Summary
The thymus is required for the development of both adaptive and innate-like T cell subsets. There is keen interest in manipulating thymic function for therapeutic purposes in circumstances of autoimmunity, immunodeficiency, and for purposes of immunotherapy. Within the thymus, thymic epithelial cells play essential roles in directing T cell development. Several transcription factors are known to be essential for thymic epithelial cell development and function, and a few transcription factors have been studied in considerable detail. However, the role of many other transcription factors is less well understood. Further, it is likely that roles exist for other transcription factors not yet known to be important in thymic epithelial cells. Recent progress in understanding of thymic epithelial cell heterogeneity has provided some new insight into transcriptional requirements in subtypes of thymic epithelial cells. However, it is unknown whether progenitors of thymic epithelial cells exist in the adult thymus, and consequently developmental relationships linking putative precursors with differentiated cell types are poorly understood. While we do not presently possess a clear understanding of stage-specific requirements for transcription factors in thymic epithelial cells, new single-cell transcriptomic and epigenomic technologies should enable rapid progress in this field. Here, we review our current knowledge of transcription factors involved in the development, maintenance, and function of thymic epithelial cells, and the mechanisms by which they act.
Keywords: Thymic Epithelial Cells, T cell development, Transcription factors
1. INTRODUCTION
The thymus is the essential organ for T cell development. Thymic epithelial cells are persistent elements of the thymus that direct the development of αβ-lineage CD4+ and CD8+ T cells, γδ-lineage T cells, invariant natural killer T (iNKT) cells, as well as other types of cells including intraepithelial lymphocytes (IELs) and some types of innate lymphocytes.1-4 In this review, we first briefly introduce the different types of thymic epithelial cells. Subsequently, we discuss transcription factors that support the development and maintenance of thymic epithelial cells, and we present our current knowledge of the mechanisms by which they act.
2. TYPES OF THYMIC EPITHELIAL CELLS
Thymic epithelial cells (TEC) are specialized cells that direct the generation of immunocompetent and self-tolerant T cells. TEC are fundamental for T cell generation and thus should not be considered as thymic stroma, because this term refers to supporting rather than essential cells. TEC cooperate with diverse other cell types including macrophages, dendritic cells, endothelial cells and fibroblast cells to coordinate T cell development.1,5
TEC are phenotypically defined as CD45−EpCAM+ cells. Based on their anatomical localization and the expression of specific markers, TEC are divided into two major subsets: cortical (c) TEC (Ly51+ UEA1−) and medullary (m) TEC (Ly51− UEA1+).3 Bipotential progenitors for cTEC and mTEC are identified in fetal mice6-8; however, the cellular basis of TEC development and maintenance in adult mice and humans is poorly understood.9-17
2.1. Cortical Thymic Epithelial Cells (cTEC).
cTEC support early steps of T cell development and positive selection of developing T cells, during which T cell precursors are assessed for productive TCR assembly by their ability to recognize self-antigens. To promote thymus homing and support intrathymic migration of T cell precursors, cTEC express CXCL12 and CCL25 chemokines.18-20 They also express the Notch ligand Delta-like 4 (DLL4) that drives Notch-dependent T cell commitment and proliferation during early steps of T cell development.21,22 In addition, cTEC produce the cytokine IL-7 which acts at several steps of thymocyte maturation.23,24 cTEC are essential drivers of positive selection as they express proteins that generate MHCI and MHCII-associated self-peptides. These include the thymus-specific proteasomal subunit β5t, and thymus-specific serine protease (TSSP).25-27
2.2. Medullary Thymic Epithelial cells (mTEC).
Subsequent to positive selection, mTEC negatively select thymocytes with dangerously high affinity to self-peptides, or divert them into the regulatory T cell (Treg) lineage.3,28 Thus, mTEC implement quality control of the T cell repertoire. Based on the expression levels of MHCII and CD80, mTEC are divided in two major populations: mTEClo (MHCIIlo and CD80lo) and mTEChi (MHCIIhi and CD80hi). mTEClo contain CCL21a-expressing cells29, whereas mTEChi express the transcription factors autoimmune regulator (Aire)30 and Fezf2.31 Recent work using single-cell RNA sequencing characterized mTEC heterogeneity in mice, and identified four mTEC subsets (mTEC I-IV).32,33 Of these, mTEC I were CCL21a-expressing cells that direct the movement of positively selected thymocytes that express CCR7 from the cortex to the medulla.32,34 mTEC II displayed promiscuous gene expression (PGE) of tissue-restricted self-antigens (TRA) and specific expression of the transcription factors Aire and Fefz2, as well as CD40, and high levels of MHC-II.32 mTEC III were identified by the expression of Krt10 and Ivl, and this population included post-Aire cells.32,33 mTEC IV were termed thymic tuft cells as the produced and expressed genes of the canonical taste transduction pathway, such as Pou2f3, Dclk1, and Trpm5.32,33
Recent single-cell RNA-sequencing (scRNA-seq) analyses of human TEC revealed TEC subpopulations that possessed transcriptional programs similar to previously identified mouse TEC, specifically cTEC, mTEClo, mTEChi, mTEC III, and mTEC IV.35,36 Significantly, new TEC clusters were identified, including immature human TEC which lacked the expression of either cTEC or mTEC genes.35 Adult human thymus showed expansion of this immature TEC population, and reduction of both mTEClo and mTEChi. Interestingly, additional rare mTEC subsets were also identified, including neuroendocrine cells, myoid cells, myelin+ cells, ciliated cells, and ionocytes, based on their transcriptional profile.35,36 Human ionocytes, tuft cells, neuroendocrine cells and myoid cells were localized close to Hassall’s corpuscles.35 The functions of these rare mTEC populations are poorly understood.
3. TRANSCRIPTIONAL REGULATORS OF TEC DEVELOPMENT AND FUNCTIONS
3.1. Foxn1
The role of TEC in thymus development was revealed in nude (hairless) mice, characterized by a rudimentary thymus and profound reduction of T cells.37 Bone marrow from nude mice repopulated thymus and peripheral T cells in lethally irradiated wild-type mice. Nude mice, by contrast, could not be reconstituted with wild-type progenitor cells.38 These results indicated that the lack of T cells in nude mice was secondary to a defect in their thymic epithelium.38
Subsequent studies have shown that in humans and mice, the nude phenotype is associated with mutations of Foxn1, a member of the forkhead/winged-helix transcription factors.39 Autosomal-recessive mutations of Foxn1 generate the nude (hairless) athymic phenotype featured by congenital absence of hair and severe combined immunodeficiency.39 Foxn1 is expressed as early as embryonic day (E) 9.5 in the third pharyngeal pouch of the mouse embryo.39 Furthermore, Foxn1 needs to be continuously expressed in TEC, as its postnatal ablation induced thymic atrophy.40,41 Autosomal recessive mutations of FOXN1 in patients result in thymic aplasia, alopecia, and nail plate dystrophy. In addition, heterozygous loss-of-function mutations of Foxn1 resulted in CD8 T cell lymphopenia during infancy and, in some cases, nail dystrophy and alopecia.42 Interestingly, two patients harboring compound heterozygous mutations of Foxn1 showed severe T cell defects but did not exhibit alopecia or nail dystrophy.43 CRISPR/ Cas9-mediated genocopy of one of these mutations in mice discovered that a stretch of 5-amino acids in the Foxn1 DNA binding domain was required in TEC but not in keratinocytes.43
Genome-wide assessments of chromatin binding and transcription identified Foxn1 gene targets in TEC. Specifically, Foxn1 regulated cTEC-specific genes necessary for early lymphoid progenitor immigration into the thymus and antigen processing and presentation (including β5t).41 Regulation of Psmb11 (the gene for β5t) by Foxn1 has been studied in more detail. Foxn1 interacts with two cis-regulatory elements in the vicinity of the transcription start site (TSS) of Psmb11. Mutation of the −80bp binding site resulted in reduced expression of Psmb11, establishing that Psmb11 receives direct regulatory inputs from Foxn1.44
Ectopic expression of Foxn1 was sufficient to partially reprogram mouse embryonic fibroblasts to display features associated with epithelial cells, such as expression of epithelial cell adhesion molecule (EpCAM) and Keratin 8.45 Remarkably, these cells were able to support T cell development in vitro. Furthermore, these Foxn1-expressing cells recapitulated thymus morphology and T cell development in vivo when mixed with embryonic TEC and grafted under the kidney capsule of adult mice.45 These data suggest that Foxn1 is a master regulator of TEC, as it is necessary for TEC development and maintenance, and appears sufficient to impose features of TEC when ectopically expressed in fibroblasts.
As mice age, the thymus undergoes involution, reducing in size and function. Thymic involution was associated with reduced expression of Foxn1.46 Ectopic overexpression of Foxn1 driven by the human keratin-14 promoter attenuated thymic involution.47 Thymic regeneration induced by sublethal total body irradiation (TBI) also was associated with induction of Foxn1 expression in TEC. Specifically, endothelial cells were found to secrete bone morphogenetic protein 4 (Bmp4) in response to TBI, and Bmp4 upregulated expression of Foxn1 in TEC.48
Several studies have examined regulation of Foxn1. Pax family genes are expressed in endoderm, and mice deficient in Pax1 have a small thymus, whereas mice deficient in Pax9 are athymic.49-51 Deficiency in transcription factors Hoxa3, Eya1, or Six1 results in thymic defects at embryonic stage (Table 1).52-55 Signaling pathways including Wnt56 and Bmp448 have also been suggested as regulators of Foxn1 expression. Further molecular characterization of the role of all these factors is needed, as it is unclear if they directly regulate Foxn1 expression, or instead may act earlier in development, as is known for Gata3.57,58 Deletion of a DNA regulatory element (RE) in the first intron of Foxn1 established that this RE is necessary for expression of Foxn1 in TEC but dispensable for Foxn1 expression in skin.59 This RE contains binding sites for many transcriptional regulators expressed in TEC that could potentially control Foxn1 expression.59
Table 1.
Transcriptional and epigenetic regulators associated with thymic development and maintenance.
