Abstract
An oncogene-centric molecular classification paradigm in non-small cell lung cancer (NSCLC) has been established. Of note, the heterogeneity within each oncogenic driver-defined subgroup may be captured by co-occurring mutations, which potentially impact response/resistance to immune checkpoint inhibitors (ICIs). We analyzed the data of 1745 NSCLCs and delineated the landscape of interaction effects of common co-mutations on ICI efficacy. Particularly in nonsquamous NSCLC, KRAS mutation remarkably interacted with its co-occurring mutations in TP53, STK11, PTPRD, RBM10, and ATM. Based on single mutation-based prediction models, adding interaction terms (referred to as inter-model) improved discriminative utilities in both training and validation sets. The scores of inter-models exhibited undifferentiated effectiveness regardless of tumor mutational burden and programmed death-ligand 1, and were identified as independent predictors for ICI benefit. Our work provides novel tools for patient selection and insights into NSCLC immunobiology, and highlights the advantage and necessity of considering interactions when developing prediction algorithms for cancer therapeutics.
Subject terms: Predictive markers, Cancer immunotherapy, Cancer genetics, Non-small-cell lung cancer
Introduction
An oncogene-centric molecular classification paradigm in non-small cell lung cancer (NSCLC) has been established by the discoveries of mutual-exclusive oncogenic drivers, e.g., KRAS, EGFR, BRAF, and ALK1. However, growing evidence points toward the biological and clinical heterogeneity within each oncogenic driver-defined subgroup1–6, which warrants further investigations into the indicators for optimizing the stratification framework in NSCLC.
Co-existing genomic aberrations in oncogenic drivers and tumor suppressor genes have emerged as the main principle of molecular diversity in NSCLC. This co-mutation pattern can capture the complexity concerning tumorigenesis, metastasis, immune microenvironment, and therapeutic vulnerabilities1,7. For instance, KRAS-driven lung adenocarcinomas (LUADs) are intrinsically heterogeneous and can be classified into three subgroups dominated, respectively, by co-mutations in TP53, STK11, and CDKN2A/B8. KRAS/TP53 co-mutations are associated with an inflamed immune microenvironment and increased tumoral programmed death-ligand 1 (PD-L1) expression8–11; However, KRAS/STK11 co-mutated LUADs appear largely “immune-inert”, characterized by a paucity of tumor-infiltrating T cells and lower PD-L1 expression8,10–12. These two different co-occurring mutations in KRAS-mutated LUADs lead to almost opposite microenvironments, demonstrating the critical impact of co-occurring mutations on the immune-related characteristics in NSCLC and the promising opportunities that interactions between co-occurring mutations may foster the improvement of prediction algorithms for cancer immunotherapy.
The predictive impacts of two co-occurring mutations may be interactive, but not simply additive, which requires further assessment of interaction effects13–15. We previously revealed an interaction of KRAS/TP53 mutations in nonsquamous NSCLC, where we observed similarly poor progression-free survival (PFS) in the patients with no or single mutation, but significant PFS advantage in the co-mutant ones16, which indicates that only when these two mutations occur simultaneously, may they predict remarkable benefits from ICIs. In a recently published 8-gene mutational signature, TP53 and KRAS mutations were calculated independently with a score associated with better response to ICIs17. In this case, the single-mutated patients had a higher score compared to the double-wildtype ones, inconsistent with their similarly poor outcomes in the actual situation. Co-mutations are rare and therefore assessing interaction effects requires a large sample size to avoid serious sampling error, which partially accounts for the omission of interaction effects in current mutational signatures for predicting immunotherapy efficacy in NSCLC17,18.
Given these, we hypothesized that co-occurring mutations may shape immune contexture and act as novel predictors for immunotherapy efficacy in NSCLC. In this study, we sought to first delineate the landscape of interaction effects separately in patients with nonsquamous or squamous NSCLC, and then to investigate whether adding significant interaction terms into prediction models could improve their performances in both the training and the validation sets. Our goal was to develop a novel tool involving interaction terms to predict ICI benefit more precisely in NSCLC and to raise the awareness of involving co-occurring genomic alterations for facilitating the refinement of prediction algorithms for cancer therapeutics.
Results
Interaction between mutational events was associated with immunotherapy efficacy
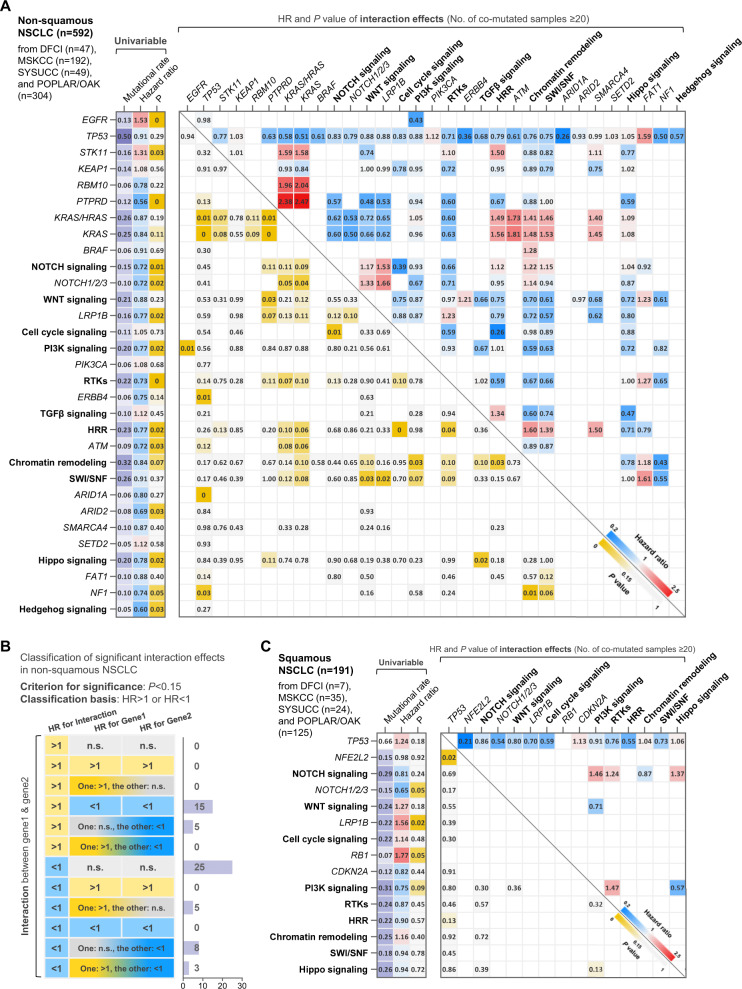
To begin with, we analyzed whether mutational events in single genes or pathways were associated with the PFS on anti-PD-(L)1 monotherapy in nonsquamous NSCLC (n = 592). The detailed information and survival data of included datasets are shown in Supplementary Table 1 and Supplementary Fig. 1, and the definitions of analyzed pathways are shown in Supplementary Table 2. Univariable analyses revealed that poorer PFS on anti-PD-(L)1 monotherapy was associated with the mutations in EGFR (P = 0.001) and STK11 (P = 0.026), and better PFS was associated with the mutations in PTPRD (P < 0.001), NOTCH pathway (P = 0.009), NOTCH1/2/3 (P = 0.025), LRP1B (P = 0.025), PI3K pathway (P = 0.022), receptor tyrosine kinases (RTKs, P = 0.004), SMAD4 (P = 0.046), homologous recombination repair (HRR) pathway (P = 0.016), ATM (P = 0.029), ATR (P = 0.049), ARID2 (P = 0.032), Hippo pathway (P = 0.023), and Hedgehog pathway (P = 0.032, Fig. 1A and Supplementary Table 3).
Fig. 1. Associations of ICI efficacy with single mutations and interactions of co-occurring mutations in NSCLC.
A Landscape of the predictive utilities of single mutations (left) and the interaction effects of common co-occurring mutations (right) in nonsquamous NSCLC. B Classification of interaction effects based on the effect sizes of single mutation terms and interaction terms. C Landscape of the predictive utilities of single mutations (left) and the interaction effects of common co-occurring mutations (right) in squamous NSCLC. Abbreviations: NSCLC nonsmall cell lung cancer.
