Abstract
Human brain organoids are generated from three-dimensional (3D) cultures of human induced pluripotent stem cells and embryonic stem cells, which partially replicate the development and complexity of the human brain. Many methods have been used to characterize the structural and molecular phenotypes of human brain organoids. Further understanding the electrophysiological phenotypes of brain organoids requires advanced electrophysiological measurement technologies to achieve long-term stable 3D recording over the time course of the organoid development with single-cell, millisecond spatiotemporal resolution. In this review, first, we briefly introduce the development, generation, and applications of human brain organoids. We then discuss the conventional methods used for characterizing the morphological, genetic, and electrical properties of brain organoids. Next, we highlight the need for characterizing electrophysiological properties of brain organoids in a minimally invasive manner. In particular, we discuss recent advances in the multi-electrode array (MEA), 3D bioelectronics, and flexible bioelectronics and their applications in brain organoid electrophysiological measurement. In addition, we introduce the recently developed cyborg organoids platform as an emerging tool for the long-term stable 3D characterization of the brain organoids electrophysiology at high spatiotemporal resolution. Finally, we discuss the perspectives of new technologies that could achieve the high-throughput, multimodal characterizations from the same brain organoids.
Graphical Abstract
Introduction
Our understanding of brain development has been mainly derived from animal models1. While these models have provided significant insight into the functionality of brain tissues, they do not fully encapsulate the molecular, structural, genetic, and functional complexity of human brains. Our current knowledge of human brain development is largely based on the analysis of postmortem of pathological specimens2,3. Although they have provided important fundamental knowledge, these tools are not amenable to experimental manipulation in real-time over the time course of development.
Recently, the development of human stem cell-derived organoids has become a major technological advance and represents a bridge between traditional two-dimensional (2D) cell cultures and in vivo animal models4–8. For example, Lancaster et al. described a novel 3D tissue model namely cerebral organoids9, which are 3D structures generated from human pluripotent stem cells (hPSCs), capable of self-organizing to form discrete regions of the human brain10–12. Neurons within these 3D assemblies can generate action potentials, display excitatory and inhibitory postsynaptic currents, and exhibit spontaneous network activity as measured by calcium imaging and extracellular field potential recordings13–15. Recent work has found that organoids can generate rhythmic activity such as synchronized activities in the delta (1–4 Hz) and gamma frequency (100–400 Hz) ranges, suggesting that the human brain organoids can partially mimic the electrical functions of the human brain16. However, technological limitations have restricted simultaneous detection of electrical activity at a high spatiotemporal resolution to small numbers of neurons over a short time. Consequently, these technical shortcomings have hindered attempts to determine electrophysiological development, evolution, and maturation among large numbers of neurons across 3D brain organoids over the time course of development for the understanding of how they collectively synchronize with rhythmic activity and study of how pharmacologic perturbations affect these electrophysiologic parameters17.
This review highlights emerging bioelectronics, capable of investigating the evolution of neural electrophysiology during human brain organoid development. In this review, we will first discuss the development and applications of brain organoids and compare different conventional technologies used to monitor their development. We will then emphasize recent advances in bioelectronics that can provide electrical monitoring at single-cell single-spike spatiotemporal resolution. In particular, we will discuss recent 2D and 3D multielectrode arrays (MEAs) for continuous measurement of brain organoids, and 3D flexible electronics for the 3D mapping of brain organoids18–21. Finally, we will discuss the further advances in tissue-like electronics that can fully integrate with brain organoids through organogenesis, which may open up new opportunities to precisely monitor and control organoid-wide development in a long-term stable manner22.
Development and applications of brain organoids
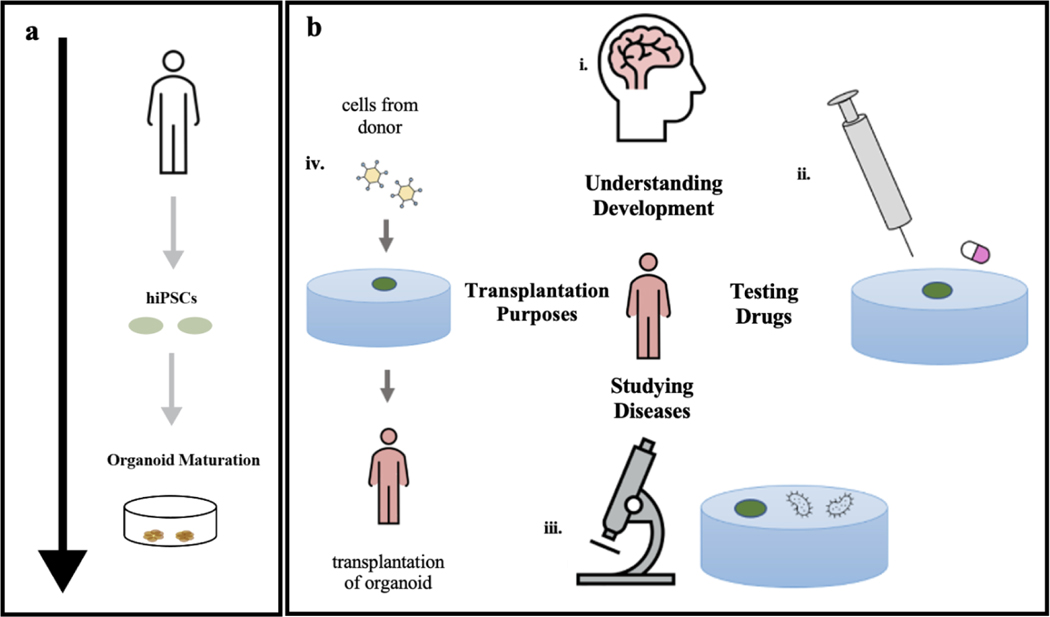
The development of brain organoids represents a major technological advance in the field of stem cell biology and regenerative medicine, a novel bridge between traditional 2D cultures and in vivo animal models, offering tremendous applications (Fig. 1). Human brain organoids are derived from human induced pluripotent stem cells (hiPSCs) and embryonic stem cells (hESCs) under 3D culture conditions with a neuronal differentiation medium (Fig. 1a). The generation of human brain organoids requires 3D self-assembly and differentiation of different types of cells (e.g., stem cells, neuron progenitor cells, astrocytes, etc.) into a functional tissue network that resembles the structure of an in vivo brain. Figure 2a introduces the timeline of developmental stages for the human brain and the corresponding brain organoid19. Brain organoids offer many opportunities for human-specific research. For example, the human brain, especially the neocortex, has evolved to be disproportionately larger compared with that of other species23–24. Brain organoids can mimic this corticogenesis process, allowing for the understanding of the mechanisms that make humans unique and aiding the translation of findings from animal models to clinical applications7 (Fig 1b. i). Moreover, brain organoids opened up new possibilities for studies of brain development across species. For example, a comparison between human, chimpanzee, and macaque iPSC-derived organoids has revealed differences in the proliferative dynamics of iPSCs that lead to different neurogenesis outputs11. In addition, brain organoids can model human diseases, which improves the efficiency of drug testing and screening (Fig. 1b ii). Notably, brain organoids have been particularly successful for recapitulating disease-related phenotypes of conditions in which structural malformations are apparent at early embryonic stages7. Therefore, brain organoids derived from patient-derived iPSCs have been extensively explored for the potential to model neurodevelopmental brain disorders and disease25–27 (Fig. 1b iii). Last, the future development of brain organoids may offer the opportunity for transplantation purposes (Fig. 1b iv). Given all the opportunities discussed above, the development of technologies that can help us to understand the spatiotemporally evolved molecular, structural, and functional properties of brain organoids is important.
Figure 1. Development and applications of human organoids.
a. Cells taken from an individual are programmed into induced pluripotent stem cells (iPSCs). iPSC colonies form embryoid bodies which are then embedded in the tissue scaffold for the 3D growth of tissue. The tissue is then cultured for several months as the organoid matures. b. (i) Organoids can be used to understand the key components of human organ development. (ii) The effectiveness of drugs against diseases can be tested on organoids. (iii) Patient-specific organoids can be used to model diseases. (iv) Organoids can be potentially used for cellular therapies. Figure 1a adapted from Mohamed et al.19 and Figure 1b adapted from Koo et al.27.