| Factor | Mouse model | Phenotype |
|---|---|---|
| Aire | Constitutive loss-of-function mutation | Aire deficiency provoked multiorgan autoimmunity.30 |
| Aff4 | Constitutive KO | Ablation of Aff4 induced a nude-like phenotype. Embryos exhibited thymic hyplopasia became that was more severe when one Foxn1 allele was defective.197 |
| Eed | Cre-Foxn1 Eedfl/fl Cre-β5t Eedfl/fl |
Deficiency of Eed resulted in thymic atrophy decrementing TCR diversity.189,191 |
| Eya1 | Constitutive KO | Eya1 deficiency compromised the formation of thymus and parathyroid structures.54,55 |
| Gata3 | Constitutive KO | Ablation of Gata3 impeded developmental progression of the thymus/parathyroid primordia.57,58 |
| HDACs | Cre-Foxn1 HDAC3fl/fl | Deficiency of HDAC3 in thymus inhibited mTEC development and TRA expression, and lead to autoimmunity.183 |
| Hoxa3 | Constitutive KO | Ablation of Hoxa3 altered the formation of the thymus, thyroid and parathyroid organs. Hoxa3 deficiency reduced Pax1 and Pax9 expression levels.52,53 |
| Fezf2 | Constitutive KO and Cre-Foxn1 Fezf2fl/− | Fezf2 regulates Aire-independent TRA expression. Knocking out Fezf2 caused autoimmunity in several peripheral tissues. 31 |
| Foxn1 | Nude mice | Deficiency of Foxn1 disrupted thymic development and hair growth. Foxn1 was also required for postnatal TEC maintenance.39-41 |
| Foxa1/2 | Cre-Foxn1 Foxa1fl/fl Foxa2fl/fl | Ablation of both Foxa1 and Foxa2 reduced thymi size, and increased the ratio of mTEC:cTEC.178 |
| Meis1 | CreERT2-K14 Meis1fl/fl | Meis1 ablation in adult mice leaded to loss of thymus size. T cell development was also compromised.173 |
| Myc | Cre-Foxn1 Mycfl/fl and Cre-Foxn1 MycTg | Myc deficiency reduced the proliferation of TEC but the differentiation to mTEC was unaltered. Ectopic Myc expression increased TEC proliferation and thymus size.166,167 |
| NF-kB | Constitutive KO of Tnfrsf11a, Traf6, Cd40, or Relb. Constitutive expression of mutated Nik (G855R) | Inhibition of NF-kB signaling resulted in thymic medullary atrophy, leading to autoimmunity.63,73,75,81,198,199 |
| p53 | Cre-Foxn1 p53fl/fl | Ablation of p53 inhibited mTEC differentiation, thymocyte maturation and compromised peripheral tolerance in 6-7 month old mice.160 |
| p63 | Constitutive KO | Deficiency in p63 resulted in thymic hyploplasia during embryonic development.91,92 |
| Pou2f3 | Constitutive KO | Pou2f3 deficiency resulted in loss of thymic tuft cells.32,33 |
| RBPJ | Cre-Foxn1 Rbpjfl/fl, CreER-Foxa2 Notchfl/fl, Cre-Foxn1 RosaN1-IC, RBPJ fl/fl Cre-Foxn1; Rosa rtTA; Teton-RBPJ-HA | Deficiency of either Notch1 or RBPJ inhibited the establishment of mTEC progenitor cells during fetal development, and reduced the mTEC compartment postnatally.179,180 |
| Six1 | Constitutive KO | Six1−/− embryos failed to generate the thymus/parathyroid common primordia. Foxn1 expression was reduced in the common primordia in Six1−/− embryos.55 |
| Tbx1 | Cre-Foxn1 R26-iTbx | Ectopic expression of Tbx1 reduced proliferation and blocked differentiation of TEC progenitors at embryonic stage. Overexpression of Tbx1 inhibited Foxn1 expression but induced targets genes of the Polycomb Repressive Complex (PRC) 2.188,189 |
| Pax1 | Constitutive KO | Pax1 ablation produced a hypoplastic thymus with deficient thymocyte development.49,200 |
| Pax9 | Constitutive KO | Organs derived from the third pharyngeal pouch, including the thymus, were compromised.50 |
3.2. NF-κB
The noncanonical NF-κB pathway responds to specific stimuli including the tumor necrosis factor (TNF) family receptors CD40, RANK, and LTβR, among others 60. Those stimuli result in a complex cascade of events that eventually activate the NF-κB inducing kinase (NIK). NIK induces proteolytic processing of p100, resulting in the generation of p52 that, in complex with RelB, is translocated to the nucleus.61,62
RANK, CD40, and LTβR each play important roles in mTEC development, Aire-dependent and -independent TRA gene expression, and establishment of self-tolerance.63-65 CD4 SP thymocytes showed low expression levels of LTα/β in mice lacking costimulatory molecules CD40, CD80 and 86. In addition, these mice exhibited reduced thymic medullary regions producing low levels of CCL21.64 Concordantly, ablation of LTβR in TEC reduced numbers of mTEC, with aberrant differentiation and reduced expression of CCL21.29,66-68 Together, the data indicate that CD80/86 interaction with CD28 mediates the expression of LTα/β by T cells, which in turn induces the production of CCL21 by mTEC via LTβR, inducing mTEC development. Ablation of both CD40 and RANK severely impaired postnatal mTEC differentiation resulting in autoimmunity.63 The inhibition of RANK signaling achieved by either knocking out Tnfrs11a (the gene for RANK) or blocking its ligand, RANKL, inhibited embryonic mTEC development.63,69 During embryonic development, γδ T cell progenitors provided RANKL prior to the appearance of αβ T cells.70 The thymus of embryonic Cd40−/− mice did not show dramatic alterations, as CD40L-expressing T cells are absent at this stage of development.71
Tumor necrosis factor receptor (TNFR)-associated factor 6 (TRAF6) is a signal transducer downstream of RANK and CD40. Traf6−/− mice displayed thymic atrophy.72 By grafting Traf6 −/− fetal thymus under the kidney capsule of athymic nude mice, it was demonstrated that cell-intrinsic TRAF6 deficiency reduces the numbers of mTEC, leading to multi-organ autoimmunity.73 In addition, RANKL-mediated mTEC development was compromised by TRAF6 ablation.63 TRAF6 is a RING domain ubiquitin E3 ligase that phosphorylates many proteins including NIK, which in turn activates the noncanonical NF-κB pathway.74 The NIKaly/aly mice strain is also known as alymphoplasia strain since it lacks lymph nodes and Peyer’s patches. These mice harbor a mutation (G855R) in the NIK gene. This mutation impedes NIK ability to bind IKK-α, and so inhibits noncanonical NF-κB activation.75 NIKaly/aly mice showed smaller medullary regions and alterations at the cortico-medullar junction of the thymus. In addition, these mice had decreased numbers of Treg cells, and autoimmune lymphocyte infiltrations in several organs.76 IKK-α is downstream of NIK, and BALB/Cnu/nu mice grafted with embryonic thymus lacking functional Ikk-α phenocopied the NIKaly/aly phenotype, as expected.77 Loss-of-function mutations in both NIK and IKK triggered autoimmunity, likely due to deficient mTEC development.63,77 By using Relb−/−, Traf6−/−, and NIKaly/aly mice, together with pharmacological inhibition of IKKβ, it was demonstrated that RANK-RANKL and CD40L-CD40 signals act to accomplish thymic self-tolerance.63
As mentioned, noncanonical NF-κB regulates the size and function of the mTEC compartment. The NF-κB family regulates Aire expression by interacting with a distal regulatory element located 3 kb upstream of the Aire gene78,79, and the Aire-deficient phenotype was recapitulated by deleting that enhancer region.78,79 Concordantly, RelB regulated Aire expression, and its ablation disrupted thymic structure and generated autoimmunity.80,81 Among mTEC, mTEC II exhibited open chromatin regions enriched for NF-κB binding sites32, consistent with the idea that many genes might be transcriptionally activated by NF-κB in addition to Aire in mTEC II. Thus, the gene regulatory program in mTEC II controlled by NF-κB remains to be better understood.
As mentioned above, RANKL, CD40L, and LTβ are required for mTEC development by activating the noncanonical NF-κB pathway. TRAF3 is an intracellular inhibitor of the noncanonical NF-κB pathway.82 Consistently, ablation of TRAF3 rescued mTEC development in Tcra−/−, Ltbr−/−/Cd40−/−, Rank−/− mice.83
Manipulation of noncanonical NF-κB signaling altered central tolerance. Osteoprotegerin (OPG) functions as a decoy receptor of the RANKL, inhibiting RANK signaling. OPG deficiency increased the number of mTEC, enlarging the thymic medulla compartment.69 Ablation of OPG in fetal thymic grafts in nude mice increased chemical-mediated tumor incidence due to increased Treg cell numbers and lower tumor-infiltrating T cells.84 Importantly, Spi-b, a transcription factor induced by RANK through NIK-mediated signaling, upregulated OPG expression in mTEC, limited mTEC development, and decreased TRA expression.84 Thus, OPG is an endogenous key regulator of the NF-κB mediated mTEC development.
Experimentally modulating mTEC via RANK has been demonstrated to modify T cell-mediated responses. Antibody-mediated blocking of RANK-RANKL signaling induced transient reduction of the Aire+ mTEC population and TRA expression. Negative selection of the thymocytes was reduced, and self-reactive T cells emerged.85 Experimental blockade of RANKL induced anti-tumor responses against a self-antigen-expressing melanoma cells, indicating that such manipulations could be useful for immunotherapy to cancers.85
3.3. p63
The p63 transcription factor, encoded by Trp63, is a protein orthologue of p53 and is highly expressed in many stratified epithelial tissues, including skin, cervix, tonsil, esophagus, urothelium, prostate and thymus.86,87 Trp63 codifies several isoforms. Alternative promotors generate two main types of isoforms: the N-terminal full transactivating (TA) p63 isoforms (N-terminal full-length) and the N-terminal truncated/Delta ΔNp63 isoforms (that lack the N-terminal transactivation domain but are still transcriptionally active).86 Alternative splicing of the C-terminus generates additional α, β, and γ isoforms.88 Mice lacking p63 exhibited severe craniofacial and limb malformations as well as profound defects in stratified epithelial tissues and derivatives such as hair follicles and breast.89,90 The epidermis of p63−/− mice was found to undergo nonregenerative differentiation and disintegration.91 Thus, mice lacking p63 die in the perinatal period due to dehydration.89,90
TEC predominantly express the ΔNp63α isoform.87 The effects of p63 deletion have been studied in the thymus of mice with p63 deleted in the germline. Loss of p63 caused thymic hypoplasia in embryos/neonatal mice, and thymocyte and TEC numbers were dramatically reduced to ~10% of wild-type numbers.91,92 However, developing thymocytes appeared normal based on surface markers such as CD4 and CD8 expression. Morphologically, the expression of TEC markers, Keratin 5 and Keratin 8, was similar between p63−/− and wild-type mice.91,92 Thus, unlike nude mice in which the thymus was almost entirely absent 39, p63−/− mice showed sufficient TEC commitment and differentiation to support T-cell development. Nevertheless, p63−/− thymi displayed biased expression towards terminally differentiated epithelial genes like involucrin, occludin and claudins.91 In addition, silencing of p63 resulted in smaller and less proliferative colonies in clonogenicity assays of TEC.91 These data suggest that p63 may be important for maintaining epithelial stem cells; thus, its ablation may result in thymic hypoplasia due to exhaustion of proliferating stem/progenitor cells.
Efforts have been made to identify p63 target genes that drive epithelial development.93 Genome-wide approaches have shown that p63 regulates epidermal proliferation and differentiation not only via direct activation or repression of target genes but also through modulating the chromatin landscape, predominantly controlling enhancers.94-97 To date, there are not genome or epigenome approaches assessing p63 regulation of TEC. However, using individual candidate gene approaches, some known p63 targets have been implicated in the TEC defect and thymic hypoplasia observed in mice lacking p63.92 Jag2 and FgfR2-IIIb expression was diminished in p63−/− whole thymi 92. Jagged (Jag) 2 is a Notch ligand expressed by TEC that supports T cell development.98,99 Fibroblast growth factor receptor 2-IIIb (FgfR2-IIIb) is essential for TEC proliferation, and FgfR2-IIIb deficiency results in a block of thymic growth, resulting in a similar phenotype as in p63 deficient mice.100 Genetic complementation with ΔNp63 in a p63−/− genetic background restored FgfR2-IIIb and Jag2 expression, suggesting that p63 may drive thymus growth at least in part via Jag2 and FgfR2IIIb.92
Chromobox homolog 4 (Cbx4), a component of the Polycomb group (PcG) multiprotein PRC1-like complex101, has been mechanistically associated with p63-mediated thymus growth.102 Like p63−/− embryos/neonatal mice, Cbx4−/− mice display decreased numbers of thymocytes and TEC but displayed normal CD4 and CD8 profile.102 Interestingly, in postnatal mice, Cbx4-deficient mice showed reduced proliferation of TEC, and blockade of T-cell development. Furthermore, Cbx4 co-immunoprecipitated with p63 in transfected HEK293T cells. Immunoassays showed co-expression of both proteins in the thymus of wild-type embryos and adult mice; however, the identity of TECs co-expressing p63 and Cbx4 was not established. These observations suggest that Cbx4 interacts with p63, contributing to the regulation of downstream gene targets.102 Notably, ex vivo embryonic skin explants assays showed that Cbx4 overexpression partially rescued the defect caused by p63 ablation.103 As previously proposed91, these studies support the hypothesis that p63 expression in TEC contributes to the self-renewal capacity of thymic epithelial stem/progenitor cells and p63 deficiency causes their premature proliferative exhaustion. The underlying mechanisms, however, are not understood, and genomic and epigenomic approaches to address how p63 shapes chromatin landscape and enhancers in TEC remain to be performed.
3. 4. Aire
As earlier noted, mTEChi is an mTEC subpopulation expressing CD80, CD86, CD40 and high levels of MHC class-II. These cells play an essential role in establishing central tolerance. They present a repertoire of tissue-restricted self-antigens (TRAs) to thymocytes, inducing the negative selection of T cells and the development of regulatory T cells.3,104 Aire is estimated to control ~40% of TRA expression31,105, and is a key controller of central tolerance as evidenced by loss-of-function mutations that cause autoimmune polyendocrine-candidiasis ectodermal dystrophy (APECED).30,106,107 APECED is characterized by autoimmunity in multiple organs and tissues including pancreas, adrenal cortex, skin, liver, and lung.30,107
Initial studies of Aire highlighted its role in the regulation of TRA expression.30,104,108 Since then, molecular mechanisms supporting Aire-mediated transcriptional activation of TRA have been elucidated. The epigenetic context in which Aire interacts with DNA regulatory regions have been characterized. Although at first glance it could be contradictory, it has been demonstrated that Aire also can restrain TRA expression, perhaps serving as a rheostat to maintain TRA gene expression at appropriate levels.109,110
3.4.1. Aire activates TRA expression
Aire harbors CARD (Caspase Recruitment Domain), SAND (Sp100, AIRE-1, NucP41/71, and DEAF-1) and two PHD (plant homeodomain) domains, as well as a nuclear localization signal (Fig. 1). The PHD1 domain interacts with DNA-PK111, which consists of Ku70 and Ku80 subunits, and is associated with DNA damage response, transcriptional regulation and cell cycle progression.112 Small molecule-mediated inhibition of DNA-PK function blocked its association with Aire and P-TEFb and inhibited the release of stalled RNA-polII.113-115
Figure 1.
