermediate strategy of transferring ubiquitin onto themselves before transferring ubiquitin to the target112,113. RBR ligases use a hybrid between the RING and HECT ligation strategies114,115. To add to the complexity of ubiquitin ligation to substrates, RING and RBR ligases have recently been shown to work in unison to mediate substrate ubiquitylation in a site-specific manner116,117. Furthermore, RING ligases are classified as Cullin–RING ligases (CRLs) and non-CRL ligases. CRLs are multi-protein complexes in which E2–Ub binding is performed by the RING domain proteins, RING-box protein 1 (RBX1) and RBX2, and target binding is performed by other subunits in the complex, held together by Cullin scaffold proteins (see Fig. 5). In such CRLs, target binding happens via substrate receptor proteins that can pair with different CRLs, as reviewed extensively elsewhere110. CRBN and VHL, for example, are substrate receptor proteins that pair with CRL4 and CRL2, respectively.
The search for new E3 ligases
The diversity and complexity of E3 ligases raises two key questions for TPD: where to start looking for new ligases, and what characteristics would make them ideal for novel PROTAC development? In our opinion, there are several exciting avenues to consider when seeking and developing a novel ligase for PROTAC development.
One practical and valuable avenue is to pursue widely applicable, ubiquitously present ligases, similar to CRBN and VHL, that could be paired with any target of choice and applied across many different therapeutic indications without limitations. Currently, all PROTACs to date use just a handful of these non-specialized ligases, collated in an online database: PROTAC-DB92.
Interestingly, however, the pairing of VHL versus CRBN for a given target can result in different degradation efficiencies49, as was first observed when targeting the fusion oncogene BCR–ABL by PROTACs118. It is therefore possible that one ligase in particular may be better than others for degrading a certain target. A recent good example, and cautionary tale, of focusing on a single ligase when targeting endogenous proteins is the development of KRAS PROTAC degraders targeting the KRASG12C variant, enabled by the revolutionary work in developing covalent KRAS(G12C) inhibitors119–121, including Amgen’s recently approved sotorasib and others that are currently in phase II trials122,123. Zeng et al.124 initially developed a covalent PROTAC (XY-4-88) based on ARS-1620, a KRAS(G12C)-binding covalent warhead, and thalidomide as the CRBN-recruiting ligand124. Although XY-4-88 was able to degrade a GFP–KRAS(G12C) fusion protein, albeit in a non-catalytic manner because it covalently bound the target, it could not degrade untagged, endogenous KRAS(G12C). Subsequent studies revealed that XY-4-88 induced the CRBN–KRAS(G12C) complex, but not its poly-ubiquitylation. By contrast, Bond et al.55 developed a covalent VHL–KRAS(G12C) PROTAC (LC-2), based on the MRTX849 KRAS(G12C)-targeting warhead and a VHL ligand. LC-2 was able to engage endogenous KRAS(G12C), induce its poly-ubiquitylation and degrade it via the 20S proteasome. The different outcomes of these two studies could arise from the different KRAS warheads, the linker between the POI and E3 ligase warheads, and/or the choice of E3 ligase. KRAS is a small protein (189 amino acids) and, when farnesylated, it is localized to the plasma membrane. Its proximity to the membrane and the large size of the Cullin 4A (CUL4A)–DDB1–CRBN–E2–Ub complex relative to the smaller CUL2–elongin B (ELOB)–ELOC–VHL–E2–Ub complex (Fig. 5) could be a contributing factor. Collectively, these findings demonstrate that KRAS(G12C) is specifically targetable via the PROTAC mechanism, but that targetability depends on the choice of E3 ligase; the findings could be extended to non-covalent PROTACS targeting other KRAS variants (for example, KRAS(G12D) or KRAS(G12D/G12V)) using non-covalent warheads and thus represent an alternative therapeutic modality for targeting KRAS-driven cancers in the future.
Similarly, building on earlier work on the degradability of kinases49,125, a recent massive kinase degradation study using various kinase inhibitors as warheads has revealed the ‘degradable kinome’ and a diversity of kinase-degrading profiles among E3 ligases — highlighting the importance that ligase choice can make in targeting kinases for degradation126. The differences in degradation profiles conferred by different ligases can be driven by several factors. One is shape complementarity between the ligase and the target. The second is the ability of the ligase to form degradation-competent ternary complexes between the ligase and POI. Ternary complex formation is a necessary step in the action of a PROTAC molecule, but the degree of ternary complex formation may differ from substrate to substrate127; that is, a ternary complex that forms too efficiently and remains tightly bound will not lead to efficient degradation. Cooperativity of the ternary complex, which is a measure of ternary complex formation in a heterobifunctional system, may be positive or negative between a ligase and a target enabled by a PROTAC molecule128 and has been shown to lead to degradation in both cases. Cooperativity plays a role in the efficiency of the VHL–BRD4 degrader MZ1 (ref.129), but is dispensable for CRBN–BRD4 degraders130. A third factor is differential subcellular localization of ligase and target; for example, a nuclear E3 ligase, DCAF16, has been shown to restrict target degradation to the nucleus131. A fourth factor is cell-type-specific expression profiles of ligase and target, which we discuss in greater detail in the next section.
Accordingly, besides the aforementioned ligases (CRBN, VHL, MDM2, IAPs, DCAF15 and DCAF16), a few additional ‘generic’ E3 ligases have been shown to be exploitable by PROTACs, using small-molecule covalent tools that specifically recruit the ligases to degrade the prototypical TPD substrate, BRD4; these ligases include ring finger protein 4 (RNF4)132, RNF114 (refs133–135), KEAP1 (refs136,137) and most recently, fem-1 homologue B (FEM1B)138. How far initial chemical matter will be developed for these newly established degrader ligases, and how widely they can and will be adopted as in vivo-compatible ligases with a broad target reach, will be the subject of intense experimentation over the next few years.
Beyond the ligases already vetted for TPD development, there are several other ubiquitin ligases, enabled by structural and/or validated substrate information, that present a path for the development of small molecules that can ultimately be converted into PROTACs, yet await development as potential PROTACable ligases. Most of these ‘low-hanging fruit’ ligases with structural data have been summarized by two excellent reviews139,140. However, it must also be mentioned that the definition of low-hanging fruit with respect to structural enablement of ligases has recently been redefined with the ground-breaking publication of artificial intelligence (AI)-driven tertiary structure prediction models from Google/DeepMind141 and RoseTTAFold142. Alphafold from DeepMind has made available all their high-quality predicted models across the proteome (https://alphafold.ebi.ac.uk/) and now serves as a major stepping stone in potentially enabling computer-aided drug discovery for many hitherto unreachable targets and ligases.
Tissue- and cell-specific E3 ligases
The discovery and development of novel E3 ligases for TPD clearly involve practical considerations, such as structural enablement, druggability and mechanistic understanding of the ligase. For example, is a validated substrate known, can that ligase–substrate relationship be exploited in development of a biochemical or cell-based binding assay to enable a screening paradigm, and is the ligase always ‘on’ or does it need to be activated by a physiological stimulus? Several ligases exist in an ‘off’ state and are auto-inhibited in the absence of an activating post-translational modification or presence of a binding partner. Co-opting such auto-inhibited ligases may pose additional challenges when considering them for degrader discovery.
However, ligases have other key characteristics, such as tissue and cell-type specificity, tumour enrichment and tumour essentiality (Fig. 6), that can offer opportunities for ‘niche’ ligase degrader development. The intersection between the practical and aspirational attributes of these niche ligases with respect to novel degrader development is often minimal at best, thus offering high risk/high reward scenarios for PROTAC discovery and development of degraders that recruit such specialized ligases. Ultimately, the wider UPS field will benefit from the intense industry interest in characterizing novel ligases for PROTACs, and from the tool molecules that will be developed.
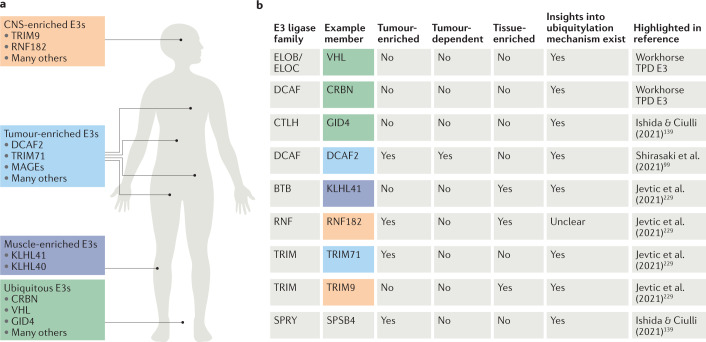
Fig. 6. Specialized E3 ligases for potential PROTAC applications.
a | Schematic representation of the human body, highlighting E3 ligases with increased tissue specificities that may be harnessed by targeted protein degradation (TPD) modalities to enable tissue- and cell-type-specific targeting of disease-causing proteins. b | Table showing representative E3 ligase examples from different mechanistic classes of E3 ligase, highlighting their various (but not exhaustive) attributes to be considered for proteolysis-targeting chimera (PROTAC) development, including tumour and tissue enrichment, tumour dependence (CERES/DepMap profile) and mechanistic understanding of ubiquitin ligation onto their substrates. The E3 examples were chosen for each E3 class from multiple E3-centric review articles80,99,139,228,229 to highlight various characteristics and the expansive choice in ligase selection for novel PROTACs. One article is referenced for each E3, but there is considerable overlap between the content of the cited references. There is no single attribute that makes or breaks an E3 ligase for PROTAC development; all attributes should be considered in totality, including the target that is being considered for degradation. CNS, central nervous system; CRBN, cereblon; DCAF2, DDB1- and CUL4-associated factor 2; ELOB, elongin B; GID4, glucose-induced degradation protein 4 homologue; KLHL40, kelch-like family member 40; MAGEs, melanoma antigen genes; RNF182, RING finger protein 182; TRIM9, tripartite motif-containing protein 9; VHL, von Hippel–Lindau.
There is high interest in academia and industry in identifying E3 ligases with unique expression profiles to enable tissue- and cell-type-specific target degradation — a concept originally proposed in the landmark PROTAC paper by Sakamoto et al.9 20 years ago. The TPD field has now matured to the point that strides are being taken to realize such specificity goals and enable precision TPD (pTPD). In our opinion, which is echoed through the TPD community, this is one of the holy grails in the field. As such, several recent reviews have analysed and categorized publicly available datasets — such as GTEx, The Cancer Genome Atlas (TCGA), ProteinAtlas and others — for ligase expression in healthy and diseased states, to highlight ligases with clear and distinct expression profiles80,99,143,144. However, as many E3 ligases are multisubunit complexes, analysing expression levels of a single subunit is only part of the whole picture.
Several such E3 ligases have been identified so far. Among the clearest examples of ligases with tissue-specific expression are kelch-like family member 40 (KLHL40) and KLHL41 in skeletal muscle145, which have a documented role in the biology of nemaline myopathy146,147. Additionally, several central nervous system (CNS)-specific E3 ligases have emerged, such as RNF182 (ref.148) and tripartite motif-containing protein 9 (TRIM9)149; these are of high interest in tackling targets in neuronal diseases, as discussed below, where CNS-specific therapeutic targeting is desirable to avoid systemic off-target and toxicity effects. There are also E3 ligases that show a certain level of enrichment — although not clear specificity — in a particular tissue or cell type, such as a non-sugar-binding member of glycan-specific substrate adaptors, F-box protein 44 (FBXO44)150 that is enriched in some tissues but not specific to any151. Despite the lack of clear tissue- or cell-specific expression, FBXO44 and other tissue- or cell-enriched ligases nonetheless have exciting potential for TPD, because the affinity of their ligands could be tuned to engineer in the specificity towards the tissue of choice, even though some basal level of ligase expression exists ubiquitously.
