Abstract
Graves’ Ophthalmopathy (GO) is an organ-specific autoimmune disease that is often characterized by infiltration of orbital tissues and is considered as the most common extra-thyroid manifestation of Graves’ disease (GD). Although genetic susceptibility has been found to be critical for the phenotype of GO, the associated risk alleles in a single gene are generally insufficient to cause the disease. Accruing evidence has shown that epigenetic disorders can act as the potentially missing link between genetic risk and clinically significant disease development. Abnormal epigenetic modifications can lead to pro-inflammatory cascades and activation of orbital fibroblasts (OFs) by promoting the various inflammatory response pathways and regulating the diverse signaling molecules that are involved in the fibrogenesis and adipogenesis, thereby leading to the significant expansion of orbital tissues, fibrosis and inflammation infiltration. Additionally, emerging evidence has shown that the gut microbiome can possibly drive the pathogenesis of GO by influencing the secretion of Thyrotropin receptor antibody (TRAb) and T-helper 17 (Th17)/regulatory T cells (Treg) imbalance. This paper describes the latest epigenetic research evidence and progress made in comprehending the mechanisms of GO development, such as DNA methylation, histone modification, non-coding RNAs, and the gut microbiome.
Keywords: Graves’ ophthalmopathy, epigenetics, DNA methylation, histone modification, noncoding RNAs, gut microbiome
Introduction
Graves’ ophthalmopathy (GO), also known as thyroid-related ophthalmopathy, is an autoimmune disease associated with Graves’ disease (GD) that involves orbital tissues. It has been found that about 25% of GD patients develop GO during their disease course (1, 2). Approximately 90% of GO develop in hyperthyroidism caused by GD, whereas GO sometimes occurs in patients with euthyroid or hypothyroid chronic autoimmune thyroiditis (3, 4). Although the ratio of female to male (F/M) has been found to vary based on the different studies, the incidence of GO is significantly higher in women than in men (5). The common clinical characteristics of GO are eyelid retraction, swelling, exophthalmos, retrobulbar pain, and even vision impairment resulting from optic neuropathy (6). Similar to GD, the autoreactive inflammatory process of orbital tissue plays a major role in the pathogenesis of GO. The extraocular muscles and connective tissue of GO patients are infiltrated by activated monocytes (such as T cells), as well as a small number of plasma cells, macrophages and mast cells (7). Activated T cells, mainly CD4+T cells, produce a large number of cytokines to amplify and maintain orbital inflammation and stimulate the proliferation of orbital fibroblasts (OFs) and the synthesis of glycosaminoglycans (GAG) (8). OFs are heterogeneous cells, and when activated, fibroblasts with a high level of thymocyte differentiation antigen 1(Thy1) surface expression (Thy1+) are more likely to form myofibroblasts, on the contrary, fibroblasts with little or no Thy1 (Thy1 -) are more likely to form adipocytes (9, 10). Accumulation of GAG leads to edema of extraocular muscles (11) and eventually leads to orbital swelling, tissue expansion and fibrosis development.
As mentioned above, cytokines and immune mechanisms play an important role in the pathogenesis of GO. CD4+T cells can be divided into auxiliary T helper 1(Th1), Th2, Th17, regulatory T (Treg) cells and other subtypes, which are considered to be related to the occurrence and development of GO (8). Studies have shown that Th1 cytokines are not only related to the progression of GO, but may play a dominant role in the orbital involvement in the early stage of GO. On the contrary, Th2 cytokines have no direct correlation with disease progression, but may play a greater role in the later stage of the disease (12–14). IL-17A secreted by Th17 cells has pro-inflammatory and pro-fibrosis effects, which further drives the clinical manifestation of GO (15). Treg cells are a subgroup of T cells with immune regulatory function, and impaired Th17/Treg balance may also be an important cause of GO (16). Furthermore, activated T cells activate the differentiation of B cells into plasma cells and the production of antibodies by secreting IL-4 (8). GD related hyperthyroidism is primarily caused by binding of TRAb to the thyrotropin receptor (TSHR) on the thyroid follicular endothelial cells, thereby stimulating the excessive secretion of the thyroid hormones. Furthermore, recent studies have reported that TRAb can effectively stimulate the proliferation of OFs, which is positively correlated with the clinical features of GO and can adversely affect the prognosis (17). Another crucial autoantigen involved in the pathogenesis of GO is insulin-like growth factor I receptor (IGF-IR), which forms a physical and functional complex with TSHR in OFs from GO patients (18). Antibodies of the IgG class generated by GD patients are able to bind to IGF-1R and initiate the signaling from the TSHR/IGF-IR protein complex. Furthermore, Teprotumumab, an IGF-IR blocking monoclonal antibody, has been approved in the US FDA for the treatment of GO and works by attenuating signaling from TSHR or IGF-IR (19). Currently, substantial progress has been made in treating GO, but the moderate-to-severe GO patients usually do not completely respond to available medical treatments, and complete restitutio ad integrum almost never occurs (5, 20). Therefore, more extensive studies are needed to explore the pathogenesis of GO and develop prompt prediction, timely referral and novel therapeutic strategies for GO.
In the past decade, increasing evidence has shown that genetic susceptibility plays a crucial role in the pathogenesis of GD and GO (21–24), but the associated risk alleles in a single gene are generally insufficient to cause disease. Moreover, several studies have performed Human leukocyte antigen (HLA), cytotoxic T lymphocyte antigen-4 (CTLA4), Interleukin (IL) - 23 receptor (IL23R), and TSHR genotyping on GO and non-GO patients in GD patients. The results show no statistical differences in allele and genotype frequency between the two groups, thereby plausibly suggesting that GO might not have significant genetic susceptibility in GD patients (25). These results suggested that epigenetics and/or environmental influences could possibly also play an important role in the pathogenesis of GO. Epigenetics as a research hotspot in recent years, researchers have found that the decreased level of ICAM1 methylation is significantly associated with exophthalmos in patients with GD (26). Similarly, micro RNAs (miRNAs) have been correlated with the level of autoimmune antibodies such as TPOAb, TgAb and TRAb (27). These findings provided us with vital background to explore the potential pathogenesis of GO. A number of previous epigenetics related studies have shown that different environmental factors can also functionally regulate gene expression and phenotypes in the disease development process without altering the DNA sequences, primarily through epigenetic mechanisms such as those of DNA methylation, histone modification, and non-coding RNAs (28). Epigenetic modifications have been found to be involved in the dysregulation of the different signaling molecules and receptors in a variety of autoimmune/inflammatory conditions, including GO (29–32). Thus, epigenetic studies could serve as interesting targets for elucidating the pathogenesis of GO. Interestingly, all environmental factors involved in the pathogenesis of GD and GO can effectively alter the balance of microorganisms located in the gut and influence the immune system, particularly the proportion of Treg and Th17 cells. A recent study demonstrated that gut dysbiosis can lead to the imbalance of Th17/Treg through the propionic acid regulated pathway, and promote the occurrence of GD (33). A number of studies in GD/GO mice model have also shown that the gut microbiome might induce the clinical manifestation of GO by influencing the levels of TRAb and Th17/Treg imbalance (34, 35). Therefore, this paper reviews the latest epigenetics landscape and progress made in deciphering the mechanisms of GO by encompassing the various aspects of DNA methylation, histone modification, noncoding RNAs and gut microbiome, with an aim to provide new effective potential therapeutic strategies for the management of GO.
Abnormal DNA methylation and GO
As one of the most common forms of epigenetic modification, DNA methylation is catalyzed by DNA methyltransferases (DNMTs) by using S-adenosyl-L-methionine (SAM) as a methyl donor, and forming 5-methylcytosine (5-MC) by the transfer of the methyl group to the five carbon atoms of the CpG dinucleotide cytosine base. Accumulating evidence has suggested that DNA methylation in the promoter regions of the CpG island is primarily linked to gene transcriptional inhibition; and the methylated CpG binding domain (MBD) family can effectively recognize and bind to methylated CpG to cause significant transcriptional inhibition (36).
