Abstract
Four Salmonella enterica serovar Virchow strains resistant to broad-spectrum cephalosporins were isolated from patients with gastroenteritis in 1997 and 1998 in Murcia and Barcelona, Spain. The isolates expressed a β-lactamase with a pI of about 8 and a positive PCR when specific primers for CTX-M-9 were used. These results suggest the presence of a CTX-M-9 β-lactamase in these strains.
Extended-spectrum β-lactamases (ESBLs) were first described in Klebsiella pneumoniae about 20 years ago (13) and now have been isolated in several species throughout the world. However, in Salmonella ESBLs have been detected only recently, mainly in the serovar Typhimurium.
Most ESBLs are derivatives of TEM-1, TEM-2, or SHV-1 enzymes, but there is an increasing number of reports of β-lactamases belonging to other families, like OXA or CTX-M, although their frequencies are still low.
This paper describes four strains of ESBL-producing Salmonella enterica serovar Virchow isolated in our laboratories in Murcia and Barcelona in 1997 and 1998. The β-lactamase belongs to the CTX-M family.
The four strains were isolated from stool samples of four patients with gastroenteritis. Three of these patients were seen at the Hospital Virgen de Arrixaca in Murcia, while the fourth was assisted at the Hospital de la Santa Creu i Sant Pau in Barcelona. Two of them were children 1 and 3 years old. One of these children required hospitalization. The other two patients were adults and were treated in the community. None of the four patients were apparently related; they lived in different towns and were seen on widely separate dates.
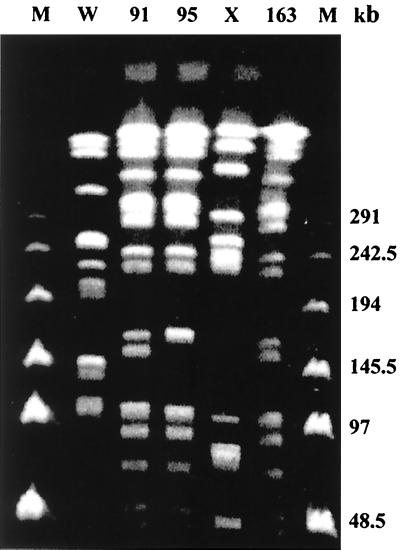
The isolates were identified biochemically by the API 20 identification system (bioMérieux S.A., Marcy-l'Etoile, France). The serogroup was determined in our laboratories and the serotype and phage type were determined in the Servicio de Enterobacterias del Centro Nacional de Microbiología, Instituto Carlos III, Majadahonda, Madrid, Spain. All four strains were identified as S. enterica serovar Virchow phage type 19. The epidemiological relationship of the three isolates from Murcia (isolates 91, 95, and 163) was investigated by pulsed-field gel electrophoresis (20) using the enzyme XbaI. The restriction patterns were identical except for one band of difference in one strain (Fig. 1). Although it is possible that a single strain had been responsible for all the cases, we do not know how discriminatory this technique is for S. enterica serovar Virchow.
FIG. 1.
Restriction pattern by pulsed-field electrophoresis of S. enterica serovar Virchow strains 91, 95, and 163. W, E. coli; X, S. enterica serovar Enteritidis; M, marker.
Susceptibility was determined by disk diffusion following the National Committee for Clinical Laboratory Standards recommendations (16). MICs were determined by E-test (AB Biodisk, Solna, Sweden) according to the manufacturer's recommendations. Escherichia coli ATCC 25922 and E. coli ATCC 35218 were used as control strains for susceptibility studies.
The four isolates were resistant to ampicillin, trimethoprim-sulfamethoxazole, tetracycline, and streptomycin, and two isolates were resistant to kanamycin. All four strains were susceptible to gentamicin, tobramycin, quinolones, and chloramphenicol. The presence of an extended-spectrum β-lactamase was suggested by the synergies detected between clavulanic acid and cefotaxime, ceftazidime, or aztreonam in a disk-diffusion assay with a typical deformation of the inhibition zone, as well as by a decreased susceptibility to third-generation cephalosporins. The MICs of the β-lactams are shown in Table 1.
TABLE 1.
