Abstract
The high-pathogenicity island (HPI) of Yersinia has been observed in 93% of 60 enteroadhesive Escherichia coli strains and 80% of E. coli strains isolated from blood samples. In the present study we investigated 671 fecal samples from patients with diarrhea in Shandong Province, China, and isolated HPI-harboring E. coli from 6.26% of the samples. The isolation rates for patients with diarrhea in three age groups, 10 to 20, 30 to 40, and 50 to 60 years, were 6.70, 12.35, and 10.81%, respectively. Therefore, HPI-harboring E. coli is the third most frequently isolated enteric pathogen from patients with diarrhea. Vomiting and abdominal pain were recorded for 33.33 and 66.67% of the patients, respectively. Stools with blood were observed for 9.52% of the patients. Twenty-four of 42 (57%) patients experienced a temperature over 37.4°C. These observations indicate that HPI-harboring E. coli is one of the major causes of diarrheal disease in China and that the clinical symptoms caused by HPI-harboring E. coli differ from those caused by enteroadhesive E. coli.
Yersinia pestis, Y. pseudotuberculosis serotype O1 to O3, and Y. enterocolitica biotype 1B strains possess a chromosomal determinant that has recently been designated the high-pathogenicity island (HPI) (3, 6, 7). In Y. pestis, the HPI comprises about 35 kb of chromosomal DNA linked to a 68-kb independent mobile pigmentation segment and includes the genes involved in iron storage and uptake, such as the irp1 and irp2 genes, which code for iron-repressible high-molecular-weight proteins HMWP1 and HMWP2, respectively, and the fyuA or psn gene (which code for ferric yersiniabactin uptake or pesticin sensitivity, respectively) (6, 8, 9, 10). The pathogenic strains of Y. enterocolitica biotype 1B carry a 45-kb stretch of chromosomal DNA comprising the irp1-irp2 and fyuA genes without a pigmentation segment, which was shown to be important for Y. pestis to block the flea proventriculus (16, 24, 25).
Diarrheagenic Escherichia coli strains have been classified into several categories, such as enterotoxinogenic E. coli (ETEC), enteroinvasive E. coli (EIEC), enteropathogenic E. coli (EPEC), enteroadhesive E. coli (EAEC), and enterohemorrhagic E. coli (EHEC). The different categories can be identified on the basis of virulence factors (20). Schubert et al. (26) reported that 93% of 60 EAEC strains and 80% of E. coli strains isolated from blood samples hybridized with an irp2 gene probe. The HPI was infrequently detected in strains of EPEC, ETEC, and EIEC. The gene for the HPI island has been proved to be absent from Shigella strains and Salmonella enterica serovars Enteritidis and Typhimurium (26). Recently, Karch et al. (18) reported that the HPI is present in 56 (27.2%) of 206 Shiga toxin-producing E. coli strains but is absent from all of 37 E. coli O157:H7 and E. coli O157:H− strains tested. Bach et al. (1) observed that, in addition to E. coli, the HPI is present in some species of Citrobacter and Klebsiella. In this study, we isolated HPI-harboring E. coli strains from patients with diarrhea and correlated the presence of HPI-harboring E. coli with clinical symptoms.
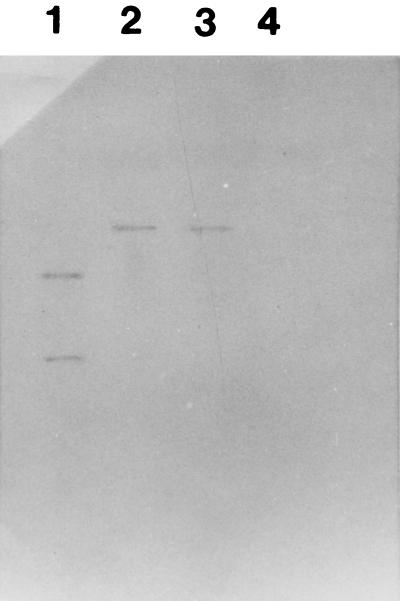
A pilot study was carried out with 43 E. coli strains isolated from patients with diarrhea in Beijing, China, in 1987, 1988, and 1989. These strains did not belong to any recognized category of diarrheagenic E. coli strains, such as EPEC, EHEC, EIEC, ETEC, or EAEC, as demonstrated by DNA probe hybridization (28). PCR analysis showed that 34.9% (15 of 43) of these strains yielded irp1, irp2, and fyuA fragments identical in size to those of Y. enterocolitica. All the positive strains were further confirmed by colony blot hybridization with irp1, irp2, and fyuA genes amplified from Y. enterocolitica as probes. Two E. coli strains harboring irp1, irp2, and fyuA genes were subjected to Southern hybridization. The purified chromosomal DNAs from the two strains were digested with EcoRI and hybridized with irp1, irp2, and fyuA DNA probes (Fig. 1). For the fyuA gene, the Southern hybridization patterns of the two E. coli strains were identical to that of Y. enterocolitica. For the irp1 and irp2 genes, the molecular sizes of the hybridized DNA fragments were different from that of Y. enterocolitica. It seems that an EcoRI restriction site of the irp1 genes was absent from E. coli strain (26).
