Dear Editor,
Programmed cell death protein 1 (PD-1; encoded by the PDCD1 gene), mostly expressed on activated T cells, is an important receptor in T-cell immunity.1–3 Together with programmed death-ligand 1 (PD-L1: encoded by the CD274 gene) PD-1 acts as an inhibitor of T cell activity under normal conditions. In addition to T-cells PD-1 is abundantly expresses in some of B-cell malignancies, including Hodgkin’s Lymphoma and Follicular lymphoma.1–3 PD-1 expression was also detected in CLL.1 Notably, circulating T-cells in CLL patients express higher PD-1 levels than T-cells of healthy donors.1–3 Interestingly, various types of cancer express high levels of PD-L1 and are able to use PD-1/PD-L1 signaling to evade T cell immunity.1–3 Moreover, interruption of immune surveillance promotes cancer cell survival by exploiting PD-1/PD-L1 signaling.1–3 In recent years, many therapeutic antibodies against PD-1 and PD-L1 have been developed and have demonstrated promising results in clinical trials for various types of cancer.1–3 The highest response rate to PD-1 blockade was achieved in classical Hodgkin lymphoma.1 In recent years anti PD-1 antibodies (Opdivo and Keytruda), as well as anti PD-L1 antibodies (Tecentric and Imfinzi) disrupting PD-1–PD-L1 interaction, were FDA approved for treatment of a number of cancers. These include melanoma, kidney cancer, bladder cancer, lung cancer, Hodgkin’s lymphoma and others.1–3 miR-15/16 is a key tumor suppressor microRNA cluster first identified as a target of 13q deletions in CLL.4 A recent report demonstrated that miR-16 regulates PD-L1 expression in prostate and other cancers.5 miR-15/16 are the first tumor suppressor microRNAs identified and alterations in miR-15/16 provided the first evidence of the involvement of noncoding RNAs in cancer pathogenesis.4 Loss of miR-15/16 is the most common genetic lesion in chronic lymphocytic leukemia, promoting overexpression of BCL2, resulting in leukemia.4 In addition to BCL2, miR-15/16 cluster targets multiple oncogenes, including ROR1, Cyclin D1, and others. Thus, miR-15/16 play an important role in many blood malignancies and solid cancers.4 For example, miR-15/16 inhibit tumor progression by directly targeting MYCN in neuroblastoma; miR-15/16 inhibit hepatocellular carcinoma progression by targeting FEAT through NF-κB signaling pathway.
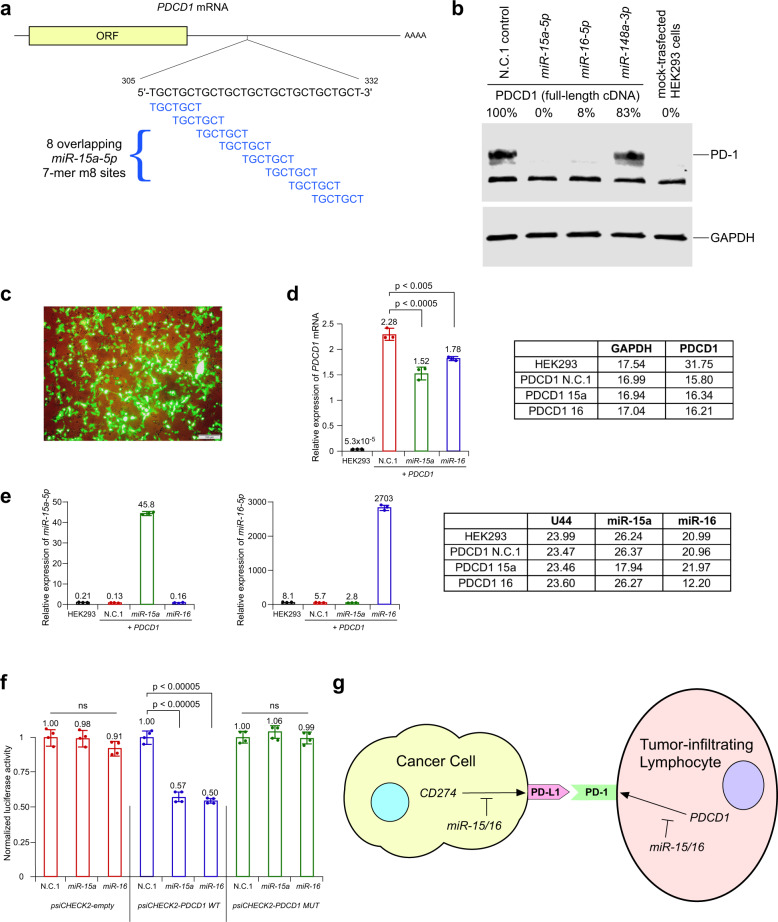
