SUMMARY
APC mutation activation of Wnt/β-catenin drives initiation of colorectal carcinogenesis (CRC). Additional factors potentiate β-catenin activation to promote CRC. Western diets are enriched in linoleic acid (LA); LA-enriched diets promote chemically induced CRC in rodents. 15-Lipoxygenase-1 (15-LOX-1), the main LA-metabolizing enzyme, is transcriptionally silenced during CRC. Whether LA and 15-LOX-1 affect Wnt/β-catenin signaling is unclear. We report that high dietary LA promotes CRC in mice treated with azoxymethane or with an intestinally targeted Apc mutation (ApcΔ580) by upregulating Wnt receptor LRP5 protein expression and β-catenin activation. 15-LOX-1 transgenic expression in mouse intestinal epithelial cells suppresses LRP5 protein expression, β-catenin activation, and CRC. 15-LOX-1 peroxidation of LA in phosphatidylinositol-3-phosphates (PI3P_LA) leads to PI3P_13-HODE formation, which decreases PI3P binding to SNX17 and LRP5 and inhibits LRP5 recycling from endosomes to the plasma membrane, thereby increasing LRP5 lysosomal degradation. This regulatory mechanism of LRP5/Wnt/β-catenin signaling could be therapeutically targeted to suppress CRC.
Graphical Abstract
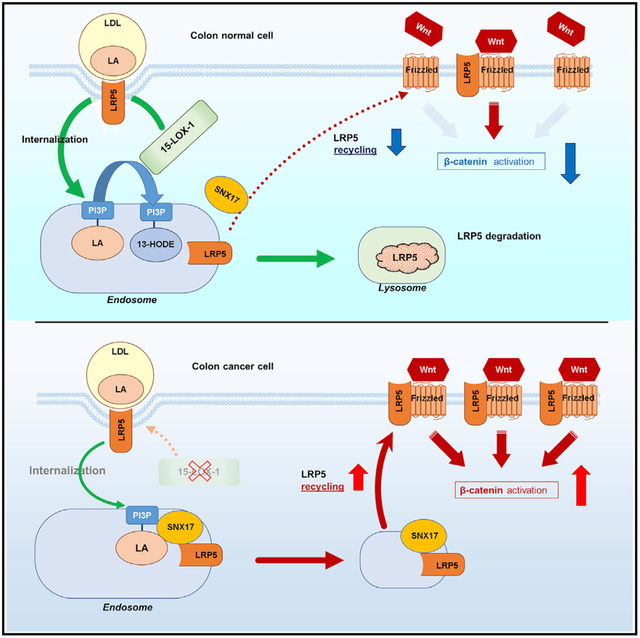
In Brief
Whether Western diet enrichment with linoleic acid promotes colorectal carcinogenesis is poorly understood. Liu et al. identify a mechanism by which 15-lipoxygenase-1 peroxidation of linoleic acid in phosphatidylinositol-3-phosphate (PI3P_LA), to induce PI3P_13-HODE formation, attenuates PI3P binding to the SNX17-LRP5 complex. This inhibits LRP5 recycling, Wnt/β-catenin activation, and colorectal carcinogenesis.
INTRODUCTION
Colorectal cancer is the third leading cause of cancer deaths in the United States (Siegel et al., 2020). APC mutations, which occur early in most colorectal cancers (Losi et al., 2005), increase β-catenin activation, driving the initiation of colorectal carcinogenesis (CRC) (Fearon and Vogelstein, 1990; Clevers and Nusse, 2012). β-catenin activation varies significantly even within the same colorectal cancer case that has the same APC mutation (Vermeulen et al., 2010; Brabletz et al., 1998). This variability has been attributed to modifiable factors that can strongly affect CRC progression via β-catenin hyperactivation (He et al., 2005; Suzuki et al., 2004; Vermeulen et al., 2010). The identification of these modifiable factors could open important therapeutic targeting opportunities, unlike APC mutations, which are un-modifiable by current therapeutic approaches.
Linoleic acid (LA) is the most commonly consumed n-6 polyunsaturated fatty acid (PUFA) in Western diets (Adam et al., 2008; Shureiqi et al., 2010) and promotes chemically induced CRC in rodents (Deschner et al., 1990; Lipkin et al., 1999). Never-theless, human diets, especially in the United States, have been increasingly enriched with LA during the past six decades, in response to the notion that LA decreases the risk for coronary artery disease (Farvid et al., 2014; Blasbalg et al., 2011). This notion has recently been challenged by new findings showing that substitution of dietary saturated fats with LA increased car-diovascular disease risk (Ramsden et al., 2013). While this debate continues, the effect of dietary LA on CRC risk remains poorly defined. Results of dietary LA studies in human CRC remain inconclusive (Jandacek, 2017), possibly because they depend on participants’ recall of dietary intake. In preclinical models, however, in which dietary factors can be strictly controlled, LA promoted azoxymethane (AOM)-induced CRC in rodents (Lipkin et al., 1999; Deschner et al., 1990). Oxidative metabolism of n-6 PUFAs such as LA influences their ability to promote AOM-induced CRC in rodents (Bull et al., 1989). LA’s oxidative metabolism occurs mainly through a pathway regulated by 15-lipoxygenase-1 (15-LOX-1) to generate 13-S-hydroxyoctadecadienoic acid (13[S]-HODE). 15-LOX-1 is downregulated in more than 60% of advanced colorectal adenomas and 100% of human invasive CRCs (Yuri et al., 2007; Shureiqi et al., 1999), and re-expression of 15-LOX-1 suppresses CRC in various preclinical models (Shureiqi et al., 2003, 2005; Nixon et al., 2004; Wu et al., 2008; Zuo et al., 2012; Mao et al., 2015).
Whether this alteration in LA oxidative metabolism via 15-LOX-1 affects aberrant Wnt/β-catenin signaling in CRC is poorly understood. The few available findings are from studies of 12/15Lox in non-cancer mouse models, and these studies have produced conflicting results, suggesting that 12/15Lox may either inhibit (Almeida et al., 2009) or stimulate (Kinder et al., 2010) β-catenin transcriptional activity. Experimental modeling of 15-LOX-1 via its murine homolog, 12/15Lox, is suboptimal because 12/15Lox is a hybrid enzyme of 15- and 12-LOX, whose products have opposing biological effects, especially in tumorigenesis (Liu et al., 1995; Müller et al., 2002). Thus, important questions remain: do LA and its oxidative metabolism by 15-LOX-1 affect Wnt/β-catenin signaling and subsequently CRC? If so, by what mechanisms? These questions are important because increasing dietary LA intake could inadvertently enhance the risk for CRC, especially when 15-LOX-1 is downregulated, as commonly occurs in CRC (Yuri et al., 2007; Shureiqi et al., 1999). The necessity of addressing these questions is further supported by the recent report that LA has an alternative 15-LOX-1-independent oxidative metabolism pathway via cytochrome P450 monooxygenases to produce 12,13-epoxyoctadecenoic acid, which promotes CRC (Wang et al., 2019).
The findings of this study address these questions: (1) high dietary LA promoted CRC by upregulating the expression of a Wnt receptor, low-density lipoprotein (LDL) receptor-related protein 5 (LRP5), and thus increasing aberrant β-catenin activation in intestinal epithelial cells (IECs), and (2) 15-LOX-1 re-expression in IECs suppressed CRC by promoting peroxidation of LA in phosphatidylinositol-3-phosphate (PI3P_LA) into PI3P_13-HODE, which decreased PI3P binding to sorting nexin 17 (SNX17) and LRP5 and subsequently inhibited LRP5 recycling from endosomes to the plasma membrane, thus increasing LRP5 lysosomal degradation.
RESULTS
15-LOX-1 Suppresses LA Promotion of AOM-Induced or Apc Mutation-Induced CRC and Aberrant β-catenin Activation in Mice
We first examined the effects of 15-LOX-1 transgenic expression (Figure S1A) on dietary LA promotion of AOM-induced CRC by feeding 15-LOX-1 mice (with 15-LOX-1 transgenic overexpression in IECs) (Zuo et al., 2012) and their wild-type (WT) littermates with standardized iso-caloric diets in which corn oil (containing 50%–55% LA) comprised either 5% of diet weight, considered low, or 20% of diet weight, which simulates the high LA content of Western diets (Deschner et al., 1990; Yang et al., 1996) (Table S1). In WT mice, AOM induced significantly more colorectal tumors per mouse in those fed 20% corn oil than in those fed 5% corn oil (Figures S1B and S1C; Table S2). In contrast, in 15-LOX-1 mice, the number of tumors per mouse was similar between those fed 20% corn oil and those fed 5% corn oil. Moreover, the 15-LOX-1 mice fed 20% corn oil had significantly fewer tumors per mouse than did the corresponding WT littermates fed the same diets (Figure S1C; Table S2).
To simulate 15-LOX-1 expression downregulation in human CRC (Shureiqi et al., 1999, 2010; Nixon et al., 2004; Yuri et al., 2007), we sought to generate a mouse model with 15-LOX-1 loss of function in IECs. The generation of a 15-LOX-1-knockout (KO) mouse model has been difficult because the 15-LOX-1 homolog in mice is 12/15Lox, which has hybrid enzymatic functions that can produce both 15-LOX-1 and 12S-LOX products (i.e., 13 [S]-HODE and 12[S]-hydroxyeicosatetraenoic [HETE]) (Müller et al., 2002; Liu et al., 1995; Grossi et al., 1989). 12/15Lox-KO (12/15LoxKO) mice have significantly reduced 13(S)-HODE and 12-HETE levels (Middleton et al., 2006). To compensate for the decreased 12-HETE levels in 12/15LoxKO mice and thus generate mice with more selective 15-LOX-1 loss of function, we bred 12/15LoxKO mice with a 12-LOX mouse model that specifically expresses 12S-LOX in IECs via villin promoter (12-LOX mice), generated by nuclear injection of a construct of villin promoter-directed human 12S-LOX expression into ICEs (Figures S1A and S1D–S1G). Breeding of 12/15LoxKO and 12-LOX mice generated 12/15LoxKO/12-LOX mice. Additionally, we bred 12/15LoxKO mice with 15-LOX-1 mice to generate 12/15LoxKO/15-LOX-1 mice (Figures S1H and S1I) to assess the effects of restoring 15-LOX-1 function in 12/15LoxKO mice on CRC.
We found that in IECs, 12/15LoxKO mice had significantly decreased 12-HETE levels compared with WT mice, and these levels were significantly increased by 12-LOX expression (12/15LoxKO/12-LOX) (Figure S1J). 12/15LoxKO mice also had significantly decreased 13-HODE levels in IECs compared with WT mice, and these levels were significantly increased by 15-LOX-1 but not 12-LOX expression (Figure S1K). 15-HETE levels were similar between WT and 12/15LoxKO mice but significantly increased by 15-LOX-1 expression (Figure S1L).
12/15LoxKO mice had significantly more tumors on the 5% corn oil diet and trended toward more tumors on the 20% corn oil diet than WT mice fed the same diets (Figure 1A). 12/15LoxKO/12-LOX mice had further increases in tumor numbers when fed 5% or 20% corn oil, and these effects were reversed in 12/15LoxKO/15-LOX-1 mice (Figures 1A–1C; Table S2). These changes were more evident for large tumors (diameter > 3 mm) (Figure 1B).
Figure 1. 15-LOX-1 Suppresses Linoleic Acid (LA) Promotion of Colorectal Carcinogenesis and Aberrant β-catenin Activation in Mice.
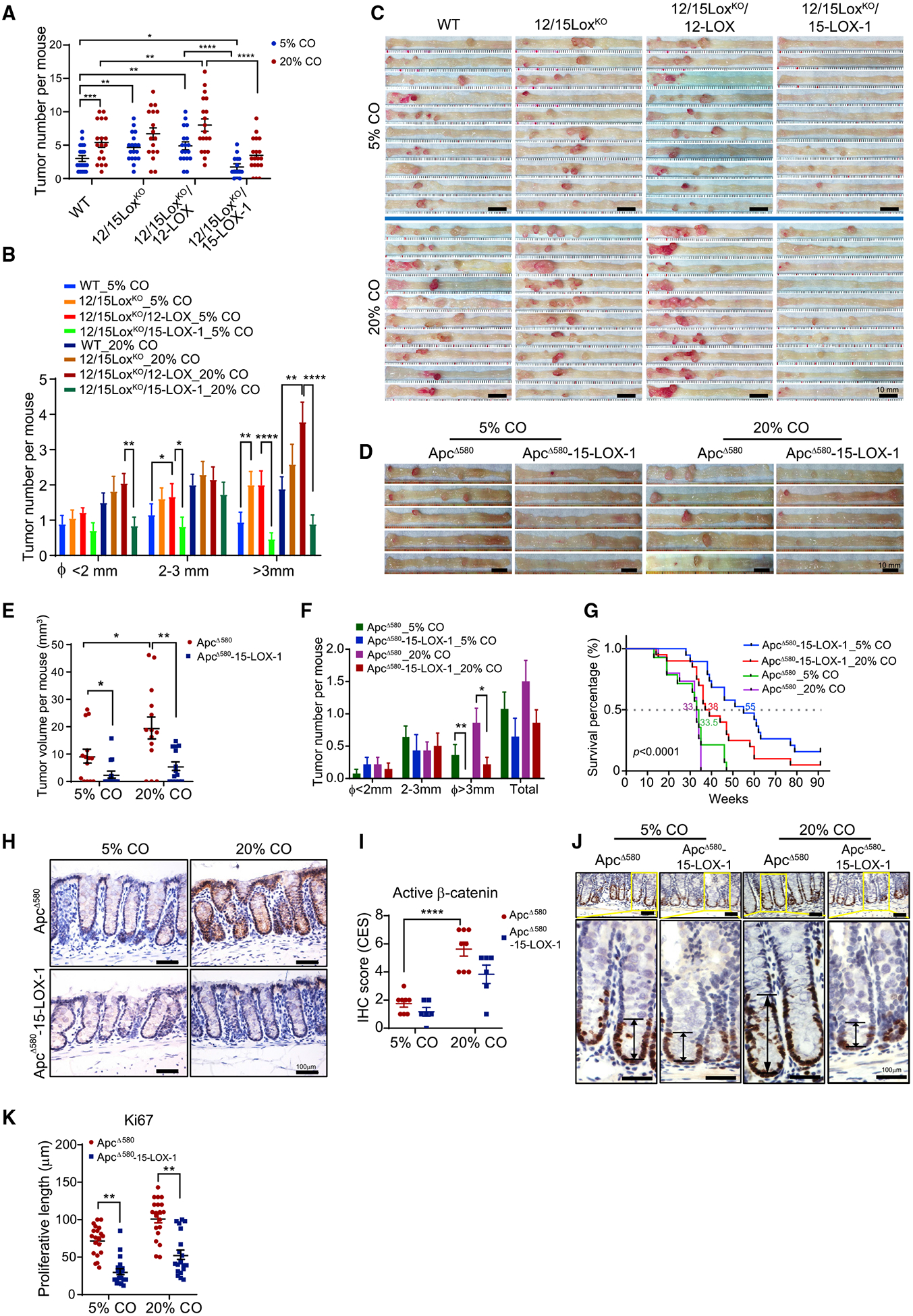
(A–C) 15-LOX-1 modulation of LA’s effects on azoxymethane (AOM)-induced colorectal carcinogenesis (CRC) in mice. FVB wild-type (WT), germline homozygous 12/15-LOX-knockout (12/15LoxKO), 12/15LoxKO plus villin promoter-driven 12-LOX expression in intestines (12/15LoxKO/12-LOX), and 12/15LoxKO plus villin promoter-driven 15-LOX-1 expression in intestines (12/15LoxKO/15-LOX-1) mice were fed either a 5% or a 20% corn oil (CO) diet and treated with AOM (10 mg/kg) via six weekly intraperitoneal injections to induce CRC. CRC was evaluated 20 weeks after completion of AOM injections. The number of tumors per mouse (A) and tumor size distributions (B) for the indicated groups (n = 17–19 per group) are shown. (C) Representative colon images are shown for the indicated mouse groups (n = 10 per group).
(D–F) ApcΔ580(+/−) (designated as ApcΔ580) and ApcΔ580 (+/−)-15-LOX-1(+/+) (designated as ApcΔ580-15-LOX-1) littermates at 4 weeks were fed either 5% or 20% CO and euthanized 10 weeks later (n = 14 mice per group). Representative colon images (D), colonic tumor volumes per mouse (E), and colonic tumor numbers and size distributions per mouse (F) of the indicated mouse groups are shown.
(G) Survival curves of ApcΔ580 and ApcΔ580-15-LOX-1 littermates at 4 weeks fed either 5% or 20% CO. The median survival age (weeks) for each group is shown (n = 14–20 mice per group). The survival time was calculated using the Kaplan-Meier method and compared between groups using the log rank test.
(H and I) Representative immunohistochemistry staining images (H) and quantitative composite expression scores (CES) (I) of active β-catenin expression and localization in normal colonic tissues from the indicated mouse groups as described in (D)–(F) (n = 6–8 mice per group).
