Abstract
Associations between brain structure and problematic alcohol use may reflect alcohol-induced toxicity and/or preexisting risk. Here, we applied a latent causal variable approach to genome-wide association study summary statistics of problematic alcohol use (n=435,563) and magnetic resonance imaging-derived brain structure phenotypes (e.g., cortical volume, cortical thickness, white matter volume; ns ranging from 17,706 to 51,665) to test whether variability in brain structure may plausibly contribute to problematic alcohol use and/or whether problematic alcohol use influences brain structure. After correction for multiple testing within each modality, we find evidence that greater volume of the pars opercularis, greater thickness of the cuneus, and lower volume of the basal forebrain may plausibly contribute to problematic alcohol use. All other nominally-significant associations identify brain structure as a potential causal contributor to problematic alcohol use; there was no evidence suggesting that problematic alcohol use may cause differences in brain structure. Collectively, these results challenge common interpretations that associations between alcohol use and brain structure reflect consequences of alcohol, instead supporting emerging work suggesting that brain structure may reflect a predispositional risk factor for alcohol involvement.
Introduction
Problematic alcohol use (PAU) has been robustly associated with smaller global and regional measures of brain structure (1). While these associations have been widely purported to arise from alcohol-induced brain atrophy, it is also possible that variability in brain structure may reflect predispositional liability (2). Mendelian Randomization (MR) approaches, which represent a form of instrumental variable analysis, have been widely used to assess whether genomic liability to one phenotype may cause another (e.g., alcohol and hypertension (3, 4)). However, MR approaches can be confounded by genetic correlations between the phenotypes. A recent method that uses a latent causal variable (LCV) approach (5) was developed to address this concern. LCV can be used to assess putatively causal relationships between pairs of phenotypes while accounting for genetic correlation and sample overlap. Here, we applied LCV to summary statistics generated from the largest GWASs of PAU and brain structure phenotypes to test whether variability in brain structure may plausibly contribute to PAU and/or whether PAU may contribute to brain structure.
Methods
Problematic Alcohol Use (PAU) genetic association summary statistics came from the Zhou et al GWAS meta-analysis of three PAU phenotypes: ICD-derived Alcohol Use Disorder (AUD) from the Million Veteran Program n= 286,202, Alcohol Use Disorder Identification Test-problem subscale (AUDIT-P) GWAS from the UK biobank, n=121,604, and DSM-III/DSM-IV alcohol dependence from the Psychiatric Genomics Consortium n=27,757; total n= 435,563)(6). The PAU GWAS had a significant heritability estimate of 0.068 (SE = 0.004).
GWAS summary statistics for global and regional magnetic resonance imaging (MRI)-derived brain structure phenotypes for cortical thickness and surface area, were obtained from analyses conducted by the Enhancing Neuro Imaging Genetics Through Meta-Analysis (ENIGMA) consortium (cortex gray matter n = 51,665 (7). Measures of gray matter volume (cortical and subcortical) and white matter volume were derived from an independent GWAS of UK biobank data (subcortical and gray matter volume n = 19,629 (8), white matter n = 17,706 (9)). N’s reflect the total number of individuals in the original study for each GWAS, with 278 imaging phenotypes total. All GWAS summary statistics include adjustments for population structure using ancestrally-informative principal components, as well as additional standard covariates (for example, sex and age).
Linkage Disequilibrium Score Regression (LDSR) (10) is commonly used to estimate the heritability of complex traits using genome-wide association study summary statistics, such as those available herein (10). As LDSR does not require raw data, this method can be extended to estimate the genetic correlation (SNP-rg) between traits measured in two different GWAS, while accounting for any sample overlap between the traits. Additionally, LDSR includes control for population structure and sample confounding.
Models built off LDSR can be parameterized to test for causality by constraining the SNP effect sizes and testing whether a latent causal variable mediates the association (5). Briefly, the GWAS summary data serve as instruments for a series of techniques that are called “genetic instrumental variable analysis”. Normally, genetic instrumental variable analysis selects the top SNPs for trait 1 and uses those as an instrumental variable “exposure”. If the SNPs that contribute to trait 1 are also associated with differences in trait 2 to a similar degree and effect size, then there is evidence for a plausibly causal association. LCV expands this analysis by using genome-wide SNPs, i.e. not only the “top” associations, as instruments. To do this, LCV models a latent “causal” variable that represents the pattern of consistency that would be observable if a causal relationship existed. LCV then tests the degree to which the latent “causal” variable mediates the correlation between both traits, so the degree of causality is measured as a ratio of sharing between the measured trait and the latent variables, giving us a two-tailed test of causality that simultaneously tests bi-directional causal effects. This ratio is known as the genetic causality proportion (GCP), an estimate of the degree to which each trait is correlated with the latent genetic variable, i.e., the extent to which each trait is potentially genetically causal for the other trait (ranging from 0 reflecting no genetic causality to |1| indicating full genetic causality). An advantage of LCV over MR is that it accounts for unknown amounts of sample overlap between the GWAS of the two traits in the model.