FIG. 1.
Southern hybridization profile of the EcoRI-digested chromosomal DNA obtained with an irp1-specific probe. Lanes: 1, Y. enterolitica O8 WA (positive control); 2, E. coli F77 (irp1, irp2, and fyuA positive); 3, E. coli E1835 (irp1, irp2, and fyuA positive); 4, E. coli HB101 (negative control).
For the PCR analysis, 30 cycles of denaturation (94°C, 1 min), extension, and annealing (at an annealing temperature [Tm] of 1 min) with one final extension step (72°C, 8 min) were performed. The sequences of the primers used for the PCRs (and the size of the amplified fragment [S], Tm, and the extension time [E] at 72°C) were as follows: (i) irp1 forward primer (HPI 1), 5′-GGCGTCTCCTCCTTTGGTATT-3′; irp1 reverse primer (HPI 2), 5′-GTGATTCCCGCTGTTGATGTT-3′ (S, 1,729 bp; Tm, 60°C; E, 2 min); (ii) irp2 forward primer (HPI 3), 5′-GCGACGGGAAGCGATGAC-3′; irp2 reverse primer (HPI 4), 5′-CGCAGTAGGCACGATGTTGTA-3′ (S, 287 bp; Tm, 62°C; E, 1 min); (iii) fyuA forward primer (HP5), 5′-GCGACGGGAAGCGATTTA-3′; fyuA reverse primer (HP6), 5′-CGCAGTAGGCACGATGTTGTA-3′ (S, 774 bp; Tm, 62°C; E, 1 min) (26). The primers were designed according to the published HPI DNA sequence (8, 9, 26). Strains of Y. enterocolitica WA (O8) and enteroaggregative E. coli O42 (EAggEC) were used as positive controls for the detection of HPI genes (2). For Southern blots, the restriction enzyme-digested genomic DNA fragments and PCR products were resolved through 0.8% agarose gels. The DNA was transferred from the gel to Zeta-Probe BT blotting membranes (Bio-Rad Laboratories, Richmond, Calif.). After prehybridization at 68°C for 2 h and addition of heat-denatured probe, the blots were incubated overnight at 68°C. Digoxigenin labeling of the probes and hybridization were performed with a DNA labeling and detection kit (Boehringer, Mannheim, Germany) according to the manufacturer's instructions.
In order to reveal the frequency of isolation of HPI-harboring E. coli strains and the relationship of the HPI-harboring E. coli from patients with diarrhea and the associated clinical symptoms, the fecal samples from patients with diarrhea visiting the outpatient units of four appointed hospitals in Shandong Province were routinely collected and cultured for enteric bacterial pathogens in 1998; the patients had not received antibiotic therapy. The enteric bacterial pathogens for which screening was performed included Vibrio cholerae, Vibrio parahaemolyticus, S. enterica serovars Typhi and Typhimurium, Shigella flexneri 2a, Shigella sonnei, Aeromonas species, Pseudomonas aeruginosa, Plesiomonas shigelloides, EPEC, EIEC, and ETEC. When the patients visited the outpatient units, the clinical symptoms were examined and recorded on diarrhea disease investigation sheets by trained doctors. All of the data for patients with culture-confirmed cases of diarrhea caused by the organisms mentioned above were then collected and analyzed.