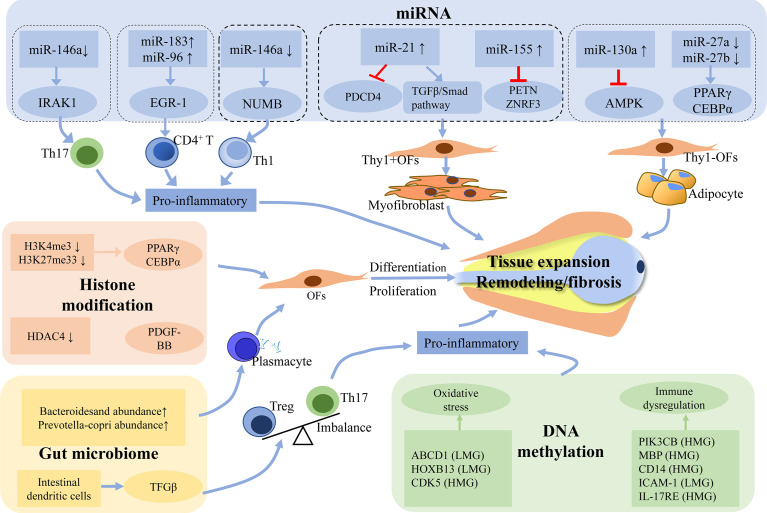
In recent years, several studies have shown that DNA methylation exhibits important functions in the pathogenesis of GO by altering the gene expression profile. Xin et al. (37) have identified 841 differentially methylated sites by sequencing the whole wide genomic DNA methylation in 6 patients with GO. It was found that 148 different genes were located near or at the methylation sites. These genes include various genes mediating immune responses such as Cluster of Differentiation 14 (CD14), interleukin 17 receptor E (IL17RE), beclin1(BECN1), cyclin dependent kinase 5 (CDK5), oxidative stress regulation genes including ATP-Binding cassette transporter superfamily D1 (ABCD1), homeobox B13 (HOXB13), thyroid function regulating gene such as dopamine receptor D4 (DRD4). The aforementioned genes IL17RE and CDK5 have been positively associated with Clinical Activity Score (CAS) and TRAb, respectively. The authors also performed enrichment of genes associated with the diverse biological processes, cellular composition, and molecular functions. To further understand the pathogenesis of GO, pathway analysis of these identified genes was carried out, and four potentially important pathways, including toxoplasmosis pathway, axon guidance pathway, focal adhesion pathway, and proteoglycans in cancer pathway, were found to be closely associated to the occurrence and development of GO. Moreover, other important pathway genes such as LDLR, CDK5, and phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit beta (PIK3CB) that play an important role in regulating inflammatory have also been found to be significantly correlated with GO phenotypes (32). These observations further confirmed the potential relationship between the DNA methylation level and its functions in the regulation of specific genes involved in the pathogenesis of GO. In another study, Shi et al. (38) carried out genome scale screening of DNA methylation in the peripheral blood samples of patients with GO and healthy controls. They next screened the different candidate genes with a good topological performance by constructing a gene regulatory network of hypermethylated and hypomethylated genes. They further expanded the sample for verification by monomethylated sequencing, thus confirming that, the methylation level of boule homolog (BOLL) was lower, and the methylation level of myelin basic protein (MBP) and CDK5 was markedly higher. BOLL, MBP, and CDK5 genes were found to be associated with increased risk of GO. The possible mechanism can be described by the fact that MBP is related to the activation of MBP specific CD8 lymphocytes and CDK5 could promote oxidative stress. Khong et al. (39) used microarray analysis to detect the gene expression profile of orbital adipose tissue obtained from GO patients. In combination with gene set enrichment analysis (GSEA), they found that compared with inactive GO, the gene enrichment of active GO showed significantly more prominent epigenetic characteristics of acute myeloid leukemia (AML). Moreover, the abnormal hypomethylation of epigenetic markers in AML was associated with immunodeficiency signaling, cytotoxic T cell-mediated apoptosis, and dysregulation of T cell receptor signaling (40). In summary, the pathogenesis of GO is driven by aberrant DNA methylation of specific genes such as ABCD1 and PIK3CB, which may cause oxidative stress and immune dysregulation leading to inflammation ( Figure 1 ).
Figure 1.
Potential mechanism of epigenetics and gut microbiome in the pathogenesis of GO. Abnormal epigenetic modifications may promote pro-inflammatory cascades and disrupt the expression of signaling molecules that are involved in the fibrogenesis and adipogenesis of orbital fibroblasts. Gut microbiome possibly drives the pathogenesis of GO by influencing secretion of TRAb and Th17/Treg imbalance. The process of all these changes will lead to the hyaluronan production and the activation of orbital fibroblasts, which eventually develop into tissue expansion, fibrosis and inflammation. LMG, lowly methylated genes; HMG, highly methylated genes; TRAb, Thyrotropin receptor antibody; Treg, regulatory T; Th17, T-helper 17.
Furthermore, OFs play a crucial role in the initiation and maintenance of the inflammatory response as well as in orbital tissue expansion and remodeling. Hu et al. (41) performed DNA methylation sequencing and orbital computer tomography (CT) measurement on the peripheral blood samples from patients with GO and healthy controls. They found that the abnormal methylation of MBP gene was significantly correlated with the ratio of the total cross-sectional area of the orbital muscles (OM) to the total orbital area in GO patients. Virakul et al. (42) similarly carried out the detection of various OFs, and further reported that OFs in active versus inactive GO patients might have different proteomic and DNA methylation characteristics, thereby indicating that OFs can show specific pathological characteristics that might be related to the disease stage during the progression of GO. These observations highlighted the potential role of OFs as a future therapeutic target in GO. These findings also suggested that DNA methylation occurs in the fibroblasts and epigenetic understanding of the transition of OFs from “inflammatory/pre-lipogenic” to “remodeling/pre-fibrotic” effector cells should be a critical area of research in the future studies.
Genetic polymorphisms of a few selected DNA methylation regulatory genes can also lead to abnormal DNA methylation and dysfunction of these genes, which can further increase the susceptibility of the host (43). For example, methylenetetra-hydrofolate reductase (MTHFR) reduces 5, 10-methylenetra-hydrofolate to 5-methylenetrahydrofol -ate in the folic acid metabolic pathway, thus extending to the methyl transport pathway and indirectly facilitating methyl groups for the DNA methylation and protein methylation through the re-methylation of homocysteine residues. Lee et al. (44) reported that MTHFR polymorphism was closely associated with GO susceptibility and that MTHFR 677 TT can serve as an independent risk factor for GO. Moreover, SNPs 677C>T of the MTHFR gene have been demonstrated to result in pronounced alteration of its enzymatic activity (45), providing indirect evidence for the important role of DNA methylation in GO pathogenesis.
Taken together, DNA methylation plays an important role in the occurrence and development of GO, but the related research is rather limited. Therefore, more studies are needed to explore the key role of DNA methylation in the pathogenesis of GO, to develop biomarkers for the rapid detection of early DNA methylation changes and establish effective GO treatment strategies.
Histone Modification and GO
The nucleosome constitutes the basic structural unit of chromosomes. It consists of an octamer composed of four different pairs of core histones (H3, H4, H2A, and H2B) and a 147bp length of the double-stranded DNA surrounding the octamer. The core histones moieties are generally spherical, and the non-structural N-terminal amino acids extended by each histone can be modified after translation. The common histone modifications events include methylation, acetylation, phosphorylation, ubiquitination, adenosine diphosphate (ADP) ribosylation, etc. (46). Currently, histone modification studies have primarily focused on the methylation and acetylation, thereby providing some evidence for the possible role of histone modification in regulating immune tolerance and autoimmune thyroid diseases (47–49).
Histone methylation can be catalyzed by histone methyltransferase (HMT) and occurs mainly on the lysine and arginine residues of H3 and H4 histones, but the transcriptional output from histone methylation primarily depends on the nature of the specific residues involved. Histone H3 methylation is one of the most well characterized epigenetic modifications and actively involved in the adipogenesis of OFs. The differentiation of OFs into adipocytes is primarily a peroxisome proliferator-activated receptor gamma (PPARγ) and CCAAT/enhancer binding protein alpha (CEBPα) dependent process (50). Khong et al. (39) conducted GSEA on the expression profile of the various differentiated genes in the active phase of GO and found that the unmethylated histone H3 in the genes with high CpG density promoters (HCP) found in embryonic fibroblast was significantly downregulated compared with the normal controls. Methylation at K4 (H3K4me3) and K27 (H3K27me33) on histone 3, which is in bivalent status on HCP has been directly related to cell lineage commitment. PPARγ often remains in a bivalent state during embryonic fibroblast differentiation (51). Matsumura et al. (52) reported that the H3K4me3 and H3K27me3 bivalent state might change to H3K4/H3K9me3 during lineage specification. The characteristics of H3K4/H3K9me3 bivalent chromatin signature can possibly maintain CEBPα and PPARγ gene expression at a very low level in lineage-committed mesenchymal stem cells (MSCs) and preadipocytes by silencing or by maintaining developmental genes to poise for subsequent activation, and they may constitute a distinct pseudo heterochromatin/euchromatin boundary which is important for adipocyte differentiation. The above studies suggested that histone H3 methylation might play an important role in the pathogenesis of GO by targeting some genes that are related to adipogenesis. However, at present, there are only few related studies, and there are no reports to establish that abnormal histone methylation might be related to the occurrence of GO. Therefore, additional studies are needed to decipher the specific mechanisms underlying these observations.
Histone acetylation and deacetylation are important components of the gene regulatory machinery. Histone acetylation is usually catalyzed by histone acetyltransferase (HAT), and addition of acetyl groups to the target histones can activate the various transcription factors which can specifically bind to the different DNA binding sites and promote gene transcription. Histone deacetylase (HDAC) can also catalyze the deacetylation of histones to make DNA more tightly encapsulated around the nucleosomes, thus inhibiting gene expression (46). Histone acetylation and deacetylation can effectively maintain a dynamic balance in the nucleus. For example, loss of HDAC expression in the thyroid and immune cells of patients with autoimmune thyroid diseases has been confirmed (53). Recently, Ekronarongchai et al. (31) reported HDAC4 mRNA upregulation and protein expression in OFs by Platelet-Derived Growth Factor-BB (PDGF-BB). It was observed that the expression of HDAC4 mRNA in the OFs of GO patients was significantly higher as compared to the healthy control group, and the lysine 9 acetylation of histone H3 (H3K9ac) was markedly decreased under PDGF-BB stimulation. It was also found that the expression of hyaluronan synthase 2 (HAS2), collagen type I alpha 1 chain (COL1A1), proliferation marker Ki67 (Ki67), and α-smooth muscle actin (α-SMA) mRNA and the production of hyaluronic acid in PDGF-BB stimulated OFs were significantly decreased upon HDAC4 silencing. These data suggested that HDAC4-induced H3K9 deacetylation could potentially exacerbate GO OFs hyperproliferation and extracellular matrix production by functionally regulating PDGF-BB, thus suggesting that HDAC4 might serve as a new target for GO therapy.