Interestingly, some ligases exhibit ‘reverse specificity’ — low expression in some tissue or cell types — that may also be advantageous for protein degraders. A remarkable example is the low expression of VHL in platelets: degradation of BCL-xL by the VHL-recruiting PROTAC DT2216 (ref.152) was spared in platelets, thus resulting in reduced platelet-driven toxicity and an improved therapeutic index compared with a BCL-xL inhibitor78,153. DT2216 is in phase I trials30 and represents an exciting step towards pTPD.
E3 ligase enrichment in diseased versus healthy tissues and cells
Another new frontier to explore for pTPD is specific targeting of a PROTAC molecule by tumour cells over adjacent, non-cancerous tissue or uptake by a given type of tumour-initiating cell, which may be achieved by targeting tumour-specific or tumour-enriched E3 ligases. Often, but not always, tumour enrichment of an E3 ligase coincides with the dependence of the tumour on expression of that ligase. These correlations can be drawn and have been shown by analysing CERES scores in DepMap98 of E3 ligases across multiple tumour cell lines. This has allowed the identification of E3 ligases and other UPS genes — some better-characterized than others, such as cell division cycle 20 (CDC20), cytosolic iron–sulfur assembly component 1 (CIAO1) and WD repeat-containing protein 82 (WDR82) — that exhibit high tumour essentiality across many different cancer cell types99. The advantage of ligases with these profiles is that tumour cells would be less able to develop ligase-based resistance to PROTACs, which has been shown to occur for some CRBN- and VHL-recruiting PROTACs, as discussed previously.
One caveat for this group of ligases is that tumour enrichment provides only a potential window of opportunity for PROTACs, which may not translate to a greatly expanded therapeutic index for pleiotropically toxic inhibitors that are converted into tumour-specific PROTACs. A second caveat is that highly tumour-enriched and tumour-dependent E3 ligases and ligase adaptors tend to be associated with the cell cycle — for example, CDC20 or S-phase kinase-associated protein 2 (SKP2) and others — and are likely to have similar profiles in tumours and rapidly dividing non-cancerous cells in the human body, such as in the bone marrow. PROTACs based on these ligases might therefore exhibit toxicities associated with classical chemotherapeutic agents, but this remains to be tested.
Finally, cancer-testis antigens (CTAs) also comprise ubiquitin ligases that have restricted expression in the normal testis but are highly overexpressed across multiple cancer types154,155. An example of such a family of ligases are the MAGE (melanoma antigen genes) family of ubiquitin E3 ligases, often referred to as the MAGE-RING ligases (MRLs), which comprise proteins that serve as substrate-recruitment modules for other RING E3 ligases156,157, adding to and/or altering the substrate specificity of their co-E3 ligases (reviewed elsewhere155,158). The exact mechanism of ubiquitylation for MRLs is not as well understood as the mechanism for CRLs, for which substrate recruitment by multiple adaptors and the molecular details of Ub transfer from E2–Ub to the target has been extensively characterized at the atomic level of an entire CRL1 complex106,117. However, MAGEs have recently received attention owing to their unique expression profiles. Although not all MAGE E3 ligases are tumour specific159 and it may not be possible to co-opt all of them for PROTACs, as some do not degrade their targets, testis-specific, tumour-enriched E3 ligases are nevertheless a space to watch for increased understanding of MRL function, biology and more.
Identifying, characterizing and developing tumour-specific E3 ligases for PROTACs by identifying small molecules that can co-opt them for TPD is only one way to potentially achieve tumour specificity and greater therapeutic indexes with PROTACs compared with classic pleiotropically toxic agents. An alternative approach is antibody-mediated tumour specificity using antibody–drug conjugates (ADCs), whereby a degrader molecule is coupled to a tumour-specific antibody. This technology has recently emerged as an exciting avenue in delivering PROTAC molecules in a tumour-directed fashion. Dragovic et al.160–162 at Genentech have pushed this modality forward and successfully demonstrated tumour-specific targeting in two different cancer types with ADCs involving VHL–BRD4 PROTACs. Although these antibody–PROTAC conjugates obviously lose the clear advantages of oral bioavailability and tunable dosing schedule of small-molecule PROTACs, the ADC approach can be valuable for potent and robust degraders that have poor physicochemical properties for parenteral administration.
Therapeutic areas beyond oncology
Degrader drugs currently on the market (the IMiD class) and the overwhelming majority of newly developed PROTACs and molecular glue compounds currently in clinical trials (Tables 1,2) target various types of cancer30,163. However, given its potential to degrade any target of choice, the reach of TPD is extending further than oncology.
Inflammation, immunity and immuno-oncology
Already in this first wave of clinical degraders, two compounds entering clinical trials could test the PROTAC modality in non-oncology indications. A BTK degrader (NX-5948) and an IRAK4 degrader (KT-474) in phase I clinical trials (Table 1), from Nurix Therapeutics and Kymera Therapeutics, respectively, could treat patients with various immuno-inflammatory diseases, such as rheumatoid arthritis.
BTK is a well-established target in inflammation, as well as in oncology, with approved drugs in the clinic164,165 and has been the subject of intense PROTAC molecule experimentation across academia and industry166–168, particularly because of emergence of the C481S resistance mutation in BTK that renders first-generation BTK inhibitors less effective169,170. Herein lies one of the great advantages of PROTACs: owing to their event-driven (versus occupancy-driven) pharmacology, PROTACs may overcome insensitivity to such resistance mutations through catalytic degradation of the target. A BTK PROTAC from Nurix Therapeutics that targets haematological malignancies (NX-2127) is currently in a phase I trial (Table 1).
By contrast, there are currently no approved IRAK4-targeting therapies, but the role of IRAK4 in inflammatory conditions, as well as in B cell lymphomas, is well established, and several inhibitors are currently in clinical trials in autoimmune indications165,171. Here, the potential scaffolding role of IRAK4 is especially intriguing as a target, as inhibitors of its kinase function may not fully address the role of IRAK4 in these conditions. Another advantage of PROTAC degraders is their ability to degrade the target completely, instead of only inhibiting its enzymatic function, which would not affect any scaffolding role that a target may have. Particularly for IRAK4, its scaffolding role in Toll-like receptor (TLR) signalling around the myddosome is established165,172, and may only be addressable by PROTAC degraders, many of which have been described to date173–175. Of these, KT-474 from Kymera Therapeutics is the first to enter clinical trials (Table 1).
Following in the footsteps of immunotherapy, small-molecule-mediated activation of the immune response against tumours is a rich area of drug development176,177. PROTACs have the potential to be first-in-class medicines in immuno-oncology as small-molecule drugs that target immune cell activation to phenocopy PD1/PDL1-directed agents178,179. Most recently, PROTACs that target mitogen-activated protein kinase kinase kinase kinase 1 (MAP4K1; also known as HPK1) have been described with promising preclinical activity180, although HPK1 inhibitors were shown to be equally effective. Here, the use of a PROTAC over an inhibitor may not be necessary or obvious, but given that PROTACs can comparatively achieve greater specificity with respect to off-target effects of some kinase inhibitors126,181, a PROTAC approach may result in a cleaner inhibitory phenotype and better tolerability as a therapeutic agent.
Neurology and neurodegeneration
A key PROTAC property is the ability to degrade proteins that are not classically targetable by small-molecule inhibitors because they lack active sites. This attribute makes targets for several neurodegenerative diseases involving toxic build-up of proteins, such as tau, α-synuclein, mutant huntingtin (mHTT), TAR RNA-binding protein (TARDBP; also known as TDP43) and FUS RNA-binding protein (FUS), potentially addressable with the PROTAC modality.
Tauopathies are good examples of diseases in which small-molecule inhibitors have disappointed but PROTACs could succeed. Tauopathies include neurological disorders, such as Alzheimer disease, frontotemporal dementia (FTD) and progressive supranuclear palsy (PSP)182, in which pathology is heavily associated with toxic accumulation of aberrant tau species, leading tau to aggregate into paired helical filaments and ultimately into neurofibrillary tangles that result in neuronal cell death183. Therefore, tau downregulation — at any node of the dynamics of tau’s transitions from monomer to oligomer to aggregate184 — is a desired therapeutic strategy. In fact, in preclinical models, decreasing tau protein levels via ASO strategies185,186 and recently by engineered zinc-finger transcription factors187 was shown to be durable and to reverse tau pathology. Importantly, induced turnover of tau by the UPS, and the degradability of tau species, has been demonstrated by a tau–kelch-like ECH-associated protein 1 (KEAP1) chimeric peptide PROTAC molecule188, as well as by a VHL–tau PROTAC189. The most advantageous point at which to interfere with the dynamics of tau aggregation and co-opt the protein for degradation, and whether degradation of various tau species is shared by the UPS and by lysosomal pathways, are areas of active investigation184.
Developing small-molecule tau-targeting PROTACs and other degraders that would combine the phenotype of knocking down tau — as ASOs do — with systemic administration and blood–brain barrier penetration, is a particularly active avenue. Tau in its monomeric form is an intrinsically disordered protein without clearly defined small-molecule binding pockets, which has made development of small-molecule inhibitors challenging. However, small-molecule probes against tau oligomeric species, such as 18F-T807, have been developed and are currently in use as clinical tau PET tracers190. In fact, one strategy has utilized such PET tracers by turning them into tool PROTAC molecules that target tau by recruiting CRBN, and these were shown to cause degradation of tau species in cell-based models of tauopathy191.
Key questions remain as to what the pathologically relevant tau species may be, and whether it is more advantageous to degrade tau generally, irrespective of its oligomeric state. Given the clear unmet medical needs represented by tauopathies and the suitability of the PROTAC approach for tackling neurodegenerative diseases, this area will see key milestones tackled over the next decade, with the potential to deliver breakthrough medicines.
Other targets in neurology where a PROTAC modality may offer clear advantages over current therapeutic agents include α-synuclein, a protein that can accumulate in the neurons of patients with Parkinson disease192. α-Synuclein and tau are both intrinsically disordered proteins with distinct conformations that probably drive their respective pathologies and for which induced proteolytic degradation could be disease-modifying184. A peptide-based α-synuclein degrader (see below) has been reported and was shown to protect neurons from α-synuclein overexpression-induced toxicity193.
In Huntington disease, poly-glutamine-expanded mHTT aggregates and accumulates194, presenting another difficult drug target that may be addressable by a PROTAC, as clearance of mHTT via the UPS pathway is thought to be beneficial for disease progression195. In fact, two small-molecule TPD strategies have been reported to lower mHTT levels: a small-molecule glue approach inducing the proximity between mHTT and microtubule-associated protein 1 light chain 3α (MAP1LC3α; also known as LC3) to target it to the autophagy pathway for degradation196; and small-molecule PROTACs that recruit mHTT aggregates to E3 ligases from the IAP family, causing the proteasomal degradation of the mutant protein197. With the recent discontinuation of the phase III trial of tominersen, an HTT-lowering ASO that it was hoped could become the first disease-modifying therapy for Huntington disease, the PROTAC modality could provide a useful alternative route to pursue this key target.
Antiviral PROTACs
Interestingly, PROTACs could emerge as potential antiviral agents as well, essentially bringing the field full circle from the viral hijacking of the UPS that inspired the TPD concept (Box 2), to turning that hijacking strategy upon viruses themselves and targeting their proteins for degradation. The feasibility of antiviral PROTACs was first established with the degradation of the hepatitis C virus (HCV) NS3/4A protease in cell-based assays by an HCV inhibitor linked to CRBN198, but has been brought to the forefront with the emergence of SARS-CoV-2, the virus responsible for the COVID-19 pandemic. A large PPI study in SARS-CoV-2-infected cells revealed several potential druggable pathways and proteins that may be exploited by repurposing inhibitors199, and that could potentially be addressed by SARS-CoV-2-targeting PROTAC molecules. One provocative paper raised the possibility of targeting the viral envelope protein200, but SARS-CoV-2 catalytic virulence factors, such as its two proteases, Mpro and PLpro201, and the RNA-dependent RNA polymerase (Rdrp), which is the target of remdesivir202, are potentially targetable by a PROTAC. Importantly, Pfizer initiated a phase I trial of its orally bioavailable SARS-CoV-2 Mpro inhibitor, PF-07321332 (NCT04756531) in healthy individuals in February 2021, followed by a phase II/III study, in which the first patient was dosed in September 2021 (NCT04960202). Pfizer has also developed an Mpro inhibitor prodrug, PF-07304814 (ref.203), which is given intravenously and which entered clinical trials in hospitalized patients with COVID-19 in 2020 (NCT04535167). These antiviral inhibitors could provide starting points for the development of antiviral PROTACs that target SARS-CoV-2.