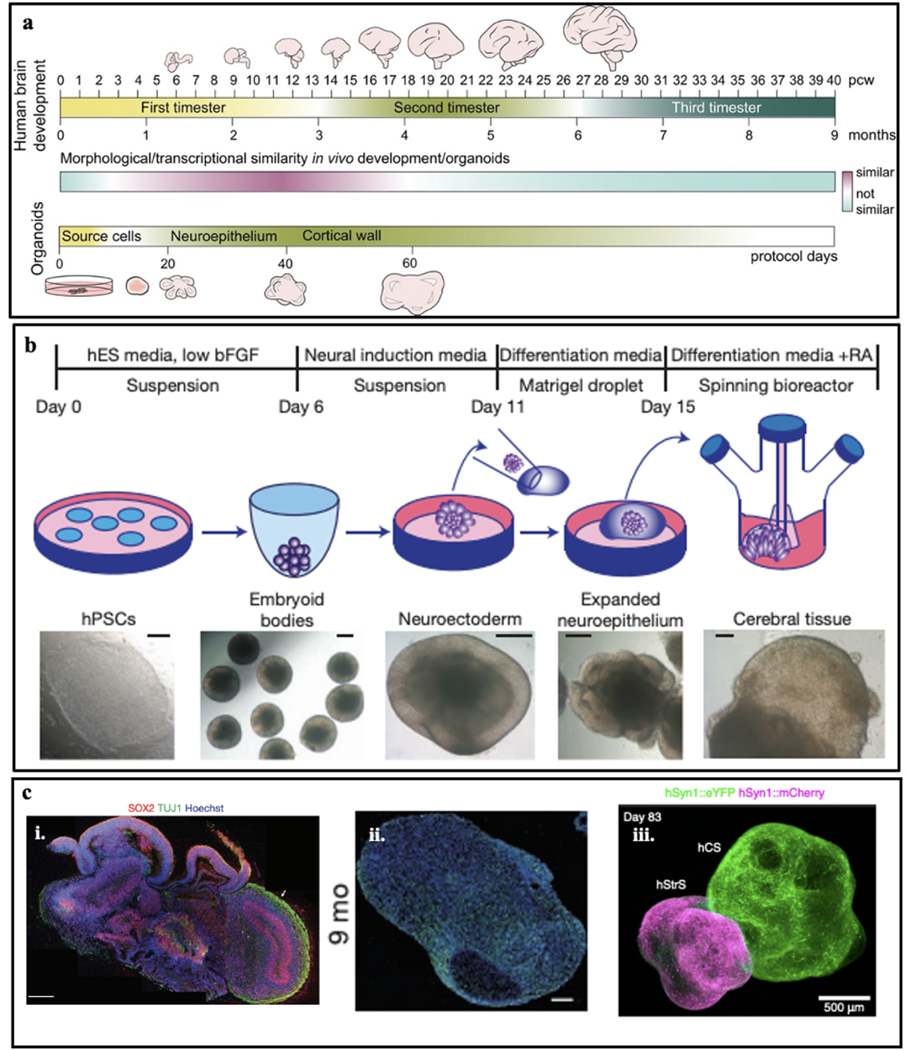
Figure 2. Human brain organoids.
a. Timeline of human brain development. A timeline showing relative similarities between human in vivo brain development and the brain organoid protocol timeline. The relative similarity (cyan-purple gradient) is based on cell-biological and transcriptomics data from several studies and is not a quantitative measure. Human developing brain and brain organoids are not to scale. b. Schematic of the typical culture system used for brain organoid generation. Example images of each stage are shown. bFGF, basic fibroblast growth factor; hESC, human embryonic stem cell; hPSCs, human pluripotent stem cells; RA, retinoic acid. c. (i) Sectioning and immunostaining revealed complex morphology with heterogeneous regions containing neural progenitors (SOX2, red) and neurons (TUJ1, green) in the human brain organoid. (ii) Immunostaining of the synaptic marker SYN1 in the human brain organoid at nine months. (iii) 3D immunostaining of clear, cortico-striatal assembloids expressing AAV-hSyn1::eYFP in human cortical spheroid and AAV-hSyn1::mCherry in human striatal spheroid. Figure 2a adapted from Kelava29, Figure 2b, 2c (i) adapted from Lancaster et al.9, Figure 2c (ii) adapted from Quadrato et al.6, and Figure 2c (iii) adapted from Miura et al.65.
Morphological and genetic characterizations of brain organoids
Morphological characterizations are the easiest way to determine the developmental stages of brain organoids (Fig. 2a). On the cellular level, imaging characterizations can reveal that brain organoids exhibit some similar features of the in vivo developing human brain in the early stages of development. For example, the progenitor ventricular zone (VZ) and subventricular zone (SVZ) can be easily recognized through bright-field (BF) optical imaging (Fig. 2b). Through labeling the stage- and cell-type-specific protein markers, fluorescence imaging can illustrate the different brain regions in the brain (Fig. 2c)9,28,29. However, human cortical development, as any other developmental process, is comprised not only of obvious cellular differences in morphology and expression of a few protein biomarkers but also employs coordinated waves of gene expression to induce all of the morphological changes and patterning cues needed to develop a functioning brain. Several technologies have recently employed gene expression analyses on neocortical cultures by microarrays30–31, RNA sequencing (RNA-seq)31–34 or single-cell RNA-seq34 and compared the gene expression of the brain organoids with the developing brain34–36. Together, these studies confirmed that in vitro brain organoids replicate well early in vivo brain development, with some reports showing that the in vitro development parallels the in vivo up to late mid-fetal period7,29,30.
Electrical characterizations of brain organoids
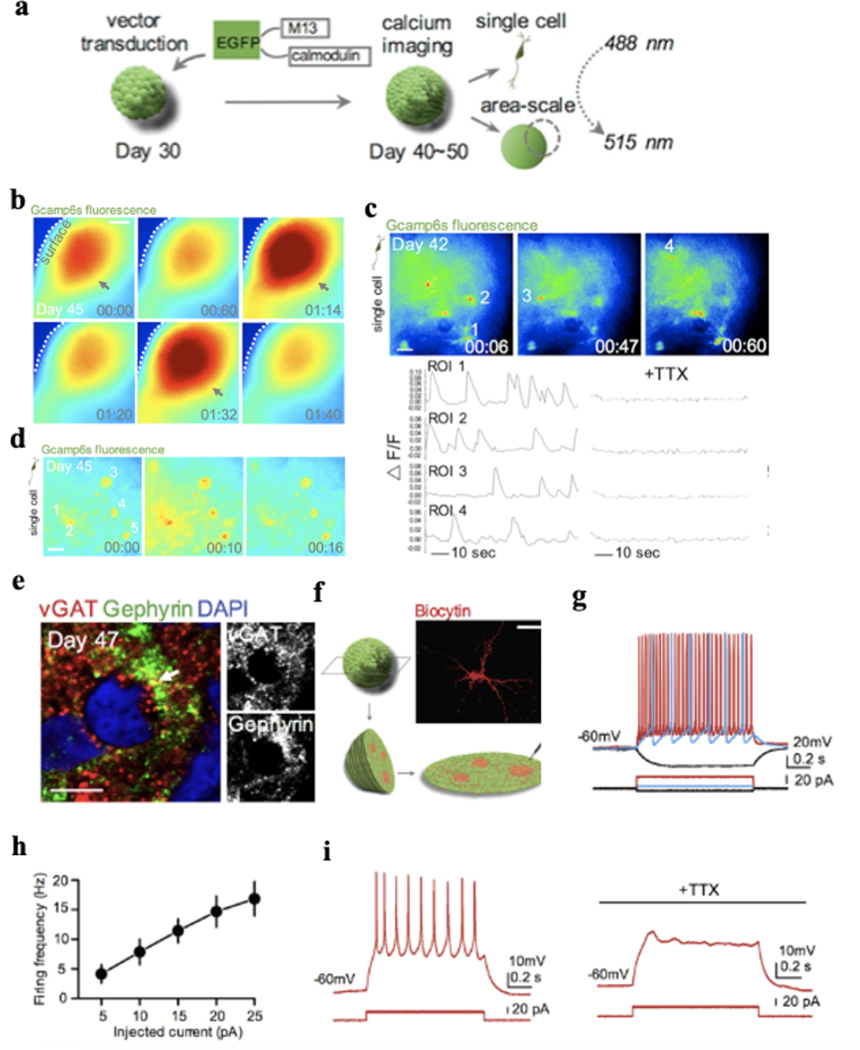
While the morphological and genetic characterizations can provide insight about the cellular and molecular level evolution over development, to examine whether the organoid culture system produces functionally active neurons and connected neural networks, it is important to determine their electrical functions during development, ideally at the single-cell single-spike spatiotemporal resolution. Figure 3 summarizes neurotechnologies developed for electrical measurement of neuronal activities and their spatiotemporal constraints. Conventional electrode technologies using rigid metal electrodes have made prominent contributions to understand the electrical activities in neural systems. However, the limited number of electrodes, the large mechanical disparity between rigid materials to soft neural tissue, and the invasive implantation process prevent them from simultaneously interrogating activities from a large number of neurons across 3D brain organoids in a long-term stable manner. Calcium imaging that employs genetically encoded indicators has been used to record the intracellular Ca2+ activity to infer the single-cell cellular electrical activity from hundreds to thousands of neurons over millimeter square areas with cell-type specificity. For example, the genetically encoded calcium indicator (GECI) GCaMP6s can be used to label neurons in human brain organoids by human synapsin I promoter (Fig. 4a–b)37. Imaging results confirmed that neurons in 3D brain organoids differentiated for 40–50 days can display calcium activities. Adding sodium channel blocker tetrodotoxin (TTX) can block the calcium activities (Fig. 4c), indicating these calcium oscillations were indeed neuronal activity-dependent signals. Moreover, when calcium imaging was performed over larger fields of view, highly synchronized calcium surges can be observed from some areas in brain organoids (Fig. 4d), suggesting the synchronization of functional neuronal networks. Further investigating the synchronized pattern of neuronal activity within the organoids by performing higher magnification imaging revealed that individual neurons displayed calcium surges simultaneously (Fig. 4d). Synchronization matrix analysis based on calcium spike patterns of individual neurons and imaging results (Fig. 4e) further revealed the presence of synchronized neuron clusters inside the brain organoids37.