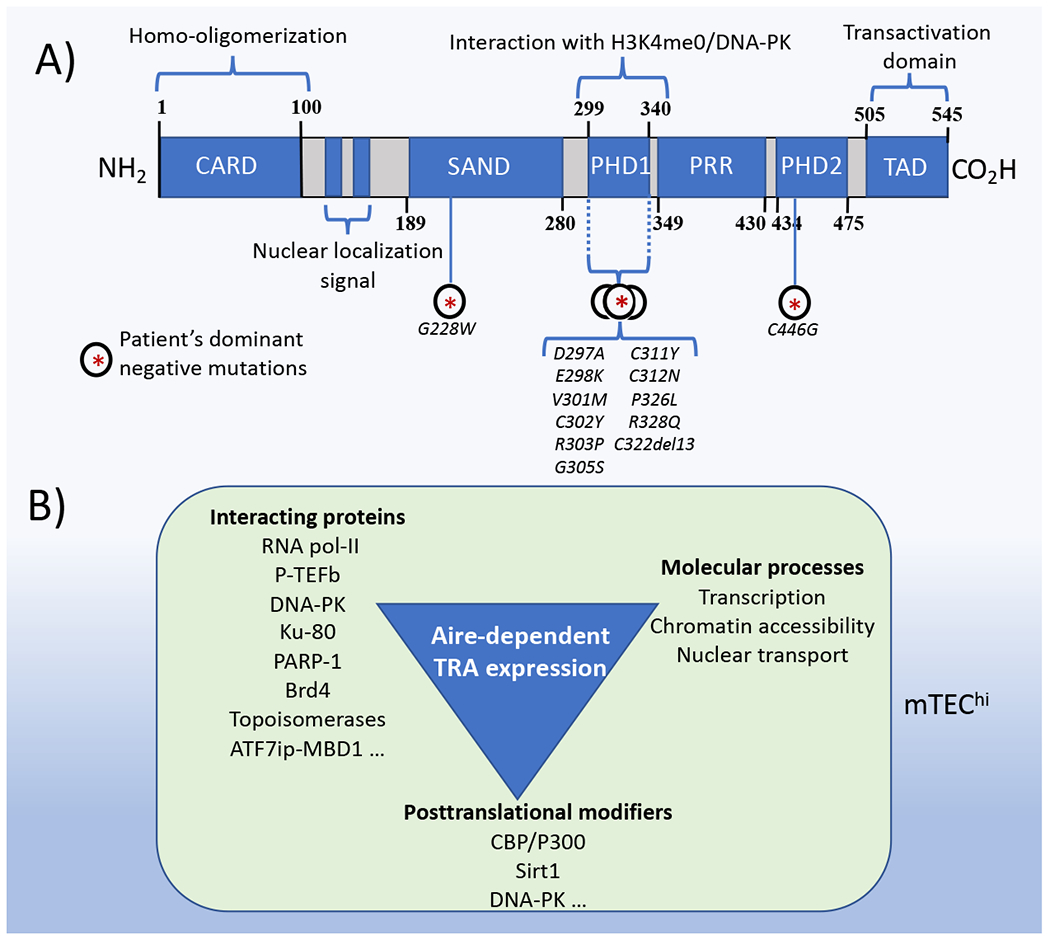
Schematic representation of the Aire protein and its functions. (A) Model of domains identified in human Aire. Known dominant-negative mutations are also indicated. (B) The diagram lists posttranslational modifiers of Aire, Aire interacting proteins, and molecular processes leading to Aire-mediated tissue-restricted antigen expression in mTEChi.
Mutations in Aire can cause autoimmune recessive as well as autosomal dominant disease in humans; however, the mechanisms are not fully understood. Aire forms homotypic interactions through its CARD domain to form multimers that bind chromatin.116,117 Mutations in the CARD domain prevented the formation of Aire multimers and showed reduced TRA expression in HEK293T cells.116 Aire-PHD1 heterozygous mutations also exhibited dominant-negative effects, inhibiting TRA expression and causing varying autoimmune phenotypes (Fig. 1).118 Aire with mutations in the PHD1 domain co-localized with wild-type Aire in nuclear speckles whereas Aire proteins with mutations in the SAND domain localized with PML bodies.116,118,119
The acetyltransferase CREB-binding protein (CBP) interacts with and acetylates Aire.120 Site-directed mutations that mimic the CBP-mediated acetylation (lysine to glutamine substitution) of Aire in a constitutive fashion impaired the ability of Aire to induce TRA gene expression.121 Sirt1 is an enzyme that removes acetyl groups from different proteins. Sirt1 is highly expressed in mTEChi. and counteracts the CBP-mediated acetylation of Aire, boosting Aire-dependent TRA expression.122 Thymic Sirt1 ablation resulted in multi-organ autoimmunity, similar to Aire-deficient mice.122
Brd4 belongs to the bromodomain and extraterminal domain (BET) family of transcriptional regulators. Brd4 interacts with P-TEFb at the transcription pre-initiation complex and promotes the transition from initiation to productive transcription elongation.123 Brd4 recognizes acetylated proteins by its bromodomain and functions as a scaffold for the recruitment of transcription factors.124 As previously mentioned, Aire influenced TRA gene expression by mediating the release of stalled RNA pol-II 115 via recruitment of P-TEFb.113 DNA-PK-mediated phosphorylation of Aire’s CARD domain triggered the association of CBP with Aire. Subsequently, CBP acetylated Aire’s CARD domain, and triggered its interaction with the Brd4 bromodomain 1.114 Inhibition of the formation of the Aire-Brd4 complex by inhibiting either CBP or DNA-PK enzymatical actions impeded the formation of the pre-initiation complex in which RNA-pol-II, topoisomerases, members of the splicing machinery, and P-TEFb are recruited.114 Concordantly, inhibition of the Aire-Brd4 interaction decreased TRA expression and led to the development of autoimmunity.114 Importantly, mutations in the Aire gene that cause APECED are also located in its CARD domain, and restricted association with Brd4.114 Mutations in the CARD domain of Aire can also compromise its ability to form self-multimers needed for the TRA expression, and so the relative contribution of each of these mechanisms to autoimmunity needs to be ascertained.
Over 40 Aire protein partners were identified by performing Aire-targeted coimmunoprecipitation coupled with mass spectrometry (MS) assays in epithelial cell lines.125 Aire was implicated in at least four nuclear processes: post-initiation RNA-PolII-mediated transcription, chromatin binding, nuclear transport, and pre-mRNA processing.125 Concordantly, RNAi-mediated downregulation of selected Aire partners impeded the nuclear location of Aire.125 Aire interacted with topoisomerases allowing the transcription of its TRA targets.125 Downregulation of TOP1 and TOP2 in HEK293T cells reduced the ability of Aire to interact with RNA-PolII, DNA-PK, Ku80, and PARP-1, among others.126 Blocking topoisomerase function repressed the transcription of Aire-responsive genes in mTEChi.126 Recent findings have shown only a limited role of Aire in alternative splicing.127,128
Aire’s protein partners were also identified in a separate study using yeast two-hybrid screening.129 Aire was found to interact with ATF7ip, a histone methylase that catalyzes H3K9me3; and with MBD1, which interacts with methylated CpG dinucleotides.129 However, the G228W mutation of the SAND domain of Aire impeded the association with the protein complex formed by ATF7ip and MBD1.129 This mutation was initially identified in patients diagnosed with APECED106,130 and is a dominant negative mutation of Aire.106,119 When Aire was ectopically expressed in HEK293T cells, it required ATF7ip and MBD1 to induce TRA expression.129 Ablation of MBD1 in mice led to autoimmunity.129
Aire localized mainly in intergenic regions instead of promoters126, highlighting a possible role in distal regulatory regions. Aire interacted with chromatin regions categorized as superenhancers (defined as those chromatin elements spanning more than 3.9 kb enriched in H3K27ac, H3K4me1, and deposition of RNA Pol-II, but not in H3K27me3).126 Aire deficiency decreased the number of superenhancer regions featured by high content of H3K27ac deposition.126
3.4.2. DNA methylation and Aire-mediated TRA expression
DNA methylation involves the addition of a methyl group to a cytosine residue by DNA methyltransferases (DNAmt). This epigenetic modification occurs predominately in CpG dinucleotides. DNA methylation is commonly associated with gene silencing and occurs in several biological processes such as development, X chromosome inactivation, genomic imprinting, among others.131
To uncover a possible association between DNA methylation in the TRA expression, the methylation profile of the casein gene locus between mTEClo and mTEChi was compared.132 This gene locus contains casein genes located in close proximity, that are expressed in mTEChi in a coregulated manner.133 Only the casein beta gene from the casein locus showed an inverse correlation between its expression levels and DNA methylation in mTEChi whereas the other casein genes remained hypermethylated regardless of their expression levels.132 These data suggest that Aire-dependent TRA expression can occur independently of the methylation status of TRA genes. Subsequent experiments corroborated these findings. eGFP was knocked into the gene for the TRA Gad67 to isolate only those mTEC expressing Gad67. There was no difference in the methylation profile of Gad67 from the GFP+mTEChi, GFP−mTEChi, mTEClo and thymocytes.132 These results suggest no obvious association between DNA methylation and TRA expression.
The number of TRA expressed in mTEC is ~4200 genes105, and genome-wide DNA methylation strategies addressed whether DNA methylation is associated to TRA expression.134 Thus, DNA methylation profiles were compared between Aire−/− and Aire+/+ mTEChi using a microarray-based screening system that contained probes to analyze tissue-dependent and differentially methylated regions.135 No major differences in the methylation patters were observed after Aire deficiency in mTEC.135 In another study using reduced representation bisulfite sequencing, no differences in the CpG methylation profiles between Aire-responsive genes versus Aire-neutral genes were observed.136 Taken together, these studies do not appear to support an evident association between the DNA methylation state of promiscuously expressed genes and their methylation. However, these studies were performed on populations of mTEChi, which are known to be heterogenous for expression of individual TRAs.105,136,137
3.4.3. Aire restrains TRA expression
In vitro studies have demonstrated that Aire, via its PHD1 domain, regulates TRA expression by binding the unmodified histone H3K4.138,139 Aire is unable to interact with the H3K4 methylated forms such as H3K4me3, an epigenetic mark associated with transcription.139 Concordantly, Aire-independent gene expression has been linked to deposition of H3K4me3.140 Aire interacts with chromatin regions enriched in repressive histone marks, including the mono-, di-, and tri-methylated forms of H3K9.138,139 Although the biological relevance of this finding has not been directly addressed yet, it has been observed that H3K9me3 is enriched close to the TSS of Aire-dependent genes in mTEChi 140, and regions close to the TSS of Aire-regulated genes are enriched in other repressive chromatin marks, with high content of H3K27me3 and low content of H3K4me3.105,140,141 Intriguingly, the same profile is present in mTEClo and tissues in which specific TRA genes are not expressed140, suggesting no association with TRA expression. Thus, these data suggest Aire acts on a preformed chromatin landscape enriched in repressive histone marks and already present in mTEClo, prior to Aire expression.