Additionally, Nurix Therapeutics has announced that it is in the discovery phase for three protein degradation chimeric targeting molecules (CTMs) against SARS-CoV-2, dubbed COVID-CTMs.
Alternative PROTAC modalities
Although PROTACs are poised to become an important modality for certain target classes, there are POIs that do not have the small-molecule binding sites needed for this approach. Even so, these targets are still susceptible to degradation by alternative PROTAC approaches, including bioPROTACs and hybrid PROTACs. Additionally, PROTACs have inspired the exploration of other classes of heterobifunctional molecules that harness non-UPS machinery to degrade POIs (Box 3), each of which offer features that sets it apart from the other TPD modalities and from small-molecule inhibitors (Table 3).
Table 3.
Characteristics of TPD approaches as therapeutic modalities
| Therapeutic modality | PROTACs/molecular glues | Small-molecule inhibitors | Gene-based strategies | bioPROTAC | oligoTAC | LYTAC | ATTEC | AUTAC | abTAC |
|---|---|---|---|---|---|---|---|---|---|
| Target scaffolding functions | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | |
| Potential to treat undruggable proteins | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | |
| Iterative mechanism of action | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ||
| Broad tissue penetration | ✓ | ✓ | ✓ | ✓ | |||||
| Orally bioavailable | ✓ | ✓ | |||||||
| Ease of manufacturing | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ||
| Preclinical validation, proof-of-concept established | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ |
| Clinical validation | ✓ (Phase II for PROTACs) | ✓ | ✓ |
PROTAC, proteolysis-targeting chimera; TPD, targeted protein degradation.
For this section, we broadly define three PROTAC categories as follows: PROTACs are composed of traditional small-molecule ligands; bioPROTACs are composed of peptide ligands; and hybrid PROTACs contain both a peptide and a traditional small-molecule warhead. This categorization redefines the original PROTAC reported in 2001 (ref.9) as a hybrid-PROTAC, as it is composed of a small-molecule warhead that interacts with METAP2 linked to a phosphopeptide that interacts with the E3 ligase β-TRCP. Each of these three forms offers powerful tool molecules for driving drug discovery, and each form has unique challenges and considerations for development as therapeutic agents. As ARV-110 and ARV-471 have demonstrated, PROTACs can be developed as oral therapeutics, but bioPROTACs and hybrid PROTACs are unlikely to be oral drugs owing to their peptide components. Indeed, bioPROTACs would most likely be delivered as mRNA or DNA constructs via a viral vector gene delivery system204–206 or in a nanoparticle207–209.
bioPROTACs and pepTACs
The bioPROTAC category can be further divided into three subgroups: bifunctional peptides (pepTACs), fusion proteins, and bispecific antibody mimics (of which several examples are provided by viruses (Box 2)). Of the three subgroups of bioPROTAC, pepTACs are the most closely related to PROTACs, in that they are composed of a peptide that binds to an E3 ligase and a peptide that binds to the POI. The two peptide warheads are typically connected with a peptide linker, and the N or C terminus typically has attached a cell-penetrating peptide (CPP). However, pepTACs composed of natural amino acids may dispense with the CPP and be delivered by transfecting cells with a DNA or mRNA transcript to synthesize the degrader in situ.
Zhou et al.210 described some of the earliest chimeric bioPROTACs, including a fusion consisting of full-length β-TRCP protein and an N-terminal peptide of the E7 protein of human papillomavirus type 16 (HPV-16) that degraded retinoblastoma protein (RB1) in a human osteosarcoma cell line. These studies demonstrated that the SCF E3 ligase could be redirected to degrade non-SCF targets.
Since then, other groups have reported fusion bioPROTACs that degraded the cancer targets human epidermal growth factor receptor 2 (HER2), MYC and KRAS and demonstrated therapeutically relevant effects in human cancer cell lines and mouse xenograft tumour models overexpressing these POIs16,17,211,212. The reported fusions involved replacing the substrate-binding domain of an E3 ligase, such as the Cbl proto-oncogene (CBL), U-box-type E3 ligase STIP1 homology and U-box-containing protein 1 (STUB1; also known as CHIP), VHL, E6AP and HIV-1 Vif, with a domain specific for the POI — or, in the case of MYC, with the MYC-associated factor X (MAX), a small protein that forms a heterodimer with MYC. Interestingly, Pan et al.212 reported a Vif-based bioPROTAC as their best KRAS degrader. However, in mice bearing xenograft pancreatic tumours, orthotopic injection of the Vif-based degrader was more efficacious than intraperitoneal injection, highlighting the need for better delivery methods to advance bioPROTACs from laboratory tools to therapeutics.
Additionally, multiple groups have described a range of pepTACs that harness SKP1/SCF F-box, VHL, KEAP1 and other E3 ligases to degrade tau or α-synuclein in primary neurons and other cell lines188,193,213, further supporting the potential of TPD to tackle Parkinson disease, Alzheimer disease and other tauopathies.
The anti-GFP bioPROTAC platform
Another approach for exploring the degradation efficiency and respective targets of chimeric E3 ligases from the CRL family is the anti-green fluorescent protein (GFP) bioPROTAC platform (anti-GFP bioPROTAC platform), developed by Partridge and colleagues214. In brief, the platform constructs bioPROTACs by replacing the natural substrate recognition domain of an adaptor protein with a GFP-binding domain, such as the anti-GFP nanobody vhhGFP4 (refs215,216); then, a doxycycline-inducible system is used to drive co-expression of the bioPROTAC and a reporter217 in cells constitutively expressing H2B–GFP218.
The platform has been used to generate GFP-degrading bioPROTACs based on various E3 ligases: the CUL1 ligases β-TRCP, F-box and WD repeat domain-containing 7 (FBW7) and SKP2; the CUL2 ligase VHL; the CUL3 ligase speckle type BTB/POZ protein (SPOP); the CUL5 ligases suppressor of cytokine signalling 2 (SOCS2) and ankyrin repeat and SOCS box-containing 1 (ASB1); and the U-box E3 ligase CHIP.
The platform also enables translation of GFP-degrading constructs into bioPROTACs that degrade a POI. For example, after demonstrating that their GFP-degrading bioPROTACs also degraded a GFP-tagged proliferating cell nuclear antigen (PCNA), Partridge and colleagues219 replaced the GFP-binding domain in one of those bioPROTACs with the 16-amino-acid Con1 peptide, which contains the conserved PIP sequence common to PCNA binding partners, to generate a Con1–SPOP bioPROTAC. This construct rapidly degraded PCNA and maintained low levels of PCNA for the 10 days of doxycycline dosing. Additionally, the platform has also been applied to the discovery of KRAS-targeting bioPROTACs.
Oligonucleotide-based PROTACs
Transcription factors and RNA-binding proteins (RBPs) are two classes of protein that are required for DNA repair, replication, transcription and many RNA-dependent processes. However, when mutated, transcription factors and RBPs give rise to multiple types of cancer, obesity and cardiovascular and neurological diseases. Despite much effort, these POIs have largely remained undrugged as many of them lack ligand-binding sites.
To address these POIs, RNA-PROTACs that target RBPs, and oligonucleotide-based PROTACs (O’PROTACs) and transcription factor-targeting chimeras (TRAFTACs) that target transcription factors have been developed.
RNA-PROTACs use an RNA consensus sequence as the RBP-binding warhead, linked to an E3 ligase-binding element220. Similarly, O’PROTACs221 and transcription factor PROTACs222 use a double-stranded DNA (dsDNA) consensus sequence as the transcription factor-binding warhead, linked to an E3 ligase-recruiting ligand. Such constructs have been shown to reduce levels of the RBP LIN28A and the transcription factors ETS transcription factor (ERG) and lymphoid enhancer-binding factor 1 (LEF1) in various human cancer cell lines.
TRAFTACs consist of a HaloPROTAC that bridges an E3 ligase and a dCas9–HT7 fusion protein, then binds a bifunctional dsDNA–CRISPR-RNA to form a transcription factor-recruiting complex. The TRAFTAC system has been used to degrade NF-κB and brachyury (T-box transcription factor T; TBXT) in human cell lines in a UPS-dependent manner that was specific to the dsDNA sequence and E3 ligase43.
The future of bioPROTACs
Although bioPROTACs, O’PROTACs and TRAFTACs have the potential to become therapeutic agents, delivery and dosing are key hurdles to overcome. Small-molecule PROTACs have the potential for oral delivery, and cellular levels can be controlled by dosing. Chimeric bioPROTACs can potentially be delivered as gene products and intracellular levels controlled by finely tuned promoters. Continued advances in nanoparticle drug delivery methods and viral vector gene delivery systems will play a major part in bringing these novel modalities to patients.
In the near term, bioPROTACs will be used as tool molecules to drive small-molecule PROTAC drug development. Like small-molecule PROTACs, bioPROTACs can be rapidly assembled to identify potent degraders if the warheads are known. For undrugged targets for which small-molecule ligands have not yet been discovered or productive small-molecule binding sites do not exist, bioPROTACs have an advantage: undrugged targets often form PPIs that are difficult to block with traditional drugs, but their protein-binding domains offer starting points for assembling bioPROTACs to initiate the drug discovery process. Therefore, bioPROTACs can be used for the following: to determine the biological benefit of complete inhibition of a protein via degradation versus partial inhibition with a small molecule; to compare the biological effect of post-expression degradation versus silencing expression with RNA interference; and to determine which face of the target protein is approachable by the E3 ligase and leads to productive lysine placement within the full E3 complex for poly-ubiquitylation.
Conclusion
Given the progress in the past two decades and the recent level of interest and investment from both academia and industry, it is clear that targeted protein degradation could become a key therapeutic modality. Witnessing its transition from intriguing idea to clinically proven concept, first via IMiDs and molecular glues and, most recently, via heterobifunctional PROTACs in phase I and phase II clinical trials, has been truly gratifying. We are excited about the discoveries in basic biology and chemistry that will further advance targeted protein degradation in the decades to come, and we believe this modality has the potential to offer patients new treatment options across diverse indications.
Acknowledgements
C.M.C. is funded by the NIH (R35CA197589) and supported by an American Cancer Research Professorship. The authors thank A. Cacace and I. Taylor for critical reading of the manuscript; M. J. Haas for providing editorial support; and the reviewers for their supportive comments during finalization of the manuscript. PROTAC is a registered trademark of Arvinas Operations, Inc.
Glossary
- Event-driven pharmacology
This refers to the mechanism of action of a small-molecule drug, whereby its function is transiently recruited to a target protein in a catalytic manner (for example, ubiquitylation by an E3 ligase recruited by a proteolysis-targeting chimera (PROTAC)), resulting in a pharmacological effect (degradation of the protein) that drives a phenotype. This contrasts with occupancy-driven pharmacology, whereby the function of a target protein is directly blocked by a small-molecule inhibitor.
- Therapeutic window
The dosage (a range of concentrations) of a drug that provides efficacious therapy and is safe (without toxic side effects).
- Scaffolding proteins
Proteins that may not have an enzymatic function on their own, but serve as protein–protein interaction hubs to recruit and orient signalling complexes.
- dTAG system
An allele-specific tagging system that uses a protein of interest (POI) fused to a FKBP12(F36V) construct that allows specific degradation of the fused POI via an FKBP12(F36V) binder linked to a cereblon (CRBN) or a von Hippel–Lindau (VHL) ligand.
- Bio-orthogonal chemistry
A chemical reaction that occurs inside a living organism without altering its biology.
