Abstract
Background:
Inflammation plays a major role in coronavirus disease (COVID-19). Factors that convey information about the status of inflammation could predict disease severity and help identify patients prone to clinical deterioration. Here, we aimed to evaluate the predictive value of inflammatory markers on the extent of lung involvement and survival of patients with COVID-19.
Materials and Methods:
Eighty patients with confirmed COVID-19 were enrolled. Demographic, clinical, and laboratory data were collected at admission. All patients underwent chest computed tomography (CT); the extent of lung involvement was assessed by a scoring system. Patients were followed up until death or discharge occurred. Logistic regression analysis was performed to evaluate the association of investigated variables with COVID-19-related death. The association between different variables and CT score was assessed using linear regression model. Receiver operator characteristic curve analysis was applied to identify the predictive value of inflammatory markers and CT score on survival.
Results:
The mean age of patients was 54.2 ± 15.2 years; 65% were male. Increased neutrophil-to-lymphocyte ratio (β =0.69, odds ratio [OR] =1.50), platelet-to-lymphocyte ratio (β =0.019, OR = 1.01), and decreased lymphocyte to C-reactive protein ratio (LCR) (β = −0.35, OR = 0.62) were significantly associated with a higher CT score and increased odds of death (P < 0.05). Lactate dehydrogenase level was also positively related with extensive lung involvement and death (β =1.15, OR = 1.52, P < 0.05). The LCR threshold for identifying survivors from nonsurvivors was 0.53 (area under curve [AUC] =0.82, 78% sensitivity and 74% specificity). Lung involvement ≥50% on chest CT was an excellent predictor of death (AUC = 0.83, 81% sensitivity and 79% specificity).
Conclusion:
Daily-performed laboratory tests that represent inflammation have great value for predicting the amount of disease burden and risk of mortality. Moreover, their cost-effectiveness and feasibility turn them into ideal prognostic markers.
Keywords: Computed tomography, COVID-19, death, inflammation, inflammatory, mortality, prognosis, survival
INTRODUCTION
Coronavirus disease (COVID-19) is a highly contagious infection caused by the novel viral pathogen, the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2).[1] This viral infection was initially identified in Wuhan, China, in a cluster of patients who mainly presented with fever, cough, and dyspnea. Nevertheless, COVID-19 encompasses a broad range of clinical manifestations from asymptomatic illness to severe respiratory distress and even death.[2]
Initial studies showed that patients with COVID-19 demonstrate elevated levels of inflammatory markers such as increased erythrocyte sedimentation rate, C-reactive protein (CRP), and lactate dehydrogenase (LDH).[3,4] Furthermore, decreased levels of lymphocytes were seen in patients with severe form of disease, which was attributed to the possible effect of SARS-CoV-2 on T-lymphocytes, subsequently leading to a reduction in CD4 and CD8 T cells.[5] Lately, Ji et al. performed an analysis on patients with COVID-19 to identify factors that are associated with disease severity. Among a multitude of investigated variables, older age and comorbidity along with inflammatory markers such as decreased lymphocyte level and increased LDH were the only independent predictors of COVID-19 progression.[6]
In recent years, novel inflammatory markers have been introduced that have the ability to convey prognostic information in a variety of diseases. For example, neutrophil to lymphocyte ratio (NLR), which can be easily measured by daily laboratory tests, has a valuable role in predicting the prognosis of a wide range of conditions from sepsis and Bell's palsy to malignancy.[7,8,9] Lymphocyte-to-CRP ratio (LCR) and platelet-to-lymphocyte ratio (PLR) are among other cost-effective inflammatory markers that are shown to be indicative of poor prognosis in a set of infectious and noninfectious diseases.[10,11] Considering these findings, several studies sought to investigate the role of these novel biomarkers in predicting the clinical severity of COVID-19. Preliminary reports showed a positive correlation between increased levels of NLR and severe disease.[7,12] Furthermore, reduced LCR and increased levels of PLR were shown to be associated with boosted inflammatory response, leading to progressive disease and unfavorable outcome.[13]
From the beginning of the COVID-19 pandemic, chest computed tomography (CT) was quickly recognized as a useful tool for early detection of the disease.[14,15] Soon after, several studies addressed the additional value of imaging findings, besides clinical and laboratory data, for follow-up and monitoring of disease course in patients with COVID-19. These studies utilized different CT scoring systems to evaluate the extent of lung involvement as a predictor of clinical severity and amount of disease burden.[16,17,18]
Since the burden associated with COVID-19 continues to remain, there is growing attention towards identification of markers of inflammation and disease severity that can help predict survival. Nevertheless, the correlation between CT findings and inflammatory biomarkers in patients with COVID-19 remains unclear. Herein, we aimed to evaluate the predictive value of several inflammatory markers on the extent of lung involvement on chest CT as well as prognosis of patients with COVID-19.
MATERIALS AND METHODS
Study population
This was a single-center cohort study conducted on consecutive patients with laboratory-confirmed COVID-19 who had been admitted to our academic hospital from February 20, 2020 to April 10, 2020. Patients aged <18 years old were excluded.
Patients' demographic and clinical data including presenting signs and symptoms, past medical history and vital signs was collected on admission. Comorbidity was defined as the presence of any of the following conditions: hypertension, diabetes, ischemic heart disease, asthma, chronic lung disease, chronic liver disease, chronic kidney disease (CKD) (defined as estimated glomerular filtration rate below 50 cc/min based on the Modification of Diet in Renal Disease equation), or immunocompromised condition. Furthermore, chest CT scan and laboratory tests were performed for all patients at admission and findings were recorded. All patients were followed until one of the study endpoints, defined as either death due to COVID-19 pneumonia or complete recovery and discharge, were reached.
Ethical considerations
The study protocol was approved by the ethics committee of our institutional review board (Ethical code: IR.SBMU.RETECH.REC.1399.035). All patients provided written informed consent prior to being enrolled in this study. All study procedures were performed in accordance with the ethical standards of the 1964 Declaration of Helsinki and its later amendments.