Since miR-15/16 target PD-L1 expression5 it is likely to regulate PD-1 – PD-L1 interaction. Thus, we thought that PDCD1 might be also regulated by miR-15/16. To determine if this is the case, we analyzed 3′ UTR of PDCD1 using TargetScan 7.2 software (http://www.targetscan.org). Remarkably we found that 3’ UTR of PDCD1 contains a 28 bp DNA fragment containing eight overlapping miR-15/16 target sites (exact match to positions 2–8 of the mature miR-15a-5p and miR-16-5p) (Fig. 1a). Thus, we proceeded to determine if miR-15/16 target PDCD1 expression. To address this, we first checked the effect of overexpressed mimics for miR-15a-5p and miR-16-5p on PD-1 in HEK293 cells. Since HEK293 cells do not express any detectable endogenous PDCD1, we co-transfected HEK293 cells with pCMV-PDCD1 (a mammalian expression vector containing full-length PDCD1 cDNA including 3’ UTR, obtained from OriGene), and set of pre-miRNA mimics from ThermoFisher (pre-miR negative control, pre-miR-15a-5p, pre-miR-16-5p and pre-miR-148a-3p). These results revealed that co-expression of PDCD1 and miR-15a-5p and miR-16-5p, but not miR-148a-3p (as a negative control) dramatically decreased PD-1 expression (Fig. 1b). In these experiments we routinely achieved ~70% transfection efficiency (Fig. 1c). To determine if these effects occur on translational level, we also measured the RNA expression of PDCD1, miR-15a, and miR-16 (Fig. 1d, e). We found that miR-15a and miR-16 expression decreased PDCD1 mRNA expression by 33 and 22%, respectively (Fig. 1d), while PDCD1 protein expression was decreased by 100 and 92% respectively (Fig. 1b). We concluded that miR-15a-5p and miR-16-5p target PDCD1 expression mostly at protein level. To determine whether PDCD1 is a direct target of miR-15a-5p and miR-16-5p, we performed luciferase reporter assays. HEK293 cells were co-transfected with constructs containing luciferase gene alone (psiCHECK2-empty) or fused with a 3’-UTR of PDCD1 containing a 28 bp DNA fragment including eight overlapping miR-15/16 target sites (psiCHECK2-PDCD1 WT) and miR negative control (N.C.1), miR-15a-5p or miR-16-5p mimics. In addition, we used psiCHECK2-PDCD1 MUT construct (psiCHECK2-PDCD1 WT, lacking 28 nt-long region that contains 8 overlapping possible binding sites for miR-15a-5p and miR-16-5p). Fig. 1f (left) shows that miR-15a-5p and miR-16-5p expression did not affect the luciferase activity of psiCHECK2 empty vector. On the other hand, miR-15a-5p and miR-16-5p expression significantly decreased the luciferase activity of the psiCHECK2-PDCD1 WT (Fig. 1f, middle). Co-transfecting miR-15a-5p and miR-16-5p and construct containing mutated form of PDCD1 3′-UTR (psiCHECK2-PDCD1 MUT) completely negated this effect (Fig. 1f, right). These results confirmed that miR-15a-5p and miR-16-5p bound directly and specifically to its target sites within the 3′-UTR of PDCD1.
Fig. 1.