Therefore, histone modification has been related to the occurrence and development of GO. Histone H3 methylation and HDAC may be potential targets for GO therapy, but the specific mechanisms are still not clear. More studies are needed to identify abnormal histone modifications associated with GO and to clarify their molecular mechanisms in GO.
Non-Coding RNAs and GO
Non-coding RNAs are a class of RNA transcripts that lack the function of coding proteins. They can be classified into miRNAs, circular RNAs (circRNAs) and long noncoding RNAs (lncRNAs), which have been reported to play an important role in the pathogenesis of the various autoimmune thyroid diseases (54).
MiRNAs and GO
MiRNAs are endogenous small non-coding RNA molecules that contain 18 to 23 nucleotides in length, and that by binding to the untranslated 3’ domain of the target gene mRNA, miRNAs can effectively mediate the degradation of mRNA or inhibit its translation, thus exerting a strong negative regulatory effect on the gene expression at the post-transcriptional level (55). MiRNAs can regulate more than 60% of protein- coding genes in the humans and have been implicated in the various biological processes such as the lineage commitment in the immune system, cell proliferation, differentiation, apoptosis, and maintenance of immune homeostasis (56, 57). An abnormal expression of miRNAs has been closely related to the pathogenesis of various autoimmune-mediated eye diseases such as GO (58). Circulating miRNAs can be detected in the peripheral blood and orbital tissues of GO patients, with different expression levels in the active disease stage of GO.
Circulating miRNAs and GO
A number of recent studies have shown that some circulating miRNAs might be directly involved in the development of GO, and have been found to be closely related to the regulation of differentiation or activation of immune cells and immune responses.
MiR-146a-5p is one of the most widely studied miRNAs and has an important role in regulating GO progression. MiR-146a is a typical multifunctional miRNA whose expression can be induced by toll-like receptor 4 (TLR4)/nuclear factor-kappa B (NF-ĸB) pathway, which acts as an important regulator of immune and inflammatory response pathways (59). By targeting tumor necrosis factor receptor-associated factor 6 (TRAF6) and IL-1 receptor-associated kinase 1 (IRAK1), miR-146a can negatively inhibit the feedback of NF-ĸB signaling cascade and modulate the signal transduction by the related receptors and subsequent cytokine release (60, 61). A few studies have shown that the peripheral blood level of miR-146a-5p in GO patients was significantly decreased as compared with their healthy counterparts as well as GD patients which lack GO phenotype (62–64). Additionally, reduced expression of miR-146a-5p has been negatively correlated with the CAS and IL-17 levels in GO patients. IL-17 levels have been positively correlated with CAS, and the downregulation of miR-146a can effectively lead to a paradoxical increase in IL-17 levels possibly due to their inability to inhibit IRAK1. This finding suggested that IRAK1 could play a key regulatory role in the differentiation of Th17 cell (62). The orbital tissues are infiltrated by activated mononuclear cell such as CD4+ T cells, which exert a key role on GO activity (7). A number of recent studies have also suggested that the down-regulated expression of miR-146a in CD4+T cells of GO patients might facilitate GO development by targeting NUMB genes, thus resulting in the development of inflammatory Th1 response, which can lead to orbital inflammation in GO patients (63, 64). In other words, the downregulation of circulating miR-146a may indirectly promote the development of GO by regulating the expression of various inflammatory mediators. Contrary to the downregulation of miR-146a, miR-155 levels have been reported to be increased in the peripheral venous blood in GO patients. In turn, both miR-155 and miR-146a have multiple target genes in the TLR4/NF-ĸB pathway, which can mutually regulate the immune response. The upregulation of miR-155 may also promote autoimmune inflammation through the targeted inhibition of cytokine signaling 1 (SOCS1) and SH2 domain-containing inositol-5-phosphatase 1 (SHIP1) (65). Other miRNAs such as miR-Let7d-5p, miR-96-5p and miR-301a-3p, were also found to be differentially expressed in the serum of GO patients versus control group, which might contribute to the regulated expression of the different inflammatory mediators in vivo and can lead to the occurrence as well as the development of GO (27). In addition, Thiel and colleagues (66) have reported an increased expression of miR-183 and miR-96 in CD4+T cells of GO patients and in activated T cells of mice. It has been found that these two kinds of miRNAs promote the activation of CD4+T cells by effectively regulating the expression of early growth response 1(EGR-1), which can directly be related to the activation of PTEN/Akt signal axis, and thus participate in the occurrence as well as the development of GO. At present, based on the special characteristic of the differential expression of miRNAs, it will be of great significance to study the role of the various influencing factors involved in the pathogenesis and to decipher the mechanisms of the transition from an active stage to static stage, to facilitate early diagnosis and treatment possibilities of GO.
The clinical utility of miRNAs in GO has also been explored. For instance, Shen et al. (67) found that the low serum miR-224-5p could be correlated with GO glucocorticoid (GC) insensitivity, and overexpression of miR-224-5p in vitro increased GC sensitivity and glucocorticoid receptor protein via targeting glycogen synthase kinase 3 beta. Moreover, the combination of baseline serum miR-224-5p and TRAb was an independent risk factor for GC insensitivity. Recently, a number of studies have analyzed the possible role of miRNAs in the assessment of GO severity and disease outcomes. One study by Martinez-Hernandez et al. (27) reported that miR-let7D-5P expression was markedly decreased in GO patients as compared with GD patients without GO and was also found to be negatively correlated with CAS. Meanwhile, Zhang et al. (68) performed miRNAs profiling of the thyroid tissues in GO patients using next-generation sequencing (NGS) and then identified their serum expression. They found that novel:19_15038 and hsa-miR-27a-3p were up-regulated in GO, while hsa-miR-22-3p was down-regulated in GO as compared to healthy controls. To sum up, the detection of the various differentially expressed miRNAs might be used as the potential biomarkers to predict the development and progression of GD into GO.
Orbital Tissue miRNAs and GO
OFs are both the target cell and effector cell in the pathogenesis process of GO. A number of recent studies have suggested that miRNA inhibitors or mimics can be used to up-regulate or down-regulate miRNAs expression in orbital tissues, thus regulating the corresponding biological behavior. Recent studies have also indicated that some miRNAs might be associated with orbital fibrosis, adipose tissue formation and inflammatory cell development.
For example, miR-146a acts as a key regulatory factor of GO orbital tissue fibrosis. To our knowledge, the level of miR-146a might be variable in the different types of tissues (63, 69). Unlike the downregulation of miR-146a in peripheral blood summarized previously, it is mostly up-regulated in orbital tissues. Wang et al. (69) reported that up-regulation of miR-146a in the orbital connective tissue of GO patients could significantly increase IL-6 level by inhibiting Notch2, thereby effectively promoting mitotic activity of fibroblasts in GO patients, inhibiting cellular apoptosis, and subsequently worsening the deterioration of GO. Woeller et al. (70) reported that TSHR signaling could induce the production of miR-146a and miR-155 on the OFs of GO patients. The expression of the different cell proliferation suppressor genes ZNRF3 and PTEN were substantially decreased, thereby indirectly promoting the proliferation response of OFs, which could possibly explain the pathological mechanisms of partial fibroplasia observed in GO. However, a few studies have also shown that miR-146a can inhibit the fibrosis process. MiR-146a also plays an active role in the anti-inflammatory and anti-fibrotic processes of OFs. Jang et al. (71, 72) have reported that inflammatory stress can significantly up-regulate miR-146a expression in OFs, but inhibit the expression of IL-6, ICAM-1, and other inflammatory proteins. It can also negatively regulate levels of transforming growth factor beta (TGF-β) -induced fibrosis markers such as fibronectin (FN), collagen I α, and α - SMA. Type I collagen is considered to be a critical marker of fibrosis, and hyaluronic acid is a glycosaminoglycan. Liu et al. (73) recently reported that miR-146a expression was markedly down-regulated in the secretion of hyaluronic acid and type I collagen in GO OFs in vitro, which can reduce the aggregation of glycosaminoglycan and collagen deposition, and thereby delay the progression of the disease. Li et al. (65) hypothesized that miR-155, in contrast to miR-146a, can enhance the development of inflammatory T cells by promoting different autoimmune responses. In particular, the levels of miR146a in circulating and orbital tissues have been found to be inconsistent, which may be because the inflammation caused by miR-146 in peripheral blood may not fully reflect the inflammation in orbit. What is more, the contradictory functions about the relationship between miR-146a and differentiation of OFs can also be difficult to interpret, mainly due to the manifestation of the different targeting effects. Furthermore, miR-146a was found to play different functions in the regulation of the diverse signaling pathways or in interactions with the different target mRNAs. However, elucidation of the detailed mechanism of miR-146 in GO requires further investigation.
