Abstract
Knockdown of patatin-like phospholipase domain-containing protein 3 (PNPLA3) increased triglycerides (TG) in primary bovine hepatocytes, suggesting that PNPLA3 plays a causal role in hepatic TG clearing. In vivo, PNPLA3 abundance across the periparturient period is inversely related to hepatic TG accumulation and circulating fatty acid (FA) concentrations. The purpose of this research was to determine if PNPLA3, as well as other lipases, transcription factors, or FA-mediated genes, are regulated by FA mimicking liver lipid accumulation (ACCUM) and liver lipid clearing (RECOV) or singular FA physiologically found in dairy cows at 0.5 mM of circulating RECOV (iRECOV). Abundance of PNPLA3 tended to decrease with ACCUM and increased quadratically with RECOV (P ≤ 0.10), differing from PNPLA3 expression, but consistent with previous in vivo research. Adipose TG lipase abundance, but not other lipase abundances, was quadratically responsive to both ACCUM and RECOV (P ≤ 0.005). Abundance of PNPLA3 and SREBP1c and expression of LXRA responded similarly to iRECOV, with C18:0 tending to decrease abundance (P ≤ 0.07). Results indicate that bovine PNPLA3 is translationally regulated by FA and although a LXRA-SREBP1c pathway mediation is possible, the mechanism warrants further investigation.
Subject terms: Lipids, Proteins, Cell biology, Mechanisms of disease
Introduction
It is well established in the dairy industry that fatty liver syndrome (FLS) occurs during the periparturient period in dairy cows1. Fatty acids (FA) are mobilized from adipose tissue via lipolysis and circulate in the bloodstream to the liver, where they are taken up for various fates including re-esterification as triglycerides (TG) for storage within hepatocytes, resulting in the onset of FLS2–4. Monetary losses of FLS were estimated to be $60 million in 20045 and likely exceed that now. While FLS remains a hinderance to cow productivity, the molecular underpinnings of FLS in dairy cows and how to mitigate it remain unknown.
Many aspects of lipid metabolism are conserved across species, and research focusing on nonalcoholic fatty liver disease (NAFLD) and steatohepatitis in humans may provide insight into the etiology of FLS in dairy cattle and vice versa. While there are a variety of differences between ruminants and nonruminants regarding physiology and digestive capabilities6–8, a key lipase involved in the development of NAFLD also plays a critical and similar role in FLS. In humans, a single nucleotide polymorphism (SNP) inactivates the lipase patatin-like phospholipase domain-containing protein 3 (PNPLA3), resulting in TG accumulation and storage in the liver9–11. An in vivo study elucidated that periparturient dairy cows with more hepatic PNPLA3 abundance had lower hepatic TG12. In conjunction with those findings, knockdown of PNPLA3 abundance using siRNA in primary bovine hepatocytes resulted in greater cellular TG content compared to cells treated with a nonspecific siRNA sequence12. These patterns are consistent with the naturally occurring inactivating SNP in humans; however, a similar pattern was not observed in mice, where knockout of PNPLA3 did not affect hepatic TG content13.
Despite the causal role of PNPLA3 in hepatic TG accumulation in both humans and bovine, the transcriptional and translational regulation of PNPLA3 remains elusive. Additionally, an understanding of the coordinated response of PNPLA3 with other lipid-related proteins is needed. Based on in vivo data, our hypothesis is that bovine PNPLA3 will be responsive to FA in a concentration and composition dependent manner, with FA composition and concentration associated with increased PNPLA3 abundance, and that this regulation may be transcription factor (TF) mediated. Thus, the objectives of this study were to 1) determine how PNPLA3 responds to FA presented in mixtures representing two distinct phases of the periparturient period [period of liver lipid accumulation (ACCUM) or period of liver lipid clearing (RECOV)], 2) determine if potential responses to FA mixtures can be attributed to individual FA [concentration of individual FA physiologically found in dairy cows present in circulating serum at 0.5 mM (iRECOV)], and 3) determine potential regulation of PNPLA3 through a targeted analysis of genes and proteins associated with lipolysis and FA-mediation.
Results
PNPLA3 cellular localization, mRNA expression, and protein abundance
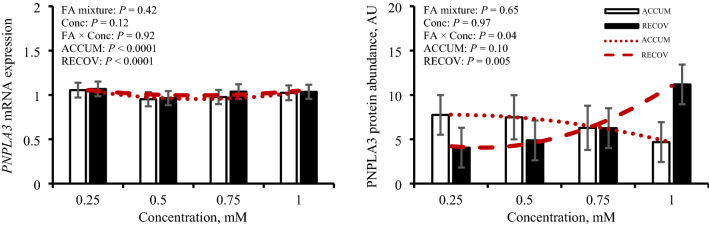
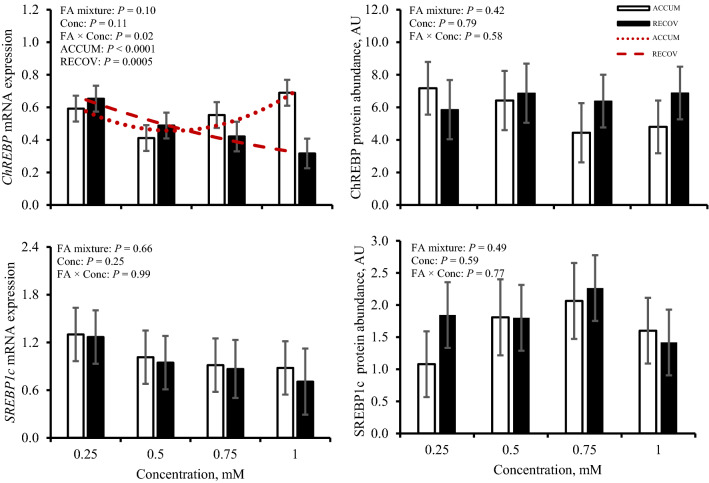
Stained primary bovine hepatocytes (Fig. 1) indicate scattered cytoplasmic localization of PNPLA3 protein within the hepatocytes, consistent with location of lipolytic activity and lipid droplets. Expression of PNPLA3 and its resulting protein abundance is in Fig. 2. An opposing quadratic relationship existed for PNPLA3 expression with ACCUM (P < 0.0001) and RECOV (P < 0.0001). Abundance of PNPLA3 was affected (P = 0.04) by concentration × FA mixture. A quadratic relationship was observed where abundance tended to decrease (P = 0.10) quadratically with greater concentrations of ACCUM, whereas PNPLA3 abundance was increased (P = 0.005) quadratically with higher concentrations of RECOV. No change in PNPLA3 expression was observed with iRECOV treatment (P = 0.48; Table 2). Conversely, abundance of PNPLA3 was affected by iRECOV (P = 0.03; Fig. 7) and tended to decrease (P = 0.07) with C18:0 compared to C18:2 n-6, and did decrease with C18:0 compared to C22:6 n-3 (P = 0.05; Fig. 7).
Figure 1.
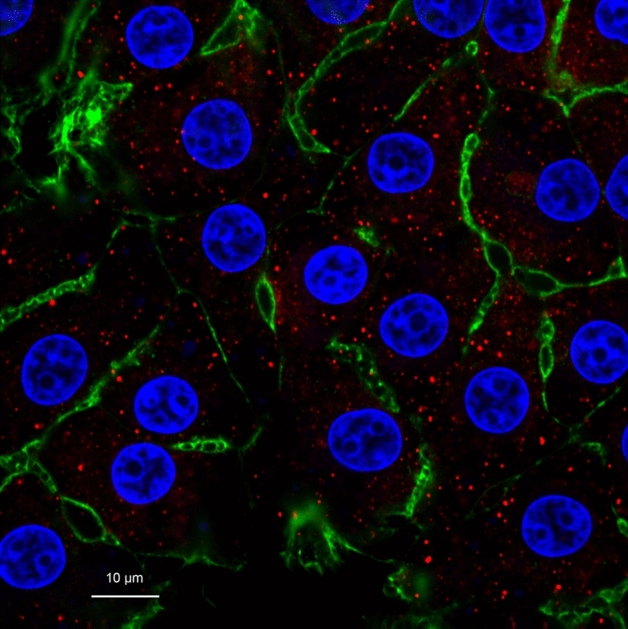
Primary bovine hepatocytes stained with DAPI (nuclei; blue), Alexa Fluor 488 Phalloidin (F-actin; green), and Cy3 for patatin-like phospholipase domain-containing protein 3 (PNPLA3; red). A common trait in hepatocytes, some hepatocytes display binucleated nuclei and some do not. The cytoskeletal protein F-actin outlines the shape of the cells. Appearance of PNPLA3 in red shows it scattered throughout the cytoplasm of the cells, where lipolytic activity occurs.
Figure 2.
Gene expression and protein abundance of patatin-like phospholipase domain-containing protein 3 (PNPLA3) in response to fatty acid (FA) mixture representing liver lipid accumulation (ACCUM; open bars and dotted line) and fatty acid mixture representing liver lipid clearing (RECOV; closed bars and dashed line) at different concentrations in primary bovine hepatocytes. Data is presented as least squares mean ± standard error of the means. Quadratic relationships are displayed if P ≤ 0.15 for a main effect.