Chromatin accessibility refers to the degree to which macromolecules are able to physically contact chromatinized DNA.142 Active DNA regulatory elements are chromatin regions accessible to transcription factors.143 Congruent with the transcriptional differences between mTEClo and mTEChi, mTEChi showed a distinctive open chromatin configuration in comparison with mTEClo, and 70% of the differentially accessible regions in mTEChi were close to TRA genes.109 Strikingly, Aire deficiency caused increased chromatin accessibility near genes upregulated in mTEChi, suggesting that Aire might establish a closed configuration in the vicinity of TRA genes. Ablation of Brg1 expression resulted in a closed chromatin configuration at these same regions. This suggests that Aire and Brg1 have opposing roles in chromatin accessibility, and that Aire contributes to closing whereas Brg1 opens chromatin regions in the vicinity of TRA genes. Interestingly, NF-κB binding sites were overrepresented in these accessible regions in mTEChi 109, consistent with the idea that NF-κB might directly regulate TRA genes. However, Aire deficiency also reduced the frequency of distant super-enhancers, and reduced chromatin accessibility at such superenhancers, in mTEChi.126 Thus, much remains to be learned about how the interplay between Aire and Brg1 regulates TRA transcription. Conceivably, new insights may be obtained using single-cell approaches, because of the limited number of mTEChi expressing any given TRA in heterogeneous mTEChi populations.105,136,137
3.4.4. Regulators of Aire gene expression
Since Aire is expressed predominantly in the thymus, whether DNA methylation regulates Aire expression was addressed. Mouse and human thymus samples showed hypomethylation in the Aire promoter.144 Treatment with 5-aza-2’-deoxycytidine, a DNAmt inhibitor, or deficiency in the DNAmts DNMT1 and DNMT3b, reactivated Aire expression in cell lines with silenced Aire expression.145,146 Tet-eleven translocation (Tet) family members display 5-methylcytosine dioxygenase activity that is associated with DNA demethylation.147 Conditional deficiency of Tet1, Tet2, and Tet3 together repressed Aire expression in mTEChi.148 The ablation of Tet caused two regions in the Aire gene that were normally demethylated to be methylated in mTEChi.148
In concordance with the prominent role of RANK-RANKL and CD40-CD40L interactions in mTEChi development63,73,76,77, transcription factors of the NF-κB family were found to regulate Aire gene expression. Mechanistically, it was shown that those factors interacted with a distal regulatory element of Aire, forming a chromatin loop to interact with the Aire promoter, thus inducing its transcriptional activation.78,79 Deletion of this enhancer was enough to phenocopy the Aire-deficient phenotype.78,79
Females are more susceptible to develop autoimmunity than males, pointing toward the existence of a potential bias in TEC-mediated T cell development.149 Aire expression was downregulated in thymic stroma in castrated and testicular feminized (ARTfm/Y) mice (which harbor a loss-of-function mutation in the androgen receptor).150 Concordantly, the number of Aire+ TEC was also decreased in castrated male mice.151 Interestingly, androgen-pretreated, male, but not female, mice were protected from autoimmune encephalitis (EAE) induced by myelin oligodendrocyte glycoprotein (MOG) immunization; this protection required Aire.150 These results support a role of Aire in resistance of male mice to autoimmune disease development.
Thymic B cells expressed Aire, and this expression required activation of CD40 signaling by CD4 cells.152 To identify transcriptional regulators of Aire, the expression profiles of Aire-expressing cells including Aire+ mTEChi, Aire+ thymic B cells, and Aire+ splenic B cells were compared to cells that do not express Aire, namely, mTEClo, cTEC, splenic T and Aire− B cells.148 Potential transcriptional regulators of Aire were identified based on the exclusively high expression levels in Aire-expressing cells.148 Of these, Tbx21, Tcf7, Irf4, and Irf8 were controllers of Aire expression in TEC, and downregulation of Aire was observed in mTEChi after ablation. Moreover, these transcription factors interacted with the Aire promoter and formed a protein complex between them.148 Notably, CTCFL contributed to the transcriptional activation of Aire by displacing CTCF from the Aire gene as observed by performing both ChIP and loss-of-function assays.148
Besides transcriptional regulation, other mechanisms control the expression levels of Aire in mTEChi. Retention of the intron 2 in the Aire pre-mRNA by the ablation of the JMJD6 introduces a premature stop codon resulting in the translation of a shorter cytoplasmic protein.153 This shorter protein interacted with the wild-type Aire isoform inducing its proteasome-mediated degradation.153 Moreover, JMJD6 KO-mediated Aire+ mTEC reduction repressed Aire-dependent TRA expression, leading to increased activated/memory CD4 T cells unleashing multi-organ autoimmunity.153
3.5. Fezf2
Aire regulates the expression of approximately 40% of TRA, indicating that additional controllers of TRA expression must exist.105,133,154 Fezf2 is a zinc finger transcription factor previously known for its role in brain development.155,156 mTEC expressed high levels of Fefz2, and gene expression analyses of mTEC isolated from Fezf2−/− mice showed that Fezf2 regulates the expression of many Aire-independent TRA genes.31 Importantly, Fezf2 and Aire were found to regulate nonoverlapping gene expression programs.31,137,154 Significantly, Aire expression and Aire-dependent TRA gene expression were not altered by loss of the Fezf2 gene. Moreover, Foxn1-Cre-mediated ablation of Fezf2 reduced numbers of mTEC and Treg cells, and increased CD4 and CD8 effector/memory T cells. Concordantly, Fezf2 deficiency led to multi-organ autoimmunity.31 Strikingly, Fezf2 ablation-mediated antibody reactivity was directed to different antigens than those generated by Aire deficiency. This suggests complementary roles of Aire and Fezf2 in regulation of TRA.31 It was initially suggested that the LTβR pathway controls Fezf2 expression in mTEC 31, however, antibody-mediated activation of LTβR did not affect expression of Fezf2. However, RANK regulated both Aire and Fezf2 expression.68 In addition, active histone marks such as H3K4me3 and H3K4ac were enriched in the TSS of Fezf2-induced genes137 whereas Aire was associated with a repressive histone profile characterized by high contents of H3K27me3.105,140
Chd4 was identified as an interacting partner of Fezf2 in Mass Spectrometry analysis of Fezf2 co-immunoprecipitated extracts. Cdh4 is an essential component of the nucleosomal remodeling and histone deacetylation complex (NuRD) which is implicated in transcriptional regulation.157 Chd4 regulated nearly one-quarter of Fezf2-regulated genes, as evident when Chd4 was ablated in TEC.137 ATAC-seq analyses from mTEC isolated from wild-type, Chd4 cKO and Fezf2 cKO mice demonstrated that approximately 50% of the Cdh4-mediated accessible-chromatin regions overlapped with those regions whose accessibility required Fezf2.137 This suggests that Chd4 and Fezf2 together regulate chromatin configuration in mTEC. Moreover, these regions are enriched in promoter sequences of genes transcriptionally activated by Fezf2 and Chd4137, and deficiency of Chd4 triggered multi-organ autoimmunity in old mice.
3. 6. p53
The tumor suppressor p53 is activated in response to several stimuli, inducing cell cycle arrest, DNA repair, or apoptosis.158 Besides these functions, p53 has also been associated with immunological responses through its transcriptional control over crucial genes involved in pathogen sensing, cytokine production and inflammation.159 By generating a mouse strain with p53 conditional deletion in TEC, it was shown that p53 regulates mTEC differentiation, thymocyte maturation, and tolerance.160 Specifically, ablation of the p53 gene in TEC reduced the number of postnatal mTEChi and decreased expression of both Aire-dependent and Aire-independent TRA genes.160 Notably, p53 was found to interact with the promoter of Tnfrsf11a (encoding RANK) inducing its transcriptional activation, which in turn increased p53 expression levels after its stimulation.160 p53 deficiency in mTEC decreased Treg cell numbers, leading to autoimmunity as evidenced by lymphocyte infiltration in organs. Moreover, immune reactivity was evident in Rag2−/− mice when reconstituted with thymocytes educated by p53 KO mTEC.160 Considering the predominant role of RANK in mTEC, altering the p53-RANK positive feedback loop may explain the phenotype evident after p53 ablation in TEC, but additional mechanisms may also exist.
3.7. Prdm1
Prdm1 was initially characterized as a repressor of Ifnb expression.161 Prdm1 plays critical roles in differentiation of B and T cells.162 In Prdm1-promoter driven YFP reporter mice, ~9 % of the mTEChi and ~2 % of the cTEC potentially expressed Prdm.163 Thymic ablation of Prdm1 using Foxn1-Cre did not appreciably alter TEC numbers or T cell development, including the development of Treg cells.163 Nevertheless, mice lacking Prdm1 in TEC developed lymphadenopathy in mandibular and accessory mandibular cervical and showed high serum levels of autoantibodies. Autoantibody production was also seen when fetal thymi lacking Prdm1 in TEC were grafted under the kidney capsule of adult nude mice, confirming that Prdm1 was needed specifically in TEC for self-tolerance.163 It remains to be determined whether Prdm1 controls the expression of some TRAs in mTEC, or acts via a distinct mechanism.
3.8. Pou2f3
Recent scRNA-seq analysis of non-hematopoietic thymic cells identified putative tuft-like cells based on the similarities with the gene expression profile of intestinal tuft cells, including expression of IL-25. mTEC IV were visualized as Dclk1+Villin+ cells by immunofluorescence imaging.32 Thymic tuft cells were also identified using an inducible Aire lineage-tracing model.33 Thymic tuft cells exhibited high expression of Dclk1 protein intracellularly, low MHC-II, and expressed transduction pathway and tuft cell markers such as IL-25 and Trpm5. Thus, mTEC IV are thymic tuft cells. Notably, thymic tuft cells expressed elevated levels of the transcription factor Pou2f3, and accessible chromatin in these cells was enriched for POU class 2 transcription factor binding sites in enhancer regions. Interestingly, ablation of Pou2f3 led to the abolition of thymic tuft cells with a concomitant increase of NKT cells32, and reduction of thymic tuft cells due to LTβR deficiency inhibited iNKT cell development.164
3.9. Myc
Myc is an essential transcription factor that controls cell growth and body size.165 The role of Myc in TEC biology has been assessed by loss- and gain-of-function approaches.166,167 TEC-specific loss of Myc expression (Foxn1-Cre-Mycfl/fl) resulted in decreased cTEC and mTEC cellularity in adult mice.166 This correlated with a reduction in the number of cycling (Ki67+) cells in both compartments, and mice with Myc-deficient TEC displayed a significantly smaller thymus. Nevertheless, Myc was not essential for the maturation of Aire+ and CD80+ mTEC; and the CD4 and CD8 profile of thymocytes as well as expression of activation and maturation markers CD69 and TCRβ were unaffected.166 Conversely, transgenic expression of Myc in TEC (Foxn1-Cre-MycTg) resulted in an enlarged thymus in adult mice.167 In addition, the total numbers of cTEC and mTEC were considerably increased, and the ratio of cTEC and mTEC was altered with a relative expansion of cTEC. Although the expansion of TEC caused a dramatic increase in thymocyte cellularity, the ratios of thymocyte populations were largely unaffected. The results were similar to previous work in which cell cycle regulator cyclin D1 was overexpressed in TEC, or the Retinoblastoma (Rb) family of genes was inactivated.168-170 Mechanistically, transcriptional profiling of Myc-transgenic TECs revealed upregulation of genes associated with ribosomal biogenesis and translation; this was distinct from mice with forced expression of cyclin D1. Taken together, these studies demonstrate that Myc coordinates a transcriptional program that determines thymus growth and size.
3.10. Meis1
Meis1 is a homeodomain transcription factor of the TALE subfamily required for embryonic development.171,172 Meis1 is primarily expressed in the mTEC compartment, in cells expressing Keratin 5 and Keratin 14 but not Keratin 8.173 Postnatal Keratin 14-CreERT2-mediated ablation of Meis1 reduced thymus size and cell numbers. Moreover, T cell development was dramatically altered, exhibiting reduction of the CD4 CD8 DP T cells in Meis1-deficient mice.173 Using Meis1-EGFP-reporter mice, Mes1high TEC were isolated and transplanted under the renal capsule of nude mice. After 4 weeks, the total number of EpCAM+ cells recovered from Mes1high TEC grafts was higher than those developed from Mes1low TEC.173 Future studies using TEC-specific ablation of Meis1 are needed to provide insight into the role of Meis1 in TEC.
3. 11. FoxA
The FoxA gene family consists of pioneer transcription factors associated with multiple stages of development.174,175 FoxA proteins mediate the emergence of enhancers through displacing linker histone H1.176 The CO2H-terminal domain of FoxA interacts with core histone proteins and is required for opening chromatin without ATP or ATP-dependent chromatin remodelers.177 Foxn1-Cre mediated ablation of both Foxa1 and Foxa2 genes generated a small thymus harboring a high ratio of mTEC to cTEC.178 Ablation of Foxa1 and Foxa2 genes induced higher MHC-I and lower MHC-II gene expression levels in both in mTEC and cTEC.178 Concordantly, T cell development was impaired after ablation of both Foxa1 and FoxA2, reducing the total number of CD4 T cells and CD8 T cells.178
3. 12. Notch-Rbpj
The Notch family is formed by four members in mammals (NOTCH1-NOTCH4). The intracellular domain (NICD) of the Notch receptor is cleaved after its activation and interacts with RBPJ, a potent DNA-binding transcription factor.22 To uncover a possible role of Notch in thymus development, Notch1 or RBPJ were ablated in TEC using Foxn1-Cre, or upstream of TEC specification using an inducible FoxA2-Cre.179,180 Cldn3,4-expressing mTEC-committed progenitors10,11 were reduced in the absence of Notch.179 Ablation of Rbpj in TEC also reduced the mTEC compartment postnatally.180 RANKL-stimulated fetal thymic organ cultures from Rbpj-deficient mice generated low numbers of mTEC compared with cells obtained from wild-type mice.180 This suggests that the Notch-mediated mTEC regulation is upstream of the NF-κB pathway. Together, these data clearly implicate canonical Notch signaling in mTEC specification during development.