- Rule of 5
A set of physicochemical property guidelines for small molecules that indicate the likelihood of a small molecule being orally bioavailable in humans. It is more of a rule of thumb than an absolute rule, and many approved drugs fall outside the rule of 5.
- Protein half-life
The time required for the amount or concentration of a protein to be reduced by 50% under physiological conditions. It is a measure of the propensity of a protein to be degraded by the ubiquitin–proteasome system (UPS). Proteins with short half-lives are rapidly degraded by the UPS (constantly being turned over), whereas proteins with long half-lives are more stable.
Competing interests
C.M.C. is a consultant and shareholder in Arvinas, Inc. and Halda Therapeutics, which support research in his lab. M.B. and D.R.L. are employees and shareholders of Arvinas, Inc, which is developing drug candidates in the targeted protein degradation space.
Footnotes
Peer review information
Nature Reviews Drug Discovery thanks John Harling, Darryl McConnell and the other, anonymous, reviewer for their contribution to the peer review of this work.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Miklós Békés, Email: miklos.bekes@arvinas.com.
David R. Langley, Email: david.langley@arvinas.com
Craig M. Crews, Email: craig.crews@yale.edu
References
- 1.Surka C, et al. CC-90009, a novel cereblon E3 ligase modulator, targets acute myeloid leukemia blasts and leukemia stem cells. Blood. 2021;137:661–677. doi: 10.1182/blood.2020008676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hansen JD, et al. Discovery of CRBN E3 ligase modulator CC-92480 for the treatment of relapsed and refractory multiple myeloma. J. Med. Chem. 2020;63:6648–6676. doi: 10.1021/acs.jmedchem.9b01928. [DOI] [PubMed] [Google Scholar]
- 3.Lu G, et al. The myeloma drug lenalidomide promotes the cereblon-dependent destruction of Ikaros proteins. Science. 2014;343:305–309. doi: 10.1126/science.1244917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kronke J, et al. Lenalidomide causes selective degradation of IKZF1 and IKZF3 in multiple myeloma cells. Science. 2014;343:301–305. doi: 10.1126/science.1244851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kronke J, et al. Lenalidomide induces ubiquitination and degradation of CK1alpha in del(5q) MDS. Nature. 2015;523:183–188. doi: 10.1038/nature14610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ito T, et al. Identification of a primary target of thalidomide teratogenicity. Science. 2010;327:1345–1350. doi: 10.1126/science.1177319. [DOI] [PubMed] [Google Scholar]
- 7.Fischer ES, et al. Structure of the DDB1-CRBN E3 ubiquitin ligase in complex with thalidomide. Nature. 2014;512:49–53. doi: 10.1038/nature13527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chamberlain PP, Hamann LG. Development of targeted protein degradation therapeutics. Nat. Chem. Biol. 2019;15:937–944. doi: 10.1038/s41589-019-0362-y. [DOI] [PubMed] [Google Scholar]
- 9.Sakamoto KM, et al. Protacs: chimeric molecules that target proteins to the Skp1-Cullin-F box complex for ubiquitination and degradation. Proc. Natl Acad. Sci. USA. 2001;98:8554–8559. doi: 10.1073/pnas.141230798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Verma R, Mohl D, Deshaies RJ. Harnessing the power of proteolysis for targeted protein inactivation. Mol. Cell. 2020;77:446–460. doi: 10.1016/j.molcel.2020.01.010. [DOI] [PubMed] [Google Scholar]
- 11.Nalawansha DA, Crews CM. PROTACs: an emerging therapeutic modality in precision medicine. Cell Chem. Biol. 2020;27:998–1014. doi: 10.1016/j.chembiol.2020.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hanzl A, Winter GE. Targeted protein degradation: current and future challenges. Curr. Opin. Chem. Biol. 2020;56:35–41. doi: 10.1016/j.cbpa.2019.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Faust TB, Donovan KA, Yue H, Chamberlain PP, Fischer ES. Small-molecule approaches to targeted protein degradation. Annu. Rev. Cancer Biol. 2021;5:181–201. [Google Scholar]
- 14.Ciechanover A, Orian A, Schwartz AL. Ubiquitin-mediated proteolysis: biological regulation via destruction. Bioessays. 2000;22:442–451. doi: 10.1002/(SICI)1521-1878(200005)22:5<442::AID-BIES6>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 15.Burslem GM, Crews CM. Proteolysis-targeting chimeras as therapeutics and tools for biological discovery. Cell. 2020;181:102–114. doi: 10.1016/j.cell.2019.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ma Y, et al. Targeted degradation of KRAS by an engineered ubiquitin ligase suppresses pancreatic cancer cell growth in vitro and in vivo. Mol. Cancer Ther. 2013;12:286–294. doi: 10.1158/1535-7163.MCT-12-0650. [DOI] [PubMed] [Google Scholar]
- 17.Hatakeyama S, Watanabe M, Fujii Y, Nakayama KI. Targeted destruction of c-Myc by an engineered ubiquitin ligase suppresses cell transformation and tumor formation. Cancer Res. 2005;65:7874–7879. doi: 10.1158/0008-5472.CAN-05-1581. [DOI] [PubMed] [Google Scholar]
- 18.Hon WC, et al. Structural basis for the recognition of hydroxyproline in HIF-1 alpha by pVHL. Nature. 2002;417:975–978. doi: 10.1038/nature00767. [DOI] [PubMed] [Google Scholar]
- 19.Min JH, et al. Structure of an HIF-1alpha-pVHL complex: hydroxyproline recognition in signaling. Science. 2002;296:1886–1889. doi: 10.1126/science.1073440. [DOI] [PubMed] [Google Scholar]
- 20.Buckley DL, et al. Small-molecule inhibitors of the interaction between the E3 ligase VHL and HIF1alpha. Angew. Chem. Int. Ed. Engl. 2012;51:11463–11467. doi: 10.1002/anie.201206231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Buckley DL, et al. Targeting the von Hippel-Lindau E3 ubiquitin ligase using small molecules to disrupt the VHL/HIF-1alpha interaction. J. Am. Chem. Soc. 2012;134:4465–4468. doi: 10.1021/ja209924v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bondeson DP, et al. Catalytic in vivo protein knockdown by small-molecule PROTACs. Nat. Chem. Biol. 2015;11:611–617. doi: 10.1038/nchembio.1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zengerle M, Chan KH, Ciulli A. Selective small molecule induced degradation of the BET bromodomain protein BRD4. ACS Chem. Biol. 2015;10:1770–1777. doi: 10.1021/acschembio.5b00216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gandhi AK, et al. Immunomodulatory agents lenalidomide and pomalidomide co-stimulate T cells by inducing degradation of T cell repressors Ikaros and Aiolos via modulation of the E3 ubiquitin ligase complex CRL4(CRBN) Br. J. Haematol. 2014;164:811–821. doi: 10.1111/bjh.12708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Han T, et al. Anticancer sulfonamides target splicing by inducing RBM39 degradation via recruitment to DCAF15. Science. 2017;356:eaal3755. doi: 10.1126/science.aal3755. [DOI] [PubMed] [Google Scholar]
- 26.Faust TB, et al. Structural complementarity facilitates E7820-mediated degradation of RBM39 by DCAF15. Nat. Chem. Biol. 2020;16:7–14. doi: 10.1038/s41589-019-0378-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Uehara T, et al. Selective degradation of splicing factor CAPERalpha by anticancer sulfonamides. Nat. Chem. Biol. 2017;13:675–680. doi: 10.1038/nchembio.2363. [DOI] [PubMed] [Google Scholar]
- 28.Du X, et al. Structural basis and kinetic pathway of RBM39 recruitment to DCAF15 by a sulfonamide molecular glue E7820. Structure. 2019;27:1625–1633 e1623. doi: 10.1016/j.str.2019.10.005. [DOI] [PubMed] [Google Scholar]
- 29.Ting TC, et al. Aryl sulfonamides degrade RBM39 and RBM23 by recruitment to CRL4-DCAF15. Cell Rep. 2019;29:1499–1510 e1496. doi: 10.1016/j.celrep.2019.09.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mullard A. Targeted protein degraders crowd into the clinic. Nat. Rev. Drug Discov. 2021;20:247–250. doi: 10.1038/d41573-021-00052-4. [DOI] [PubMed] [Google Scholar]
- 31.Wu T, et al. Targeted protein degradation as a powerful research tool in basic biology and drug target discovery. Nat. Struct. Mol. Biol. 2020;27:605–614. doi: 10.1038/s41594-020-0438-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chamberlain PP, et al. Evolution of cereblon-mediated protein degradation as a therapeutic modality. ACS Med. Chem. Lett. 2019;10:1592–1602. doi: 10.1021/acsmedchemlett.9b00425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Salami J, et al. Androgen receptor degradation by the proteolysis-targeting chimera ARCC-4 outperforms enzalutamide in cellular models of prostate cancer drug resistance. Commun. Biol. 2018;1:100. doi: 10.1038/s42003-018-0105-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Neklesa TK, Winkler JD, Crews CM. Targeted protein degradation by PROTACs. Pharmacol. Ther. 2017;174:138–144. doi: 10.1016/j.pharmthera.2017.02.027. [DOI] [PubMed] [Google Scholar]
- 35.Flanagan JJ, Neklesa TK. Targeting nuclear receptors with PROTAC degraders. Mol. Cell Endocrinol. 2019;493:110452. doi: 10.1016/j.mce.2019.110452. [DOI] [PubMed] [Google Scholar]
- 36.Sievers QL, et al. Defining the human C2H2 zinc finger degrome targeted by thalidomide analogs through CRBN. Science. 2018;362:eaat0572. doi: 10.1126/science.aat0572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Donovan KA, et al. Thalidomide promotes degradation of SALL4, a transcription factor implicated in Duane radial ray syndrome. eLife. 2018;7:e38430. doi: 10.7554/eLife.38430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McDonnell DP, Wardell SE, Norris JD. Oral selective estrogen receptor downregulators (SERDs), a breakthrough endocrine therapy for breast cancer. J. Med. Chem. 2015;58:4883–4887. doi: 10.1021/acs.jmedchem.5b00760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ariazi EA, Ariazi JL, Cordera F, Jordan VC. Estrogen receptors as therapeutic targets in breast cancer. Curr. Top. Med. Chem. 2006;6:181–202. [PubMed] [Google Scholar]
- 40.Petrylak DP, et al. First-in-human phase I study of ARV-110, an androgen receptor (AR) PROTAC degrader in patients (pts) with metastatic castrate-resistant prostate cancer (mCRPC) following enzalutamide (ENZ) and/or abiraterone (ABI) J. Clin. Oncol. 2020;38:3500–3500. [Google Scholar]
- 41.Snyder, L. B. et al. The discovery of ARV-471, an orally bioavailable estrogen receptor degrading PROTAC for the treatment of patients with breast cancer. In Proc. 112th Annual Meeting of the American Association for Cancer Research 1116 (AACR, 2021).