In addition to miR-146a, miR-21 has also been reported to play an important role in orbital muscle fibrogenesis. Lee et al. (74) found that in human OFs, PDGF-BB can significantly inhibit the expression of Programmed cell death factor 4 (PDCD4) by up-regulating miR-21, and thus promoting the proliferation of OFs. It is consistent with the study that the translation of PDCD4 was inhibited by miR-21 by Young et al. (75). Therefore, PDGF-BB/miRNA-21/PDCD4 pathway might form the basis of novel strategies for developing therapeutic interventions against GO. Tong et al. (76) reported that expression of miR-21-5p in GO eye fibroblasts was markedly higher than that of the control group and that it could enhance TGF-β 1-induced expression of total type I collagen and mRNA level of type I collagen. In addition, anti-miR-21 not only blocked decapentaplegic3 (Smad3) phosphorylation but can also mimic activated Smad3 phosphorylation. These results suggested that miR-21 could substantially enhance Smad3 phosphorylation and activate TGF-β/Smad signaling pathway to participate in orbital muscle fibrogenesis. In summary, these studies indicated that miR-21 may provide new targets for the treatment of GO.
Adipogenesis has been found to be closely related to the degree of ocular protruding (77). Therefore, efforts are being made to find effective ways to block the adipogenesis pathway as a new therapeutic option. Hammond et al. (78) reported that when Thy1- OFs were accumulated in GO, the miR-130a level was significantly increased, resulting in the decreased AMPK activity and promotion of the fat accumulation in orbit. Moreover, the expression level of miR-27a and miR-27b in the orbital adipose tissue of GO patients was significantly lower than that of the controls, which explains the inability to negatively regulate the mRNA expression of the various adipogenic genes such as PPARγ and CEBPβ in GO fibroblasts, thus indirectly causing the adipogenic differentiation of OFs (79). Therefore, blocking orbital muscle fibrogenesis and adipose generating pathways through modulating miRNA expression could provide novel therapeutic options.
In summary, multiple miRNAs can play an important role in the occurrence as well as the development of GO, and especially miR-146a may serve as an important target ( Table 1 ). However, the therapeutic applications of miRNA have not been fully developed, and hence more studies are needed to clarify the possible effects of miRNAs on other target genes in GO or its pathogenesis. In the future, miRNAs are expected to aid significantly in the precise diagnosis and targeted therapy of GO.
Table 1.
Non-coding RNAs in the pathogenesis of GO.
| Noncoding-RNAs | Samples/cells | Expression change | Function | Effects in GO | References |
|---|---|---|---|---|---|
| miR-146a | Plasma | Downregulation | Promote the differentiation of Th17 cell | Pro-inflammatory | (62) |
| CD4+ T | Downregulation | Increase Th1 response | Pro-inflammatory | (63, 64) | |
| Orbital tissue | Upregulation | Increase the IL-6 level | Exacerbate GO | (69) | |
| Orbital tissue | Upregulation | Decreases FN, collagen Iα, and α -SMA | Inhibit fibrosis process | (72, 73) | |
| miR-183 and miR-96 | CD4+ T | Upregulation | Contribute to the activation of CD4+ T cells | Pro-inflammatory | (66) |
| miR-21-5P | Orbital tissue | Upregulation | Promote collagen Iα and total collagen production | Promote fibrogenesis | (76) |
| miR-146a and miR-155 | Orbital tissue | Upregulation | Reducing the expression of PTEN and ZNRF3 | Promote proliferation | (70) |
| m iR-27a and miR-27b | Orbital tissue | Downregulation | Cause the adipogenic differentiation of OFs | Promote adipogenesis | (79) |
| m iR-130a | Orbital tissue | Upregulation | Enhance lipid accumulation | Fatty tissue accumulation | (78) |
| circRNA_14940 | Orbital tissue | Upregulation | Regulate the Wnt signaling pathway, ECM receptor interaction and PIK3-AKT signaling pathway | Participate in the pathogenesis of GO | (80) |
Other Noncoding RNAs and GO
CircRNAs are endogenous entities formed by the covalent binding of 3’ and 5’ end reverse splicing, which can display important biological functions in acting as miRNA sponge adsorption, attaching to the different RNA-binding proteins, and participating in protein translation (81, 82). However, the role of circRNAs in GO development is still in its infancy. For example, Wu et al. identified (80) 163 differentially expressed circRNAs from the orbital fat/connective tissues of GO patients. Through circRNA-mRNA co-expression and circRNA-mRNA interaction analysis, possible crosstalk of circRNA_14940 with down-regulated mRNA tenascin XB (TNXB) and up-regulated mRNA cyclin D1(CCND1) was analyzed and it was observed that abnormal regulation of the Wnt signaling pathway, PI3K-Akt signaling cascade, and extracellular matrix (ECM) receptor might also be involved in the pathogenesis of GO. CircRNA_10135 can also interact with prostaglandin F receptor (PTGFR) through modulating the calcium signaling pathway and participating in the adipogenesis process of GO. Meanwhile, circRNA_14936 has been found to be correlated with up-regulated mRNA TNF receptor superfamily member 19 (TNFRSF19), and hsa-miR-10392-3p that may potentially affect the occurrence and development of GO by regulating the interactions between circRNA_14936 and TNFRSF19 which can influence B cell survival. Therefore, circRNAs might play an important role in the pathogenesis and progression of GO ( Table 1 ). However, there are only few studies conducted to evaluate the role of circRNAs in the pathogenicity of GO, so the functions of most circRNAs are not clear and their molecular mechanism remains to be further elucidated.
LncRNAs are an important class of non-coding RNAs with a length of more than 200 nucleotides. LncRNAs can regulate the gene expression through affecting various mechanisms, including those related to epigenetics, transcriptional, post-transcriptional, and miRNAs regulation. A number of previous studies have reported that lncRNAs may play an important role in the pathogenesis of GD (83). For instance, Christensen et al. (84) reported the first lncRNA Heg to be associated with GD and showed that was negatively correlated with monocyte CD14 mRNA and serum TRAb level in GD patients. Moreover, anti-thyroid therapy could not change lncRNA Heg level in GD patients. The intimate temporal relationship between the onset of GD and GO also clearly suggests that these two diseases may have the same underlying etiology. Xia et al. (85) identified a novel lncRNA named Lnc-Smad3, which can maintain the tight chromatin structure of Smad3 promoter through direct interaction with HDAC1, thereby inhibiting Smad3 transcription and affecting inducible Treg (iTreg) cell polarization through modulating the TGFβ/Smad3 signaling pathway.
LncRNAs are also involved in the development of Treg cells, which provides a new direction to further study of the possible relationship between Lnc-Smad3 and GO. Although there are some difficulties in screening and studying lncRNAs, identifying their role and mechanism in GO may be a necessary direction for future research.
Gut Microbiome and GO
The human gut microbiome consists of about 100 trillion different gut microbial cells and serves as the main entry point for bacteria. In a symbiotic manner, the human host primarily provides the gut microbiota with crucial nutrients for promoting growth. In contrast, the gut microbes play an important role in the food digestion, detoxification, protection from pathogens, and the regulation of the host immune system. In fact, the highest concentration of T cells in the human body is located in the intestinal mucosa, where lymphocytes are primed to respond to the various microorganisms. Furthermore, the intestinal microbiome can also effectively regulate the local intestinal immune system and the systemic immune response (86, 87).
Moreover, multiple human and animal studies have shown that gut microbiome have been closely related to a variety of autoimmune diseases, such as inflammatory colitis (IBD) (88), rheumatoid arthritis (RA) (89), type I diabetes (T1D) (90), and systemic lupus erythematosus (SLE) (91). For instance, Jiang et al. (92) sequenced the fecal samples of patients with GD and healthy controls, and found that the richness of intestinal microflora in patients with GD decreased significantly. This alteration was characterized by a significant increase in Bacteroides, which damaged intestinal barrier function, resulting in the release of a large number of pro-inflammatory factors outside the intestine, thus resulting in immune dysfunction. In addition, it was also found that changes in thyroid hormone levels can lead to an increase in the number of Lactobacillus. It may play a harmful role in GD by activating NF- κB signaling pathway and autoimmune response to thyroid gland. At present, there are only few studies on intestinal flora in GO. Several reports have suggested that there is a marked decrease in bacterial community diversity in GO. Shi et al. (93) sequenced the fecal samples of patients with GO and healthy controls and found that compared with the control group, the proportion of Bacteroidetes and Prevotellaceae in patients with GO at different levels increased significantly, while the proportion of Blautia and Fusicatenibacter decreased significantly. The proportion of the Succinivibrionaceae and Subdoligranulum was positively correlated with TRAb, and there was a negative correlation between Parabacteroides_disasonis and TRAb. Further analysis showed that Prevotellaceae could clearly distinguish GO patients from the control group. The level of s_Prevotea_copri was reported to be positively correlated with the level of serum TRAb, which may be related to active orbital inflammation (94). Thereafter, the researchers conducted a comparative analysis of GD, GO and healthy people for the first time. They found that the proportion of Deinococcus-Thermus and Chloroflexi in patients with GO was significantly lower than that in GD (95). Interestingly, after transplanting the normal human feces into type 2 diabetic mice, researchers found that the abundance of Bacteroides and some bacteria decreased and blood glucose levels recovered. This led to the speculation that it may be possibly increasing the diversity of intestinal microflora that can affect the different types of microorganisms in the intestines of mice to achieve the effect of treatment (96). Therefore, whether fecal microbiome transplantation can also have a therapeutic effect on GO can be an important direction for future research. Additional in-depth studies about the role of intestinal microorganisms in GO will possibly provide new methods and ideas for the treatment of GO. In addition, some studies have reported that Salmonella that can cause enteritis possesses an antigenic cluster similar to the thyroid stimulating hormone receptor protein, where antibodies against the thyrotropin receptor proteins might be produced after infection (96). These results laid a sound foundation for exploring the interaction between intestinal microorganisms and TRAb. However, the current findings may also be related to thyroid autoimmunity rather than GO, and might not be entirely specific for GO.