MICs of β-lactams for S. enterica serovar Virchow strains 91, 95, 163, and 144Ma and the respective transconjugants (MSP498, MSP499, MSP500, and MSP501)
| β-lactam agent | MIC (μg/ml) required
|
|||||||
|---|---|---|---|---|---|---|---|---|
| 91 | MSP498 | 95 | MSP499 | 163 | MSP500 | 144Ma | MSP501 | |
| Amoxicillin-clavulanic acid (2:1) | 4 | 1.5 | 4 | 2 | 4 | 2 | 4 | 2 |
| Cefuroxime | >256 | 48 | >256 | >256 | >256 | 48 | >256 | 128 |
| Cefotaxime | 4 | 1 | 8 | 4 | 4 | 1 | 6 | 1.5 |
| Cefotaxime-clavulanic acid (2:1) | 0.023 | ≤0.016 | 0.023 | ≤0.016 | 0.032 | ≤0.016 | 0.023 | ≤0.016 |
| Ceftazidime | 1 | 0.094 | 1 | 0.19 | 1 | 0.064 | 0.75 | 0.125 |
| Ceftazidime-clavulanic acid (2:1) | ≤0.064 | ≤0.064 | ≤0.064 | ≤0.064 | ≤0.064 | ≤0.064 | ≤0.064 | ≤0.064 |
| Cefepime | 2 | 0.094 | 2 | 0.5 | 1 | 0.094 | 1 | 0.19 |
| Aztreonam | 0.5 | 0.032 | 1 | 0.5 | 0.5 | 0.047 | 1 | 0.125 |
The β-lactamase extracts and isoelectric focusing were performed as described by Barthélémy et al. (1), with a pH gradient from 4 to 11 (Servalyt 4-9 T, 9-11 T; Serva, Heidelberg, Germany). The enzyme activities were revealed by the iodometric method. The acrylamide gel was overlaid with agar gel containing iodine and ceftriaxone in order to detect activity against ceftriaxone. Afterward, a gel containing iodine with penicillin G was applied to detect those β-lactamases without ceftriaxone activity. The four isolates produced two β-lactamases, one with an isoelectric point (pI) of 5.4 and the other with a pI of about 8. The latter β-lactamase showed activity against ceftriaxone, whereas the pI-5.4 β-lactamase did not reveal any activity. From the total DNA obtained directly from a colony of each of these four strains, a positive PCR was obtained when a pair of specific primers for TEM-type β-lactamases were used as previously described (19). These results suggest that the pI-5.4 β-lactamase is consistent with TEM-1.
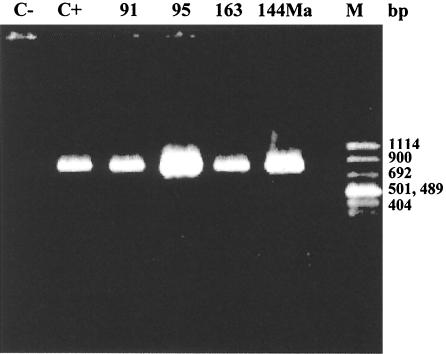
The fact that the strains were more resistant to cefotaxime than to ceftazidime, the presence of a β-lactamase with a pI of about 8, and negative PCR results when specific primers for SHV-type β-lactamases were used (17) suggested the existence of a β-lactamase belonging to the recently reported CTX-M type and more probably the CTX-M-9 type (18). To confirm this possibility, a PCR assay was performed using the primers 5′-GTG ACA AAG AGA GTG CAA CGG-3′ and 5′-ATG ATT CTC GCC GCT GAA GCC-3′, which comprise the positions 4 to 24 and 860 to 840, respectively, with respect to the CTX-M-9 translational starting point (GenBank accession number AF174129). PCR was performed with the four isolates, and an 856-bp fragment was obtained (Fig. 2).
FIG. 2.
PCR products of the four S. enterica serovar Virchow strains (91, 95, 163, and 144Ma) by using CTX-M-9 primers (C− and C+). M, marker.
The transconjugants MSP498, MSP499, MSP500, and MSP501 were obtained for the four isolates, 91, 95, 163, and 144Ma, respectively, presenting a similar resistance pattern (Table 1) and expressing β-lactamases with pIs of 5.4 and about 8, similar to the donors.
CTX-M-type β-lactamase-producing strains have been identified incidentally in different geographical areas: CTX-M-1 in Germany in 1989 (4); MEN-1 in France from an Italian patient in 1989 (2, 6); CTX-M-2 in Argentina in 1990 (3, 5); CTX-M-3 in Poland in 1996 (11); CTX-M-4 in Russia in 1996 (8, 10); CTX-M-5 in Latvia in 1991 (7); CTX-M-5 in Greece in 1996 (9, 20); CTX-M-6 in Greece in 1997 (9, 20); Toho-1 (1993) and Toho-2 (1995) in Japan (12, 14); CTX-M-7 in Brazil (GenBank accession number AF189721); and CTX-M-9 in Barcelona (18). In addition to these initial findings, the CTX-M-type β-lactamases have been described in several species of the Enterobacteriaceae family in widely differing geographical areas, with all of the β-lactamases being plasmid encoded. The divergence of the amino acid sequences (between 70 and 98% sequence homology), as well as the temporal and geographical dispersion of strains carrying this β-lactamase, makes any assumption about the origin of this plasmid-mediated β-lactamase difficult.