- 42.Luh LM, et al. Prey for the proteasome: targeted protein degradation-a medicinal chemist’s perspective. Angew. Chem. Int. Ed. Engl. 2020;59:15448–15466. doi: 10.1002/anie.202004310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Samarasinghe KTG, et al. Targeted degradation of transcription factors by TRAFTACs: TRAnscription Factor TArgeting Chimeras. Cell Chem. Biol. 2021 doi: 10.1016/j.chembiol.2021.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Farnaby W, Koegl M, McConnell DB, Ciulli A. Transforming targeted cancer therapy with PROTACs: A forward-looking perspective. Curr. Opin. Pharmacol. 2021;57:175–183. doi: 10.1016/j.coph.2021.02.009. [DOI] [PubMed] [Google Scholar]
- 45.Hopkins AL, Groom CR. The druggable genome. Nat. Rev. Drug Discov. 2002;1:727–730. doi: 10.1038/nrd892. [DOI] [PubMed] [Google Scholar]
- 46.Guharoy M, Bhowmick P, Sallam M, Tompa P. Tripartite degrons confer diversity and specificity on regulated protein degradation in the ubiquitin-proteasome system. Nat. Commun. 2016;7:10239. doi: 10.1038/ncomms10239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Davis C, Spaller BL, Matouschek A. Mechanisms of substrate recognition by the 26S proteasome. Curr. Opin. Struct. Biol. 2021;67:161–169. doi: 10.1016/j.sbi.2020.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mares A, et al. Extended pharmacodynamic responses observed upon PROTAC-mediated degradation of RIPK2. Commun. Biol. 2020;3:140. doi: 10.1038/s42003-020-0868-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bondeson DP, et al. Lessons in PROTAC design from selective degradation with a promiscuous warhead. Cell Chem. Biol. 2018;25:78–87 e75. doi: 10.1016/j.chembiol.2017.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Alabi S, et al. Mutant-selective degradation by BRAF-targeting PROTACs. Nat. Commun. 2021;12:920. doi: 10.1038/s41467-021-21159-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Farnaby W, et al. BAF complex vulnerabilities in cancer demonstrated via structure-based PROTAC design. Nat. Chem. Biol. 2019;15:672–680. doi: 10.1038/s41589-019-0294-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bensimon A, et al. Targeted degradation of SLC transporters reveals amenability of multi-pass transmembrane proteins to ligand-induced proteolysis. Cell Chem. Biol. 2020;27:728–739 e729. doi: 10.1016/j.chembiol.2020.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nabet B, et al. The dTAG system for immediate and target-specific protein degradation. Nat. Chem. Biol. 2018;14:431–441. doi: 10.1038/s41589-018-0021-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang Y, et al. In vitro and in vivo degradation of programmed cell death ligand 1 (PD-L1) by a proteolysis targeting chimera (PROTAC) Bioorg. Chem. 2021;111:104833. doi: 10.1016/j.bioorg.2021.104833. [DOI] [PubMed] [Google Scholar]
- 55.Bond MJ, Chu L, Nalawansha DA, Li K, Crews CM. Targeted degradation of oncogenic KRAS(G12C) by VHL-recruiting PROTACs. ACS Cent. Sci. 2020;6:1367–1375. doi: 10.1021/acscentsci.0c00411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sletten EM, Bertozzi CR. Bioorthogonal chemistry: fishing for selectivity in a sea of functionality. Angew. Chem. Int. Ed. Engl. 2009;48:6974–6998. doi: 10.1002/anie.200900942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ray S, Murkin AS. New electrophiles and strategies for mechanism-based and targeted covalent inhibitor design. Biochemistry. 2019;58:5234–5244. doi: 10.1021/acs.biochem.9b00293. [DOI] [PubMed] [Google Scholar]
- 58.Lossouarn A, Renard PY, Sabot C. Tailored bioorthogonal and bioconjugate chemistry: a source of inspiration for developing kinetic target-guided synthesis strategies. Bioconjug Chem. 2021;32:63–72. doi: 10.1021/acs.bioconjchem.0c00568. [DOI] [PubMed] [Google Scholar]
- 59.Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 2001;46:3–26. doi: 10.1016/s0169-409x(00)00129-0. [DOI] [PubMed] [Google Scholar]
- 60.Doak BC, Over B, Giordanetto F, Kihlberg J. Oral druggable space beyond the rule of 5: insights from drugs and clinical candidates. Chem. Biol. 2014;21:1115–1142. doi: 10.1016/j.chembiol.2014.08.013. [DOI] [PubMed] [Google Scholar]
- 61.Shultz MD. Two decades under the influence of the rule of five and the changing properties of approved oral drugs. J. Med. Chem. 2019;62:1701–1714. doi: 10.1021/acs.jmedchem.8b00686. [DOI] [PubMed] [Google Scholar]
- 62.Young RJ, Leeson PD. Mapping the efficiency and physicochemical trajectories of successful optimizations. J. Med. Chem. 2018;61:6421–6467. doi: 10.1021/acs.jmedchem.8b00180. [DOI] [PubMed] [Google Scholar]
- 63.Scott JS, Waring MJ. Practical application of ligand efficiency metrics in lead optimisation. Bioorg. Med. Chem. 2018;26:3006–3015. doi: 10.1016/j.bmc.2018.04.004. [DOI] [PubMed] [Google Scholar]
- 64.Johnson TW, Gallego RA, Edwards MP. Lipophilic efficiency as an important metric in drug design. J. Med. Chem. 2018;61:6401–6420. doi: 10.1021/acs.jmedchem.8b00077. [DOI] [PubMed] [Google Scholar]
- 65.Han X, et al. Strategies toward discovery of potent and orally bioavailable proteolysis targeting chimera degraders of androgen receptor for the treatment of prostate cancer. J. Med. Chem. 2021;64:12831–12854. doi: 10.1021/acs.jmedchem.1c00882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Poongavanam V, Kihlberg J. PROTAC cell permeability and oral bioavailability: a journey into uncharted territory. Future Med. Chem. 2021 doi: 10.4155/fmc-2021-0208. [DOI] [PubMed] [Google Scholar]
- 67.Troup RI, Fallan C, Baud MGJ. Current strategies for the design of PROTAC linkers: a critical review. Explor. Target. Antitumor Ther. 2020;1:273–312. doi: 10.37349/etat.2020.00018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Atilaw Y, et al. Solution conformations shed light on PROTAC cell permeability. ACS Med. Chem. Lett. 2021;12:107–114. doi: 10.1021/acsmedchemlett.0c00556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cyrus K, et al. Impact of linker length on the activity of PROTACs. Mol. Biosyst. 2011;7:359–364. doi: 10.1039/c0mb00074d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schneider M, et al. The PROTACtable genome. Nat. Rev. Drug Discov. 2021;20:789–797. doi: 10.1038/s41573-021-00245-x. [DOI] [PubMed] [Google Scholar]
- 71.Snyder, L. B. et al. Discovery of ARV-110, a first in class androgen receptor degrading PROTAC for the treatment of men with metastatic castration resistant prostate cancer. In Proc. 112th Annual Meeting of the American Association for Cancer Research 1115 (AACR, 2021).
- 72.Chamberlain PP, Cathers BE. Cereblon modulators: low molecular weight inducers of protein degradation. Drug Discov. Today Technol. 2019;31:29–34. doi: 10.1016/j.ddtec.2019.02.004. [DOI] [PubMed] [Google Scholar]
- 73.Matyskiela ME, et al. A cereblon modulator (CC-220) with improved degradation of Ikaros and Aiolos. J. Med. Chem. 2018;61:535–542. doi: 10.1021/acs.jmedchem.6b01921. [DOI] [PubMed] [Google Scholar]
- 74.Petzold G, Fischer ES, Thoma NH. Structural basis of lenalidomide-induced CK1alpha degradation by the CRL4(CRBN) ubiquitin ligase. Nature. 2016;532:127–130. doi: 10.1038/nature16979. [DOI] [PubMed] [Google Scholar]
- 75.Matyskiela ME, et al. A novel cereblon modulator recruits GSPT1 to the CRL4(CRBN) ubiquitin ligase. Nature. 2016;535:252–257. doi: 10.1038/nature18611. [DOI] [PubMed] [Google Scholar]
- 76.Ege N, Bouguenina H, Tatari M, Chopra R. Phenotypic screening with target identification and validation in the discovery and development of E3 ligase modulators. Cell Chem. Biol. 2021;28:283–299. doi: 10.1016/j.chembiol.2021.02.011. [DOI] [PubMed] [Google Scholar]
- 77.He Y, et al. DT2216-a Bcl-xL-specific degrader is highly active against Bcl-xL-dependent T cell lymphomas. J. Hematol. Oncol. 2020;13:95. doi: 10.1186/s13045-020-00928-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhang X, et al. Utilizing PROTAC technology to address the on-target platelet toxicity associated with inhibition of BCL-XL. Chem. Commun. 2019;55:14765–14768. doi: 10.1039/c9cc07217a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Oh E, Akopian D, Rape M. Principles of ubiquitin-dependent signaling. Annu. Rev. Cell Dev. Biol. 2018;34:137–162. doi: 10.1146/annurev-cellbio-100617-062802. [DOI] [PubMed] [Google Scholar]
- 80.Schapira M, Calabrese MF, Bullock AN, Crews CM. Targeted protein degradation: expanding the toolbox. Nat. Rev. Drug. Discov. 2019;18:949–963. doi: 10.1038/s41573-019-0047-y. [DOI] [PubMed] [Google Scholar]
- 81.Fukuoka K, et al. Mechanisms of action of the novel sulfonamide anticancer agent E7070 on cell cycle progression in human non-small cell lung cancer cells. Invest. New Drugs. 2001;19:219–227. doi: 10.1023/a:1010608317361. [DOI] [PubMed] [Google Scholar]
- 82.Owa T, et al. Discovery of novel antitumor sulfonamides targeting G1 phase of the cell cycle. J. Med. Chem. 1999;42:3789–3799. doi: 10.1021/jm9902638. [DOI] [PubMed] [Google Scholar]
- 83.Bussiere DE, et al. Structural basis of indisulam-mediated RBM39 recruitment to DCAF15 E3 ligase complex. Nat. Chem. Biol. 2020;16:15–23. doi: 10.1038/s41589-019-0411-6. [DOI] [PubMed] [Google Scholar]
- 84.Lv L, et al. Discovery of a molecular glue promoting CDK12-DDB1 interaction to trigger cyclin K degradation. eLife. 2020;9:e59994. doi: 10.7554/eLife.59994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Slabicki M, et al. The CDK inhibitor CR8 acts as a molecular glue degrader that depletes cyclin K. Nature. 2020;585:293–297. doi: 10.1038/s41586-020-2374-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Mayor-Ruiz C, et al. Rational discovery of molecular glue degraders via scalable chemical profiling. Nat. Chem. Biol. 2020;16:1199–1207. doi: 10.1038/s41589-020-0594-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Dieter SM, et al. Degradation of CCNK/CDK12 is a druggable vulnerability of colorectal cancer. Cell Rep. 2021;36:109394. doi: 10.1016/j.celrep.2021.109394. [DOI] [PubMed] [Google Scholar]
- 88.Asatsuma-Okumura T, Ito T, Handa H. Molecular mechanisms of cereblon-based drugs. Pharmacol. Ther. 2019;202:132–139. doi: 10.1016/j.pharmthera.2019.06.004. [DOI] [PubMed] [Google Scholar]
- 89.Nguyen KM, Busino L. Targeting the E3 ubiquitin ligases DCAF15 and cereblon for cancer therapy. Semin. Cancer Biol. 2020;67:53–60. doi: 10.1016/j.semcancer.2020.03.007. [DOI] [PubMed] [Google Scholar]