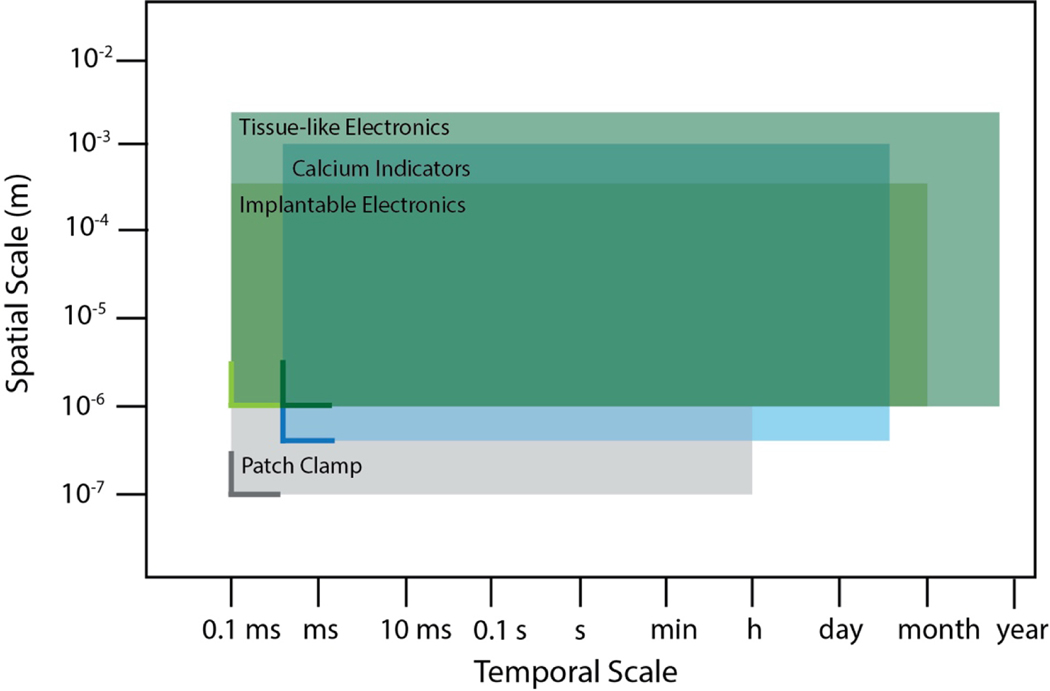
Figure 3. Spatiotemporal constraints of electrical recording technologies for brain organoid measurement.
The spatiotemporal constraints of various electrical recording technologies have been developed in the field of neuroengineering. The horizontal and vertical solid lines represent resolution in temporal and spatial scales, respectively, whereas the extent of the rectangular shade represents the spatiotemporal span of each technology. Figure 3 adapted from Hong et al.66.
Figure 4. Calcium Imaging.
a. Schematic view of the methods for calcium imaging of intact brain organoids. b. Representative image of area-scale calcium imaging in intact 45-day-old brain organoids. The synchronized calcium surges are indicated with arrows. Time is shown in mins. Scale bar: 100 μm. c. Representative image showing cells expressing hSyn-GCaMP6s in an intact 42-day-old brain organoid. The single-cell tracings of calcium transient (region of interest [ROI] indicated on top) are shown, which are blocked by the application of tetrodotoxin (TTX) (1μM). Scale bar: 25 μm. d. Calcium imaging of synchronized area (b) at single-cell level. ROIs are indicated. Time is shown as mins. Scale bar: 25 μm. e. Immunostaining for pre-synaptic protein vesicular GABA transporter (VGAT) and postsynaptic protein gephyrin in a 47-day-old brain organoid section. Scale bar: 5 μm. f-g. Diagram showing slice patch-clamp and identification of the neuronal morphology of the recorded cell by filling with biocytin. Scale bar: 25 μm. h. Graph depicting the firing frequency of the recorded cells from brain organoids plotted against injected current (n = 7 cells). Mean ± SE is shown. i. Representative image of action potentials of a cell in brain organoid slice before and during application of TTX (1μM). Figure 4 adapted from Xiang et al.37.
Calcium imaging, however, has limited temporal resolution and penetration depth (Fig. 3). Thus, it cannot resolve the individual action potential from the neurons at millisecond temporal resolution and cannot capture activities of neurons inside the brain organoid. Whole-cell patch-clamp recording can characterize the electrophysiological properties of individual neurons in brain organoids at millisecond temporal resolution (Fig. 3, Fig. 4f–i)37. Patch-clamp allows researchers to record the cellular membrane properties and intracellular action potential from individual neurons in brain organoids, which provides detailed electrophysiological analysis of neurons20,22,30,38. The high temporal resolution is also useful for determining neuronal responses to pharmacological treatment or optogenetic stimulation21. However, patch-clamp can only assay one cell at a time with membrane disruption. Thus, little-to-no information on network connectivity and neuronal population dynamics can be provided, which are important to regional and global organoid function. To increase spatial coverage and analyze network activity, calcium imaging has been typically combined with patch-clamp9,37,39 to overcome these limitations, while the long-term stable measure of the electrical properties of brain organoids is still a challenge.
Emergent bioelectronics for electrophysiological activity of organoids
2D MEAs
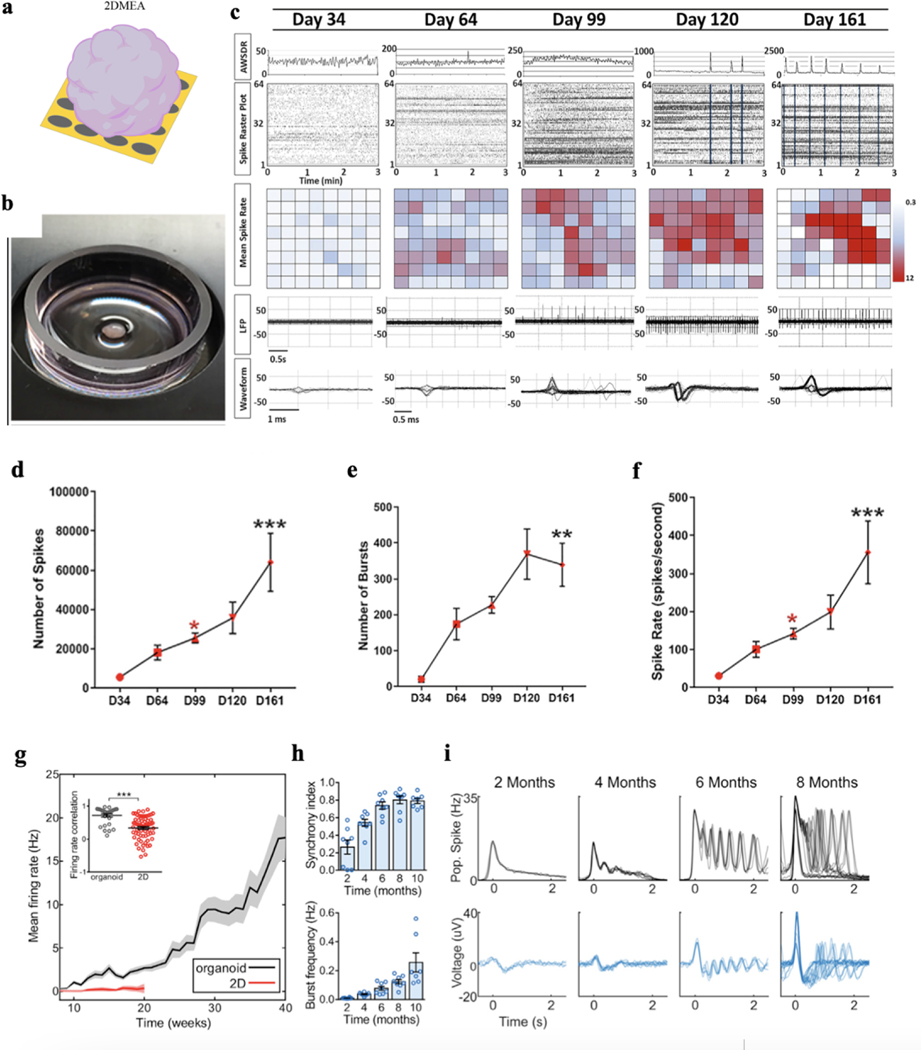
To measure the electrical activities from brain organoids at millisecond resolution over development, MEAs have been used to enable a non-invasive and stable long-term bioelectronic interface with brain organoids. Previously, 2D MEA has been widely used to long-term stably monitor the electrical properties of 2D cultured neurons and neural networks either from tissue-harvested or stem cell-derived neurons40. Brain organoids can be placed on the top of the microelectrode array on a silicon or glass substrate inside a bioreactor or multi-well plate for measurement. The microelectrode array was connected through encapsulated metallic interconnects to the amplification system for continuous recording. Action potentials can be detected by the microelectrode array from the neurons at the bottom of the brain organoids. Field potential can also be detected from the brain organoids. When the neurons form an interconnected network, synchronized action potentials can be detected as organized bursts of action potentials across multiple electrodes on the array. These temporal and spatial organization and synchronization of action potentials can thus provide crucial insights into the healthiness and maturation of brain organoids.