As for the interaction effects between co-occurring genetic aberrations, the co-mutations existing in more than 20 patients were included in the analysis to reduce the impact of serious sampling error. We found 20 significant interactions with a P value below 0.05, and 41 interactions with a P value between 0.05 and 0.15 (Fig. 1A). Among these 61 interactions, 28 (45.9%) had effect sizes opposite to the trends of the effect sizes of single mutations (e.g., PTPRD*KRAS pathway: hazard ratio [HR] = 2.47, 95% confidence interval (CI) 1.33-4.61, P = 0.004; PTPRD: HR = 0.43, 95% CI 0.30-0.62, P < 0.001; KRAS: HR = 0.76, 95% CI 0.60-0.95, P = 0.018, Fig. 1B and Supplementary Table 4). These results indicate that gene mutations may exhibit different associations with immunotherapy efficacy based on their co-occurring mutations in nonsquamous NSCLC.
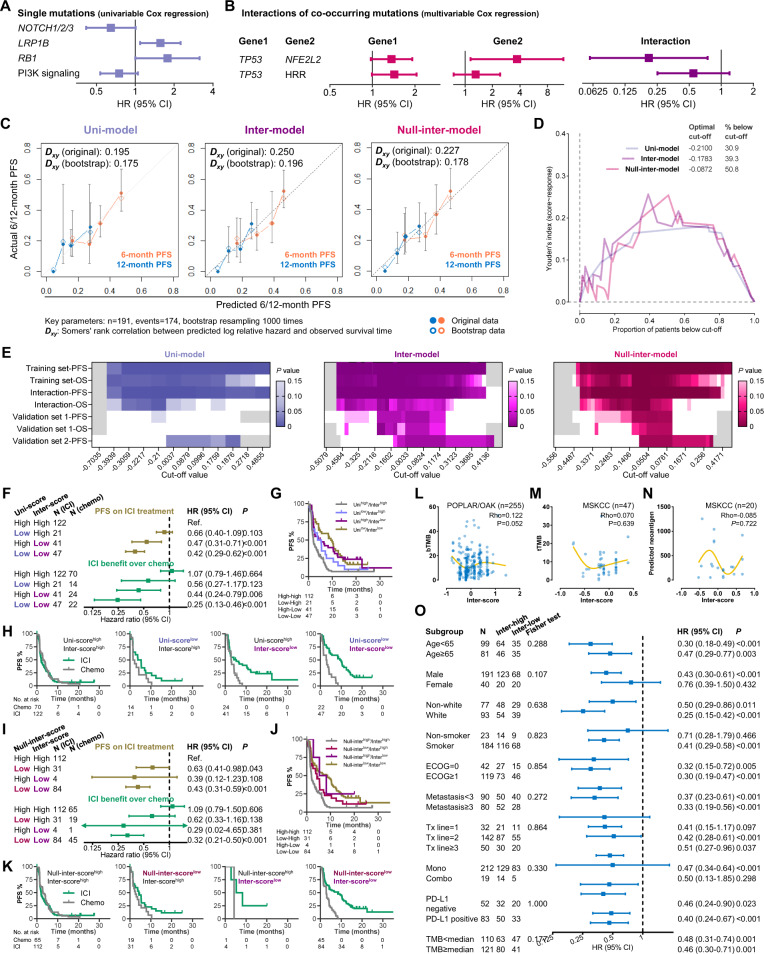
In the patients with squamous NSCLC (n = 191), the relatively small sample size limits the exploration of significant effects to a certain extent, and therefore we relaxed the requirement for p value from 0.05 to 0.15 in the following analysis. In terms of individual gene mutations, the mutations in NOTCH1/2/3 (P = 0.053) and PI3K pathway (P = 0.092) were associated with longer PFS on anti-PD-(L)1 monotherapy and the mutations in LRP1B (P = 0.016) and RB1 (P = 0.051) were associated with worse PFS (Fig. 1C and Supplementary Table 5). For the interaction effects, three remarkable interactions were discovered (TP53*NFE2L2, TP53*HRR pathway, and PI3K pathway*Hippo pathway, Fig. 1C and Supplementary Table 6). Two interactions in squamous NSCLC (TP53*NFE2L2 and TP53*HRR pathway) had effect sizes opposite to the trends of the effect sizes of single mutations (e.g., TP53*NFE2L2: HR = 0.21, 95% CI 0.06–0.74, P = 0.016; TP53: HR = 1.37, 95% CI 0.97–1.92, P = 0.071; NFE2L2: HR = 3.71, 95% CI 1.15–12.00, P = 0.029), highlighting the importance of investigating interaction effects of co-occurring mutations.
Development of a model involving interactions for anti-PD-(L)1 therapy in nonsquamous NSCLC
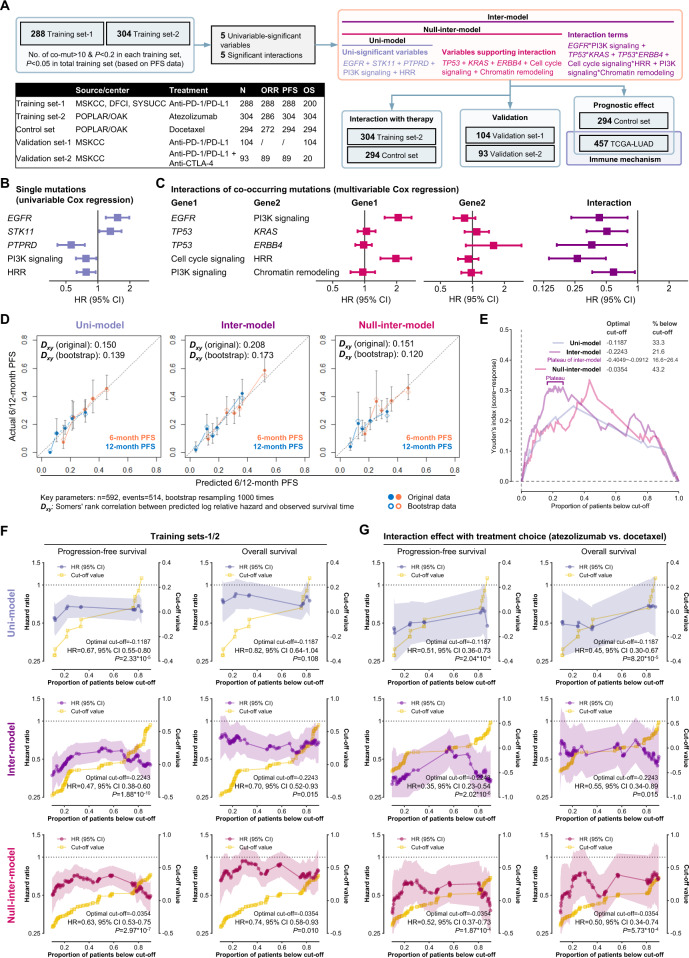
As shown in Fig. 2A, of the total 1083 patients with nonsquamous NSCLC, 288 anti-programmed death-(ligand)1 (anti-PD-(L)1) monotherapy-treated patients with PFS data from Sun Yat-Sen University Cancer Center (SYSUCC)19, Dana Farber Cancer Institute (DFCI)20, and Memorial Sloan-Kettering Cancer Center (MSKCC)21–23 were included in the training set-1, and 304 patients treated with atezolizumab and 294 patients treated with docetaxel from the POPLAR/OAK cohort were included in the training set-2 and the control set, respectively24.
Fig. 2. Performances of the three models in the training sets of nonsquamous NSCLC.
A Workflow of developing and validating three models in nonsquamous NSCLC. B, C The single mutations (B) and interaction effects of co-mutations (C) selected for model development. D Calibration curves of the three models in the training sets-1/2. E Youden’s index based on the receiver operating characteristic curve of the three inter-scores and response to anti-PD-(L)1 monotherapy in the training sets-1/2. F Performances of the three models on discriminating the PFS and OS on anti-PD-(L)1 monotherapy in the training sets-1/2. G Performances of the three models on predicting the PFS and OS benefit from atezolizumab over docetaxel in the POPLAR/OAK cohort. Abbreviations: DFCI Dana Farber Cancer Institute, MSKCC Memorial Sloan-Kettering Cancer Center, NSCLC nonsmall cell lung cancer, OS overall survival, PD-1 programmed death-1, PD-L1 programmed death-ligand 1, PFS progression-free survival, SYSUCC Sun Yat-Sen University Cancer Center.