Among a collection of 671 fecal samples, 448 yielded recognized enteric pathogens, with Shigella species being the most frequently isolated pathogen, while 176 yielded pure but not recognized diarrheagenic E. coli strains in diagnostic antiserum kits. However, 42 strains of E. coli hybridized with DNA probes specific for the irp2 gene, of which one strain also hybridized with the ipaB probe specific for EIEC. None of them hybridized with an EAggEC-specific DNA probe (2). By PCR analysis, the irp2 and fyuA genes of HPI were confirmed to be present in these strains. The HPI has recently been reported to be present in as many as 90% of the EAEC strains tested (26). HPI-harboring E. coli has now identified in diarrheal patients of Shandong Province, China. The rate of isolation (6.26%) of HPI-harboring E. coli appears to be higher than those for ETEC, EIEC, or EPEC in this study. Therefore, it was the third most frequently isolated enteric bacterial pathogens studied, after V. parahaemolyticus and Shigella species (Table 1). It should be mentioned that the 42 isolates of HPI-harboring E. coli could not be considered EAggEC since they did not hybridize with the EAggEC-specific DNA probe (27). By HEp-2 cell adherence assay, many strains can be identified as EAggEC. However, some E. coli strains could adhere to HEp-2 cell in an aggregative pattern, but do not hybridize to the EAggEC-specific probe. Moreover, not all EAggEC isolates harbor the HPI (2, 20).
TABLE 1.
Bacterial pathogens from patients with diarrhea in Shandong Province
| Bacterial species | No. of strains isolated | Isolation rate (%) |
|---|---|---|
| Shigella flexneri 2a | 161 | 23.99 |
| Shigella sonnei | 10 | 1.49 |
| Vibrio parahaemolyticus | 58 | 11.24 |
| ETEC | 26 | 3.87 |
| EPEC | 14 | 2.09 |
| EIEC | 19 | 2.83 |
| HPI-harboring Escherichia coli | 42 | 6.26 |
| Atypical Escherichia coli | 134 | 19.99 |
| Salmonella enterica serovar Typhimurium | 15 | 2.29 |
| Yersinia enterocolitica | 9 | 1.34 |
| Pseudomonas aeruginosa | 9 | 1.34 |
| Citrobacter freundii | 22 | 3.28 |
| Proteus | 22 | 3.28 |
| Klebsiella | 11 | 1.64 |
| Enterobacter cloacae | 13 | 1.94 |
| Enterobacter aerogenes | 1 | 0.25 |
| Enterobacter agglomerans | 2 | 0.30 |
| Morganella morganii | 4 | 0.60 |
| Serratia liquefaciens | 3 | 0.45 |
| Providencia | 6 | 0.89 |
| Hafnia spp. | 1 | 0.15 |
| Total (exclusion of atypical E. coli) | 448 | 66.8 |
Nataro and Kaper (20) referred to EAEC as EAggEC. EAggEC does not secrete an enterotoxin such as the heat-labile or heat-stable toxin and has the ability to adhere to HEp-2 cells in an aggregative pattern (20). Baudry et al. (2) had developed a 1.0-kb plasmid-derived fragment as an EAggEC-specific diagnostic probe. In a prospective study of 513 Venezuelan infants with diarrhea and 241 age-matched controls, EAEC strains were found in 26.9% of diarrheal patients and 15% of control (11, 14). Several studies have suggested the association of EAEC with diarrhea in pediatric patients, especially those with persistent diarrhea (4, 5, 17, 20). With the data from outbreaks, sporadic cases, and the volunteer study, it has been suggested that EAEC causes a watery, mucoid, secretory diarrheal illness with low-grade fever and little to no vomiting (20). In volunteers infected with EAEC, the stools were generally mucoid and low volume without occult blood or fecal leukocytes. It appears that EAEC infection may be accompanied by a subtle form of mucosal inflammation (20).