Table 2.
Least squares means and standard error of the means (SE) of the gene expression of genes of interest in primary bovine hepatocytes treated with individual fatty acids physiologically found circulating in dairy cows at an in vivo mixture at 0.5 mM1 during liver lipid clearing (iRECOV) with significant differences between means denoted with different superscript letters.
| Gene | iRECOV2 | SE | P-value | |||||
|---|---|---|---|---|---|---|---|---|
| C14:0 | C16:0 | C18:0 | C18:1 | C18:2 | C22:6 | |||
| ChREBP | 0.91 | 0.89 | 0.88 | 0.91 | 0.90 | 0.90 | 0.04 | 0.85 |
| CPT1A | 0.80 | 1.11 | 0.70 | 0.78 | 0.52 | 0.80 | 0.27 | 0.64 |
| CPT2 | 0.98 | 0.97 | 0.98 | 0.89 | 0.96 | 0.98 | 0.05 | 0.08 |
| FASN | 0.75 | 0.67 | 0.66 | 0.69 | 0.73 | 0.75 | 0.10 | 0.87 |
| FOXO1 | 1.05 | 0.89 | 0.89 | 1.00 | 1.08 | 0.89 | 0.09 | 0.33 |
| LXRA | 0.80a | 0.60b | 0.57b | 0.72ab | 0.68ab | 0.73ab | 0.10 | 0.03 |
| LXRB | 0.97 | 0.94 | 0.92 | 0.86 | 0.54 | 0.64 | 0.19 | 0.30 |
| PGC1A | 0.93 | 0.94 | 0.95 | 0.95 | 0.95 | 0.95 | 0.03 | 0.87 |
| PNPLA3 | 0.99 | 0.99 | 1.04 | 1.01 | 1.06 | 1.06 | 0.07 | 0.48 |
| PPARA | 0.95 | 0.95 | 1.00 | 1.00 | 1.00 | 0.91 | 0.07 | 0.21 |
| SIRT1 | 0.90 | 0.91 | 0.93 | 0.94 | 0.94 | 0.93 | 0.03 | 0.60 |
| SIRT3 | 0.40 | 0.23 | 0.21 | 0.29 | 0.18 | 0.37 | 0.12 | 0.44 |
| SREBP1 | 1.94 | 2.60 | 2.27 | 1.84 | 0.84 | 1.49 | 0.62 | 0.34 |
| UCP2 | 0.40 | 0.23 | 0.21 | 0.29 | 0.18 | 0.37 | 0.19 | 0.60 |
1Concentratoin of C14:0, C16:0, C18:0, C18:1 n-9, C18:2 n-6, and C22:6 n-3 was treated at 0.009, 0.175, 0.279, 0.021, and 0.011 mM, respectively.
2C14:0, C16:0, C18:0, C18:1 n-9, C18:2 n-6, C20:5 n-3, and C22:6 n-3 are abbreviated as C14:0, C16:0, C18:0, C18:1, C18:2, C20:5 and C22:6 in the table for conciseness.
Figure 7.
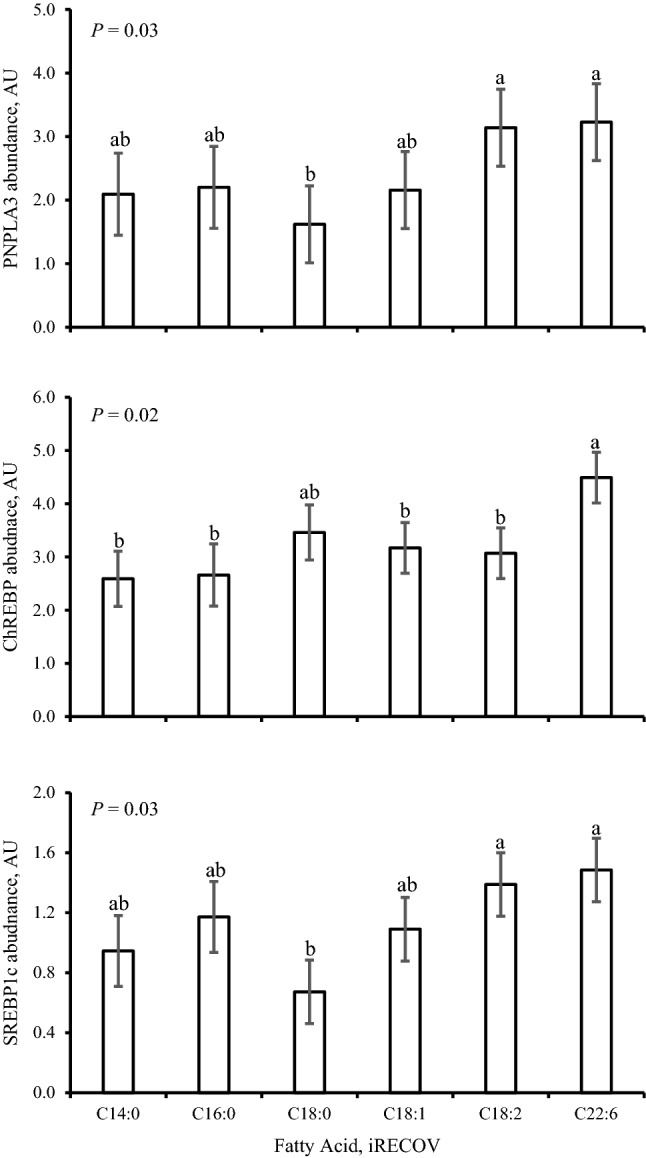
Protein abundance of patatin-like phospholipase domain-containing protein 3 (PNPLA3), carbohydrate response element binding protein (ChREBP), and sterol regulatory element-binding transcription factor 1c (SREBP1c) in response to individual fatty acids at the concentration physiologically found circulating in serum in dairy cows at 0.5 mM of circulating liver lipid clearing profile (iRECOV) in primary bovine hepatocytes. C14:0, C16:0, C18:0, C18:1 n-9, C18:2 n-6, and C22:6 n-3 are abbreviated as C14:0, C16:0, C18:0, C18:1, C18:2, and C22:6 in the figure for conciseness. Data is presented as least squares mean ± standard error of the means. Differences between individual fatty acids are indicated as alphabetic superscripts; when superscripts do not share the same letter, differences between the fatty acids were found (P ≤ 0.07).
Protein abundance of lipases and gene expression of FA-mediated genes and TFs
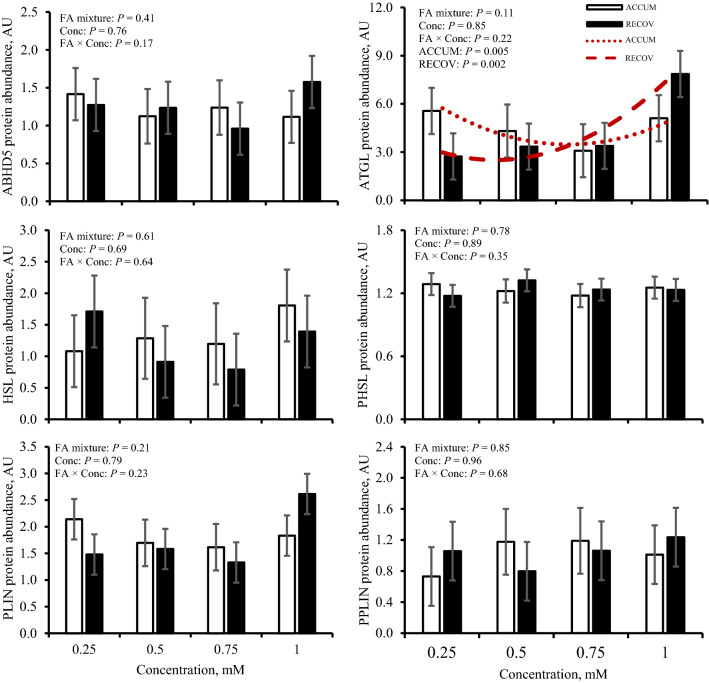
An opposing quadratic relationship for adipose TG lipase (ATGL) existed in which ATGL decreased with ACCUM (P = 0.005) and increased with RECOV (P = 0.002; Fig. 3). No change was observed on abundance of abhydrolase domain containing 5 (ABHD5), hormone sensitive lipase (HSL), phosphorylated HSL (PHSL), perilipin (PLIN), and phosphorylated PLIN (PPLIN) in response to FA mixtures at varying concentrations (P ≥ 0.17; Fig. 3).
Figure 3.
Protein abundance of abhydrolase domain containing 5 (ABHD5), adipose triglyceride lipase (ATGL), hormone sensitive lipase (HSL), phosphorylated HSL (PHSL), perilipin (PLIN), and phosphorylated PLIN (PPLIN) in response to fatty acid mixture representing liver lipid accumulation (ACCUM; open bars and dotted line) and fatty acid mixture representing liver lipid clearing (RECOV; closed bars and dashed line) at different concentrations in primary bovine hepatocytes. Data is presented as least squares mean ± standard error of the means. Quadratic relationships are displayed if P ≤ 0.15 for a main effect.
