Abstract
Uncertainty surrounding the role of Dientamoeba fragilis in human disease could be due in part to the existence of pathogenic and nonpathogenic variants. Evidence for two genetically distinct forms was obtained using PCR-restriction fragment length polymorphism analysis of ribosomal genes. Future studies in humans will need to take D. fragilis diversity into account.
Dientamoeba fragilis is a little-studied ameboid inhabitant of the human large bowel related to trichomonad flagellates (13). Its role, if any, in human disease is unclear, but several reports have linked its presence to gastrointestinal symptoms (see, for example, references 1, 6, 8, 10, 12, and 14). The prevalence of D. fragilis is not well known: in some surveys it was not reported, while in others a prevalence of 5% or more has been found. Diagnosis requires fresh feces since trophozoites degenerate within a few hours of stool passage, and a cyst form has never been demonstrated. Identification of two nuclei with fragmented chromatin in the stained trophozoite is the primary diagnostic characteristic. D. fragilis has the reputation of being difficult to maintain in culture and has never been cultivated axenically.
In several other gut protozoa, including Entamoeba species, Cryptosporidium parvum, and Blastocystis hominis, significant genetic diversity is present in the absence of morphological variation (2, 4, 7, 9). Uncertainty surrounding the role of D. fragilis in disease arises from the existence of asymptomatic infections (8) and from the fact that the reported symptoms could have many etiologies. However, it is possible that two or more genetically distinct organisms are being called D. fragilis but only one is pathogenic. If true, the effects of the pathogen could be masked in population surveys by the existence of the commensal. To address this possibility, we investigated genetic variation in D. fragilis by riboprinting (3)-restriction fragment length polymorphism analysis of PCR-amplified small-subunit rRNA genes.
All fecal specimens sent to the East Surrey Hospital Department of Microbiology from August 1998 to August 1999 (4,280 samples, all from symptomatic individuals) were examined microscopically in saline wet mounts, as well as by standard microbiological culture. The 17 samples containing trophozoites were stained with Leishman stain, a rapid method for identifying nuclei. The 10 samples with binucleate trophozoites were inoculated into Robinson's medium (11) with rice starch and incubated at 37°C; they were also fixed in sodium acetate-acetic acid-formalin for later iron-hematoxylin staining. Cultures were examined for D. fragilis every 2 days. Phagocytosis of rice starch proved a simple method for distinguishing D. fragilis from B. hominis, since the latter does not ingest it while the former does so voraciously. After 5 to 6 days, cultures were pelleted and lysed in 0.25 ml of 0.25% sodium dodecyl sulfate–0.1 M EDTA (pH 8), and DNA was extracted as previously described (5). PCR amplification with Taq DNA polymerase was done under standard conditions and with primers TRD5 (GATACTTGGTTGATCCTGCCAAGG) and TRD3 (GATCCAACGGCAGGTTCACCTACC) (13), which amplify only trichomonad small-subunit rRNA genes. Amplification was achieved by 30 cycles of 94°C for 1 min, 55°C for 1.5 min, and 72°C for 2 min. The 1.7-kbp PCR products were digested with 10 restriction enzymes, and the fragments were separated in 2.4% agarose gels (NuSieve 3:1 Agarose: FMC BioProducts) with appropriate size markers.
All nine stool samples positive for D. fragilis gave rise to short-term cultures of the ameba. Only 1 of 10 samples in which binucleate trophozoites were seen with Leishman stain did not contain D. fragilis in the iron-hematoxylin-stained preparation (it contained Endolimax nana). All of the D. fragilis-positive patients had diarrhea, and five of nine patients reported abdominal pain. Seven had other parasites (five had B. hominis, one had Entamoeba coli, and one had Endolimax nana), and one was coinfected with Campylobacter spp. Infection with D. fragilis in our population appears to be short-lived. Five of the nine infected patients provided follow-up stool samples approximately 2 weeks after initial detection. None of the samples were positive in saline wet mounts or by culture, although one was found to be positive after iron-hematoxylin staining. The presence of D. fragilis was reported to the physician, but only the patient with the concurrent Campylobacter infection was treated, with ciprofloxacin.