In our study, HPI-harboring E. coli has been isolated from all age groups. It was isolated from 6.70% (14 of 209) of patients with diarrhea under age 10 years. The isolation rates for other age groups are as follows: 3.60% (4 of 111) for those ages 10 to 19 years, 4.43% (7 of 158) for those ages 20 to 29 years, 12.35% (10 of 81) for those ages 30 to 39 years, 3.85% (2 of 52) for those ages 40 to 49 years, 10.81% (4 of 37) for those ages 50 to 59 years, and 4.35% (1 of 23) for those >60 years of age. It seems that HPI-harboring E. coli can cause diarrhea in all age groups and is most frequently detected in association with diarrhea in children under age 10 years (14 of 44 bacteriologically confirmed cases). Among the patients with HPI-harboring E. coli-related diarrhea, vomiting was observed in 33.33% of the patients; a normal temperature, low-grade fever, and high-grade fever were observed in 40.48, 28.57, and 28.57% of the patients, respectively. A total of 66.67% of the patients experienced abdominal pain; mucoid, watery, and liquid green stools were observed in 54.76, 19.05, and 11.90% of the patients, respectively. Stools with blood were observed in 9.52% of the patients. In the patients with diarrhea related to atypical E. coli isolates, 35.1 and 41.8% of the patients had watery and mucoid feces, respectively; 89.1% of the patients experienced a low, medium, or high temperature (Table 2). Only 23.9% (13 of 134) of the patients had a normal body temperature. These observations suggested that the diarrhea related to HPI-harboring E. coli is a watery, mucoid illness (31 of 42 patients [73.81%]) with no fever or a low-grade fever (30 of 42 patients [71.43%]). A total of 78.57% (33 of 42) patients had diarrhea more than six times a day (Table 2). The remarkable differences observed between the clinical symptoms related to HPI-harboring E. coli and those related to atypical E. coli are stool forms and body temperature.
TABLE 2.
Clinical symptoms of 42 patients with diarrhea related to HPI-harboring E. coli and 134 patients with diarrhea related to atypical E. coli
| Symptoms | No. (%) of patients with the symptom
|
|
|---|---|---|
| HPI-harboring E. coli | Other atypical E. coli | |
| Nausea | 14 (33.33) | 44 (32.8) |
| Vomiting | 11 (26.19) | 40 (29.9) |
| Diarrhea the following no. of times/day: | ||
| 3–5 | 9 (21.43) | 26 (19.4) |
| 6–8 | 16 (38.10) | 36 (26.9) |
| 9–10 | 12 (28.57) | 48 (35.8) |
| >10 | 5 (11.90) | 24 (17.9) |
| Form of feces | ||
| Mucous | 23 (54.76) | 47 (35.1) |
| Watery | 8 (19.05) | 56 (41.8) |
| With blood | 4 (9.52) | 18 (13.4) |
| Unformed | 7 (16.67) | 12 (9.0) |
| Inappetence | 26 (61.90) | 54 (40.3) |
| Abdominal pain | 28 (66.67) | 40 (29.9) |
| Vapidity | 21 (50) | 41 (30.6) |
| Headache | 10 (23.8) | 23 (17.2) |
| Abdominal movement | 8 (19.05) | 14 (10.4) |
| Temp | ||
| Normal | 18 (42.86) | 32 (23.9) |
| 37.4–37.9°C | 12 (28.57) | 45 (33.6) |
| 38–38.9°C | 8 (19.05) | 43 (32.1) |
| >39°C | 4 (9.52) | 14 (10.4) |
The gene products of the fyuA-irp gene cluster of the HPI island may benefit E. coli pathotypes (22, 25). The yersiniabactin has a possible cytotoxic effect on T cells (12). Pyochelin, the yersiniabactin-like siderophore of P. aeruginosa, could promote damage to endothelial cells by formation of free radicals (21, 26). The irp2 gene product, HMWP2, has extensive similarity to a superfamily of adenylate-forming enzymes involved in the nonribosomal peptide synthesis of not only siderophores but also peptide antibiotics (19, 23, 24, 26). By analogy with yersiniae, it has been suggested that the fyuA-irp gene cluster may contribute to the virulence of certain pathogenic E. coli strains, such as EAEC and septicemia-causing E. coli strains (26). Since the reason for the Yersinia HPI among EAEC strains is unknown, more studies are needed to clarify its clinical importance and the pathogenic role that it plays.
Acknowledgments
We are grateful to James B. Kaper for kindly providing E. coli strain O42 and the EAggEC-specific DNA probe. We appreciate Jun Yu for critical reading of the manuscript.
This work was supported by the Basic Research Program (grant G1999054101 to J.X.) from the Ministry of Science and Technology and by an Outstanding Young Scientist Award (award 39625001 to J.X.) from the Chinese National Natural Sciences Foundation, Beijing, China.