2D MEAs enable minimally invasive, long-term electrical measurement. Using 2D MEAs, Fair et al. evaluated the cortical organoid electrical functionality at a mesoscopic level by recording spontaneous neuronal electrical activity on a weekly basis18 (Fig. 5a). Analyzing the gene expression and electrophysiological profiles of cortical organoids, they learned about the long-term network development in brain organoids (Fig. 5b). Specifically, using a 64-channel MEA, the earliest time point at which electrical activity emerges in cortical organoids was captured at day 34 of development, which was expected since this time point is correlated to a developmental window of the rapid expansion of neuroepithelium and the beginning of early neurogenesis within cortical organoids that precedes neuronal maturation. After 64 days of development, the authors found that cortical organoids exhibited increases in both mean spike rate and field spike amplitude, with both properties continuing to increase thereafter into day 99 of development (Fig. 5d–f). Furthermore, 2D MEA can detect characteristic spontaneous action potentials with discrete depolarizations that coincided with the observed expansion of astrocytes. At day 120 of development, as indicated by a significant increase in the array-wide spike detection rate, cortical organoids exhibited synchronized burst firings, which are prominent characteristic features of functional cortical neural networks in the developing mammalian cortex and a hallmark of synaptic signal propagation. After day 161 of development, cortical organoids showed further increases in the number of synchronized burst firings, which suggests the cortical organoids formed highly interconnected neural networks. This developmental link between synchronization and development supports their previously described role in the establishment and control of neural network activity40,41.
Figure 5. 2D multielectrode array (MEA).
a-b. Schematic (a) and optical photograph (b) showing a brain organoid placed on the top of a 2D MEA. c. Detection of spontaneous electrical activity in day 34 brain organoids and approximately every month thereafter shows a progressive increase in and complexity of electrophysiological properties. Top: Graphical representation of the array-wide spike detection rate (AWSDR) above every spike raster plot with a 3-min recording interval. Synchronized burst firing (SBF) activity is observed in late-stage brain organoid cultures and is indicated by light blue columns within raster plots of days 120 and 161. Note the corresponding peaks in the AWSDR above each SBF. Middle: Heatmap plots of the mean spike rates of neurons within brain organoids recorded by each individual microelectrode. Bottom: Local field potential (LFP) traces and a corresponding waveform taken from a shorter time interval showing a progressive increase in voltage amplitude and spiking rate at successive developmental time points. Note the characteristic action potential waveforms shown in days 99, 120, and 161. d-f. Number of spikes (d), number of bursts (e), and spike rate (f) of developing brain organoids at approximately monthly intervals initiating at day 34 (n = 4/time point). *, P<0.05, ** P< 0.01, and *** P<0.001 One-way ANOVA with Tukey’s multiple comparisons test. g. Brain organoids show elevated and continuously increasing mean firing rate compared to 2D monolayer neurons (n = 8 for organoid cultures and n = 12 for 2D neurons). Inset: correlation of the firing rate vector over 12 weeks of differentiation (from 8 to 20) between pairs of cultures showing reduced variability among organoid replicates. ***P<0.001, t-test. h. Temporal evolution of brain organoid network activity. i. Time series of population spiking and LFP during network events in brain organoid development. Each overlaid trace represents a single event during the same recording session. Figure 5a adapted from Passaro et al.21; Figures 5b-f adapted from Fair et al.18, and Figures 5g-i adapted from Trujillo et al.16.
A similar study conducted by Trujillo et al. used 2D MEA to analyze single-channel and population firing characteristics derived from channel-wise spike times and the local field potential16. These spikes from each channel do not represent putative single-unit action potentials but represent multi-unit activity which is mainly due to the limited spatial and temporal resolution of 2D MEA used in the study. Therefore, single-unit spike trains cannot be isolated. Over the course of 10 months, 2D MEA recording showed that cortical organoids exhibited consistent increases in electrical activity, as parametrized by channel-wise firing rate, burst frequency, and synchrony (Fig. 5g and 5h), which indicates a continually evolving neural network. Additionally, using the 2D MEA to measure organoids and iPSC-derived neurons in monolayer cultures, their results showed that the variability between organoids over 40 weeks of differentiation was significantly lower compared to 2D neuron cultures (Fig. 5g). But the brain organoids showed a robust pattern of activity, switching between long periods of quiescence and short bursts of spontaneous network-synchronized spiking. Notably, the authors found that these network events were periodic (~0.05 Hz) but infrequent early in development (~2 months), occurring roughly every 20 s and decayed monotonically after the initial onset (Fig. 5i). Further analyzing the characteristics of the data, they concluded that the burst dynamics recorded from brain organoids by MEA are very similar to the brain activity from a human infant recorded by EEG devices, suggesting the capability of brain organoids to resemble the electrical functions in early human development16.
3D MEAs
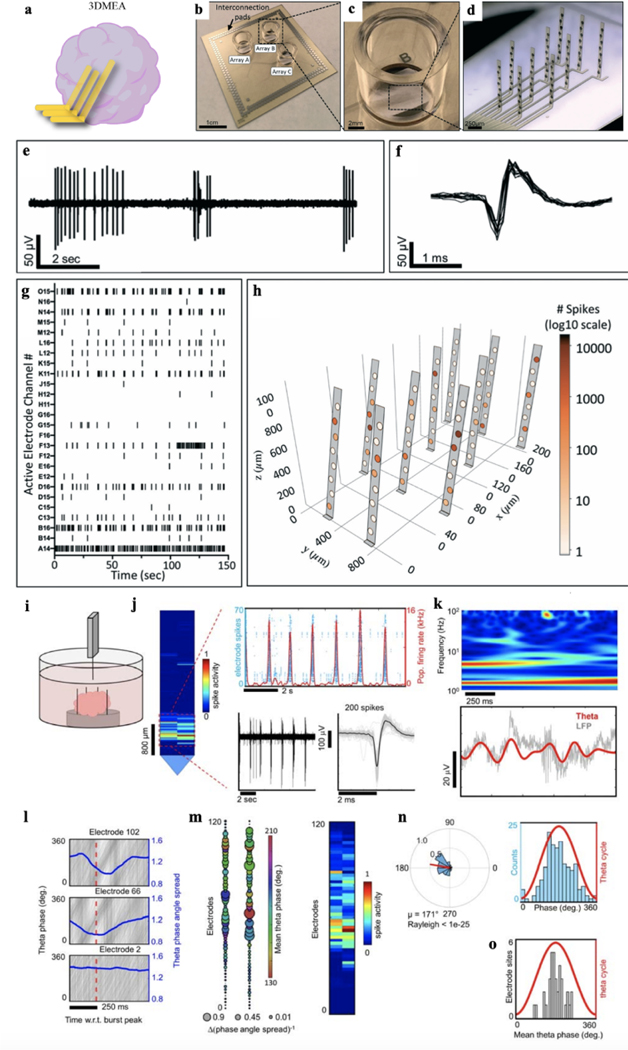
While the 2D MEAs can measure the activities from the neurons located on the bottom of brain organoids, the 3D structure of brain organoids still limits their accessibility to the majority of neurons in the organoid. To overcome this issue, 3D MEAs are developed for brain organoid measurement20,21 (Fig. 6a–d). For example, Soscia et al. developed an electrode array using polyimide as the encapsulating layer and metal as interconnects20. After releasing the electrode array from the substrate, they used mechanical force to fold the electrodes into 3D. To assess the ability of the 3D MEA to monitor the electrical activity of a 3D neuronal culture system, recordings were performed on human iPSC-derived neuronal cultures over 38 days of in vitro culture. By 15 days electrodes began detecting electrophysiological activity from the cells. Across the 3 arrays from the device examined, there were 70 active electrodes out of the total 80 electrodes in arrays that exhibited activity at 38 days. The bursting activity was also present, which is expected for functional and mature networks (Fig. 6e). An example overlay of action potential waveforms captured by one electrode is shown in Fig. 6f. Neuronal activity was observed up to 45 days of culture. The electrophysiological activity was distributed throughout the 3D MEA and was able to be visually represented in 3D plots based on the location of the electrodes calculated from the probe actuation angles measured at termination of the cell culture20 (Fig. 6g–h).
