Abstract
We have demonstrated, by PCR and restriction enzyme analysis of the PCR product, the presence of bovine herpesvirus 4 (BoHV-4) DNA in the cell fraction of milk from dairy cattle with a history of BoHV-4 infection. We next evaluated the infectious nature of BoHV-4 DNA in those cells. Cocultivation of a BoHV-4-sensitive cell line with BoHV-4 DNA-positive milk cell samples produced cytopathic effects. The same result was obtained from frozen and thawed milk cell fraction coming from the cell milk fraction PCR-positive cows, ensuring that cells were killed and only infectious virus could be recovered after cocultivation with sensitive cells. This report shows that infectious BoHV-4 can be present in milk cells and that therefore nursing may be one of the transmission routes of BoHV-4.
Bovine herpesvirus 4 (BoHV-4) is a herpesvirus originally included in the Betaherpesvirinae subfamily and referred to as bovine cytomegalovirus, primarily because its biological properties in tissue culture most closely resembled those of human cytomegaloviruses (19). However, molecular evidence has accumulated, indicating that BoHV-4 is genetically more closely related to members of the Gammaherpesvirinae subfamily. This evidence includes large blocks of homologous genes arranged in the same order which are shared among BoHV-4 and two gammaherpesviruses, Epstein-Barr virus and herpesvirus saimiri (2). BoHV-4 was first isolated in Europe from animals with respiratory and ocular diseases by Bartha and colleagues (1) and later in the United States by Mohanty and colleagues (16). Subsequently, distinct BoHV-4 isolates were obtained, either in Europe or in the United States (7, 14, 17, 19). BoHV-4 has been isolated from a variety of samples and cells (8) from healthy cattle and from cattle with abortion, metritis, pneumonia, diarrhea, respiratory infection, and mammary pustular dermatitis (reviewed by Bartha et al. [1] and Thiry et al. [21]). Although BoHV-4 has been demonstrated in many tissues, accumulated evidence suggests that one site of persistence in both the natural and experimental host is cells of the monocyte/macrophage lineage (7, 17, 18), nothing else is known about BoHV-4 persistent infection. However, the pathogenic role of BoHV-4 remains unclear; the direct correlation between particular strains of BoHV-4 with variable disease conditions is a delicate question, unsolved even through experimental infection.
Only few investigators have successfully produced experimental disease (reviewed by Thiry et al. [21]), and direct inoculation of the natural host only occasionally elicited respiratory and genital disease (3, 22). Notwithstanding, no direct correlation can at present be demonstrated between BoHV-4 and specific lesions. In vivo distribution of BoHV-4 was examined by testing nasal and conjunctival exudates, peripheral blood leukocytes, and various organs of experimentally infected calves (10, 17). However, little information about excretion and transmission of BoHV-4 from naturally infected cattle has been generated. In this report, we describe the detection of infectious BoHV-4 in the cell fraction of milk from dairy cattle with a history of natural BoHV-4 infection and speculate that nursing may be one of the potential transmission routes of BoHV-4.
Herd history and sampling strategy.
A herd of 100 dairy cows experiencing a high incidence of postpartum metritis, abortion, and infertility was positive by indirect fluorescent antibody testing (IFAT) (4) for BoHV-4 antibodies. BoHV-4 antibodies were detected in the sera of 19 of 100 cows tested, and 7 of 19 were repeatedly serologically positive at three samplings, carried out at 2-month intervals during a period of 6 months. The blood and milk of these seven cows were sampled for BoHV-4 DNA.
Cell lines.
A bovine arterial endothelial cell line (BAE-7372) (obtained from Stefano Grolli, Veterinary Biochemistry Institute, Parma University, Italy) was used, due to the high sensitivity of bovine arterial endothelial cells towards BoHV-4 and other bovine herpesviruses (12). Cells were grown at 37°C in minimum essential medium (Gibco-BRL, Paisley, United Kingdom) supplemented with 10% heat-inactivated fetal calf serum, penicillin (100 IU/ml), and streptomycin (100 μg/ml) in a humidified atmosphere containing 5% CO2.
DNA preparation. (i) Blood samples.