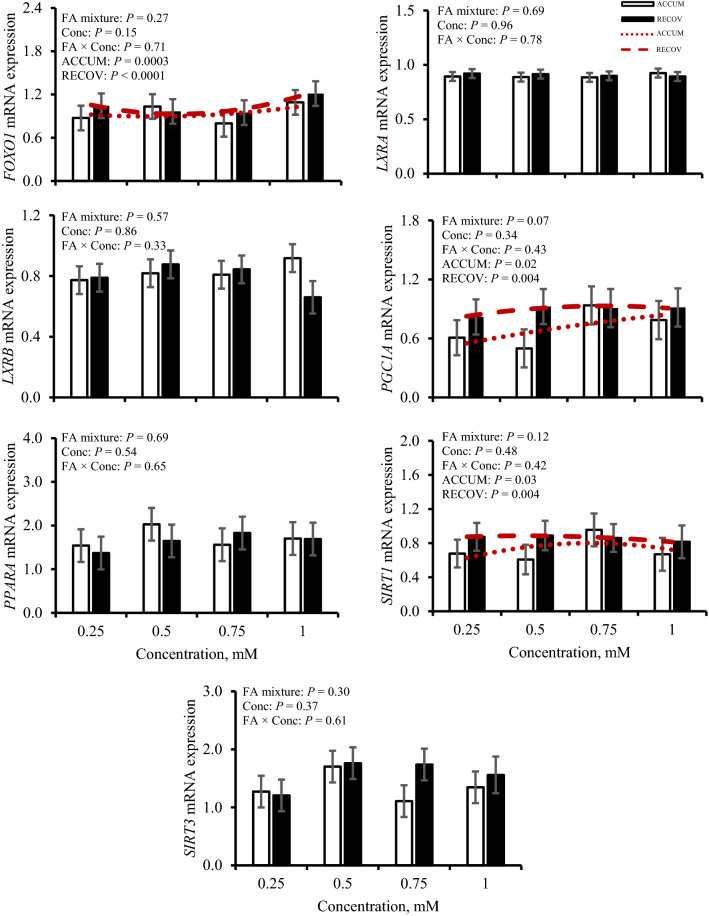
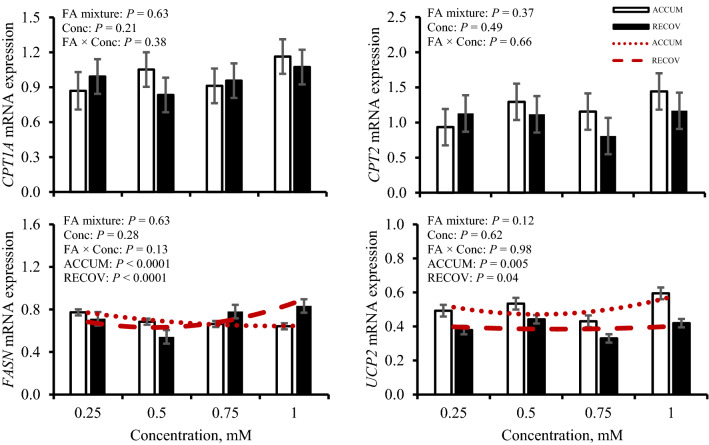
Responses of expression of FA-mediated genes and TFs regarding FA mixture and concentration treatment are in Figs. 4 and 5, respectively, and responses to iRECOV are in Table 2. Expression of FA-transport genes carnitine palmitoyltransferase 1A (CPT1A), carnitine palmitoyltransferase 2 (CPT2), and peroxisome proliferator-activator α (PPARA) were not altered by FA mixture nor concentration (P ≥ 0.21). However, CPT2 expression tended to change with treatment of iRECOV (P = 0.08). Neither CPT1A nor PPARA were affected by iRECOV (P ≥ 0.21). An opposing quadratic relationship of fatty acid synthase (FASN) expression was observed with ACCUM (P < 0.0001) and RECOV (P < 0.0001); however, FASN expression was not altered by iRECOV (P = 0.87). A quadratic relationship was observed for sirtuin 1 (SIRT1) expression to change with both ACCUM (P = 0.03) and RECOV (P = 0.004), yet expression was not altered by iRECOV (P = 0.60). Expression of sirtuin 3 (SIRT3) was not altered by any FA treatment (P ≥ 0.30). A quadratic relationship was observed for uncoupling protein 2 (UCP2) expression in response to both ACCUM (P = 0.005) and RECOV (P = 0.04), yet expression was not altered by iRECOV (P = 0.60).
Figure 4.
Gene expression of transcription factors forkhead box O1 (FOXO1), liver X receptor α (LXRA), liver X receptor β (LXRB), peroxisome proliferator-activated receptor gamma coactivator 1-alpha (PGC1A), peroxisome proliferator-activated receptor α (PPARA), sirtuin 1 (SIRT1), and sirtuin 3 (SIRT3) in response to fatty acid mixture representing liver lipid accumulation (ACCUM; open bars and dotted line) and fatty acid mixture representing liver lipid clearing (RECOV; closed bars and dashed line) at different concentrations in primary bovine hepatocytes. Data is presented as least squares mean ± standard error of the means. Quadratic relationships are displayed if P ≤ 0.15 for a main effect.
Figure 5.
Gene expression of fatty acid-mediated genes carnitine palmitoyltransferase 1A (CPT1A), carnitine palmitoyltransferase 2 (CPT2), fatty acid synthase (FASN), and uncoupling protein 2 (UCP2) in response to fatty acid mixture representing liver lipid accumulation (ACCUM; open bars and dotted line) and fatty acid mixture representing liver lipid clearing (RECOV; closed bars and dashed line) at different concentrations in primary bovine hepatocytes. Data is presented as least squares mean ± standard error of the means. Quadratic relationships are displayed if P ≤ 0.15 for a main effect.
Expression of liver X receptor α (LXRA) and liver X receptor β (LXRB) were not affected by FA mixture nor concentration (P ≥ 0.33); however, expression of LXRA was affected by iRECOV (P = 0.03). Expression decreased (P = 0.03) with C18:0 compared to C14:0 and tended to decrease (P = 0.06) with C16:0 compared to C14:0. No evidence was observed for LXRB to alter expression in response to iRECOV (P = 0.30). Expression of peroxisome proliferator- activated receptor gamma coactivator 1 α (PGC1A) tended to be affected by FA mixture (P = 0.07); expression increased quadratically with increasing concentration of ACCUM (P = 0.02) and RECOV (P = 0.004), but expression was not affected by iRECOV (P = 0.87). A quadratic relationship for forkhead box O1 (FOXO1) expression existed with ACCUM (P = 0.003) and RECOV (P < 0.0001) but expression was not affected by iRECOV (P = 0.33).
ChREBP and SREBP1c mRNA expression and protein abundance
Expression of carbohydrate response element binding protein (ChREBP) and sterol regulatory element-binding transcription factor 1c (SREBP1c) and resulting protein abundances are shown in Fig. 6. Expression of ChREBP was affected (P = 0.02) by concentration × FA mixture. A quadratic relationship was observed where expression was affected (P < 0.0001) by greater concentrations of ACCUM, whereas with higher concentrations of RECOV, ChREBP expression was quadratically increased (P = 0.0005). No evidence was observed for ChREBP expression to be altered with iRECOV (P = 0.85). Abundance of ChREBP was not affected (P ≥ 0.42) by concentration nor FA mixture, but ChREBP abundance was affected by iRECOV (P = 0.02; Fig. 7). Abundance of ChREBP tended to decrease with C18:0 (P = 0.08) and C18:2 n-6 (P = 0.06) compared to C22:6 n-3 and did decrease (P ≤ 0.05) with C14:0 and C16:0 compared to C22:6 n-3.
Figure 6.
Gene expression and protein abundance of carbohydrate response element binding protein (ChREBP) and sterol regulatory element-binding transcription factor 1c (SREBP1c) in response to fatty acid mixture representing liver lipid accumulation (ACCUM; open bars and dotted line) and fatty acid mixture representing liver lipid clearing (RECOV; closed bars and dashed line) at different concentrations in primary bovine hepatocytes. Data is presented as least squares mean ± standard error of the means. Quadratic relationships are displayed if P ≤ 0.15 for a main effect.
No evidence was observed for expression of SREBP1c to be affected by concentration or FA mixture (P ≥ 0.25). Expression of SREBP1c was not affected by iRECOV (P = 0.34). Abundance of SREBP1c was not affected (P ≥ 0.49) by concentration nor FA mixture, but SREBP1c abundance was affected by iRECOV (P = 0.03; Fig. 7). Abundance of SREBP1c tended to decrease with C18:0 (P = 0.06) compared to C18:2 n-6 and did decrease (P = 0.03) with C18:0 compared to C22:6 n-3.
