Abstract
Epicardial cells (EpiCs) are necessary for myocardium formation, yet little is known about crosstalk between EpiCs and cardiomyocytes (CMs) during development and the potential impact of EpiCs on CM maturation. To investigate the effects of EpiCs on CM commitment and maturation, we differentiated human pluripotent stem cells (hPSCs) to cardiac progenitor cells (CPCs) and EpiCs, and cocultured EpiCs and CPCs for two weeks. When EpiCs were allowed to form epicardial-derived cells, we observed increased expression of cTnI in developing CMs. In the presence of the TGFβ inhibitor A83–01, EpiCs remained in the epicardial state and induced CM proliferation, increased MLC2v expression, and led to less organized sarcomeres. These effects were not observed if CPCs were treated with EpiC-conditioned medium or if CPCs were indirectly cocultured with EpiCs. Finally, single cell RNA sequencing identified that EpiC-CPC coculture had bi-directional effects on transcriptional programs in EpiCs and CMs, and biased EpiC lineages from a SFRP2-enriched population to a DLK1- or C3-enriched population. This work suggests important crosstalk between EpiCs and CMs during differentiation can be used to influence cell fate and improve the ability to generate cardiac cells and tissues for in vitro models and development of cardiac cellular therapies.
Keywords: hPSC, cardiomyocyte, epicardial cell, proliferation, coculture
Graphical Abstract
1. Introduction
Human pluripotent stem cells (hPSCs) provide a proliferative and scalable cardiac cell source by which heart function may eventually be restored via cell therapy. Under protocols developed by our lab and others, hPSC-cardiomyocytes (CMs) begin spontaneously beating in culture after 1.5 to 3 weeks and can be maintained and matured in culture for many months or years [1,2]. However, a major barrier to widespread use of hPSC-CMs for in vitro and in vivo applications is that they are immature compared to human adult CMs [3,4]. When implanted into a porcine model following an induced myocardial infarction, cardiac patches including hPSC-CMs improved ejection fraction, capillary density in the border region, and decreased the total fibrotic area [5]. Another group demonstrated a similar improvement in ejection fraction in a macaque myocardial infarction model treated with direct injection of hPSC-CMs; however, in a subset of animals hPSC-CM grafts caused ventricular arrhythmias, likely due to their immaturity [6].
hPSC-CM maturation has been measured by a myriad of metrics with many assays probing specification and sarcomere maturation. During development and differentiation, ventricular CMs undergo a transition in which the cells switch from primarily expressing MLC2a to MLC2v [7]. This isoform shift has been widely used to benchmark hPSC-CM maturation and assess ventricular specification [2,8,9]. Additionally, adult CMs have higher cTnT expression compared to embryonic CMs, and Troponin I switches from the slow skeletal (ssTnI) to the cardiac isoform (cTnI) and has been suggested as a later metric for hPSC-CM maturation than the MLC2a to MLC2v switch [10–12]. Lastly, developing CMs elongate, become larger, and form organized sarcomere structures during development [13].
To create more mature cardiac tissues, we and others have begun to include cell types other than CMs in hPSC-derived cardiac tissues [14–16]. Epicardial cells (EpiCs) are of particular interest because they are one of the first cell types present in the developing vertebrate heart and represent a key signaling center during cardiac development. This multipotent epithelial cell population lines the outer surface of the heart then migrates and undergoes epithelial-to-mesenchymal transition (EMT) to give rise to epicardial-derived cells (EPDCs), a collection of smooth muscle cells (SMCs), pericytes, and nearly 80% of the fibroblasts (FBs) in the adult mouse heart [17]. Ablation or knockout of EPDCs demonstrated that they play important roles in development of the compact ventricular wall, coronary arteries, atrioventricular cushions and valves, and formation of Purkinje fibers [18]. Furthermore, the epicardium is required for normal cardiac muscle regeneration in zebrafish and plays a role in heart fibrosis, remodeling, and repair in mammals [19].
Several studies have explored interactions between EpiCs or EPDCs on CM maturation. One report analyzed the effects of coculturing quail EPDCs with mouse CMs [20]. The cocultured CMs began beating earlier and expressed higher levels of electrical, mechanical, and sarcomere proteins, including connexin 43, N-cadherin, cardiac troponin I, and α-actinin. Additionally, the authors showed that direct contact of dispersed EpiCs amongst CMs was needed for this maturation. Indirect coculture, conditioned media, and paracrine signaling were not sufficient.