Peripheral blood mononuclear cells (PBMC) were separated by Ficoll-Paque (Pharmacia-Biotech, Uppsala, Sweden) gradient centrifugation. They were washed with phosphate-buffered saline (PBS), and aliquots of 2 × 106 cells were made.
(ii) Milk samples.
Each milk sample was centrifuged at 400 × g for 10 min, and the supernatant was collected. Subsequently, the sedimented cells were washed once with PBS and resuspended in 3 ml of PBS. Aliquots of 2 × 106 cells were made. Cell-free milk supernatant (CFMS) was centrifuged at 3,500 × g for 30 min to remove cellular debris and ultracentrifuged at 90,000 × g for 2 h to collect the virus if present. Nucleic acids from PBMC aliquots, milk cell aliquots, and pellets from ultracentrifuged CFMS were extracted as suggested by standard methods (15).
PCR and restriction enzyme analysis.
PCR, restriction enzyme analysis, probe preparation, and Southern hybridization were essentially performed as previously described by Donofrio et al. (5). A 1-μl sample of DNA was amplified during 30 cycles, with each cycle consisting of denaturation at 94°C for 1 min, primer annealing at 55°C for 1 min, and chain elongation with 1 U of Taq polymerase (Boehringer Diagnostics, Milan, Italy) at 72°C for 2 min. PCR amplification was performed in a final volume of 50 μl of 10 mM Tris-hydrochloride, pH 8.3, containing 0.2 mM deoxynucleoside triphosphate, 3 mM MgCl2, 50 mM KCl, and a 0.25 μM concentration of each primer. In the first cycle, the samples were denatured at 94°C for 5 min, and in the last cycle the extension step was increased to 7 min. The primers used for amplification were selected from the published sequences of BoHV-4 (13) (GenBank accession number S49773) and compared with the base sequences of the genes from different bovine herpesviruses. The primers were checked for self-complementarity by the method of Innis et al. (11). The oligonucleotides were designated α (5′-CGAATTATAGTCTAAAGTCATCCTC-3′) and β (5′- GTAAGGACCTTTCACACTCTTAAGC-3′), and amplification led to a 2,538-bp fragment which includes the 3′ end of open reading frame 1 (ORF1) (homologous to the EBV BVRF1 gene), ORF2 (homologous to the Epstein-Barr virus BXRF1 gene), the thymidine kinase gene (ORF3), and the 5′ end of the glycoprotein H gene (ORF4). The PCR product was electrophoresed in 1% agarose gel and visualized after ethidium bromide staining. The expected, amplified 2,538-bp fragment was extracted from the agarose gel, digested with HindIII restriction endonuclease, and analyzed on 1.5% agarose gel in 1× TAE buffer (40 mM Tris-acetate, 1 mM EDTA) containing ethidium bromide for DNA staining. The gel was run for 2.5 h in 1× TAE buffer. The specificity of the PCR product was determined by sequencing, using a ThermoSequenase kit (Amersham International, Amersham, United Kingdom). The resulting sequence was checked with the corresponding sequence in the GenBank database, under accession number S49773 (13).
Infection assay and immunostaining.
Infection assay and immunostaining were performed as previously described by Donofrio et al. (6).
Results and discussion.