Masetti et al. (34) used intramuscular injection of eukaryotic expression plasmid (thyroid stimulating receptor) in two distinct experimental centers in two different countries (Center 1 and 2). The GD/GO disease model of BALB/C female mice was established by immunological method of TSHR. The results showed that only BALB/C mice from Center 2 displayed hyperthyroidism and apparent changes in orbital tissues. Interestingly, it was found that the abundance and species composition of intestinal flora were significantly different between BALB/C mice tested from the two centers by 16S rRNA gene sequencing and traditional microbial culture. In Center 2, the intestinal flora structure of different intervention groups (TSHR group, β Gal group) was analyzed by 16S rRNA gene sequencing and it was found that the intestinal flora of TSHR group changed significantly (increased relative abundance of Lactobacillus, decreased relative abundance of bacteroides, etc.). These alterations were also associated with some clinical features such as thyroxine levels (fT4), TRAb, orbital adipogenesis, and muscular atrophy. The same group constructed another GD/GO model by using BALB/c and C57BL/6J mice. The results indicated that the C57BL/6 mice had higher TRAb, but compared with BALB/c mice, the spleen T cells of C57BL/6 mice exhibited poor proliferation of TSHR response, and secreted mainly anti-inflammatory cytokines such as IL-10, without elevated thyroxine levels or any orbital lesions, while resembling immunized BALB/C mice showed orbital pathology. These observations suggested that there was a different pattern of correlation between the microbiomes and disease (endocrine and immune) characteristics between the two different mouse strains (97). In addition, Shi et al. (94) found that operational taxonomic units (OTUs) derived from the bacteroidesstercoris species were markedly associated with the thyroglobulin autoantibodies, whereas OTUs from the bacteroides genus were associated with CAS. Masetti et al. (98) deciphered that the effect of intestinal microorganisms on the immune system can be primarily revealed by maintaining a balance between the Tregs and pro-inflammatory Th17 cells and Th1 cells. When the microbiome might not be ideal, intestinal dendritic cells can secrete TGF-β, which can cause the Th17/Treg imbalance, thus leading to the loss of immune tolerance. In individuals with a particular genetic predisposition, the result can potentially lead to GD and GO. These results clearly suggested that the gut microbiome could play a critical role in shaping the immune response that might significantly influence the TRAb levels and immune tolerance, thereby influencing thyroid and eye disease phenotypes.
The above studies have provided a novel perspective research pathway to clarify the potential mechanisms of intestinal microbiome disorders observed in GO. Hence, understanding that how the composition of the gut bacterial microbiome and the abundance of specific bacterial strains might affect the autoimmune response in GD and GO remains the next important aspect.
Conclusion
With the emergence of novel bioinformatics tools and high-throughput gene sequencing technologies, the important role of epigenetics and gut microbiomes in GO has been gradually elucidated ( Figure 1 ). Still, several crucial mechanisms especially from GD to GO remain unclear. At present, the study of GO epigenetics focuses primarily on only one kind of epigenetic modifications, but the role of its regulatory network in controlling molecular mechanisms of GO remains unknown. Therefore, there is an unmet need to conduct a combined analysis of the varying epigenetic modifications that can further explore the possible roles of various less explored epigenetic factors that have been implicated in the pathogenesis of GO.
Epigenetic inheritance can link environmental factors and diseases. How environmental factors can lead to the progression of GO by facilitating epigenetic inheritance and intestinal microbial dysregulation in GO remains to be elucidated. To date, GO is clinically diagnosed by an ophthalmologist with an adjunct imaging such as CT or magnetic resonance imaging (MRI). A few studies have shown that some epigenetic markers and intestinal microflora might be different in GD patients with or without GO. However, it is still difficult to predict which GD patients will develop into GO or severe GO, and which GD patients will be responsive or unresponsive to the different types of treatment. Further studies should focus on the current challenges in the future.
Author Contributions
YW and H-SY participated in the conceptualization and writing. X-MM, XW, XS, L-JW, X-QL, and X-YL participated in the review and editing. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by National Key R&D Program of China (grant number: 2018YFC1004300), National Natural Science Foundation Project of China (grant number: 82160154, 81670844), the Key Project of Guizhou Provincial Science and Technology Department (grant number: QKH-JC-2019-1464), the Excellent Talent Support Program of Guizhou Provincial Education Department (grant number: QJH-KY-2017-077), the Science and Technology Foundation of Guizhou Province (grant number: QKH-PTRC-2018-5772-042), the Science and Technology Project of Zunyi (grant number: ZSKH-HZ-2020-35), and the Program for Excellent Young Talents of Zunyi Medical University (grant number: 18-ZY-001).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
- 1. Edmunds MR, Boelaert K. Knowledge of Thyroid Eye Disease in Graves’ Disease Patients With and Without Orbitopathy. Thyroid (2019) 29(4):557–62. doi: 10.1089/thy.2018.0665 [DOI] [PubMed] [Google Scholar]
- 2. Tanda ML, Piantanida E, Liparulo L, Veronesi G, Lai A, Sassi L, et al. Prevalence and Natural History of Graves’ Orbitopathy in a Large Series of Patients With Newly Diagnosed Graves’ Hyperthyroidism Seen at a Single Center. J Clin Endocrinol Metab (2013) 98(4):1443–9. doi: 10.1210/jc.2012-3873 [DOI] [PubMed] [Google Scholar]
- 3. Bahn RS. Graves’ Ophthalmopathy. N Engl J Med (2010) 362(8):726–38. doi: 10.1056/NEJMra0905750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bartley GB. The Epidemiologic Characteristics and Clinical Course of Ophthalmopathy Associated With Autoimmune Thyroid Disease in Olmsted County, Minnesota. Trans Am Ophthalmol Soc (1994) 92:477–588. [PMC free article] [PubMed] [Google Scholar]
- 5. Bartalena L, Piantanida E, Gallo D, Lai A, Tanda ML. Epidemiology, Natural History, Risk Factors, and Prevention of Graves’ Orbitopathy. Front Endocrinol (Lausanne) (2020) 11:615993. doi: 10.3389/fendo.2020.615993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bartalena L, Baldeschi L, Boboridis K, Eckstein A, Kahaly GJ, Marcocci C, et al. The 2016 European Thyroid Association/European Group on Graves’ Orbitopathy Guidelines for the Management of Graves’ Orbitopathy. Eur Thyroid J (2016) 5(1):9–26. doi: 10.1159/000443828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Rotondo Dottore G, Torregrossa L, Lanzolla G, Mariotti S, Menconi F, Piaggi P, et al. Role of the Mononuclear Cell Infiltrate in Graves’ Orbitopathy (GO): Results of a Large Cohort Study. J Endocrinol Invest (2021). doi: 10.1007/s40618-021-01692-4 [DOI] [PubMed] [Google Scholar]
- 8. Huang Y, Fang S, Li D, Zhou H, Li B, Fan X. The Involvement of T Cell Pathogenesis in Thyroid-Associated Ophthalmopathy. Eye (Lond) (2019) 33(2):176–82. doi: 10.1038/s41433-018-0279-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kuriyan AE, Woeller CF, O’Loughlin CW, Phipps RP, Feldon SE. Orbital Fibroblasts From Thyroid Eye Disease Patients Differ in Proliferative and Adipogenic Responses Depending on Disease Subtype. Invest Ophthalmol Vis Sci (2013) 54(12):7370–7. doi: 10.1167/iovs.13-12741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Koumas L, Smith TJ, Phipps RP. Fibroblast Subsets in the Human Orbit: Thy-1+ and Thy-1- Subpopulations Exhibit Distinct Phenotypes. Eur J Immunol (2002) 32(2):477–85. doi: [DOI] [PubMed] [Google Scholar]