Imaging studies
As part of national COVID-19 guidelines, all patients underwent low-dose noncontrast chest CT scan at admission.[19] All chest CT scans were performed using a 64-slice scanner (Siemens sensation; Siemens Healthineers, Erlangen, Germany) in supine position during end-inspiration without contrast medium injection and the following scanning parameters were applied: gantry rotation time of 0.5 s, 0.625 mm × 64 detector array, pitch of 1.4, table speed of 45.2 mm/rotation, 20 mAs, 120 kVp, and a 300 × 300 matrix. CARE Dose4D and CARE kV scanning parameters were off. For the purpose of reconstruction (sagittal and coronal), 1 mm slice thickness and 1 mm reconstruction intervals were used. After each scan, disinfection with ethanol and didecyldimethylammonium chloride as well as passive air ventilation was performed.
Two expert radiologists with 9 and 18 years of experience interpreted the images independently. In case of disagreement between the two readers, images were reassessed in order to reach consensus. All the CT scans were reviewed in axial, sagittal and coronal planes. Lung zones were classified as follows: above the carina region as the upper zone, the area between the carina and inferior pulmonary vein as the middle zone and below the inferior pulmonary vein as the lower zone.
The predominant pattern of involvement was assessed and categorized as ground-glass opacity, consolidation, reticular or mixed. The distribution of lesions (peripheral, central or both) was also recorded. In addition, the presence of other imaging features including airway thickening, crazy paving, reverse halo sign, dilated vessels, airway dilatation, air bronchogram, lymphadenopathy (defined as a lymph node with a short axis > 10 mm) and pleural or pericardial effusions was assessed. Extent of lung involvement was evaluated using the following scoring system: 0: no involvement, 1: <25%, 2: 26%–50%, 3: 51%–75% and 4: >75%.[20] The individual specific zone score of one lung was summed up with that of the other lung to calculate bilateral zonal score. In addition, total lung involvement score was measured by summation of all of the zonal scores (maximum score = 24).
Laboratory procedures
On admission, real-time polymerase chain reaction (DAAN gene Co. Ltd) was performed on the throat swab samples of all patients with suspected COVID-19 infection. Routine laboratory tests including complete blood count (CBC) with cell differentiation and serum biochemistry (including renal function test, Troponin, CRP, LDH, creatine phosphokinase) were performed for every patient at admission. Also, data regarding NLR, LCR and PLR was calculated and recorded.
CRP levels were measured using the Rondox essay kit with immunoturbidimetric techniques. To evaluate the NLR and other inflammatory markers, venous blood samples were collected in potassium-ethylene diamine tetraacetic acid tubes (dipotassium EDTA tubes) and the Sysmex-XE 2000i automated blood cell analyzer (Sysmex, Kobe, Japan) was then used to measure the CBC within an hour. This duration is standard for the laboratory of our hospital and helps prevent EDTA-induced swelling.
Statistical analysis
Continuous variables are reported as mean ± standard deviation normally distributed continuous variables were compared between the two groups (death vs. discharge) using independent sample t-test. Skewed continuous data are reported as median (quartile1–quartile3) and Mann–Whitney U-test was utilized to compare this data between the two groups. Categorical variables are presented as frequency (percentage) and Chi-square test with exact P value was applied for comparison between groups. Normality assumption was tested using the Kolmogorov–Smirnov test. Binary logistic regression models were applied to determine the potential risk factors that could be utilized to predict COVID-19-related mortality. The odds ratio (OR) and its 95% confidence intervals were calculated to analyze the intensity and direction of the relationship between risk factors and death. Receiver operator characteristic (ROC) curve analysis was used to evaluate the value of laboratory markers and CT score in predicting death or discharge; optimal cutoff points were determined and their sensitivity and specificity was calculated. Multivariate linear regression method was used for modeling the association between laboratory parameters and total lung score. These scores of involvement were used in multivariate analysis simultaneously and considered as a matrix of dependent variables. The assumptions of errors variance consistency and normality of residuals were checked in regression models and if applicable, appropriate transformation was made to meet the criteria. Also, to handle the problem of collinearity, resulting from highly correlated independent variables, the variance inflation factor (VIF) was calculated for all determinants and covariates with high VIF were removed from analysis, and analysis was continued with stepwise regression methods. All statistical analysis was performed by SPSS version 24 (IBM Corp, Illinois, USA). P <0.05 was considered statistically significant.
RESULTS
Patient's demographic and radio-clinical characteristics
The mean age of patients was 54.2 ± 15.2 years old; 65% were male. Patients' baseline demographic and clinical data is presented in Table 1 in detail. On follow-up, 67 patients (83.7%) were discharged and 13 (16.3%) died. As shown in Table 1, patients' age and sex were not significantly different between discharged and deceased patients (P > 0.05). CKD and immunocompromised condition were the only comorbidities that were significantly more prevalent in those who died (P = 0.01 and P = 0.02). We did not observe a significant difference in the presenting signs and symptoms of patients in either group (P > 0.05). Furthermore, oxygen saturation on room air did not differ significantly between the two groups at the time of admission (P = 0.24).
Table 1.