4. OTHER EPIGENETIC REGULATORS IN TEC
Below, we discuss enzymatic activities that modify chromatin, and play essential roles in TEC (Table 1), but the transcription factors they work with remain to be established.
4. 1. HDACS
Histone deacetylases (HDACs) remove the acetyl group from previously acetylated lysine residues at histone tails.181 This promotes the formation of a compacted chromatin structure and represses gene expression. There are eighteen highly conserved genes encoding HDAC, that are classified in 4 subclasses. HDAC1, HDAC2 and HDAC3 are members of the HDAC class I. These HDACs are recruited by protein complexes such as NuRD, CoREST and SMRT.182
Foxn1-Cre-mediated ablation of HDAC1 and HDAC2 genes did not show dramatic structural changes in the thymus of 6-week-old mice.183 However, conditional deficiency of HDAC3 in TEC greatly reduced the mTEC compartment, inhibited both Aire-dependent and Aire-independent TRA expression, and reduced the frequency of Treg.183 Interestingly, Aire gene expression was not altered, suggesting that deacetylation might influence the ability of Aire to interact with histones, as previously observed.139 HDAC3 deficiency generated autoimmunity in old mice. Although HDAC3 ablation generated a phenotype similar to that displayed by altering the NF-κB pathway, NIKaly/aly mice revealed that Hdac3 deficiency is not associated with alterations in NF-κB.183 Specifically, fetal thymic organ cultures prepared from NIKaly/aly embryos, but not HDAC3-deficient embryos, were rescued by RANKL. To further understand the phenotype presented by HDAC3 ablation in mTEC, the transcriptional programs were compared between HDAC3 KO mTEC and their normal counterparts. HDAC3 was found to positively regulate expression of key mTEC genes such as Fezf2, SpiB and Pou2f3.183 In addition, ablation of HDAC3 repressed genes linked to Notch signaling.183
HDAC3 forms a repressor complex with NCoR and SMART. However, conditional ablation of these proteins in the thymus did not alter thymus structure or TEC subpopulations.183
4.2. PRC1 complex
Polycomb group repressive complex (PRC) 1 represses transcription by inducing H2A-K119 monoubiquitylation. PRC1 complex is formed by members of the CBXs, PCGFs, PHCs and RING E3 ligases. Bmi1 is a member of the PCGF proteins and plays a key role in the PRC1 complex by regulating H2A ubiquitination and mediating protein-protein interactions. In addition, Bmi1 interacts with RING E3 ligases of the PRC1 complex to monoubiquitinate histone H2A at lysine 119 (present in silenced regions).184 Interestingly, fetal Bmi1−/− thymic lobes produced smaller grafts than control thymic lobes when implanted under the kidney capsule.185
Cbx4 is another member of the PRC1 complex that harbors both Polycomb and non-Polycomb SUMO E3 ligase activities (which are necessary for the control differentiation programs).103 As mentioned above, Cbx4 is able to interact with p63 and its ablation generated smaller thymi, which was attributed to reduced proliferation and delayed maturation of embryonic TEC.102
4.3. PRC2 complex
PRC2 establishes H3K27me3 by the activity of methyltransferases EZH1 and EZH2. The core components of PRC2 are EZH1/ EZH2, EED, SUZ12, And RbAp26 or RbAp48. Individual ablation of these components causes embryonic lethality186, highlighting their essential role in life. PRC2 is recruited to CpG-rich DNA elements to maintain gene silencing.187
Insight into the importance of PRC2 in the thymus came from experiments uncovering the role the transcription factor Tbx1 in the thymus.188 Ectopic expression of Tbx1 blocked thymus organogenesis.188 Mechanistically, Tbx1 overexpression resulted in upregulation of PRC2 dependent genes.188 These data associated the thymus defects with impaired activity of PRC2.188,189 In a subsequent study, an EED cKO mouse was generated to elucidate the role of PRC2 in the thymus. EED recognizes the H3K27me3 epigenetic mark inducing allosteric activation of the methyltransferase EZH2, thus propagating a repressive chromatin configuration.190 Foxn1-cre-mediated EED ablation caused a reduction in H3K27me3 in TEC, and a reduction in thymus size in four-week-old mice.189 Similar observations were made using β5t-cre-mediated EED ablation.191 Notably, thymus cellularity, TEC numbers, and mTEC frequency were lower in EED ablated neonatal mice.189,191 TEC proliferation, survival, and differentiation were also altered.189,191 Importantly, transcriptomic analyses showed that Foxn1 and its gene targets were downregulated in the EED ablated embryos, suggesting that PRC2 mediated repression of negative regulators of Foxn1.189 Interestingly, EED ablation activated transcriptional programs not associated with TEC development such as neural and mesenchymal differentiation.189 In concordance with this, it has been described that loss of H3K27me3 provokes aberrant accumulation of CBP/p300-mediated H3K27 acetylation.190 Extensive characterization of thymocyte education by Eed−/− TEC with showed a reduced TCR diversity and increased generation of Treg with higher suppressive capacity.191
5. CONLUSIONS
Perhaps half of all known transcription factors are expressed in any cell type.192 Of these, a core set is expressed in a relatively cell-type specific manner, and important for cell identity.193 Thus, many transcriptional regulators remain to be identified that are important for development and maintenance of the thymus, and the different types of TEC within it. Application of high-throughput single-cell technologies to dissociated thymus preparations have boosted our knowledge of TEC subsets. However, there is the possibility that such isolation methods may miss some TEC populations194. Thus, spatial transcriptomics methods may allow us to identify new TEC subsets.195,196 Additional work is needed to identify relevant transcriptional controllers and understand the mechanisms by which they act. Deciphering the cellular and molecular basis of TEC development and maintenance is essential for understanding how TEC perturbations result in autoimmunity or immunodeficiency, and for devising TEC-based therapies.
Acknowledgements
We thank Susannah Shissler and Yongge Zhao for essential insight and critical comments. GUMR is supported by grants from National Council of Science and Technology (CONACyT-México; A1-S-16997) and National Autonomous University of México (PAPIIT-UNAM; IA204820). AMS is supported by the patronage of the Children’s Hospital of Mexico. AB is supported by the Intramural Research Program of the National Cancer Institute, Center for Cancer Research, National Institutes of Health.
Footnotes
Conflict of interest
The authors have declared that no conflict of interest exist.
References
- 1.Han J, Zuniga-Pflucker JC. A 2020 View of Thymus Stromal Cells in T Cell Development. J Immunol. 2021;206(2):249–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abramson J, Anderson G. Thymic Epithelial Cells. Annu Rev Immunol. 2017;35:85–118. [DOI] [PubMed] [Google Scholar]
- 3.Anderson G, Takahama Y. Thymic epithelial cells: working class heroes for T cell development and repertoire selection. Trends Immunol. 2012;33(6):256–263. [DOI] [PubMed] [Google Scholar]
- 4.Takahama Y, Ohigashi I, Baik S, Anderson G. Generation of diversity in thymic epithelial cells. Nat Rev Immunol. 2017;17(5):295–305. [DOI] [PubMed] [Google Scholar]
- 5.James KD, Jenkinson WE, Anderson G. Non-Epithelial Stromal Cells in Thymus Development and Function. Front Immunol. 2021;12:634367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ucar A, Ucar O, Klug P, et al. Adult thymus contains FoxN1(−) epithelial stem cells that are bipotent for medullary and cortical thymic epithelial lineages. Immunity. 2014;41(2):257–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bleul CC, Corbeaux T, Reuter A, Fisch P, Monting JS, Boehm T. Formation of a functional thymus initiated by a postnatal epithelial progenitor cell. Nature. 2006;441(7096):992–996. [DOI] [PubMed] [Google Scholar]
- 8.Rossi SW, Jenkinson WE, Anderson G, Jenkinson EJ. Clonal analysis reveals a common progenitor for thymic cortical and medullary epithelium. Nature. 2006;441(7096):988–991. [DOI] [PubMed] [Google Scholar]
- 9.Depreter MG, Blair NF, Gaskell TL, et al. Identification of Plet-1 as a specific marker of early thymic epithelial progenitor cells. Proc Natl Acad Sci U S A. 2008;105(3):961–966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hamazaki Y Adult thymic epithelial cell (TEC) progenitors and TEC stem cells: Models and mechanisms for TEC development and maintenance. Eur J Immunol. 2015;45(11):2985–2993. [DOI] [PubMed] [Google Scholar]
- 11.Hamazaki Y, Sekai M, Minato N. Medullary thymic epithelial stem cells: role in thymic epithelial cell maintenance and thymic involution. Immunol Rev. 2016;271(1):38–55. [DOI] [PubMed] [Google Scholar]
- 12.Jenkinson WE, Bacon A, White AJ, Anderson G, Jenkinson EJ. An epithelial progenitor pool regulates thymus growth. J Immunol. 2008;181(9):6101–6108. [DOI] [PubMed] [Google Scholar]
- 13.Ohigashi I, Kozai M, Takahama Y. Development and developmental potential of cortical thymic epithelial cells. Immunol Rev. 2016;271(1):10–22. [DOI] [PubMed] [Google Scholar]
- 14.Ohigashi I, Zuklys S, Sakata M, et al. Adult Thymic Medullary Epithelium Is Maintained and Regenerated by Lineage-Restricted Cells Rather Than Bipotent Progenitors. Cell Rep. 2015;13(7):1432–1443. [DOI] [PubMed] [Google Scholar]
- 15.Ulyanchenko S, O’Neill KE, Medley T, et al. Identification of a Bipotent Epithelial Progenitor Population in the Adult Thymus. Cell Rep. 2016;14(12):2819–2832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miyao T, Miyauchi M, Kelly ST, et al. Integrative analysis of scRNAs-seq and scATAC-seq revealed transit-amplifying thymic epithelial cells expressing autoimmune regulator. bioRxiv. 2021:2021.2010.2004.463004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wells KL, Miller CN, Gschwind AR, et al. Combined transient ablation and single-cell RNA-sequencing reveals the development of medullary thymic epithelial cells. Elife. 2020;9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lucas B, White AJ, Parnell SM, Henley PM, Jenkinson WE, Anderson G. Progressive Changes in CXCR4 Expression That Define Thymocyte Positive Selection Are Dispensable For Both Innate and Conventional alphabetaT-cell Development. Sci Rep. 2017;7(1):5068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kadakia T, Tai X, Kruhlak M, et al. E-protein-regulated expression of CXCR4 adheres preselection thymocytes to the thymic cortex. J Exp Med. 2019;216(8):1749–1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wurbel MA, Philippe JM, Nguyen C, et al. The chemokine TECK is expressed by thymic and intestinal epithelial cells and attracts double- and single-positive thymocytes expressing the TECK receptor CCR9. Eur J Immunol. 2000;30(1):262–271. [DOI] [PubMed] [Google Scholar]
- 21.Gonzalez-Garcia S, Garcia-Peydro M, Alcain J, Toribio ML. Notch1 and IL-7 receptor signalling in early T-cell development and leukaemia. Curr Top Microbiol Immunol. 2012;360:47–73. [DOI] [PubMed] [Google Scholar]
- 22.Maillard I, Fang T, Pear WS. Regulation of lymphoid development, differentiation, and function by the Notch pathway. Annu Rev Immunol. 2005;23:945–974. [DOI] [PubMed] [Google Scholar]
- 23.Boudil A, Matei IR, Shih HY, et al. IL-7 coordinates proliferation, differentiation and Tcra recombination during thymocyte beta-selection. Nat Immunol. 2015;16(4):397–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hong C, Luckey MA, Park JH. Intrathymic IL-7: the where, when, and why of IL-7 signaling during T cell development. Semin Immunol. 2012;24(3):151–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Murata S, Sasaki K, Kishimoto T, et al. Regulation of CD8+ T cell development by thymus-specific proteasomes. Science. 2007;316(5829):1349–1353. [DOI] [PubMed] [Google Scholar]
- 26.Sasaki K, Takada K, Ohte Y, et al. Thymoproteasomes produce unique peptide motifs for positive selection of CD8(+) T cells. Nat Commun. 2015;6:7484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gommeaux J, Gregoire C, Nguessan P, et al. Thymus-specific serine protease regulates positive selection of a subset of CD4+ thymocytes. Eur J Immunol. 2009;39(4):956–964. [DOI] [PubMed] [Google Scholar]