By cross-validation in training sets-1/2 (detailed criteria were shown in Fig. 2A: [i] number of mutation or co-mutation>10 and P < 0.2 in training set-1 [n = 288] and -2 [n = 304]; [ii] P < 0.05 in total training sets [n = 592]), five single mutational events (EGFR, STK11, PTPRD, PI3K pathway, and HRR pathway, Fig. 2B), and five interactions (EGFR*PI3K pathway, TP53*KRAS, TP53*ERBB4, cell cycle pathway*HRR pathway, and PI3K pathway*chromatin remodeling pathway, Fig. 2C) were selected to develop three different prediction models. The first model only includes the five single mutational events (termed uni-model); The second model consists of the five single mutations, five interactions, and five terms that support the valid calculation of interaction effects (termed inter-model); The third model was designed to be a control model between the uni-model and the inter-model, contains five single mutations and five supporting terms, but no interaction terms (termed null-inter-model, Fig. 2A). By comparing the performances of these three models (coefficients: Supplementary Table 8, nomograms: Supplementary Fig. 2), it was possible to evaluate whether adding interaction terms into the prediction model with only individual mutations could improve its predictive effectiveness.
In the training sets-1/2, lower scores of the inter-model were associated with better PFS, and the inter-model showed numerically better discriminative performance (bootstrap Dxy = 0.173) and predictive utility for the response to anti-PD-(L)1 monotherapy (area under the curve of receiver operating characteristic [AUROC] = 0.690 [P = 7.0*10−]) compared to the uni-model (bootstrap Dxy = 0.139, AUROC = 0.655 [P = 4.7*10−7]) and the null-inter-model (bootstrap Dxy = 0.120, AUROC = 0.673 [P = 2.0*10−8], Fig. 2D–E and Supplementary Fig. 3A). As genomic features likewise, the AUROCs of tissue tumor mutational burden (tTMB, 0.603, P = 0.026) and blood TMB (bTMB, 0.547, P = 0.445) were lower than all three models (Supplementary Fig. 3A). The potentially optimal cut-off of each model was identified when Youden’s index reached the maximum (Fig. 2E). In addition to the optimal cut-off, other cut-off values were also taken into consideration. We compare the survival data of the patients with a score below each cut-off with those with a score above this cut-off. By this methodology, we aimed to comprehensively evaluate the robustness of predictive effectiveness for each model.
Lower scores were associated with longer survival on anti-PD-1/PD-L1 monotherapy (Fig. 2F), but poorer survival on docetaxel and prognosis in The Cancer Genomic Atlas (TCGA)-LUAD cohort (Supplementary Fig. 4). By directly calculating the interaction effect between treatment (atezolizumab vs. docetaxel) and each score in the POPLAR/OAK cohort, all three models exhibited excellent discriminative effectiveness in predicting PFS benefit from atezolizumab over docetaxel. However, when it comes to predicting OS benefit, only the uni-model and the inter-model showed good discriminative effectiveness (Fig. 2G), indicating their predictive utility.
Outperformance and robustness of the inter-model in predicting immunotherapy efficacy in nonsquamous NSCLC
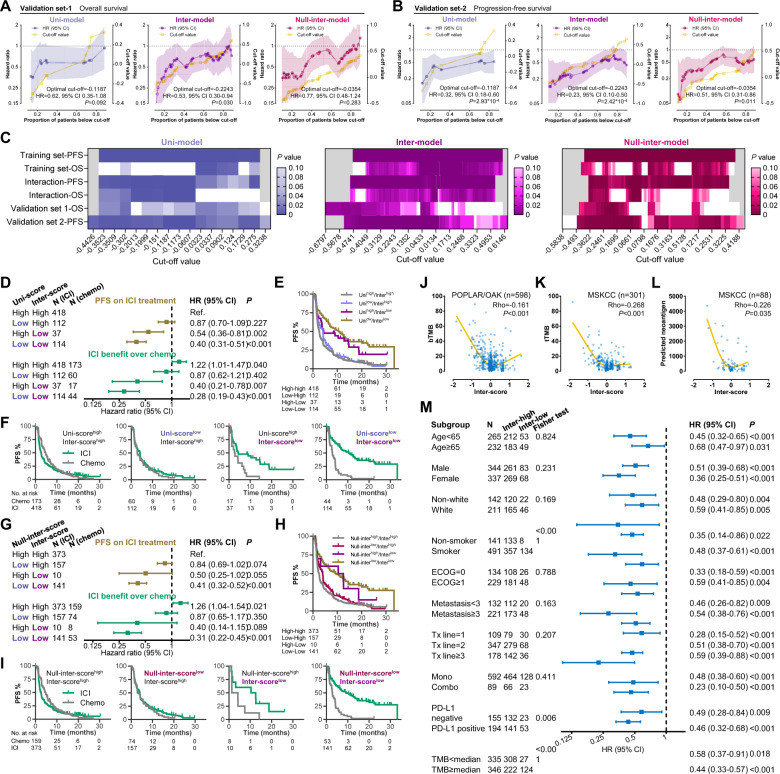
First, two cohorts were employed to validate the models for immunotherapy in nonsquamous NSCLC. As mentioned in Fig. 2A, the validation set-1 consists of 104 patients with only OS data, who had received anti-PD-(L)1 monotherapy, and the validation set-2 includes 93 patients with objective response rate (ORR) and PFS data treated with combination therapy with anti-cytotoxic T lymphocyte antigen-4 (anti-CTLA-4). In the validation set-1, the inter-model outperformed the other two models on account of (1) wider range of applicable cut-off values and (2) the significant result at optimal cut-off (Fig. 3A). In the validation set-2, all three models showed good discriminative effectiveness. However, compared to the other two models, the inter-model exhibited a numerically higher AUROC of response (inter-model: 0.816, P = 2.0*10−6; uni-model: 0.692, P = 0.003; null-inter-model: 0.708, P = 0.002, Supplementary Fig. 3B), and a lower HR value at optimal cut-off (Fig. 3B).
Fig. 3. Validation and comparison of the three models and potential immune-related mechanism of the inter-score in nonsquamous NSCLC.
A, B Performances of the three models on discriminating the OS on anti-PD-(L)1 monotherapy in the validation set-1 (A) and the PFS on combination therapy with anti-CTLA-4 in the validation set-2 (B). C Summary of the performances of the three models in all training/validation sets. D–F Comparison between the uni-model and the inter-model when two results are inconsistent (D). KM curves illustrating PFS on ICI treatment (E) and PFS benefit from ICI therapy over docetaxel (F). G–I Comparison between the null-inter-model and the inter-model when two results are inconsistent (G). KM curves illustrating PFS on ICI treatment (H) and PFS benefit from ICI therapy over docetaxel (I). J–L Association of the inter-score with blood TMB (J), tissue TMB (K), and neoantigen load (L). M Effectiveness of the inter-score in patients with different clinicopathological features, TMB, and PD-L1. Abbreviations: CTLA-4 cytotoxic T lymphocyte antigen-4, ICI immune checkpoint inhibitor, KM Kaplan-Meier, NSCLC nonsmall cell lung cancer, OS overall survival, PD-1 programmed death-1, PD-L1 programmed death-ligand 1, PFS progression-free survival, TMB tumor mutational burden.
Second, we sought to compare the utility of three models by summarizing the P values at all cut-offs in both training and validation sets (Fig. 3C). The inter-score exhibited consistent discriminative effectiveness ranging from −0.4059 to 0.0163 (approximately from 15th percentile to 55th percentile), outperforming the other two scores. To further compare the inter-model with the other two models, we classified all patients with nonsquamous NSCLC by the optimal cut-offs and compared their survival outcomes (all HR and 95% CI results below are summarized in Fig. 3D). To start with, we compared the inter-model with the uni-model. As shown in Fig. 3E illustrating the PFS on ICI treatment, the uni-scorehigh/inter-scorehigh subgroup had the shortest PFS. Compared to this subgroup, slightly longer PFS was observed in the uni-scorelow/inter-scorehigh subgroup (HR = 0.87, 95% CI 0.70–1.29, P = 0.227), but markedly longer PFS was revealed in the uni-scorehigh/inter-scorelow subgroup (HR = 0.54, 95% CI 0.36–0.81, P = 0.002). As shown in Fig. 3F, compared to chemotherapy, ICI treatment decreased the hazard of progression or death by only 13% in the uni-scorelow/inter-scorehigh subgroup (HR = 0.87, 95% CI 0.62–1.21, P = 0.402), and by higher proportion as 60% in the uni-scorehigh/inter-scorelow subgroup (HR = 0.40, 95% CI 0.21–0.78, P = 0.007). These results demonstrate that when the results of the two models are inconsistent, the inter-model may be more accurate in predicting ICI benefit. We further compared the inter-model with the null-inter-model, and similar superiority of the inter-model was observed (Fig. 3G–I). Taken together, the inter-model involving interaction effects of co-occurring genomic alterations outperformed the other two control models, indicating the necessity of adding interaction terms into the prediction model for optimizing its discriminative utility.