Unlike the other CTX-M-type β-lactamases described to date in Salmonella, including CTX-M-2 (3, 5), CTX-M-4 (8, 10), CTX-M-5 (7), CTX-M-5 (9, 21), and CTX-M-6 (9, 21), which all present high resistances to cefotaxime, our isolates require much more moderate MICs (between 4 and 8 μg/ml).
The first and, to date, the only report of an extended-spectrum β-lactamase of Salmonella in Spain was that described by Morosini et al. (15). It was the TEM-27 β-lactamase and was isolated in a nosocomial outbreak produced by S. enterica serovar Othmarschen. Our isolates represent the first description of a β-lactamase of the CTX-M family in salmonellae in Spain and also the first description of an ESBL in S. enterica serovar Virchow.
The establishment and spread of S. enterica serovar Virchow organisms resistant to therapeutically important broad-spectrum β-lactams are cause for concern.
Acknowledgments
We thank Guillermo Prats for critical reading of the manuscript and M. A. Usera, A. Echeita, and A. Aladueña from Servicio de Enterobacterias del Centro Nacional de Microbiología, Instituto Carlos III, Majadahonda, Madrid, Spain, for determination of the serotype and phage type of Salmonella. We are indebted to Graham C. Arnold for his assistance in the preparation of the manuscript.
This work was supported by grant 98/1293 from the Fondo de Investigaciones Sanitarias de la Seguridad Social de España.
REFERENCES
- 1.Barthélémy M, Guionie M, Labia R. Beta-lactamases: determination of their isoelectric points. Antimicrob Agents Chemother. 1978;13:695–698. doi: 10.1128/aac.13.4.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barthélémy M, Peduzzi J, Bernard H, Tancrède C, Labia R. Close amino acid sequence relationship between the new plasmid-mediated extended-spectrum β-lactamase MEN-1 and chromosomally encoded enzymes of Klebsiella oxytoca. Biochim Biophys Acta. 1992;1122:15–22. doi: 10.1016/0167-4838(92)90121-s. [DOI] [PubMed] [Google Scholar]
- 3.Bauernfeind A, Casellas J M, Goldberg M, Holley M, Jungwirth R, Mangold P, Röhnisch T, Schweighart S, Wilhelm R. A new plasmidic cefotaximase from patients infected with Salmonella typhimurium. Infection. 1992;20:158–163. doi: 10.1007/BF01704610. [DOI] [PubMed] [Google Scholar]
- 4.Bauernfeind A, Grimm H, Schweighart S. A new plasmidic cefotaximase in a clinical isolate of Escherichia coli. Infection. 1990;18:294–298. doi: 10.1007/BF01647010. [DOI] [PubMed] [Google Scholar]
- 5.Bauernfeind A, Stemplinger I, Jungwirth R, Ernst S, Casellas J M. Sequence of β-lactamase genes encoding CTX-M-1 (MEN-1) and CTX-M-2 and relationship of their amino acid sequences with those of other β-lactamases. Antimicrob Agents Chemother. 1996;40:509–513. doi: 10.1128/aac.40.2.509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bernard H, Tancrede C, Livrelli V, Morand A, Barthélémy M, Labia R. A novel plasmid-mediated extended-spectrum β-lactamase not derived from TEM- or SHV-type enzymes. J Antimicrob Chemother. 1992;29:590–592. doi: 10.1093/jac/29.5.590. [DOI] [PubMed] [Google Scholar]
- 7.Bradford P A, Yang Y, Sahm D, Grope I, Gardovska D, Storch G. CTX-M-5, a novel cefotaxime-hydrolyzing β-lactamase from an outbreak of Salmonella typhimurium in Latvia. Antimicrob Agents Chemother. 1998;42:1980–1984. doi: 10.1128/aac.42.8.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gazouli M, Sidorenko S V, Tzelepi E, Kozlova N S, Gladin D P, Tzouvelekis L S. A plasmid-mediated β-lactamase conferring resistance to cefotaxime in a Salmonella typhimurium clone found in St. Petersburg, Russia. J Antimicrob Chemother. 1998;41:119–121. doi: 10.1093/jac/41.1.119. [DOI] [PubMed] [Google Scholar]
- 9.Gazouli M, Tzelepi E, Markogiannakis A, Legakis N J, Tzouvelekis L S. Two novel plasmid-mediated cefotaxime-hydrolyzing β-lactamases (CTX-M-5 and CTX-M-6) from Salmonella typhimurium. FEMS Microbiol Lett. 1998;165:289–293. doi: 10.1111/j.1574-6968.1998.tb13159.x. [DOI] [PubMed] [Google Scholar]