Patient’s demographic and radio-clinical characteristics at the time of admission
| Total (n=80) | Discharged (n=67) | Deceased (n=13) | P | |
|---|---|---|---|---|
| Age (years) | 54.29±15.21 | 53.85±15.64 | 56.54±13.13 | 0.56 |
| Sex | ||||
| Male | 52 (65.0) | 41 (61.1) | 11 (84.6) | 0.12 |
| Female | 28 (35.0) | 26 (38.8) | 2 (13.4) | |
| Duration of hospitalization (days) | 10.73±7.9 | 9.34±7.42 | 16.38±7.5 | 0.007 |
| O2 saturation (%) | 89.15±5.59 | 89.48±5.34 | 87.46±6.70 | 0.24 |
| Signs and symptoms | ||||
| Fever | 50 (62.5) | 42 (62.7) | 8 (61.5) | 0.94 |
| Cough | 57 (71.3) | 49 (73.1) | 8 (61.5) | 0.51 |
| Sore throat | 10 (12.5) | 9 (13.4) | 1 (7.7) | 0.99 |
| Dyspnea | 51 (63.7) | 44 (65.7) | 7 (53.8) | 0.53 |
| Chilling | 14 (17.5) | 13 (19.4) | 1 (7.7) | 0.45 |
| Headache | 10 (12.5) | 9 (13.4) | 1 (7.7) | 0.99 |
| Myalgia | 21 (26.3) | 18 (26.9) | 3 (23.1) | 0.99 |
| Nausea | 8 (10.0) | 7 (10.4) | 1 (7.7) | 0.99 |
| Abdominal pain | 9 (11.3) | 7 (10.4) | 2 (15.3) | 0.63 |
| Diarrhea | 8 (10.0) | 6 (8.9) | 2 (15.3) | 0.61 |
| Comorbidity status | ||||
| Asthma | 7 (8.8) | 6 (8.9) | 1 (7.7) | 0.99 |
| DM | 12 (15.0) | 11 (16.4) | 1 (7.7) | 0.68 |
| IHD | 15 (18.8) | 11 (16.4) | 4 (30.8) | 0.25 |
| HTN | 20 (25.0) | 18 (26.9) | 2 (15.3) | 0.50 |
| CKD | 19 (23.8) | 12 (17.9) | 7 (53.8) | 0.01 |
| Liver disease | 1 (1.3) | 1 (1.5) | - | 0.99 |
| Immunocompromised condition | 12 (15.0) | 7 (10.4) | 5 (38.5) | 0.02 |
| Comorbidity* | 45 (56.3) | 35 (52.2) | 1 (7.7) | 0.10 |
| Involvement pattern | ||||
| Ground glass opacity | 53 (66.3) | 45 (67.1) | 8 (61.5) | 0.31 |
| Consolidation | 13 (16.3) | 9 (13.4) | 4 (30.7) | |
| Reticular | 8 (10.0) | 8 (11.9) | - | |
| Mixed | 5 (7.5) | 5 (83.3) | 1 (16.7) | |
| Lesion distribution | ||||
| Peripheral | 50 (73.8) | 50 (74.6) | 9 (69.2) | 0.78 |
| Central | 8 (10.0) | 7 (10.4) | 1 (7.7) | |
| Diffuse | 13 (16.2) | 10 (14.9) | 3 (23.1) | |
| Upper zone score | 2 (1-3) | 2 (0-3) | 3 (2-5) | 0.006 |
| Middle zone score | 3 (2-5) | 3 (2-4) | 6 (5-6.5) | <0.001 |
| Lower zone score | 4 (2-6) | 3 (2-5) | 7 (4.5-8) | <0.001 |
| Total score | 9.76±5.73 | 8.67±5.28 | 15.38±4.25 | <0.001 |
| Abnormal imaging findings | ||||
| Airway thickening | 62 (77.5) | 50 (80.6) | 12 (19.4) | 0.28 |
| Crazy paving | 9 (11.3) | 8 (11.9) | 1 (7.7) | 0.99 |
| Reverse halo | 1 (1.3) | 1 (1.5) | - | 0.99 |
| Lymph node | 4 (5.0) | 3 (4.4) | 1 (7.6) | 0.52 |
| Dilated vessels | 55 (68.8) | 42 (62.6) | 13 (100) | 0.007 |
| Airway dilatation | 37 (46.3) | 28 (41.8) | 9 (69.2) | 0.11 |
| Air bronchogram | 25 (31.3) | 18 (26.9) | 7 (53.8) | 0.09 |
| Septal thickening | 11 (13.8) | 9 (13.4) | 2 (15.3) | 0.99 |
| Pericardial effusion | 15 (18.8) | 12 (17.9) | 3 (23.1) | 0.70 |
| Pleural effusion | 14 (17.5) | 11 (16.4) | 3 (23.1) | 0.69 |
| Laboratory parameters | ||||
| WBC count (×109/L) | 5.3 (4.1-7.3) | 5.2 (4.1-6.9) | 6.4 (4.0-8.6) | 0.44 |
| Hb (g/dl) | 13.72±2.60 | 13.94±2.58 | 12.61±2.52 | 0.09 |
| Platelet (×109/L) | 189 (139-241) | 197 (144-251) | 171 (128.8-208) | 0.18 |
| Neutrophil count (×109/L) | 7.2 (6.5-7.9) | 7.2 (6.5-7.7) | 7.5 (6.8-8.9) | 0.09 |
| Lymphocyte count (×109/L) | 1.1 (0.9-1.7) | 1.2 (0.9-1.8) | 1.0 (0.7-1.2) | 0.02 |
| NLR | 2.9 (2.2-4.0) | 2.8 (2.0-3.9) | 3.8 (2.5-11.2) | 0.04 |
| PLR | 76.0 (52.8-119.7) | 74.3 (52.7-113.5) | 93.5 (41.5-202.7) | 0.02 |
| Eosin (%) | 2 (1-2) | 2 (1-2) | 2 (1-3) | 0.67 |
| Troponin (ng/ml×103) | 2 (1-5) | 2 (1-4) | 4 (1-8) | 0.15 |
| Cr (mg/dl) | 1.3 (1.0-1.9) | 1.2 (1.0-1.5) | 2.3 (1.5-4.1) | 0.002 |
| BUN (mg/dl) | 30 (18-50.5) | 28 (16-40) | 93 (40-153.5) | <0.001 |
| CRP (mg/l) | 37.5 (16.3-50) | 36 (14.5-48) | 49.0 (34-57) | 0.04 |
| CPK (IU/l) | 141.5 (52-386) | 153 (55-398) | 94 (43-321) | 0.68 |
| LDH (IU/l) | 434.5 (332-570) | 390 (321-501) | 578 (455-1316) | 0.002 |
| LCR | 0.61 (0.42-1.6) | 0.68 (0.46-2.8) | 0.36 (0.22-0.55) | <0.001 |
Data are represented as mean±SD, median (Q1-Q3) and frequency (percentage). *Defined as the presence of any co-existing morbidity including hypertension, diabetes, ischemic heart disease, asthma, chronic lung disease, chronic liver disease, CKD, or immunocompromised condition. Mean and median differences were tested using independent t-test and Mann-Whitney U test, respectively. The distribution of categorical data was compared by Chi-square test (with exact P value). O2 saturation=Oxygen saturation; DM=Diabetes mellitus; IHD=Ischemic heart disease; HTN=Hypertension; CKD=Chronic kidney disease; WBC=White blood cell; Hb=Hemoglobin; NLR=Neutrophil-to-lymphocyte ratio; PLR=Platelet to lymphocyte ratio; Cr=Creatinine; BUN=Blood urea nitrogen; CRP=C-reactive protein; CPK=Creatine phosphokinase; LDH=Lactate dehydrogenase; LCR=Lymphocyte-to-C-reactive protein; SD=Standard deviation
In terms of laboratory parameters, our results showed that mean serum creatinine and blood urea nitrogen (BUN) were significantly higher among deceased patients (P < 0. 001 and P = 0.002, respectively). Also, we observed that lymphocyte count (P = 0.02) and LCR (P < 0.001) were significantly lower in deceased patients while NLR (P = 0.04), PLR (P = 0.02), CRP (P = 0.04) and LDH (P = 0.002) were markedly elevated in these patients compared with those who survived.