Using a combination of Wt1CreERT2/+, Rosa26mTmG/+ mice, Zhou et al. tagged all cells that progressed through an epicardial progenitor and isolated EPDCs via GFP expression [21]. EPDC-conditioned medium administered immediately after induced myocardial infarction in mice led to reduced cardiac remodeling after 9 weeks but did not significantly improve ejection fraction. The group identified proangiogenic factors highly enriched in EPDCs including Vegfa, Angpt1, Ang, Fgf1, Fgf2, Fgf9, Pdgfa, Pdgfc, Pdgfd, Adamts1, Tgfb2, Jag1, Hgf1, Sdf1, Mcp1, and Il6.
In another study, mouse embryonic stem cell-derived CMs were either directly cocultured with epicardial mesothelial cells or treated with medium conditioned by these cells [22]. In both cases, coculture increased the number of α-actinin+ CMs and the expression of Myh6, Mlc2v, and Mlc2a in the CMs. Their work further suggested that epicardial-derived follistatin-like 1 (FSTL1), a glycoprotein implicated in suppression of inflammation, could evoke CM division and have pro-regenerative effects.
Finally, hPSC-EpiCs have been incorporated in hPSC-CM-containing microtissues and implanted into rat hearts immediately after induced myocardial infarction [23]. At 14 and 28 days post-transplantation, microtissues incorporating both CMs and EpiCs contained FB-like cells expressing S100A4 and demonstrated improved engraftment, enhanced microvessel density, and enhanced hPSC-CM proliferation compared to microtissues containing only CMs. Functionally, this resulted in increased fractional shortening and left-ventricular end-systolic dimension at day 28, which led to persistent improvements in cardiac pumping capability for more than 12 weeks. Taken together, these prior studies demonstrate that hPSC-EpiCs may interact with hPSC-CMs to influence mechanisms of development or repair.
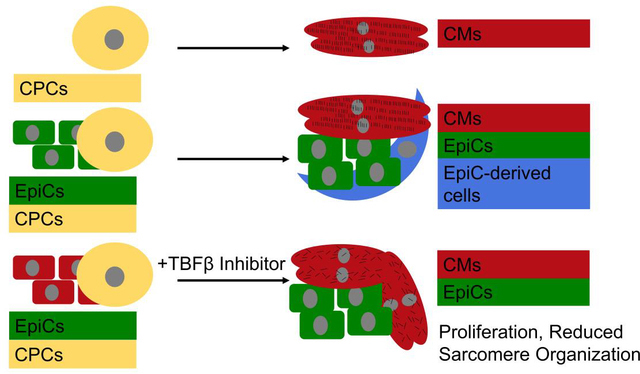
EpiCs are present early in the developing mouse heart, surround the myocardium by E8.5, and undergo EMT and differentiate to EPDCs around E14.5 [24]. Thus, we initiated coculture of hPSC-EpiCs with differentiating CMs at the cardiac progenitor cell (CPC) stage to test the effects of EpiCs on CM differentiation and maturation. Previous studies from our group and others have demonstrated that hPSC-CMs can receive microenvironmental cues, including signals from hPSC-endothelial cells or mechanical pacing, which direct maturation during this early differentiation window, whereas at later timepoints (e.g. after CM specification) these cues may have little to no effect [25,26]. To investigate if EpiCs or EPDCs had the capability to impact CM maturation or proliferation, we cultured hPSC-EpiCs with hPSC-CPCs in dispersed direct coculture at a 1:1 ratio in LaSR basal medium with or without the TGFβ inhibitor A83–01 for 2 weeks with medium change every two days, and compared the coculture to hPSC-CPC monocultures. We refer to EpiC/CPC coculture in LaSR basal medium supplemented with A83–01 as LAEC (LaSR A83–01 EpiC/CPC), CPC monoculture in LaSR basal medium supplemented with A83–01 as LAC (LaSR A83–10 CPC), EpiC/CPC coculture in LaSR basal medium as LEC (LaSR EpiC/CPC), and CPC monoculture in LaSR basal medium as LC (LaSR CPC) as shown in Figure 1A. We performed additional coculture studies to investigate the dose response in LAEC cocultures and compare the effects of LAEC conditioned medium or indirect cocultures to direct cocultures.