Third, we proposed to examine the applicability of the inter-model in the nonsquamous NSCLCs with different clinical characteristics. As for TMB and PD-L1, lower inter-score was weakly associated with higher TMB and neoantigen load (Rho < 0.30, Fig. 3J–L) and PD-L1 positivity (69.7% vs. 51.6%, P = 0.006). However, consistent predictive effectiveness was revealed in the TMB < median (HR = 0.58, 95% CI 0.37–0.91, P = 0.018), the TMB ≥ median (HR = 0.44, 95% CI 0.33–0.57, P < 0.001), the PD-L1negative (HR = 0.49, 95% CI 0.28–0.84, P = 0.009), and the PD-L1positive nonsquamous NSCLCs (HR = 0.46, 95% CI 0.32–0.68, P < 0.001, Fig. 3M). Moreover, the inter-score showed undifferentiated predictive value regardless of other key clinicopathological features (age, sex, race, smoking, Eastern Cooperative Oncology Group [ECOG], number of metastasis sites, treatment lines, and ICI regimen, Fig. 3M). These results demonstrate the robustness of the inter-model in nonsquamous NSCLC with different clinical characteristics.
Immune-related mechanisms of the inter-score in nonsquamous NSCLC
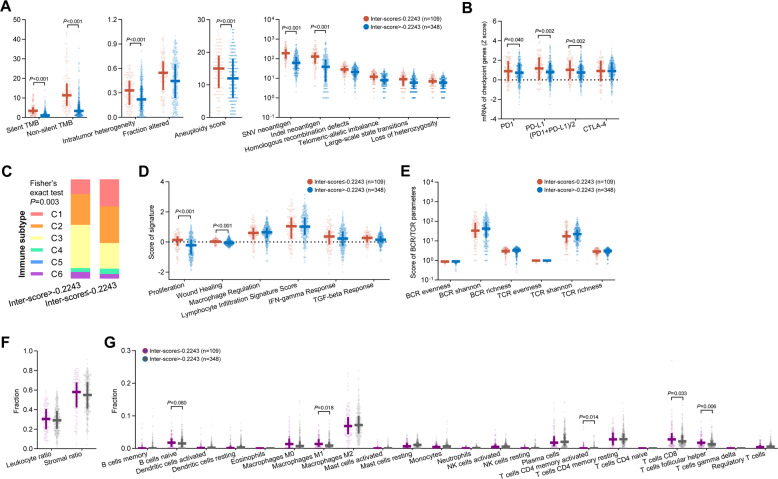
The TCGA-LUAD cohort with genomic, transcriptomic, and survival data was used for exploring the immune-related mechanisms of the inter-score. Similar to the previous result about TMB and PD-L1 expression, higher TMB, neoantigen load, aneuploidy score, intratumoral heterogeneity, PD-1 mRNA, and PD-L1 mRNA were observed in the inter-scorelow group (Fig. 4A, B).
Fig. 4. Immune correlates of the inter-score in the TCGA-LUAD cohort.
A–G Associations of the inter-score with TMB, intratumor heterogeneity, fraction altered, aneuploidy score, neoantigen load, homologous recombination defects (A), mRNA of immune checkpoint genes (B), immune subtype (C), signatures supporting immune subtyping (D), and BCR/TCR parameters (E), leukocyte/stromal ratio (F), and tumor-infiltrating immune cells (G). Abbreviations: BCR B cell receptor, LUAD lung adenocarcinoma, SNV single-nucleotide variation, TCGA The Cancer Genomic Atlas, TCR T cell receptor, TMB tumor mutational burden.
Thorsson et al. presented immunogenomics analyses of more than 10,000 tumors in TCGA, identifying 6 immune subtypes (C1: wound healing, C2: IFN-γ dominant, C3: inflammatory, C4: lymphocyte depleted, C5: immunologically quiet, C6: TGF-β dominant) that encompass 33 cancer types based on six key signatures25. Higher prevalence of C1 and C2 and lower prevalence of C3 were observed in the inter-scorelow group (P = 0.003, Fig. 4C, D). Both C1 and C2 subtypes had low Th1/Th2 ratio, high proliferation rate, high intratumoral heterogeneity, and C2 had the highest M1/M2 macrophage polarization, and a strong CD8 signal25. In addition, no significant difference of the features of B and T cell receptors (BCR/TCR) were observed (Fig. 4E). Regarding immune cell infiltration, despite the similar fractions of leukocyte and stromal area in both groups (Fig. 4F), greater infiltration of naïve B cell (P = 0.060), M1 macrophage (P = 0.018), activated memory CD4+ T cell (P = 0.014), CD8+ T cell (P = 0.033), and follicular helper T cell (P = 0.006) was found in the inter-scorelow group (Fig. 4G).
These immune-related associations in addition to TMB and PD-L1 may serve as the foundation explaining why its predictive utility for immunotherapy efficacy was undifferentiated in the patients with different levels of TMB and PD-L1 expression (Fig. 3M). To further confirm this independence, we performed multivariable analysis including the inter-score, TMB, PD-L1, and key clinicopathological features in the patients with valid TMB and PD-L1 data (Table 1). Lower inter-score, rather than high TMB and PD-L1 positivity, was independently associated with longer PFS on ICI treatment (inter-score: multivariable HR = 0.45, 95% CI 0.31–0.67, P < 0.001) rather than docetaxel (multivariable HR = 1.16, 95% CI 0.80–1.68, P = 0.425). More importantly, the interaction effect between the inter-score and treatment choice (ICI vs. chemotherapy) was remarkably significant (multivariable HR = 0.38, 95% CI = 0.23–0.61, P < 0.001). These findings demonstrate that the inter-score, rather than TMB and PD-L1, may be a robust and independent predictor for favorable benefit from ICIs in nonsquamous NSCLC.
Table 1.
Univariable and multivariable analyses of PFS in patients with nonsquamous NSCLC or squamous NSCLC.