Regarding imaging finds, the involvement pattern did not differ significantly across the two groups (P = 0.31); however, lung involvement scores (upper, middle, lower zone and total) were significantly higher in the deceased group compared with discharged patients [Table 1]. Vessel dilatation was observed in more than 62% of discharged patients while it was detected in all of the deceased patients (P = 0.007).
Association of computed tomography score and laboratory findings with the odds of COVID-19-related death
By applying logistic regression analysis, we aimed to identify the impact of laboratory findings and CT involvement scores on patients' survival status. Table 2 shows the crude and adjusted ORs of different variables. According to the adjusted analysis (that was adjusted for age, gender and comorbidity), greater extent of lung involvement was significantly correlated with increased odds of death as one-point increase in the total lung score raised the odds of death by 26% (P = 0.002) [Table 2].
Table 2.
Adjusted and unadjusted univariate logistic regression analysis to identify factors predictive of coronavirus disease-19-related death
| Crude analysis | Adjusted analysis* | |||
|---|---|---|---|---|
|
|
|
|||
| OR (95% CI) | P | OR (95% CI) | P | |
| Total CT score | 1.27 (1.10-1.45) | 0.001 | 1.26 (1.09-1.45) | 0.002 |
| WBC count | 1.07 (0.88-1.31) | 0.49 | 1.02 (0.82-1.28) | 0.84 |
| Hb | 0.83 (0.66-1.04) | 0.10 | 0.81 (0.64-1.01) | 0.06 |
| Platelet | 0.99 (0.98-1.01) | 0.17 | 0.99 (0.98-1.00) | 0.16 |
| Neutrophil count | 1.76 (0.90-3.42) | 0.09 | 1.70 (0.88-3.30) | 0.12 |
| Lymphocyte count | 0.30 (0.07-1.17) | 0.08 | 0.28 (0.07-1.22) | 0.09 |
| NLR | 1.50 (1.12-2.02) | 0.007 | 1.50 (1.16-1.95) | 0.002 |
| PLR | 1.01 (1.01-1.02) | 0.016 | 1.01 (1.01-1.02) | 0.019 |
| Eosin | 1.03 (0.78-1.36) | 0.84 | 0.96 (0.71-1.28) | 0.76 |
| Troponin | 1.02 (0.99-1.05) | 0.19 | 1.01 (0.98-1.05) | 0.38 |
| Cr | 1.66 (1.13-2.44) | 0.009 | 1.49 (1.01-2.22) | 0.049 |
| BUN | 1.02 (1.01-1.04) | 0.001 | 1.02 (1.01-1.03) | 0.007 |
| CRP | 1.03 (1.01-1.07) | 0.04 | 1.03 (1.00-1.07) | 0.07 |
| CPK | 0.96 (0.78-1.18) | 0.68 | 0.95 (0.75-1.22) | 0.70 |
| LDH | 1.48 (1.13-1.96) | 0.005 | 1.52 (1.13-2.05) | 0.005 |
| LCR | 0.60 (0.44-0.82) | 0.001 | 0.62 (0.45-0.85) | 0.003 |
*Adjusted for age, sex, and presence of comorbidity. WBC=White blood cell; Hb=Hemoglobin; NLR=Neutrophil-to-lymphocyte ratio; PLR=Platelet-to-lymphocyte ratio; Cr=Creatinine; BUN=Blood urea nitrogen; CRP=C-reactive protein; CPK=Creatine phosphokinase; LDH=Lactate dehydrogenase; LCR=Lymphocyte-to-C-reactive protein; CT=Computed tomography; CI=Confidence interval; OR=Odds ratio
As for laboratory parameters, elevated levels of NLR (OR = 1.5, P = 0.002), PLR (OR = 1.01, P = 0.019), serum creatinine (OR = 1.49, P = 0.049), BUN (OR = 1.02, P = 0.007), and LDH (OR = 1.52, P = 0.005) significantly increased the odds of death whereas one-point increase in LCR reduced the odds of death by 38% (P = 0.003). After performing stepwise logistic model and excluding creatinine due to the collinearity issues, the positive associations of the NLR (OR = 1.35, P = 0.04), BUN (OR = 1.02, P = 0.01) and LDH (OR = 1.40, P = 0.03) with increased odds of death remained significant. The negative association of LCR and odds of death was not significant after stepwise model results (OR = 0.98, P = 0.31).