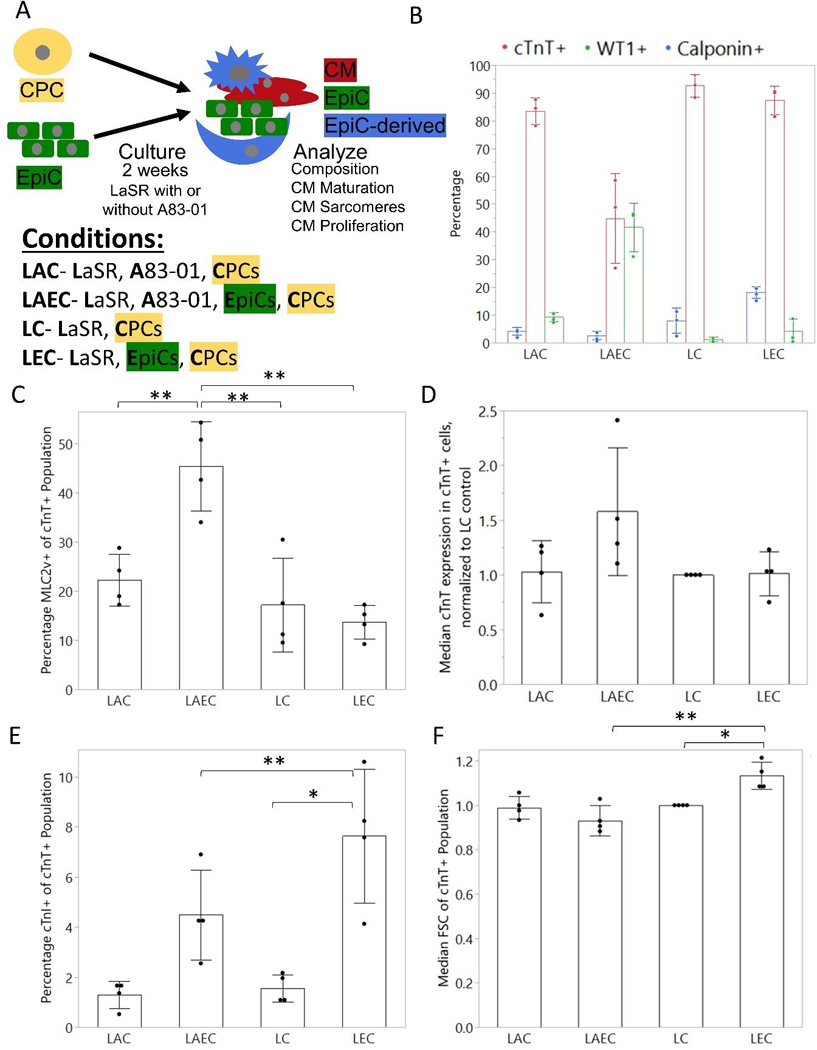
Figure 1:
Description and analysis of EpiC and CPC coculture composition. (A) Schematic of coculture experiments. Cardiac progenitor cells (CPCs) were seeded with epicardial cells (EpiCs) at a 1:1 ratio and cultured for two weeks with or without TGFβ inhibitor A83–01 in direct contact coculture. (B) Percentage of cTnT+, WT1+, and Calponin+ cells at the end of the two week cocultures assessed by flow cytometry. (C) Percentage MLC2v+ of the cTnT+ population. (D) Mean fluorescence intensity (MFI) of cTnT expression in cTnT+ cells, normalized to EpiC samples to control for variability between flow cytometry experiments. (E) Percentage cTnI+ cells in cTnT+ population (F) Normalized median FSC-A of cTnT+ population. Dots represent the average of one well from 3–4 independent differentiations. Bars represent the average across 3–4 differentiations and error bars represent the standard deviation. Statistics are ANOVA with Tukey’s post hoc test using raw data where * is p<0.05 and ** is p<0.01.
Here, we demonstrate that coculture of hPSC-EpiCs with developing hPSC-CMs for two weeks increased CM proliferation, increased expression of MLC2v, and decreased sarcomere organization. When EpiCs were allowed to undergo EMT, they increased expression of cTnI but did not retain the potential to alter CM proliferation and sarcomere organization. EpiC-conditioned medium or indirect coculture did not induce the same level of proliferation as direct coculture. Furthermore, CMs altered EpiCs lineage bias from a SFRP2-enriched population to a DLK1- or C3-enriched cocultured population. Overall, this suggests the importance of EpiC and CM bi-directional crosstalk during hPSC-CM differentiation and maturation which provides crucial insight for the design of cardiac tissue engineering and regenerative medicine strategies.