Discussion
Understanding the etiology of the development and subsequent recovery of FLS in periparturient dairy cows is key to mitigating FLS and may provide mechanistic insights into understanding similar metabolic disorders in other species. Administering FA mixtures mimicking the physiological states during FLS development and determining if individual FA within the mixture is responsible for regulating a wide variety of genes and proteins involved in lipolysis, FA-transport, or are FA-mediated is a key piece of furthering our understanding. Therefore, the primary objective was understanding the regulation of PNPLA3 due to its impact on FLS in dairy cows. In humans and mice, PNPLA3 is localized in the cytoplasm and on the lipid droplet9,14,15. Likewise, bovine PNPLA3 appears to be localized in a similar pattern (Fig. 1). Expression of PNPLA3 and its resulting protein abundance did not exhibit similar responses, suggesting that regulation, at least in response to FA, is primarily at the translational level.
To elucidate which singular FA was responsible for this shift, iRECOV treatments were administered. Given the opposing quadradic response of PNPLA3 abundance to increasing concentrations RECOV and ACCUM (Fig. 2), individual FA most likely to be responsible for the regulation would be those with differential inclusion proportions between the two physiological mixtures, specifically C18:0, C18:1 n-9, or C20:5 n-3 (Table 1). In bovine, C18:0 originates from rumen biohydrogenation of dietary polyunsaturated FA and adipose tissue stores16,17, making it a key component of circulating FA profile during mobilization (ie, the physiological phase represented by ACCUM). C18:0 can also be stored in hepatocytes16,17 and thus could potentially be released from hepatic TG by lipolysis during periods of liver TG recovery (ie. the physiological phase represented by RECOV). Treatment with C18:0 inhibited PNPLA3 when presented alone; however, greater contribution of C18:0 to the RECOV mixture would imply that increasing concentration of that mixture would decrease PNPLA3, which was not observed. Although C18:0 has been demonstrated to have inhibitory regulatory effects in isolation, these effects are often blighted when cells are treated in mixtures that reflect physiological states18,19. In contrast, C18:1 n-9 and C18:2 n-6 are found in lower concentration in RECOV than ACCUM and both resulted in increased PNPLA3 abundance compared with C18:0 and maintained abundance compared with C14:0 and C16:0 (Fig. 7). These findings suggest that C18:1 n-9 and C18:2 n-6 could be inhibiting PNPLA3 abundance, as evidenced by the decreased abundance during treatment with ACCUM but increased abundance with RECOV (containing less of these FA) and exposure to iRECOV. This is in agreement with human hepatocytes treated with C18:1 n-9 and C18:2 n-6, where PNPLA3 abundance increased, even though treatment was not at a physiologically relevant concentration (400 µM)20. Since hepatocytes are never exposed to single fatty acids in vivo, it is not surprising that the combination of FA is an important component of regulation. Ultimately, the pattern observed within is in agreement with prior in vivo work12.
Table 1.
Treatment composition of fatty acids (FA) of physiologically representative mixtures present in circulating serum [liver lipid accumulation (ACCUM) and liver lipid clearing (RECOV)] and concentration of individual FA physiologically present in circulating serum in dairy cows at 0.5 mM of RECOV (iRECOV).
| FAa | Mixture composition, % | iRECOV, mM | ||
|---|---|---|---|---|
| ACCUM | RECOV | |||
| Myristic | C14:0 | 2.63 | 1.81 | 0.009 |
| Palmitic | C16:0 | 33.01 | 34.93 | 0.175 |
| Stearic | C18:0 | 43.39 | 55.81 | 0.279 |
| Oleic | C18:1 | 16.4 | 4.2 | 0.021 |
| Linoleic | C18:2 | 1.3 | 0.95 | 0.005 |
| EPA | C20:5 | 0.9 | – | – |
| DHA | C22:6 | 2.38 | 2.26 | 0.011 |
aC14:0, C16:0, C18:0, C18:1 n-9, C18:2 n-6, C20:5 n-3, and C22:6 n-3 are abbreviated as C14:0, C16:0, C18:0, C18:1, C18:2, C20:5 and C22:6 in the table for conciseness.
Expression of PNPLA3 and abundance of PNPLA3 were not congruent in this in vitro study, which is in agreement with previous findings12. It may be possible that the differences seen in PNPLA3 expression and resulting protein both in vitro and in vivo are due to regulation at both the transcriptional and translational levels. Energy status is known to affect PNPLA3 protein in rodents and humans20, and a growing body of evidence is in agreement that bovine PNPLA3 mRNA and its resulting protein are also affected by energy status12,21. Although cell culture models cannot fully recapitulate physiological energy status, a recent in vivo study demonstrated the relationships between energy status, liver TG, and PNPLA3 protein abundance12. Given that hepatic TG accumulation is responsive to energy status during the periparturient period, the lower abundance of PNPLA3 in this study in response to ACCUM, yet higher abundance in response to RECOV, further supports the importance of PNPLA3 abundance on clearing TG in vivo and in vitro12.
The TF SREBP1c is a known regulator of lipogenesis22 and growing evidence suggests SREBP1c may play a role in NAFLD23. Abundance of SREBP1c protein responded similarly to iRECOV as PNPLA3 protein abundance did (Fig. 7). Both PNPLA3 and SREBP1c were decreased with C18:0 compared to C22:6 n-3, while ChREBP protein abundance was not affected by C18:0. The findings presented here suggest that transcriptional and translational levels of bovine PNPLA3 are differentially regulated, and similar to human PNPLA3, bovine PNPLA3 protein may be regulated by SREBP1c20,24,25 although further research is needed to elucidate the direct role of SREBP1c on PNPLA3 abundance in bovine.
Although the rate limiting lipase, ATGL decreased quadratically with ACCUM and increased quadratically with RECOV, its coactivtor, ABHD526, was not affected by FA mixture nor concentration. This finding suggests that the role ATGL has in FLS within hepatocytes may not be as prominent as the role it has in lipolysis in adipose tissue27–29. This is further supported by the lack of change we observed in hepatocyte ATGL abundance during PNPLA3 knockdown in bovine hepatocytes12. Even in rodents, knockout of ATGL in hepatocytes reduces, but does not dissipate, TG hydrolysis because other lipases compensate for the lack of ATGL29. Across species, the working hypothesis is that PLIN encapsulates the lipid droplet and when the AMPK pathway is activated, both HSL and PLIN are phosphorylated, allowing ATGL, ABHD5, and PHSL access to the lipid droplet26,28,30. Previous work focused on lipolysis in adipose tissue in dairy cows found that ATGL, PHSL, and PPLIN abundance were affected by the periparturient period but ABHD5, HSL, and PLIN were not28. This further implicates the role PNPLA3 has on FLS.
Across species, CPT1A is regulated by PPARA and transports FA into the mitochondria31 followed by transport into the mitochondrial matrix for β-oxidation by CPT2, which was recently identified as the rate-limiting step of β-oxidation in cancerous human cell lines32,33. Multiple bovine studies have conflicting evidence on if CPT1A expression varies over the periparturient period34 or remains constant31,35, where these studies focus on various treatment effects (i.e., excess dietary energy, nutritional intervention, etc.) and the effect of time on expression. In the current work, there was no response of CPT1A and CPT2 to ACCUM nor RECOV, although CPT2 tended to be responsive to iRECOV (Table 2). Previous work in dairy cows found that both CPT1A and CPT2 expression were both lower with hyperketonemia, a common peripartum metabolic disorder defined as high concentrations (≥ 1.2 mM) of circulating β-hydroxybutyrate, but expression was only measured at one time point during the study36. The lack of consistent evidence in the literature prevents us from drawing conclusions on CPT expression responsiveness in bovine.
Sirtuins are a family of genes involved in deacetylation that are well conserved across species37. Divided into 4 classes, SIRT1 and SIRT3 are categorized into class I, which primarily encompasses involvement in metabolic diseases and FA oxidation37. While SIRT1 is located in the nucleus and cytoplasm, the location of SIRT3 is in the mitochondria and cytoplasm37,38. A previous study in which dairy cows were identified as having FLS (defined as hepatic TG content of ≥ 5% wet weight) had lower SIRT3 expression in response to higher concentrations of circulating FA39. While this is a similar pattern observed in mice40, these findings are contrary to ours. It should be noted that SIRT3 can regulate PPARA in humans38. Both SIRT1 and PGC1A exhibited similar patterns of expression with administration of FA mixture and concentration. In human pluripotent stem cells, SIRT1 knockdown significantly decreased PGC1A, resulting in a decrease in lipolysis41. Previous work in mice determined that SIRT1 successfully induced complete FA oxidation via deacetylation of PGC1A with treatment of C18:1 n-942 and that deacetylation of PGC1A increases gluconeogenesis43. However, with PGC1A expression tending to be greater with RECOV, it is possible that this TF may regulate PNPLA3 protein in FLS (Fig. 4). Evidence in humans suggests that SIRT1-mediated pathways aided in resolving NAFLD by increasing NAD + in the liver, but knockdown of SIRT3 does not change liver status38. With SIRT1 and PGC1A having a quadratic response to ACCUM and RECOV but SIRT3 and PPARA having no response, it may be possible that the SIRT1-mediated pathway may aid in resolving FLS in bovine. More work on clarifying the relationships between SIRT1, PGC1A, and PNPLA3 mRNA and its resulting protein in FLS and primary bovine hepatocytes is warranted29,42,44.