- 28.Kyewski B, Klein L. A central role for central tolerance. Annu Rev Immunol. 2006;24:571–606. [DOI] [PubMed] [Google Scholar]
- 29.Lkhagvasuren E, Sakata M, Ohigashi I, Takahama Y. Lymphotoxin beta receptor regulates the development of CCL21-expressing subset of postnatal medullary thymic epithelial cells. J Immunol. 2013;190(10):5110–5117. [DOI] [PubMed] [Google Scholar]
- 30.Anderson MS, Venanzi ES, Klein L, et al. Projection of an immunological self shadow within the thymus by the aire protein. Science. 2002;298(5597):1395–1401. [DOI] [PubMed] [Google Scholar]
- 31.Takaba H, Morishita Y, Tomofuji Y, et al. Fezf2 Orchestrates a Thymic Program of Self-Antigen Expression for Immune Tolerance. Cell. 2015;163(4):975–987. [DOI] [PubMed] [Google Scholar]
- 32.Bornstein C, Nevo S, Giladi A, et al. Single-cell mapping of the thymic stroma identifies IL-25-producing tuft epithelial cells. Nature. 2018;559(7715):622–626. [DOI] [PubMed] [Google Scholar]
- 33.Miller CN, Proekt I, von Moltke J, et al. Thymic tuft cells promote an IL-4-enriched medulla and shape thymocyte development. Nature. 2018;559(7715):627–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kozai M, Kubo Y, Katakai T, et al. Essential role of CCL21 in establishment of central self-tolerance in T cells. J Exp Med. 2017;214(7):1925–1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bautista JL, Cramer NT, Miller CN, et al. Single-cell transcriptional profiling of human thymic stroma uncovers novel cellular heterogeneity in the thymic medulla. Nat Commun. 2021;12(1):1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Park JE, Botting RA, Dominguez Conde C, et al. A cell atlas of human thymic development defines T cell repertoire formation. Science. 2020;367(6480). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brissette JL, Li J, Kamimura J, Lee D, Dotto GP. The product of the mouse nude locus, Whn, regulates the balance between epithelial cell growth and differentiation. Genes Dev. 1996;10(17):2212–2221. [DOI] [PubMed] [Google Scholar]
- 38.Wortis HH, Nehlsen S, Owen JJ. Abnormal development of the thymus in “nude” mice. J Exp Med. 1971;134(3 Pt 1):681–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nehls M, Pfeifer D, Schorpp M, Hedrich H, Boehm T. New member of the winged-helix protein family disrupted in mouse and rat nude mutations. Nature. 1994;372(6501):103–107. [DOI] [PubMed] [Google Scholar]
- 40.Cheng L, Guo J, Sun L, et al. Postnatal tissue-specific disruption of transcription factor FoxN1 triggers acute thymic atrophy. J Biol Chem. 2010;285(8):5836–5847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zuklys S, Handel A, Zhanybekova S, et al. Foxn1 regulates key target genes essential for T cell development in postnatal thymic epithelial cells. Nat Immunol. 2016;17(10):1206–1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bosticardo M, Yamazaki Y, Cowan J, et al. Heterozygous FOXN1 Variants Cause Low TRECs and Severe T Cell Lymphopenia, Revealing a Crucial Role of FOXN1 in Supporting Early Thymopoiesis. Am J Hum Genet. 2019;105(3):549–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Du Q, Huynh LK, Coskun F, et al. FOXN1 compound heterozygous mutations cause selective thymic hypoplasia in humans. J Clin Invest. 2019;129(11):4724–4738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Uddin MM, Ohigashi I, Motosugi R, et al. Foxn1-beta5t transcriptional axis controls CD8(+) T-cell production in the thymus. Nat Commun. 2017;8:14419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bredenkamp N, Ulyanchenko S, O’Neill KE, Manley NR, Vaidya HJ, Blackburn CC. An organized and functional thymus generated from FOXN1-reprogrammed fibroblasts. Nat Cell Biol. 2014;16(9):902–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bredenkamp N, Nowell CS, Blackburn CC. Regeneration of the aged thymus by a single transcription factor. Development. 2014;141(8):1627–1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zook EC, Krishack PA, Zhang S, et al. Overexpression of Foxn1 attenuates age-associated thymic involution and prevents the expansion of peripheral CD4 memory T cells. Blood. 2011;118(22):5723–5731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wertheimer T, Velardi E, Tsai J, et al. Production of BMP4 by endothelial cells is crucial for endogenous thymic regeneration. Sci Immunol. 2018;3(19). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wallin J, Eibel H, Neubuser A, Wilting J, Koseki H, Balling R. Pax1 is expressed during development of the thymus epithelium and is required for normal T-cell maturation. Development. 1996;122(1):23–30. [DOI] [PubMed] [Google Scholar]
- 50.Peters H, Neubuser A, Kratochwil K, Balling R. Pax9-deficient mice lack pharyngeal pouch derivatives and teeth and exhibit craniofacial and limb abnormalities. Genes Dev. 1998;12(17):2735–2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yamazaki Y, Urrutia R, Franco LM, et al. PAX1 is essential for development and function of the human thymus. Sci Immunol. 2020;5(44). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Manley NR, Capecchi MR. The role of Hoxa-3 in mouse thymus and thyroid development. Development. 1995;121(7):1989–2003. [DOI] [PubMed] [Google Scholar]
- 53.Manley NR, Capecchi MR. Hox group 3 paralogs regulate the development and migration of the thymus, thyroid, and parathyroid glands. Dev Biol. 1998;195(1):1–15. [DOI] [PubMed] [Google Scholar]
- 54.Xu PX, Zheng W, Laclef C, et al. Eya1 is required for the morphogenesis of mammalian thymus, parathyroid and thyroid. Development. 2002;129(13):3033–3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zou D, Silvius D, Davenport J, Grifone R, Maire P, Xu PX. Patterning of the third pharyngeal pouch into thymus/parathyroid by Six and Eya1. Dev Biol. 2006;293(2):499–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Balciunaite G, Keller MP, Balciunaite E, et al. Wnt glycoproteins regulate the expression of FoxN1, the gene defective in nude mice. Nat Immunol. 2002;3(11):1102–1108. [DOI] [PubMed] [Google Scholar]
- 57.Grigorieva IV, Mirczuk S, Gaynor KU, et al. Gata3-deficient mice develop parathyroid abnormalities due to dysregulation of the parathyroid-specific transcription factor Gcm2. J Clin Invest. 2010;120(6):2144–2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wei Q, Condie BG. A focused in situ hybridization screen identifies candidate transcriptional regulators of thymic epithelial cell development and function. PLoS One. 2011;6(11):e26795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Larsen BM, Cowan JE, Wang Y, et al. Identification of an Intronic Regulatory Element Necessary for Tissue-Specific Expression of Foxn1 in Thymic Epithelial Cells. J Immunol. 2019;203(3):686–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sen R, Smale ST. Selectivity of the NF-{kappa}B response. Cold Spring Harb Perspect Biol. 2010;2(4):a000257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dainichi T, Matsumoto R, Mostafa A, Kabashima K. Immune Control by TRAF6-Mediated Pathways of Epithelial Cells in the EIME (Epithelial Immune Microenvironment). Front Immunol. 2019;10:1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Taniguchi K, Karin M. NF-kappaB, inflammation, immunity and cancer: coming of age. Nat Rev Immunol. 2018;18(5):309–324. [DOI] [PubMed] [Google Scholar]
- 63.Akiyama T, Shimo Y, Yanai H, et al. The tumor necrosis factor family receptors RANK and CD40 cooperatively establish the thymic medullary microenvironment and self-tolerance. Immunity. 2008;29(3):423–437. [DOI] [PubMed] [Google Scholar]
- 64.Williams JA, Zhang J, Jeon H, et al. Thymic medullary epithelium and thymocyte self-tolerance require cooperation between CD28-CD80/86 and CD40-CD40L costimulatory pathways. J Immunol. 2014;192(2):630–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Williams JA, Tai X, Hodes RJ. CD28-CD80/86 and CD40-CD40L Interactions Promote Thymic Tolerance by Regulating Medullary Epithelial Cell and Thymocyte Development. Crit Rev Immunol. 2015;35(1):59–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wu W, Shi Y, Xia H, et al. Epithelial LTbetaR signaling controls the population size of the progenitors of medullary thymic epithelial cells in neonatal mice. Sci Rep. 2017;7:44481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Boehm T, Scheu S, Pfeffer K, Bleul CC. Thymic medullary epithelial cell differentiation, thymocyte emigration, and the control of autoimmunity require lympho-epithelial cross talk via LTbetaR. J Exp Med. 2003;198(5):757–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cosway EJ, Lucas B, James KD, et al. Redefining thymus medulla specialization for central tolerance. J Exp Med. 2017;214(11):3183–3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hikosaka Y, Nitta T, Ohigashi I, et al. The cytokine RANKL produced by positively selected thymocytes fosters medullary thymic epithelial cells that express autoimmune regulator. Immunity. 2008;29(3):438–450. [DOI] [PubMed] [Google Scholar]
- 70.Roberts NA, White AJ, Jenkinson WE, et al. Rank signaling links the development of invariant gammadelta T cell progenitors and Aire(+) medullary epithelium. Immunity. 2012;36(3):427–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Desanti GE, Cowan JE, Baik S, et al. Developmentally regulated availability of RANKL and CD40 ligand reveals distinct mechanisms of fetal and adult cross-talk in the thymus medulla. J Immunol. 2012;189(12):5519–5526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Naito A, Azuma S, Tanaka S, et al. Severe osteopetrosis, defective interleukin-1 signalling and lymph node organogenesis in TRAF6-deficient mice. Genes Cells. 1999;4(6):353–362. [DOI] [PubMed] [Google Scholar]
- 73.Akiyama T, Maeda S, Yamane S, et al. Dependence of self-tolerance on TRAF6-directed development of thymic stroma. Science. 2005;308(5719):248–251. [DOI] [PubMed] [Google Scholar]
- 74.Liu T, Zhang L, Joo D, Sun SC. NF-kappaB signaling in inflammation. Signal Transduct Target Ther. 2017;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Miyawaki S, Nakamura Y, Suzuka H, et al. A new mutation, aly, that induces a generalized lack of lymph nodes accompanied by immunodeficiency in mice. Eur J Immunol. 1994;24(2):429–434. [DOI] [PubMed] [Google Scholar]
- 76.Kajiura F, Sun S, Nomura T, et al. NF-kappa B-inducing kinase establishes self-tolerance in a thymic stroma-dependent manner. J Immunol. 2004;172(4):2067–2075. [DOI] [PubMed] [Google Scholar]
- 77.Kinoshita D, Hirota F, Kaisho T, et al. Essential role of IkappaB kinase alpha in thymic organogenesis required for the establishment of self-tolerance. J Immunol. 2006;176(7):3995–4002. [DOI] [PubMed] [Google Scholar]
- 78.Haljasorg U, Bichele R, Saare M, et al. A highly conserved NF-kappaB-responsive enhancer is critical for thymic expression of Aire in mice. Eur J Immunol. 2015;45(12):3246–3256. [DOI] [PubMed] [Google Scholar]
- 79.LaFlam TN, Seumois G, Miller CN, et al. Identification of a novel cis-regulatory element essential for immune tolerance. J Exp Med. 2015;212(12):1993–2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zuklys S, Balciunaite G, Agarwal A, Fasler-Kan E, Palmer E, Hollander GA. Normal thymic architecture and negative selection are associated with Aire expression, the gene defective in the autoimmune-polyendocrinopathy-candidiasis-ectodermal dystrophy (APECED). J Immunol. 2000;165(4):1976–1983. [DOI] [PubMed] [Google Scholar]
- 81.Burkly L, Hession C, Ogata L, et al. Expression of relB is required for the development of thymic medulla and dendritic cells. Nature. 1995;373(6514):531–536. [DOI] [PubMed] [Google Scholar]