Figure 6. 3D MEA.
a. Schematic depicting a 3D MEA inserted into a brain organoid for electrical recording. b-d. Photographs of 3D MEAs. e-g. Representative voltage traces (e), spikes (f), and raster plot (g) of neuronal spiking data from the 3D MEA. h. 3D visualization plot showing spike activity from a single array of the 3D MEA device throughout 3D space. The number of spikes recorded from each electrode is represented by shading of electrodes on a log10 scale. i. A Neuropixels high-density CMOS shank was attached to a custom-made mount and controlled by a micromanipulator to be inserted into a cerebral organoid kept in media. j. Representative spikes detected near the tip of the Neuropixels shank. k. Top: spectrogram plot of LFP from an electrode illustrates dominant oscillation power in the theta and delta bands. Bottom, raw LFP (gray line) from the same electrode overlaid with the theta-filtered band (red line). l. Theta phase traces (gray lines) shown relative to population burst peak events (red dotted line) reveal phase coherence as illustrated by a drop in the phase angle spread (blue line). The bottom plot shows an electrode site displaying no phase coherence relative to population burst events. m. Left, a spatial map of the change in theta phase angle spread shown relative to burst peak time across the Neuropixels shank. The bubble size indicates the inverse of the phase angle spread relative to the burst peak for each electrode. Notice the overlap with phase coherent sites and the spatial region registering spiking activity on the right. n. Left, circular distribution of theta-spike phase angles measured from a single electrode site. The direction of the mean spike angle (μ) relative to the theta phase and magnitude (mean resultant length) are shown as polar plots. The Rayleigh criteria for non-uniformity was used to determine if spikes were distributed non-uniformly over the theta cycle (0°, 360°). Right, distributions of theta-spike angles are shown relative to the theta cycle. o. Mean theta phase angle of spike phase-locked electrodes across the Neuropixels probe satisfying the Rayleigh criterion (P< 0.05). Figure 6a adapted from Passaro et al.21; Figures 6b-h adapted from Soscia et al.20; Figures 6i-o adapted from Sharf et al.17.
Meanwhile, the conventional Michigan probes has been employed as 3D MEA to measure the brain organoids. Quadrato et al. applied silicon electrode array to the brain organoid at 8 months of culture6. Single unit action potentials can be isolated from seven of eight brain organoids. A high-density 3D MEA platform can allow for more precise spatial mapping of active neurons21,42. To increase the number and density of the electrodes, the Neuopixels probe that integrates the complementary metal-oxide-semiconductor (CMOS) multiplexing circuit with MEA, which shares the similar design as the Michigan probe, has been developed for the in vivo activity measurement. Sharf et al. applied the Neuropixels probe to 3D brain organoid recording by inserting the probe into brain organoids17. Acute electrophysiology recordings were performed from the intact brain organoids with multiple Neuropixels probes each containing a densely tiled 960 electrode CMOS shank with 20 μm inter-electrode pitch. Neuropixel probe was inserted by using a motorized, low-drift micromanipulator. Single-unit spiking activity that extended hundreds of microns across the organoid tissue and spontaneous, synchronized population bursts can be observed (Fig. 6i, j). Analyzing these mapping data, the authors found that the amplitude of theta oscillations increased when synchronized with neuronal population bursts. Neuropixels probe offers an advantage to examine the relationship between theta oscillations with neuronal spikes by simultaneously recording a greater number of neuronal activities comparing to other techniques. To confirm the synchronization of theta oscillations to neuronal population bursts, the authors performed simultaneous extra-cellular field potential recordings alongside action potential recording across the whole organoids through three Neuropixels probes. They found that theta phase alignment was indeed synchronized with population bursts and co-localized with spatial regions where spiking activity was observed (Fig. 6k–m, right). Meanwhile, theta phases caused deconstructivity in regions where the organoid showed no signs of spiking activity. In addition, the ultra-high-bandwidth recordings from the entire brain organoids confirmed the presence phase-locking of spikes to theta oscillation (Fig. 6n) simultaneously across multiple sites (Fig. 6o) in the z-plane of the organoid17. Specifically, they found that phase-locked sites form an interconnected ensemble composed of higher relative connectivity strengths when compared against the larger constituent connectivity network in which they reside17. Therefore, their ultra-high-bandwidth measurements of electrical activity generated by brain organoids provided strong evidence that theta oscillation is synchronized over temporal epochs of increased neuronal activity within the time window of population bursts.
3D flexible bioelectronics
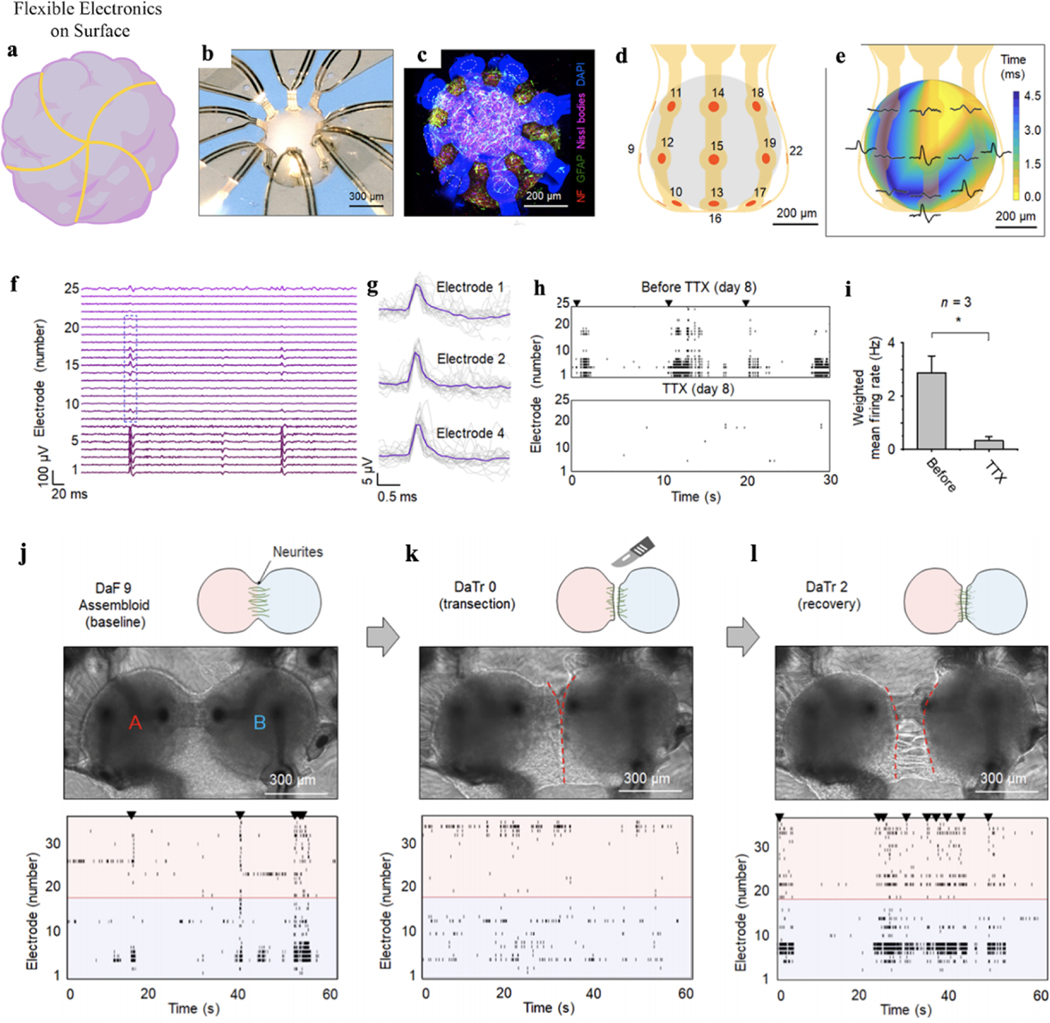
3D MEA can measure the electrical activity across the brain organoids, but the insertion process introduces substantial damages to the neural network. The recent development of flexible bioelectronics provides soft, shape-matched electronics in 3D geometries that can gently envelop the 3D brain organoids for the electrical interface (Fig. 7a). Initial work in this direction includes a thin-film flexible microelectrode array to wrap cardiac cells43 and cylindrical electrode arrays to contact cortical spheroids44. Controlling the mechanical buckling of the thin-film electronics, Rogers, et al. developed a series method to precisely fold the 2D flexible electronics into a designed 3D structure, which allows multifunctional electronics to be integrated with the 3D bioelectronics45,46. They recently applied these techniques to interface with 3D brain organoids. Through reverse engineering and considering shapes, sizes, and geometries of brain organoids, they built the 3D flexible bioelectronics that precisely matches the morphology of the brain organoids. For example, in a typical device, Park et al. integrated an array of gold microelectrodes encapsulated by polyimide into a 3D flexible bioelectronics buckled into a shape matching the geometry of cortical spheroids47. Figure 7b shows that a hiPSC cortical spheroid has been inserted into the 3D flexible bioelectronic device. Confocal microscopic imaging showed the intimate contact between the 