2. Results
2.1. Coculture of EpiCs and CPCs
We cocultured developing CPCs and EpiCs at a 1:1 ratio with or without A83–01, a TGFβ inhibitor, for two weeks (Figure 1A). Prior to analysis, brightfield images were taken from each monoculture and coculture condition (Figure S1A). In the monocultures, the CPCs formed a thick layer of cells which, in the LC sample, began to detach from the plate after spontaneous contraction of the CMs began. In the LEC coculture, the CPCs formed dense contracting areas surrounded by noncontracting cells. Similarly, in the LAEC coculture, the beating areas were exclusively found within the web-like network or spheroid-like clusters of cells, and the contracting clusters were surrounded by cells that exhibited an epithelial cobblestone morphology similar to EpiCs. We did not observe cTnT+ cells in monocultured EpiCs treated with or without A83–01 (Figure S1B). Cellular localization was verified through immunocytochemistry for cTnT to identify CM sarcomeres and WT1 to identify EpiCs (Figure S1C).
Next, we compared the resulting populations from two-week monocultures and cocultures using flow cytometry for cTnT to mark CMs, WT1 to mark EpiCs, and Calponin to mark smooth muscle-like cells (Figure 1B). Example flow cytometry gating plots are shown in Figure S2. Monoculture CMs in LaSR medium with and without A83–01 contained over 80% cTnT+ cells with small numbers of EpiCs or smooth muscle-like cells as byproducts of the CM differentiation. This is consistent with a single cell sequencing analysis which identified EpiCs, SMCs, endothelial cells, and FBs in CM differentiations [27]. When 1:1 EpiCs:CPCs were cocultured in the presence of A83–01 (LAEC), we observed approximately 50% WT1+ cells, much higher than <5% WT1+ cells in the CPC monoculture control (LAC). In the absence of A83–01 (LEC), less than 5% of the 1:1 EpiC:CPC coculture population was WT1+ and approximately 20% of cells expressed Calponin, much higher than <5% WT1+ cells or Calponin+ cells in the CPC monoculture control (LC), suggesting that the EpiCs in coculture underwent EMT and at least some differentiated into smooth muscle-like cells. Comparing LAEC and LEC coculture conditions, the higher percentage of non-CMs in the LAEC condition may result from differential proliferative capacity of EpiCs and EPDCs.
2.2. Cardiac protein expression and cell size are altered in CPCs cocultures with EpiCs and EpiCs undergoing EMT
To assess the effects of EpiCs on the maturation of cocultured CMs, the cocultured cells were analyzed via flow cytometry for the expression of sarcomere proteins that have been linked to maturation. By co-labeling the cells with cTnT and MLC2v, the percentages of MLC2v+ CMs in two-week cocultures and monocultures were determined via flow cytometry (Figure 1C). A significant increase in MLC2v+ CMs was found in the LAEC coculture in comparison to the LAC monoculture. No change was found between the LC and LAC monocultures, nor between the LEC coculture and LC monoculture, suggesting that EpiCs increased expression of MLC2v in co-cultured CMs but EPDCs did not.
The median fluorescence intensity (MFI) of cTnT was analyzed within cTnT+ CMs in Figure 1D. The MFI was higher in the LAEC coculture condition compared to the LAC monoculture condition. No significant change was found between the LC or LAC monocultures nor between the LC monoculture and LEC coculture.
To determine if troponin I isoform expression is impacted by the coculture, the percentage of cTnI+ cTnT+ CMs was quantified in cocultures and monocultures (Figure 1E). Very few of the CMs expressed cTnI in any of the conditions. However, there was a slight but significant increase in cTnI+ cells in the LEC coculture, which is consistent with previous reports of CM maturation by EPDCs [20].
Using the forward scatter from flow cytometry, a semi-quantitative measurement of the cell volume can be determined (Figure 1F). The cTnT+ CMs from the 1:1 EpiC:CPC coculture in the absence of A83–01 (LEC) had significantly higher forward scatter than the CM monoculture (LC).
Together, these coculture experiments demonstrate that the EpiCs, when treated with A83–01 to prevent EMT (LAEC), increased both cTnT and MLC2v expression in the CMs derived from the CPCs, a few key markers of structural maturation. In the presence of EPDCs (LEC), CMs had increased cell area and cardiac troponin I expression, suggesting differential molecular maturation effects specific to EpiC or EPDC cocultures.