To evaluate our results for evidence of correlation and causality, we evaluate both the direction of effect of the SNP-rg and the direction of effect of the GCP. We can evaluate the direction of the LDSR genetic correlation as we would any other correlation measure, with a positive SNP-rg providing evidence of a positive relationship between the traits, and a negative SNP-rg suggesting an inverse correlation. If the GCP is positive, it suggests that the first trait in the model is causal for trait 2; if the GCP is negative, it means the second trait is causal for trait 1. For example, if trait 1 in our model is PAU and trait 2 is volume of a brain region, a negative genetic correlation would imply that lower volume for the brain region is correlated with greater risk for problematic alcohol use; furthermore, a negative GCP would imply that volume of the brain region has a causal effect on problematic alcohol use.
Latent causal variable (LCV) analyses (5) between PAU and the 278 MRI imaging phenotypes were conducted using the MASSIVE pipeline (https://view.genoma.io/) (11). For each of the 278 imaging phenotypes, we tested for putatively causal associations and genetic correlations with PAU; we corrected for multiple testing using Bonferroni correction within each modality. The major histocompatibility complex (MHC) region was removed for all analyses.
Results
Three GCP estimates were significant after multiple corrections. Increased cortical volume of the left pars operculais (GCP = −0.643(0.173)) was significant after multiple corrections within modality (P =0.00019, N regions= 62, Bonferroni threshold = 0.00081). Enigma cortical surface metrics were averaged across the hemisphere, and greater thickness of the cuneus (GCP −0.226(0.066), P= 0.00059) was significant when correcting within modality (N regions = 35, Bonferroni threshold= 0.001429). Finally, lower subcortical brain volume of the left basal forebrain (GCP = −0.489(0.144)) was implicated when correcting within number of subcortical brain areas (p = 0.00067, N regions = 35, Bonferroni Threshold = 0.001429). No results were significant after a conservative correction for all brain regions (0.05/278, correction = 0.00018). All nominally significant results were in line with deviations in brain areas causing PAU, rather than PAU causing changes in brain area (Table 1 and Figure 1).
Table 1.
Bonferroni Corrected and Nominally Significant Results from Latent Causal Variable Analyses of Brain Structure and Problematic Alcohol Use
| Modality | Region | GCP | GCP SE | GCP P | SNP-rg | h2 |
|---|---|---|---|---|---|---|
| Cortical Volume | Left Pars Opercularis | −0.643 | 0.173 | 0.00019 | 0.062 | 0.172 |
| Cortical Thickness | Cuneus | −0.226 | 0.066 | 0.00059 | 0.113 |
0.103
|
| Subcortical Volume | Left Basal Forebrain | −0.489 | 0.144 | 0.00067 | -0.067 | 0.103 |
| Cortical Thickness | Precuneus | −0.447 | 0.162 | 0.00584 | 0.100 | 0.124 |
| Cortical Volume | Right Pars Triangularis | −0.584 | 0.243 | 0.01613 | −0.104 | 0.147 |
| Cortical Thickness | Pericalcarine Gyrus | −0.343 | 0.143 | 0.01641 | 0.109 | 0.061 |
| Cortical Thickness | Postcentral Gyrus | −0.600 | 0.260 | 0.02116 | 0.103 | 0.101 |
| Cortical Volume | Right Superior Frontal Gyrus | −0.555 | 0.250 | 0.02629 | −0.186 | 0.333 |
| Subcortical Volume | Right Vessel | −0.563 | 0.259 | 0.02977 | −0.182 | 0.233 |
| Cortical Volume | Right Lateral Occipital Gyrus | −0.553 | 0.258 | 0.03227 | −0.092 | 0.242 |
| Cortical Thickness | Lingual Gyrus | −0.481 | 0.227 | 0.03380 | 0.183 | 0.101 |
| Cortical Thickness | Mean Thickness | −0.567 | 0.270 | 0.03553 | −0.216 | 0.260 |
| Cortical Thickness | Medial Orbitofrontal Cortex | −0.522 | 0.264 | 0.04790 | −0.117 | 0.029 |
Note: Nominal significance was determined by GCP estimates with P < .05 before multiple correction. GCP=genetic causality proportion, SE = Standard error of GCP, GCP P = GCP p-value, SNP-rg = genetic correlation, h2 = SNP heritability (calculated by the original study). Bold represents phenotypes that are significant after Bonferroni correction. For ENGIMA cortical surface phenotypes, only mean across hemispheres was available. All results from the UKBiobank (gray and white matter volume) use bilateral parcellations. The h2 is from the original study for each phenotype.