3D flexible electronics with the cortical spheroid (Fig. 7c). The dashed circles indicate the locations of microelectrodes designed for electrophysiological monitoring. The results highlight soft and close proximity contacts (separations of less than ∼60 μm) to the surfaces of the spheroid, across most of its surface area (Fig. 7d), without any noticeable physical damage or deformation of the tissues. Using the 3D organoid-contacted MEA, the authors can successfully record field potentials (Fig. 7e–g) at a 200-μm resolution. The 3D flexible MEA can record the 3D spatial propagation of wave spreading and firing and bursting events across cortical spheroid (Fig. 7e), which is similar to the electrical activities recorded from 2D cell cultures on 2D MEAs. They can also record the uniform potential waveform with an average duration of ∼0.5 ms and a peak-to-peak amplitude of ∼15 μV. The signals can be abolished by applying TTX into the culture media, confirming as single-unit action potentials from the cortical spheroid47 (Fig. 7h–i). Remarkably, as the 3D flexible MEA can be designed in different shapes, the authors designed the devices to record the activity from the assembloids to reveal electrical activities that are associated with growth, transection, and regrowth of the neurite bridge between these spheroids. An experiment that involved electrophysiological recordings before and after cutting the neurite bridge that forms between two spheroids illustrated processes of neural recovery for days (Fig. 7j–l). They first recorded the neural activities across both spheroids until their activities are synchronous after the formation of the bridge (Fig. 7j) and then cut the bridge showing synchrony declines immediately (Fig. 7k). One day after the cutting, the neurite bridge reestablished, and synchronous firing resumed (Fig. 7l). They believed that the model presented here reproduces neurite neural injury and recovery in a 3D human system and provides direct electrical evidence of functional reintegration. Finally, another unique capability of this platform is that advanced semiconductor-based planar electronic and optoelectronic components can be easily integrated to provide various types of neural interfaces. For example, microscale inorganic light-emitting diodes, electrochemical sensors, thermal actuators, and precision temperature gauges have been integrated with flexible 3D bioelectronics, capable of enabling multimodal interrogation of the brain organoid physiology47–51.
Figure 7. Flexible electronics.
a. Schematic depicting flexible electronics on the surface of an organoid. b. Optical image of a cortical spheroid enclosed in a 3D flexible bioelectronics designed for electrophysiological recording. c. Confocal microscope image of the spheroid in a similar 3D mesostructure, neurofilament (red), GFAP (green), Nissl (magenta), DAPI (blue), and autofluorescence from the parylene-C in bioelectronics (blue). Dashed circles indicate the approximate positions of microelectrodes in a corresponding functional system. d. 3D illustration of the positions of the microelectrodes across the surface of the spheroid. e. 3D plot of time latency associated with traces in the blue box of (f). f. Representative field potentials were recorded from all 25 microelectrodes in the system. g. Overlaid plots of 30 spikes from channels 1, 2, and 4. h. Network activity of day 8 and suppressed behavior resulting from the application of TTX. i. Weighted mean firing rate (the spike rate multiplied by the number of active electrodes) is representative of 2 min at each condition. n = 3 spheroids; *P < 0.05; two-tailed paired t-test; means ± SEM. j-l, Illustrations, optical images, and raster plots (j) before and (k) after transection of the neurite bridge that joins the spheroids of this assembloid and (l) recovery after transection. Figure 7a adapted from Passaro et al.21, Figures 7b-l adapted from Park et al.47.
Cyborg organoids
Despite the remarkable progress in the development and application of 3D flexible electronics into brain organoid measurement, long-term stable 3D recording of single-cell electrophysiology in developing brain organoids is still a challenge. The recording technology not only needs to form stable electrical interfaces with brain organoids but also needs to accommodate the rapid volume change occurring during the organoid organogenesis. Recently, through the convergence of soft materials and nanoelectronics, tissue-like electronics have been introduced52–55. The incorporation of tissue-like properties such as tissue-level flexibility, subcellular feature size, and mesh-like networks allows the nanoelectronics to seamlessly integrate throughout 3D tissue and record multimodal activities at single-cell resolution in a chronically stable manner (Fig. 3 and Fig. 8a).
Figure 8. Cyborg organoids.
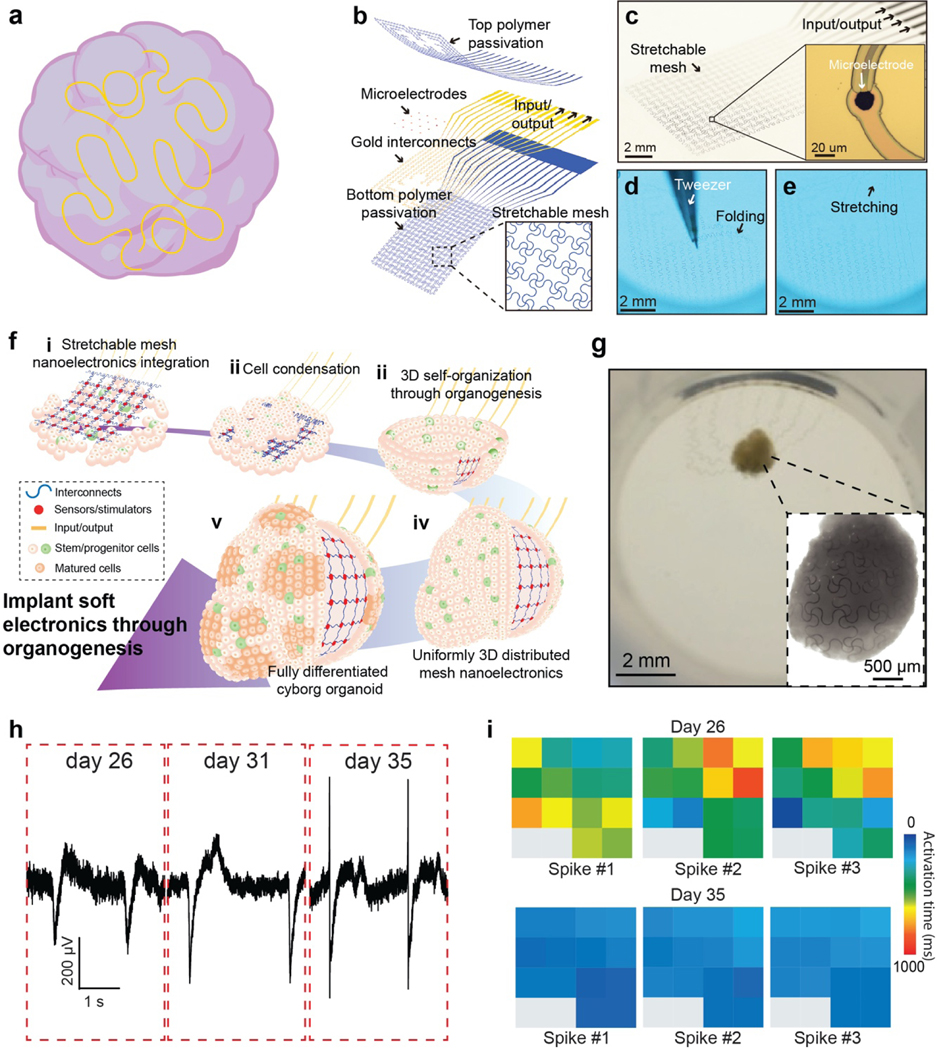
a. Schematic illustrating the cyborg organoid platform that stretchable mesh nanoelectronics fully distributed across the 3D volume of organoids for electrical recording. b-e. Schematics (b) and optical images of stretchable mesh nanoelectronics (c) and their mechanical properties (d-e). For a typical dimension, the bending stiffness of the mesh is less than 0.1 nN•m capable of being bucked and stretched by the force at the level of 1 μN, which is one order of magnitude smaller than the cell-cell attraction force from tissue development. f. (i) Lamination of stretchable mesh nanoelectronics onto a continuous sheet of Matrigel seeded with hiPSCs or hiPSCs-derived progenitor cells. (ii) Aggregation of the cell sheet into a cell-dense plate through cell proliferation and migration induced by cell−cell attractive forces, embedding the stretchable mesh nanoelectronics into the cell plate and folding them into a tightly packed structure. (iii) Organogenetic 2D-to-3D self-organization folds the 2D cell plate/nanoelectronics hybrid into a 3D structure with a bowl-like geometry. (iv) Organogenesis unfolds the closely packed nanoelectronics and distributes their structure across the entire 3D organoid. (v) Further development and differentiation of stem/progenitor cells into different cell types with their electrophysiological behaviors continuously monitored by 3D embedded nanoelectronics. g. Optical image shows fully connected cyborg organoids and organoid-wide distributed mesh nanoelectronics (inset). h-i. Long-term stable electrophysiological mapping of tissue development showed changes in spike dynamics (h) and related synchronization (i) at different stages of cardiac organoids. Figure 8a adapted from Passaro et al.21, Figures 8b-i adapted from Li et al.22.