(J and K) Representative immunohistochemistry staining images for cell proliferation marker Ki-67 (J) and corresponding proliferation zone lengths (K) in normal colonic tissues from the indicated mouse groups as described in (D)–(F) (30 crypts from three mice per group were counted).
Values represent mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001; two-sided Poisson (A, B, and F) and two-way ANOVA (E, I, and K).
To ensure that LA promotion of CRC and its modulation by 15-LOX-1 are not limited to the AOM carcinogenesis model, we examined the effects of 15-LOX-1 and the same low and high corn oil diets on Apc mutation-driven CRC using ApcΔ580 mice without and with transgenic 15-LOX-1 expression in IECs (Apc Δ580-15-LOX-1 mice) (Figures S2A–S2C). In ApcΔ580 mice, increasing the corn oil content from 5% to 20% significantly increased tumor volume per mouse at age 14 weeks (Figures 1D and 1E; Table S2). In contrast, ApcΔ580-15-LOX-1 mice fed either 5% or 20% corn oil had a lower tumor volume per mouse and fewer large tumors (diameter > 3 mm) per mouse than did the ApcΔ580 mice on the same diets (Figures 1E and 1F), despite a body weight increase in those fed 20% corn oil compared with those fed 5% corn oil (Figure S2D). In a longitudinal follow-up survival experiment, ApcΔ580-15-LOX-1 mice lived significantly longer than did the corresponding ApcΔ580 littermates fed the same diets; at 47 weeks, none of the ApcΔ580 mice were alive, whereas 53% of the ApcΔ580-15-LOX-1 mice fed 5% corn oil and 25% of the ApcΔ580-15-LOX-1 mice fed 20% corn oil were still alive (Figure 1G). To determine whether 15-LOX-1 impacts β-catenin and CRC even in the absence of dietary LA variation, we fed ApcΔ580-15-LOX-1 mice and their ApcΔ580 littermates the same 7% corn oil diet. At age 25 weeks, ApcΔ580-15-LOX-1 mice had markedly higher 13-HODE levels but lower active β-catenin levels and lower colorectal tumor multiplicity, especially for large tumors (diameter > 3 mm) (Figures S2E–S2H; Table S2), than did ApcΔ580 littermates.
ApcΔ580 mutation induces aberrant β-catenin activation to initiate CRC in ApcΔ580 mice (Hinoi et al., 2007). We therefore examined whether dietary LA and 15-LOX-1 further modulated β-catenin activation. β-catenin levels in both normal-appearing mucosa (Figures 1H, 1I, 2A, and 2C) and tumor mucosa (Figures S3A and S3B) were increased by high dietary corn oil but decreased by 15-LOX-1 transgenic expression in IECs. 15-LOX-1 expression also decreased active β-catenin levels in AOM-induced CRC (Figures S3C and S3E). Apc mutations have been reported to expand the colonic crypt proliferative zones as an important mechanism to drive CRC (Barthold and Beck, 1980; Boman and Fields, 2013); in our study, colonic crypt proliferative zones as measured using Ki-67 immunohistochemical analyses were increased by high dietary corn oil but decreased by transgenic 15-LOX-1 expression in IECs (Figures 1J and 1K). Together, these data suggest that 15-LOX-1 suppressed β-catenin-driven CRC promotion, especially in relation to high dietary LA.
Figure 2. 15-LOX-1 Suppresses Linoleic Acid (LA) Upregulation of LRP5 and Active β-catenin Expression in Intestinal Epithelial Cells (IECs) of ApcΔ580 Mice and in Human Colorectal Cancer Cells.
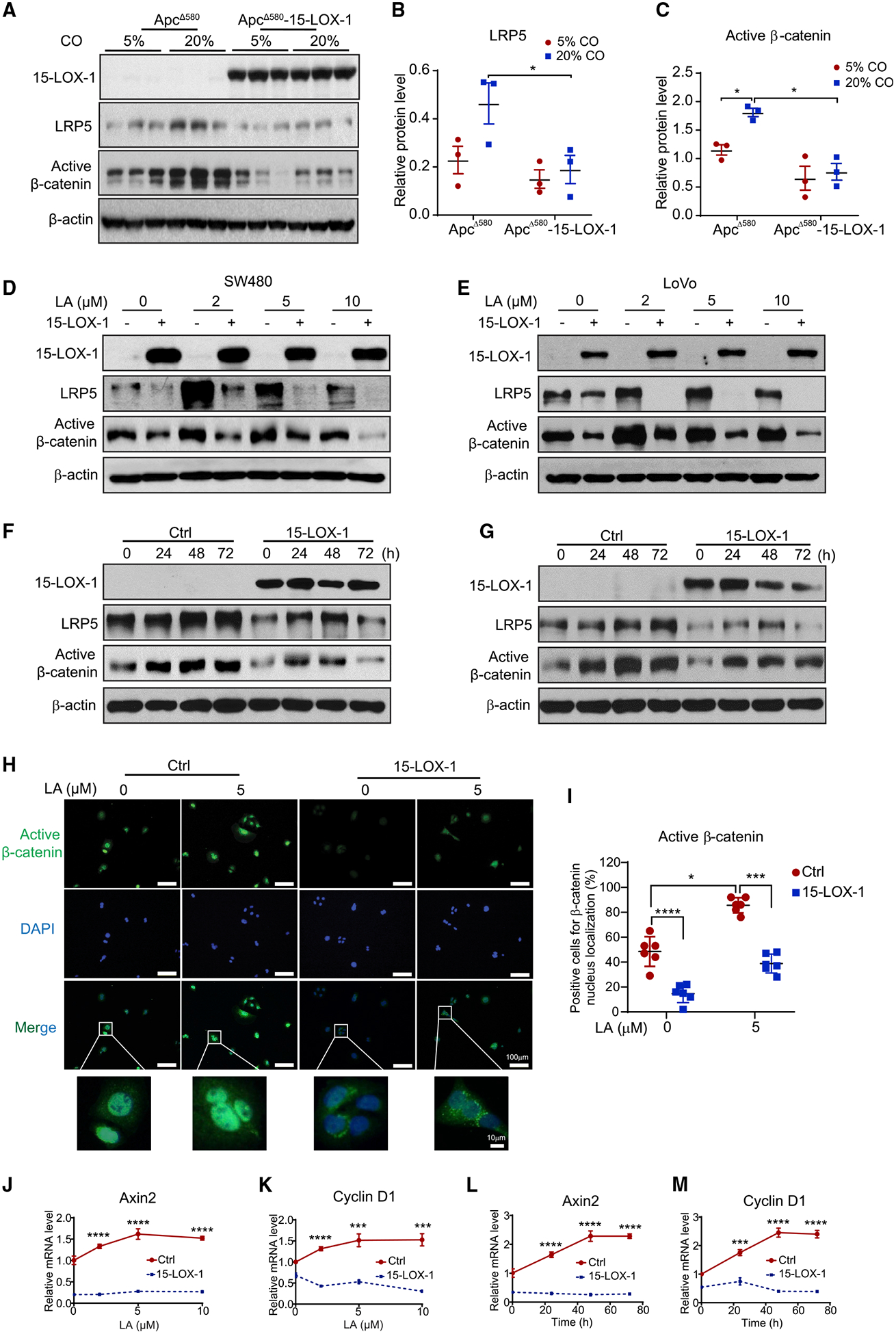
(A) Western blot images showing 15-LOX-1, LRP5, and active β-catenin protein expression in normal IECs from the indicated mouse groups as described in Figures 1D–1F (n = 3 per group).
(B and C) Band density ratios of LRP5 (B) and active β-catenin (C) to β-actin corresponding to (A), measured using ImageJ. Values in (B) and (C) are mean ± SEM.
(D–G) Active β-catenin and LRP5 protein levels as measured using western blot analysis in SW480 and LoVo cells stably transduced with either control (Ctrl) or 15-LOX-1 lentivirus and treated with LA at the indicated concentrations in culture media for 72 h (D and E) or treated with 5 μM LA at the indicated time points (F and G).
(H and I) Representative immunofluorescence staining images of active β-catenin (H) and quantification of cells positive for active β-catenin nuclear localization (I) in SW480 cells treated with 5 μM LA for 48 h. n = 3 repeats with similar results. Values are mean ± SD.
(J–M) Axin2 and cyclin D1 mRNA expression in SW480 cells stably transduced with either control (Ctrl) or 15-LOX-1 lentivirus and treated with LA at different concentrations in culture medium supplemented with 5% dialyzed fetal bovine serum for 48 h (J and K) or treated with 5 μM LA at the indicated time points (L and M), measured using reverse transcription real-time quantitative polymerase chain reaction (n = 3 replicates/condition).
Values are mean ± SD. *p < 0.05, ***p < 0.001, and ****p < 0.0001; two-way ANOVA (B, C, I, and J–M).
15-LOX-1 Suppresses LA Upregulation of LRP5 and Active β-catenin in IECs of ApcΔ580 Mice and in Human Colorectal Cancer Cells
LRPs have dual receptor roles: (1) LA-enriched LDL endocytosis and subsequent degradation (Spiteller and Spiteller, 2000) and (2) transduction of Wnt signals to activate β-catenin (Tortelote et al., 2017). In their LDL-related role, LRPs recruit 12/15-LOX to the cell membrane to oxidize LDL (Zhu et al., 2003). We therefore examined the effects of LA and 15-LOX-1 on LRPs.
LRP5 and active β-catenin levels were lower in ApcΔ580-15-LOX-1 mice than in ApcΔ580 mice at age 6–8 weeks, which precedes the development of CRC in these mice (Figure S3D). In contrast, LRP6 protein levels were not significantly changed by 15-LOX-1 transgenic expression (Figure S3D); thus, our further studies were focused on LRP5. LRP5 and active β-catenin levels were higher in 12/15LoxKO mice compared with WT mice, and further increased in 12/15LoxKO/12-LOX mice, but decreased in 12/15LoxKO/15-LOX-1 mice; however, cleaved PARP levels were altered in the opposite direction (Figure S3E). Similarly, LRP5 and active β-catenin levels in ApcΔ580 mice at age 14 weeks were upregulated by the LA-enriched high corn oil diet and downregulated by transgenic 15-LOX-1 expression in IECs (Figures 2A–2C). Also, mRNA levels of β-catenin downstream targets (Axin2 and cyclin D1) were increased by higher LA dietary content in ApcΔ580 mice but decreased by 15-LOX-1 transgenic expression in IECs (Figures S3F and S3G).
We next sought to determine whether our mouse findings were applicable to human CRC by re-expressing 15-LOX-1 via lentivirus infection in SW480 and LoVo cells, which, like other colorectal cancer cells, lack 15-LOX-1 expression (Moussalli et al., 2011). In the control SW480 and LoVo cells, LA supplementation increased protein levels of LRP5 and active β-catenin, which peaked at 2 and 5 μM LA, respectively, after 48–72 h (Figures 2D–2G). β-catenin nuclear localization (Figures 2H, 2I, S4A, and S4B) and Axin2 and cyclin D1 mRNA levels increased after LA supplementation in a time- and concentration-dependent manner (Figures 2J–2M and S4C–S4F). All these effects were inhibited by 15-LOX-1 re-expression in SW480 and LoVo cells (Figures 2D–2M and S4A–S4F). These observations implicate 15-LOX-1 as a negative modulator of LA-increased LRP5 protein expression and β-catenin activation.
15-LOX-1 Inhibits Colonic Stem Cell Regeneration in Organoids Derived from ApcΔ580 Mice and Human Colorectal Cancers
We assessed the biological significance of 15-LOX-1’s suppression of LRP5 and β-catenin signaling by examining how 15-LOX-1 affects colonic tumor stem cell self-renewal, which is strongly enhanced by aberrant β-catenin activation (Schwitalla et al., 2013). Colonic organoids derived from IECs of ApcΔ580-15-LOX-1 mice had significantly lower expression of LRP5, active β-catenin, Axin2, and cyclin D1 (Figures 3A–3C) and yielded fewer primary and secondary organoid numbers, especially primitive spheroids, than did the organoids derived from IECs of ApcΔ580 mice (Figures 3D–3F). More important, 15-LOX-1 re-expression via lentiviral infection in human organoids derived from patients’ colorectal cancer tissues reduced LRP5, active β-catenin, Axin1, and cyclin D1 protein levels (Figures 3G and 3H); decreased Axin2 and cyclin D1 mRNA expression (Figures 3I and 3J); and repressed the organoids’ self-renewal, as measured using organoid formation assay (Figures 3K and 3L).
Figure 3. 15-LOX-1 Inhibits Colonic Stem Cell Self-Renewal in Intestinal Epithelial Cells of ApcΔ580 Mice and Human Colorectal Cancer Cells.
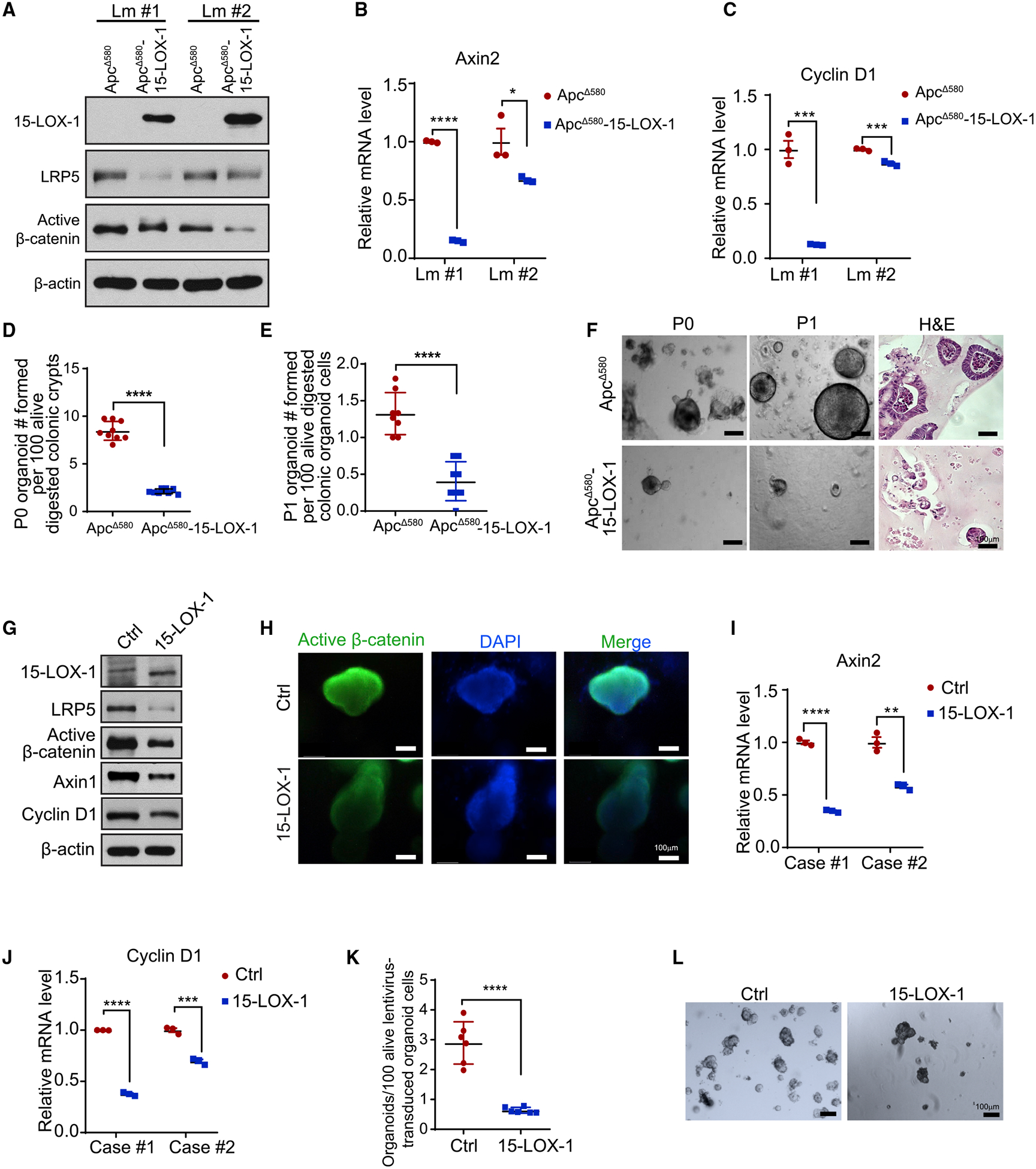
(A–C) ApcΔ580 and ApcΔ580-15-LOX-1 littermates were euthanized at 6 weeks and isolated colonic mucosa cells from two littermates (Lm) of the indicated mice were cultured for primary organoids for 8 days and then analyzed for LRP5, active β-catenin, and 15-LOX-1 protein expression levels (A), as well as Axin2 (B) and cyclin D1 (C) mRNA expression.
(D–F) Primary (P0) (D) and secondary (P1) (E) organoid formation efficacies at 8 days of culture. (F) Representative bright-field light microscopic (P0 and P1) and hematoxylin and eosin (H&E) staining of images (P1) of organoids for the indicated mice.