Figure 1. Latent Causal Variable Estimates and Genetic Correlations Between Brain Structure Phenotypes and Problematic Alcohol Use (n=278).
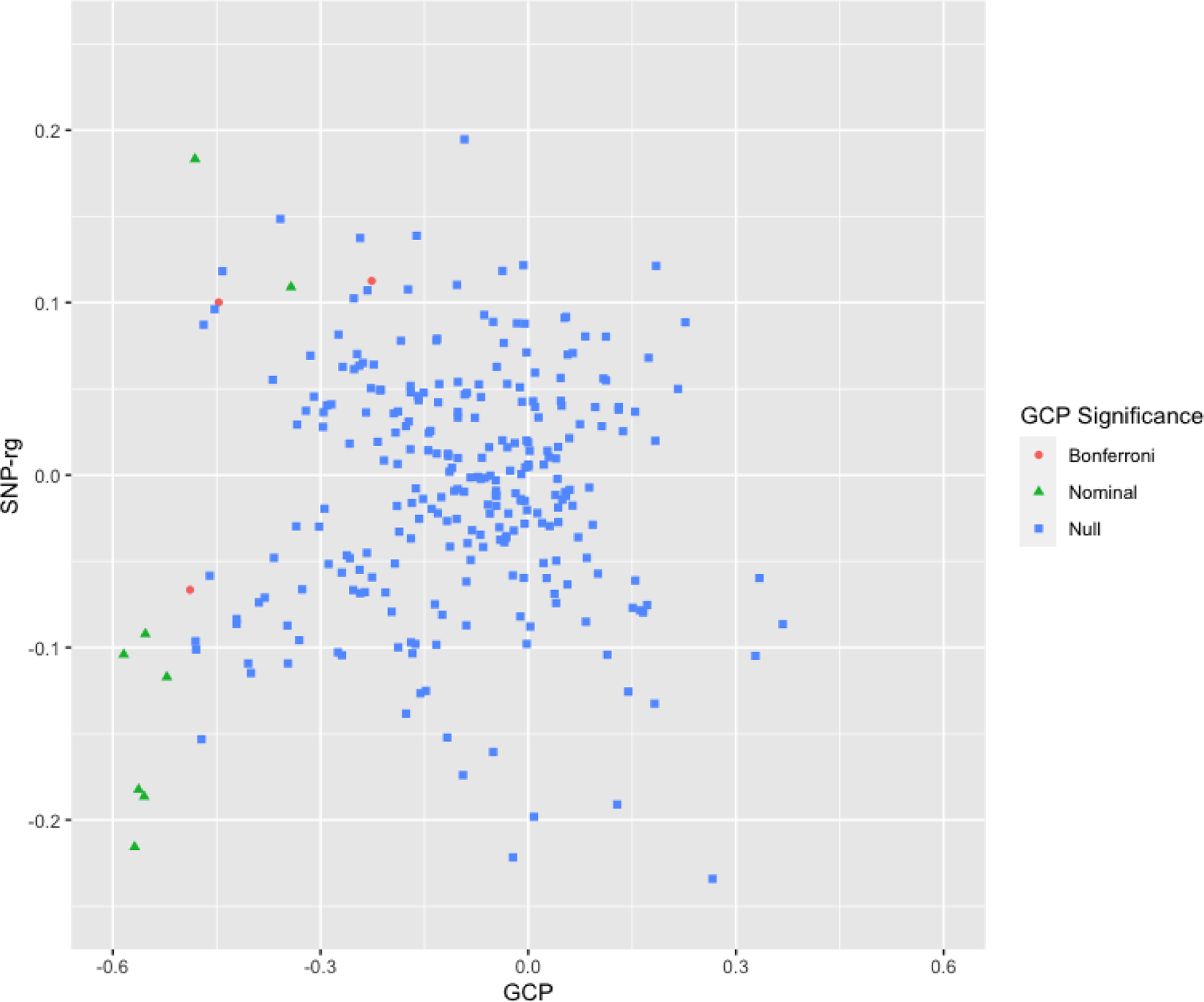
Latent causal variable estimates for brain structure phenotypes (n=278) and problematic alcohol use plotted by Genetic Causality Proportion (GCP; X axis) and Genetic Correlation (SNP-rg Y Axis). Coloration and shape represent GCP significance; blue squares represent non-significant GCPs, green triangles represent nominally-significant GCPs (p < .05), orange squares represent significant GCPs following Bonferroni correction (correction within modality). All Bonferroni and nominally significant GCPs were negative and the spread of the scatter plot is greater on the negative side. This is suggestive of genetic liability for brain structure putatively causing liability for problematic alcohol use.