- 90.Fang Y, Liao G, Yu B. Small-molecule MDM2/X inhibitors and PROTAC degraders for cancer therapy: advances and perspectives. Acta Pharm. Sin. B. 2020;10:1253–1278. doi: 10.1016/j.apsb.2020.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Naito M, Ohoka N, Shibata N, Tsukumo Y. Targeted protein degradation by chimeric small molecules, PROTACs and SNIPERs. Front. Chem. 2019;7:849. doi: 10.3389/fchem.2019.00849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Weng G, et al. PROTAC-DB: an online database of PROTACs. Nucleic Acids Res. 2021;49:D1381–D1387. doi: 10.1093/nar/gkaa807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wang Y, Jiang X, Feng F, Liu W, Sun H. Degradation of proteins by PROTACs and other strategies. Acta Pharm. Sin. B. 2020;10:207–238. doi: 10.1016/j.apsb.2019.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Buckley DL, et al. HaloPROTACS: use of small molecule PROTACs to induce degradation of halotag fusion proteins. ACS Chem. Biol. 2015;10:1831–1837. doi: 10.1021/acschembio.5b00442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Nabet B, et al. Rapid and direct control of target protein levels with VHL-recruiting dTAG molecules. Nat. Commun. 2020;11:4687. doi: 10.1038/s41467-020-18377-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Roth S, Fulcher LJ, Sapkota GP. Advances in targeted degradation of endogenous proteins. Cell Mol. Life Sci. 2019;76:2761–2777. doi: 10.1007/s00018-019-03112-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Prozzillo Y, et al. Targeted protein degradation tools: overview and future perspectives. Biology (Basel) 2020;9:421. doi: 10.3390/biology9120421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Pacini C, et al. Integrated cross-study datasets of genetic dependencies in cancer. Nat. Commun. 2021;12:1661. doi: 10.1038/s41467-021-21898-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Shirasaki R, et al. Functional genomics identify distinct and overlapping genes mediating resistance to different classes of heterobifunctional degraders of oncoproteins. Cell Rep. 2021;34:108532. doi: 10.1016/j.celrep.2020.108532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zhang L, Riley-Gillis B, Vijay P, Shen Y. Acquired resistance to BET-PROTACs (proteolysis-targeting chimeras) caused by genomic alterations in core components of E3 ligase complexes. Mol. Cancer Ther. 2019;18:1302–1311. doi: 10.1158/1535-7163.MCT-18-1129. [DOI] [PubMed] [Google Scholar]
- 101.Ottis P, et al. Cellular resistance mechanisms to targeted protein degradation converge toward impairment of the engaged ubiquitin transfer pathway. ACS Chem. Biol. 2019;14:2215–2223. doi: 10.1021/acschembio.9b00525. [DOI] [PubMed] [Google Scholar]
- 102.Gooding S, et al. Multiple cereblon genetic changes are associated with acquired resistance to lenalidomide or pomalidomide in multiple myeloma. Blood. 2021;137:232–237. doi: 10.1182/blood.2020007081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Barrio S, et al. IKZF1/3 and CRL4(CRBN) E3 ubiquitin ligase mutations and resistance to immunomodulatory drugs in multiple myeloma. Haematologica. 2020;105:e237–e241. doi: 10.3324/haematol.2019.217943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Zimmerman ES, Schulman BA, Zheng N. Structural assembly of cullin-RING ubiquitin ligase complexes. Curr. Opin. Struct. Biol. 2010;20:714–721. doi: 10.1016/j.sbi.2010.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Duda DM, et al. Structural regulation of cullin-RING ubiquitin ligase complexes. Curr. Opin. Struct. Biol. 2011;21:257–264. doi: 10.1016/j.sbi.2011.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Baek K, et al. NEDD8 nucleates a multivalent cullin-RING-UBE2D ubiquitin ligation assembly. Nature. 2020;578:461–466. doi: 10.1038/s41586-020-2000-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Baek K, Scott DC, Schulman BA. NEDD8 and ubiquitin ligation by cullin-RING E3 ligases. Curr. Opin. Struct. Biol. 2020;67:101–109. doi: 10.1016/j.sbi.2020.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Yau R, Rape M. The increasing complexity of the ubiquitin code. Nat. Cell Biol. 2016;18:579–586. doi: 10.1038/ncb3358. [DOI] [PubMed] [Google Scholar]
- 109.Komander D, Rape M. The ubiquitin code. Annu. Rev. Biochem. 2012;81:203–229. doi: 10.1146/annurev-biochem-060310-170328. [DOI] [PubMed] [Google Scholar]
- 110.Zheng N, Shabek N. Ubiquitin ligases: structure, function, and regulation. Annu. Rev. Biochem. 2017;86:129–157. doi: 10.1146/annurev-biochem-060815-014922. [DOI] [PubMed] [Google Scholar]
- 111.Liu L, et al. UbiHub: a data hub for the explorers of ubiquitination pathways. Bioinformatics. 2019;35:2882–2884. doi: 10.1093/bioinformatics/bty1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Zhang W, et al. System-wide modulation of HECT E3 ligases with selective ubiquitin variant probes. Mol. Cell. 2016;62:121–136. doi: 10.1016/j.molcel.2016.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kamadurai HB, et al. Mechanism of ubiquitin ligation and lysine prioritization by a HECT E3. eLife. 2013;2:e00828. doi: 10.7554/eLife.00828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Yuan L, Lv Z, Atkison JH, Olsen SK. Structural insights into the mechanism and E2 specificity of the RBR E3 ubiquitin ligase HHARI. Nat. Commun. 2017;8:211. doi: 10.1038/s41467-017-00272-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Dove KK, et al. Structural studies of HHARI/UbcH7~Ub reveal unique E2~Ub conformational restriction by RBR RING1. Structure. 2017;25:890–900 e895. doi: 10.1016/j.str.2017.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Scott DC, et al. Two distinct types of E3 ligases work in unison to regulate substrate ubiquitylation. Cell. 2016;166:1198–1214 e1124. doi: 10.1016/j.cell.2016.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Horn-Ghetko D, et al. Ubiquitin ligation to F-box protein targets by SCF-RBR E3-E3 super-assembly. Nature. 2021;590:671–676. doi: 10.1038/s41586-021-03197-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Lai AC, et al. Modular PROTAC design for the degradation of oncogenic BCR-ABL. Angew. Chem. Int. Ed. Engl. 2016;55:807–810. doi: 10.1002/anie.201507634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Ostrem JM, Peters U, Sos ML, Wells JA, Shokat KM. K-Ras(G12C) inhibitors allosterically control GTP affinity and effector interactions. Nature. 2013;503:548–551. doi: 10.1038/nature12796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Janes MR, et al. Targeting KRAS mutant cancers with a covalent G12C-specific inhibitor. Cell. 2018;172:578–589 e517. doi: 10.1016/j.cell.2018.01.006. [DOI] [PubMed] [Google Scholar]
- 121.Moore AR, Rosenberg SC, McCormick F, Malek S. RAS-targeted therapies: is the undruggable drugged? Nat. Rev. Drug Discov. 2020;19:533–552. doi: 10.1038/s41573-020-0068-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Hallin J, et al. The KRAS(G12C) inhibitor MRTX849 provides insight toward therapeutic susceptibility of KRAS-mutant cancers in mouse models and patients. Cancer Discov. 2020;10:54–71. doi: 10.1158/2159-8290.CD-19-1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Canon J, et al. The clinical KRAS(G12C) inhibitor AMG 510 drives anti-tumour immunity. Nature. 2019;575:217–223. doi: 10.1038/s41586-019-1694-1. [DOI] [PubMed] [Google Scholar]
- 124.Zeng M, et al. Exploring targeted degradation strategy for oncogenic KRAS(G12C) Cell Chem. Biol. 2020;27:19–31 e16. doi: 10.1016/j.chembiol.2019.12.006. [DOI] [PubMed] [Google Scholar]
- 125.Huang HT, et al. A chemoproteomic approach to query the degradable kinome using a multi-kinase degrader. Cell Chem. Biol. 2018;25:88–99 e86. doi: 10.1016/j.chembiol.2017.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Donovan KA, et al. Mapping the degradable kinome provides a resource for expedited degrader development. Cell. 2020;183:1714–1731 e1710. doi: 10.1016/j.cell.2020.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Rodriguez-Rivera FP, Levi SM. Unifying catalysis framework to dissect proteasomal degradation paradigms. ACS Cent. Sci. 2021;7:1117–1125. doi: 10.1021/acscentsci.1c00389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Hughes SJ, Ciulli A. Molecular recognition of ternary complexes: a new dimension in the structure-guided design of chemical degraders. Essays Biochem. 2017;61:505–516. doi: 10.1042/EBC20170041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Gadd MS, et al. Structural basis of PROTAC cooperative recognition for selective protein degradation. Nat. Chem. Biol. 2017;13:514–521. doi: 10.1038/nchembio.2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Nowak RP, et al. Plasticity in binding confers selectivity in ligand-induced protein degradation. Nat. Chem. Biol. 2018;14:706–714. doi: 10.1038/s41589-018-0055-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Zhang X, Crowley VM, Wucherpfennig TG, Dix MM, Cravatt BF. Electrophilic PROTACs that degrade nuclear proteins by engaging DCAF16. Nat. Chem. Biol. 2019;15:737–746. doi: 10.1038/s41589-019-0279-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Ward CC, et al. Covalent ligand screening uncovers a RNF4 E3 ligase recruiter for targeted protein degradation applications. ACS Chem. Biol. 2019;14:2430–2440. doi: 10.1021/acschembio.8b01083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Spradlin JN, et al. Harnessing the anti-cancer natural product nimbolide for targeted protein degradation. Nat. Chem. Biol. 2019;15:747–755. doi: 10.1038/s41589-019-0304-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Luo M, et al. Chemoproteomics-enabled discovery of covalent RNF114-based degraders that mimic natural product function. Cell Chem. Biol. 2021;28:559–566. doi: 10.1016/j.chembiol.2021.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Tong B, et al. A nimbolide-based kinase degrader preferentially degrades oncogenic BCR-ABL. ACS Chem. Biol. 2020;15:1788–1794. doi: 10.1021/acschembio.0c00348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Tong B, et al. Bardoxolone conjugation enables targeted protein degradation of BRD4. Sci. Rep. 2020;10:15543. doi: 10.1038/s41598-020-72491-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Wei J, et al. Harnessing the E3 Ligase KEAP1 for targeted protein degradation. J. Am. Chem. Soc. 2021;143:15073–15083. doi: 10.1021/jacs.1c04841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Henning NJ, et al. Discovery of a covalent FEM1B recruiter for targeted protein degradation applications. bioRxiv. 2021 doi: 10.1101/2021.04.15.439993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Ishida T, Ciulli A. E3 ligase ligands for PROTACs: how they were found and how to discover new ones. SLAS Discov. 2021;26:484–502. doi: 10.1177/2472555220965528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Ramachandran S, Ciulli A. Building ubiquitination machineries: E3 ligase multi-subunit assembly and substrate targeting by PROTACs and molecular glues. Curr. Opin. Struct. Biol. 2020;67:110–119. doi: 10.1016/j.sbi.2020.10.009. [DOI] [PubMed] [Google Scholar]