2.3. EpiC coculture with CPCs induces CM proliferation and reduces structural maturation
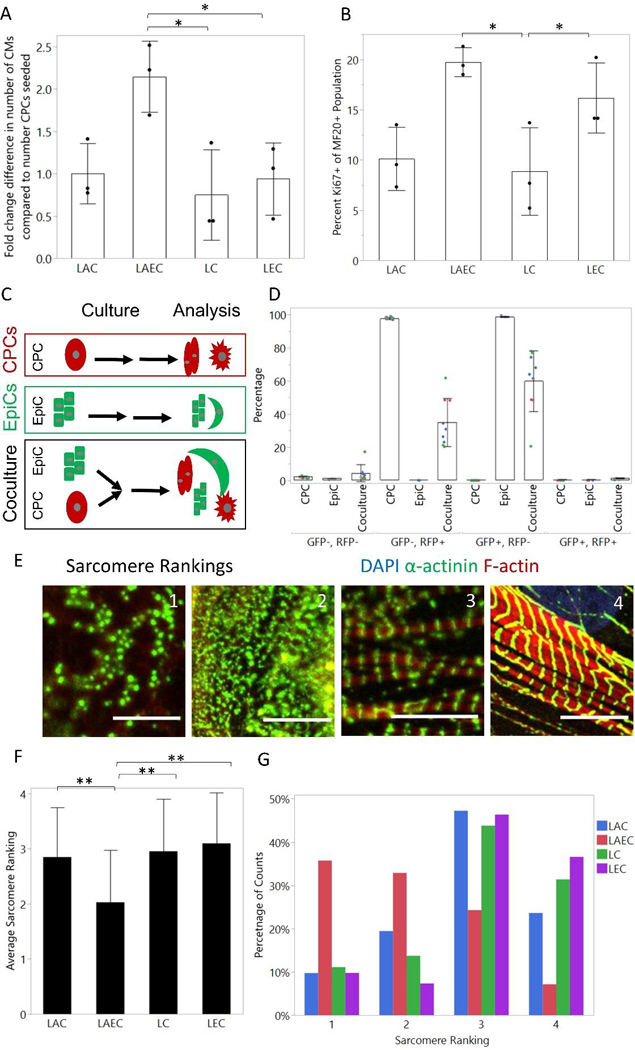
When comparing cocultures to monocultures, we noticed that there were more total cells in the CPC-EpiC coculture containing A83–01 (LAEC) samples compared to monocultures and the LEC coculture. To determine if this represented an increase in the number of CMs or was due to an increase in the number of EpiCs, we quantified the ratio of cTnT+ cells at the end of the coculture compared to the number of CPCs seeded at the beginning of the experiment. As shown in Figure 2A, there were approximately two-fold more CMs per CPC seeded in the LAEC coculture than in the LEC coculture or monoculture conditions, suggesting the EpiC coculture induced CM proliferation. To further investigate CM proliferation, we stained for Ki67 and MF20, which binds cardiac myosin heavy chain, and quantified the percentage of MF20+ cells that were Ki67+ in each condition (Figure 2B). We identified a significantly higher percentage of Ki67+ CMs in the LAEC coculture compared to the LAC and LC monocultures. Ki67 is unable to distinguish between cell division, endoreduplication, and poly-nucleation [13], so we also stained for Vybrant DyeCycle Green DNA dye and observed an increased fraction of cycling cells in the cTnT+ population of the LAEC coculture samples (Figure S3A–B). Since across all conditions approximately 8% of D20 hPSC-CMs were binuclear, and the LAEC samples with higher Ki67 in MF20+ cells also contained many more CMs, so it is likely the Ki67 expression and DNA dye incorporation is indicative of proliferation of CPCs and CMs between D6 and D20. We next performed cocultures using WTC-CAAX-RFP CPCs and WTC-LMNB1-eGFP EpiCs to determine if cell fusion had occurred (Figure 2C, Figure S4). In the LAC coculture, we observed less than 1% GFP+ RFP+ double positive cells by flow cytometry, suggesting that cell fusion is likely not the cause of an increased number of CMs compared to number of CPCs seeded (Figure 2D).
Figure 2: LAEC cocultures cause CM proliferation and reduce sarcomere organization.