(G–L) LRP5, active β-catenin, 15-LOX-1, Axin1, and cyclin D1 protein expression levels, measured using western blot analysis (G); immunofluorescence staining images of active β-catenin (H); and mRNA expression of Axin2 (I) and cyclin D1 (J) measured using reverse transcription real-time quantitative polymerase chain reaction performed in triplicate in human colorectal cancer tissue-derived organoid cells transduced with control (Ctrl) or 15-LOX-1 lentivirus for at least five passages. (K) Organoid formation efficacies and (L) representative bright-field light microscopic images of human colorectal cancer tissue-derived organoids transduced with either control or 15-LOX-1 lentivirus (n = 6 cases).
Values are mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001; unpaired t test (B–E, I–K).
15-LOX-1 Downregulates LRP5 Protein Expression via 13-HODE
To identify the molecular mechanisms by which 15-LOX-1 downregulates LRP5 expression, we first examined the oxidative lipid profiles of IECs via liquid chromatography-tandem mass spectrometry to determine the relationship between 15-LOX-1 enzymatic products and their suppression of CRC in ApcΔ580 mice. In ApcΔ580-15-LOX-1 mice, the dominant oxidative lipid products were 13-HODE and 15-HETE acid (Figure 4A), whereas in ApcΔ580 mice, the dominant products were prostaglandin E2 and 12-HETE (Figures 4A and S5A). Lipoxin A4 (LXA4) and lipoxin B4 (LXB4) levels were also significantly higher in ApcΔ580-15-LOX-1 mice than in ApcΔ580 mice (Figure 4A). Levels of these 15-LOX-1 enzymatic products (13-HODE, 15-HETE, LXA4, and LXB4) were significantly negatively correlated with tumor volumes in both ApcΔ580 and ApcΔ580-15-LOX-1 mice (Figures 4B–4E). In contrast, levels of non-15-LOX-1 enzymatic products (prostaglandin E2, 5-HETE, 12-HETE, and LTB4) did not significantly correlate with tumor volumes (Figures S5B–S5E).
Figure 4. 15-LOX-1 Downregulates LRP5 Protein Expression via 13-HODE.
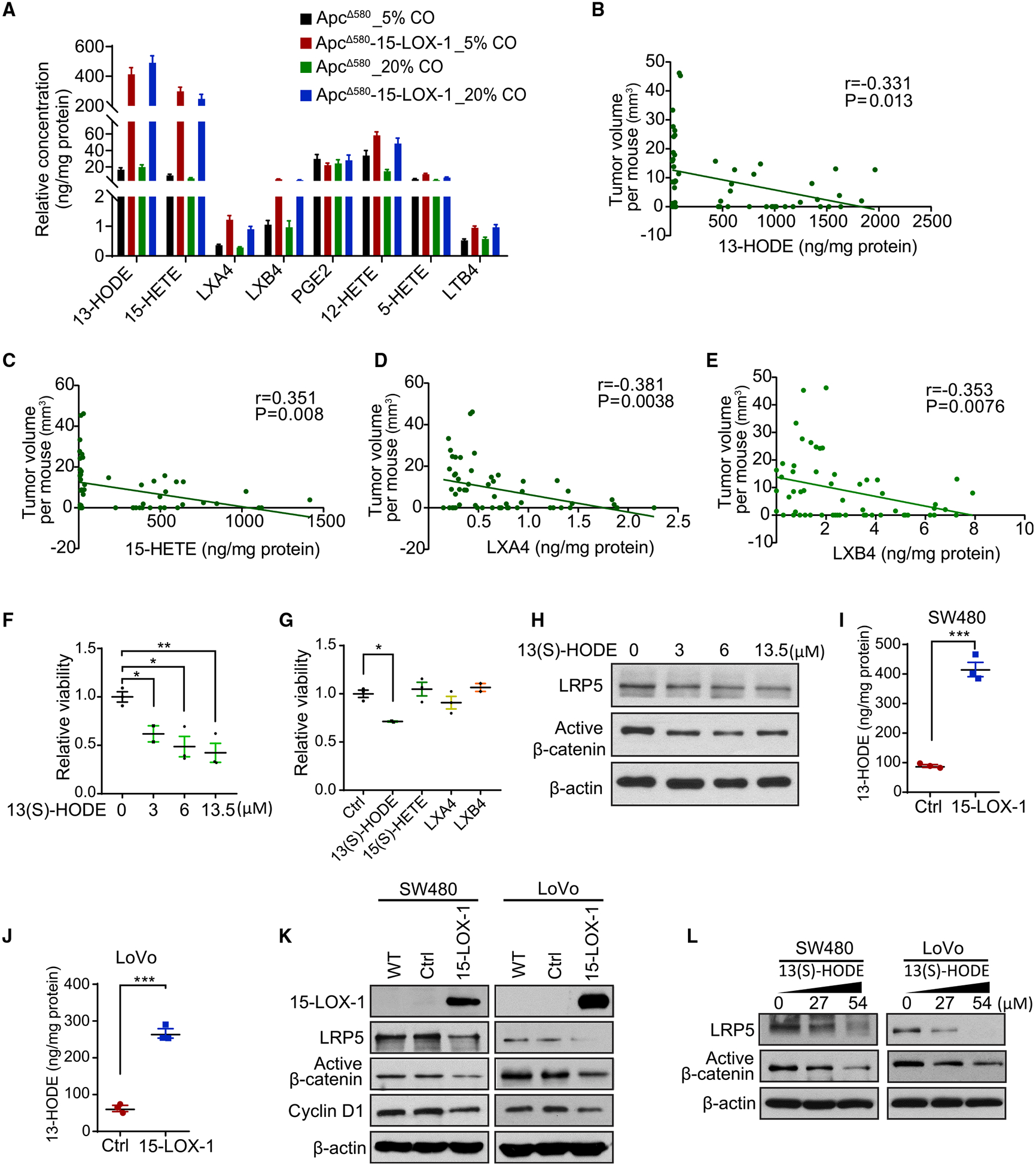
(A) Eicosanoid metabolite profile of the IECs from ApcΔ580 and ApcΔ580-15-LOX-1 mice, as described in Figures 1D–1F, measured using liquid chromatography-tandem mass spectrometry (LC-MS/MS) (n = 14 mice per group). Values are mean ± SEM.
(B–E) Spearman correlation analysis of colonic tumor volumes and levels of 13-HODE (B), 15-HETE (C), LXA4 (D), and LXB4 (E) in IECs from the mice as described in (A) (n = 56 for all four groups).
(F and G) Cell viability of colonic organoids derived from the IECs of ApcΔ580 mice treated with 13(S)-HODE at 0, 3, 6, or 13.5 μM (F) or treated with 13(S)-HODE (3 μM), 15(S)-HETE (2.15 μM), LXA4 (8.86 nM), or LXB4 (36.24 nM) for 6 days (G), measured using CellTiter-Glo Luminescent Cell Viability Assay (n = 3 repeats). Values are mean ± SEM. *p < 0.05 and **p < 0.01 (one-way ANOVA).
(H) LRP5 and active β-catenin protein expression levels in organoid cells treated with 13(S)-HODE at the indicated concentrations for 6 days, as described in (F).
(I and J) Levels of 13-HODE in SW480 (I) and LoVo (J) cells stably transduced with either control (Ctrl) or 15-LOX-1 lentivirus, measured using LC-MS/MS in triplicated measurements. Values are mean ± SD. ***p < 0.001 (unpaired t test).
(K) 15-LOX-1, LRP5, and active β-catenin protein levels in SW480 and LoVo cells transduced with either Ctrl or 15-LOX-1 lentivirus, measured using western blot analysis.
(L) LRP5 and active β-catenin protein levels measured using western blot analysis in SW480 and LoVo cells supplemented with 13(S)-HODE at the indicated concentrations in cell culture media for 48 h.
We then determined the mechanistic relevance of those correlations to the previously observed effects of 15-LOX-1 on stem cell regeneration (Figures 3D–3F). Treatment with 13(S)-HODE inhibited regeneration of ApcΔ580 IEC-derived organoids in a concentration-dependent manner (Figure 4F). In contrast, other 15-LOX-1 enzymatic products (15[S]-HETE, LXA4, and LXB4), administered at concentrations proportional to their levels relative to 13-HODE levels in IECs of the ApcΔ580 mice as measured using liquid chromatography-tandem mass spectrometry (Figure 4A), had no effect on regeneration of colonic organoids from IECs of ApcΔ580 mice (Figure 4G). Furthermore, treatment with 13(S)-HODE at the concentrations that repressed organoid regeneration also decreased LRP5 and active β-catenin protein levels in ApcΔ580 IEC-derived organoids (Figure 4H).
Next, we investigated the relevance of our mouse findings to human CRC. Re-expression of 15-LOX-1 in SW480 and LoVo cells via lentiviral infection significantly increased 13-HODE levels and inhibited LRP5 and active β-catenin protein levels (Figures 4I–4K). Treatment of SW480 and LoVo cells with 13(S)-HODE, at the same concentrations that inhibited cell proliferation (Figures S5F and S5G), also downregulated LRP5 and active β-catenin protein levels (Figure 4L). In contrast, treatment with other 15-LOX-1 products, such as 15(S)-HETE, LXA4, and LXB4, did not affect LRP5 or active β-catenin protein levels in either SW480 or LoVo cells (Figures S5H and S5I). Together, these results suggest that 13-HODE mediates 15-LOX-1-induced downregulation of LRP5 protein expression.
15-LOX-1 Regulates LRP5 Expression in the Cell Membrane
Membranous LRPs are tightly regulated at the protein stability level, as their fate is balanced during their endocytosis with LDL between lysosomal degradation and recycling to the cell membrane (van Kerkhof et al., 2005). When we tested 15-LOX-1’s effects on LRP5 protein stability in SW480 and LoVo cells, 15-LOX-1 re-expression decreased LRP5 protein stability following treatment with cycloheximide (Figures 5A and 5B; Table S3). Because LRP5 promotes 15-LOX-1 membranous translocation to oxidize LA-enriched LDL (Zhu et al., 2003), we investigated whether LA supplementation to LoVo and SW480 cells is sufficient to trigger 15-LOX-1 membranous re-localization and whether this re-localization affects LRP5 protein internalization and subsequently its membranous abundance. LA supplementation to LoVo and SW480 cells increased 15-LOX-1 protein distribution from the cytoplasm to the cell membrane, when 15-LOX-1 was re-expressed in cancer cells; this 15-LOX-1 redistribution was associated with increased LRP5 protein localization from the cell membrane to the cytoplasm (Figures 5C–5F, S6A, and S6B). To examine whether this observed LRP5 redistribution is due to increased LRP5 internalization, we measured LRP5 internalization using imaging flow cytometry with immunofluorescence-labeled LRP5 protein. The number of cells with LRP5 internalization was larger in SW480 cells stably transduced with 15-LOX-1 lentivirus than in those transduced with control lentivirus with the same LA treatment (Figures 5G and 5H; Table S4).
Figure 5. 15-LOX-1 Decreases Cell Membranous LRP5 Levels.
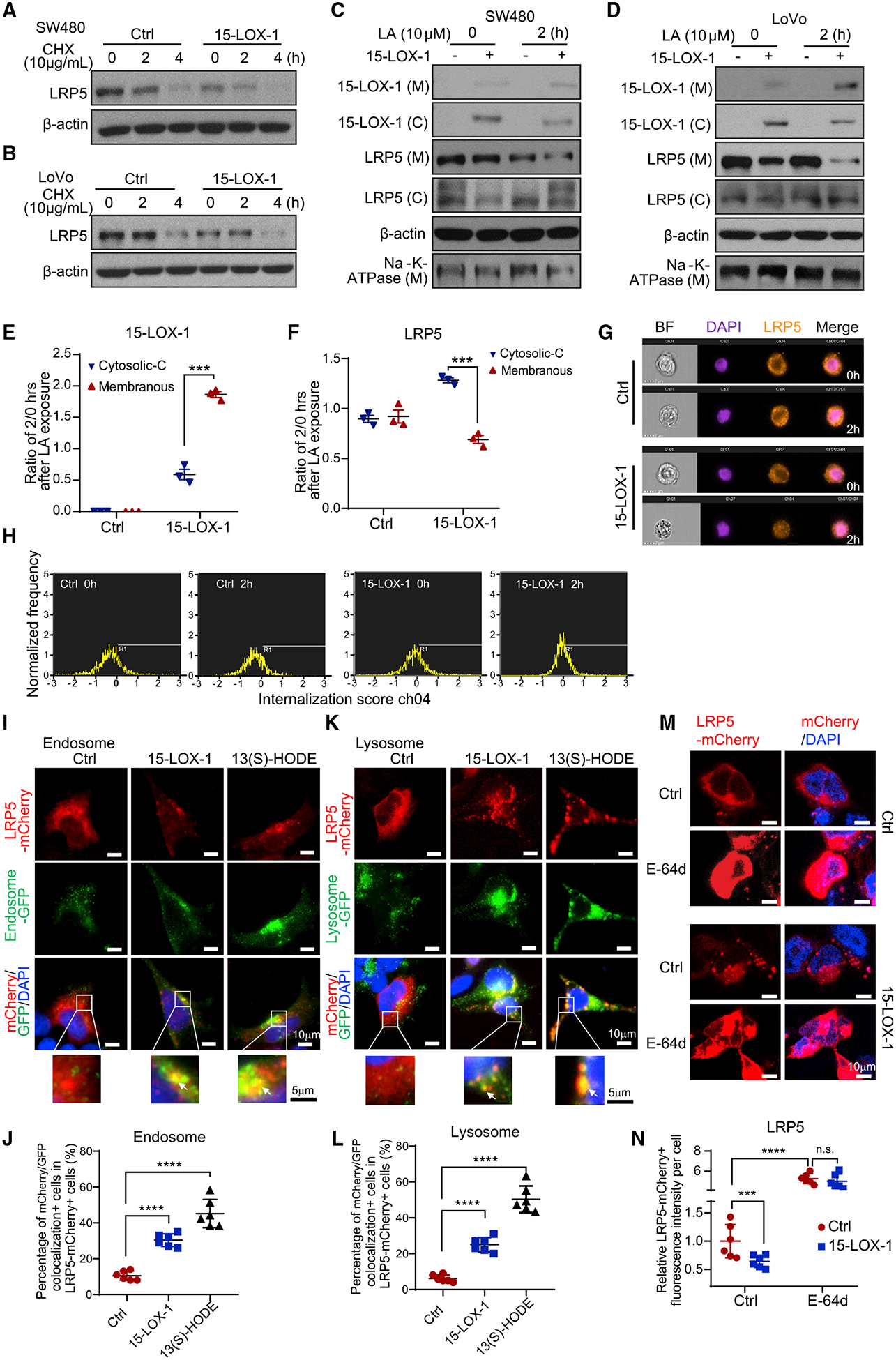
(A and B) LRP5 protein expression in SW480 (A) and LoVo (B) cells stably transduced with control (Ctrl) or 15-LOX-1 lentivirus and treated with 10 μg/mL cycloheximide (CHX) for 0, 2, and 4 h. The whole-cell lysates were analyzed using western blot.
(C and D) Cytoplasmic (C) and membranous (M) LRP5 and 15-LOX-1 protein expression in SW480 (C) and LoVo (D) cells stably transduced with control (Ctrl) or 15-LOX-1 lentivirus and treated with 10 μM linoleic acid (LA) for 2 h. The cells were processed into cytoplasmic and membranous protein fractions and then analyzed using western blot.
(E and F) Two hour to 0 h ratios of cytoplasmic protein band densities normalized to β-actin and ratios of membranous protein band densities normalized to Na+-K+-ATPase for 15-LOX-1 (E) and LRP5 (F) expression in SW480 cells transduced with Ctrl or 15-LOX-1 lentivirus, corresponding to (C) (n = 3). Values are mean ± SEM. ***p < 0.001 (two-way ANOVA).
(G and H) SW480 cells stably transduced with Ctrl or 15-LOX-1 lentivirus were treated with 10 μM LA for 2 h. The cells were analyzed for LRP5 internalization using imaging flow cytometry assay. Representative images (G) and cell internalization distribution (H) of 5,000 examined cells (cells with internalization score R1 > 0 were counted as internalization positive) are shown (n = 3 repeats with similar results).
(I–L) 293T cells stably transfected with Tet-on 15-LOX-1 inducible vector were transfected with mCherry-tagged LRP5 expression vector and treated with (15-LOX-1) or without (Ctrl) doxycycline (2 μg/mL) or treated with 13(S)-HODE (27 μM) for 24 h and then incubated with CellLight Early Endosomes-GFP, BacMam 2.0 or CellLight Lysosomes-GFP, BacMam 2.0 for another 24 h. Representative microphotographs and quantitative results of mCherry-labeled LRP5 colocalization with GFP-traced cytoplasmic endosomes (I and J) or lysosomes (K and L) are shown. Quantification was evaluated as percentages of the cells that had co-localization of mCherry and GFP fluorescence out of mCherry-positive cells from six random fields under a confocal microscope. Arrows indicate colocalization. (M and N) 293T cells were co-transfected with mCherry-tagged LRP5 expression plasmid with pCMV5-IRES-GFP (Ctrl) or with pCMV5–15-LOX-1-IRES-GFP (15-LOX-1) for 48 h and then treated with E-64d (10 μM) or dissolvent (Ctrl) for 1 h. Representative microphotographs (M) and quantitative results (N) of LRP5 trans-localization traced with mCherry fluorescence protein from six random fields under a confocal microscope are shown.