Association between laboratory parameters and total computed tomography score
As shown in Table 3, linear regression analysis revealed that neutrophil count (β =1.29, P = 0.02), NLR (β =1.29, P = 0.02), PLR (β =0.02, P = 0.02), BUN (β =0.05, P = 0.001), CRP (β =0.13, P < 0.001) and LDH (β =1.15, P < 0.001) were positively correlated with total CT score while LCR demonstrated a negative relationship (β = −0.35, P < 0.00) [Figure 1]. After performing stepwise linear regression analysis, the positive association of age (β =0.09, P = 0.005), NLR (β =0.31, P = 0.04), BUN (β =0.03, P = 0.02), and LDH (β =1.08, P < 0.001) remained significant.
Table 3.
Linear regression analysis to identify predictive factors of extent of lung involvement
| Total CT score | ||
|---|---|---|
|
| ||
| β (SE) | P | |
| Age | 0.07 (0.04) | 0.08 |
| Sex (male) | 1.61 (1.34) | 0.23 |
| Comorbidity* | 1.71 (1.28) | 0.19 |
| WBC count | 0.14 (0.23) | 0.56 |
| Hb | −0.47 (0.24) | 0.06 |
| Platelet | −0.006 (0.007) | 0.34 |
| Neutrophil count | 1.29 (0.53) | 0.02 |
| Lymphocyte count | −0.89 (0.55) | 0.11 |
| NLR | 0.69 (0.20) | 0.001 |
| PLR | 0.019 (0.008) | 0.02 |
| Eosin | −0.22 (0.33) | 0.51 |
| Troponin | −0.005 (0.04) | 0.89 |
| Cr | 0.27 (0.48) | 0.57 |
| BUN | 0.046 (0.013) | 0.001 |
| CRP | 0.13 (0.03) | <0.001 |
| CPK | 0.08 (0.23) | 0.73 |
| LDH | 1.15 (0.18) | <0.001 |
| LCR | −0.35 (0.09) | <0.001 |
*Defined as the presence of any co-existing morbidity including hypertension, diabetes, ischemic heart disease, asthma, chronic lung disease, chronic liver disease, CKD or immunocompromised condition. WBC=White blood cell; Hb=Hemoglobin; NLR=Neutrophil-to-lymphocyte ratio; PLR=Platelet-to-lymphocyte ratio; Cr=Creatinine; BUN=Blood urea nitrogen; CRP=C-reactive protein; CPK=Creatine phosphokinase; LDH=Lactate dehydrogenase; LCR=Lymphocyte-to-C-reactive protein; CT=Computed tomography; SE=Standard error
Figure 1.
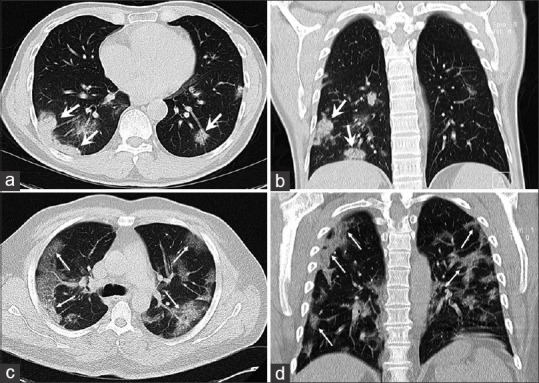
(a and b) A 49-year-old male patient with COVID-19 presenting with fever, dyspnea, sore throat and a lymphocyte count within the normal range (neutrophil to lymphocyte ratio: 1.0, lymphocyte to C-reactive protein ratio: 2.8). A computed tomography scan obtained 4 days after the onset of symptoms showed bilateral peripheral multifocal patchy consolidations predominantly in the lower lobes with subtle air bronchogram (thick arrows). (c and d) 52-year-old man with positive reverse transcription polymerase chain reaction for severe acute respiratory syndrome coronavirus 2 manifesting with initial symptoms of fever, cough, dyspnea, myalgia and lymphocytopenia (neutrophil-to-lymphocyte ratio: 11.1, lymphocyte-to-C-reactive protein ratio: 0.23). A computed tomography scan obtained 4 days after the onset of symptoms showed bilateral diffused peripheral ground glass opacities with crazy-paving pattern (thin arrows)
Optimal threshold of inflammatory markers and computed tomography score for predicting survival
The ROC curve analysis of NLR, PLR, LCR and total CT score for predicting survivors from nonsurvivors are shown in Figure 2. The area under curve (AUC) for NLR, PLR, LCR and total CT score was 0.70, 0.62, 0.82 and 0.83, respectively. Since the AUC for PLR was not statistically significant (P = 0.19), PLR was not included in further analysis. The optimal threshold value of NLR, LCR and total CT score was ≥3.25, ≤0.53, and ≥12, respectively. The specificity and sensitivity of the determined cut-off points are reported in Table 4. Among the investigated variables, total CT score had the best performance for predicting survival with 81% sensitivity and 79% specificity.
Figure 2.
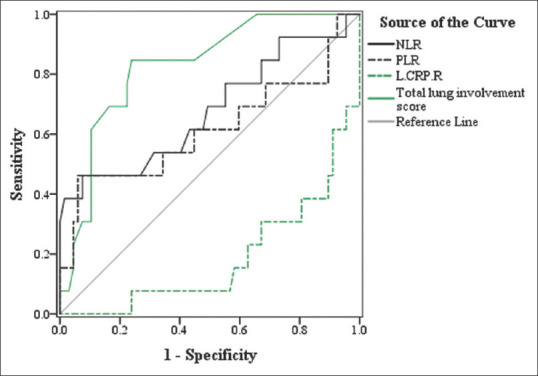
Receiver operator characteristic curve analysis of neutrophil to lymphocyte ratio, platelet to lymphocyte ratio, lymphocyte to C-reactive protein ratio, and total computed tomography score for discriminating deceased from discharged patients
Table 4.