(A) Fold change difference in number of CMs at day 20 compared to seeding measured by total number of cTnT+ cells at end of coculture divided by the number of CPCs seeded. (B) Percentage Ki67+ cells of MF20+ population as measured by flow cytometry. Dots represent the average of one well from three differentiations. Bars represent the average across 3 differentiations and error bars represent the standard deviation. Statistics are one-way ANOVA with Tukey’s post-hoc test where * is p<0.05 and ** is p<0.01. (C) Schematic of cell fusion experiment. CPCs were differentiated in the WTC-CAAX-RFP line and EpiCs were differentiated in the WTC-LMNB1-eGFP line. After the two-week coculture, samples were analyzed by flow cytometry. (D) Percentage GFP−/RFP−; GFP−/RFP+; GFP+/RFP−; and GFP+/RFP+. Dots represent well replicates (n=3) and colors represent 3 independent differentiations. Bars represent the average across 3 differentiations and error bars represent the standard deviation. (E) Example images of sarcomere rankings. Scale bar is 10 μm. (F) Average sarcomere rating from blinded image analysis of CMs in coculture and monoculture conditions. Images were from 3 independent CM differentiations with 3 wells per differentiation and a total of at least 40 cells per condition. Bar graph represents the average across all differentiations and the error bars represent the standard deviation. Statistics are ANOVA two-way test with Tukey’s post-hoc test, where * is p<0.05 and ** is p<0.01. (G) Histogram of sarcomere ranking scores for each monoculture and coculture condition across all differentiations.
To determine if EpiC cocultures affected CM sarcomere organization, we analyzed CMs for the presence of H zones and Z lines and quantified sarcomere length in replated CMs by immunofluorescent staining for α-actinin and phalloidin (F-actin). We then imaged individual cells via confocal microscopy and ranked blinded images on the following scale: 1) no visible sarcomere structure, 2) some sarcomere organization, 3) H zones and Z lines visible in areas with some sarcomere organization, and 4) near perfect sarcomeres with clear H zones, Z lines, and thick myofibrils [26]. Example images of each of these scores are shown in Figure 2E. We observed a lower average sarcomere rating and a distinctly lower distribution of sarcomere ratings in the CPC-EpiC coculture in the presence of A83–01 (LAEC) compared to all other conditions (Figure 2F–G). We did not observe any difference in sarcomere length between coculture conditions; sarcomere lengths were approximately 1.4 μm in all conditions, similar to other reports for immature hPSC-CMs (Figure S3C) [23]. We also performed automated sarcomere analysis using SarcTrack2 [28] and did not observe a difference in mean sarcomere length or angle between sarcomeres (Figure S3D–F).
We next stained for cTnT, thresholded images, and performed cell profile analysis to further discover differences in CM structural maturation resulting from EpiC coculture (Figure S5) [13]. We observed decreased CM area in LAEC cocultures compared to the LAC monoculture control. Interestingly, this measurement of cell spreading contrasts the previous forward scatter analysis where we had observed an increase in LEC coculture average forward scatter, which may be due to the complex metric of forward scatter which is influenced by cell volume and shape rather than spread area. This suggests that EpiC coculture induces CM proliferation, reduces CM cell area, and decreases sarcomere organization.