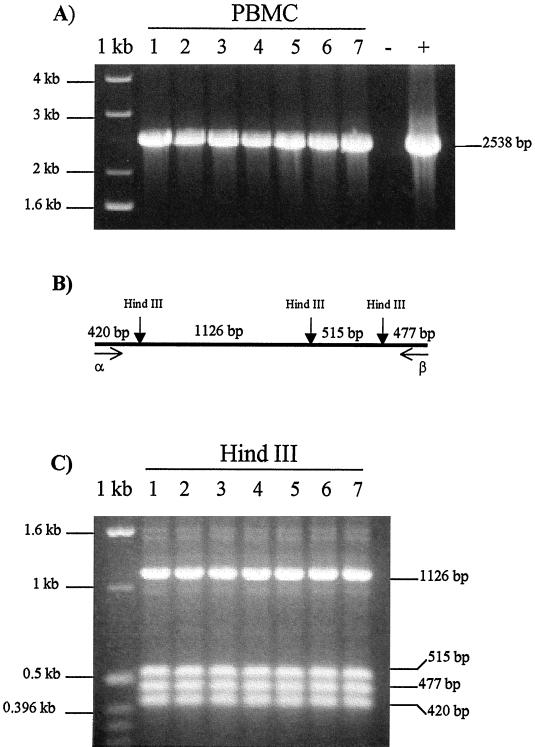
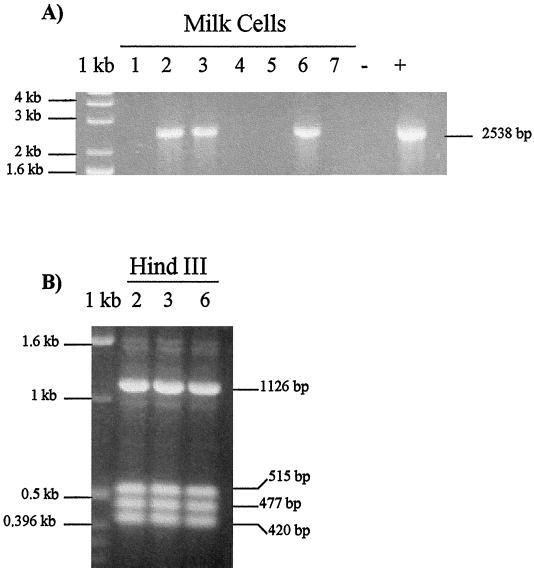
A herd of 100 dairy cattle was tested by IFAT for BoHV-4 antibodies, and 19 of 100 cows tested were found positive. To assess the BoHV-4 infection of the 19 BoHV-4 IFAT-positive cows, IFAT was repeated three times at 2-month intervals during a period of 6 months. Seven of 19 cows were found repeatedly IFAT positive. Blood and milk of these seven cows were collected for BoHV-4 DNA detection. PCR allowed us to detect BoHV-4 DNA in the PBMC of all seven cows tested (Fig. 1A) and in three of the seven milk cell fractions, too (Fig. 2A), but none was detected in the CFMS (data not shown). The identity of the amplicon was confirmed by HindIII restriction enzyme digestion (Fig. 1C); the same fragments predicted from the published sequence were detected in the PCR products of all seven PBMC samples (Fig. 1B) and three milk fraction samples (Fig. 2B). Then, to demonstrate the infectious nature of the virus in the milk cell fraction, we cocultured the milk cell fraction with a BoHV-4-sensitive cell line. Cocultures with each of the three samples developed cytopathic effects. To confirm the specificity of the cytopathic effects, the resulting plaques were stained with an anti-BoHV-4 hyperimmune serum, and a specific positive staining was obtained (data not shown). To determine whether the milk cell fraction was productively infected or if the virus obtained after cocultivation with sensitive cells was just the reactivation of latent BoHV-4 genomes, we froze and thawed the milk cell fraction coming from the three cell milk fraction PCR-positive cows (samples 2, 3, and 6). That procedure ensured that cells were killed and only infectious virus could be recovered after cocultivation with sensitive cells. BoHV-4 was recovered from all three cell fraction samples (data not shown). We concluded that complete viral particles had been assembled in all three milk cell fraction samples. This result indicates a productive infection.
FIG. 1.
(A) Specific amplification of BoHV-4 DNA fragment from PBMC samples from BoHV-4 IFAT-positive cows. PCR amplified a 2,538-bp fragment of the BoHV-4 genome containing ORF1 overlapping ORF2 gene, ORF3 corresponding to the thymidine kinase gene, and ORF4 homologous to the herpes simplex virus type 1 glycoprotein H gene. Lanes 1 to 7 correspond to PBMC samples; lanes − and + correspond to negative (DNA from PBMC of BoHV-4 seronegative cow) and positive controls, respectively. 1 kb, molecular size marker. (B) Predicted location of HindIII restriction sites and respective expected restriction fragment sizes (not shown to scale). (C) Ethidium bromide-stained gel of HindIII-digested 2,538-bp BoHV-4 amplified sequence from PBMC DNA samples (lanes 1 to 7). 1 kb, molecular size marker.
FIG. 2.