- 82.Vallabhapurapu S, Matsuzawa A, Zhang W, et al. Nonredundant and complementary functions of TRAF2 and TRAF3 in a ubiquitination cascade that activates NIK-dependent alternative NF-kappaB signaling. Nat Immunol. 2008;9(12):1364–1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Jenkinson SR, Williams JA, Jeon H, et al. TRAF3 enforces the requirement for T cell cross-talk in thymic medullary epithelial development. Proc Natl Acad Sci U S A. 2013;110(52):21107–21112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Akiyama N, Shinzawa M, Miyauchi M, et al. Limitation of immune tolerance-inducing thymic epithelial cell development by Spi-B-mediated negative feedback regulation. J Exp Med. 2014;211(12):2425–2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Khan IS, Mouchess ML, Zhu ML, et al. Enhancement of an anti-tumor immune response by transient blockade of central T cell tolerance. J Exp Med. 2014;211(5):761–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yang A, Kaghad M, Wang Y, et al. p63, a p53 homolog at 3q27-29, encodes multiple products with transactivating, death-inducing, and dominant-negative activities. Mol Cell. 1998;2(3):305–316. [DOI] [PubMed] [Google Scholar]
- 87.Di Como CJ, Urist MJ, Babayan I, et al. p63 expression profiles in human normal and tumor tissues. Clin Cancer Res. 2002;8(2):494–501. [PubMed] [Google Scholar]
- 88.Fisher ML, Balinth S, Mills AA. p63-related signaling at a glance. J Cell Sci. 2020;133(17). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yang A, Schweitzer R, Sun D, et al. p63 is essential for regenerative proliferation in limb, craniofacial and epithelial development. Nature. 1999;398(6729):714–718. [DOI] [PubMed] [Google Scholar]
- 90.Mills AA, Zheng B, Wang XJ, Vogel H, Roop DR, Bradley A. p63 is a p53 homologue required for limb and epidermal morphogenesis. Nature. 1999;398(6729):708–713. [DOI] [PubMed] [Google Scholar]
- 91.Senoo M, Pinto F, Crum CP, McKeon F. p63 Is essential for the proliferative potential of stem cells in stratified epithelia. Cell. 2007;129(3):523–536. [DOI] [PubMed] [Google Scholar]
- 92.Candi E, Rufini A, Terrinoni A, et al. DeltaNp63 regulates thymic development through enhanced expression of FgfR2 and Jag2. Proc Natl Acad Sci U S A. 2007;104(29):11999–12004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kouwenhoven EN, van Bokhoven H, Zhou H. Gene regulatory mechanisms orchestrated by p63 in epithelial development and related disorders. Biochim Biophys Acta. 2015;1849(6):590–600. [DOI] [PubMed] [Google Scholar]
- 94.Qu J, Tanis SEJ, Smits JPH, et al. Mutant p63 Affects Epidermal Cell Identity through Rewiring the Enhancer Landscape. Cell Rep. 2018;25(12):3490-3503 e3494. [DOI] [PubMed] [Google Scholar]
- 95.Bao X, Rubin AJ, Qu K, et al. A novel ATAC-seq approach reveals lineage-specific reinforcement of the open chromatin landscape via cooperation between BAF and p63. Genome Biol. 2015;16:284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Rinaldi L, Datta D, Serrat J, et al. Dnmt3a and Dnmt3b Associate with Enhancers to Regulate Human Epidermal Stem Cell Homeostasis. Cell Stem Cell. 2016;19(4):491–501. [DOI] [PubMed] [Google Scholar]
- 97.Kouwenhoven EN, Oti M, Niehues H, et al. Transcription factor p63 bookmarks and regulates dynamic enhancers during epidermal differentiation. EMBO Rep. 2015;16(7):863–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Anderson G, Pongracz J, Parnell S, Jenkinson EJ. Notch ligand-bearing thymic epithelial cells initiate and sustain Notch signaling in thymocytes independently of T cell receptor signaling. Eur J Immunol. 2001;31(11):3349–3354. [DOI] [PubMed] [Google Scholar]
- 99.Felli MP, Maroder M, Mitsiadis TA, et al. Expression pattern of notch1, 2 and 3 and Jagged1 and 2 in lymphoid and stromal thymus components: distinct ligand-receptor interactions in intrathymic T cell development. Int Immunol. 1999;11(7):1017–1025. [DOI] [PubMed] [Google Scholar]
- 100.Revest JM, Suniara RK, Kerr K, Owen JJ, Dickson C. Development of the thymus requires signaling through the fibroblast growth factor receptor R2-IIIb. J Immunol. 2001;167(4):1954–1961. [DOI] [PubMed] [Google Scholar]
- 101.Schwartz YB, Pirrotta V. Polycomb complexes and epigenetic states. Curr Opin Cell Biol. 2008;20(3):266–273. [DOI] [PubMed] [Google Scholar]
- 102.Liu B, Liu YF, Du YR, et al. Cbx4 regulates the proliferation of thymic epithelial cells and thymus function. Development. 2013;140(4):780–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Mardaryev AN, Liu B, Rapisarda V, et al. Cbx4 maintains the epithelial lineage identity and cell proliferation in the developing stratified epithelium. J Cell Biol. 2016;212(1):77–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Mathis D, Benoist C. Aire. Annu Rev Immunol. 2009;27:287–312. [DOI] [PubMed] [Google Scholar]
- 105.Sansom SN, Shikama-Dorn N, Zhanybekova S, et al. Population and single-cell genomics reveal the Aire dependency, relief from Polycomb silencing, and distribution of self-antigen expression in thymic epithelia. Genome Res. 2014;24(12):1918–1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ilmarinen T, Eskelin P, Halonen M, et al. Functional analysis of SAND mutations in AIRE supports dominant inheritance of the G228W mutation. Hum Mutat. 2005;26(4):322–331. [DOI] [PubMed] [Google Scholar]
- 107.Nagamine K, Peterson P, Scott HS, et al. Positional cloning of the APECED gene. Nat Genet. 1997;17(4):393–398. [DOI] [PubMed] [Google Scholar]
- 108.Anderson MS, Venanzi ES, Chen Z, Berzins SP, Benoist C, Mathis D. The cellular mechanism of Aire control of T cell tolerance. Immunity. 2005;23(2):227–239. [DOI] [PubMed] [Google Scholar]
- 109.Koh AS, Miller EL, Buenrostro JD, et al. Rapid chromatin repression by Aire provides precise control of immune tolerance. Nat Immunol. 2018;19(2):162–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Bortnick A, Murre C. mTECs Aire on the side of caution. Nat Immunol. 2018;19(2):100–101. [DOI] [PubMed] [Google Scholar]
- 111.Liiv I, Rebane A, Org T, et al. DNA-PK contributes to the phosphorylation of AIRE: importance in transcriptional activity. Biochim Biophys Acta. 2008;1783(1):74–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Mohiuddin IS, Kang MH. DNA-PK as an Emerging Therapeutic Target in Cancer. Front Oncol. 2019;9:635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Oven I, Brdickova N, Kohoutek J, Vaupotic T, Narat M, Peterlin BM. AIRE recruits P-TEFb for transcriptional elongation of target genes in medullary thymic epithelial cells. Mol Cell Biol. 2007;27(24):8815–8823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Yoshida H, Bansal K, Schaefer U, et al. Brd4 bridges the transcriptional regulators, Aire and P-TEFb, to promote elongation of peripheral-tissue antigen transcripts in thymic stromal cells. Proc Natl Acad Sci U S A. 2015;112(32):E4448–4457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Giraud M, Yoshida H, Abramson J, et al. Aire unleashes stalled RNA polymerase to induce ectopic gene expression in thymic epithelial cells. Proc Natl Acad Sci U S A. 2012;109(2):535–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Huoh YS, Wu B, Park S, et al. Dual functions of Aire CARD multimerization in the transcriptional regulation of T cell tolerance. Nat Commun. 2020;11(1):1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Kumar PG, Laloraya M, Wang CY, et al. The autoimmune regulator (AIRE) is a DNA-binding protein. J Biol Chem. 2001;276(44):41357–41364. [DOI] [PubMed] [Google Scholar]
- 118.Oftedal BE, Hellesen A, Erichsen MM, et al. Dominant Mutations in the Autoimmune Regulator AIRE Are Associated with Common Organ-Specific Autoimmune Diseases. Immunity. 2015;42(6):1185–1196. [DOI] [PubMed] [Google Scholar]
- 119.Su MA, Giang K, Zumer K, et al. Mechanisms of an autoimmunity syndrome in mice caused by a dominant mutation in Aire. J Clin Invest. 2008;118(5):1712–1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Pitkanen J, Doucas V, Sternsdorf T, et al. The autoimmune regulator protein has transcriptional transactivating properties and interacts with the common coactivator CREB-binding protein. J Biol Chem. 2000;275(22):16802–16809. [DOI] [PubMed] [Google Scholar]
- 121.Saare M, Rebane A, Rajashekar B, Vilo J, Peterson P. Autoimmune regulator is acetylated by transcription coactivator CBP/p300. Exp Cell Res. 2012;318(14):1767–1778. [DOI] [PubMed] [Google Scholar]
- 122.Chuprin A, Avin A, Goldfarb Y, et al. The deacetylase Sirt1 is an essential regulator of Aire-mediated induction of central immunological tolerance. Nat Immunol. 2015;16(7):737–745. [DOI] [PubMed] [Google Scholar]
- 123.Devaiah BN, Singer DS. Two faces of brd4: mitotic bookmark and transcriptional lynchpin. Transcription. 2013;4(1):13–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Wu SY, Chiang CM. The double bromodomain-containing chromatin adaptor Brd4 and transcriptional regulation. J Biol Chem. 2007;282(18):13141–13145. [DOI] [PubMed] [Google Scholar]
- 125.Abramson J, Giraud M, Benoist C, Mathis D. Aire’s partners in the molecular control of immunological tolerance. Cell. 2010;140(1):123–135. [DOI] [PubMed] [Google Scholar]
- 126.Bansal K, Yoshida H, Benoist C, Mathis D. The transcriptional regulator Aire binds to and activates super-enhancers. Nat Immunol. 2017;18(3):263–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Jansen K, Shikama-Dorn N, Attar M, et al. RBFOX splicing factors contribute to a broad but selective recapitulation of peripheral tissue splicing patterns in the thymus. Genome Res. 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Danan-Gotthold M, Guyon C, Giraud M, Levanon EY, Abramson J. Extensive RNA editing and splicing increase immune self-representation diversity in medullary thymic epithelial cells. Genome Biol. 2016;17(1):219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Waterfield M, Khan IS, Cortez JT, et al. The transcriptional regulator Aire coopts the repressive ATF7ip-MBD1 complex for the induction of immunotolerance. Nat Immunol. 2014;15(3):258–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Cetani F, Barbesino G, Borsari S, et al. A novel mutation of the autoimmune regulator gene in an Italian kindred with autoimmune polyendocrinopathy-candidiasis-ectodermal dystrophy, acting in a dominant fashion and strongly cosegregating with hypothyroid autoimmune thyroiditis. J Clin Endocrinol Metab. 2001;86(10):4747–4752. [DOI] [PubMed] [Google Scholar]
- 131.Greenberg MVC, Bourc’his D. The diverse roles of DNA methylation in mammalian development and disease. Nat Rev Mol Cell Biol. 2019;20(10):590–607. [DOI] [PubMed] [Google Scholar]
- 132.Tykocinski LO, Sinemus A, Rezavandy E, et al. Epigenetic regulation of promiscuous gene expression in thymic medullary epithelial cells. Proc Natl Acad Sci U S A. 2010;107(45):19426–19431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Derbinski J, Gabler J, Brors B, et al. Promiscuous gene expression in thymic epithelial cells is regulated at multiple levels. J Exp Med. 2005;202(1):33–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Yong WS, Hsu FM, Chen PY. Profiling genome-wide DNA methylation. Epigenetics Chromatin. 2016;9:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Wu G, Hirabayashi K, Sato S, et al. DNA methylation profile of Aire-deficient mouse medullary thymic epithelial cells. BMC Immunol. 2012;13:58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Meredith M, Zemmour D, Mathis D, Benoist C. Aire controls gene expression in the thymic epithelium with ordered stochasticity. Nat Immunol. 2015;16(9):942–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Tomofuji Y, Takaba H, Suzuki HI, et al. Chd4 choreographs self-antigen expression for central immune tolerance. Nat Immunol. 2020;21(8):892–901. [DOI] [PubMed] [Google Scholar]
- 138.Org T, Chignola F, Hetenyi C, et al. The autoimmune regulator PHD finger binds to non-methylated histone H3K4 to activate gene expression. EMBO Rep. 2008;9(4):370–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Koh AS, Kuo AJ, Park SY, et al. Aire employs a histone-binding module to mediate immunological tolerance, linking chromatin regulation with organ-specific autoimmunity. Proc Natl Acad Sci U S A. 2008;105(41):15878–15883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Handel AE, Shikama-Dorn N, Zhanybekova S, et al. Comprehensively Profiling the Chromatin Architecture of Tissue Restricted Antigen Expression in Thymic Epithelial Cells Over Development. Front Immunol. 2018;9:2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Org T, Rebane A, Kisand K, et al. AIRE activated tissue specific genes have histone modifications associated with inactive chromatin. Hum Mol Genet. 2009;18(24):4699–4710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Klemm SL, Shipony Z, Greenleaf WJ. Chromatin accessibility and the regulatory epigenome. Nat Rev Genet. 2019;20(4):207–220. [DOI] [PubMed] [Google Scholar]