2.4. CM proliferation and maturation is dose dependent on the EpiC:CPC ratio
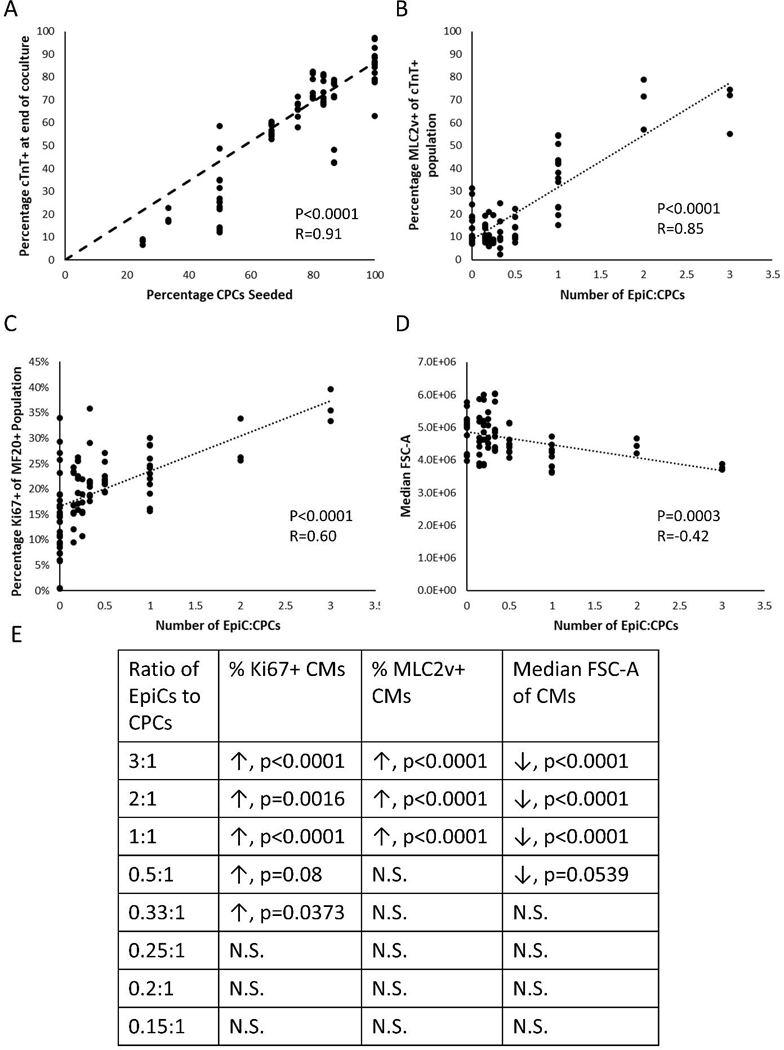
To ascertain if CM proliferation was dose dependent on the relative number of EpiCs to CPCs in coculture, we performed LAEC cocultures at various seeding ratios of EpiCs:CPCs (3:1, 2:1, 1:1, 0.5:1, 0.33:1, 0.25:1, 0.2:1, 0.15:1), keeping the total number of cells in the coculture constant. First, we observed an approximately linear correlation between the ratio of CPCs seeded in the cocultures and the ratio of CMs in the cell population after the two-week cocultures (Figure 3A). The ratio of EpiCs:CPCs also strongly correlated with the percentage of MLC2v+ cells in the cTnT+ population and the percentage of Ki67+ cells in the MF20+ population (Figure 3B–C). Consistent with previous results, we observed a negative correlation between CM cell size measured by forward scatter and the ratio of EpiCs:CPCs (Figure 3D). Furthermore, we observed statistically significant differences in these properties when directly comparing LAEC coculture at various seeding ratios with LAC monoculture above a ratio of approximately 0.5:1 EpiCs:CPCs (Figure 3E). These results demonstrate that EpiCs impart proliferative effects and acquisition of MLC2v expression in CMs in a dose-dependent manner.
Figure 3: Dose response of EpiCs on CM proliferation in coculture.
Flow cytometry analysis of LAC monoculture and LAEC coculture after two weeks. Three well replicates from at least three independent differentiations were analyzed. (A) Percentage of cTnT+ CMs at end of two-week cocultures as a function of CPCs seeded. (B) Percentage of MLC2v+ cells of cTnT+ population as a function of EpiCs:CPCs initially seeded. (C) Percentage of Ki67+ cells of MF20+ population as a function of EpiCs:CPCs initially seeded. (D) Median FSC-A of cTnT+ population as a function of EpiCs:CPCs initially seeded. Each dot represents a single well. Analysis includes data from 8 independent differentiations. Statistics are F-test where null hypothesis is slope equal to zero. (E) Table shows statistics from a two-way ANOVA with Dunnett’s post-hoc comparison to LAC monoculture control. Arrows indicate if coculture was higher or lower than monoculture CMs and N.S. represents p>0.1.