- 141.Tunyasuvunakool K, et al. Highly accurate protein structure prediction for the human proteome. Nature. 2021;596:590–596. doi: 10.1038/s41586-021-03828-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Baek M, et al. Accurate prediction of protein structures and interactions using a three-track neural network. Science. 2021;373:871–876. doi: 10.1126/science.abj8754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Khan S, et al. PROteolysis TArgeting Chimeras (PROTACs) as emerging anticancer therapeutics. Oncogene. 2020;39:4909–4924. doi: 10.1038/s41388-020-1336-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.He Y, et al. Proteolysis targeting chimeras (PROTACs) are emerging therapeutics for hematologic malignancies. J. Hematol. Oncol. 2020;13:103. doi: 10.1186/s13045-020-00924-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Ehrlich KC, Baribault C, Ehrlich M. Epigenetics of muscle- and brain-specific expression of KLHL family genes. Int. J. Mol. Sci. 2020;21:8394. doi: 10.3390/ijms21218394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Gupta VA, et al. Identification of KLHL41 mutations implicates BTB-kelch-mediated ubiquitination as an alternate pathway to myofibrillar disruption in nemaline myopathy. Am. J. Hum. Genet. 2013;93:1108–1117. doi: 10.1016/j.ajhg.2013.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Garg A, et al. KLHL40 deficiency destabilizes thin filament proteins and promotes nemaline myopathy. J. Clin. Invest. 2014;124:3529–3539. doi: 10.1172/JCI74994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Liu QY, Lei JX, Sikorska M, Liu R. A novel brain-enriched E3 ubiquitin ligase RNF182 is up regulated in the brains of Alzheimer’s patients and targets ATP6V0C for degradation. Mol. Neurodegener. 2008;3:4. doi: 10.1186/1750-1326-3-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Menon S, et al. The TRIM9/TRIM67 neuronal interactome reveals novel activators of morphogenesis. Mol. Biol. Cell. 2021;32:314–330. doi: 10.1091/mbc.E20-10-0622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Kumanomidou T, et al. The structural differences between a glycoprotein specific F-box protein Fbs1 and its homologous protein FBG3. PLoS ONE. 2015;10:e0140366. doi: 10.1371/journal.pone.0140366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Glenn KA, Nelson RF, Wen HM, Mallinger AJ, Paulson HL. Diversity in tissue expression, substrate binding, and SCF complex formation for a lectin family of ubiquitin ligases. J. Biol. Chem. 2008;283:12717–12729. doi: 10.1074/jbc.M709508200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Khan S, et al. A selective BCL-XL PROTAC degrader achieves safe and potent antitumor activity. Nat. Med. 2019;25:1938–1947. doi: 10.1038/s41591-019-0668-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Zhang X, et al. Discovery of PROTAC BCL-XL degraders as potent anticancer agents with low on-target platelet toxicity. Eur. J. Med. Chem. 2020;192:112186. doi: 10.1016/j.ejmech.2020.112186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Wei R, et al. Cancer testis antigens in sarcoma: expression, function and immunotherapeutic application. Cancer Lett. 2020;479:54–60. doi: 10.1016/j.canlet.2019.10.024. [DOI] [PubMed] [Google Scholar]
- 155.Weon JL, Potts PR. The MAGE protein family and cancer. Curr. Opin. Cell Biol. 2015;37:1–8. doi: 10.1016/j.ceb.2015.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Yang SW, et al. A cancer-specific ubiquitin ligase drives mRNA alternative polyadenylation by ubiquitinating the mRNA 3′ end processing complex. Mol. Cell. 2020;77:1206–1221 e1207. doi: 10.1016/j.molcel.2019.12.022. [DOI] [PubMed] [Google Scholar]
- 157.Pineda CT, et al. Degradation of AMPK by a cancer-specific ubiquitin ligase. Cell. 2015;160:715–728. doi: 10.1016/j.cell.2015.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Lee AK, Potts PR. A comprehensive guide to the MAGE family of ubiquitin ligases. J. Mol. Biol. 2017;429:1114–1142. doi: 10.1016/j.jmb.2017.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Tacer KF, Potts PR. Cellular and disease functions of the Prader-Willi syndrome gene MAGEL2. Biochem. J. 2017;474:2177–2190. doi: 10.1042/BCJ20160616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Pillow TH, et al. Antibody conjugation of a chimeric BET degrader enables in vivo activity. ChemMedChem. 2020;15:17–25. doi: 10.1002/cmdc.201900497. [DOI] [PubMed] [Google Scholar]
- 161.Dragovich PS, et al. Antibody-mediated delivery of chimeric BRD4 degraders. Part 2: improvement of in vitro antiproliferation activity and in vivo antitumor efficacy. J. Med. Chem. 2021;64:2576–2607. doi: 10.1021/acs.jmedchem.0c01846. [DOI] [PubMed] [Google Scholar]
- 162.Dragovich PS, et al. Antibody-mediated delivery of chimeric BRD4 degraders. Part 1: exploration of antibody linker, payload loading, and payload molecular properties. J. Med. Chem. 2021;64:2534–2575. doi: 10.1021/acs.jmedchem.0c01845. [DOI] [PubMed] [Google Scholar]
- 163.Jan M, Sperling AS, Ebert BL. Cancer therapies based on targeted protein degradation — lessons learned with lenalidomide. Nat. Rev. Clin. Oncol. 2021;18:401–417. doi: 10.1038/s41571-021-00479-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Dobrovolsky D, et al. Bruton tyrosine kinase degradation as a therapeutic strategy for cancer. Blood. 2019;133:952–961. doi: 10.1182/blood-2018-07-862953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Zarrin AA, Bao K, Lupardus P, Vucic D. Kinase inhibition in autoimmunity and inflammation. Nat. Rev. Drug Discov. 2021;20:39–63. doi: 10.1038/s41573-020-0082-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Schiemer J, et al. Snapshots and ensembles of BTK and cIAP1 protein degrader ternary complexes. Nat. Chem. Biol. 2021;17:152–160. doi: 10.1038/s41589-020-00686-2. [DOI] [PubMed] [Google Scholar]
- 167.Zorba A, et al. Delineating the role of cooperativity in the design of potent PROTACs for BTK. Proc. Natl Acad. Sci. USA. 2018;115:E7285–E7292. doi: 10.1073/pnas.1803662115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Tinworth CP, et al. PROTAC-mediated degradation of bruton’s tyrosine kinase is inhibited by covalent binding. ACS Chem. Biol. 2019;14:342–347. doi: 10.1021/acschembio.8b01094. [DOI] [PubMed] [Google Scholar]
- 169.Buhimschi AD, et al. Targeting the C481S ibrutinib-resistance mutation in Bruton’s tyrosine kinase using PROTAC-mediated degradation. Biochemistry. 2018;57:3564–3575. doi: 10.1021/acs.biochem.8b00391. [DOI] [PubMed] [Google Scholar]
- 170.Sun Y, et al. PROTAC-induced BTK degradation as a novel therapy for mutated BTK C481S induced ibrutinib-resistant B-cell malignancies. Cell Res. 2018;28:779–781. doi: 10.1038/s41422-018-0055-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Wiese MD, Manning-Bennett AT, Abuhelwa AY. Investigational IRAK-4 inhibitors for the treatment of rheumatoid arthritis. Expert Opin. Investig. Drugs. 2020;29:475–482. doi: 10.1080/13543784.2020.1752660. [DOI] [PubMed] [Google Scholar]
- 172.Qin J, Jiang Z, Qian Y, Casanova JL, Li X. IRAK4 kinase activity is redundant for interleukin-1 (IL-1) receptor-associated kinase phosphorylation and IL-1 responsiveness. J. Biol. Chem. 2004;279:26748–26753. doi: 10.1074/jbc.M400785200. [DOI] [PubMed] [Google Scholar]
- 173.Zhang J, et al. Assessing IRAK4 functions in ABC DLBCL by IRAK4 kinase inhibition and protein degradation. Cell Chem. Biol. 2020;27:1500–1509 e1513. doi: 10.1016/j.chembiol.2020.08.010. [DOI] [PubMed] [Google Scholar]
- 174.Nunes J, et al. Targeting IRAK4 for degradation with PROTACs. ACS Med. Chem. Lett. 2019;10:1081–1085. doi: 10.1021/acsmedchemlett.9b00219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Chen Y, et al. Design, synthesis, and biological evaluation of IRAK4-targeting PROTACs. ACS Med. Chem. Lett. 2021;12:82–87. doi: 10.1021/acsmedchemlett.0c00474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.van der Zanden SY, Luimstra JJ, Neefjes J, Borst J, Ovaa H. Opportunities for small molecules in cancer immunotherapy. Trends Immunol. 2020;41:493–511. doi: 10.1016/j.it.2020.04.004. [DOI] [PubMed] [Google Scholar]
- 177.Kerr WG, Chisholm JD. The next generation of immunotherapy for cancer: small molecules could make big waves. J. Immunol. 2019;202:11–19. doi: 10.4049/jimmunol.1800991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Wang Y, Deng S, Xu J. Proteasomal and lysosomal degradation for specific and durable suppression of immunotherapeutic targets. Cancer Biol. Med. 2020;17:583–598. doi: 10.20892/j.issn.2095-3941.2020.0066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Shifrut E, et al. Genome-wide CRISPR screens in primary human T cells reveal key regulators of immune function. Cell. 2018;175:1958–1971 e1915. doi: 10.1016/j.cell.2018.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Si J, et al. Hematopoietic progenitor kinase1 (HPK1) mediates T cell dysfunction and is a druggable target for T cell-based immunotherapies. Cancer Cell. 2020;38:551–566 e511. doi: 10.1016/j.ccell.2020.08.001. [DOI] [PubMed] [Google Scholar]
- 181.Burslem GM, et al. The advantages of targeted protein degradation over inhibition: an RTK case study. Cell Chem. Biol. 2018;25:67–77 e63. doi: 10.1016/j.chembiol.2017.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Wang Y, Mandelkow E. Tau in physiology and pathology. Nat. Rev. Neurosci. 2016;17:5–21. doi: 10.1038/nrn.2015.1. [DOI] [PubMed] [Google Scholar]
- 183.Goedert M. Tau protein and neurodegeneration. Semin. Cell Dev. Biol. 2004;15:45–49. doi: 10.1016/j.semcdb.2003.12.015. [DOI] [PubMed] [Google Scholar]
- 184.Chang CW, Shao E, Mucke L. Tau: enabler of diverse brain disorders and target of rapidly evolving therapeutic strategies. Science. 2021;371:eabb8255. doi: 10.1126/science.abb8255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.DeVos SL, et al. Tau reduction prevents neuronal loss and reverses pathological tau deposition and seeding in mice with tauopathy. Sci. Transl. Med. 2017;9:eaag0481. doi: 10.1126/scitranslmed.aag0481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Yiannopoulou KG, Papageorgiou SG. Current and future treatments in Alzheimer disease: an update. J. Cent. Nerv. Syst. Dis. 2020;12:1179573520907397. doi: 10.1177/1179573520907397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Wegmann S, et al. Persistent repression of tau in the brain using engineered zinc finger protein transcription factors. Sci. Adv. 2021;7:eabe1611. doi: 10.1126/sciadv.abe1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Lu M, et al. Discovery of a Keap1-dependent peptide PROTAC to knockdown Tau by ubiquitination-proteasome degradation pathway. Eur. J. Med. Chem. 2018;146:251–259. doi: 10.1016/j.ejmech.2018.01.063. [DOI] [PubMed] [Google Scholar]
- 189.Wang W, et al. A novel small-molecule PROTAC selectively promotes tau clearance to improve cognitive functions in Alzheimer-like models. Theranostics. 2021;11:5279–5295. doi: 10.7150/thno.55680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Chien DT, et al. Early clinical PET imaging results with the novel PHF-tau radioligand [F-18]-T807. J. Alzheimers Dis. 2013;34:457–468. doi: 10.3233/JAD-122059. [DOI] [PubMed] [Google Scholar]
- 191.Silva MC, et al. Targeted degradation of aberrant tau in frontotemporal dementia patient-derived neuronal cell models. eLife. 2019;8:e45457. doi: 10.7554/eLife.45457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Kotzbauer PT, Trojanowsk JQ, Lee VM. Lewy body pathology in Alzheimer’s disease. J. Mol. Neurosci. 2001;17:225–232. doi: 10.1385/jmn:17:2:225. [DOI] [PubMed] [Google Scholar]
- 193.Qu J, et al. Specific knockdown of alpha-synuclein by peptide-directed proteasome degradation rescued its associated neurotoxicity. Cell Chem. Biol. 2020;27:763. doi: 10.1016/j.chembiol.2020.05.012. [DOI] [PubMed] [Google Scholar]
- 194.MacDonald M. E. A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington’s disease chromosomes. Cell. 1993;72:971–983. doi: 10.1016/0092-8674(93)90585-e. [DOI] [PubMed] [Google Scholar]
- 195.Sap KA, Reits EA. Strategies to investigate ubiquitination in Huntington’s disease. Front. Chem. 2020;8:485. doi: 10.3389/fchem.2020.00485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Li Z, et al. Allele-selective lowering of mutant HTT protein by HTT-LC3 linker compounds. Nature. 2019;575:203–209. doi: 10.1038/s41586-019-1722-1. [DOI] [PubMed] [Google Scholar]
- 197.Tomoshige S, Nomura S, Ohgane K, Hashimoto Y, Ishikawa M. Discovery of small molecules that induce the degradation of huntingtin. Angew. Chem. Int. Ed. Engl. 2017;56:11530–11533. doi: 10.1002/anie.201706529. [DOI] [PubMed] [Google Scholar]