- 11. Wiersinga WM, Prummel MF. Pathogenesis of Graves’ Ophthalmopathy–Current Understanding. J Clin Endocrinol Metab (2001) 86(2):501–3. doi: 10.1210/jcem.86.2.7338 [DOI] [PubMed] [Google Scholar]
- 12. Wakelkamp IM, Bakker O, Baldeschi L, Wiersinga WM, Prummel MF. TSH-R Expression and Cytokine Profile in Orbital Tissue of Active vs. Inactive Graves’ Ophthalmopathy Patients. Clin Endocrinol (Oxf) (2003) 58(3):280–7. doi: 10.1046/j.1365-2265.2003.01708.x [DOI] [PubMed] [Google Scholar]
- 13. Aniszewski JP, Valyasevi RW, Bahn RS. Relationship Between Disease Duration and Predominant Orbital T Cell Subset in Graves’ Ophthalmopathy. J Clin Endocrinol Metab (2000) 85(2):776–80. doi: 10.1210/jcem.85.2.6333 [DOI] [PubMed] [Google Scholar]
- 14. Xia N, Zhou S, Liang Y, Xiao C, Shen H, Pan H, et al. CD4+ T Cells and the Th1/Th2 Imbalance Are Implicated in the Pathogenesis of Graves’ Ophthalmopathy. Int J Mol Med (2006) 17(5):911–6. [PubMed] [Google Scholar]
- 15. Fang S, Huang Y, Wang S, Zhang Y, Luo X, Liu L, et al. IL-17a Exacerbates Fibrosis by Promoting the Proinflammatory and Profibrotic Function of Orbital Fibroblasts in TAO. J Clin Endocrinol Metab (2016) 101(8):2955–65. doi: 10.1210/jc.2016-1882 [DOI] [PubMed] [Google Scholar]
- 16. Lv M, Shen J, Li Z, Zhao D, Chen Z, Wan H, et al. [Role of Treg/Th17 Cells and Related Cytokines in Graves’ Ophthalmopathy]. Nan Fang Yi Ke Da Xue Xue Bao = J South Med Univ (2014) 34(12):1809–13. [PubMed] [Google Scholar]
- 17. Eckstein AK, Plicht M, Lax H, Neuhäuser M, Mann K, Lederbogen S, et al. Thyrotropin Receptor Autoantibodies Are Independent Risk Factors for Graves’ Ophthalmopathy and Help to Predict Severity and Outcome of the Disease. J Clin Endocrinol Metab (2006) 91(9):3464–70. doi: 10.1210/jc.2005-2813 [DOI] [PubMed] [Google Scholar]
- 18. Tsui S, Naik V, Hoa N, Hwang CJ, Afifiyan NF, Sinha Hikim A, et al. Evidence for an Association Between Thyroid-Stimulating Hormone and Insulin-Like Growth Factor 1 Receptors: A Tale of Two Antigens Implicated in Graves’ Disease. J Immunol (2008) 181(6):4397–405. doi: 10.4049/jimmunol.181.6.4397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Smith TJ. Insulin-Like Growth Factor Pathway and the Thyroid. Front Endocrinol (Lausanne) (2021) 12:653627. doi: 10.3389/fendo.2021.653627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bartalena L, Kahaly G, Baldeschi L, Dayan C, Eckstein A, Marcocci C, et al. The 2021 European Group on Graves' Orbitopathy (EUGOGO) Clinical Practice Guidelines for the Medical Management of Graves' Orbitopathy. Eur J Endocrinol (2021) 185(4):G43–g67. doi: 10.1530/eje-21-0479 [DOI] [PubMed] [Google Scholar]
- 21. Hadj-Kacem H, Rebuffat S, Mnif-Feki M, Belguith-Maalej S, Ayadi H, Peraldi-Roux S. Autoimmune Thyroid Diseases: Genetic Susceptibility of Thyroid-Specific Genes and Thyroid Autoantigens Contributions. Int J Immunogenet (2009) 36(2):85–96. doi: 10.1111/j.1744-313X.2009.00830.x [DOI] [PubMed] [Google Scholar]
- 22. Yin X, Latif R, Bahn R, Tomer Y, Davies TF. Influence of the TSH Receptor Gene on Susceptibility to Graves’ Disease and Graves’ Ophthalmopathy. Thyroid (2008) 18(11):1201–6. doi: 10.1089/thy.2008.0098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Du J, Wang X, Tan G, Wei W, Zhou F, Liang Z, et al. Predisposition to Graves’ Disease and Graves’ Ophthalmopathy by Genetic Variants of IL2RA. J Mol Med (Berl) (2021) 99(10):1487–95. doi: 10.1007/s00109-021-02111-0 [DOI] [PubMed] [Google Scholar]
- 24. Zhou F, Liang Z, Wang X, Tan G, Wei W, Zheng G, et al. The VDR Gene Confers a Genetic Predisposition to Graves’ Disease and Graves’ Ophthalmopathy in the Southwest Chinese Han Population. Gene (2021) 793:145750. doi: 10.1016/j.gene.2021.145750 [DOI] [PubMed] [Google Scholar]
- 25. Yin X, Latif R, Bahn R, Davies TF. Genetic Profiling in Graves’ Disease: Further Evidence for Lack of a Distinct Genetic Contribution to Graves’ Ophthalmopathy. Thyroid (2012) 22(7):730–6. doi: 10.1089/thy.2012.0007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Shalaby SM, Mackawy AMH, Atef DM, Atef RM, Saeed J. Promoter Methylation and Expression of Intercellular Adhesion Molecule 1 Gene in Blood of Autoimmune Thyroiditis Patients. Mol Biol Rep (2019) 46(5):5345–53. doi: 10.1007/s11033-019-04990-6 [DOI] [PubMed] [Google Scholar]
- 27. Martinez-Hernandez R, Sampedro-Nunez M, Serrano-Somavilla A, Ramos-Levi AM, de la Fuente H, Trivino JC, et al. A MicroRNA Signature for Evaluation of Risk and Severity of Autoimmune Thyroid Diseases. J Clin Endocrinol Metab (2018) 103(3):1139–50. doi: 10.1210/jc.2017-02318 [DOI] [PubMed] [Google Scholar]
- 28. Feil R, Fraga MF. Epigenetics and the Environment: Emerging Patterns and Implications. Nat Rev Genet (2012) 13(2):97–109. doi: 10.1038/nrg3142 [DOI] [PubMed] [Google Scholar]
- 29. Wang B, Shao X, Song R, Xu D, Zhang JA. The Emerging Role of Epigenetics in Autoimmune Thyroid Diseases. Front Immunol (2017) 8:396. doi: 10.3389/fimmu.2017.00396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mazzone R, Zwergel C, Artico M, Taurone S, Ralli M, Greco A, et al. The Emerging Role of Epigenetics in Human Autoimmune Disorders. Clin Epigenet (2019) 11(1):34. doi: 10.1186/s13148-019-0632-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ekronarongchai S, Palaga T, Saonanon P, Pruksakorn V, Hirankarn N, van Hagen PM, et al. Histone Deacetylase 4 Controls Extracellular Matrix Production in Orbital Fibroblasts From Graves’ Ophthalmopathy Patients. Thyroid (2021) 31(10):1566–76. doi: 10.1089/thy.2020.0948 [DOI] [PubMed] [Google Scholar]
- 32. Xin Z, Hua L, Yang YL, Shi TT, Liu W, Tuo X, et al. A Pathway Analysis Based on Genome-Wide DNA Methylation of Chinese Patients With Graves’ Orbitopathy. BioMed Res Int (2019) 2019:9565794. doi: 10.1155/2019/9565794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Su X, Yin X, Liu Y, Yan X, Zhang S, Wang X, et al. Gut Dysbiosis Contributes to the Imbalance of Treg and Th17 Cells in Graves’ Disease Patients by Propionic Acid. J Clin Endocrinol Metab (2020) 105(11):dgaa511. doi: 10.1210/clinem/dgaa511 [DOI] [PubMed] [Google Scholar]
- 34. Masetti G, Moshkelgosha S, Kohling HL, Covelli D, Banga JP, Berchner-Pfannschmidt U, et al. Gut Microbiota in Experimental Murine Model of Graves’ Orbitopathy Established in Different Environments may Modulate Clinical Presentation of Disease. Microbiome (2018) 6(1):97. doi: 10.1186/s40168-018-0478-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Moshkelgosha S, Verhasselt HL, Masetti G, Covelli D, Biscarini F, Horstmann M, et al. Modulating Gut Microbiota in a Mouse Model of Graves’ Orbitopathy and Its Impact on Induced Disease. Microbiome (2021) 9(1):45. doi: 10.1186/s40168-020-00952-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zhu H, Wang G, Qian J. Transcription Factors as Readers and Effectors of DNA Methylation. Nat Rev Genet (2016) 17(9):551–65. doi: 10.1038/nrg.2016.83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Xin Z, Hua L, Shi TT, Tuo X, Yang FY, Li Y, et al. A Genome-Wide DNA Methylation Analysis in Peripheral Blood From Patients Identifies Risk Loci Associated With Graves’ Orbitopathy. J Endocrinol Invest (2018) 41(6):719–27. doi: 10.1007/s40618-017-0796-6 [DOI] [PubMed] [Google Scholar]