The TF FOXO1 plays a role in insulin signaling, regulates lipid metabolism and gluconeogenesis in NAFLD patients, and regulates SREBP1c in mice45. Expression of both FOXO1 (Fig. 4) and UCP2 (Fig. 5), a gene involved in insulin signaling, had a quadratic relationship with ACCUM and RECOV, but the pattern of expression of both genes differed with RECOV. In vitro mouse adipocyte research observed that lower FOXO1 expression appeared to cause lower UCP2 expression46, and that in goat mammary epithelial cells, knockdown of FOXO1 increased both FASN expression and TG synthesis47. Fatty acid synthase is key in synthesizing FA, promotes lipid storage, and has previously been shown to be upregulated in NAFLD patients48. In our study, the quadratic relationship of FASN expression increased with RECOV, although FASN was not responsive to individual FA. Expression of PNPLA3 (Fig. 2) and of FOXO1 (Fig. 4) did not exhibit the same expression patterns with FA mixtures across concentrations. The relationships between these three genes and proteins are unclear and should be further explored.
Transcription factor LXRA regulates biochemical pathways involved in regulating inflammatory responses, glycolysis, and lipid metabolism22,49 with primary action in the liver, compared to the isoform LXRB which is ubiquitously expressed in tissues across species49,50. Downstream targets of LXRA are ChREBP and SREBP1c, both regulators of glycolysis and lipogenesis, respectively22,48, and known regulators of PNPLA3 in mice and humans. Surprisingly, although LXRA expression was responsive to iRECOV, ChREBP and SREBP1c expression were not; however, LXRA expression and SREBP1c abundance both decreased with C18:0 iRECOV treatment (Table 2, Fig. 7). Since LXRA expression, SREBP1c abundance, and PNPLA3 abundance exhibited similar patterns in response to iRECOV, this finding may further provide evidence that the LXRA-SREBP1c pathway is regulating PNPLA3 abundance.
Conclusions
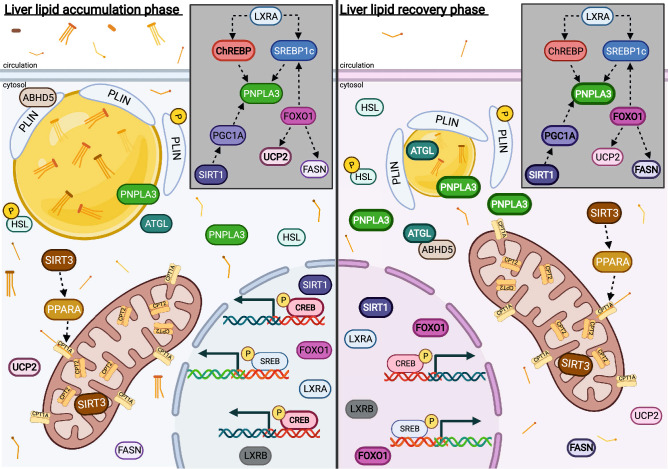
Bovine PNPLA3 appears to be differentially regulated at the transcriptional and translational level, and the importance of mimicking physiological states via FA mixtures rather than singular FA provides a more complete picture of regulation. Although bovine PNPLA3 abundance is clearly responsive to FA in a manner that is consistent between primary bovine hepatocytes herein, and previously published in vivo research, the relationships between the gene expression of SIRT1, PGC1A, SIRT3, PPARA, and PNPLA3 and its resulting protein remain unclear and warrant further investigation into which pathway aids in FLS recovery. A working hypothesis of associations between these key components is shown in Fig. 8, based on published research across species and the current data. Based on the current work, it is possible that PNPLA3 regulation is mediated by SREBP1c via LXRA, similar to PNPLA3 regulation in humans, due to the similar pattern of response to iRECOV. Further research elucidating the role of SREBP1c and LXRA on bovine PNPLA3 regulation could provide valuable insight leading to both bovine targeted interventions and potential use of bovine models to further understanding of human PNPLA3 and NAFLD.
Figure 8.
Working hypothesis of lipid related genes and proteins in primary bovine hepatocytes during the liver lipid accumulation phase (left panel) and the liver lipid recovery phase (right panel). The gray box insert within each panel represents the associations between PNPLA3, transcription factors, and genes where potential for regulatory influence exists. Relative abundance between panels are shown visually with line weight and quantity. Arrows between genes and proteins indicate associative relationships based on published research across species and the current data and do not denote directionality of regulation. Created with BioRendeder.com.
Materials and methods
All animal use and handling protocols were approved by the University of Wisconsin-Madison College of Agricultural and Life Sciences Animal Care and Use Committee and were carried out in accordance with relevant guidelines and regulations. Materials and methods presented follow the ARRIVE guidelines.
Isolation and cell culture
Primary bovine hepatocytes were isolated from 4 Holstein bull calves (5 ± 2 days; calf as biological replicate) as described previously12 and all hepatocyte isolation preparations were used in this experiment (n = 4). The caudate process was excised and perfused using collagenase at 37° C and hepatocytes were isolated as previously described51–53. Cells were plated on 35-mm tissue treated dishes (Eppendorf, Hauppauge, NY) in sterile Dulbecco’s Modified Eagle’s Medium (D2902, Sigma-Aldrich, St. Louis, MO) with added cell culture grade HEPES and sodium bicarbonate (DMEM; Sigma-Aldrich) supplemented with 20% fetal bovine serum and 1% antibiotic, antimycotic solution (Sigma-Aldrich) at a density of approximately 2 million cells per dish. Media was refreshed with sterile DMEM supplemented with 10% fetal bovine serum and 1% antibiotic, antimycotic solution 4 h after initial plating. Cells were maintained in monolayer cultures for 24 h and were at least 80% confluent.
Fatty acid treatments
Wells were randomly assigned to treatment in triplicate. Treatment media comprised of sterile DMEM with the addition of 1% bovine serum albumin (BSA; EMD Millipore, Burlington, MA) and 1% antibiotic, antimycotic solution plus either individual FA (iRECOV) or FA mixtures. Cell culture grade C14:0, C16:0, C18:0, C18:1 n-9, C18:2 n-6, C20:5 n-3, and C22:6 n-3 were independently bound to BSA (9% solution) to achieve individual 8 mM stock FA solutions as described previously54. Two FA mixtures were comprised of varying percentages of each stock FA (Table 1) to represent circulating serum FA in vivo55 reflective of two distinct postpartum periods (1) accumulation of liver lipids (ACCUM) and (2) recovery from fatty liver (RECOV). Cells were treated with 0.25, 0.5, 0.75, or 1 mM of ACCUM or RECOV, respectively.
RNA isolation and RT-qPCR analysis
For genes with known strong correlations between expression and protein abundance50,56–58, only gene expression was quantified. An unabridged version of RNA isolation and real-time qPCR (RT-qPCR) methods has been published previously59. As described therein, cells were harvested in TRIzol reagent (Invitrogen, Carlsbad, CA), RNA extracted using a phenol–chloroform extraction60 (Life Technologies), and RNA pooled and purified using the Aurum Total RNA 96 Kit61 (732–6800; Bio-Rad Laboratories, Hercules, CA). Quantified (Synergy H1 Hybrid Spectrophotometer; BioTek, Winooski, VT) and quality assured (ratio absorbances between 1.9 and 2.152,59) RNA samples were reverse transcribed to cDNA using 0.5 µg RNA and iScript Reverse Transcription Supermix (170–8840; Bio-Rad Laboratories) in a C1000 Touch Thermo Cycler (Bio-Rad Laboratories). Standard curves and controls were as described previously59 and cDNA samples were diluted 1:20 for qPCR.
Expression of genes ChREBP, CPT1A, CPT2, FASN, FOXO1, LXRA, LXRB, PGC1A, PNPLA3, PPARA, SIRT1, SIRT3, SREBP1c, and UCP2 was quantified using RT-qPCR in a CFX-384 Real-Time System (Bio-Rad Laboratories, Hercules, CA) with SsoAdvanced SYBR (172–5270, Bio-Rad Laboratories). Gene expression of reference genes ribosomal 18S (18S), glyceraldehyde 3-phosphate dehydrogenase (GAPDH), ribosomal protein L32 (RPL32), and ribosomal protein S9 (RPS9) were quantified as described above and the stability of reference gene normalization using the geometric mean of all four reference genes (M = 0.06) instead of a single gene or other gene combination (M ≥ 0.09) was confirmed via NormFinder62. All primers (Table 3) were optimized and either validated previously or verified within as the single product using melt-peak analysis. Standards, controls, and samples were amplified in triplicate for all genes as follows: 1 cycle at 95° C for 3 min, 45 cycles of 95° C for 15 s and 55° C for 5 s, then a melt curve from 65° C to 95° C at increasing increments of 0.5° C for 3 s. Primers for PNPLA3, SIRT3, and SREBP1c used annealing temperatures of 55.3, 54.3, and 58.4° C instead of 55° C, respectively.