Values are mean ± SD. ***p < 0.001, ****p < 0.0001, n.s., no significant difference; one-way ANOVA (J and L), two-way ANOVA (N). n = 3 repeats with similar results.
To investigate whether this 15-LOX-1-induced increase in LRP5 internalization augments LRP5 lysosomal degradation, we transfected 293T cells with mCherry-labeled LRP5 expression vector to trace LRP5 distribution in endosomes and lysosomes labeled with green fluorescent protein (GFP) via incubation with CellLightEarly Endosomes-GFP BacMam 2.0 or CellLight Lysosomes-GFP BacMam 2.0. Doxycycline-induced 15-LOX-1 expression or treatment with 13(S)-HODE increased LRP5 redistribution to the cell endosomes and lysosomes (Figures 5I–5L). Furthermore, treatment with E-64d, an irreversible broad-spectrum cysteine protease inhibitor that suppresses lysosomal protein degradation, increased both membranous and cytoplasmic LRP5 protein levels in 293T cells transfected with control vector. In contrast, transfection with 15-LOX-1 vector in 293T cells reduced LRP5 levels, especially in the membranous compartment; E-64d blocked that effect (Figures 5M and 5N). Similar results were obtained in SW480 cells (Figures S6C and S6D). These findings demonstrate that 15-LOX-1 downregulates LRP5 protein expression levels by increasing LRP5 cell internalization and subsequent LRP5 lysosomal degradation.
15-LOX-1 Inhibits SNX17-Mediated LRP5 Recycling to the Cell Membrane
SNX17 binds membranous phosphatidylinositol 3-phosphate (PI3P) for anchorage to direct LRPs away from lysosomal degradation and recycle them into their membranous pools (van Kerkhof et al., 2005). We sought to examine whether 15-LOX-1 modulates SNX17’s role in rescuing LRP5 from lysosomal degradation to be recycled to the membrane. We first examined whether SNX17 affects LRP5 and subsequently active β-catenin protein and found that SNX17 overexpression increased LRP5 and active β-catenin protein levels in 293T and SW480 cells (Figure 6A). In complementary experiments, SNX17 downregulation by small interfering RNA (siRNA) reduced LRP5 and active β-catenin protein levels (Figure 6B). Rescue experiments, to ensure that the effects of SNX17 siRNA were not a result of off-target effects, showed that overexpression of the mutant SNX17 sequence, which is resistant to degradation by SNX17 siRNA (McNally et al., 2017), blocked LRP5 downregulation by SNX17 siRNA (Figures S6E and S6F). Next, we used immunoprecipitation and immunoblotting to determine whether LRP5 binds to SNX17 to promote LRP5 protein recycling. When Myc-tagged LRP5 was expressed in 293T and SW480 cells, SNX17 was detected by immunoblotting in immunoprecipitated products with anti-c-Myc antibody (Figure 6C). Additionally, when FLAG-tagged SNX17 was expressed in 293T and SW480 cells, LRP5 was also detected by immunoblotting in immunoprecipitated products with anti-FLAG antibody (Figure 6D). These findings demonstrate the ability of LRP5 to bind SNX17.
Figure 6. 15-LOX-1 Reduces Membranous LRP5 Levels by Inhibiting SNX17-Mediated LRP5 Recycling to the Cell Membrane.
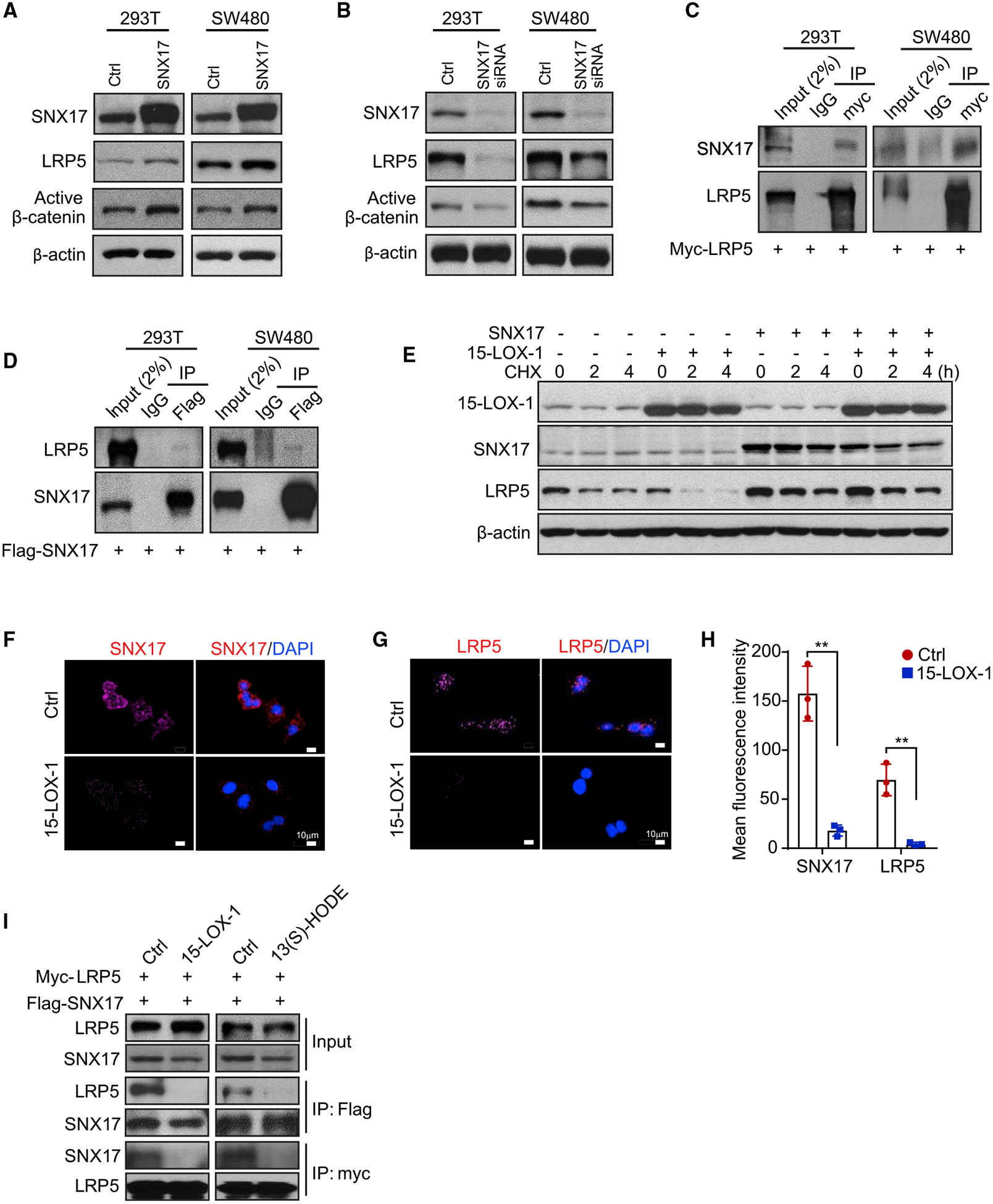
(A) SNX17, LRP5, and active β-catenin expression in 293T or SW480 cells transfected with control vector (Ctrl) or FLAG-tagged SNX17 expression vector for 48 h, then processed for western blot analysis.
(B) SNX17, LRP5, and active β-catenin expression levels in 293T or SW480 cells transfected with control (Ctrl) or pooled SNX17 siRNA for 72 h, measured using western blot analysis.
(C) SNX17 and LRP5 interaction assessment in 293T cells or SW480 cells transfected with Myc-tagged LRP5 expression vector for 48 h. Cell lysates were immunoprecipitated (IP) by anti-Myc antibody or rabbit immunoglobulin G (IgG) and then analyzed using immunoblotting with anti-SNX17 or anti-LRP5 antibody.
(D) LRP5 and SNX17 interaction assessment in 293T cells or SW480 cells transfected with FLAG-tagged SNX17 expression plasmid for 48 h. Cell lysates were immunoprecipitated by anti-FLAG antibody or rabbit IgG and then analyzed using immunoblotting with anti-LRP5 or anti-SNX17 antibody.
(E) SW480 cells stably transduced with control (−) or 15-LOX-1 (+) lentivirus were transfected with either SNX17 expression vector (+) or control vector (−) and treated with 5 μM linoleic acid (LA) for 48 h, followed by treatment with cycloheximide (CHX; 10 μg/mL) for 0, 2, and 4 h. SNX17, LRP5, and active β-catenin expression levels were measured by western blot analysis.
(F–H) SW480 cells stably transduced with control (Ctrl) or 15-LOX-1 lentivirus were treated with 100 μM alkynyl LA for 36 h, and the cells were then analyzed using proximity ligation assay using the click chemistry reaction of alkynyl LA with SNX17 or with LRP5. Representative click microphotographs for SNX17 (F) and LRP5
(G) and their corresponding quantitative intensity (H) are shown. Values in (H) are mean ± SEM. **p < 0.01 (unpaired t test).
(I) Effects of 15-LOX-1 or 13(S)-HODE on LRP5 binding to SNX17. SW480 cells stably transduced with control (Ctrl) or 15-LOX-1 lentivirus were co-transfected with Myc-tagged LRP5 expression plasmid and FLAG-tagged SNX17 expression plasmid for 48 h, then treated with 10 μM LA for an additional 24 h (left). Wild-type SW480 cells were co-transfected with LRP5 and SNX17 expression plasmids for 48 h, then treated with 27 μM 13(S)-HODE for another 24 h (right). Cell lysates were immunoprecipitated by anti-Myc antibody or anti-FLAG antibody and then analyzed using immunoblotting with anti-SNX17 or anti-LRP5 antibody.
To examine whether SNX17 affects 15-LOX-1’s modulation of LRP5 protein stability, we performed an LRP5 cycloheximide pulse-chase assay with SNX17 and 15-LOX-1 overexpression. SNX17 overexpression reduced LRP5 degradation following treatment with cycloheximide and, furthermore, blocked the augmentation of LRP5 degradation by 15-LOX-1 re-expression in SW480 cells with LA supplementation (Figure 6E). We next examined whether 15-LOX-1 affects LRP5 or SNX17 binding to LA using a chemical ligation proximity assay. 15-LOX-1 re-expression markedly decreased the colocalization of alkynyl LA with SNX17 or with LRP5 in SW480 cells (Figures 6F–6H). In co-immunoprecipitation studies, 15-LOX-1 re-expression in SW480 cells with LA supplementation or treatment of SW480 cells with 13(S)-HODE inhibited LRP5 binding to SNX17 (Figure 6I), suggesting that 15-LOX-1 conversion of LA to 13(S)-HODE suppresses SNX17 binding to LRP5.
15-LOX-1-Mediated Peroxidation of PI3P_LA to Generate PI3P_13-HODE Inhibits PI3P Binding to SNX17 and thus Suppresses LRP5 Recycling to the Cell Membrane
Within endosomes, PI3P anchors SNX17 to recycle LRPs (van Kerkhof et al., 2005); we therefore examined the role of PI3Ps in LRP5 recycling by SNX17 using a specific PI3P inhibitor, VPS34-IN-1. VPS34-IN-1 specifically inhibits vacuolar protein sorting 34 (VPS34), which phosphorylates endosomal phosphatidylinositol to generate PI3P (van Kerkhof et al., 2005; Bago et al., 2014). VPS34-IN-1 decreased LRP5 protein expression, especially when 15-LOX-1 was re-expressed, in SW480 and LoVo cells (Figures 7A and 7B), suggesting that PI3P increases LRP5 protein expression and that 15-LOX-1 might inhibit these PI3P’s effects. Given that PI3P-SNX17 plays a critical role in mediating the trafficking of LRP5 by endosomes and the fate of LRP5 (van Kerkhof et al., 2005), we tracked LRP5 protein expression using mCherry red fluorescence in 293T and SW480 cells treated with VPS34-IN-1. LA increased whereas VPS34-IN-1 or 13(S)-HODE decreased LRP5 levels (Figures 7C and 7D). Additionally, compared with LA, VPS34-IN-1 increased LRP5 lysosomal localization (Figures S7A and S7B), which further supported the significance of PI3P in rescuing LRP5 from being trafficked for degradation in the lysosomes.
Figure 7. 15-LOX-1 Increases Phosphatidylinositol 3-Phosphate (PI3P) 13-HODE Production, which Inhibits PI3P’s Binding to SNX17 in Endosomes and Subsequently Suppresses LRP5 Recycling to the Cell Membrane.
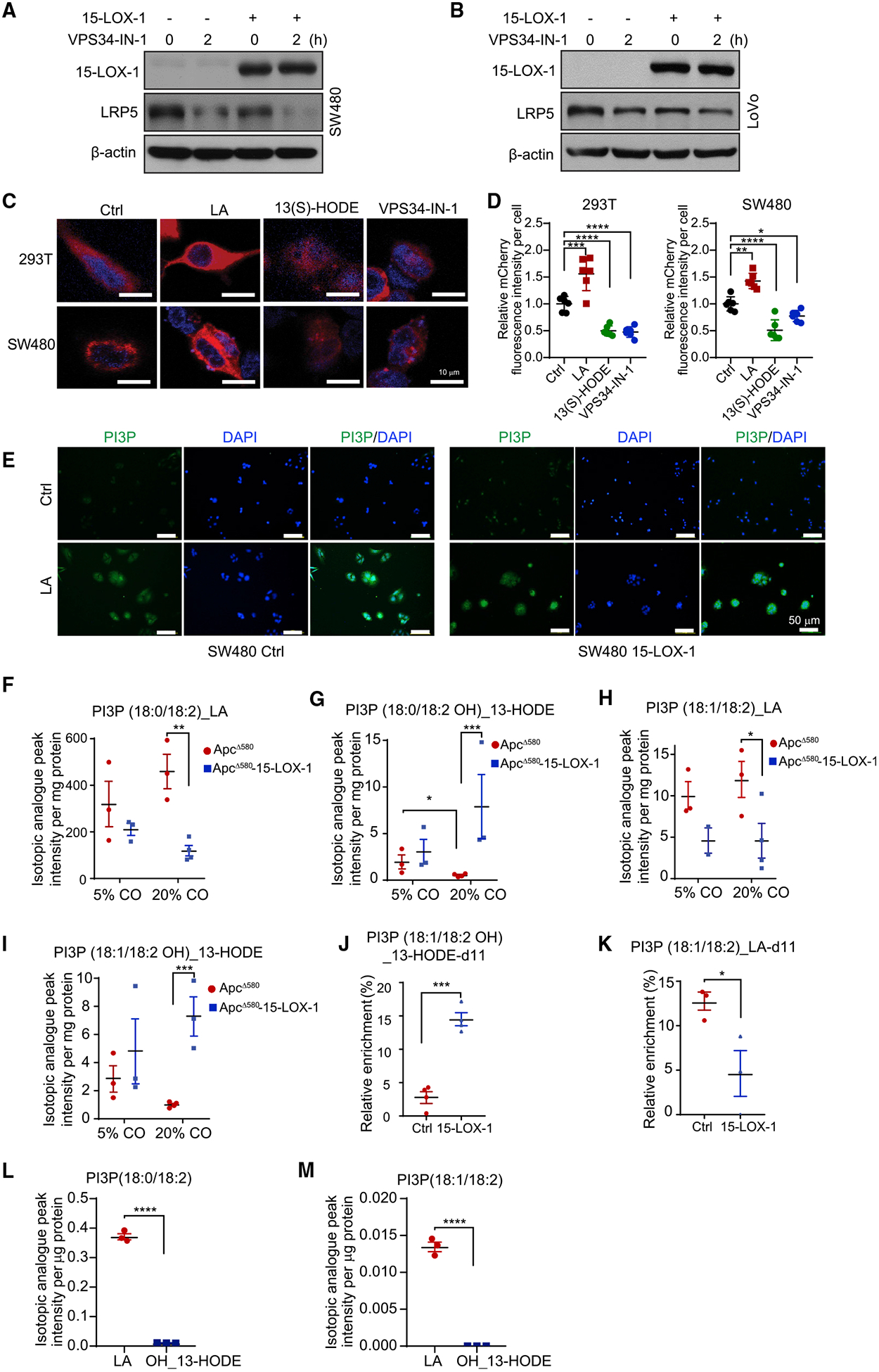
(A and B) SW480 (A) or LoVo (B) cells stably transduced with control (Ctrl) or 15-LOX-1 lentivirus were treated with the PI3P biosynthesis inhibitor VPS34-IN-1 (1 μM) for 2 h and processed for the indicated protein expression using western blot analysis.