Discussion
We used LCV analyses to estimate putative causal relationships between brain structure and problematic alcohol use (PAU). In contrast to speculation that neuroimaging-derived brain structure correlates of PAU reflect neurotoxic consequences of alcohol (12), our analyses revealed evidence that brain structure phenotypes may, at least partially, contribute to PAU (Table 1; Figure 1). We found no evidence that PAU contributes to brain structure. After correction for multiple testing, there was evidence that lower basal forebrain volume as well as greater volume of the pars opercularis and thickness of the cuneus may genetically contribute to PAU.
Reduced volume of the basal forebrain has been linked to chronic alcohol use disorder as well as working memory performance (13). Alongside evidence from non-human animal models that binge drinking can reduce basal forebrain volume (14) this has led to speculation that heavy alcohol use may cause these reductions, and, in turn, underlie behavioral impairments associated with PAU (e.g., executive control (13)). Our findings provide a counterpoint to this interpretation and suggest that lower basal forebrain volume may, at least partially, represent a genetically associated predisposing risk factor for problematic alcohol use. Such findings are consistent with emerging research suggesting that genomic liability to executive function is shared with PAU (15).
In contrast to evidence linking reduced inferior frontal gyrus volume to AUD, we find evidence that greater pars opercularis volume is associated with PAU. It is likely that Pars Opercularis is playing a role through language (16). Better language ability may contribute to initial escalations in alcohol use that provide the foundation for the development of alcohol use disorder (17). We are unaware of any prior findings linking cortical thickness of the cuneus to alcohol, though cuneus functional activation has been related to impulsive choice in AUD cases (18).
Some limitations are worth noting. First, as the GWAS were conducted in individuals of European ancestry, findings may not generalize to other ancestral groups. Second, we were unable to evaluate whether our findings replicate using independent GWAS summary statistics. As most GWASs gather and meta-analyze the largest possible datasets for discovery, there are limited opportunities to use well-powered GWAS summary statistics generated from independent samples for replication. Nonetheless, related approaches assessing the plausibility of causality (e.g., longitudinal data, discordant twin/sibling designs) may be leveraged to assess convergence of results (2). Limitations notwithstanding, our estimation of putatively causal bidirectional genetic relationships between brain structure and PAU using GWAS data yielded evidence that brain structure may contribute to the development of PAU, but that PAU may not contribute to brains structure.
Acknowledgments.
The Million Veterans Project summary statistics were accessed via dbGaP (phs001672.v1.p1) as part of #24806: Neurobiological bases of psychiatric traits. The authors thank Million Veteran Program (MVP) staff, researchers, and volunteers, who have contributed to MVP, and especially participants who previously served their country in the military and now generously agreed to enroll in the study. (See https://www.research.va.gov/mvp/ for more details).The citation for MVP is Gaziano, J.M. et al. Million Veteran Program: A mega-biobank to study genetic influences on health and disease. J Clin Epidemiol 70, 214–23 (2016). This research is based on data from the Million Veteran Program, Office of Research and Development, Veterans Health Administration, and was supported by the Veterans Administration (VA) Cooperative Studies Program (CSP) award #G002. Data were also accessed via the Enhancing Neuro Imaging Genetics Through Meta-Analysis (ENIGMA) Consortium. More on ENIGMA can be found here: http://enigma.ini.usc.edu/. Data are made available via the MASSIVE pipeline here: https://view.genoma.io/.