The results of receiver operator characteristics curve analysis of inflammatory biomarkers and total lung involvement score for discriminating deceased from discharged patients
| AUC (95% CI) | P | Cutoff point | Sensitivity | Specificity | |
|---|---|---|---|---|---|
| NLR | 0.70 (0.51-0.86) | 0.040 | ≥3.25 | 0.62 | 0.60 |
| PLR | 0.62 (0.41-0.82) | 0.19 | - | - | - |
| LCR | 0.82 (0.69-0.94) | <0.001 | ≤0.53 | 0.78 | 0.74 |
| Total CT score | 0.83 (0.71-0.94) | <0.001 | ≥12 | 0.81 | 0.79 |
AUC=Area under curve; CI=Confidence interval; NLR=Neutrophil-to-lymphocyte ratio; PLR=Platelet-to-lymphocyte ratio; LCR=Lymphocyte-to-C-reactive protein; CT=Computed tomography
DISCUSSION
Inflammation significantly contributes to the pathogenesis and prognosis of COVID-19.[21] During severe course of this disease, patients might experience an exaggerated and uncontrollable inflammatory response, known as the “cytokine storm,” which increases the chance of death.[22] Therefore, factors that convey information about the status of inflammation and immunity could be used as potential predictors of disease severity and can help identify patients who are at higher risk of rapid clinical deterioration; this will promote timely and efficient patient management and optimize allocation of healthcare resources. In the present study, we showed that inflammatory markers such as NLR, PLR, LCR and LDH are related with the odds of COVID-19-related death. Interestingly, these factors were positively associated with more severe lung involvement on imaging. Moreover, we observed that the extent of lung involvement (assessed by a CT scoring system) is an excellent predictor of survival, with high sensitivity and specificity and that higher CT scores are associated with increased chance of mortality.
In general, the immune system responds rapidly to a viral infection by initially activating the components of the innate immunity including Type I interferons, monocytes, macrophages and neutrophils. If necessary, a delayed but more complex adaptive immunity that is mainly mediated by lymphocytes, steps into action to eliminate the pathogen and develop long-lasting immune response.[23] Experience from past pandemics caused by SARS-CoV and middle east respiratory syndrome (MERS)-CoV infection shows that white blood cells have an undeniable role in manipulating inflammatory response and subsequently, altering the risk of disease progression. Similarly, in COVID-19, hyper-activated neutrophils and monocytes/macrophages have been shown to play a major role in sustained inflammation and triggering the so-called “cytokine storm.” Furthermore, decreased lymphocyte count has been implicated as an important risk factor for ARDS and mortality in patients with COVID-19.[24] Thus, it is evident that laboratory parameters that represent alterations in the hematological system should have great value for predicting disease burden and assessing the risk of mortality. Moreover, their cost-effectiveness and feasibility, due to being performed in everyday clinical practice, turns them into ideal prognostic markers.
In this aspect, several studies have shown that absolute neutrophil and lymphocyte count as well as NLR, which represents the proportion of these cells, and LCR are strong indicators of advanced disease in patients with COVID-19.[25,26] In line with these findings, in our cohort of patients, those who experienced death presented with significantly lower lymphocyte count and LCR and increased NLR at admission. We also observed that after adjusted analysis, higher values of NLR and decreased LCR were associated with an increase in the odds of death. It is proposed that the decrease in lymphocyte count could be due to the cytokine storm which causes SARS-CoV-2-induced lysis and atrophy of lymphoid organs, including the spleen and lymph nodes.[27,28] Although the relationship between inflammatory cells and biomarkers with the progression of pneumonia has not been studied in the context of COVID-19, previous studies in patients with MERS-CoV suggested that lymphocytopenia and elevated CRP level are predictive of pneumonia development and its progression into ARDS.[29] Interestingly, increased pro-inflammatory cells and cytokines in the sera of patients with aberrant pulmonary inflammation and extensive lung damage were also reported in SARS-CoV.[30] The results of our study approved that increased levels of circulatory inflammatory biomarkers such as NLR and neutrophil are associated with a greater extent of lung involvement on chest CT scan. Also, decreased values of LCR were seen in patients with higher total CT score. By using ROC curve analysis, we also assessed the predictive value of LCR and NLR for distinguishing between survivors and nonsurvivors of COVID-19. At a cut-off point of 3.25, NLR had 60% sensitivity and 62% specificity for predicting survival. Since a similar optimal threshold has been reported in many previous studies,[14,31,32] our finding supports existing evidence to implement NLR along with this cut-off point as an important prognostic indicator in daily clinical practice as well as in future management guidelines. Nevertheless, we observed that LCR performs better in predicting survival with an AUC of 0.82 (95% confidence interval [CI]: 0.69–0.94, P < 0.00), 78% sensitivity and 74% specificity at a cut-off point of 0.52. To our knowledge, no study has yet investigated the optimal threshold of this biomarker for predicting survival of patients with COVID-19. Thus, further studies are recommended to shed light on our finding.
Besides neutrophils and lymphocytes, thrombocytopenia has also been reported in patients with severe COVID-19.[33,34] Despite this, our study failed to show significant difference in the platelet count of patients who died and those who were discharged. In addition, we did not find any association between platelet count and increased odds of death or more severe lung involvement. However, PLR is another novel biomarker that has been evidenced to be prognostic for COVID-19.[31,35] Due to the interactions between platelets and lymphocytes, PLR indicates both aggregation and inflammation and might be a more sensitive marker for the intensity of systemic inflammation rather than platelets or lymphocytes alone. Consistent with this, the findings of our study showed that PLR is an indicator of both increased odds of death and higher CT score.
Severe infections result in cytokine-mediated tissue damage and release of LDH.[36] LDH is largely present in lung tissue, thus, as like with other respiratory infections such as the MERS, patients with severe COVID-19 infection are also likely to have increased amounts of LDH.[37] Many previous reports have demonstrated an association between elevated LDH values and poor outcome in patients with COVID-19.[2,38,39] It has been reported that LDH can result in 16-fold increase in the odds of COVID-19-related mortality.[40] We also showed that LDH levels were significantly higher in deceased patient. Furthermore, LDH was predictive of mortality, with a one-unit rise in the level of LDH increasing the chance of death by approximately 50%. We also observed a significant relation between higher levels of LDH and greater extent of lung involvement.
One of the limitations of our study was that this was a single center study. Another limitation was the relatively small sample size. In addition, assessment of laboratory data at different time points during the disease course will provide further detailed insight. Thus, we recommend future investigations to overcome these limitations to obtain optimal results.