(C and D) 293T or SW480cells were transfected with mCherry-tagged LRP5 expression plasmid for 48 h, followed by treatment with linoleic acid (LA; 100 μM), 13(S)-HODE (27 μM), or VPS34-IN-1 (1 μM) for another 4 h. Representative microphotographs (C) and quantitative results (D) of mCherry-tagged LRP5 localization from six random fields under a confocal microscope are shown. Values are mean ± SD; one-way ANOVA (D). n = 3 repeats with similar results.
(E) SW480 cells stably transduced with control (Ctrl) or 15-LOX-1 lentivirus were treated with either vehicle solvent (Ctrl) or 5 μM LA for 48 h. PI3P levels in those cells were analyzed by immunofluorescence staining. Representative microphotographs are shown.
(F–I) ApcΔ580 and ApcΔ580-15-LOX-1 littermates at age 4 weeks fed 5% or 20% corn oil (CO) were euthanized at age 14 weeks. The IECs of these mice were scraped and examined for PI3P incorporated with (18:0/18:2)_LA (F), (18:0/18:2 OH)_13-HODE (G), (18:1/18:2)_LA (H), and (18:1/18:2 OH)_13-HODE (I) using LC-HRMS (n = 3 or 4 mice per group).
(J and K) SW480 cells stably transduced with control (Ctrl) or 15-LOX-1 lentivirus were treated with 100 μM LA-d11 for 36 h in culture medium containing 5% dialyzed fetal bovine serum. PI3P profiling was traced with d11 labeling by LC-HRMS. The percentages of PI3P (18:1/18:2 OH)_13-HODE-d11 over total PI3P (18:1/18:2 OH)_13-HODE (J) and PI3P (18:1/18:2)_LA-d11 over total PI3P (18:1/18:2)_LA (K) were calculated as relative enrichment (%) (n = 3 repeats).
(L and M) SW480 cells stably transduced with control (Ctrl) or 15-LOX-1 lentivirus were transfected with FLAG-tagged SNX17 expression plasmid for 48 h, followed by treatment with 100 μM LA for another 24 h. Cell lysates were immunoprecipitated by anti-FLAG antibody-coated magnetic beads, then analyzed using LC-HRMS to examine the levels of PI3P incorporated with (18:0/18:2)_LA or (18:0/18:2 OH)_13-HODE (L) or with (18:1/18:2)_LA or (18:1/18:2 OH)_13-HODE (M) (n = 3 repeats).
Values are mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001; two-way ANOVA (F–I), unpaired t test (J–M).
PI3P contains fatty acid residues that include LA, which could influence the biological functions of PI3P (Gu et al., 2013). We found that LA increased cellular PI3P production in SW480 cells, and this was not altered by 15-LOX-1 re-expression (Figures 7E and S7C), suggesting that 15-LOX-1 does not change PI3P abundance to regulate LRP5. To evaluate whether 15-LOX-1 instead alters PI3P composition, we measured PI3P_LA and PI3P_13-HODE relative abundance in IECs of ApcΔ580 mice using liquid chromatography-high-resolution mass spectrometry (LC-HRMS). PI3P(18:0/18:2)_LA levels showed an upward trend with high dietary LA (20% versus 5% corn oil), and this trend was decreased by 15-LOX-1 transgenic expression in IECs (Figure 7F). In contrast, PI3P (18:0/18:2 OH)_13-HODE levels decreased with high dietary LA but were increased by 15-LOX-1 transgenic expression in IECs (Figure 7G). Similar changes to those in the PI3P(18:0/18:2)_LA species were seen in PI3P (18:1/18:2)_LA (Figure 7H), and similar changes to those in the PI3P (18:0/18:2 OH)_13-HODE species were seen in PI3P (18:1/18:2 OH)_13-HODE (Figure 7I) and PI3P (16:0/18:2 OH)_13-HODE (Figure S7D) in IECs of ApcΔ580 and ApcΔ580-15-LOX-1 mice.
To examine the human relevance of the effect of 15-LOX-1 on PI3P species that contain LA, we performed LA-d11 metabolic tracing analyses in SW480 cells using LC-HRMS. We found that 15-LOX-1 re-expression considerably increased PI3P (18:1/18:2 OH)_13-HODE-d11 levels (Figure 7J) and decreased PI3P (18:1/18:2)_LA-d11 levels (Figure 7K), but these effects were not significant for PI3P (18:0/18:2)_LA-d11 and PI3P (18:0/18:2 OH)_13-HODE-d11 production (Figures S7E and S7F).
To elucidate whether PI3P_13-HODE has differential binding affinity to SNX17 compared with PI3P_LA, we examined using LC-HRMS the levels of PI3P species that were bound to immunoprecipitated FLAG-tagged SNX17 protein ectopically overexpressed in SW480 cells with or without 15-LOX-1 re-expression. The relative enrichments of PI3P (18:0/18:2)_LA and PI3P (18:1/18:2)_LA as measurements of their binding abilities to SNX17 were significantly higher than those of PI3P (18:0/18:2 OH)_13-HODE and PI3P (18:1/18:2 OH)_13-HODE (Figures 7L and 7M). Together, our results demonstrate that 15-LOX-1 mediates conversion of PI3P_LA to PI3P_13-HODE, which decreases PI3P binding to SNX17 and, consequently, inhibits LRP5 recycling from lysosomes to the cell membrane.
DISCUSSION
We found that (1) 15-LOX-1 suppressed LA promotion of CRC induced by AOM or by ApcΔ580 mutation in mice; (2) 15-LOX-1 downregulated LRP5 protein expression and β-catenin activation in ApcΔ580 mouse IECs, ApcΔ580 IEC-derived organoid cells, human colorectal cancer cells, and human colorectal cancer tissue-derived organoid cells; (3) 15-LOX-1 suppressed CRC stem cell self-renewal in mice and humans; (4) 13(S)-HODE, the primary 15-LOX-1 metabolic product, downregulated LRP5 protein expression and β-catenin activation in ApcΔ580 IECs and human colorectal cancer cells; and (5) 15-LOX-1 promoted PI3P_LA conversion to PI3P_13-HODE, which markedly reduced PI3P binding to SNX17 and subsequently inhibited LRP5 recycling from lysosomal degradation to the cytoplasmic membrane pool. These findings identify a mechanism by which 15-LOX-1 oxidative metabolism of LA residue within PI3P decreases LRP5 protein recycling, which inhibits Wnt/β-catenin signaling and subsequently CRC.
High dietary LA promoted AOM-induced and ApcΔ580 mutation-driven CRC in mice, which was inhibited by IECs’ 15-LOX-1 transgenic expression. High dietary LA, which is typical of Western-type diets (Yang et al., 1996), has been reported to promote AOM-induced CRC in mice (Deschner et al., 1990). Our findings show that LA promotion of CRC is not limited to CRC induction by AOM but also applies to CRC induction by other experimental approaches, such as the intestinally targeted ApcΔ580 mutation in mice, which closely mimics human CRC (Hinoi et al., 2007), further confirming the deleterious effects of excess dietary LA. More important, these data identify LA enhancement of aberrant β-catenin activation as a mechanism to promote CRC, which can be strongly suppressed by LA’s oxidative metabolism via 15-LOX-1. Furthermore, our experimental simulation in mice of 15-LOX-1 downregulation in human CRC shows how this 15-LOX-1 loss enhances CRC promotion, especially when coupled with excess dietary LA. 15-LOX-1’s repression of β-catenin activation was associated with enhanced apoptosis in IECs. These findings are consistent with prior reports showing that aberrant β-catenin activation represses, whereas 15-LOX-1 enhances, apoptosis in colonic cells (He et al., 2005; Shureiqi et al., 2003).
That 15-LOX-1 suppressed CRC progression beyond its initiation by ApcΔ580 mutation in mice is supported by the following observations. First, 15-LOX-1 transgenic expression repressed primarily large colorectal tumor formation, which is associated with CRC progression (Oshima et al., 2015). Second, 15-LOX-1 repressed intestinal crypt proliferative zone expansion, which is also associated with CRC progression (Wong et al., 2002; van de Wetering et al., 2002). Finally, 15-LOX-1 repressed colorectal cancer stem cell self-renewal, a critical phenotypic feature by which colorectal cancer stem cells promote CRC progression (Brabletz et al., 2005). These results demonstrated that 15-LOX-1 suppression of β-catenin signaling significantly modulates CRC progression beyond CRC initiation by APC mutations.
15-LOX-1 inhibited Wnt/β-catenin signaling via downregulation of LRP5. LRP5 is a key component of the LRP5/LRP6/Frizzled co-receptor group that acts as a Wnt receptor, which when activated stabilizes cytoplasmic β-catenin to potentiate its downstream signaling (MacDonald and He, 2012; Tortelote et al., 2017). LRP5 also functions as an endocytic receptor of LDL to mediate LDL internalization for intracellular processing via the endosomal/lysosomal system (Krieger and Herz, 1994; Herz et al., 2009; Tortelote et al., 2017). Cells tightly control LRPs’ membrane abundance, and subsequently Wnt signaling, by balancing their lysosomal degradation and recycling to the cell membrane through SNX proteins (e.g., SNX17) (van Kerkhof et al., 2005). LDL binding to LRPs has also been demonstrated to promote 15-LOX-1 translocation from the cytoplasm to the cell membrane to oxidize LA incorporated in complex lipids to form 13(S)-HODE (Zhu et al., 2003). Our results demonstrate that 15-LOX-1 translocation and oxidization of LA to 13-HODE affect LRP5 membranous abundance and subsequently Wnt/β-catenin signaling. LA upregulated LRP5 in colorectal cancer cells, which agrees with a previous report of LA upregulating LRP5 protein expression in peritoneal macrophages (Schumann et al., 2015). Our present findings, however, demonstrate that LA upregulation of LRP5 augments aberrant β-catenin activation beyond its activation by APC mutations in mouse models and human colorectal cancer cells. More important, these findings identify 15-LOX-1 as a negative regulator of LRP5 augmentation of aberrant β-catenin activation.
15-LOX-1 downregulated LRP5 via 13-HODE. 15-LOX-1 transgenic expression in mouse IECs increased 13-HODE as the predominant oxidative metabolite compared with 15-LOX-1’s other known metabolites of arachidonic acid (15-HETE, LXA4, and LXB4) (Brash, 1999; Serhan et al., 2008). Although the levels of these various 15-LOX-1 products negatively correlated with the tumor burden in ApcΔ580 mice, 13(S)-HODE was the only one among these metabolites to suppress ApcΔ580 mutant colonic stem cell self-renewal and to downregulate LRP5 and β-catenin activation. These findings indicate the specific mechanistic contribution of 13(S)-HODE to 15-LOX-1’s downregulation of LRP5 to suppress hyperactivation of Wnt/β-catenin signaling.
15-LOX-1 re-expression inhibited LRP5 protein recycling to the cell membrane via PI3P_13-HODE. LRPs bind to SNX17, and SNX17 is anchored via PIPs, especially PI3P in early endosomes, to promote subsequent LRP recycling to the cell membrane (van Kerkhof et al., 2005). PI3Ps are critical regulators of cell signaling, particularly via membrane trafficking events that affect cell health and disease (e.g., cancer) status. Our data uncover the biological significance of oxidative metabolism of LA, as a part of PI3Ps, which are membrane-bound complex lipids. Most of the research attention in studying the effects of LA and other n-6 PUFA oxidative products on cell signaling has been focused on the free form of these products (e.g., PGE2). 15-LOX-1 is unique among mammalian lipoxygenases in its ability to catalyze lipid peroxidation even when the substrate is bound by an ester linkage within a complex biological structure (Kuhn et al., 1990; Schewe et al., 1975). Our findings demonstrate that modulation of LA as a PI3P lipid residue via 15-LOX-1 to form PI3P_13-HODE reduced SNX17 binding to LRP5, shifting LRP5 fate to lysosomal degradation and thus reducing its membranous recycling and subsequently suppressing Wnt signaling and CRC. This 15-LOX-1 regulatory mechanism of LRP5 membranous abundance in normal colonic cells prevents excessive activation of Wnt/β-catenin signaling. In contrast, cancer cells ubiquitously silence 15-LOX-1 expression (Moussalli et al., 2011; Il Lee et al., 2011), which could increase LRP5 membranous abundance to hyperactivate Wnt/β-catenin signaling, especially in the presence of excess dietary LA, to drive CRC progression.
In summary, our findings demonstrate that increasing dietary LA intake promotes CRC by increasing Wnt/β-catenin activation via increased LRP5 recycling to the cell membrane and that 15-LOX-1 represses LRP5 membranous recycling by promoting the conversion of PI3P_LA to PI3P_13-HODE. This previously unknown regulation of Wnt/β-catenin signaling via 15-LOX-1 metabolism of PI3P_LA represents a modifiable mechanism, beyond APC mutations, by which Wnt/β-catenin signaling can be therapeutically targeted for CRC prevention and treatment.
STAR★METHODS
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Imad Shureiqi (ishureiqi@mdanderson.org).
Materials Availability
Unique reagents and materials generated in this study are available after completing an institutionally required Material Transfer Agreement.
Data and Code Availability
The reported studies did not generate any unique datasets or code.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cell culture
Human colon cancer cell lines SW480 and LoVo and human embryonic kidney cell line 293T were purchased from ATCC and were maintained in McCoy 5A, RPMI 1640, and Dulbecco modified Eagle (high-glucose) media, respectively. All culture media were supplemented with 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin, and all cell lines were cultured in a humidified atmosphere with 5% CO2 at 37°C. The cell lines were authenticated by short tandem repeat analyses and routinely tested for mycoplasma.
Animals
Mouse care and experimental protocols were approved by and conducted in accordance with the guidelines of the Animal Care and Use Committee of The University of Texas MD Anderson Cancer Center. We generated the two mouse models using targeted human 15-lipoxygenase-1 (15-LOX-1) or 12S-lipoxygenase (12-LOX) overexpression in IECs via a villin promoter, designated as 15-LOX-1 mice as described previously (Zuo et al., 2012) and 12-LOX mice, respectively. B6.129S2-Alox15tm1Fun/J homozygous mice (12/15Lox(−/−), designated as 12/15LoxKO, #002778) and B6.Cg-Tg (CDX2-Cre) 101Erf/J mice (CDX2-Cre, #009350) were purchased from Jackson Laboratory. 12/15LoxKO mice with the C57/BL6 background were crossbred into the FVB/N background by breeding with FVB/N WT mice for at least 10 generations before they were used for the experiments. ApcΔ580-flox mice, in which Apc exon 14 is flanked with loxP sites, were a gift from Dr. Kenneth E. Hung at Tufts Medical Center (Hung et al., 2010). Breeding of ApcΔ580-flox mice with Cre recombinase–expressing mice (CDX2-Cre) generated ApcΔ580-flox(+/−); CDX2-Cre (+/−) mice (ApcΔ580(+/−), designated as ApcΔ580 mice in this work), in which one allele has an Apc codon 580 frameshift mutation (Hung et al., 2010, Liu et al., 2019).
METHOD DETAILS
Mouse intestinal tumorigenesis evaluation and survival experiments
1) Four-week-old 15-LOX-1 mice and their WT littermates were fed diets containing 5% or 20% corn oil (Envigo) for 4 weeks and then treated with AOM via intraperitoneal injection (7.5 mg/kg) once per week for 6 weeks (n = 6–10 mice per group). The mice were sacrificed at 20 weeks after the last injection of AOM. 2) Several steps of breeding 12/15LoxKO mice with 12-LOX or 15-LOX-1 mice were done to generate 12/15LoxKO/12-LOX or 12/15LoxKO/15-LOX-1, respectively. Four-week-old WT, 12/15LoxKO, 12/15LoxKO/12-LOX, or 12/15LoxKO/15-LOX-1 mice were fed diets containing 5% or 20% corn oil (Envigo) for 4 weeks and then treated with AOM via intraperitoneal injection (10 mg/kg) once per week for 6 weeks (n = 17–19 mice per group). The mice were sacrificed at 20 weeks after the last injection of AOM. 3) Several steps of breeding ApcΔ580 mice with 15-LOX-1 mice were done to produce ApcΔ580 (+/−);15-LOX-1(+/+) mice, designated as ApcΔ580–15-LOX-1 mice. Four-week-old ApcΔ580 and ApcΔ580–15-LOX-1 littermates were fed diets containing 5% or 20% corn oil (Envigo) for 10 consecutive weeks (n = 14 mice per group). The mice were sacrificed at age 14 weeks. The colons from the rectum to the cecum were removed and washed with phosphate-buffered saline (PBS) and photographed, tumors were counted, and tumor sizes were measured. The tumor volumes were calculated using the formula volume = (length × width2)/2 (Szot et al., 2018). The distal one-third of colons were collected in 10% neutral formalin for hematoxylin and eosin staining and immunohistochemical staining, and the other two-thirds of colons were scraped to harvest colonic epithelial cells for further analyses (e.g., RNA, protein, and mass spectrometry) as described in the corresponding Supplementary Methods sections.