| Nonsquamous NSCLC | ICI treatment (n = 349) | Chemotherapy (n = 224) | Interaction effect (n = 573) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Parameter | Univariable analysis | Multivariable analysis | Univariable analysis | Multivariable analysis | Univariable analysis | Multivariable analysis | ||||||
| HR (95%CI) | P value | HR (95%CI) | P value | HR (95%CI) | P value | HR (95%CI) | P value | HR (95%CI) | P value | HR (95%CI) | P value | |
| Age (≥65 vs. <65) | 1.06 (0.81–1.39) | 0.659 | 0.94 (0.72–1.24) | 0.680 | 1.00 (0.82–1.21) | 0.970 | ||||||
| Sex (male vs. female) | 0.81 (0.64–1.02) | 0.069 | 0.79 (0.58–1.08) | 0.144 | 1.02 (0.78–1.35) | 0.872 | 0.88 (0.74–1.05) | 0.169 | ||||
| Race (white vs. nonwhite) | 0.81 (0.62–1.04) | 0.099 | 1.02 (0.75–1.39) | 0.900 | 0.99 (0.74–1.33) | 0.954 | 0.88 (0.72–1.06) | 0.184 | ||||
| Smoking (smoker vs. non-smoker) | 0.72 (0.55–0.95) | 0.022 | 0.80 (0.55–1.16) | 0.245 | 1.00 (0.71–1.42) | 0.984 | 0.80 (0.64–0.99) | 0.040 | 0.83 (0.65–1.06) | 0.142 | ||
| ECOG (≥1 vs. 0) | 1.21 (0.94–1.56) | 0.145 | 1.40 (1.05–1.86) | 0.021 | 1.35 (1.01–1.80) | 0.040 | 1.26 (1.04–1.53) | 0.016 | 1.49 (1.21–1.84) | <0.001 | ||
| Metastatic sites (≥3 vs. <3) | 1.51 (1.16–1.97) | 0.002 | 1.61 (1.19–2.17) | 0.002 | 1.55 (1.15–2.08) | 0.004 | 1.40 (1.04–1.89) | 0.027 | 1.49 (1.23–1.81) | <0.001 | 1.49 (1.21–1.84) | <0.001 |
| Treatment lines (≥3 vs. 2 vs. 1) | 1.25 (1.06–1.47) | 0.007 | 0.73 (0.53–1.00) | 0.048 | 0.72 (0.53–0.98) | 0.036 | 0.72 (0.53–0.98) | 0.037 | 1.15 (1.01–1.32) | 0.035 | 0.70 (0.56–0.87) | 0.001 |
| PD-L1 (positive vs. negative) | 0.73 (0.58–0.92) | 0.007 | 0.82 (0.61–1.09) | 0.173 | 0.93 (0.71–1.23) | 0.624 | 0.94 (0.71–1.24) | 0.660 | 0.79 (0.67–0.95) | 0.010 | 0.89 (0.73–1.08) | 0.240 |
| TMB ( ≥ median vs. <median) | 0.79 (0.63–1.00) | 0.046 | 1.56 (1.14–2.13) | 0.005 | 1.74 (1.32–2.31) | <0.001 | 1.59 (1.17–2.17) | 0.003 | 1.00 (0.84-1.19) | 0.977 | 1.57 (1.25-1.96) | <0.001 |
| Inter-score (≤cut-off vs. >cut-off) | 0.45 (0.33–0.61) | <0.001 | 0.45 (0.30–0.67) | <0.001 | 1.42 (1.02–1.98) | 0.039 | 1.16 (0.80–1.68) | 0.425 | 1.35 (0.97–1.88) | 0.077 | 1.13 (0.80–1.59) | 0.494 |
| Therapy (ICI vs. chemotherapy) | 1.08 (0.89–1.32) | 0.440 | 1.17 (0.94–1.46) | 0.158 | ||||||||
| Interaction: inter-score*therapy | 0.31 (0.20–0.49) | <0.001 | 0.38 (0.23–0.61) | <0.001 | ||||||||
| Squamous NSCLC | ICI treatment (n = 135) | Chemotherapy (n = 91) | Interaction effect (n = 226) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Parameter | Univariable analysis | Multivariable analysis | Univariable analysis | Multivariable analysis | Univariable analysis | Multivariable analysis | ||||||
| HR (95%CI) | P value | HR (95%CI) | P value | HR (95%CI) | P value | HR (95%CI) | P value | HR (95%CI) | P value | HR (95%CI) | P value | |
| Age (≥65 vs. <65) | 0.88 (0.57–1.36) | 0.570 | 1.63 (1.05–2.53) | 0.028 | 1.56 (1.00–2.42) | 0.049 | 1.12 (0.83–1.51) | 0.455 | ||||
| Sex (male vs. female) | 0.79 (0.46–1.36) | 0.398 | 1.40 (0.84–2.32) | 0.195 | 1.03 (0.72–1.49) | 0.861 | ||||||
| Race (white vs. nonwhite) | 1.00 (0.67–1.50) | 0.981 | 1.32 (0.79–2.21) | 0.283 | 1.16 (0.85–1.57) | 0.349 | ||||||
| Smoking (smoker vs. non-smoker) | 0.91 (0.44–1.89) | 0.805 | 0.66 (0.16-2.73) | 0.568 | 0.96 (0.51-1.83) | 0.908 | ||||||
| ECOG (≥1 vs. 0) | 1.40 (0.87–2.26) | 0.166 | 1.26 (0.78–2.02) | 0.344 | 1.28 (0.92–1.79) | 0.145 | ||||||
| Metastatic sites (≥3 vs. <3) | 1.30 (0.88–1.93) | 0.193 | 0.75 (0.48–1.15) | 0.185 | 1.11 (0.83–1.48) | 0.484 | ||||||
| Treatment lines (≥3 vs. 2 vs. 1) | 1.41 (1.06–1.87) | 0.019 | 1.53 (1.16–2.03) | 0.003 | 0.88 (0.46-1.67) | 0.697 | 1.35 (1.06–1.72) | 0.016 | 1.40 (1.09–1.78) | 0.007 | ||
| PD-L1 (positive vs. negative) | 0.99 (0.67–1.45) | 0.954 | 1.11 (0.75–1.64) | 0.599 | 1.27 (0.82–1.95) | 0.280 | 1.17 (0.76–1.80) | 0.483 | 1.04 (0.79–1.38) | 0.775 | 1.15 (0.87–1.54) | 0.328 |
| TMB ( ≥ median vs. <median) | 0.83 (0.57–1.21) | 0.340 | 0.69 (0.47–1.01) | 0.054 | 1.58 (1.02–2.44) | 0.040 | 1.52 (0.98–2.36) | 0.064 | 1.00 (0.76–1.32) | 1.000 | 0.97 (0.73–1.29) | 0.815 |
| Inter-score (≤cut-off vs. >cut-off) | 0.43 (0.29–0.64) | <0.001 | 0.37 (0.24–0.56) | <0.001 | 1.12 (0.71–1.78) | 0.616 | 1.14 (0.72–1.81) | 0.565 | 1.08 (0.69–1.70) | 0.741 | 1.07 (0.68-1.68) | 0.779 |
| Therapy (ICI vs. chemotherapy) | 1.03 (0.73–1.46) | 0.850 | 1.09 (0.77–1.56) | 0.619 | ||||||||
| Interaction: inter-score*therapy | 0.37 (0.20–0.69) | 0.001 | 0.35 (0.19–0.65) | <0.001 | ||||||||
CI confidence interval, ECOG Eastern Cooperative Oncology Group, HR hazard ratio, PD-L1 programmed death-ligand 1, TMB tumor mutational burden.
Involving interaction effects in prediction model improved predictive utility in squamous NSCLC
A refinement of the prediction model was observed in nonsquamous NSCLC by considering the interaction effects of co-occurring mutations. We hereby performed the same analyses in squamous NSCLC to investigate whether this improvement could occur in squamous NSCLC as well. To avoid repetition, we shall briefly describe the results. The available data of the training sets and the validation sets are shown in Supplementary Fig. 5. Four single mutational events (mutations in NOTCH1/2/3, LRP1B, RB1, and PI3K pathway) and two interactions (TP53*NFE2L2 and TP53*HRR pathway) were involved for developing the three models (Fig. 5A, B and Supplementary Table 9). The coefficients and the nomograms of these three models are illustrated in Supplementary Table 10 and Supplementary Fig. 6.
Fig. 5. Outperformance of the inter-model in squamous NSCLC.
A, B The single mutations (A) and interaction effects of co-mutations (B) selected for model development. C Calibration curves of the three-models in the training sets-1/2. D Youden’s index based on the receiver operating characteristic curve of the three inter-scores and response to anti-PD-(L)1 monotherapy in the training sets-1/2. E Summary of the performances of the three models in all training/validation sets. F–H Comparison between the uni-model and the inter-model when two results are inconsistent (F). KM curves illustrating PFS on ICI treatment (G) and PFS benefit from ICI therapy over docetaxel (H). I–K Comparison between the null-inter-model and the inter-model when two results are inconsistent (I). KM curves illustrating PFS on ICI treatment (J) and PFS benefit from ICI therapy over docetaxel (K). L–N Association of the inter-score with blood TMB (L), tissue TMB (M), and neoantigen load (N). O Effectiveness of the inter-score in patients with different clinicopathological features, TMB, and PD-L1. Abbreviations: ICI immune checkpoint inhibitor, KM Kaplan-Meier, NSCLC nonsmall cell lung cancer, OS overall survival, PD-1 programmed death-1, PD-L1 programmed death-ligand 1, PFS progression-free survival, TMB tumor mutational burden.
In the training sets-1/2, the inter-model showed numerically better discriminative performance (Fig. 5C) and predictive utility on response to anti-PD-(L)1 (Fig. 5D and Supplementary Fig. 7A), PFS/OS on anti-PD-(L)1 (Supplementary Fig. 8A), and survival benefit from atezolizumab over docetaxel (Supplementary Fig. 8B). Similar advantages were seen in validation sets-1/2 (response: Supplementary Fig. 7B-C, PFS/OS: Supplementary Fig. 9). P values at all cut-offs of the analyses in the training sets and the validation sets were summarized in Fig. 5E. The inter-score exhibited consistent discriminative utility (criterion: P < 0.15) ranging from −0.1528 to 0.1312 (approximately from 41st percentile to 61st percentile), outperforming the other two scores (uni-model: none; null-inter-model: from −0.0709 [50th percentile] to 0.1094 [61st percentile]). In addition, lower scores were associated with better immunotherapy efficacy rather than longer survival on docetaxel and better prognosis in the TCGA-lung squamous carcinoma (LUSC) cohort (Supplementary Fig. 10), indicating their predictive, but not prognostic utilities.
When the result of the inter-score was opposite to the one of the uni-score (Fig. 5F–H) or the null-inter-score (Fig. 5I–K), the inter-model performed numerically better than the other two models in predicting the PFS on ICI treatment and the PFS benefit from immunotherapy over chemotherapy. These findings demonstrate the necessity of adding interaction terms into the prediction model for optimizing its discriminative utility.