Table 3.
Primers used for real time-quantitative PCR organized by function: reference, fatty acid (FA)-mediated, and transcription factor (TFs) genes.
| Gene1 | Accession no. | Position2 | Sequence (5ʹ–3ʹ) | Source |
|---|---|---|---|---|
| References | ||||
| 18S | NR_036642.1 | F | ACCCATTCGAACGTCTGCCCTATT | 52 |
| R | TCCTTGGATGTGGTAGCCGTTTCT | |||
| GAPDH | NM_001034034.2 | F | AAGGTCGGAGTGAACGGATTC | 65 |
| R | ATGGCGACGATGTCCACTTT | |||
| RPL32 | NM_001034783.2 | F | AGACCCCTCGTGAAGCCTAA | 65 |
| R | CCGCCAGTTCCGCTTGATTT | |||
| RPS9 | NM_001101152.2 | F | CCTCGACCAAGAGCTGAA | 53 |
| R | CCTCCAGACCTCACGTTT | |||
| FA-mediated | ||||
| CPT1A | NM_001304989.1 | F | TTCGCGATGGACTTGCTGTA | 35 |
| R | TTTCCTCCCGGTCCAGTTTG | |||
| CPT2 | NM_001045889.1 | F | TGAACATCCTCTCCATCTGG | 39 |
| R | GGTCAACAGCAACTACTACG | |||
| FASN | NM_001012669.1 | F | CAGCTTTGTGTTGGCAGAGAAG | 39 |
| R | AGCGAGCTGTCCAGGTTGAC | |||
| PNPLA3 | XM_005207459.4 | F | ACGAAGGGTTCACCAAGCTC | 12 |
| R | GCATTAACAGCGACCGGAAC | |||
| UCP2 | NM_001033611.2 | F | ACCTAACACAGCCGGTCTC | Verified within |
| R | GCAACCTCCCAGGAGATGAA | |||
| TFs | ||||
| ChREBP | TC350579 | F | GCTGCAGCATGGCAATGTACT | 66 |
| R | GGAGCAGAAGAGGCGTTCAA | |||
| FOXO1 | XM_025000053.1 | F | AAGCGAGCAAGCAGGCTACA | Verified within |
| R | GGACTGCTTCTCTCAGTTCCTG | |||
| LXRA | NM_001014861.1 | F | CATGCCTACGTCTCCATCCA | 66 |
| R | TCACCAGTTTCATCAGCATCCT | |||
| LXRB | NM_001014883.1 | F | TGCAGCTCGGTCGTGAAG | 66 |
| R | CAGCAGCATGATCTCGATGGT | |||
| PGC1A | NM_177945.3 | F | CAGAAGGCAATTGAAGAGCG | Verified within |
| R | CCTCAGTTCTGTCCGTGTTGT | |||
| PPARA | NM_001034036.1 | F | ACAAAGCCTCTGGCTACCAC | 31 |
| R | AGCTTCAGCCGAATCGTTCT | |||
| SIRT1 | NM_001192980.3 | F | AAGACCTCGGATAGGTCCAT | Verified within |
| R | GAATTGTTCGAGGATCTGTGCC | |||
| SIRT3 | XM_005200933.4 | F | CAACCTGGAGAAATACCGTCTT | 67 |
| R | CAGTCCTTTTTCCTTCAGCAG | |||
| SREBP1c | NM_001113302.1 | F | CGACACCACCAGCATCAACCACG | 68 |
| R | GCAGCCCATTCATCAGCCAGACC | |||
Protein isolation and Western Blot analysis
For genes subject to translational regulation, protein abundance was quantified28,29. Molecular grade ethanol was added to the phenol-intermediate phase for precipitation of DNA and tubes were centrifuged at 2000×g for 5 min at 4 °C and supernatant saved for protein isolation via ethanol addition per the manufacturer’s protocol (Life Technologies). The resulting protein pellet was re-suspended in a modified lysis buffer63 with addition of Halt Protease Inhibitor Cocktail (78445; Pierce BioTechnology, ThermoFisher, Rockford, IL) and then warmed at 55 °C until the pellet solubilized as described previously63. Samples were centrifuged at 10,000×g for 10 min at 4 °C to remove impurities (Life Technologies), and supernatants collected and pooled across technical replicates. As done previously12, a protein pool comprised of equal volumes of samples was created as a quality control standard for downstream analysis via Western Blot, herein referred to as the pool.
Protein concentration of samples and the pool was determined by BCA assay per the manufacturer’s protocol (23227, Pierce, ThermoFisher, Rockford, IL). Any samples that did not fall within the standard curve were concentrated using molecular weight concentrators following the manufacturer’s protocol (88502; Pierce, ThermoFisher, Rockford, IL) and samples re-analyzed via BCA assay to determine concentration with coefficient of variations never exceeding 10%.
Samples were prepared for Western Blot analysis as previously described12,64. All proteins of interest were probed on Western Blots loaded with 25 µg of protein per lane and were heated at 37 °C for 30 min, except for ChREBP which was analyzed on Western Blots loaded with 50 µg of protein per lane and were heated at 100 °C for 5 min. After blocking, blots were probed with primary antibody for 1 h (ABHD5, ab59488, Abcam, Cambridge, MA; PHSL, 4139S, Cell Signaling Technologies, Danvers, MA; PLIN and PPLIN, AB10200, EMD Millipore Sigma, Darmstadt, Germany; PNPLA3, ab81874, Abcam; SREBP1c, LSB-93, LS-Bio, Seattle, WA) or overnight at 4 °C (ATGL, ab99532, Abcam; ChREBP, sc-515922, Santa Cruz Biotechnologies; HSL; 4107S, Cell Signaling Technologies; SREBP1c, LS-93; Life Science Biology). Before blocking and probing for ChREBP abundance, blots were blocked with an endogenous biotin blocking kit (E-21390; Thermo Fisher Scientific) for 20 min for each reagent and diluted in extra-pure water.
Hepatocyte imaging of PNPLA3
Bovine neonatal hepatocytes were stained with DAPI (MP01306; Invitrogen, ThermoFisher Scientific) and Alexa Fluor 488 Phalloidin (A12379; Invitrogen, ThermoFisher Scientific) for nuclei and F-actin staining, respectively, following the manufacturer’s protocols (Invitrogen, ThermoFisher Scientific). Cells were then exposed to incubation with the primary antibody used for Western Blot analysis for PNPLA3 but at a 1:400 dilution (ab81874, Abcam). After primary incubation overnight at room temperature, a secondary anti-rabbit Cy3 antibody (711-165-152, Jackson Research Laboratories, West Grove, PA) was administered at 1:100 for a 2 h incubation period. Images were taken on each respective channel, the three images merged, and the final image was altered minimally to reduce background noise.
Statistical analysis
Data was checked for normality using PROC UNIVARIATE in SAS 9.4 (SAS Institute Inc., Cary, NC) and transformed, when necessary, based on Shapiro-Wilks. Normality was achieved using root transformation, or square root of the multiplicative inverse, and the appropriate transformation determined and applied on an individual gene or protein basis. Data was analyzed using PROC MIXED (SAS 9.4). Separate mixed models were built for FA mixture treatments and iRECOV treatments. For FA mixture treatments, models contained the fixed effect of FA mixture treatment, concentration, their interaction, and random effect of calf. For iRECOV treatments, models contained the fixed effect of individual FA and random effect of calf. Significance was declared at P ≤ 0.05 and tendencies at 0.05 < P ≤ 0.10. If a main effect of FA mixture, concentration, or the interaction for FA mixture and concentration treatments for either gene expression or protein abundance was P ≤ 0.15, quadratic relationships were explored. When the interaction of FA mixture × concentration was significant, means were separated by Tukey–Kramer adjustment. All data reported are least square means ± standard error of the mean (LSM ± SEM).
Acknowledgements
The authors acknowledge the support of D. Rieman, manager at the UW-Madison Dairy Cattle Center, and the UW-Madison Dairy Cattle Instruction and Research Center staff (University of Wisconsin-Madison, Madison, WI). Additionally, the authors would like to acknowledge S. J. Bertics, R. S. Pralle, R. Caputo Oliveira, K. A. Weld, and H. T. Holdorf for their expertise, experience, and support. This project was supported by the Agricultural Food Research Initiative of the National Institute of Food and Agriculture, USDA, Grant # 2016-67015-24573 and the USDA Hatch Grant # WIS01877.