Survival experiments were also performed for ApcΔ580 and ApcΔ580–15-LOX-1 mice (n = 14–20 mice per group) on a diet containing 5% or 20% corn oil. The mice were followed until they required euthanasia on the basis of at least one of the following preset criteria: (1) persistent rectal bleeding for 3 consecutive days and (2) weight loss of more than 20%.
Immunohistochemical analysis
Immunohistochemical staining was performed as described previously (Zuo et al., 2017, Liu et al., 2019). Colon tissues from the indicated experimental mice were fixed in 10% buffered formalin, embedded in paraffin, and cut into 5-μm sections. The tissue sections were deparaffinized and rehydrated, and antigen retrieval was performed with antigen unmasking solution (Vector Laboratories), and then the sections were incubated in blocking buffer (PBS with 1.5% goat serum and 0.3% Triton X-100) for 1 hour at room temperature and incubated with primary antibodies in a humidified chamber at 4°C overnight. For immunohistochemistry staining, the following primary antibodies were used: active β-catenin (#8814; Cell Signaling Technology) and Ki-67 (#RM-9106-S1; Invitrogen). Subsequently, the tissue sections were incubated with biotinylated secondary antibodies (VECTASTAIN ABC kit; Vector Laboratories) for 1 hour, followed by incubation with avidin-coupled peroxidase (Vector Laboratories) for 30 minutes. 3,3′-diaminobenzidine (DAB; Agilent Dako) was used as the chromogen, and the slides were counterstained with Mayer’s hematoxylin (Agilent Dako). Composite expression scores for immunohistochemical quantification were recorded as described previously (Zuo et al., 2017).
Cell immunofluorescence assay
Cells were seeded on coverslips in a 6-well or 12-well plate and cultured for 24 hours. The cells were fixed in cold 4% paraformaldehyde for 15 minutes and then simply rinsed with PBS. The cells were incubated with 0.1% Triton X-100 PBS on ice for 10 minutes, blocked in blocking buffer of 5% goat serum PBS containing 0.1% Triton X-100 at 37°C for 30 minutes, and then incubated with primary antibody at 4°C overnight. Subsequently, the cells were incubated with fluorochrome-conjugated secondary antibody at room temperature for 2 hours and then stained and mounted by ProLong Gold Antifade Mountant with 4′,6-diamidino-2-phenylindole (DAPI) (Thermo Fisher Scientific). For fluorescence image quantitative analysis, fluorescence intensities were measured from six random areas under a fluorescence microscope (NIS-Element software, Nikon), and then relative fluorescence intensity per cell was calculated. Cells with higher fluorescence in the nucleus than in the cytoplasm were evaluated as cells positive for active β-catenin nuclear localization and counted from six random fields under a fluorescence microscope. For quantitative analysis of colocalization fluorescence images, the cells that had mCherry and GFP fluorescence co-localization from six random areas were counted under a confocal microscope, and then the percentages of dual-fluorescence co-localization–positive cells in mCherry cells were calculated.
RNA isolation and reverse transcription real-time quantitative polymerase chain reaction (RT-qPCR) assay
Total RNAs from the cultured cells, IECs from colons, or three-dimensionally cultured colonic organoids were extracted using TRIzol reagent (Invitrogen), and then cDNA was synthesized using the Bio-Rad cDNA Synthesis Kit (Bio-Rad Laboratories). RT-qPCR was performed using FastStart Universal Probe Master (ROX) (Roche) and the StepOnePlus PCR system (Applied Biosystems) as described previously (Liu et al., 2019). An HPRT probe for human cells and β-actin probe for mouse cells were used as endogenous controls, and the mRNA expression levels of indicated targets were calculated using the comparative cycle threshold method (ddCt). All probes were purchased from Applied Biosystems. All experiments were performed in triplicate.
Western blot analysis
Western blot analysis was performed as described previously (Zuo et al., 2017). Briefly, the cultured cells or the IECs scraped from the mice colons were lysed in the lysis buffer (Zuo et al., 2017) with sonication in ice water. The blots were probed with primary and secondary antibodies. The blots were then imaged using enhanced chemiluminescence (Thermo Fisher Scientific).
Eicosanoid metabolite profiling of IECs in the mice
Four-week-old ApcΔ580 and ApcΔ580-15-LOX-1 littermates were fed diets containing 5% or 20% corn oil (Envigo) for 10 consecutive weeks. The mice (n = 14 mice per group) were euthanized at age 14 weeks. The colon crypts were scraped for eicosanoid metabolite profiling by liquid chromatography–tandem mass spectrometry measurements as described previously (Shureiqi et al., 2010).
Generation of stable 15-LOX-1–overexpressing cell lines
Precision LentiORF 15-LOX-1 in pLOC plasmid (#OHS5898–224632216, clone ID: PLOHS_ccsbBEn_05805; Dharmacon) and Precision LentiORF RFP in pLOC vector (#OHS5832; Dharmacon) were packaged into lentivirus particles by MD Anderson’s shRNA and ORFeome Core Facility. LoVo and SW480 cells were transduced with lentiviral particles (10 MOIs) with 8 μg/mL hexadimethrine bromide. Twelve hours later, the culture medium was replaced with fresh medium containing blasticidin (15 μg/mL for SW480 cells, 20 μg/mL for LoVo cells). The medium was changed every 72 hours. The cells after blasticidin selection for 2 weeks were collected and expanded under blasticidin selection for further analyses.
Generation of stable Tet-on 15-LOX-1 cell line using Tet-One system
The Tet-One Inducible Expression system was purchased from Clontech (#634303). The stable Tet-One inducible 15-LOX-1 overexpression clones were generated according to the manufacturer’s instructions. Briefly, 15-LOX-1 full-length cDNA was subcloned into pTetOne vector, and then the subcloned 15-LOX-1 construct was co-transfected with a linear puromycin marker (supplied in the kit) into 293T cells. Forty-eight hours later, cells were selected using 1 μg/mL puromycin. The medium was changed every 72 hours. Single cell clones were collected after puromycin selection for 2 weeks and expanded under puromycin selection. The positive clones were screened by treatment with doxycycline (2 μg/mL) for 48 hours and examined for 15-LOX-1 expression by western blot analysis.
Plasmid constructions
Flag-tagged SNX17 expression plasmid (pECE-M2-Snx17) was purchased from Addgene (#69811). Myc-tagged LRP5 expression plasmid (pcDNA3.1-Lrp5-myc-His) was a gift from Dr. Matthew Warman at Boston Children’s Hospital. pAdenoVator-CMV5-IRES-GFP and pAdenoVator-CMV5–15-LOX-1-IRES-GFP were obtained as described previously (Shureiqi et al., 2003). To subclone the mCherry coding sequence into the C terminus of pcDNA3.1-Lrp5-myc-His vector, we performed fusion PCR methods as follows: a pair of primers (Lrp5-F: 5′-CTCCCTCGAGACCAATAACAAC-3′; Lrp5-R: 5′-CTCGCCCTTGCTCACGGATGAGTCCGTG CAGGGGG-3′) was used to amplify the C terminus of Lrp5, and another pair of primers (mCherry-F: 5′-CTGCACGGACTCATCCGT GAGCAAGGGCGAGGAGGA-3′; mCherry-R: 5′-CCAGTTTAAACTTACTTGTACAGCTCGTCCATGCC-3′) was used to amplify mCherry cDNA. The fusion PCR products were then amplified by primers of Lrp5-F and mCherry-R using C terminus of Lrp5 and mCherry cDNA fragments as the PCR templates. The fusion PCR products were constructed into pcDNA3.1-Lrp5-Myc-His using XhoI and PmeI restriction sites to generate the pcDNA3.1-Lrp5-mCherry plasmid.
An SNX17 expression plasmid that is resistant to an individual SNX17 siRNA (Dharmacon, #J-013427-12-0002) was constructed by modifying SNX17 cDNA from 1359-CAGT-1362 to 1359-ATCC-1362 in the pECE-M2-SNX17 plasmid using GeneArt Site-Directed Mutagenesis PLUS System (A14604; Thermo Fisher Scientific) using methods similar to those described previously (McNally et al., 2017). To construct this plasmid (siRNA resistant M2-SNX17), we used the following two primers: SNX17-MF 5′-CAGATGCCAGTGCATCCGATGTCCACGGCAATTTCGC-3′, SNX17-MR 5′-TGCCGTGGACATCGGATGCACTGGCATCTGTGCTGGG-3′.
siRNA transient transfection
Cells were seeded and cultured to 60% confluence and then transiently transfected with 100nM ON-TARGETplus siRNA SMARTpool [human SNX17 (Dharmacon #L-013427-01-0005) or control/non-targeting (Dharmacon #D-001810-10-05)] using Lipofectamine RNAiMax (#13778030, Thermo Fisher Scientific) transfection reagent following the manufacturer’s protocol. The cells were further analyzed 48–72 hours after transfection.
Cell proliferation and viability assay
The cells were seeded into a 96-well plate (3 × 103/well). Twenty-four hours later, the cells were treated with 13(S)-HODE (#38610, Cayman) or dissolvent in 5% dialyzed FBS of the medium. The cell proliferation and viability were analyzed at 0, 24, 48, and 72 hours after treatment unless otherwise specified using MTT assay.
15-LOX-1 and LRP5 translocation analysis
SW480 and LoVo cells stably transduced with 15-LOX-1 or control lentivirus were seeded in a six-well plate (5 × 105/well). Twenty-four hours later, the cells were cultured in serum-free medium for another 24 hours, followed by treatment with 10μM LA dissolved in 5% dialyzed FBS of the medium for 2 hours. Cells were lysated in chilled lysis buffer (50mM HEPES [4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid], 150mM NaCl, 3mM MgCl2 in PBS, pH 7.5) with proteinase inhibitor cocktail (#11697498001; Roche). After the pellets were discarded by spinning down at 1000g for 3 minutes, the supernatants were centrifuged at 4°C at 21,460g for 1 hour. The supernatants/cytoplasmic proteins were added with a final 1% sodium dodecyl sulfate, and membrane/cytoskeleton protein pellets were then re-suspended with 1% sodium dodecyl sulfate in the same lysis buffer as mentioned above. Cytoplasmic and cell membranous 15-LOX-1 and LRP5 protein expression levels were measured by western blot analysis.
Organoid culture for mouse colon epithelial cells and human CRC tissues and organoid cell viability assay
Six-week-old ApcΔ580 and ApcΔ580–15-LOX-1 littermates (n = 6 per group) were sacrificed, and their colons were harvested and digested for three-dimensional organoid culture using the method described by Clevers and colleagues, with some modifications (Sato et al., 2011, Liu et al., 2019). Human CRC tissues for three-dimensional organoid culture were obtained from CRC surgery from patients at MD Anderson, with Institutional Review Board approval. Briefly, the glands were isolated with 10mM ethylenediaminete-traacetic acid by incubation for 10 minutes at room temperature. Equal amounts of glands (500 glands/well) were seeded in growth factor–reduced Matrigel (Corning) with the organoid culture medium (Sato et al., 2011) in 24 wells of plates. Mouse primary organoid cells were digested into single cells with TrypLE (Thermo Fisher Scientific), seeded onto Matrigel, and cultured using the same method as described above. On day 8 of the culture, primary and secondary organoids were imaged, counted, and harvested for further analyses. In addition, mouse organoid cells were treated with 13(S)-HODE (0μM, 3μM, 6μM, and 13.5μM), 15-HETE (2.15μM), LXA4 (8.86nM), and LXB4 (36.24nM) in the organoid culture medium, and the medium was changed every 3 days. On day 7 of treatment, organoid cell viability was measured using CellTiter-Glo Luminescent Cell Viability Assay kit (Promega) according to the manufacturer’s manual.
Human CRC organoids were passaged at least five generations and then digested into single cells with TrypLE (Thermo Fisher Scientific) and plated in 24-well plates with the Ultra-Low attachment surface (Corning). The cells were then transduced with 15-LOX-1 or control lentivirus particles. Twelve hours after lentivirus transduction, the organoid cells were seeded onto Matrigel and cultured using the same method as described above. On day 7 of culture, the organoids transduced with 15-LOX-1 or control lentivirus were imaged, counted, and harvested for further analyses.
Image flow cytometry for LRP5 internalization analysis
SW480 cells stably transduced with 15-LOX-1 or control lentivirus were seeded in 6-cm dishes (1 × 106 per dish). Twenty-four hours later, the cells were starved in a culture of serum-free medium for another 24 hours, then treated with 10μM LA in 5% dialyzed FBS of the medium for 2 hours. Then, the cells were digested and fixed in 4% paraformaldehyde on ice for 15 minutes, followed by incubation with blocking buffer with 0.1% Triton X-100 at room temperature for 1 hour. Subsequently, cells were incubated with anti-LRP5 primary antibody (1:200, Novopro) for 1 hour and Alexa Fluor 594 secondary antibody (1:250, #A-11012; Invitrogen) for 30 minutes at room temperature. Before the analysis, the cells were stained with DAPI (5 μg/mL) (#D9542, Sigma-Aldrich) for 5 minutes. LRP5 internalization analysis was performed using Amnis FlowSight and ImageStreamX (Luminex) according to the manufacturer’s instructions.
Immunoprecipitation and co-immunoprecipitation assays
293T cells or SW480 cells were seeded in six-well plates (5 × 105/well). Twenty-four hours later, the cells were transiently transfected with Flag-tagged SNX17 expression plasmid (pECE-M2-Snx17-Flag), Myc-tagged LRP5 expression plasmid (pcDNA3.1-Lrp5-myc-His), or their control plasmids using jetPRIME transfection reagent (#114–07, Polyplus-transfection) and were harvested 48 hours after transfection for immunoprecipitation or co-immunoprecipitation assays. These assays were performed using anti-Flag M2-tagged magnetic beads (#M8823; Sigma-Aldrich) and/or Pierce Myc–tagged magnetic IP/Co-IP Kit (#88844; Thermo Fisher Scientific) according to the manufacturer’s protocol instructions. The same amount of the lysates was incubated with magnetic bead–conjugated mouse immunoglobulin G (#5873, Cell Signaling Technology) as a control. The subsequently precipitated proteins were immunoblotted using the indicated antibodies as described in the corresponding figure legends.
Click chemistry assay
A click chemistry assay was performed as described previously (Gao and Hannoush, 2016), except LA alkyne (Alk-LA) was used instead of Alk-C16, and anti-SNX17 or anti-LRP5 antibody was used instead of anti–Wnt-3a antibody. Briefly, SW480 cells stably transduced with 15-LOX-1 or control lentivirus were seeded on micro coverslips (#10026–136, VWR) in a 12-well plate and cultured in 10% FBS of McCoy 5A medium. The cells were treated with 100 μM Alk-LA for 36 hours. Then, the cells were fixed with pre-chilled methanol at −20°C for 10 minutes, followed by incubation with 0.1% Triton X-100 PBS for 5 minutes at room temperature. The mixture solution of 5mM biotin-azide, 50mM Tris (2-carboxyethyl) phosphine hydrochloride (Sigma-Aldrich), and 50mM CuSO4 in PBS was added to the cells and incubated at room temperature for 1 hour. The cells were then incubated with blocking buffer (5% bovine serum albumin and 0.3% Triton X-100 in PBS) at room temperature for 1 hour, probed with either anti-SNX17 antibody (1:200; Proteintech) or anti-LRP5 antibody (1:200; NovoPro) at 4°C overnight, then probed with anti-biotin antibody (1:3000, #ab36406; Abcam) at room temperature for 1 hour, and then incubated with the oligonucleotide-conjugated secondary antibody solution by mixing Duolink In Situ PLA Probe Anti-Rabbit PLUS and Anti-Goat MINUS (#DUO92002 and #DUO92006, respectively; Sigma-Aldrich) together in blocking buffer at 37°C for 1 hour. Finally, the cells were incubated with ligation solution (#DUO92013; Sigma-Aldrich) in water, then with amplification solution (#DUO92013, Sigma-Aldrich) at 37°C for 100 minutes in a dark area, and then the cells were mounted with ProLong Gold Antifade Mountant with DAPI (Thermo Fisher Scientific).
Treatment with cycloheximide for protein degradation analysis
SW480 and LoVo cells stably transduced with 15-LOX-1 and control lentivirus were seeded in six-well plates (6 × 105 per well). The cells were starved with serum-free medium for another 24 hours before 10 μg/mL cycloheximide (#14126, Cayman Chemical) dissolved in dimethyl sulfoxide was added. The cells were harvested at 0, 2, and 4 hours after treatment for further analyses by western blot.
In vitro PI3-kinase VPS34 inhibition assay
SW480 and LoVo cells stably transduced with 15-LOX-1 and control lentivirus were seeded in 6-well plates (3 × 105 per well). Twenty-four hours later, the cells were treated with 10μM LA in 5% FBS of the cell culture medium or an equal amount of dissolvent (dimethyl sulfoxide) for 48 hours, then treated with 1μM class III phosphoinositide 3-kinase vacuolar protein sorting 34 inhibitor 1 (VPS34-IN-1) in serum-free medium for 2 hours. Then, the cells were harvested and measured for LRP5 protein expression by western blot analysis.