- 38. Shi TT, Hua L, Xin Z, Li Y, Liu W, Yang YL. Identifying and Validating Genes With DNA Methylation Data in the Context of Biological Network for Chinese Patients With Graves’ Orbitopathy. Int J Endocrinol (2019) 2019:6212681. doi: 10.1155/2019/6212681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Khong JJ, Wang LY, Smyth GK, McNab AA, Hardy TG, Selva D, et al. Differential Gene Expression Profiling of Orbital Adipose Tissue in Thyroid Orbitopathy. Invest Ophthalmol Vis Sci (2015) 56(11):6438–47. doi: 10.1167/iovs.15-17185 [DOI] [PubMed] [Google Scholar]
- 40. Figueroa ME, Lugthart S, Li Y, Erpelinck-Verschueren C, Deng X, Christos PJ, et al. DNA Methylation Signatures Identify Biologically Distinct Subtypes in Acute Myeloid Leukemia. Cancer Cell (2010) 17(1):13–27. doi: 10.1016/j.ccr.2009.11.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hu YF, Hua L, Tuo X, Shi TT, Yang YL, Liu YF, et al. Preliminary Evidence of the Association Between DNAm and Orbital Volumetry in GO. Endocr Connect (2020) 9(7):617–26. doi: 10.1530/EC-20-0147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Virakul S, Somparn P, Pisitkun T, van der Spek PJ, Dalm V, Paridaens D, et al. Integrative Analysis of Proteomics and DNA Methylation in Orbital Fibroblasts From Graves’ Ophthalmopathy. Front Endocrinol (Lausanne) (2020) 11:619989. doi: 10.3389/fendo.2020.619989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kato N, Loh M, Takeuchi F, Verweij N, Wang X, Zhang W, et al. Trans-Ancestry Genome-Wide Association Study Identifies 12 Genetic Loci Influencing Blood Pressure and Implicates a Role for DNA Methylation. Nat Genet (2015) 47(11):1282–93. doi: 10.1038/ng.3405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lee JY, Kim NK, Cho YW, Lew H. Association Between Methylenetetrahydrofolate Reductase (MTHFR) Polymorphisms and Susceptibility to Graves’ Ophthalmopathy. Mol Med Rep (2016) 14(3):2276–82. doi: 10.3892/mmr.2016.5458 [DOI] [PubMed] [Google Scholar]
- 45. Mao R, Fan Y, Zuo L, Geng D, Meng F, Zhu J, et al. Association Study Between Methylenetetrahydrofolate Reductase Gene Polymorphisms and Graves’ Disease. Cell Biochem Funct (2010) 28(7):585–90. doi: 10.1002/cbf.1694 [DOI] [PubMed] [Google Scholar]
- 46. Kouzarides T. Chromatin Modifications and Their Function. Cell (2007) 128(4):693–705. doi: 10.1016/j.cell.2007.02.005 [DOI] [PubMed] [Google Scholar]
- 47. Yan N, Zhou JZ, Zhang JA, Cai T, Zhang W, Wang Y, et al. Histone Hypoacetylation and Increased Histone Deacetylases in Peripheral Blood Mononuclear Cells From Patients With Graves’ Disease. Mol Cell Endocrinol (2015) 414:143–7. doi: 10.1016/j.mce.2015.05.037 [DOI] [PubMed] [Google Scholar]
- 48. Stefan M, Wei C, Lombardi A, Li CW, Concepcion ES, Inabnet WB, 3rd, et al. Genetic-Epigenetic Dysregulation of Thymic TSH Receptor Gene Expression Triggers Thyroid Autoimmunity. Proc Natl Acad Sci U S A (2014) 111(34):12562–7. doi: 10.1073/pnas.1408821111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Limbach M, Saare M, Tserel L, Kisand K, Eglit T, Sauer S, et al. Epigenetic Profiling in CD4+ and CD8+ T Cells From Graves’ Disease Patients Reveals Changes in Genes Associated With T Cell Receptor Signaling. J Autoimmun (2016) 67:46–56. doi: 10.1016/j.jaut.2015.09.006 [DOI] [PubMed] [Google Scholar]
- 50. Jung SJ, Choi YJ, Park TK, Woo SE, Kim BY, Yoon JS, et al. Wnt Signalling Inhibits Adipogenesis in Orbital Fibroblasts From Patients With Graves’ Orbitopathy. Br J Ophthalmol (2021). doi: 10.1136/bjophthalmol-2020-316898 [DOI] [PubMed] [Google Scholar]
- 51. Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, et al. Genome-Wide Maps of Chromatin State in Pluripotent and Lineage-Committed Cells. Nature (2007) 448(7153):553–60. doi: 10.1038/nature06008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Matsumura Y, Nakaki R, Inagaki T, Yoshida A, Kano Y, Kimura H, et al. H3K4/H3K9me3 Bivalent Chromatin Domains Targeted by Lineage-Specific DNA Methylation Pauses Adipocyte Differentiation. Mol Cell (2015) 60(4):584–96. doi: 10.1016/j.molcel.2015.10.025 [DOI] [PubMed] [Google Scholar]
- 53. Sacristan P, Serrano-Somavilla A, Gonzalez-Amaro R, Martinez-Hernandez R, Marazuela M. Analysis of Expression of Different Histone Deacetylases in Autoimmune Thyroid Disease. J Clin Endocrinol Metab (2021) 106(11):3213–27. doi: 10.1210/clinem/dgab526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Yin L, Zeng C, Yao J, Shen J. Emerging Roles for Noncoding RNAs in Autoimmune Thyroid Disease. Endocrinology (2020) 161(8):bqaa053. doi: 10.1210/endocr/bqaa053 [DOI] [PubMed] [Google Scholar]
- 55. Mehta A, Baltimore D. MicroRNAs as Regulatory Elements in Immune System Logic. Nat Rev Immunol (2016) 16(5):279–94. doi: 10.1038/nri.2016.40 [DOI] [PubMed] [Google Scholar]
- 56. Friedman RC, Farh KK, Burge CB, Bartel DP. Most Mammalian mRNAs Are Conserved Targets of microRNAs. Genome Res (2009) 19(1):92–105. doi: 10.1101/gr.082701.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Chen JQ, Papp G, Szodoray P, Zeher M. The Role of microRNAs in the Pathogenesis of Autoimmune Diseases. Autoimmun Rev (2016) 15(12):1171–80. doi: 10.1016/j.autrev.2016.09.003 [DOI] [PubMed] [Google Scholar]
- 58. Wei Y, Li N, Zhao L, Yang C, Ma B, Li X, et al. MicroRNAs and Autoimmune-Mediated Eye Diseases. Front Cell Dev Biol (2020) 8:818. doi: 10.3389/fcell.2020.00818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Ma X, Becker Buscaglia LE, Barker JR, Li Y. MicroRNAs in NF-kappaB Signaling. J Mol Cell Biol (2011) 3(3):159–66. doi: 10.1093/jmcb/mjr007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Jiang W, Kong L, Ni Q, Lu Y, Ding W, Liu G, et al. miR-146a Ameliorates Liver Ischemia/Reperfusion Injury by Suppressing IRAK1 and TRAF6. PLoS One (2014) 9(7):e101530. doi: 10.1371/journal.pone.0101530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Taganov KD, Boldin MP, Chang KJ, Baltimore D. NF-kappaB-Dependent Induction of microRNA miR-146, an Inhibitor Targeted to Signaling Proteins of Innate Immune Responses. Proc Natl Acad Sci USA (2006) 103(33):12481–6. doi: 10.1073/pnas.0605298103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Wei H, Guan M, Qin Y, Xie C, Fu X, Gao F, et al. Circulating Levels of miR-146a and IL-17 Are Significantly Correlated With the Clinical Activity of Graves’ Ophthalmopathy. Endocrine J (2014) 61(11):1087–92. doi: 10.1507/endocrj.ej14-0246 [DOI] [PubMed] [Google Scholar]
- 63. Hu ZJ, He JF, Li KJ, Chen J, Xie XR. Decreased microRNA-146a in CD4+T Cells Promote Ocular Inflammation in Thyroid-Associated Ophthalmopathy by Targeting NUMB. Eur Rev Med Pharmacol Sci (2017) 21(8):1803–9. [PubMed] [Google Scholar]
- 64. Yang WJ, Ma PF, Li SP, Su H, Liu YJ. MicroRNA-146a Contributes to CD4(+) T Lymphocyte Differentiation in Patients With Thyroid Ophthalmopathy. Am J Trans Res (2017) 9(4):1801–9. [PMC free article] [PubMed] [Google Scholar]
- 65. Li K, Du Y, Jiang BL, He JF. Increased microRNA-155 and Decreased microRNA-146a may Promote Ocular Inflammation and Proliferation in Graves’ Ophthalmopathy. Med Sci Monit (2014) 20:639–43. doi: 10.12659/MSM.890686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Thiel J, Alter C, Luppus S, Eckstein A, Tan S, Fuhrer D, et al. MicroRNA-183 and microRNA-96 Are Associated With Autoimmune Responses by Regulating T Cell Activation. J Autoimmun (2019) 96:94–103. doi: 10.1016/j.jaut.2018.08.010 [DOI] [PubMed] [Google Scholar]
- 67. Shen L, Huang F, Ye L, Zhu W, Zhang X, Wang S, et al. Circulating microRNA Predicts Insensitivity to Glucocorticoid Therapy in Graves’ Ophthalmopathy. Endocrine (2015) 49(2):445–56. doi: 10.1007/s12020-014-0487-4 [DOI] [PubMed] [Google Scholar]