No significant correlation of the inter-score with mutational burden, neoantigen load, and PD-L1 positivity was observed (Fig. 5L–O and Supplementary Fig. 11A–B). As shown in Supplementary Fig. 11, the immune mechanism of the inter-score may include higher mRNA expressions of PD-1 and CTLA-4, BCR richness and Shannon index, the fraction of stromal area and infiltration of naïve B cell, monocyte, M1 macrophage, activated NK cell, resting memory CD4+ T cell, and follicular helper T cell. These may be part of the mechanisms underlying its predictive function for ICI treatment and explain why the inter-score showed undifferentiated predictive utility in the patients with different levels of TMB and PD-L1 expression (Fig. 5O).
The inter-score showed concordant associations with PFS in the patients with different TMB, PD-L1, and clinical characteristics, indicating wide applicability. We further performed multivariable analyses (including TMB, PD-L1, and clinical characteristics) and identified lower inter-score as an independent indicator of longer PFS on ICI treatment (inter-score: multivariable HR = 0.37, 95% CI 0.24–0.56, P < 0.001) rather than chemotherapy (multivariable HR = 1.14, 95% CI 0.72–1.81, P = 0.565), and larger PFS benefit from ICI treatment to chemotherapy (multivariable HR = 0.35, 95% CI = 0.19–0.65, P < 0.001).
Altogether, the outperformance of the inter-model over the other two models in squamous NSCLC demonstrates the improvement by adding interaction terms into prediction models. Moreover, good performances in subgroup analysis and multivariable analysis indicate the effectiveness and robustness of the inter-model in predicting immunotherapy efficacy in squamous NSCLC.
Discussion
In this retrospective study involving 1745 patients from eight cohorts, we delineated the landscape of interaction effects between co-occurring mutations on ICI efficacy and we further developed and validated two cost-effective mutational signatures involving interaction terms to predict ICI benefit more precisely in nonsquamous and squamous NSCLC. Taken together, our comprehensive analysis demonstrates the advantage and necessity of involving interaction effects when developing a prediction algorithm.
Despite abundant studies exploring the mutational biomarkers for ICI treatment, most of these focused on the impact of a single gene (e.g., EGFR) or a group of genes with similar functions (e.g., NOTCH1/2/3 and EPHA receptors)19,20,23,26–28. Unlike previous studies in nonsquamous NSCLC29, KEAP1 mutation was not associated with poorer ICI efficacy in the training datasets and thereby was not included in the prediction models. Given the inconsistent predictive effects of KEAP1 mutation in different cohorts (Supplementary Fig. 12A), further investigations of the association between KEAP1 and ICI efficacy are warranted. In the studies aiming at single genes, whether their predictive utilities are influenced by other mutations was not investigated, partially due to the small sample size of co-mutated tumors which could lead to serious sampling error. We hereby collected the PFS data of nearly 800 NSCLC patients treated with anti-PD-(L)1 monotherapy and evaluated the interaction effects with more than 20 co-mutated samples separately in nonsquamous and squamous NSCLC.
Within KRAS-mutant LUADs, previous studies have extensively explored the impact of co-occurring mutations (e.g., TP53, STK11, CDKN2A/B) on pathogenesis. In terms of immune-related features, KRAS/TP53 co-mutated LUADs exhibit high TMB, CD8A mRNA, and PD-L1 protein expression8,9, while KRAS/STK11 co-mutations are associated with PD-L1 negativity and T cell suppressive properties, despite an intermediate-to-high TMB12. In keeping with their prominent role in shaping immune-related characteristics, co-occurring mutations further impacted survival outcome with ICIs in the present study (KRAS*TP53, HR = 0.51, P = 0.003; KRAS*STK11, HR = 1.58, P = 0.082). In addition to TP53 and STK11 mutations, we also revealed remarkable interactions of KRAS mutations with co-occurring genomic alterations in PTPRD (HR = 2.47, P = 0.004), RBM10 (HR = 2.04, P = 0.094), NOTCH1/2/3 (HR = 0.50, P = 0.036), ATM (HR = 1.81, P = 0.062), RTKs (HR = 0.63, P = 0.097), and switch/sucrose non-fermentable complex (SWI/SNF, HR = 1.53, P = 0.079). The immune-related mechanisms of these co-mutations are largely unknown, and our results provide new perspectives for the basic research into tumor immunobiology and immune subtyping in KRAS-driven LUADs.
Of note, in a single-center retrospective analysis, the NSCLC patients with STK11/KRAS co-mutations (n = 36) exhibited longer OS on ICI treatment compared to the ones with STK11 mutation only (n = 37)30, inconsistent with the previous and our results12. The reason for this discord may include that the STK11/KRAS co-mutated patients in their cohort were older at diagnosis, more likely to have received nivolumab, and more likely to have longer smoking histories, compared with the STK11mut/KRASWT patients30. This highlights the importance of balancing key clinical characteristics before investigating biomarkers, especially ICI usage and treatment lines.
Among LUSCs, both TP53 and NFE2F2 mutations are frequent31. A recent study suggested the potential relationship between them by uncovering an NFE2L2-mediated increase in tumor growth seen in Keap1/Tp53-double-deleted LUSC cells32. We observed a substantial interaction effect between these two mutations impacting the PFS on anti-PD-(L)1 monotherapy (HR = 0.21, P = 0.016), which introduces a possibility of the interactive effect of TP53/NFE2L2 co-mutations on immune phenotypes in LUSC.
The advantage of adding key interaction terms into prediction models to improve discriminative effectiveness in both training and validation sets was revealed by comparing three models (uni-model, inter-model, and null-inter-model) in nonsquamous and squamous NSCLC respectively. In the combined analysis, we further assessed the benefit from ICIs over docetaxel when the results of different models are inconsistent, and the inter-model exhibited better accuracy in predicting ICI benefit, compared to the other two models involving no interaction terms.
Importantly, our inter-models held great promise by their broad applicability, for the equivalent predictive value for ICI treatment regardless of age, sex, race, smoking history, ECOG, metastasis, treatment lines, combination with anti-CTLA-4, TMB level, and PD-L1 expression. Even in the patients with high tissue TMB (≥10) or tumoral PD-L1 ( ≥ 50%), the inter-score can also successfully discriminate the survival outcome with ICI therapy (Supplementary Fig. 13). TMB is not a validated predictive biomarker of survival benefit to ICIs in NSCLC, lacking unified cut-off in several analyses and not predicting survival benefit in some large phase 3 trials (e.g., CheckMate 227 and KEYNOTE-189)24,33–36. As genomic features likewise, the inter-model exhibited higher AUROC in predicting response to anti-PD-(L)1 therapy in nonsquamous and squamous NSCLCs. Furthermore, we also noticed that the inter-score was able to identify responders with TMB-low or PD-L1-negative NSCLC, making it a meaningful work for patient selection. The inter-score rather than TMB and PD-L1 is an independent predictor in our multivariable analysis, indicating that the inter-score might in some way cover TMB and PD-L1 with more predictive power. This speculation is partly supported by the association between the inter-score and the immune-related features other than TMB/PD-L1 (e.g., BCR/TCR diversity, mRNA expression of immune checkpoints, and quantity of tumor-infiltrating immune cells). In addition, we assessed the performance at other cut-off values besides the optimal one derived from the ORR-based receiver operating characteristic (ROC) curve (nonsquamous: 15th–55th percentile; squamous: 41st–61st percentile), indicating the robustness of our inter-models.
As for limitation, first, KRASG12C mutations are not reported in the OAK/POPLAR cohort. In the present study, the TP53/KRAS co-mutation was associated with better immunotherapy efficacy in nonsquamous NSCLCs, and the interaction term was included in the inter-model. In the datasets other than the POPLAR/OAK cohort, we analyzed the PFS on ICIs of the nonsquamous NSCLC patients with TP53/KRASG12C co-mutation, which was relatively longer than the ones without co-mutation (P = 0.067, Supplementary Fig. 12B). This result indicates that the predictive effect of KRASG12C on ICI efficacy may be similar to the one of other KRAS mutations. Given this, the lack of KRASG12C assessment in the POPLAR/OAK cohort may not affect the main findings of our study. Second, the present study is strong at clinical analysis and relatively weak at exploring mechanism. Investigations into the biological mechanism of co-mutations are warranted in the future studies. Third, the validation cohorts are heterogenous in terms of clinical characteristics, ICIs given (especially anti-PD(L)1 monotherapy vs. combination with anti-CTLA-4), mutational status, treatment lines, and etc., which might affect the results. Of note, the training sets merely include patients treated with monotherapy. Despite that the inter-models performed well in the validation sets consisting of patients undergoing combination therapy, there may be room for improvement. Moreover, further validations by other cohorts are required. Fourth, the retrospective setting of our study may introduce biases, but this limitation can be greatly minimized by large sample size, multi-cohort methodology, and implementation of subgroup analysis and multivariable analysis, by which experimental features could be balanced (e.g., race, ICI regimen, treatment lines, and the platform/panel/used samples of next-generation sequencing [NGS] testing) and the possibility of confounding impact from these variables might be excluded to some extent.