(A) Specific amplification, performed with primers α and β, of DNA from milk cell fractions of milk coming from PBMC PCR-positive cows and producing a 2,538-bp product. Lanes 1 to 7 correspond to milk cell fraction samples; lanes − and + correspond to negative and positive controls respectively. 1 kb, molecular size marker. (B) Ethidium bromide-stained gel of HindIII-digested 2,538-bp BoHV-4 amplified sequence from PBMC DNA samples (2, 3, and 6). 1 kb, molecular size marker.
This study does not define the type of cell carrying the virus into the milk. However, the demonstration that BoHV-4 can establish a persistent infection in lymphoid tissues and a prolonged viremia associated with the PBMC fraction (17, 9, 10) could explain the recovery of the virus in the milk as a consequence of transport through lymphatic and circulatory system routes.
We have examined BoHV-4 only from the milk of animals serologically positive for the virus and positive for the presence of the virus in PBMC. BoHV-4 presence or prevalence in milk of other populations has not been determined. Longitudinal studies of subjects with known serostatus will be required to address shedding and transmissibility of BoHV-4 by milk.
Bovine herpesviruses can generally be transmitted by horizontal and vertical routes. The horizontal transmission occurs by close contact with moist contaminated surfaces, but droplet infections are also common. Vertical transmission via fetal infection occurs during parturition (14). Because bovine herpesviruses are highly labile once shed from the body and are readily inactivated by sunlight or drying, milk represents a good candidate as a vehicle for BoHV-4 shedding and transmission. The virus is protected by cell lipid membranes, and this could increase the probability of BoHV-4 infection of nursing animals through the oral mucosa surface. In addition, the humoral immune response following BoHV-4 infection in cattle is characterized by the production of low-avidity neutralizing antibodies (17, 22). The lack of neutralizing antibodies in milk and colostrum from infected cows could favor the transmission of BoHV-4 to nursing calves. Whether contact with infectious milk plays a role in the transmission of the virus or the stage at which shedding of BoHV-4 in milk occurs is unknown. Although this report demonstrates the potential infectivity of BoHV-4 in milk, the importance of BoHV-4 shedding in milk in the transmission of the virus remains to be determined. However, this report emphasizes that infectious BoHV-4 can be present in milk, and nursing may be one of the transmission routes of BoHV-4.
Acknowledgments
This work was supported by internal funding of Parma University.
We thank V. Van Santen, Department of Pathobiology, Auburn University, Auburn, Ala., for helpful discussion and reading of the manuscript and L. Gandolfi for technical support.
REFERENCES
- 1.Bartha A, Juhasz M, Liebermann H. Isolation of a bovine herpesvirus from calves with respiratory disease and keratoconjuntivitis. Acta Vet Acad Sci Hung. 1966;16:357–358. [PubMed] [Google Scholar]
- 2.Bublot M, Lomonte P, Lequarre A S, Albrecht J C, Nicholas J, Fleckenstein B, Pastoret P P, Thiry E. Genetic relationships between bovine herpesvirus 4 and the gammaherpesviruses Epstein-Barr virus and herpesvirus saimiri. Virology. 1992;190:654–655. doi: 10.1016/0042-6822(92)90903-3. [DOI] [PubMed] [Google Scholar]
- 3.Castrucci G, Frigeri F, Ferrari M, Ranucci S, Aldrovandi V, Cilli V, Rampichini L, Gatti R. Experimental infection of calves with strain of bovid herpesvirus-4. Comp Immunol Microbiol Inf Dis. 1987;10:41–49. doi: 10.1016/0147-9571(87)90039-7. [DOI] [PubMed] [Google Scholar]