CONCLUSION
Inflammation plays a major role in the progression of COVID-19. We showed that increased level of inflammatory biomarkers such as NLR, PLR, and LDH and decreased LCR are associated with more extensive lung involvement and increased risk of COVID-19-related death. In addition, lung involvement ≥50% on chest CT was an excellent predictor of death (AUC = 0.83, 95% CI: 0.71–0.94) with 81% sensitivity and 79% specificity. These findings suggest that routinely-performed, inexpensive laboratory data that are markers of inflammation have great value for risk-stratification of patients with COVID-19.
Financial support and sponsorship
Nil.
Conflicts of interest
There are no conflicts of interest.
Acknowledgments
We would like to thank all of the patients who participated in this study.
REFERENCES
- 1.Li Q, Guan X, Wu P, Wang X, Zhou L, Tong Y, et al. Early transmission dynamics in Wuhan, China, of novel coronavirus-infected pneumonia. N Engl J Med. 2020;382:1199–207. doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhou F, Yu T, Du R, Fan G, Liu Y, Liu Z, et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: A retrospective cohort study. Lancet. 2020;395:1054–62. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Roxburgh CS, McMillan DC. Role of systemic inflammatory response in predicting survival in patients with primary operable cancer. Future Oncol. 2010;6:149–63. doi: 10.2217/fon.09.136. [DOI] [PubMed] [Google Scholar]
- 4.Viganó A, Bruera E, Jhangri GS, Newman SC, Fields AL, Suarez-Almazor ME. Clinical survival predictors in patients with advanced cancer. Arch Intern Med. 2000;160:861–8. doi: 10.1001/archinte.160.6.861. [DOI] [PubMed] [Google Scholar]
- 5.Gao Y, Li T, Han M, Li X, Wu D, Xu Y, et al. Diagnostic utility of clinical laboratory data determinations for patients with the severe COVID-19. J Med Virol. 2020;92:791–6. doi: 10.1002/jmv.25770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ji D, Zhang D, Xu J, Chen Z, Yang T, Zhao P, et al. Prediction for progression risk in patients with COVID-19 pneumonia: The CALL score. Clin Infect Dis. 2020;71:1393–9. doi: 10.1093/cid/ciaa414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu X, Shen Y, Wang H, Ge Q, Fei A, Pan S. Prognostic Significance of Neutrophil-to-Lymphocyte Ratio in Patients with Sepsis: A Prospective Observational Study. Mediators Inflamm. 2016;2016:8191254. doi: 10.1155/2016/8191254. doi: 10.1155/2016/8191254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bucak A, Ulu S, Oruc S, Yucedag F, Tekin MS, Karakaya F, et al. Neutrophil-to-lymphocyte ratio as a novel-potential marker for predicting prognosis of bell palsy. Laryngoscope. 2014;124:1678–81. doi: 10.1002/lary.24551. [DOI] [PubMed] [Google Scholar]
- 9.Templeton AJ, McNamara MG, Šeruga B, Vera-Badillo FE, Aneja P, Ocaña A, et al. Prognostic role of neutrophil-to-lymphocyte ratio in solid tumors: A systematic review and meta-analysis. J Natl Cancer Inst. 2014;106:dju124. doi: 10.1093/jnci/dju124. [DOI] [PubMed] [Google Scholar]
- 10.Shen Y, Huang X, Zhang W. Platelet-to-lymphocyte ratio as a prognostic predictor of mortality for sepsis: Interaction effect with disease severity-a retrospective study. BMJ Open. 2019;9:e022896. doi: 10.1136/bmjopen-2018-022896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheng CB, Zhang QX, Zhuang LP, Sun JW. Prognostic value of lymphocyte-to-C-reactive protein ratio in patients with gastric cancer after surgery: A multicentre study. Jpn J Clin Oncol. 2020;50:1141–9. doi: 10.1093/jjco/hyaa099. [DOI] [PubMed] [Google Scholar]
- 12.Lagunas-Rangel FA. Neutrophil-to-lymphocyte ratio and lymphocyte-to-C-reactive protein ratio in patients with severe coronavirus disease 2019 (COVID-19): A meta-analysis. J Med Virol. 2020;92:1733–4. doi: 10.1002/jmv.25819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Qu R, Ling Y, Zhang YH, Wei LY, Chen X, Li XM, et al. Platelet-to-lymphocyte ratio is associated with prognosis in patients with coronavirus disease-19. J Med Virol. 2020;92:1533–41. doi: 10.1002/jmv.25767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yufei Y, Mingli L, Xuejiao L, Xuemei D, Yiming J, Qin Q, et al. Utility of the neutrophil-to-lymphocyte ratio and C-reactive protein level for coronavirus disease 2019 (COVID-19) Scand J Clin Lab Invest. 2020;80:536–40. doi: 10.1080/00365513.2020.1803587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fang Y, Zhang H, Xie J, Lin M, Ying L, Pang P, et al. Sensitivity of chest CT for COVID-19: Comparison to RT-PCR. Radiology. 2020;296:E115–7. doi: 10.1148/radiol.2020200432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Colombi D, Bodini FC, Petrini M, Maffi G, Morelli N, Milanese G, et al. Well-aerated lung on admitting chest CT to predict adverse outcome in COVID-19 pneumonia. Radiology. 2020;296:E86–96. doi: 10.1148/radiol.2020201433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wynants L, Van Calster B, Collins GS, Riley RD, Heinze G, Schuit E, et al. Prediction models for diagnosis and prognosis of COVID-19 infection: Systematic review and critical appraisal. BMJ. 2020;369:m1328. doi: 10.1136/bmj.m1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Francone M, Iafrate F, Masci GM, Coco S, Cilia F, Manganaro L, et al. Chest CT score in COVID-19 patients: correlation with disease severity and short-term prognosis. Eur Radiol. 2020;30:6808–17. doi: 10.1007/s00330-020-07033-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Radpour A, Bahrami-Motlagh H, Taaghi MT, Sedaghat A, Karimi MA, Hekmatnia A, et al. COVID-19 evaluation by low-dose high resolution CT scans protocol. Acad Radiol. 