To our knowledge, this is the first study delineating the landscape of interaction effects between co-occurring mutations on the ICI efficacy in NSCLC, which provides novel insights for basic research into the interactive impacts of co-occurring genomic alterations on tumor immunobiology and immune contexture. Furthermore, we developed and validated two cost-effective and quantitative prediction models involving key interaction terms to predict favorable benefit from ICI treatment in nonsquamous and squamous NSCLC more precisely, compared to the models without interaction terms. Our comparative analysis highlights the advantage and necessity of involving co-occurring genomic alterations for facilitating the refinement of prediction algorithms for cancer therapeutics.
Methods
Patients
Eight cohorts of 1745 NSCLC patients treated with ICI were analyzed19–24,26,37, from National Cancer Center (NCC), SYSUCC, DFCI, MSKCC, and the POPLAR/OAK trial, and TCGA database. ICI agent, setting, number of patients, treatment lines, outcome, PD-L1 antibody, NGS technique, and survival data are shown in Supplementary Table 1 and Supplementary Fig. 1. Of note, overlapped patients were identified among the separately published four MSKCC cohorts by the patient identifier (e.g., P-0003869). We merged these cohorts and then classified them into two subsets based on the used ICI agents (anti-PD-(L)1 monotherapy or combination therapy with anti-CTLA-4). The written consents were received from all the participated patients. Besides, the TCGA-LUAD and -LUSC datasets, and the results of immunogenomics analysis from Thorsson et al.38 were analyzed to explore the immune-related mechanism. Outcomes, analyzed mutations, and definition of TMB > median and PD-L1 positivity are in Supplemental Methods and Supplementary Table 2.
Our study is following the principles of the Declaration of Helsinki and approved by the Institution Review Board of Chinese PLA General Hospital (2015L01380). This report follows the Strengthening the Reporting of Observational Studies in Epidemiology (STROBE) reporting guideline.
Statistical analysis
To assess the between-group difference, we performed (i) Fisher exact test and Chi-square test for categorical variables, (ii) Mann-Whitney tests for continuous variables, and (iii) Kaplan-Meier (KM) method, Log-rank method, and Cox regression (hazard ratio [HR] and 95% confidence interval [CI]) for survival variables. The variables with p value below 0.10 in the univariable analyses were included in the following multivariable analyses.
The area under the curve (AUC) of the ROC curve was calculated to estimate the discriminative performance. Multivariable Cox regression was performed to develop prediction models, and the scores were calculated for each patient using a formula derived from the mutation status (1 or 0) weighted by their regression coefficient: . The coefficients of the models for nonsquamous and squamous NSCLC are displayed in Supplementary Table 8 and Supplementary Table 10, respectively. The nomograms are displayed in Supplementary Fig. 2 and Supplementary Fig. 6, respectively. The potential optimal cut-off was determined by the ROC curve of objective response in the training set. Calibration curves were drawn via bootstrap resampling 1000 times, and Dxy was calculated by Somers’ rank correlation between predicted log relative hazard and PFS. All statistical analyses mentioned above were performed using IBM SPSS Statistics 22 or R 3.4.2. The nominal level of significance was set as 5%, and all 95% CIs were 2-sided.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Acknowledgements
This work was supported by National Natural Science Foundation of China (81770204, 81773056), The Young Talent of PLA General Hospital Program and Clinical Medical Data Research of PLA General Hospital (2019XXMBD-010), Program for Young Talents of Science and Technology in PLA high-level innovative talents, Project of National Commission capacity building and Continuing Education Center (GWJJ2021100304), Special key project of military health care (20BJZ37), Military logistics scientific research project (BEP19J005), Special key project of military health care (18BJZ28), Medical big data and artificial intelligence development fund of Chinese PLA General Hospital (2019MBD-049). We thank Dizai Shi (Stitch) for his emotional support, the patients included and their family members for their participation, and the researchers in NCC, SYSUCC, DFCI, MSKCC, and POPLAR/OAK team who shared treatment data for public use.
Author contributions
Conception and design: F.Z., J.W., T.L., Y.X., Y.H., and L.Z. Development of methodology: Y.X. Acquisition of data: F.Z., J.W., T.L., Y.X., Y.H., and L.Z. Analysis and interpretation of data: Y.X., C.L., and G.W. Writing, review, and/or revision of the manuscript: All authors Administrative, technical, or material support: S.C., Y.H., and L.Z. Study supervision: Y.H., and L.Z. Final approval of manuscript: All authors.
Data availability
The authors declare that relevant data supporting the findings of this study are available within the paper and its Supplementary files. The source of the data (hyperlinks and DOIs) of all the included datasets are shown in Supplementary Table 1. The data of the NCC and SYSUCC cohorts are obtained through sending requests to the corresponding authors. Due to ethical and privacy concerns, we are sorry that we are unable to publish their full data in our study. Readers may contact the corresponding authors for the access of individual patient-level data for non-commercial purposes.
Code availability
We used IBM SPSS Statistics 25 and R version 4.1.0 for data analysis. The packages we performed in R include “ggplot2”, “ggpubr”, “magrittr”, “survival”, “survminer”, “reshape2”, “forcats”, “rms”, “foreign”, “caret”, “PresenceAbsence”, “riskRegression”, and “prodlim”.
Competing interests
The authors declare no potential competing interest, except the employment of Yu Xu, Shangli Cai, Guoqiang Wang, Chengcheng Li in Burning Rock Biotech.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Fan Zhang, Jinliang Wang, Yu Xu, Shangli Cai.
Contributor Information
Lei Zhao, Email: zhaolei301@126.com.
Yi Hu, Email: huyi301zlxb@sina.com.
Supplementary information
The online version contains supplementary material available at 10.1038/s41698-021-00243-7.