- 143.Thurman RE, Rynes E, Humbert R, et al. The accessible chromatin landscape of the human genome. Nature. 2012;489(7414):75–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Kont V, Murumagi A, Tykocinski LO, et al. DNA methylation signatures of the AIRE promoter in thymic epithelial cells, thymomas and normal tissues. Mol Immunol. 2011;49(3):518–526. [DOI] [PubMed] [Google Scholar]
- 145.Murumagi A, Vahamurto P, Peterson P. Characterization of regulatory elements and methylation pattern of the autoimmune regulator (AIRE) promoter. J Biol Chem. 2003;278(22):19784–19790. [DOI] [PubMed] [Google Scholar]
- 146.Klamp T, Sahin U, Kyewski B, Schwendemann J, Dhaene K, Tureci O. Expression profiling of autoimmune regulator AIRE mRNA in a comprehensive set of human normal and neoplastic tissues. Immunol Lett. 2006;106(2):172–179. [DOI] [PubMed] [Google Scholar]
- 147.Zhao H, Chen T. Tet family of 5-methylcytosine dioxygenases in mammalian development. J Hum Genet. 2013;58(7):421–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Herzig Y, Nevo S, Bornstein C, et al. Transcriptional programs that control expression of the autoimmune regulator gene Aire. Nat Immunol. 2017;18(2):161–172. [DOI] [PubMed] [Google Scholar]
- 149.Ngo ST, Steyn FJ, McCombe PA. Gender differences in autoimmune disease. Front Neuroendocrinol. 2014;35(3):347–369. [DOI] [PubMed] [Google Scholar]
- 150.Zhu ML, Bakhru P, Conley B, et al. Sex bias in CNS autoimmune disease mediated by androgen control of autoimmune regulator. Nat Commun. 2016;7:11350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Dragin N, Bismuth J, Cizeron-Clairac G, et al. Estrogen-mediated downregulation of AIRE influences sexual dimorphism in autoimmune diseases. J Clin Invest. 2016;126(4):1525–1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Yamano T, Nedjic J, Hinterberger M, et al. Thymic B Cells Are Licensed to Present Self Antigens for Central T Cell Tolerance Induction. Immunity. 2015;42(6):1048–1061. [DOI] [PubMed] [Google Scholar]
- 153.Yanagihara T, Sanematsu F, Sato T, et al. Intronic regulation of Aire expression by Jmjd6 for self-tolerance induction in the thymus. Nat Commun. 2015;6:8820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Takaba H, Takayanagi H. The Mechanisms of T Cell Selection in the Thymus. Trends Immunol. 2017;38(11):805–816. [DOI] [PubMed] [Google Scholar]
- 155.Shimizu T, Nakazawa M, Kani S, et al. Zinc finger genes Fezf1 and Fezf2 control neuronal differentiation by repressing Hes5 expression in the forebrain. Development. 2010;137(11):1875–1885. [DOI] [PubMed] [Google Scholar]
- 156.Kwan KY. Transcriptional dysregulation of neocortical circuit assembly in ASD. Int Rev Neurobiol. 2013;113:167–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.O’Shaughnessy A, Hendrich B. CHD4 in the DNA-damage response and cell cycle progression: not so NuRDy now. Biochem Soc Trans. 2013;41(3):777–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Levine AJ. p53: 800 million years of evolution and 40 years of discovery. Nat Rev Cancer. 2020;20(8):471–480. [DOI] [PubMed] [Google Scholar]
- 159.Munoz-Fontela C, Mandinova A, Aaronson SA, Lee SW. Emerging roles of p53 and other tumour-suppressor genes in immune regulation. Nat Rev Immunol. 2016;16(12):741–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Rodrigues PM, Ribeiro AR, Perrod C, et al. Thymic epithelial cells require p53 to support their long-term function in thymopoiesis in mice. Blood. 2017;130(4):478–488. [DOI] [PubMed] [Google Scholar]
- 161.Keller AD, Maniatis T. Identification and characterization of a novel repressor of beta-interferon gene expression. Genes Dev. 1991;5(5):868–879. [DOI] [PubMed] [Google Scholar]
- 162.Nutt SL, Fairfax KA, Kallies A. BLIMP1 guides the fate of effector B and T cells. Nat Rev Immunol. 2007;7(12):923–927. [DOI] [PubMed] [Google Scholar]
- 163.Roberts NA, Adams BD, McCarthy NI, et al. Prdm1 Regulates Thymic Epithelial Function To Prevent Autoimmunity. J Immunol. 2017;199(4):1250–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Lucas B, White AJ, Cosway EJ, et al. Diversity in medullary thymic epithelial cells controls the activity and availability of iNKT cells. Nat Commun. 2020;11(1):2198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Trumpp A, Refaeli Y, Oskarsson T, et al. c-Myc regulates mammalian body size by controlling cell number but not cell size. Nature. 2001;414(6865):768–773. [DOI] [PubMed] [Google Scholar]
- 166.Hauri-Hohl M, Zuklys S, Hollander GA, Ziegler SF. A regulatory role for TGF-beta signaling in the establishment and function of the thymic medulla. Nat Immunol. 2014;15(6):554–561. [DOI] [PubMed] [Google Scholar]
- 167.Cowan JE, Malin J, Zhao Y, et al. Myc controls a distinct transcriptional program in fetal thymic epithelial cells that determines thymus growth. Nat Commun. 2019;10(1):5498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Robles AI, Larcher F, Whalin RB, et al. Expression of cyclin D1 in epithelial tissues of transgenic mice results in epidermal hyperproliferation and severe thymic hyperplasia. Proc Natl Acad Sci U S A. 1996;93(15):7634–7638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Klug DB, Crouch E, Carter C, Coghlan L, Conti CJ, Richie ER. Transgenic expression of cyclin D1 in thymic epithelial precursors promotes epithelial and T cell development. J Immunol. 2000;164(4):1881–1888. [DOI] [PubMed] [Google Scholar]
- 170.Garfin PM, Min D, Bryson JL, et al. Inactivation of the RB family prevents thymus involution and promotes thymic function by direct control of Foxn1 expression. J Exp Med. 2013;210(6):1087–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Azcoitia V, Aracil M, Martinez AC, Torres M. The homeodomain protein Meis1 is essential for definitive hematopoiesis and vascular patterning in the mouse embryo. Dev Biol. 2005;280(2):307–320. [DOI] [PubMed] [Google Scholar]
- 172.Hisa T, Spence SE, Rachel RA, et al. Hematopoietic, angiogenic and eye defects in Meis1 mutant animals. EMBO J. 2004;23(2):450–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Hirayama T, Asano Y, Iida H, Watanabe T, Nakamura T, Goitsuka R. Meis1 is required for the maintenance of postnatal thymic epithelial cells. PLoS One. 2014;9(3):e89885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Kaestner KH. The FoxA factors in organogenesis and differentiation. Curr Opin Genet Dev. 2010;20(5):527–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Zaret KS, Carroll JS. Pioneer transcription factors: establishing competence for gene expression. Genes Dev. 2011;25(21):2227–2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Iwafuchi-Doi M, Donahue G, Kakumanu A, et al. The Pioneer Transcription Factor FoxA Maintains an Accessible Nucleosome Configuration at Enhancers for Tissue-Specific Gene Activation. Mol Cell. 2016;62(1):79–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Swinstead EE, Paakinaho V, Presman DM, Hager GL. Pioneer factors and ATP-dependent chromatin remodeling factors interact dynamically: A new perspective: Multiple transcription factors can effect chromatin pioneer functions through dynamic interactions with ATP-dependent chromatin remodeling factors. Bioessays. 2016;38(11):1150–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Lau CI, Yanez DC, Solanki A, Papaioannou E, Saldana JI, Crompton T. Foxa1 and Foxa2 in thymic epithelial cells (TEC) regulate medullary TEC and regulatory T-cell maturation. J Autoimmun. 2018;93:131–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Li J, Gordon J, Chen ELY, et al. NOTCH1 signaling establishes the medullary thymic epithelial cell progenitor pool during mouse fetal development. Development. 2020;147(12). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Liu D, Kousa AI, O’Neill KE, et al. Canonical Notch signaling controls the early thymic epithelial progenitor cell state and emergence of the medullary epithelial lineage in fetal thymus development. Development. 2020;147(12). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Bannister AJ, Kouzarides T. Regulation of chromatin by histone modifications. Cell Res. 2011;21(3):381–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Delcuve GP, Khan DH, Davie JR. Roles of histone deacetylases in epigenetic regulation: emerging paradigms from studies with inhibitors. Clin Epigenetics. 2012;4(1):5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Goldfarb Y, Kadouri N, Levi B, et al. HDAC3 Is a Master Regulator of mTEC Development. Cell Rep. 2016;15(3):651–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Wang H, Wang L, Erdjument-Bromage H, et al. Role of histone H2A ubiquitination in Polycomb silencing. Nature. 2004;431(7010):873–878. [DOI] [PubMed] [Google Scholar]
- 185.Guo Y, Miyazaki M, Itoi M, et al. Polycomb group gene Bmi1 plays a role in the growth of thymic epithelial cells. Eur J Immunol. 2011;41(4):1098–1107. [DOI] [PubMed] [Google Scholar]
- 186.Margueron R, Reinberg D. The Polycomb complex PRC2 and its mark in life. Nature. 2011;469(7330):343–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Riising EM, Comet I, Leblanc B, Wu X, Johansen JV, Helin K. Gene silencing triggers polycomb repressive complex 2 recruitment to CpG islands genome wide. Mol Cell. 2014;55(3):347–360. [DOI] [PubMed] [Google Scholar]
- 188.Reeh KA, Cardenas KT, Bain VE, et al. Ectopic TBX1 suppresses thymic epithelial cell differentiation and proliferation during thymus organogenesis. Development. 2014;141(15):2950–2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Singarapu N, Ma K, Reeh KAG, et al. Polycomb Repressive Complex 2 is essential for development and maintenance of a functional TEC compartment. Sci Rep. 2018;8(1):14335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Margueron R, Justin N, Ohno K, et al. Role of the polycomb protein EED in the propagation of repressive histone marks. Nature. 2009;461(7265):762–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Barthlott T, Handel AE, Teh HY, et al. Indispensable epigenetic control of thymic epithelial cell development and function by polycomb repressive complex 2. Nat Commun. 2021;12(1):3933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Vaquerizas JM, Kummerfeld SK, Teichmann SA, Luscombe NM. A census of human transcription factors: function, expression and evolution. Nat Rev Genet. 2009;10(4):252–263. [DOI] [PubMed] [Google Scholar]
- 193.D’Alessio AC, Fan ZP, Wert KJ, et al. A Systematic Approach to Identify Candidate Transcription Factors that Control Cell Identity. Stem Cell Reports. 2015;5(5):763–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Sakata M, Ohigashi I, Takahama Y. Cellularity of Thymic Epithelial Cells in the Postnatal Mouse. J Immunol. 2018;200(4):1382–1388. [DOI] [PubMed] [Google Scholar]
- 195.Petukhov V, Xu RJ, Soldatov RA, et al. Cell segmentation in imaging-based spatial transcriptomics. Nature Biotechnology. 2021. [DOI] [PubMed] [Google Scholar]
- 196.Dries R, Chen J, Del Rossi N, Khan MM, Sistig A, Yuan GC. Advances in spatial transcriptomic data analysis. Genome Res. 2021;31(10):1706–1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Li J, Lee YK, Fu W, et al. Modeling by disruption and a selected-for partner for the nude locus. EMBO Rep. 2021;22(3):e49804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Weih F, Carrasco D, Durham SK, et al. Multiorgan inflammation and hematopoietic abnormalities in mice with a targeted disruption of RelB, a member of the NF-kappa B/Rel family. Cell. 1995;80(2):331–340. [DOI] [PubMed] [Google Scholar]
- 199.Shinkura R, Kitada K, Matsuda F, et al. Alymphoplasia is caused by a point mutation in the mouse gene encoding Nf-kappa b-inducing kinase. Nat Genet. 1999;22(1):74–77. [DOI] [PubMed] [Google Scholar]
- 200.Su DM, Manley NR. Hoxa3 and pax1 transcription factors regulate the ability of fetal thymic epithelial cells to promote thymocyte development. J Immunol. 2000;164(11):5753–5760. [DOI] [PubMed] [Google Scholar]