- 198.de Wispelaere M, et al. Small molecule degraders of the hepatitis C virus protease reduce susceptibility to resistance mutations. Nat. Commun. 2019;10:3468. doi: 10.1038/s41467-019-11429-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Gordon DE, et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020;583:459–468. doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Martinez-Ortiz W, Zhou MM. Could PROTACs protect us from COVID-19? Drug Discov. Today. 2020;25:1894–1896. doi: 10.1016/j.drudis.2020.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Ghosh AK, Brindisi M, Shahabi D, Chapman ME, Mesecar AD. Drug development and medicinal chemistry efforts toward SARS-coronavirus and Covid-19 therapeutics. ChemMedChem. 2020;15:907–932. doi: 10.1002/cmdc.202000223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Yin W, et al. Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir. Science. 2020;368:1499–1504. doi: 10.1126/science.abc1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Boras B, et al. Discovery of a novel inhibitor of coronavirus 3CL protease as a clinical candidate for the potential treatment of COVID-19. bioRxiv. 2020 doi: 10.1101/2020.09.12.293498. [DOI] [Google Scholar]
- 204.De Haan P, Van Diemen FR, Toscano MG. Viral gene delivery vectors: the next generation medicines for immune-related diseases. Hum. Vaccin. Immunother. 2021;17:14–21. doi: 10.1080/21645515.2020.1757989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Sung YK, Kim SW. Recent advances in the development of gene delivery systems. Biomater. Res. 2019;23:8. doi: 10.1186/s40824-019-0156-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Ghosh S, Brown AM, Jenkins C, Campbell K. Viral vector systems for gene therapy: a comprehensive literature review of progress and biosafety challenges. Appl. Biosaf. 2020;25:7–18. doi: 10.1177/1535676019899502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Yang KC, et al. Nanotechnology advances in pathogen- and host-targeted antiviral delivery: multipronged therapeutic intervention for pandemic control. Drug Deliv. Transl. Res. 2021;11:1420–1437. doi: 10.1007/s13346-021-00965-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Steinhauff D, Ghandehari H. Matrix mediated viral gene delivery: a review. Bioconjug Chem. 2019;30:384–399. doi: 10.1021/acs.bioconjchem.8b00853. [DOI] [PubMed] [Google Scholar]
- 209.Editorial Let’s talk about lipid nanoparticles. Nat. Rev. Mater. 2021;6:99. [Google Scholar]
- 210.Zhou P, Bogacki R, McReynolds L, Howley PM. Harnessing the ubiquitination machinery to target the degradation of specific cellular proteins. Mol. Cell. 2000;6:751–756. doi: 10.1016/s1097-2765(00)00074-5. [DOI] [PubMed] [Google Scholar]
- 211.Li X, et al. Degradation of HER2 by Cbl-based chimeric ubiquitin ligases. Cancer Res. 2007;67:8716–8724. doi: 10.1158/0008-5472.CAN-06-3731. [DOI] [PubMed] [Google Scholar]
- 212.Pan T, et al. A recombinant chimeric protein specifically induces mutant KRAS degradation and potently inhibits pancreatic tumor growth. Oncotarget. 2016;7:44299–44309. doi: 10.18632/oncotarget.9996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Chu TT, et al. Specific knockdown of endogenous tau protein by peptide-directed ubiquitin-proteasome degradation. Cell Chem. Biol. 2016;23:453–461. doi: 10.1016/j.chembiol.2016.02.016. [DOI] [PubMed] [Google Scholar]
- 214.Lim S, et al. bioPROTACs as versatile modulators of intracellular therapeutic targets including proliferating cell nuclear antigen (PCNA) Proc. Natl Acad. Sci. USA. 2020;117:5791–5800. doi: 10.1073/pnas.1920251117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Fulcher LJ, et al. An affinity-directed protein missile system for targeted proteolysis. Open Biol. 2016;6:160255. doi: 10.1098/rsob.160255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Saerens D, et al. Identification of a universal VHH framework to graft non-canonical antigen-binding loops of camel single-domain antibodies. J. Mol. Biol. 2005;352:597–607. doi: 10.1016/j.jmb.2005.07.038. [DOI] [PubMed] [Google Scholar]
- 217.Caussinus E, Kanca O, Affolter M. Fluorescent fusion protein knockout mediated by anti-GFP nanobody. Nat. Struct. Mol. Biol. 2011;19:117–121. doi: 10.1038/nsmb.2180. [DOI] [PubMed] [Google Scholar]
- 218.Shin YJ, et al. Nanobody-targeted E3-ubiquitin ligase complex degrades nuclear proteins. Sci. Rep. 2015;5:14269. doi: 10.1038/srep14269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Lim S, et al. Exquisitely specific anti-KRAS biodegraders inform on the cellular prevalence of nucleotide-loaded states. ACS Cent. Sci. 2021;7:274–291. doi: 10.1021/acscentsci.0c01337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Ghidini A, Clery A, Halloy F, Allain FHT, Hall J. RNA-PROTACs: Degraders of RNA-binding proteins. Angew. Chem. Int. Ed. Engl. 2021;60:3163–3169. doi: 10.1002/anie.202012330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Shao J, et al. Destruction of DNA-binding proteins by programmable O’PROTAC: oligonucleotide-based PROTAC. bioRxiv. 2021 doi: 10.1101/2021.03.08.434493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Liu J, et al. TF-PROTACs enable targeted degradation of transcription factors. J. Am. Chem. Soc. 2021;143:8902–8910. doi: 10.1021/jacs.1c03852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Angers S, et al. Molecular architecture and assembly of the DDB1-CUL4A ubiquitin ligase machinery. Nature. 2006;443:590–593. doi: 10.1038/nature05175. [DOI] [PubMed] [Google Scholar]
- 224.Page RCP, Amick JJN, Klevit RE, Misra S. Structural Insights into the conformation and oligomerization of E2∼ubiquitin conjugates. Biochemistry. 2012;51:4175–4187. doi: 10.1021/bi300058m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Faull SV, et al. Structural basis of Cullin 2 RING E3 ligase regulation by the COP9 signalosome. Nat. Commun. 2019;10:3814. doi: 10.1038/s41467-019-11772-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Zhang M, et al. Chaperoned ubiquitylation–crystal structures of the CHIP U box E3 ubiquitin ligase and a CHIP-Ubc13-Uev1a complex. Mol. Cell. 2005;20:525–538. doi: 10.1016/j.molcel.2005.09.023. [DOI] [PubMed] [Google Scholar]
- 227.Kiss L, et al. A tri-ionic anchor mechanism drives Ube2N-specific recruitment and K63-chain ubiquitination in TRIM ligases. Nat. Commun. 2019;10:4502. doi: 10.1038/s41467-019-12388-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Kannt A, Dikic I. Expanding the arsenal of E3 ubiquitin ligases for proximity-induced protein degradation. Cell Chem. Biol. 2021;28:1014–1031. doi: 10.1016/j.chembiol.2021.04.007. [DOI] [PubMed] [Google Scholar]
- 229.Jevtic P, Haakonsen DL, Rape M. An E3 ligase guide to the galaxy of small-molecule-induced protein degradation. Cell Chem. Biol. 2021;28:1000–1013. doi: 10.1016/j.chembiol.2021.04.002. [DOI] [PubMed] [Google Scholar]
- 230.Chen YN, et al. Allosteric inhibition of SHP2 phosphatase inhibits cancers driven by receptor tyrosine kinases. Nature. 2016;535:148–152. doi: 10.1038/nature18621. [DOI] [PubMed] [Google Scholar]
- 231.Liu J, et al. Calcineurin is a common target of cyclophilin-cyclosporin A and FKBP-FK506 complexes. Cell. 1991;66:807–815. doi: 10.1016/0092-8674(91)90124-h. [DOI] [PubMed] [Google Scholar]
- 232.Schreiber SL. The rise of molecular glues. Cell. 2021;184:3–9. doi: 10.1016/j.cell.2020.12.020. [DOI] [PubMed] [Google Scholar]
- 233.Deshaies RJ. Multispecific drugs herald a new era of biopharmaceutical innovation. Nature. 2020;580:329–338. doi: 10.1038/s41586-020-2168-1. [DOI] [PubMed] [Google Scholar]
- 234.Simonetta KR, et al. Prospective discovery of small molecule enhancers of an E3 ligase-substrate interaction. Nat. Commun. 2019;10:1402. doi: 10.1038/s41467-019-09358-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Koduri V, et al. Targeting oncoproteins with a positive selection assay for protein degraders. Sci. Adv. 2021;7:eabd6263. doi: 10.1126/sciadv.abd6263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Lou Z, Wang S. E3 ubiquitin ligases and human papillomavirus-induced carcinogenesis. J. Int. Med. Res. 2014;42:247–260. doi: 10.1177/0300060513506655. [DOI] [PubMed] [Google Scholar]
- 237.Planelles V, Barker E. Roles of Vpr and Vpx in modulating the virus-host cell relationship. Mol. Asp. Med. 2010;31:398–406. doi: 10.1016/j.mam.2010.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Yan J, et al. HIV-1 Vpr reprograms CLR4(DCAF1) E3 ubiquitin ligase to antagonize exonuclease 1-mediated restriction of HIV-1 infection. mBio. 2018;9:e01732-18. doi: 10.1128/mBio.01732-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Dharmasiri N, Dharmasiri S, Estelle M. The F-box protein TIR1 is an auxin receptor. Nature. 2005;435:441–445. doi: 10.1038/nature03543. [DOI] [PubMed] [Google Scholar]
- 240.Kepinski S, Leyser O. Auxin-induced SCFTIR1-Aux/IAA interaction involves stable modification of the SCFTIR1 complex. Proc. Natl Acad. Sci. USA. 2004;101:12381–12386. doi: 10.1073/pnas.0402868101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Thines B, et al. JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature. 2007;448:661–665. doi: 10.1038/nature05960. [DOI] [PubMed] [Google Scholar]
- 242.Larrieu A, Vernoux T. Comparison of plant hormone signalling systems. Essays Biochem. 2015;58:165–181. doi: 10.1042/bse0580165. [DOI] [PubMed] [Google Scholar]
- 243.Guo Y, et al. Structural basis for hijacking CBF-beta and CUL5 E3 ligase complex by HIV-1 Vif. Nature. 2014;505:229–233. doi: 10.1038/nature12884. [DOI] [PubMed] [Google Scholar]
- 244.Wu Y, et al. The DDB1-DCAF1-Vpr-UNG2 crystal structure reveals how HIV-1 Vpr steers human UNG2 toward destruction. Nat. Struct. Mol. Biol. 2016;23:933–940. doi: 10.1038/nsmb.3284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Martinez-Zapien D, et al. Structure of the E6/E6AP/p53 complex required for HPV-mediated degradation of p53. Nature. 2016;529:541–545. doi: 10.1038/nature16481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Ding Y, Fei Y, Lu B. Emerging new concepts of degrader technologies. Trends Pharmacol. Sci. 2020;41:464–474. doi: 10.1016/j.tips.2020.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Kastl JM, Davies G, Godsman E, Holdgate GA. Small-molecule degraders beyond PROTACs-challenges and opportunities. SLAS Discov. 2021;26:524–533. doi: 10.1177/2472555221991104. [DOI] [PubMed] [Google Scholar]
- 248.Alabi S, Crews C. Major advances in targeted protein degradation: PROTACs, LYTACs, and MADTACs. J. Biol. Chem. 2021;296:100647. doi: 10.1016/j.jbc.2021.100647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Takahashi D, et al. AUTACs: cargo-specific degraders using selective autophagy. Mol. Cell. 2019;76:797–810 e710. doi: 10.1016/j.molcel.2019.09.009. [DOI] [PubMed] [Google Scholar]
- 250.Banik SM, et al. Lysosome-targeting chimaeras for degradation of extracellular proteins. Nature. 2020;584:291–297. doi: 10.1038/s41586-020-2545-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Cotton AD, Nguyen DP, Gramespacher JA, Seiple IB, Wells JA. Development of antibody-based PROTACs for the degradation of the cell-surface immune checkpoint protein PD-L1. J. Am. Chem. Soc. 2021;143:593–598. doi: 10.1021/jacs.0c10008. [DOI] [PMC free article] [PubMed] [Google Scholar]