References
- 1.Skoulidis F, Heymach JV. Co-occurring genomic alterations in non-small-cell lung cancer biology and therapy. Nat. Rev. Cancer. 2019;19:495–509. doi: 10.1038/s41568-019-0179-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen J, et al. Genomic landscape of lung adenocarcinoma in East Asians. Nat. Genet. 2020;52:177–186. doi: 10.1038/s41588-019-0569-6. [DOI] [PubMed] [Google Scholar]
- 3.Xu JY, et al. Integrative Proteomic Characterization of Human Lung Adenocarcinoma. Cell. 2020;182:245–261 e217. doi: 10.1016/j.cell.2020.05.043. [DOI] [PubMed] [Google Scholar]
- 4.Gillette MA, et al. Proteogenomic Characterization Reveals Therapeutic Vulnerabilities in Lung Adenocarcinoma. Cell. 2020;182:200–225 e235. doi: 10.1016/j.cell.2020.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stewart PA, et al. Proteogenomic landscape of squamous cell lung cancer. Nat. Commun. 2019;10:3578. doi: 10.1038/s41467-019-11452-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nahar R, et al. Elucidating the genomic architecture of Asian EGFR-mutant lung adenocarcinoma through multi-region exome sequencing. Nat. Commun. 2018;9:216. doi: 10.1038/s41467-017-02584-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arbour KC, et al. Effects of Co-occurring Genomic Alterations on Outcomes in Patients with KRAS-Mutant Non-Small Cell Lung Cancer. Clin. Cancer Res. 2018;24:334–340. doi: 10.1158/1078-0432.CCR-17-1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Skoulidis F, et al. Co-occurring genomic alterations define major subsets of KRAS-mutant lung adenocarcinoma with distinct biology, immune profiles, and therapeutic vulnerabilities. Cancer Disco. 2015;5:860–877. doi: 10.1158/2159-8290.CD-14-1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dong ZY, et al. Potential Predictive Value of TP53 and KRAS Mutation Status for Response to PD-1 Blockade Immunotherapy in Lung Adenocarcinoma. Clin. Cancer Res. 2017;23:3012–3024. doi: 10.1158/1078-0432.CCR-16-2554. [DOI] [PubMed] [Google Scholar]
- 10.Jiao X, Qin B, Xu Y, Gong F, Zang Y. 1987P Discordant genomic correlates of PD-L1 expression in lung adenocarcinoma among multiple cohorts using dissimilar PD-L1 testing techniques. Ann. Oncol. 2020;31:S1113. doi: 10.1016/j.annonc.2020.08.1293. [DOI] [Google Scholar]
- 11.Schoenfeld AJ, et al. Clinical and molecular correlates of PD-L1 expression in patients with lung adenocarcinomas. Ann. Oncol. 2020 doi: 10.1016/j.annonc.2020.01.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Skoulidis F, et al. STK11/LKB1 Mutations and PD-1 Inhibitor Resistance in KRAS-Mutant Lung Adenocarcinoma. Cancer Disco. 2018;8:822–835. doi: 10.1158/2159-8290.CD-18-0099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hayes AF, Matthes J. Computational procedures for probing interactions in OLS and logistic regression: SPSS and SAS implementations. Behav. Res. Methods. 2009;41:924–936. doi: 10.3758/BRM.41.3.924. [DOI] [PubMed] [Google Scholar]
- 14.Altman DG, Bland JM. Interaction revisited: the difference between two estimates. BMJ. 2003;326:219. doi: 10.1136/bmj.326.7382.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vatcheva KP, Lee M, McCormick JB, Rahbar MH. The Effect of Ignoring Statistical Interactions in Regression Analyses Conducted in Epidemiologic Studies: An Example with Survival Analysis Using Cox Proportional Hazards Regression Model. Epidemiology (Sunnyvale) 2015;6:1000216. doi: 10.4172/2161-1165.1000216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li X, Xu Y, Wang G, Li L. OA07.06 Interdependence of KRAS and TP53 Mutations in Predicting ICI Efficacy in EGFR/ALKWT Non-Squamous NSCLC: Results From 1129 Patient-Level Data. J. Thorac. Oncol. 2021;16:S117. doi: 10.1016/j.jtho.2021.01.297. [DOI] [Google Scholar]
- 17.Bai X, et al. Development and validation of a genomic mutation signature to predict response to PD-1 inhibitors in non-squamous NSCLC: a multicohort study. J. Immunother. Cancer. 2020;8:000381. doi: 10.1136/jitc-2019-000381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pan D, Hu AY, Antonia SJ, Li CY. A Gene Mutation Signature Predicting Immunotherapy Benefits in Patients With NSCLC. J. Thorac. Oncol. 2021;16:419–427. doi: 10.1016/j.jtho.2020.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fang W, et al. Comprehensive Genomic Profiling Identifies Novel Genetic Predictors of Response to Anti-PD-(L)1 Therapies in Non-Small Cell Lung Cancer. Clin. Cancer Res. 2019;25:5015–5026. doi: 10.1158/1078-0432.CCR-19-0585. [DOI] [PubMed] [Google Scholar]
- 20.Miao D, et al. Genomic correlates of response to immune checkpoint blockade in microsatellite-stable solid tumors. Nat. Genet. 2018;50:1271–1281. doi: 10.1038/s41588-018-0200-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rizvi NA, et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science. 2015;348:124–128. doi: 10.1126/science.aaa1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Samstein RM, et al. Tumor mutational load predicts survival after immunotherapy across multiple cancer types. Nat. Genet. 2019;51:202–206. doi: 10.1038/s41588-018-0312-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rizvi H, et al. Molecular Determinants of Response to Anti-Programmed Cell Death (PD)-1 and Anti-Programmed Death-Ligand 1 (PD-L1) Blockade in Patients With Non-Small-Cell Lung Cancer Profiled With Targeted Next-Generation Sequencing. J. Clin. Oncol. 2018;36:633–641. doi: 10.1200/JCO.2017.75.3384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gandara DR, et al. Blood-based tumor mutational burden as a predictor of clinical benefit in non-small-cell lung cancer patients treated with atezolizumab. Nat. Med. 2018;24:1441–1448. doi: 10.1038/s41591-018-0134-3. [DOI] [PubMed] [Google Scholar]
- 25.Thorsson V, et al. The Immune Landscape of Cancer. Immunity. 2018;48:812–830 e814. doi: 10.1016/j.immuni.2018.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hellmann MD, et al. Genomic Features of Response to Combination Immunotherapy in Patients with Advanced Non-Small-Cell Lung Cancer. Cancer Cell. 2018;33:843–852 e844. doi: 10.1016/j.ccell.2018.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bai H, et al. EPHA mutation as a predictor of immunotherapeutic efficacy in lung adenocarcinoma. J. Immunother. Cancer. 2020;8:001315. doi: 10.1136/jitc-2020-001315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang K, et al. Identification of deleterious NOTCH mutation as novel predictor to efficacious immunotherapy in NSCLC. Clin Cancer Res. 2020;26:3649–3661. doi: 10.1158/1078-0432.CCR-19-3976. [DOI] [PubMed] [Google Scholar]
- 29.Marinelli D, et al. KEAP1-driven co-mutations in lung adenocarcinoma unresponsive to immunotherapy despite high tumor mutational burden. Ann. Oncol. 2020;31:1746–1754. doi: 10.1016/j.annonc.2020.08.2105. [DOI] [PubMed] [Google Scholar]
- 30.Basher F, Saravia D, Fanfan D, Cotta JA, Lopes G. Impact of STK11 and KRAS co-mutations on outcomes with immunotherapy in non-small cell lung cancer. J. Clin. Oncol. 2020;38:e15135–e15135. doi: 10.1200/JCO.2020.38.15_suppl.e15135. [DOI] [Google Scholar]
- 31.Herbst RS, Morgensztern D, Boshoff C. The biology and management of non-small cell lung cancer. Nature. 2018;553:446–454. doi: 10.1038/nature25183. [DOI] [PubMed] [Google Scholar]
- 32.Jeong Y, et al. Role of KEAP1/NRF2 and TP53 Mutations in Lung Squamous Cell Carcinoma Development and Radiation Resistance. Cancer Disco. 2017;7:86–101. doi: 10.1158/2159-8290.CD-16-0127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hellmann MD, et al. Nivolumab plus Ipilimumab in Advanced Non–Small-Cell Lung Cancer. N. Eng. J. Med. 2019;381:2020–2031. doi: 10.1056/NEJMoa1910231. [DOI] [PubMed] [Google Scholar]
- 34.Rizvi NA, et al. Durvalumab With or Without Tremelimumab vs Standard Chemotherapy in First-line Treatment of Metastatic Non-Small Cell Lung Cancer: The MYSTIC Phase 3 Randomized Clinical Trial. JAMA Oncol. 2020;6:661–674. doi: 10.1001/jamaoncol.2020.0237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Garassino M, et al. OA04.06 Evaluation of TMB in KEYNOTE-189: Pembrolizumab Plus Chemotherapy vs Placebo Plus Chemotherapy for Nonsquamous NSCLC. J. Thorac. Oncol. 2019;14:S216–S217. doi: 10.1016/j.jtho.2019.08.427. [DOI] [Google Scholar]
- 36.Carbone DP, et al. First-Line Nivolumab in Stage IV or Recurrent Non-Small-Cell Lung Cancer. N. Engl. J. Med. 2017;376:2415–2426. doi: 10.1056/NEJMoa1613493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang Z, et al. Assessment of Blood Tumor Mutational Burden as a Potential Biomarker for Immunotherapy in Patients With Non-Small Cell Lung Cancer With Use of a Next-Generation Sequencing Cancer Gene Panel. JAMA Oncol. 2019;5:696–702. doi: 10.1001/jamaoncol.2018.7098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Newman AM, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods. 2015;12:453–457. doi: 10.1038/nmeth.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors declare that relevant data supporting the findings of this study are available within the paper and its Supplementary files. The source of the data (hyperlinks and DOIs) of all the included datasets are shown in Supplementary Table 1. The data of the NCC and SYSUCC cohorts are obtained through sending requests to the corresponding authors. Due to ethical and privacy concerns, we are sorry that we are unable to publish their full data in our study. Readers may contact the corresponding authors for the access of individual patient-level data for non-commercial purposes.
We used IBM SPSS Statistics 25 and R version 4.1.0 for data analysis. The packages we performed in R include “ggplot2”, “ggpubr”, “magrittr”, “survival”, “survminer”, “reshape2”, “forcats”, “rms”, “foreign”, “caret”, “PresenceAbsence”, “riskRegression”, and “prodlim”.