Author contributions
S.J.E., T.L.C., and H.M.W executed in vitro experiments. T.L.C. generated Fig. 1. S.J.E. analyzed samples and data included in all other figures and tables. S.J.E. and H.M.W. responsible for data analysis and writing. All authors reviewed the manuscript. H.M.W. secured funding for the research.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Grummer RR. Etiology of lipid-related metabolic disorders in periparturient dairy cows. J. Dairy Sci. 1993;76:3882–3896. doi: 10.3168/jds.S0022-0302(93)77729-2. [DOI] [PubMed] [Google Scholar]
- 2.White, H. M. Invited Review: Foundation Scholar Review. Influencing hepatic metabolism: can nutrient partitioning be modulated to optimize metabolic health in the transition dairy cow? J. Dairy Sci. 103, 6741–6750 (2020). [DOI] [PubMed]
- 3.Contreras GA, Strieder-Barboza C, Raphael W. Adipose tissue lipolysis and remodeling during the transition period of dairy cows. J. Anim. Sci. Biotechnol. 2017;8:41–52. doi: 10.1186/s40104-017-0174-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.White H. The role of TCA cycle anaplerosis in ketosis and fatty liver in periparturient dairy cows. Animals. 2015;5:793–802. doi: 10.3390/ani5030384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bobe G, Young JW, Beitz DC. Invited review: pathology, etiology, prevention, and treatment of fatty liver in dairy cows. J. of Dairy Sci. 2004;87:3105–3124. doi: 10.3168/jds.S0022-0302(04)73446-3. [DOI] [PubMed] [Google Scholar]
- 6.Glover J, Duthie DW. The apparent digestibility of crude protein by non-ruminants and ruminants. J. Agric. Sci. 1958;51:289–293. [Google Scholar]
- 7.Keys JE, Jr, Van Soest PJ, Young EP. Comparative study of the digestibility of forage cellulose and hemicellulose in ruminants and nonruminants. J. Anim. Sci. 1969;29:11–15. doi: 10.2527/jas1969.29111x. [DOI] [PubMed] [Google Scholar]
- 8.Mosher GA, Krishnamurti CR, Kitts WD. Metabolism of the toxic principle of astragalas serotinas in ruminants and nonruminants. Can. J. Anim. Sci. 1971;51:475–480. [Google Scholar]
- 9.Huang Y, Cohen JC, Hobbs HH. Expression and characterization of a PNPLA3 protein isoform (I148M) associated with nonalcoholic fatty liver disease. J. Biol. Chem. 2011;286:37085–37093. doi: 10.1074/jbc.M111.290114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bruschi FV, Tardelli M, Herac M, Claudel T, Trauner M. Metabolic regulation of hepatic PNPLA3 expression and severity of liver fibrosis in patients with NASH. Liver Int. 2020;40:1098–1110. doi: 10.1111/liv.14402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Speliotes EK, Butler JL, Palmer CD, Voight BF, Hirschhorn JN. PNPLA3 variants specifically confer increased risk for histologic nonalcoholic fatty liver disease but not metabolic disease. Hepatology. 2010;52:904–912. doi: 10.1002/hep.23768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pralle RS, Erb SJ, Holdorf HT, White HM. Greater liver PNPLA3 protein abundance in vivo and in vitro supports lower triglyceride accumulation in dairy cows. Sci. Rep. 2021;11:2839. doi: 10.1038/s41598-021-82233-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Basantani MK, et al. Pnpla3/Adiponutrin deficiency in mice does not contribute to fatty liver disease or metabolic syndrome. J. Lipid Resear. 2011;52:318–329. doi: 10.1194/jlr.M011205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.BasuRay S, Wang Y, Smagris E, Cohen JC, Hobbs HH. Accumulation of PNPLA3 on lipid droplets is the basis of associated hepatic steatosis. Proc. Natl. Acad. Sci. 2019;116:201901974. doi: 10.1073/pnas.1901974116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smagris E, et al. Pnpla3I148M knockin mice accumulate PNPLA3 on lipid droplets and develop hepatic steatosis. Hepat. Baltim. Md. 2014;61:108–118. doi: 10.1002/hep.27242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weld, K. A., Oliveira, R. C., Bertics, S. J., Erb, S. J. & White, H. M. Hepatic pyruvate carboxylase expression differed prior to hyperketonemia onset in transition dairy cows. Plos One 15, e0241929 (2020). [DOI] [PMC free article] [PubMed]
- 17.Rukkwamsuk T, Kruip TA, Meijer GA, Wensing T. Hepatic fatty acid composition in periparturient dairy cows with fatty liver induced by intake of a high energy diet in the dry period. J. Dairy Sci. 1999;82:280–287. doi: 10.3168/jds.S0022-0302(99)75234-3. [DOI] [PubMed] [Google Scholar]
- 18.White HM, Koser SL, Donkin SS. Characterization of bovine pyruvate carboxylase promoter 1 responsiveness to serum from control and feed-restricted cows. J. Anim. Sci. 2011;89:1763–1768. doi: 10.2527/jas.2010-3407. [DOI] [PubMed] [Google Scholar]
- 19.White HM, Koser SL, Donkin SS. Gluconeogenic enzymes are differentially regulated by fatty acid cocktails in Madin-Darby bovine kidney cells. J. Dairy Sci. 2012;95:1249–1256. doi: 10.3168/jds.2011-4644. [DOI] [PubMed] [Google Scholar]
- 20.Huang Y, et al. A feed-forward loop amplifies nutritional regulation of PNPLA3. Proceed. Natl. Acad. Sci. USA. 2010;107:7892–7897. doi: 10.1073/pnas.1003585107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McCann CC, Viner ME, Donkin SS, White HM. Hepatic patatin-like phospholipase domain-containing protein 3 sequence, single nucleotide polymorphism presence, protein confirmation, and responsiveness to energy balance in dairy cows. J. Dairy Sci. 2014;97:5167–5175. doi: 10.3168/jds.2014-7910. [DOI] [PubMed] [Google Scholar]
- 22.Iizuka K, Horikawa Y. ChREBP: A glucose-activated transcription factor involved in the development of metabolic syndrome. Endocr. J. 2008;55:617–624. doi: 10.1507/endocrj.k07e-110. [DOI] [PubMed] [Google Scholar]
- 23.Ferré P, Foufelle F. Hepatic steatosis: A role for de novo lipogenesis and the transcription factor SREBP-1c. Diabet. Obes. Met. 2010;12:83–92. doi: 10.1111/j.1463-1326.2010.01275.x. [DOI] [PubMed] [Google Scholar]
- 24.Dubuquoy C, et al. Distinct regulation of adiponutrin/PNPLA3 gene expression by the transcription factors ChREBP and SREBP1c in mouse and human hepatocytes. J. Hepat. 2011;55:145–153. doi: 10.1016/j.jhep.2010.10.024. [DOI] [PubMed] [Google Scholar]
- 25.Liang H, et al. The SRE motif in the human PNPLA3 promoter (−97 to −88 bp) mediates transactivational effects of SREBP-1c. J. Cell. Physiol. 2015;230:2224–2232. doi: 10.1002/jcp.24951. [DOI] [PubMed] [Google Scholar]
- 26.Yu L, Li Y, Grisé A, Wang H. Lipid transfer in lipoprotein metabolism and cardiovascular disease. Adv. Exp. Med. Biol. 2020;1276:197–222. doi: 10.1007/978-981-15-6082-8_13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koltes DA, Spurlock ME, Spurlock DM. Adipose triglyceride lipase protein abundance and translocation to the lipid droplet increase during leptin-induced lipolysis in bovine adipocytes. Domest. Anim. Endo. 2017;61:62–76. doi: 10.1016/j.domaniend.2017.06.001. [DOI] [PubMed] [Google Scholar]
- 28.Koltes DA, Spurlock DM. Coordination of lipid droplet-associated proteins during the transition period of Holstein dairy cows. J. Dairy Sci. 2011;94:1839–1848. doi: 10.3168/jds.2010-3769. [DOI] [PubMed] [Google Scholar]
- 29.Schreiber, R., Xie, H. & Schweiger, M. Of mice and men: The physiological role of adipose triglyceride lipase (ATGL). Biochim. Biophys. Acta BBA Mol. Cell Biol. Lipids.1864, 880–899 (2019). [DOI] [PMC free article] [PubMed]
- 30.Wang S, et al. Lipolysis and the integrated physiology of lipid energy metabolism. Mol. Genet. Metab. 2008;95:117–126. doi: 10.1016/j.ymgme.2008.06.012. [DOI] [PubMed] [Google Scholar]
- 31.Caprarulo V, et al. The effects of prepartum energy intake and peripartum rumen-protected choline supplementation on hepatic genes involved in glucose and lipid metabolism. J. Dairy Sci. 2020;103:11439–11448. doi: 10.3168/jds.2020-18840. [DOI] [PubMed] [Google Scholar]