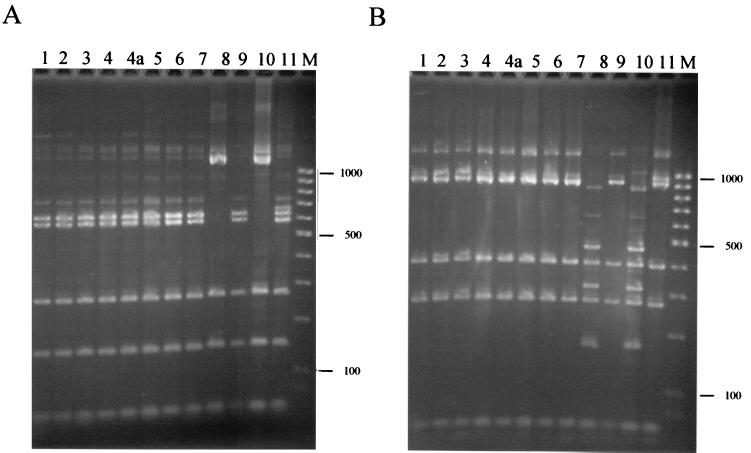
Small-subunit rRNA genes were amplified successfully from D. fragilis in all cases. Two control samples were used: D. fragilis strain Bi/PA (ATCC 30948), for which the small-subunit rRNA gene sequence is available (13), and a short-term D. fragilis culture from the London School of Hygiene and Tropical Medicine. Restriction patterns were identical for all isolates with eight enzymes (Sau3AI, Sau96I, HaeIII, HhaI, AluI, MspI, HinfI, and TaqI), but two enzyme patterns (RsaI and DdeI) were distinct in two isolates, one of which was Bi/PA. The patterns of the latter were exactly those predicted from the gene sequence. The two groups within the 12 isolates examined differed by only three restriction enzyme sites in their small-subunit rRNA genes, a finding which extrapolates to approximately 2% sequence divergence.
The number of isolates examined was small; however, the less common of the two variants is present in both Europe and North America since Bi/PA (isolated in Illinois in 1948) shares the same pattern as one of our isolates (Fig. 1, lane 8). The latter came from a 2-year-old child with no history of travel. We cannot exclude the existence of more variants within the species at present, as many more samples of geographically diverse origins are needed.
FIG. 1.
Ribosomal gene sequence variation among D. fragilis isolates. PCR-amplified small-subunit rRNA genes from 12 isolates of D. fragilis were digested with restriction enzyme RsaI (A) or DdeI (B), and the fragments were separated in 2.4% agarose gels before staining them with ethidium bromide. The size marker (M) is a 100-bp ladder. The faint bands represent incomplete restriction digestion products. Lanes 1 to 9, isolates from East Surrey Hospital; lane 10, isolate Bi/PA; lane 11, isolate from the London School of Hygiene and Tropical Medicine. Lanes 4 and 4a are duplicate samples from cultures inoculated using the same stool sample.
The existence of genetic variants in D. fragilis does not necessarily indicate that they have distinct effects in the human host, but it does show that organisms currently being reported as D. fragilis represent at least two significantly different genetic entities. The degree of divergence between them is comparable to that between Entamoeba histolytica and Entamoeba dispar, and it could be argued that they represent distinct species. We believe such a conclusion is premature, but our data do indicate that future studies of the role of D. fragilis in human disease need to take into account the existence of genetic diversity.
(This study formed part of an M.Sc. Thesis at the University of Surrey [J.A.J.])
Acknowledgments
We thank the Department of Microbiology at East Surrey Hospital for use of their facilities and John E. Williams of the London School of Hygiene and Tropical Medicine for advice, media, and one of the samples.