REFERENCES
- 1.Bach S, de Almeida A, Carniel E. The Yersinia high-pathogenicity island is present in different members of the family Enterobacteriaceae. FEMS Microbiol Lett. 2000;183:289–294. doi: 10.1111/j.1574-6968.2000.tb08973.x. [DOI] [PubMed] [Google Scholar]
- 2.Baudry B, Savarino S J, Vial P, Kaper J B, Levine M M. A sensitive and specific DNA probe to identify enteroaggregative Escherichia coli, a recently discovered diarrheal pathogen, J. Infect Dis. 1990;161:1249–1251. doi: 10.1093/infdis/161.6.1249. [DOI] [PubMed] [Google Scholar]
- 3.Bearden S W, Fetherston J D, Perry R D. Genetic organization of the yersiniabactin biosynthetic region and construction of avirulent mutants in Yersinia pestis. Infect Immun. 1997;65:1659–1668. doi: 10.1128/iai.65.5.1659-1668.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bhan M K, Khoshoo V, Sommerfelt H, Raj P, Sazawal S, Srivastava R. Enteroaggregative Escherichia coli and Salmonella associated with nondysenteric persistent diarrhea. Pediatr Infect Dis J. 1989;8:499–502. doi: 10.1097/00006454-198908000-00005. [DOI] [PubMed] [Google Scholar]
- 5.Bhan M K, Raj P, Levine M M, Kaper J B, Bhandari N, Srivastava R, Kumar R, Sazawal S. Enteroaggregative Escherichia coli associated with persistent diarrhea in a cohort of rural children in India. J Infect Dis. 1989;159:1061–1064. doi: 10.1093/infdis/159.6.1061. [DOI] [PubMed] [Google Scholar]
- 6.Buchrieser C, Prentice M, Carniel E. The 102-kilobase unstable region of Yersinia pestis comprises a high-pathogenicity island linked to a pigmentation segment which undergoes internal rearrangement. J Bacteriol. 1998;180:2321–2329. doi: 10.1128/jb.180.9.2321-2329.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buchrieser C, Brosch R, Bach S, Guiyoule A, Carniel E. The high-pathogenicity island of Yersinia pseudotuberculosis can be inserted into any of the three chromosomal and tRNA genes. Mol Microbiol. 1998;30:965–978. doi: 10.1046/j.1365-2958.1998.01124.x. [DOI] [PubMed] [Google Scholar]
- 8.Carniel E, Mercereau-Puijalon O, Bonnefoy S. The gene coding for the 190,000-dalton iron-regulated protein of Yersinia species is present only in the highly pathogenic strains. Infect Immun. 1989;57:1211–1217. doi: 10.1128/iai.57.4.1211-1217.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carniel E, Guiyoule A, Guilvout L, Mercereau-Puijalon O. Molecular cloning, iron-regulation and mutagenesis of the irp2 gene encoding HMWP2, a protein specific for the highly pathogenic Yersinia. Mol Microbiol. 1992;6:379–388. doi: 10.1111/j.1365-2958.1992.tb01481.x. [DOI] [PubMed] [Google Scholar]
- 10.Carniel E, Guilvout I, Prentice M. Characterization of a large chromosomal “high-pathogenicity island” in biotype 1B Yersinia enterocolitica. J Bacteriol. 1996;178:6743–6751. doi: 10.1128/jb.178.23.6743-6751.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cravioto A, Tello A, Navarro A, Ruiz J, Villafan H, Uribe F, Eslava C. Association of Escherichia coli HEp-2 adherence patterns with type and duration of diarrhoea. Lancet. 1991;337:262–264. doi: 10.1016/0140-6736(91)90868-p. [DOI] [PubMed] [Google Scholar]
- 12.de Almeida A M, Guiyoule A, Guilvout I, Iteman I, Baranton G, Carniel E. Chromosomal irp2 gene in Yersinia: distribution, expression, deletion and impact on virulence. Microb Pathog. 1993;14:9–21. doi: 10.1006/mpat.1993.1002. [DOI] [PubMed] [Google Scholar]
- 13.Fetherston J D, Schuetze P, Perry R D. Loss of the pigmentation phenotype in Yersinia pestis is due to the spontaneous deletion of 102 kb of chromosomal DNA which is flanked by a repetitive element. Mol Microbiol. 1992;6:2693–2704. doi: 10.1111/j.1365-2958.1992.tb01446.x. [DOI] [PubMed] [Google Scholar]