Liu et al. previously developed 3D soft, mesh-like nanoelectronics that fully mimics the physicochemical properties of tissue scaffolds and possesses tissue-like bending stiffness52–54. The mesh nanoelectronics was fabricated by photolithographic patterning of low impedance Pt microelectrodes, metal interconnections, and polymer passivation with nanoscale thickness and microscale width in a fully integrated mesh network. Li et al. further implemented a stretchable design using serpentine structures in the mesh (Fig. 8b)22. These elastic interconnections and the resulting tissue-like mechanical properties allow the nanoelectronics to be readily stretched and re-organized by forces generated during tissue morphogenesis (Fig. 8c–e). Importantly, all the microelectrodes are individually addressable to an external recording set-up for continuous measurement.
The stretchable mesh nanoelectronics was then engineered to grow into and migrate with a 2D stem cell layer (Fig. 8f). During the endogenous 2D-to-3D reconfiguration of organoid development, the tissue itself folds the 2D stretchable mesh nanoelectronics into the appropriate 3D structure. The stretchable microelectrodes on the mesh are distributed uniformly across the entire 3D volume of the organoids (Fig. 8g) as “cyborg organoids”. The seamless integration with tissue and intimate contact with cells enables the recording of the cellular activity at single-cell resolution and millisecond resolution over the entire developmental time course (Fig. 8h). The electrodes allow for simultaneous, multisite electrophysiological recording, enabling a systematic mapping of physiological of the 3D tissue network. Toxicity, metabolic and immunostaining characterizations confirmed the long-term biocompatibility for the mesh nanoelectronics in the tissue culture. Li et al. demonstrated the feasibility of chronic electrophysiological recording over the entire course of human cardiac organoid development22. Multi-channel electrophysiological recording across the 3D volume of the cardiac organoids was obtained. These recordings illustrated heterogeneous activity patterns with clear time latencies, including the tissue-wide propagation of local field potentials. Chronic recordings at millisecond temporal resolution during cardiac organoid maturation were sufficient to reveal changes (Fig. 8i). We envision that further application of the cyborg organoid technology to brain organoids will enable the long-term stable, tissue-wide electrophysiology over the entire time course of brain organoid development and maturation56.
Conclusion
The capacity of brain organoids to differentiate, self-organize, and form distinct, complex, biologically relevant structures makes them ideal in vitro models of development, disease pathogenesis, and platforms for drug screening. They also hold the promise of better relevance for understanding human brain development and disease than current rodent models. However, to take full advantage of this potential, technologies need to be developed to probe the structural, genetic, and functional properties and heterogeneities from brain organoids across time. While the current development of bioelectronics has offered the unique bioelectrical interface with organoids to long-term stably record single-cell activities at millisecond temporal resolution over the time course of development, the electrical recording still cannot probe the neuronal activities with cell-type specificity and simultaneously record from a large number of neurons across the 3D volume of brain organoids. Further development of multimodal recording that combines in situ sequencing or cell-type-specific imaging with electrical recording may further relate the electrical signals with cell-type-specific information57,58. Integration of CMOS multiplexing circuits with flexible electronics will also enable a scalable platform to simultaneously record activities from a large number of neurons in brain organoids59–62. Last, integration of multifunctional sensors and stimulators into these bioelectronic platforms may introduce the multimodal in situ mapping of brain organoid activities beyond the electrical signals and provide closed-loop feedback to control the developmental trajectories or accelerate the maturation of brain organoids63,64.
Research Highlights:
Brief introduction of the development, generation, and applications of human brain organoids, and conventional methods used for characterizing the morphological, genetic, and electrical properties of brain organoids.
Highlight of the need for characterizing electrophysiological properties of brain organoids in a minimally invasive manner.
Introduction of recent advances in the multi-electrode array (MEA), 3D bioelectronics, and flexible bioelectronics and their applications in brain organoid electrophysiological measurement.
Introduction of the recently developed cyborg organoids platform as an emerging tool for the long-term stably 3D characterization of the brain organoids electrophysiology at high spatiotemporal resolution.
Perspectives of new technologies that could achieve the high-throughput, cell-type-specific and multimodal characterizations from the same brain organoids.
Acknowledgements
We acknowledge the helpful discussion and assistance from Qiang Li and Paul Le Floch. We acknowledge the support from the NIH/NIMH 1RF1MH123948 and the William F. Milton Fund.
Footnotes
Competing interests
The authors declare no competing interests.
Kazi Tasnim and Jia Liu: Conceptualization, Writing- Original draft preparation and Writing- Reviewing and Editing.