REFERENCES
- 1.Ayadi A, Bahri I. Dientamoeba fragilis: flagelle pathogène? Bull Soc Pathol Exot. 1999;92:299–301. [PubMed] [Google Scholar]
- 2.Clark C G. Extensive genetic diversity in Blastocystis hominis. Mol Biochem Parasitol. 1997;87:79–83. doi: 10.1016/s0166-6851(97)00046-7. [DOI] [PubMed] [Google Scholar]
- 3.Clark C G. Riboprinting: a tool for the study of genetic diversity in microorganisms. J Eukaryot Microbiol. 1997;44:277–283. doi: 10.1111/j.1550-7408.1997.tb05667.x. [DOI] [PubMed] [Google Scholar]
- 4.Clark C G, Diamond L S. The Laredo strain and other Entamoeba histolytica-like amoebae are Entamoeba moshkovskii. Mol Biochem Parasitol. 1991;46:11–18. doi: 10.1016/0166-6851(91)90194-b. [DOI] [PubMed] [Google Scholar]
- 5.Clark C G, Diamond L S. Intraspecific variation and phylogenetic relationships in the genus Entamoeba as revealed by riboprinting. J Eukaryot Microbiol. 1997;44:142–154. doi: 10.1111/j.1550-7408.1997.tb05951.x. [DOI] [PubMed] [Google Scholar]
- 6.Cuffari C, Oligny L, Seidman E G. Dientamoeba fragilis masquerading as allergic colitis. J Pediatr Gastroenterol Nutr. 1998;26:16–20. doi: 10.1097/00005176-199801000-00003. [DOI] [PubMed] [Google Scholar]
- 7.Diamond L S, Clark C G. A redescription of Entamoeba histolytica Schaudinn, 1903 (Emended Walker, 1911) separating it from Entamoeba dispar Brumpt, 1925. J Eukaryot Microbiol. 1993;40:340–344. doi: 10.1111/j.1550-7408.1993.tb04926.x. [DOI] [PubMed] [Google Scholar]
- 8.Grendon J H, DiGiacomo R F, Frost F J. Descriptive features of Dientamoeba fragilis infections. J Trop Med Hyg. 1995;98:309–315. [PubMed] [Google Scholar]
- 9.Morgan U M, Xiao L, Fayer R, Lal A A, Thompson R C A. Variation in Cryptosporidium: towards a taxonomic revision of the genus. Int J Parasitol. 1999;29:1733–1751. doi: 10.1016/s0020-7519(99)00109-5. [DOI] [PubMed] [Google Scholar]
- 10.Preiss U, Ockert G, Broemme S, Otto A. On the clinical importance of Dientamoeba fragilis infections in childhood. J Hyg Epidemiol Microbiol Immunol. 1991;35:27–34. [PubMed] [Google Scholar]
- 11.Robinson G L. The laboratory diagnosis of human parasitic amoebae. Trans R Soc Trop Med Hyg. 1968;62:285–294. doi: 10.1016/0035-9203(68)90170-3. [DOI] [PubMed] [Google Scholar]
- 12.Sawangjaroen N, Luke R, Prociv P. Diagnosis by faecal culture of Dientamoeba fragilis infections in Australian patients with diarrhoea. Trans R Soc Trop Med Hyg. 1993;87:163–165. doi: 10.1016/0035-9203(93)90472-3. [DOI] [PubMed] [Google Scholar]
- 13.Silberman J D, Clark C G, Sogin M L. Dientamoeba fragilis shares a recent common evolutionary history with the trichomonads. Mol Biochem Parasitol. 1996;76:311–314. doi: 10.1016/0166-6851(95)02516-2. [DOI] [PubMed] [Google Scholar]
- 14.Windsor J J, Rafay A M, Shenoy A K, Johnson E H. Incidence of Dientamoeba fragilis in faecal samples submitted for routine microbiological analysis. Br J Biomed Sci. 1998;55:172–175. [PubMed] [Google Scholar]