- 14.Fetherston J D, Perry R D. The pigmentation locus of Yersinia pestis KIM6+ is flanked by an insertion sequence and includes the structural genes for pesticin sensitivity and HMWP2. Mol Microbiol. 1994;13:697–708. doi: 10.1111/j.1365-2958.1994.tb00463.x. [DOI] [PubMed] [Google Scholar]
- 15.Hare J M, Wagner A K, McDonough K A. Independent acquisition and insertion into different chromosomal locations of the same pathogenicity island in Yersinia pestis and Yersinia pseudotuberculosis. Mol Microbiol. 1999;31:291–303. doi: 10.1046/j.1365-2958.1999.01172.x. [DOI] [PubMed] [Google Scholar]
- 16.Hinnebusch B J, Perry R D, Schwan T G. Role of the Yersinia pestis hemin storage (hms) locus in the transmission of plague by fleas. Science. 1996;273:367–370. doi: 10.1126/science.273.5273.367. [DOI] [PubMed] [Google Scholar]
- 17.Huppertz H, Rutkowski S, Aleksic S, Karch H. Acute and chronic diarrhoea and abdominal colic associated with enteroaggregative Escherichia coli in young children living in western Europe. Lancet. 1997;349:1660–1662. doi: 10.1016/S0140-6736(96)12485-5. [DOI] [PubMed] [Google Scholar]
- 18.Karch H, Schubert S, Zhang D, Zhang W, Schmidt H, Olschlager T, Hacker J. A genomic island, termed high-pathogenicity island, is present in certain non-O157 Shiga toxin-producing Escherichia coli clonal lineages. Infect Immun. 1999;67:5994–6001. doi: 10.1128/iai.67.11.5994-6001.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lucier T S, Fetherston J D, Brubaker R R, Perry R D. Iron uptake and iron-repressible polypeptides in Yersinia pestis. Infect Immun. 1996;64:3023–3031. doi: 10.1128/iai.64.8.3023-3031.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nataro J P, Kaper J B. Diarrheagenic Escherichia coli. Clin Microbiol Rev. 1998;11:142–201. doi: 10.1128/cmr.11.1.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pendrak M L, Perry R D. Proteins essential for expression of the Hms+ phenotype of Yersinia pestis. Mol Microbiol. 1993;8:857–864. doi: 10.1111/j.1365-2958.1993.tb01632.x. [DOI] [PubMed] [Google Scholar]
- 22.Perry R D, Lucier T S, Sikkema D J, Brubaker R R. Storage reservoirs of hemin and inorganic iron in Yersinia pestis. Infect Immun. 1993;61:32–39. doi: 10.1128/iai.61.1.32-39.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rakin A, Heesemann J. Virulence-associated fyuA/irp2 gene cluster of Yersinia enterocolitica biotype 1B carries a novel insertion sequence IS1328. FEMS Microbiol Lett. 1995;129:287–292. doi: 10.1111/j.1574-6968.1995.tb07594.x. [DOI] [PubMed] [Google Scholar]
- 24.Rakin A, Saken E, Harmsen D, Heesemann J. The pesticin receptor of Yersinia enterocolitica: a novel virulence factor with dual function. Mol Microbiol. 1994;13:253–263. doi: 10.1111/j.1365-2958.1994.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 25.Rakin A, Urbitsch P, Heesemann J. Evidence for two evolutionary lineages of highly pathogenic Yersinia species. J Bacteriol. 1995;177:2292–2298. doi: 10.1128/jb.177.9.2292-2298.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schubert S, Rakin A, Karch H, Carniel E, Heesemann J. Prevalence of the “high-pathogenicity island” of Yersinia species among Escherichia coli strains that are pathogenic to humans. Infect Immun. 1998;66:480–485. doi: 10.1128/iai.66.2.480-485.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vial P A, Robins Browne R, Lior H, Prado V, Kaper J B, Nataro J P, Maneval D, Elsayed A, Levine M M. Characterization of enteroadherent-aggregative Escherichia coli, a putative agent of diarrheal disease. J Infect Dis. 1988;158:70–79. doi: 10.1093/infdis/158.1.70. [DOI] [PubMed] [Google Scholar]
- 28.Xu J-G, Cheng B Q, Wu Y P, Huang L B, Lai X H, Liu B Y, Lo X Z, Li H F. Adherence patterns and DNA probe types of Escherichia coli isolated from diarrheal patients in China. Microbiol Immunol. 1996;40:89–97. doi: 10.1111/j.1348-0421.1996.tb03318.x. [DOI] [PubMed] [Google Scholar]