Analysis of PI3P profile by LC–HRMS
The following experiments were performed for LC-HRMS. 1) Colonic epithelial cells were scraped from ApcΔ580 and ApcΔ580-15-LOX-1 mice at 14 weeks fed with diets containing either 5% or 20% corn oil for 10 weeks (n = 3–4 mice per group). 2) For the tracing-combined assay, SW480 cells stably transduced with 15-LOX-1 and control lentivirus were seeded in 10-cm dishes to reach 80% confluence, then treated with 100 μM LA–d11 (Cayman Chemical) in 5% dialyzed FBS of McCoy 5A medium for 24 hours. The cells were quickly washed with pre-chilled PBS to remove medium completely and then harvested. 3) For the immunoprecipitation-combined assay, SW480 cells stably transduced with 15-LOX-1 and control lentivirus were seeded in 10-cm dishes and transfected with Flag-tagged SNX17 expression plasmid (pECE-M2-Snx17-Flag) using jetPRIME for 48 hours. The cells were then treated in 100 μM LA for another 24 hours and harvested for processing immunoprecipitation using anti-Flag M2 magnetic beads. Then, precipitated samples were collected.
The samples collected from the three experiments described above were extracted for lipid metabolites using cold chloroform/methanol/water acidified with hydrochloric acid (Kim et al., 2010). The extracted samples were centrifuged at 4°C at 17,000g for 5 minutes, and organic layers were transferred to clean tubes followed by evaporation to dryness under nitrogen. Samples were reconstituted in 90/10 acetonitrile/water and injected into a Thermo Fisher Scientific Vanquish liquid chromatography system containing a SeQuant ZIC-pHILIC 4.6 × 100 mm column with a 5-μm particle size. Mobile phase A was acetonitrile containing 95/5 of acetonitrile/200mM ammonium acetate, pH 5.8. Mobile phase B (MPB) was 90/5/5 of water/acetonitrile/200mM ammonium acetate, pH 5.8. The flow rate was 300 μL/minute at 35°C, and the gradient conditions were initially 15% MPB, increased to 95% MPB at 10 minutes, held at 95% MPB for 5 minutes, and returned to initial conditions and equilibrated for 5 minutes. The total run time was 20 minutes. Data were acquired using a Thermo Fisher Scientific Orbitrap Fusion Tribrid mass spectrometer with electrospray ionization negative mode at a resolution of 240,000. Then, the raw files were imported to TraceFinder software for final analysis (Thermo Fisher Scientific). The fractional abundance of each isotopologue is calculated by the peak area of the corresponding isotopologue normalized by the sum of all isotopologue areas. The isotopic relative enrichments of PI3Ps incorporated with LA or OH_13-HODE were calculated by comparing the peak areas normalized by the standard concentration of PI3P (18:1/18:1) and protein levels of each sample.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical significance was determined by the unpaired Student t test, Poisson test, or analysis of variance (one-way or two-way) with Bonferroni adjustments for all multiple comparisons. The significance of correlations was determined by the Spearman correlation coefficient. Survival time was calculated using the Kaplan-Meier method and compared between groups using the log-rank test. All tests were two-sided, and significance was defined as p < 0.05. Data were analyzed using SAS software, version 9.4 (SAS Institute) or GraphPad Prism 7.01 (GraphPad Software).
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse polyclonal anti–β-actin (WB) | Santa Cruz Biotechnology | Cat# sc-47778; RRID:AB_626632 |
| Rabbit monoclonal anti–active β-catenin (WB/IHC/IF) | Cell Signaling Technology | Cat# 8814; RRID:AB_11127203 |
| Rabbit monoclonal anti-LRP5 (WB/IP) | Cell Signaling Technology | Cat# 5731, RRID:AB_10705602 |
| Rabbit polyclonal anti-LRP5 (IF/CCA) | NovoPro | Cat# 112323 |
| Rabbit polyclonal anti-15-LOX-1 (WB) | Generated by our laboratory | N/A |
| Rabbit polyclonal anti-SNX17 (WB/CCA) | Proteintech | Cat# 10275-1-AP; RRID:AB_2192394 |
| Rabbit monoclonal anti–Na-K-ATPase (WB) | Cell Signaling Technology | Cat# 3010; RRID:AB_2060983 |
| Rabbit polyclonal anti-PI3P (IF) | Echelon Biosciences | Cat# Z-P003; RRID:AB_427221 |
| Rabbit monoclonal anti–Ki-67 (IHC) | Lab Vision | Cat# RM-9106-S1; RRID:AB_149792 |
| Rabbit monoclonal anti-cyclin D1 (WB) | Cell Signaling Technology | Cat# 2978; RRID:AB_2259616 |
| Rabbit monoclonal anti-Axin1 (WB) | Cell Signaling Technology | Cat# 2087; RRID:AB_2274550 |
| Anti-mouse immunoglobulin G (IgG), horseradish peroxidase (HRP)–linked antibody | Cell Signaling Technology | Cat# 7076S; RRID:AB_330924 |
| Anti-rabbit IgG, HRP-linked antibody | Cell Signaling Technology | Cat# 7074S; RRID:AB_2099233 |
| Biotinylated goat anti-rabbit IgG antibody | Vector Laboratories | Cat# BA-1000; RRID:AB_2313606 |
| Goat anti-rat IgG (H+L) secondary antibody, Alexa Fluor 594 conjugated | Thermo Fisher Scientific | Cat# A-11007; RRID:AB_10561522 |
| Goat anti-rabbit IgG (H+L) secondary antibody, Alexa Fluor 488 conjugated | Thermo Fisher Scientific | Cat# A-11034; RRID:AB_2576217 |
| Goat anti-rabbit IgG (H+L) secondary antibody, Alexa Fluor 594 conjugated | Thermo Fisher Scientific | Cat# A-11012; RRID:AB_141359 |
| Goat polyclonal anti-biotin (CCA) | Sigma-Aldrich | Cat# B3640; RRID:AB_258552 |
| Mouse polyclonal anti–Myc (IF) | OriGene | Cat# TA150121; RRID:AB_2622266 |
| Anti-rabbit PLUS (CCA) | Sigma-Aldrich | Cat# DUO92002; RRID:AB_2810940 |
| Anti-goat MINUS (CCA) | Sigma-Aldrich | Cat# DUO92006 |
| Biological Samples | ||
| Human colon cancer tissues | MD Anderson Cancer Center tissue bank | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| 13(S)-HODE | Cayman Chemical | Cat# 38610 |
| Linoleic acid | Cayman Chemical | Cat# 90150 |
| Alkynyl linoleic acid | Kerafast | Cat# EVU112 |
| 15(S)-HETE | Cayman Chemical | Cat# 34720 |
| Lipoxin A4 | Cayman Chemical | Cat# 90410 |
| Lipoxin B4 | Cayman Chemical | Cat# 90420 |
| Linoleic acid d-11 | Cayman Chemical | Cat# 9002193 |
| Chlorhexidine | Cayman Chemical | Cat# 17343 |
| E-64d | Cayman Chemical | Cat# 13533 |
| VPS34-IN1 | Cayman Chemical | Cat# 17392 |
| PI3P (18:1/18:1) | Avanti Polar Lipids | Cat# 850150 |
| 10% buffered formalin | Thermo Fisher Scientific | Cat# 23-245-685 |
| 4% paraformaldehyde solution | Thermo Fisher Scientific | Cat# AAJ19943K2 |
| Advanced Dulbecco modified Eagle medium | Thermo Fisher Scientific | Cat# 12634010 |
| (DMEM)/Ham F-12 medium | ||
| RPMI 1640 medium | Thermo Fisher Scientific | Cat# 11875119 |
| McCoy 5A (modified) medium | Thermo Fisher Scientific | Cat# 16600082 |
| Antigen unmasking solution | Vector Laboratories | Cat# H3300 |
| Collagenase, type IV | Sigma-Aldrich | Cat# C5138 |
| DMEM medium | Thermo Fisher Scientific | Cat# 11995073 |
| Dimethyl sulfoxide | Sigma-Aldrich | Cat# D2650 |
| ECL western blotting substrate | Thermo Fisher Scientific | Cat# PI32209 |
| Eosin-Y | Thermo Fisher Scientific | Cat# 7111 |
| Ethylenediaminetetraacetic acid (EDTA) | Sigma-Aldrich | Cat# E9884 |
| Fetal bovine serum | Atlanta Biologicals | Cat# S12450H |
| GlutaMAX (100 ×) | Thermo Fisher Scientific | Cat# 35050061 |
| Normal goat serum | Cell Signaling Technology | Cat# 5425S |
| Hexadimethrine bromide (Polybrene) | Santa Cruz Biotechnology | Cat# SC-134220 |
| Hydrogen peroxide | Sigma-Aldrich | Cat# H3410 |
| Matrigel | Corning | Cat# 356231 |
| Mayer hematoxylin | Agilent Dako | Cat# S330930-2 |
| Nonidet P-40 | Sigma-Aldrich | Cat# 74385 |
| N-acetyl-L-cysteine | Sigma-Aldrich | Cat# A7250 |
| Penicillin-streptomycin-glutamine (100 ×) | Thermo Fisher Scientific | Cat# 10378016 |
| Phenylmethylsulfonyl fluoride | Sigma-Aldrich | Cat# P7626 |
| Recombinant murine Wnt-3a | EMD Millipore | Cat# GF154 |
| TRIzol | Thermo Fisher Scientific | Cat# 15596018 |
| Trypsin-EDTA | Thermo Fisher Scientific | Cat# 25300062 |
| Recombinant human Wnt-3a protein | R&D Systems | Cat# 5036-WN-010 |
| cOmplete protease inhibitor cocktail | Roche | Cat# 11697498001 |
| Critical Commercial Assays | ||
| iScript cDNA Synthesis Kit | Bio-Rad | Cat# 1708891BUN |
| FastStart Universal Probe Master (ROX) | Roche | Cat# 4914058001 |
| CellTiter-Glo Luminescent Cell Viability Assay kit | Promega | Cat# G7570 |
| Mouse IgG-Ab magnetic beads | Cell Signaling Technology | Cat# 5873 |
| Pierce Myc-Tag Magnetic IP/Co-IP Kit | Thermo Fisher Scientific | Cat# 88844 |
| Detection Reagents FarRed Kit | Sigma-Aldrich | Cat# DUO92013 |
| Anti-FLAG M2 magnetic beads | Sigma-Aldrich | Cat# M8823 |
| ProLong Gold Antifade Mountant with DAPI | Thermo Fisher Scientific | Cat# P36935 |
| Lipofectamine RNAiMax | Thermo Fisher Scientific | Cat# 13778030 |
| jetPRIME | Polyplus-transfection | Cat# 114-07 |
| CellLight Lysosomes-GFP, BacMam 2.0 | Thermo Fisher Scientific | Cat# C10507 |
| CellLight Early Endosomes-GFP, BacMam 2.0 | Thermo Fisher Scientific | Cat# C10586 |
| Experimental Models: Cell Lines | ||
| Human: LoVo | ATCC | CCL-229 |
| Human: SW480 | ATCC | CCL-228 |
| Human: HEK293T/17 | ATCC | CRL-11268 |
| Experimental Models: Organisms/Strains | ||
| Mouse: B6.Cg-Tg (CDX2-Cre)101Erf/J | Jackson Laboratory | Stock# 009350 |
| Mouse: C57BL/6J (B6) | Jackson Laboratory | Stock# 664 |
| Mouse: FVB/N | Charles River Laboratories | Strain Code 207 |
| Mouse: ApcΔ580-flox | Donated by Dr. Kenneth E. Hung | N/A |
| Oligonucleotides | ||
| Human HPRT1 (VIC/MGB) endogenous control | Thermo Fisher Scientific | 4Cat# 326321E |
| Human Axin2 (FAM/MGB) | Thermo Fisher Scientific | Hs00610344_m1 |
| Human cyclin D1 (FAM/MGB) | Thermo Fisher Scientific | Hs00765553_m1 |
| Mouse β-actin (FAM/MGB) endogenous control | Thermo Fisher Scientific | Mm02619580_g1 |
| Mouse Axin2 (FAM/MGB) | Thermo Fisher Scientific | Mm00443610_m1 |
| Mouse cyclin D1 (FAM/MGB) | Thermo Fisher Scientific | Mm00432359_m1 |
| ON-TARGETplus human SNX17 siRNA SMARTpool | Dharmacon | Cat# L-013427-01-0005 |
| Individual ON-TARGETplus human SNX17 siRNA | Dharmacon | Cat# J-013427-12-0002 |
| Control/non-targeting siRNA | Dharmacon | Cat# D-001810-10-05 |
| Recombinant DNA | ||
| pcDNA3.1-LRP5-Myc-His | Donated by Dr. Matthew Warman in Boston Children’s Hospital | N/A |
| pcDNA3.1-Lrp5-mCherry | This paper | N/A |
| p12.4Kvill-12-S-LOX | This paper | N/A |
| Precision LentiORF 15-LOX-1 in pLOC | Dharmacon | OCat# HS5898-224632216 |
| Precision LentiORF RFP in pLOC | Dharmacon | Cat# OHS5832 |
| pECE-M2-SNX17 | Addgene | 6Cat# 9811 |
| M2-SNX17 (siRNA resistant) | This paper | N/A |
| pAdenoVator-CMV5-IRES-GFP | This paper | N/A |
| pAdenoVator-CMV5-15-LOX-1-IRES-GFP | This paper | N/A |
| Tet-One Inducible Expression System | Clontech | Cat# 634301 |
| Software and Algorithms | ||
| Thermo Trace Finder | Thermo Fisher Scientific | N/A |
| GraphPad Prism 7 | GraphPad Software Inc. | SCR_002798 |
| ImageJ | NIH: Open source image processing software | SCR_003070 |
| SAS software 9.4 | SAS Institute | https://www.sas.com/genome.html |
| Adobe Illustrator CC 2017 | Adobe Inc. | SCR_010279 |
Highlights.