miR-15/16 target PDCD1 (PD-1) expression. a A 28 bp DNA fragment containing eight overlapping miR-15/16 target sites in the 3′ UTR of PDCD1. b miR-15/16 inhibit PD-1 protein expression. miR-15a-5p and miR-16-5p expression suppress the PDCD1 expression by targeting its 3’-UTR. Western blot data showed the protein level of overexpressed PDCD1 in HEK293 cells co-transfected with scrambled pre-miR negative control, pre-mir-15a-5p, pre-miR-16-5p and pre-miR-148a-3p mimics. GAPDH served as a loading control. c An example of transformation efficiency in experiments in HEK293 cells for western blot and Luciferase assays (as shown in b, f). d, e Results of real-time PCR experiments on RNAs isolated from HEK293 cells co-transfected with PDCD1 construct and miR mimics (same as in b). d Results of real-time PCR using TaqMan probe for human PDCD1 transcript. Average CT numbers are shown (right). e Results of real-time PCR using TaqMan probes for human miR-15a-5p (left) and miR-16-5p (middle). Average CT numbers are shown (right). f PDCD1 is a direct target of miR-15/16. Renilla luciferase reporter assay showing the reporter expression in HEK293 cells co-transfected with wild-type 3′-UTR of PDCD1 and mutant 3′-UTR of PDCD1 along with scrambled negative control 1, miR-15a and miR-16 mimics. Renilla luciferase activity was normalized to firefly luciferase activity. The normalized luciferase activities in HEK293 cells transfected with different psiCHECK2 constructs and scrambled negative control 1 (pre-miR-N.C.1) were set at 1 and relative luciferase activities of HEK293 cells co-transfected with each psiCHECK2 construct and miR-15a or miR-16 mimics are shown. Two independent experiments were carried out in duplicates and data were presented as mean ± SD. g miR-15/16 are key regulators of PD-1–PD-L1 interaction
PD-1, together with its ligand PD-L1 functions as a negative regulator of T-cell response in immune system. PD-1–PD-L1 interaction is a critical mechanism utilized by many tumor types to avoid T-cell response.1–3 The disruption of this interaction was targeted by many drug companies. Several immunotherapy antibodies (such as Opdivo, Keytruda, Tecentric, and Imfinzi) disrupting this interaction were FDA approved in recent years for treatment of Hodgkin’s lymphoma, melanoma, lung cancer, kidney cancer, bladder and other cancers.1–3 Previously we identified miR-15/16, key tumor suppressor microRNAs, as targets of 13q deletions in CLL.4 Loss of miR-15/16 is the most common genetic lesion in CLL, promoting overexpression of BCL2 and causing CLL development. Since it was recently reported that miR-16 target PD-L1 expression5 and regulate PD-1–PD-L1 interaction, we thought that PDCD1 might be also be under miR-15/16 control. Here we identified a 28 bp DNA fragment containing eight overlapping miR-15/16 target sites in the 3’ UTR of PDCD1. Using luciferase assay and western blot analysis we demonstrated that miR-15/16 target PDCD1 expression. Since miR-15/16 regulate PD-1 and PD-L1 expression, our results suggest that miR-15/16 are critical in the regulation of PD-1–PD-L1 interaction, a critical mechanism utilized by malignant cells to avoid T-cell immunity (Fig. 1g). Restoration of miR-15/16 activity in both, T-cells and tumor cells can be a promising opportunity in cancer therapy.
Supplementary information
Acknowledgements
This work was supported by the R35CA197706 grant from the National Institutes of Health to CMC.
Author contributions
A.P., L.Ts., L.To., and Y.P. designed the study, performed the research, analyzed data, and wrote the manuscript; C.M.C. designed research, analyzed data, and wrote the manuscript.
Data availability
The datasets used and/or analyzed during this study are available from the corresponding author on reasonable request.
Competing interests
The authors have no conflicts of interest to declare. Carlo M. Croce is one of the Editors-in-Chief of Signal Transduction and Targeted Therapy, but he has not been involved in the process of the manuscript handling.
Contributor Information
Yuri Pekarsky, Email: Yuri.Pekarsky@osumc.edu.
Carlo M. Croce, Email: Carlo.Croce@osumc.edu
Supplementary information
The online version contains supplementary material available at 10.1038/s41392-021-00832-9.
References
- 1.Xu-Monette ZY, Zhou J, Young KH. PD-1 expression and clinical PD-1 blockade in B-cell lymphomas. Blood. 2018;131:68–83. doi: 10.1182/blood-2017-07-740993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berger KN, Pu JJ. PD-1 pathway and its clinical application: a 20 year journey after discovery of the complete human PD-1 gene. Gene. 2018;638:20–25. doi: 10.1016/j.gene.2017.09.050. [DOI] [PubMed] [Google Scholar]
- 3.Cha JH, Chan LC, Li CW, Hsu JL, Hung MC. Mechanisms controlling PD-L1 expression in cancer. Mol. Cell. 2019;76:359–370. doi: 10.1016/j.molcel.2019.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pekarsky Y, Balatti V, Croce CM. BCL2 and miR-15/16: from gene discovery to treatment. Cell Death Differ. 2018;25:21–26. doi: 10.1038/cdd.2017.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tao Z, et al. MiR-195/-16 family enhances radiotherapy via T cell activation in the tumor microenvironment by blocking the PD-L1 immune checkpoint. Cell Physiol. Biochem. 2018;48:801–814. doi: 10.1159/000491909. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during this study are available from the corresponding author on reasonable request.