- 4.Cavirani S, Martelli P, Cabassi C S, Lavazza A, Allegri G, Flammini C F. Proceedings of the XIX World Buiatric Congress. 1996. Isolation of bovide herpesvirus 4 (BHV-4) from dairy cows with digital dermatitis; p. 121. [Google Scholar]
- 5.Donofrio, G., S. Cavirani, C. S. Flammini, and F. Scatozza. Molecular typing of a BHV-4 (bovine herpesvirus 4) isolate. Vet. Res. Commun., in press. [DOI] [PubMed]
- 6.Donofrio G, Cavirani S, van Santen V L. Establishment of a cell line persistently infected with recombinant BHV-4. J Gen Virol. 2000;81:1807–1814. doi: 10.1099/0022-1317-81-7-1807. [DOI] [PubMed] [Google Scholar]
- 7.Dubuisson J, Thiry E, Thalasso F, Bublot M, Pastoret P P. Biological and biochemical comparison of bovid herpesvirus-4 strains. Vet Microbiol. 1988;16:339–349. doi: 10.1016/0378-1135(88)90015-6. [DOI] [PubMed] [Google Scholar]
- 8.Egyed L. Replication of bovine herpesvirus type 4 in human cells in vitro. J Clin Microbiol. 1998;36:2109–2111. doi: 10.1128/jcm.36.7.2109-2111.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Egyed L, Bartha A. PCR studies on the potential sites for latency of BHV-4 in calves. Vet Res Commun. 1998;22:209–216. doi: 10.1023/a:1006029523226. [DOI] [PubMed] [Google Scholar]
- 10.Egyed L, Ballagy-Pordany A, Bartha A, Belak S. Studies of in vivo distribution of bovine herpesvirus type 4 in the natural host. J Clin Microbiol. 1996;34:1091–1095. doi: 10.1128/jcm.34.5.1091-1095.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Innis A M, Gelfand H D, Sninsky J J, White J. PCR protocols: a guide to methods and applications. New York, N.Y: Academic Press; 1990. [Google Scholar]
- 12.Lin T M, Shi G Y, Tsai C F, Janny Su H, Leon Guo Y L, Wu H L. Susceptibility of endothelial cells to bovine herpesvirus type 4 (BHV-4) J Virol Methods. 1997;63:219–225. doi: 10.1016/s0166-0934(96)02132-5. [DOI] [PubMed] [Google Scholar]
- 13.Lomonte P, Bublot M, Pastoret P, Thiry E. Location and characterization of the bovine herpes virus type 4 thymidine kinase gene; comparison with thymidine kinase genes of other herpesviruses. Arch Virol. 1992;127:327–337. doi: 10.1007/BF01309595. [DOI] [PubMed] [Google Scholar]
- 14.Ludwig H. Bovine herpesviruses. In: Roizman B, editor. The herpesviruses. Vol. 2. New York, N.Y: Plenum Press; 1983. pp. 135–214. [Google Scholar]
- 15.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- 16.Mohanty S B, Hammond R C, Lillie M G. A new bovine herpesvirus and its effect on experimentally infected calves. Arch Gesamte Virusforsch. 1971;34:394–395. doi: 10.1007/BF01254696. [DOI] [PubMed] [Google Scholar]
- 17.Osorio F A, Rock D L, Reed D E. Studies on the pathogenesis of a bovine cytomegalo-like virus in an experimental host. J Gen Virol. 1985;66:1941–1951. doi: 10.1099/0022-1317-66-9-1941. [DOI] [PubMed] [Google Scholar]
- 18.Osorio F A, Reed D E, Van Der Maaten M J, Metz C A. Comparison of the herpesviruses of cattle by restriction endonuclease analysis and serologic analysis. Am J Vet Res. 1985;46:2104–2109. [PubMed] [Google Scholar]
- 19.Storz J, Ehlers B, Todd V J, Ludwig H. Bovine cytomegaloviruses: identification and differential properties. J Gen Virol. 1984;65:697–706. doi: 10.1099/0022-1317-65-4-697. [DOI] [PubMed] [Google Scholar]
- 20.Sunil-Chandra N P, Efstathious S, Nash A A. Murine gammaherpesvirus 68 establishes a latent infection in mouse B lymphocyte in vivo. J Gen Virol. 1992;73:3275–3279. doi: 10.1099/0022-1317-73-12-3275. [DOI] [PubMed] [Google Scholar]
- 21.Thiry E, Bublot M, Dubuisson J, Pastoret P P. Bovine herpesvirus-4 (BHV-4) infection in cattle. In: Wittmann G, editor. Herpesvirus diseases of cattle, horses and pigs. Boston, Mass: Kluwer; 1989. pp. 96–115. [Google Scholar]
- 22.Wellemans G, Van Opdenbosh E, Mammerickx M. Inoculation expérimental du virus LVR 140 (herpes bovin IV) à des vaches gestantes et non-gestantes. Ann Rech Vet. 1986;17:89–94. [PubMed] [Google Scholar]