Colonic 15-LOX-1 expression inhibits colon cancer promotion by linoleic acid in mice
Dietary linoleic acid upregulates LRP5 to enhance colonic Wnt/β-catenin activation
15-LOX-1 peroxidation of linoleic acid in PI3P promotes PI3P_13-HODE formation
Low affinity of PI3P_13-HODE for SNX17 inhibits LRP5 recycling
ACKNOWLEDGMENTS
This work was supported by the National Cancer Institute (R01-CA195686 and R01-CA206539 to I.S.) and the Cancer Prevention and Research Institute of Texas (RP150195 to I.S.). This study made use of the MD Anderson Cancer Center Genetically Engineered Mouse Facility, Functional Genomics Core, Flow Cytometry and Cellular Imaging Facility, Sequencing and Microarray Facility, and Research Animal Support Facility—Smithville Laboratory Animal Genetic Services, supported by Cancer Center Support Grant P30CA016672, and the MD Anderson Cancer Center metabolomics facility service, supported in part by Cancer Prevention and Research Institute of Texas grant RP130397 and National Institutes of Health grant 1S10OD012304-01. We thank Dr. Jae-Il Park at The University of Texas MD Anderson Cancer Center for his feedback on this work. We thank Ms. Sarah J. Bronson and Ms. Erica Goodoff in Scientific Publications, Research Medical Library at MD Anderson for editing the article. We thank Dr. Matthew Warman in Boston Children’s Hospital for donating the LRP5 plasmid.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2020.108049.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Adam O, Tesche A, and Wolfram G (2008). Impact of linoleic acid intake on arachidonic acid formation and eicosanoid biosynthesis in humans. Prostaglandins Leukot. Essent. Fatty Acids 79, 177–181. [DOI] [PubMed] [Google Scholar]
- Almeida M, Ambrogini E, Han L, Manolagas SC, and Jilka RL (2009). Increased lipid oxidation causes oxidative stress, increased peroxisome proliferator-activated receptor-g expression, and diminished pro-osteogenic Wnt signaling in the skeleton. J. Biol. Chem 284, 27438–27448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bago R, Malik N, Munson MJ, Prescott AR, Davies P, Sommer E, Shpiro N, Ward R, Cross D, Ganley IG, and Alessi DR (2014). Characterization of VPS34-IN1, a selective inhibitor of Vps34, reveals that the phosphatidylinositol 3-phosphate-binding SGK3 protein kinase is a downstream target of class III phosphoinositide 3-kinase. Biochem. J 463, 413–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barthold SW, and Beck D (1980). Modification of early dimethylhydrazine carcinogenesis by colonic mucosal hyperplasia. Cancer Res. 40, 4451–4455. [PubMed] [Google Scholar]
- Blasbalg TL, Hibbeln JR, Ramsden CE, Majchrzak SF, and Rawlings RR (2011). Changes in consumption of omega-3 and omega-6 fatty acids in the United States during the 20th century. Am. J. Clin. Nutr 93, 950–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boman BM, and Fields JZ (2013). An APC:WNT counter-current-like mechanism regulates cell division along the colonic crypt axis: a mechanism that explains how APC mutations induce proliferative abnormalities that drive colon cancer development. Front. Oncol 3, 244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brabletz T, Jung A, Hermann K, Günther K, Hohenberger W, and Kirchner T (1998). Nuclear overexpression of the oncoprotein beta-catenin in colorectal cancer is localized predominantly at the invasion front. Pathol. Res. Pract 194, 701–704. [DOI] [PubMed] [Google Scholar]
- Brabletz T, Jung A, Spaderna S, Hlubek F, and Kirchner T (2005). Opinion: migrating cancer stem cells - an integrated concept of malignant tumour progression. Nat. Rev. Cancer 5, 744–749. [DOI] [PubMed] [Google Scholar]
- Brash AR (1999). Lipoxygenases: occurrence, functions, catalysis, and acquisition of substrate. J. Biol. Chem 274, 23679–23682. [DOI] [PubMed] [Google Scholar]
- Bull AW, Bronstein JC, and Nigro ND (1989). The essential fatty acid requirement for azoxymethane-induced intestinal carcinogenesis in rats. Lipids 24, 340–346. [DOI] [PubMed] [Google Scholar]
- Clevers H, and Nusse R (2012). Wnt/β-catenin signaling and disease. Cell 149, 1192–1205. [DOI] [PubMed] [Google Scholar]
- Deschner EE, Lytle JS, Wong G, Ruperto JF, and Newmark HL (1990). The effect of dietary omega-3 fatty acids (fish oil) on azoxymethanol-induced focal areas of dysplasia and colon tumor incidence. Cancer 66, 2350–2356. [DOI] [PubMed] [Google Scholar]
- Farvid MS, Ding M, Pan A, Sun Q, Chiuve SE, Steffen LM, Willett WC, and Hu FB (2014). Dietary linoleic acid and risk of coronary heart disease: a systematic review and meta-analysis of prospective cohort studies. Circulation 130, 1568–1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fearon ER, and Vogelstein B (1990). A genetic model for colorectal tumorigenesis. Cell 61, 759–767. [DOI] [PubMed] [Google Scholar]
- Gao X, and Hannoush RN (2016). Visualizing Wnt palmitoylation in single cells. Methods Mol. Biol 1481, 1–9. [DOI] [PubMed] [Google Scholar]
- Grossi IM, Fitzgerald LA, Umbarger LA, Nelson KK, Diglio CA, Taylor JD, and Honn KV (1989). Bidirectional control of membrane expression and/or activation of the tumor cell IRGpIIb/IIIa receptor and tumor cell adhesion by lipoxygenase products of arachidonic acid and linoleic acid. Cancer Res. 49, 1029–1037. [PubMed] [Google Scholar]
- Gu Z, Wu J, Wang S, Suburu J, Chen H, Thomas MJ, Shi L, Edwards IJ, Berquin IM, and Chen YQ (2013). Polyunsaturated fatty acids affect the localization and signaling of PIP3/AKT in prostate cancer cells. Carcinogenesis 34, 1968–1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He B, Reguart N, You L, Mazieres J, Xu Z, Lee AY, Mikami I, McCormick F, and Jablons DM (2005). Blockade of Wnt-1 signaling induces apoptosis in human colorectal cancer cells containing downstream mutations. Oncogene 24, 3054–3058. [DOI] [PubMed] [Google Scholar]
- Herz J, Chen Y, Masiulis I, and Zhou L (2009). Expanding functions of lipoprotein receptors. J. Lipid Res 50 (Suppl), S287–S292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinoi T, Akyol A, Theisen BK, Ferguson DO, Greenson JK, Williams BO, Cho KR, and Fearon ER (2007). Mouse model of colonic adenoma-carcinoma progression based on somatic Apc inactivation. Cancer Res. 67, 9721–9730. [DOI] [PubMed] [Google Scholar]
- Hung KE, Maricevich MA, Richard LG, Chen WY, Richardson MP, Kunin A, Bronson RT, Mahmood U, and Kucherlapati R (2010). Development of a mouse model for sporadic and metastatic colon tumors and its use in assessing drug treatment. Proc. Natl. Acad. Sci. U S A 107, 1565–1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Il Lee S, Zuo X, and Shureiqi I (2011). 15-Lipoxygenase-1 as a tumor suppressor gene in colon cancer: is the verdict in? Cancer Metastasis Rev. 30, 481–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jandacek RJ (2017). Linoleic acid: a nutritional quandary. Healthcare (Basel) 5, 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Shanta SR, Zhou LH, and Kim KP (2010). Mass spectrometry based cellular phosphoinositides profiling and phospholipid analysis: a brief review. Exp. Mol. Med 42, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinder M, Wei C, Shelat SG, Kundu M, Zhao L, Blair IA, and Puré E (2010). Hematopoietic stem cell function requires 12/15-lipoxygenase-dependent fatty acid metabolism. Blood 115, 5012–5022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieger M, and Herz J (1994). Structures and functions of multiligand lipoprotein receptors: macrophage scavenger receptors and LDL receptor-related protein (LRP). Annu. Rev. Biochem 63, 601–637. [DOI] [PubMed] [Google Scholar]
- Kuhn H, Belkner J, Wiesner R, and Brash AR (1990). Oxygenation of biological membranes by the pure reticulocyte lipoxygenase. J. Biol. Chem 265, 18351–18361. [PubMed] [Google Scholar]
- Lipkin M, Reddy B, Newmark H, and Lamprecht SA (1999). Dietary factors in human colorectal cancer. Annu. Rev. Nutr 19, 545–586. [DOI] [PubMed] [Google Scholar]
- Liu B, Khan WA, Hannun YA, Timar J, Taylor JD, Lundy S, Butovich I, and Honn KV (1995). 12(S)-hydroxyeicosatetraenoic acid and 13(S)-hydroxyoctadecadienoic acid regulation of protein kinase C-alpha in melanoma cells: role of receptor-mediated hydrolysis of inositol phospholipids. Proc. Natl. Acad. Sci. U S A 92, 9323–9327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Deguchi Y, Tian R, Wei D, Wu L, Chen W, Xu W, Xu M, Liu F, Gao S, et al. (2019). Pleiotropic effects of PPARD accelerate colorectal tumorigenesis, progression, and invasion. Cancer Res. 79, 954–969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losi L, Di Gregorio C, Pedroni M, Ponti G, Roncucci L, Scarselli A, Genuardi M, Baglioni S, Marino M, Rossi G, et al. (2005). Molecular genetic alterations and clinical features in early-onset colorectal carcinomas and their role for the recognition of hereditary cancer syndromes. Am. J. Gastroenterol 100, 2280–2287. [DOI] [PubMed] [Google Scholar]
- MacDonald BT, and He X (2012). Frizzled and LRP5/6 receptors for Wnt/β-catenin signaling. Cold Spring Harb. Perspect. Biol 4, a007880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao F, Xu M, Zuo X, Yu J, Xu W, Moussalli MJ, Elias E, Li HS, Watowich SS, and Shureiqi I (2015). 15-Lipoxygenase-1 suppression of colitis-associated colon cancer through inhibition of the IL-6/STAT3 signaling pathway. FASEB J. 29, 2359–2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNally KE, Faulkner R, Steinberg F, Gallon M, Ghai R, Pim D, Langton P, Pearson N, Danson CM, Nägele H, et al. (2017). Retriever is a multiprotein complex for retromer-independent endosomal cargo recycling. Nat. Cell Biol 19, 1214–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middleton MK, Zukas AM, Rubinstein T, Jacob M, Zhu P, Zhao L, Blair I, and Puré E (2006). Identification of 12/15-lipoxygenase as a suppressor of myeloproliferative disease. J. Exp. Med 203, 2529–2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moussalli MJ, Wu Y, Zuo X, Yang XL, Wistuba II, Raso MG, Morris JS, Bowser JL, Minna JD, Lotan R, and Shureiqi I (2011). Mechanistic contribution of ubiquitous 15-lipoxygenase-1 expression loss in cancer cells to terminal cell differentiation evasion. Cancer Prev. Res. (Phila.) 4, 1961–1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller K, Siebert M, Heidt M, Marks F, Krieg P, and Fürstenberger G (2002). Modulation of epidermal tumor development caused by targeted overexpression of epidermis-type 12S-lipoxygenase. Cancer Res. 62, 4610–4616. [PubMed] [Google Scholar]
- Nixon JB, Kim KS, Lamb PW, Bottone FG, and Eling TE (2004). 15-Lipoxygenase-1 has anti-tumorigenic effects in colorectal cancer. Prostaglandins Leukot. Essent. Fatty Acids 70, 7–15. [DOI] [PubMed] [Google Scholar]
- Oshima H, Nakayama M, Han TS, Naoi K, Ju X, Maeda Y, Robine S, Tsuchiya K, Sato T, Sato H, et al. (2015). Suppressing TGFb signaling in regenerating epithelia in an inflammatory microenvironment is sufficient to cause invasive intestinal cancer. Cancer Res. 75, 766–776. [DOI] [PubMed] [Google Scholar]
- Ramsden CE, Zamora D, Leelarthaepin B, Majchrzak-Hong SF, Faurot KR, Suchindran CM, Ringel A, Davis JM, and Hibbeln JR (2013). Use of dietary linoleic acid for secondary prevention of coronary heart disease and death: evaluation of recovered data from the Sydney Diet Heart Study and updated meta-analysis. BMJ 346, e8707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T, Stange DE, Ferrante M, Vries RG, Van Es JH, Van den Brink S, Van Houdt WJ, Pronk A, Van Gorp J, Siersema PD, and Clevers H (2011). Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett’s epithelium. Gastroenterology 141, 1762–1772. [DOI] [PubMed] [Google Scholar]
- Schewe T, Halangk W, Hiebsch C, and Rapoport SM (1975). A lipoxygenase in rabbit reticulocytes which attacks phospholipids and intact mitochondria. FEBS Lett. 60, 149–152. [DOI] [PubMed] [Google Scholar]
- Schumann T, Adhikary T, Wortmann A, Finkernagel F, Lieber S, Schnitzer E, Legrand N, Schober Y, Nockher WA, Toth PM, et al. (2015). Deregulation of PPARβ/δ target genes in tumor-associated macrophages by fatty acid ligands in the ovarian cancer microenvironment. Oncotarget 6, 13416–13433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwitalla S, Fingerle AA, Cammareri P, Nebelsiek T, Göktuna SI, Ziegler PK, Canli O, Heijmans J, Huels DJ, Moreaux G, et al. (2013). Intestinal tumorigenesis initiated by dedifferentiation and acquisition of stem-cell-like properties. Cell 152, 25–38. [DOI] [PubMed] [Google Scholar]
- Serhan CN, Chiang N, and Van Dyke TE (2008). Resolving inflammation: dual anti-inflammatory and pro-resolution lipid mediators. Nat. Rev. Immunol 8, 349–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shureiqi I, Wojno KJ, Poore JA, Reddy RG, Moussalli MJ, Spindler SA, Greenson JK, Normolle D, Hasan AA, Lawrence TS, and Brenner DE (1999). Decreased 13-S-hydroxyoctadecadienoic acid levels and 15-lipoxygenase-1 expression in human colon cancers. Carcinogenesis 20, 1985–1995. [DOI] [PubMed] [Google Scholar]
- Shureiqi I, Jiang W, Zuo X, Wu Y, Stimmel JB, Leesnitzer LM, Morris JS, Fan HZ, Fischer SM, and Lippman SM (2003). The 15-lipoxygenase-1 product 13-S-hydroxyoctadecadienoic acid down-regulates PPAR-delta to induce apoptosis in colorectal cancer cells. Proc. Natl. Acad. Sci. USA 100, 9968–9973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shureiqi I, Wu Y, Chen D, Yang XL, Guan B, Morris JS, Yang P, Newman RA, Broaddus R, Hamilton SR, et al. (2005). The critical role of 15-lipoxygenase-1 in colorectal epithelial cell terminal differentiation and tumorigenesis. Cancer Res. 65, 11486–11492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shureiqi I, Chen D, Day RS, Zuo X, Hochman FL, Ross WA, Cole RA, Moy O, Morris JS, Xiao L, et al. (2010). Profiling lipoxygenase metabolism in specific steps of colorectal tumorigenesis. Cancer Prev. Res. (Phila.) 3, 829–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel RL, Miller KD, and Jemal A (2020). Cancer statistics, 2020. CA Cancer J. Clin 70, 7–30. [DOI] [PubMed] [Google Scholar]
- Spiteller D, and Spiteller G (2000). Oxidation of linoleic acid in low-density lipoprotein: an important event in atherogenesis. Angew. Chem. Int. Ed. Engl 39, 585–589. [DOI] [PubMed] [Google Scholar]
- Suzuki H, Watkins DN, Jair K-W, Schuebel KE, Markowitz SD, Chen WD, Pretlow TP, Yang B, Akiyama Y, Van Engeland M, et al. (2004). Epigenetic inactivation of SFRP genes allows constitutive WNT signaling in colorectal cancer. Nat. Genet 36, 417–422. [DOI] [PubMed] [Google Scholar]
- Szot C, Saha S, Zhang XM, Zhu Z, Hilton MB, Morris K, Seaman S, Dunleavey JM, Hsu KS, Yu GJ, et al. (2018). Tumor stroma-targeted antibody-drug conjugate triggers localized anticancer drug release. J. Clin. Invest 128, 2927–2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tortelote GG, Reis RR, de Almeida Mendes F, and Abreu JG (2017). Complexity of the Wnt/β-catenin pathway: Searching for an activation model. Cell. Signal 40, 30–43. [DOI] [PubMed] [Google Scholar]
- van de Wetering M, Sancho E, Verweij C, de Lau W, Oving I, Hurlstone A, van der Horn K, Batlle E, Coudreuse D, Haramis AP, et al. (2002). The beta-catenin/TCF-4 complex imposes a crypt progenitor phenotype on colorectal cancer cells. Cell 111, 241–250. [DOI] [PubMed] [Google Scholar]
- van Kerkhof P, Lee J, McCormick L, Tetrault E, Lu W, Schoenfish M, Oorschot V, Strous GJ, Klumperman J, and Bu G (2005). Sorting nexin 17 facilitates LRP recycling in the early endosome. EMBO J. 24, 2851–2861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeulen L, De Sousa E Melo F, van der Heijden M, Cameron K, de Jong JH, Borovski T, Tuynman JB, Todaro M, Merz C, Rodermond H, et al. (2010). Wnt activity defines colon cancer stem cells and is regulated by the microenvironment. Nat. Cell Biol 12, 468–476. [DOI] [PubMed] [Google Scholar]
- Wang W, Yang J, Edin ML, Wang Y, Luo Y, Wan D, Yang H, Song C-Q, Xue W, Sanidad KZ, et al. (2019). Targeted metabolomics identifies the cytochrome P450 monooxygenase eicosanoid pathway as a novel therapeutic target of colon tumorigenesis. Cancer Res. 79, 1822–1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong WM, Mandir N, Goodlad RA, Wong BC, Garcia SB, Lam SK, and Wright NA (2002). Histogenesis of human colorectal adenomas and hyperplastic polyps: the role of cell proliferation and crypt fission. Gut 50, 212–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Fang B, Yang XQ, Wang L, Chen D, Krasnykh V, Carter BZ, Morris JS, and Shureiqi I (2008). Therapeutic molecular targeting of 15-lipoxygenase-1 in colon cancer. Mol. Ther 16, 886–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang K, Fan K, Newmark H, Leung D, Lipkin M, Steele VE, and Kelloff GJ (1996). Cytokeratin, lectin, and acidic mucin modulation in differentiating colonic epithelial cells of mice after feeding Western-style diets. Cancer Res. 56, 4644–4648. [PubMed] [Google Scholar]
- Yuri M, Sasahira T, Nakai K, Ishimaru S, Ohmori H, and Kuniyasu H (2007). Reversal of expression of 15-lipoxygenase-1 to cyclooxygenase-2 is associated with development of colonic cancer. Histopathology 51, 520–527. [DOI] [PubMed] [Google Scholar]
- Zhu H, Takahashi Y, Xu W, Kawajiri H, Murakami T, Yamamoto M, Iseki S, Iwasaki T, Hattori H, and Yoshimoto T (2003). Low density lipoprotein receptor-related protein-mediated membrane translocation of 12/15-lipoxygenase is required for oxidation of low density lipoprotein by macrophages. J. Biol. Chem 278, 13350–13355. [DOI] [PubMed] [Google Scholar]
- Zuo X, Peng Z, Wu Y, Moussalli MJ, Yang XL, Wang Y, Parker-Thornburg J, Morris JS, Broaddus RR, Fischer SM, and Shureiqi I (2012). Effects of gut-targeted 15-LOX-1 transgene expression on colonic tumorigenesis in mice. J. Natl. Cancer Inst 104, 709–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo X, Xu W, Xu M, Tian R, Moussalli MJ, Mao F, Zheng X, Wang J, Morris JS, Gagea M, et al. (2017). Metastasis regulation by PPARD expression in cancer cells. JCI Insight 2, e91419. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The reported studies did not generate any unique datasets or code.