Authors received funding support from NIH: Dr. Hatoum (DA007261–17), Dr. Johnson (F32AA027435), Dr. Agrawal (MH109532 and K02DA032573), Dr. Bogdan (AG052564, AA027827, DA046224). ASH, ECJ, AA, and RB developed the research questions. ASH conducted analyses. ASH and RB drafted the manuscript. ECJ and AA provided critical revision of the manuscript for important intellectual content. ASH had full access to all data in the study and take responsibility for the integrity of the data and accuracy of the data analyses. Conflict of interest disclosures: No disclosures were reported.
References
- 1.Durazzo TC, et al. , Cortical thickness, surface area, and volume of the brain reward system in alcohol dependence: Relationships to relapse and extended abstinence. Alcohol. Clin. Exp. Res 35, 1187–1200 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baranger DAA, et al. , Convergent Evidence for Predispositional Effects of Brain Gray Matter Volume on Alcohol Consumption. Biol. Psychiatry (2019) 10.1016/j.biopsych.2019.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen L, Davey Smith G, Harbord RM, Lewis SJ, Alcohol Intake and Blood Pressure: A Systematic Review Implementing a Mendelian Randomization Approach. PLoS Med. 5, e52 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smith GD, Ebrahim S, What can mendelian randomisation tell us about modifiable behavioural and environmental exposures? Br. Med. J 330, 1076–1079 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.O’Connor LJ, Price AL, Distinguishing genetic correlation from causation across 52 diseases and complex traits. Nat. Genet 50, 1728–1734 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhou H, et al. , Genome-wide meta-analysis of problematic alcohol use in 435,563 individuals yields insights into biology and relationships with other traits. Nat. Neurosci, 1–10 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grasby KL, et al. , The genetic architecture of the human cerebral cortex. Science (80-. ) 367 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhao B, et al. , Genome-wide association analysis of 19,629 individuals identifies variants influencing regional brain volumes and refines their genetic co-architecture with cognitive and mental health traits. Nat. Genet 51, 1637–1644 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhao B, et al. , Large-scale GWAS reveals genetic architecture of brain white matter microstructure and genetic overlap with cognitive and mental health traits (n = 17,706). Mol. Psychiatry, 1–13 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bulik-Sullivan BK, et al. , LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat. Genet 47, 291–295 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cuellar-Partida G, et al. , Complex-Traits Genetics Virtual Lab: A community-driven web platform for post-GWAS analyses. bioRxiv, 518027 (2019). [Google Scholar]
- 12.Anstey KJ, et al. , Weekly Alcohol Consumption, Brain Atrophy, and White Matter Hyperintensities in a Community-Based Sample Aged 60 to 64 Years. Psychosom. Med 68, 778–785 (2006). [DOI] [PubMed] [Google Scholar]
- 13.Sullivan EV, Deshmukh A, De Rosa E, Rosenbloom MJ, Pfefferbaum A, Striatal and forebrain nuclei volumes: Contribution to motor function and working memory deficits in alcoholism. Biol. Psychiatry 57, 768–776 (2005). [DOI] [PubMed] [Google Scholar]
- 14.Coleman LG, He J, Lee J, Styner M, Crews FT, Adolescent Binge Drinking Alters Adult Brain Neurotransmitter Gene Expression, Behavior, Brain Regional Volumes, and Neurochemistry in Mice. Alcohol. Clin. Exp. Res 35, 671–688 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hatoum A, et al. , GWAS of Over 427,000 Individuals Establishes GABAergic and Synaptic Molecular Pathways as Key for Cognitive Executive Functions. GWAS Over 427,000 Individ. Establ. GABAergic Synaptic Mol. Pathways as Key Cogn. Exec. Funct, 674515 (2019). [Google Scholar]
- 16.Woodward KE, et al. , Childhood language development and later alcohol use behaviors. Drug Alcohol Depend. 198, 95–99 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Latvala A, Rose RJ, Pulkkinen L, Dick DM, Kaprio J, Childhood Verbal Development and Drinking Behaviors from Adolescence to Young Adulthood: A Discordant Twin-Pair Analysis. Alcohol. Clin. Exp. Res 38, 457–465 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Claus ED, Kiehl KA, Hutchison KE, Neural and Behavioral Mechanisms of Impulsive Choice in Alcohol Use Disorder. Alcohol. Clin. Exp. Res 35, 1209–1219 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]