- 10.Gazouli M, Tzelepi E, Sidorenko S V, Tzouvelekis L S. Sequence of the gene encoding a plasmid-mediated cefotaxime-hydrolyzing class A β-lactamase (CTX-M-4): involvement of serine 237 in cephalosporin hydrolysis. Antimicrob Agents Chemother. 1998;42:1259–1262. doi: 10.1128/aac.42.5.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gniadkowski M, Schneider I, Palucha A, Jungwirth R, Mikiewicz B, Bauernfeind A. Cefotaxime-resistant Enterobacteriaceae isolates from a hospital in Warsaw, Poland: identification of a new CTX-M-3 cefotaxime-hydrolyzing β-lactamase that is closely related to the CTX-M-1/MEN-1 enzyme. Antimicrob Agents Chemother. 1998;42:827–832. doi: 10.1128/aac.42.4.827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ishii Y, Ohno A, Taguchi H, Imajo S, Ishiguro M, Matsuzawa H. Cloning and sequence of the gene encoding a cefotaxime-hydrolyzing class A β-lactamase isolated from Escherichia coli. Antimicrob Agents Chemother. 1995;39:2269–2275. doi: 10.1128/aac.39.10.2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Knothe H, Shah P, Kromery V, Antal M, Mitsuashi S. Transferable resistance to cefotaxime, cefoxitin, cefamandole and cefuroxime in clinical isolates of Klebsiella pneumoniae and Serratia marcescens. Infection. 1983;11:315–317. doi: 10.1007/BF01641355. [DOI] [PubMed] [Google Scholar]
- 14.Ma L, Ishii Y, Ishiguro M, Matsuzawa H, Yamaguchi K. Cloning and sequencing of the gene encoding Toho-2, a class A β-lactamase preferentially inhibited by tazobactam. Antimicrob Agents Chemother. 1998;42:1181–1186. doi: 10.1128/aac.42.5.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morosini M I, Blazquez J, Negri M C, Canton R, Loza E, Baquero F. Characterization of a nosocomial outbreak involving an epidemic plasmid encoding for TEM-27 in Salmonella enterica subspecies enterica serotype Othmarschen. J Infect Dis. 1996;174:1015–1020. doi: 10.1093/infdis/174.5.1015. [DOI] [PubMed] [Google Scholar]
- 16.National Committee for Clinical Laboratory Standards. Performance standards for antimicrobial susceptibility testing, 8th informational suppl. M100–S8. Wayne, Pa: National Committee for Clinical Laboratory Standards; 1998. [Google Scholar]
- 17.Nüesch-Inderbinen M T, Hächler H, Kayser F H. Detection of genes coding for extended-spectrum SHV beta-lactamases in clinical isolates by a molecular genetic method, and comparison with E-test. Eur J Clin Microbiol Infect Dis. 1996;15:398–402. doi: 10.1007/BF01690097. [DOI] [PubMed] [Google Scholar]
- 18.Sabaté M, Tarragó R, Navarro F, Miró E, Vergés C, Barbé J, Prats G. Cloning and sequence of the gene encoding a novel cefotaxime-hydrolyzing β-lactamase (CTX-M-9) from Escherichia coli in Spain. Antimicrob Agents Chemother. 2000;44:1970–1973. doi: 10.1128/aac.44.7.1970-1973.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sabaté M, Vergés C, Miró E, Mirelis B, Navarro F, del Rio E, Prats G. Incidencia de betalactamasas de espectro ampliado en Escherichia coli en un hospital universitario durante 1994–1996. Enferm Infecc Microbiol Clin. 1999;17:401–404. [PubMed] [Google Scholar]
- 20.Smith C L, Klco S R, Cantor C R. Pulsed-field gel electrophoresis and the technology of large DNA molecules. In: Davies K, editor. Genome analysis: a practical approach. Oxford, United Kingdom: IRL Press; 1988. pp. 41–72. [Google Scholar]
- 21.Tzouvelekis L S, Gazouli M, Markogiannakis A, Paraskaki E, Legakis N J, Tzelepi E. Emergence of resistance to third-generation cephalosporins amongst Salmonella typhimurium isolates in Greece: report of the first three cases. J Antimicrob Chemother. 1998;42:273–275. doi: 10.1093/jac/42.2.273. [DOI] [PubMed] [Google Scholar]