- 32.Zhang X, et al. CPT2 down-regulation promotes tumor growth and metastasis through inducing ROS/NFκB pathway in ovarian cancer. Transl. Oncol. 2021;14:1023. doi: 10.1016/j.tranon.2021.101023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lin M, et al. Downregulation of CPT2 promotes tumorigenesis and chemoresistance to cisplatin in hepatocellular carcinoma. Oncotargets Ther. 2018;11:3101–3110. doi: 10.2147/OTT.S163266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Khan MJ, et al. Overfeeding dairy cattle during late-pregnancy alters hepatic PPARα-regulated pathways including hepatokines: Impact on metabolism and peripheral insulin sensitivity. Gene Reg. Syst. Biol. 2014;8:97–111. doi: 10.4137/GRSB.S14116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chandler TL, White HM. Choline and methionine differentially alter methyl carbon metabolism in bovine neonatal hepatocytes. PLoS ONE. 2017;12:1080. doi: 10.1371/journal.pone.0171080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li P, et al. Alterations of fatty acid β-oxidation capability in the liver of ketotic cows. J. Dairy Sci. 2012;95:1759–1766. doi: 10.3168/jds.2011-4580. [DOI] [PubMed] [Google Scholar]
- 37.Zhang J, et al. Mitochondrial sirtuin 3: New emerging biological function and therapeutic target. Theranostics. 2020;10:8315–8342. doi: 10.7150/thno.45922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dall M, Hassing AS, Treebak JT. NAD+ and NAFLD– caution, causality and careful optimism. J. Physiol. 2021;1:1–20. doi: 10.1113/JP280908. [DOI] [PubMed] [Google Scholar]
- 39.Liu L, et al. Sirtuin 3 improves fatty acid metabolism in response to high nonesterified fatty acids in calf hepatocytes by modulating gene expression. J. Dairy Sci. 2020;103:6557–6568. doi: 10.3168/jds.2019-17670. [DOI] [PubMed] [Google Scholar]
- 40.Kendrick AA, et al. Fatty liver is associated with reduced SIRT3 activity and mitochondrial protein hyperacetylation. Biochem. J. 2011;433:505–514. doi: 10.1042/BJ20100791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.l’Hortet, A. C. de et al. Generation of human fatty livers using custom-engineered induced pluripotent stem cells with modifiable SIRT1 metabolism. Cell Metab. 30, 385–401 (2019). [DOI] [PMC free article] [PubMed]
- 42.Lim J-H, et al. Oleic acid stimulates complete oxidation of fatty acids through protein kinase A-dependent activation of SIRT1-PGC1α Complex. J. Biol. Chem. 2013;288:7117–7126. doi: 10.1074/jbc.M112.415729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang T, Fu M, Pestell R, Sauve AA. SIRT1 and endocrine signaling. Trends Endocrinol. Metab. 2006;17:186–191. doi: 10.1016/j.tem.2006.04.002. [DOI] [PubMed] [Google Scholar]
- 44.Lin Y-C, Chang P-F, Chang M-H, Ni Y-H. A common variant in the peroxisome proliferator-activated receptor-γ coactivator-1α gene is associated with nonalcoholic fatty liver disease in obese children. Amer. J. Clin. Nutr. 2012;97:326–331. doi: 10.3945/ajcn.112.046417. [DOI] [PubMed] [Google Scholar]
- 45.Deng X, et al. FoxO1 inhibits sterol regulatory element-binding protein-1c (SREBP-1c) gene expression via transcription factors Sp1 and SREBP-1c. J. Biol. Chem. 2012;287:20132–20143. doi: 10.1074/jbc.M112.347211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu L, et al. FoxO1 interacts with transcription factor EB and differentially regulates mitochondrial uncoupling proteins via autophagy in adipocytes. Cell Death Discov. 2016;2:16066. doi: 10.1038/cddiscovery.2016.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.He Q, et al. FoxO1 knockdown promotes fatty acid synthesis via modulating SREBP1 activities in the dairy goat mammary epithelial cells. J. Agr. Food Chem. 2020;68:12067–12078. doi: 10.1021/acs.jafc.0c05237. [DOI] [PubMed] [Google Scholar]
- 48.Higuchi N, et al. Liver X receptor in cooperation with SREBP-1c is a major lipid synthesis regulator in nonalcoholic fatty liver disease. Hepatol. Res. 2008;38:1122–1129. doi: 10.1111/j.1872-034X.2008.00382.x. [DOI] [PubMed] [Google Scholar]
- 49.Patel M, Oza N, Anand I, Deshpande S, Patel C. Liver X Receptor: A novel therapeutic target. Indian J. Pharm. Sci. 2008;70:135–144. doi: 10.4103/0250-474X.41445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cariello M, Piccinin E, Moschetta A. Transcriptional regulation of metabolic pathways via lipid-sensing nuclear receptors. Cell Mol. Gastroent. Hepat. 2021;11:1519–1539. doi: 10.1016/j.jcmgh.2021.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Donkin SS, Armentano LE. Preparation of extended in vitro cultures of bovine hepatocytes that are hormonally responsive. J. Anim. Sci. 1993;71:2218–2227. doi: 10.2527/1993.7182218x. [DOI] [PubMed] [Google Scholar]
- 52.Chandler TL, White HM. Glucose metabolism is differentially altered by choline and methionine in bovine neonatal hepatocytes. PLoS ONE. 2019;14:e0217160–e217217. doi: 10.1371/journal.pone.0217160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang, Q. & White, H. M. Regulation of inflammation, antioxidant production, and methyl-carbon metabolism during methionine supplementation in lipopolysaccharide-challenged neonatal bovine hepatocytes. J. Dairy Sci. 1–13 (2017). [DOI] [PubMed]
- 54.Berry, M. N., Edwards, A. M. & Barritt, G. J. Isolated hepatocytes: Preparation, properties and applications (1991).
- 55.Rukkwamsuk T, Geelen MJ, Kruip TA, Wensing T. Interrelation of fatty acid composition in adipose tissue, serum, and liver of dairy cows during the development of fatty liver postpartum. J. Dairy Sci. 2000;83:52–59. doi: 10.3168/jds.S0022-0302(00)74854-5. [DOI] [PubMed] [Google Scholar]
- 56.ijcep0003-0505.pdf.
- 57.Yamazaki, M. et al. Maternal fructose consumption down-regulates LXRa expression via miR-206-mediated regulation. J. Nutr. Biochem.82, 108386 (2020). [DOI] [PubMed]
- 58.Whitney KD, et al. Liver X Receptor (LXR) regulation of the LXRα gene in human macrophages. J. Biol. Chem. 2001;276:43509–43515. doi: 10.1074/jbc.M106155200. [DOI] [PubMed] [Google Scholar]
- 59.Oliveira RC, et al. Postpartum supplementation with fermented ammoniated condensed whey altered nutrient partitioning to support hepatic metabolism. J. Dairy Sci. 2020;103:7055–7067. doi: 10.3168/jds.2019-17790. [DOI] [PubMed] [Google Scholar]
- 60.Chomczynski P. A reagent for the single-step simultaneous isolation of RNA, DNA and proteins from cell and tissue samples. Biotechniques. 1993;15:532–536. [PubMed] [Google Scholar]
- 61.Miller A, Spagnuolo RA, Baras V, Pyles R. High-throughput automated extraction of RNA using the AurumTM Total RNA 96 kit. BioRad. Tech Note. 2010;6018:1–6. [Google Scholar]
- 62.Andersen CL, Jensen JL, Ørntoft TF. Normalization of real-time quantitative reverse transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004;64:5245–5250. doi: 10.1158/0008-5472.CAN-04-0496. [DOI] [PubMed] [Google Scholar]
- 63.Kopec AM, Rivera PD, Lacagnina MJ, Hanamsagar R, Bilbo SD. Optimized solubilization of TRIzol-precipitated protein permits Western blotting analysis to maximize data available from brain tissue. J. Neurosci. Meth. 2017;280:64–76. doi: 10.1016/j.jneumeth.2017.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Caputo Oliveira, R. et al. Postpartum fermented ammoniated condensed whey supplementation altered nutrient partitioning to support hepatic metabolism. J. Dairy Sci. (2020). [DOI] [PubMed]
- 65.Weld KA, Erb SJ, White HM. Short communication: Effect of manipulating fatty acid profile on gluconeogenic gene expression in bovine primary hepatocytes. J. Dairy Sci. 2019;102:7576–7582. doi: 10.3168/jds.2018-16150. [DOI] [PubMed] [Google Scholar]
- 66.Harvatine KJ, Boisclair YR, Bauman DE. Liver x receptors stimulate lipogenesis in bovine mammary epithelial cell culture but do not appear to be involved in diet-induced milk fat depression in cows. Physiol. Rep. 2014;2:266. doi: 10.1002/phy2.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gui L, Hong J, Raza SHA, Zan L. Genetic variants in SIRT3 transcriptional regulatory region affect promoter activity and fat deposition in three cattle breeds. Mol. Cell. Probe. 2017;32:40–45. doi: 10.1016/j.mcp.2016.12.002. [DOI] [PubMed] [Google Scholar]
- 68.Li X, et al. Insulin suppresses the AMPK signaling pathway to regulate lipid metabolism in primary cultured hepatocytes of dairy cows. J. Dairy Res. 2018;85:157–162. doi: 10.1017/S002202991800016X. [DOI] [PubMed] [Google Scholar]