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References:
- 1.Spear LP (2004). Adolescent brain development and animal models. Annals of the New York Academy of Sciences, 1021, 23–26. [DOI] [PubMed] [Google Scholar]
- 2.Lewis D. (2002). The human brain revisited opportunities and challenges in postmortem studies of psychiatric disorders, Neuropsychopharmacology, 26, 143–154. [DOI] [PubMed] [Google Scholar]
- 3.Giridharan V, et al. , (2020). Postmortem evidence of brain inflammatory markers in bipolar disorder: a systematic review. Mol. Psych, 2, 94–113. [DOI] [PubMed] [Google Scholar]
- 4.Di Lullo E, et al. , (2017). The use of brain organoids to investigate neural development and disease. Nat. Rev. Neurosci, 18, 573–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.AlFatah Mansour A, et al. , (2018). An in vivo model of functional and vascularized human brain organoids. Nat. Biotechnol, 36, 432–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Quadrato G, et al. , (2017). Cell diversity and network dynamics in photosensitive human brain organoids. Nature, 545, 48–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Qian X, Song H. & Ming GL, (2019). Brain organoids: advances, applications and challenges. Development, 146, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pasca SP, (2019). Assembling human brain organoids. Science, 363. 126–127. [DOI] [PubMed] [Google Scholar]
- 9.Lancaster MA, et al. , (2013). Cerebral organoids model human brain development and microcephaly. Nature, 501, 373–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pollen AA, et al. , (2015). Molecular identity of human outer radial glia during cortical development. Cell, 163, 55–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thomsen ER, et al. , (2016). Fixed single-cell transcriptomic characterization of human radial glial diversity. Nat. Methods, 13, 87–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Close JL, et al. , (2017). Single-cell profiling of an in vitro model of human interneuron development reveals temporal dynamics of cell type production and maturation. Neuron, 93, 1035–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Durens M, et al. , (2020). High-throughput screening of human induced pluripotent stem cell-derived brain organoids. J. Neurosci. Methods, 335, 108627–108627. [DOI] [PubMed] [Google Scholar]
- 14.Mansour A, et al. , (2018). An in vivo model of functional and vascularized human brain organoids. Nat. Biotechnol, 36, 432–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Trujillo CA, et al. , (2018). Brain organoids and the study of neurodevelopment. Trends. Mol. Med, 24, 982–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Trujillo CA, et al. , (2019). Complex oscillatory waves emerging from cortical organoids model early human brain network development. Cell Stem Cell, 25, 558–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sharf T, et al. , (2021). Intrinsic network activity in human brain organoids. BioRxiv. [Preprint] 10.1101/2021.01.28.428643. [DOI] [Google Scholar]
- 18.Fair SR, et al. , (2020). Electrophysiological maturation of cerebral organoids correlates with dynamic morphological and cellular development. Stem Cell Rep, 1, 855–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mohamed NV, et al. , (2021). Generation of human midbrain organoids from induced pluripotent stem cells. MNI Open Research, 3, 1–1. [Google Scholar]
- 20.Soscia DA, et al. , (2020). A flexible 3-dimensional microelectrode array for in vitro brain models. Lab Chip, 20, 901–911. [DOI] [PubMed] [Google Scholar]
- 21.Passaro AP & Stice SL, (2021). Electrophysiological analysis of brain organoids: current approaches and advancements. Front. Neurosci, 14, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li Q, et al. , (2019). Cyborg organoids: implantation of nanoelectronics via organogenesis for tissue-wide electrophysiology. Nano Lett, 19, 5781–5789. [DOI] [PubMed] [Google Scholar]
- 23.Rakic P, (2009). Evolution of the neocortex: a perspective from developmental biology. Nat. Rev. Neurosci, 10, 724–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sousa AMM, et al. , (2017). Molecular and cellular reorganization of neural circuits in the human lineage. Science, 358, 1027–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Otani T, et al. , (2016). 2D and 3D stem cell models of primate cortical development identify species-specific differences in progenitor behavior contributing to brain size. Cell Stem Cell, 18, 467–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rossi G, Manfrin A. & Lutolf MP, (2018). Progress and potential in organoid research. Nat. Rev. Genet, 19, 671–687. [DOI] [PubMed] [Google Scholar]
- 27.Koo B, et al. , (2019). Past, present, and future of brain organoid technology. Mol. Cells, 42, 617–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smart HM, et al. , (1991). Unique morphological features of the proliferative zones and postmitotic compartments of the neural epithelium giving rise to striate and extrastriate cortex in the monkey. Cereb, 12, 37–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kelava I, Lancaster MA, (2016). Dishing out mini-brains: current progress and future prospects in brain organoid research. Dev. Biol, 420, 199–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pasca AM, et al. , (2015). “Functional cortical neurons and astrocytes from human pluripotent stem cells in 3D culture. Nat. Methods, 12, 671–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mariani J, et al. , (2012). Modeling human cortical development in vitro using induced pluripotent stem cells. Proc. Natl. Acad. Sci. USA, 109, 12770–12775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Qian X, et al. , (2016). Brain-region-specific organoids using mini-bioreactors for modeling ZIKV exposure. Cell, 165, 1238–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van de Leemput J, et al. , (2014). CORTECON: a temporal transcriptome analysis of in vitro human cerebral cortex development from human embryonic stem cells. Neuron, 83, 51–68. [DOI] [PubMed] [Google Scholar]
- 34.Camp JG, et al. , (2015). Human cerebral organoids recapitulate gene expression programs of fetal neocortex development. Proc. Natl. Acad. Sci. USA, 112, 15672–15677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kang HJ, et al. , (2011). Spatio-temporal transcriptome of the human brain. Nature, 478, 483–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miller JA, et al. , (2014). Transcriptional landscape of the prenatal human brain. Nature, 508, 199–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xiang Y, et al. , (2017). Fusion of regionally specified hPSC-derived organoids models human brain development and interneuron migration. Cell Stem Cell, 21, 383–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sakaguchi H, et al. , (2019). Self-organized synchronous calcium transients in a cultured human neural network derived from cerebral organoids. Stem Cell Rep, 13, 458–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee SY, et al. , (2019). Optogenetic control of iPS cell-derived neurons in 2D and 3D culture systems using channelrhodopsin-2 expression driven by the synapsin-1 and calcium-calmodulin kinase II promoters. J. Tissue. Eng. Regen. Med, 13, 369–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kuijlaars J, et al. , (2016). Sustained synchronized neuronal network activity in a human astrocyte co-culture system. Sci. Rep, 6, 36529–36529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mann EO, et al. , (2007). “Role of GABAergic inhibition in hippocampal network oscillations”. Trends Neurosci, 30, 343–349. [DOI] [PubMed] [Google Scholar]
- 42.Yuan X, et al. , (2020). Versatile live-cell activity analysis platform for characterization of neuronal dynamics at single-cell and network level. Nat. Commun, 11, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cools J, et al. , (2018). A micropatterned multielectrode shell for 3D spatiotemporal recording from live cells. Adv. Sci, 5, 1700731–1700731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kalmykov A. et al. , (2020). Bioelectrical interfaces with cortical spheroids in three-dimensions. BioRxiv [Preprint] 10.1101/2020.11.29. [DOI] [PubMed] [Google Scholar]
- 45.Xu S, et al. , (2015). Assembly of micro/nanomaterials into complex, three-dimensional architectures by compressive buckling. Science, 347, 154–159. [DOI] [PubMed] [Google Scholar]
- 46.Zhang Y, et al. , (2015). A mechanically driven form of Kirigami as a route to 3D mesostructures in micro/nanomembranes. Proc. Natl. Acad. Sci. USA, 112, 11757–11764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Park Y, et al. , (2021). Three-dimensional, multifunctional neural interfaces for cortical spheroids and engineered assembloids. Sci. Adv, 7,1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fenno L, et al. , (2011). The development and application of optogenetics. Sci. Adv, 7,1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ryu H, et al. , (2021). Transparent, compliant 3D mesostructures for precise evaluation of mechanical characteristics of organoids”. Adv. Mater, 2100026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dahl-Jensen S, et al. , (2017). The physics of organoids: a biophysical approach to understanding organogenesis. Development, 144, 946–951. [DOI] [PubMed] [Google Scholar]
- 51.Nasr B, et al. , (2018). Self-organized nanostructure modified microelectrode for sensitive electrochemical glutamate detection in stem cells-derived brain organoids. Biosensors, 8, 14–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu J, et al. , (2015). Syringe-injectable electronics. Nat. Nanotechnol, 10, 629–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu J, et al. , (2020). Intrinsically stretchable electrode array enabled in vivo electrophysiological mapping of atrial fibrillation at cellular resolution”. Proc. Natl. Acad. Sci. USA, 117, 14769–14778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xie C, et al. , (2015). Three-dimensional macroporous nanoelectronic networks as minimally invasive brain probes. Nat. Mater, 14, 1286–1292. [DOI] [PubMed] [Google Scholar]
- 55.Liu Y, et al. , (2019). Soft and elastic hydrogel-based microelectronics for localized low-voltage neuromodulation. Nat. Biomed. Eng, 3, 58–68. [DOI] [PubMed] [Google Scholar]
- 56.Le Floch P. et al. , (2021). A method for three-dimensional single-cell chronic electrophysiology from developing brain organoids. BioRxiv, 10.1101/2021.06.22.449502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang X, et al. , (2018). Three-dimensional intact-tissue sequencing of single-cell transcriptional states. Science, 361, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li Q, et al. , (2021). In situ electro-sequencing in three-dimensional tissues. BioRxiv, 10.1101/2021.04.22.440941. [DOI] [Google Scholar]
- 59.Müller J, et al. , (2015). High-resolution CMOS MEA platform to study neurons at subcellular, cellular, and network levels. Lab Chip, 15, 2767–2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dipalo M, et al. , (2018). Plasmonic meta-electrodes allow intracellular recordings at network level on high-density CMOS-multi-electrode arrays. Nat. Nanotechnol, 13, 965–971. [DOI] [PubMed] [Google Scholar]
- 61.Bakkum DJ, et al. , (2013). Tracking axonal action potential propagation on a high-density microelectrode array across hundreds of sites. Nat. Commun, 4, 2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shein-Idelson M, et al. , (2017). Large-scale mapping of cortical synaptic projections with extracellular electrode arrays. Nat. Methods, 14, 882–890. [DOI] [PubMed] [Google Scholar]
- 63.Takebe T. & Well JM, (2019). Organoids by design. Science, 364, 956–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jgamadze D, et al. , (2020). Modeling traumatic brain injury with human brain organoids. Curr. Opin. Biomed, 14, 52–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Miura Y, et al. , (2020). Generation of human striatal organoids and cortico-striatal assembloids from human pluripotent stem cells. Nat. Biotechnol, 38, 1421–1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hong G, et al. , (2019). Novel electrode technologies for neural recordings. Nat. Rev. Neurosci, 20, 330–345. [DOI] [PMC free article] [PubMed] [Google Scholar]