- 68. Zhang L, Masetti G, Colucci G, Salvi M, Covelli D, Eckstein A, et al. Combining Micro-RNA and Protein Sequencing to Detect Robust Biomarkers for Graves’ Disease and Orbitopathy. Sci Rep (2018) 8(1):8386. doi: 10.1038/s41598-018-26700-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Wang N, Chen FE, Long ZW. Mechanism of MicroRNA-146a/Notch2 Signaling Regulating IL-6 in Graves Ophthalmopathy. Cell Physiol Biochem (2017) 41(4):1285–97. doi: 10.1159/000464430 [DOI] [PubMed] [Google Scholar]
- 70. Woeller CF, Roztocil E, Hammond C, Feldon SE. TSHR Signaling Stimulates Proliferation Through PI3K/Akt and Induction of miR-146a and miR-155 in Thyroid Eye Disease Orbital Fibroblasts. Invest Ophthalmol Vis Sci (2019) 60(13):4336–45. doi: 10.1167/iovs.19-27865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Jang SY, Chae MK, Lee JH, Lee EJ, Yoon JS. Role of miR-146a in the Regulation of Inflammation in an In Vitro Model of Graves’ Orbitopathy. Invest Ophthalmol Vis Sci (2016) 57(10):4027–34. doi: 10.1167/iovs.16-19213 [DOI] [PubMed] [Google Scholar]
- 72. Jang SY, Park SJ, Chae MK, Lee JH, Lee EJ, Yoon JS. Role of microRNA-146a in Regulation of Fibrosis in Orbital Fibroblasts From Patients With Graves’ Orbitopathy. Br J Ophthalmol (2018) 102(3):407–14. doi: 10.1136/bjophthalmol-2017-310723 [DOI] [PubMed] [Google Scholar]
- 73. Liu W, Ma C, Li HY, Chen L, Yuan SS, Li KJ. MicroRNA-146a Downregulates the Production of Hyaluronic Acid and Collagen I in Graves’ Ophthalmopathy Orbital Fibroblasts. Exp Ther Med (2020) 20(5):38. doi: 10.3892/etm.2020.9165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Lee JY, Yun M, Paik JS, Lee SB, Yang SW. PDGF-BB Enhances the Proliferation of Cells in Human Orbital Fibroblasts by Suppressing PDCD4 Expression Via Up-Regulation of microRNA-21. Invest Ophthalmol Vis Sci (2016) 57(3):908–13. doi: 10.1167/iovs.15-18157 [DOI] [PubMed] [Google Scholar]
- 75. Young MR, Santhanam AN, Yoshikawa N, Colburn NH. Have Tumor Suppressor PDCD4 and Its Counteragent Oncogenic miR-21 Gone Rogue? Mol Interventions (2010) 10(2):76–9. doi: 10.1124/mi.10.2.5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Tong BD, Xiao MY, Zeng JX, Xiong W. MiRNA-21 Promotes Fibrosis in Orbital Fibroblasts From Thyroid-Associated Ophthalmopathy. Mol Vision (2015) 21:324–34. [PMC free article] [PubMed] [Google Scholar]
- 77. Nishida Y, Tian S, Isberg B, Hayashi O, Tallstedt L, Lennerstrand G. Significance of Orbital Fatty Tissue for Exophthalmos in Thyroid-Associated Ophthalmopathy. Graefes Arch Clin Exp Ophthalmol (2002) 240(7):515–20. doi: 10.1007/s00417-002-0498-3 [DOI] [PubMed] [Google Scholar]
- 78. Hammond CL, Roztocil E, Gonzalez MO, Feldon SE, Woeller CF. MicroRNA-130a Is Elevated in Thyroid Eye Disease and Increases Lipid Accumulation in Fibroblasts Through the Suppression of AMPK. Invest Ophthalmol Vis Sci (2021) 62(1):29. doi: 10.1167/iovs.62.1.29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Jang SY, Chae MK, Lee JH, Lee EJ, Yoon JS. MicroRNA-27 Inhibits Adipogenic Differentiation in Orbital Fibroblasts From Patients With Graves’ Orbitopathy. PLoS One (2019) 14(8):e0221077. doi: 10.1371/journal.pone.0221077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Wu L, Zhou R, Diao J, Chen X, Huang J, Xu K, et al. Differentially Expressed Circular RNAs in Orbital Adipose/Connective Tissue From Patients With Thyroid-Associated Ophthalmopathy. Exp Eye Res (2020) 196:108036. doi: 10.1016/j.exer.2020.108036 [DOI] [PubMed] [Google Scholar]
- 81. Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, et al. Natural RNA Circles Function as Efficient microRNA Sponges. Nature (2013) 495(7441):384–8. doi: 10.1038/nature11993 [DOI] [PubMed] [Google Scholar]
- 82. Bolisetty MT, Graveley BR. Circuitous Route to Transcription Regulation. Mol Cell (2013) 51(6):705–6. doi: 10.1016/j.molcel.2013.09.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Wu GC, Pan HF, Leng RX, Wang DG, Li XP, Li XM, et al. Emerging Role of Long Noncoding RNAs in Autoimmune Diseases. Autoimmun Rev (2015) 14(9):798–805. doi: 10.1016/j.autrev.2015.05.004 [DOI] [PubMed] [Google Scholar]
- 84. Christensen NJ, Habekost G. Bratholm P. A RNA Transcript (Heg) in Mononuclear Cells is Negatively Correlated With CD14 mRNA and TSH Receptor Autoantibodies. Clin Exp Immunol (2008) 154(2):209–15. doi: 10.1111/j.1365-2249.2008.03744.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Xia M, Liu J, Liu S, Chen K, Lin H, Jiang M, et al. Ash1l and lnc-Smad3 Coordinate Smad3 Locus Accessibility to Modulate Itreg Polarization and T Cell Autoimmunity. Nat Commun (2017) 8:15818. doi: 10.1038/ncomms15818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Covelli D, Ludgate M. The Thyroid, the Eyes and the Gut: A Possible Connection. J Endocrinol Invest (2017) 40(6):567–76. doi: 10.1007/s40618-016-0594-6 [DOI] [PubMed] [Google Scholar]
- 87. Kataoka K. The Intestinal Microbiota and Its Role in Human Health and Disease. J Med Investigation: JMI (2016) 63(1-2):27–37. doi: 10.2152/jmi.63.27 [DOI] [PubMed] [Google Scholar]
- 88. Conrad MA, Wu GD, Kelsen JR. The Gut Microbiota and Inflammatory Bowel Disease. Pediatric Inflammatory Bowel Disease. Springer International Publishing; (2017) 45–54. doi: 10.1007/978-3-319-49215-5_4 [DOI] [Google Scholar]
- 89. Zhang X, Zhang D, Jia H, Feng Q, Wang D, Liang D, et al. The Oral and Gut Microbiomes Are Perturbed in Rheumatoid Arthritis and Partly Normalized After Treatment. Nat Med (2015) 21(8):895–905. doi: 10.1038/nm.3914 [DOI] [PubMed] [Google Scholar]
- 90. Kugelberg E. Microbiota: Diet can Protect Against Type 1 Diabetes. Nat Rev Immunol (2017) 17(5):279. doi: 10.1038/nri.2017.40 [DOI] [PubMed] [Google Scholar]
- 91. Edwards CJ, Costenbader KH. Epigenetics and the Microbiome: Developing Areas in the Understanding of the Aetiology of Lupus. Lupus (2014) 23(6):505–6. doi: 10.1177/0961203314531636 [DOI] [PubMed] [Google Scholar]
- 92. Jiang W, Yu X, Kosik RO, Song Y, Qiao T, Tong J, et al. Gut Microbiota May Play a Significant Role in the Pathogenesis of Graves’ Disease. Thyroid (2021) 31(5):810–20. doi: 10.1089/thy.2020.0193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Shi TT, Xin Z, Hua L, Zhao RX, Yang YL, Wang H, et al. Alterations in the Intestinal Microbiota of Patients With Severe and Active Graves’ Orbitopathy: A Cross-Sectional Study. J Endocrinol Invest (2019) 42(8):967–78. doi: 10.1007/s40618-019-1010-9 [DOI] [PubMed] [Google Scholar]
- 94. Shi TT, Hua L, Wang H, Xin Z. The Potential Link Between Gut Microbiota and Serum TRAb in Chinese Patients With Severe and Active Graves’ Orbitopathy. Int J Endocrinol (2019) 2019:9736968. doi: 10.1155/2019/9736968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Shi TT, Xin Z, Hua L, Wang H, Zhao RX, Yang YL, et al. Comparative Assessment of Gut Microbial Composition and Function in Patients With Graves’ Disease and Graves’ Orbitopathy. J Endocrinol Invest (2021) 44(2):297–310. doi: 10.1007/s40618-020-01298-2 [DOI] [PubMed] [Google Scholar]
- 96. Han X, Wang Y, Zhang P, Zhu M, Li L, Mao X, et al. Kazak Faecal Microbiota Transplantation Induces Short-Chain Fatty Acids That Promote Glucagon-Like Peptide-1 Secretion by Regulating Gut Microbiota in Db/Db Mice. Pharm Biol (2021) 59(1):1077–87. doi: 10.1080/13880209.2021.1954667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Moshkelgosha S, Masetti G, Berchner-Pfannschmidt U, Verhasselt HL, Horstmann M, Diaz-Cano S, et al. Gut Microbiome in BALB/c and C57BL/6J Mice Undergoing Experimental Thyroid Autoimmunity Associate With Differences in Immunological Responses and Thyroid Function. Horm Metab Res (2018) 50(12):932–41. doi: 10.1055/a-0653-3766 [DOI] [PubMed] [Google Scholar]
- 98. Masetti G, Ludgate M. Microbiome and Graves’ Orbitopathy. Eur Thyroid J (2020) 9(Suppl 1):78–85. doi: 10.1159/000512255 [DOI] [PMC free article] [PubMed] [Google Scholar]