2020;27:901. doi: 10.1016/j.acra.2020.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li K, Fang Y, Li W, Pan C, Qin P, Zhong Y, et al. CT image visual quantitative evaluation and clinical classification of coronavirus disease (COVID-19) Eur Radiol. 2020;30:4407–16. doi: 10.1007/s00330-020-06817-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Feng X, Li S, Sun Q, Zhu J, Chen B, Xiong M, et al. Immune-inflammatory parameters in COVID-19 cases: A systematic review and meta-analysis. Front Med (Lausanne) 2020;7:301. doi: 10.3389/fmed.2020.00301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ragab D, Salah Eldin H, Taeimah M, Khattab R, Salem R. The COVID-19 cytokine storm; what we know so far. Front Immunol. 2020;11:1446. doi: 10.3389/fimmu.2020.01446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ortiz-Prado E, Simbaña-Rivera K, Gómez-Barreno L, Rubio-Neira M, Guaman LP, Kyriakidis NC, et al. Clinical, molecular, and epidemiological characterization of the SARS-CoV-2 virus and the coronavirus disease 2019 (COVID-19), a comprehensive literature review. Diagn Microbiol Infect Dis. 2020;98:115094. doi: 10.1016/j.diagmicrobio.2020.115094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ponti G, Maccaferri M, Ruini C, Tomasi A, Ozben T. Biomarkers associated with COVID-19 disease progression. Crit Rev Clin Lab Sci. 2020;57:389–99. doi: 10.1080/10408363.2020.1770685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qin C, Zhou L, Hu Z, Zhang S, Yang S, Tao Y, et al. Dysregulation of immune response in patients with coronavirus 2019 (COVID-19) in Wuhan, China. Clin Infect Dis. 2020;71:762–8. doi: 10.1093/cid/ciaa248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Słomka A, Kowalewski M, Żekanowska E. Coronavirus Disease 2019 (COVID-19): A Short Review on Hematological Manifestations. Pathogens. 2020;9:493. doi: 10.3390/pathogens9060493. doi: 10.3390/pathogens9060493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Soy M, Keser G, Atagündüz P, Tabak F, Atagündüz I, Kayhan S. Cytokine storm in COVID-19: Pathogenesis and overview of anti-inflammatory agents used in treatment. Clin Rheumatol. 2020;39:2085–94. doi: 10.1007/s10067-020-05190-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang W, Zhao Y, Zhang F, Wang Q, Li T, Liu Z, et al. The use of anti-inflammatory drugs in the treatment of people with severe coronavirus disease 2019 (COVID-19): The perspectives of clinical immunologists from China. Clin Immunol (Orlando, Fla) 2020;214:108393. doi: 10.1016/j.clim.2020.108393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ko JH, Park GE, Lee JY, Lee JY, Cho SY, Ha YE, et al. Predictive factors for pneumonia development and progression to respiratory failure in MERS-CoV infected patients. J Infect. 2016;73:468–75. doi: 10.1016/j.jinf.2016.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lam CW, Chan MH, Wong CK. Severe acute respiratory syndrome: clinical and laboratory manifestations. Clin Biochem Rev. 2004;25:121–32. [PMC free article] [PubMed] [Google Scholar]
- 31.Yang AP, Liu JP, Tao WQ, Li HM. The diagnostic and predictive role of NLR, d-NLR and PLR in COVID-19 patients. Int Immunopharmacol. 2020;84:106504. doi: 10.1016/j.intimp.2020.106504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Eid M, Al-Kaisy M, Regeia W, Jiwa Khan H. The Prognostic Accuracy of Neutrophil-Lymphocyte Ratio in COVID-19 Patients. Front Emerg Med. 5:e8. [Google Scholar]
- 33.Lippi G, Plebani M, Henry BM. Thrombocytopenia is associated with severe coronavirus disease 2019 (COVID-19) infections: A meta-analysis. Clin Chim Acta. 2020;506:145–8. doi: 10.1016/j.cca.2020.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bomhof G, Mutsaers PG, Leebeek FW, Te Boekhorst PA, Hofland J, Croles FN, et al. COVID-19-associated immune thrombocytopenia. Br J Haematol. 2020;190:e61–4. doi: 10.1111/bjh.16850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Allegra A, Di Gioacchino M, Tonacci A, Musolino C, Gangemi S. Immunopathology of SARS-CoV-2 Infection: Immune Cells and Mediators, Prognostic Factors, and Immune-Therapeutic Implications? Int J Mol Sci. 2020;21:4782. doi: 10.3390/ijms21134782. doi: 10.3390/ijms21134782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martinez-Outschoorn UE, Prisco M, Ertel A, Tsirigos A, Lin Z, Pavlides S, et al. Ketones and lactate increase cancer cell “stemness,” driving recurrence, metastasis and poor clinical outcome in breast cancer: Achieving personalized medicine via Metabolo-Genomics. Cell Cycle. 2011;10:1271–86. doi: 10.4161/cc.10.8.15330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Assiri A, Al-Tawfiq JA, Al-Rabeeah AA, Al-Rabiah FA, Al-Hajjar S, Al-Barrak A, et al. Epidemiological, demographic, and clinical characteristics of 47 cases of Middle East respiratory syndrome coronavirus disease from Saudi Arabia: A descriptive study. Lancet Infect Dis. 2013;13:752–61. doi: 10.1016/S1473-3099(13)70204-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guan W, Ni Z, Hu Y, Liang W, Ou C, He J, et al. Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med. 2020;382:1708–20. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Henry BM, Aggarwal G, Wong J, Benoit S, Vikse J, Plebani M, et al. Lactate dehydrogenase levels predict coronavirus disease 2019 (COVID-19) severity and mortality: A pooled analysis. Am J Emerg Med. 2020;38:1722–6. doi: 10.1016/j.ajem.2020.05.073. [DOI] [PMC free article] [PubMed] [Google Scholar]