2.5. EpiC conditioned medium and indirect coculture does not induce CM proliferation
Crosstalk between EpiCs and CPCs could be through soluble factors or be mediated through cell-cell signaling that requires close contact. To determine if CM proliferation was induced via soluble factors secreted by EpiCs, we performed conditioned medium studies by treating CPCs every two days with fresh LA (LaSR A83–01) medium or 1:1 fresh LA medium to LA medium that had been conditioned by EpiCs for two days. After two weeks of conditioned medium culture, we analyzed the CMs for changes in proliferation and structural maturation. Over four independent differentiations, we did not observe any statistically-significant differences in percentage of MLC2v+ CMs, percentage of Ki67+ CMs, cell size as measured by forward scatter, or an increase in the total number of CMs measure by total cTnT+ cells, comparing unconditioned medium to conditioned medium (Figure S6A–D). Structurally, we observed no difference in cell area, perimeter, circularity, or aspect ratio in CMs cultured in EpiC conditioned medium over the course of ten independent differentiations (Figure S6E–H). Additionally, we did not observe any differences in sarcomere organization between CMs cultured in unconditioned and conditioned medium in four independent differentiations (Figure S6I–J). Automated SarcTrack2 analysis identified slightly higher sarcomere angles in the conditioned medium samples compared to the unconditioned medium sample and no change in the average sarcomere length or standard deviation of the sarcomere angle (Figure S6K–M). Together these data suggest that EpiC-conditioned medium was not capable of replicating the effects of direct EpiC coculture on CM proliferation, MLC2v gene expression, and sarcomere organization indicating that EpiCs impart pro-proliferative signals through cell-cell interactions.
We next performed indirect cocultures by seeding CPCs across from CPCs or EpiCs separated by a cell-free region with fresh medium changes every two days (Figure S7A). After the two-week coculture, we did not observe any difference in forward scatter, percentage Ki67+ cells in the MF20+ population, or percentage cycling cells in the cTnT+ population (Figure S7B–D) between LAC monoculture and indirect coculture. Additionally, we observed a slight change in the average sarcomere ranking using manual blinded analysis, however this was not to the same degree as the direct cocultures (Figure S7E–G). SarcTrack2 analysis also identified a slight increase in the average sarcomere length in the indirect coculture and no change in the sarcomere angle (Figure S7H–J). Together, these results suggest that indirect coculture reduces the effect of EpiC crosstalk with developing CMs compared to direct EpiC coculture.
2.6. Single cell transcriptomics identifies bi-direction crosstalk between EpiCs and differentiating CMs
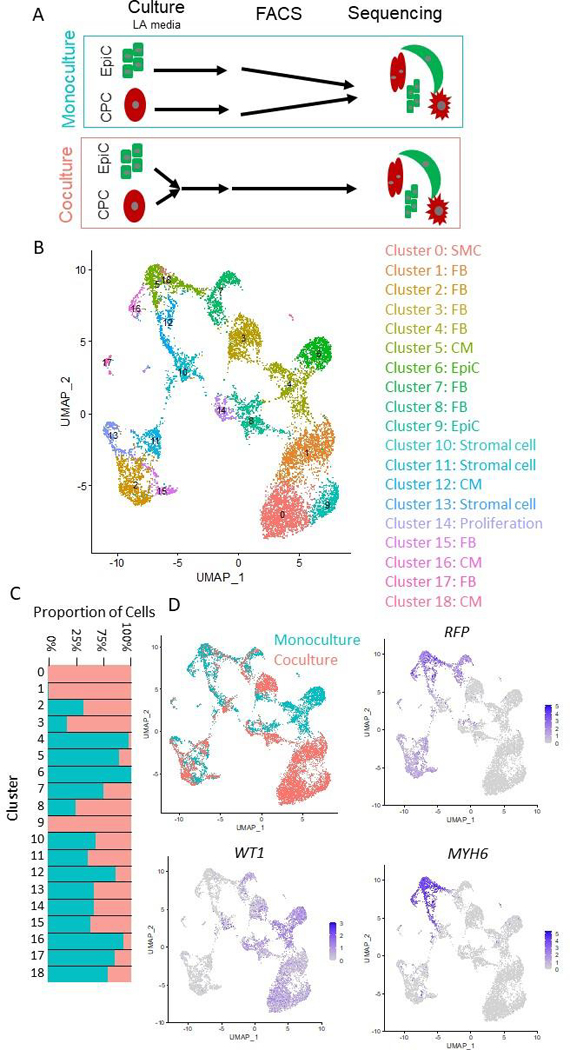
To further investigate how coculture of EpiCs with differentiating CMs affects the resulting cell populations, we performed single cell RNA-sequencing (Figure 4A). First, we cocultured CPCs differentiated in the WTC11-CAAX-RFP iPSC line with EpiCs differentiated in the WTC11-LMNB1-eGFP iPSC line for two weeks in the presence of A83–01. We chose these lines as they have the same parental background to minimize genetic variation. Additionally, expression of RFP and GFP allowed us to determine cell types that arose from CPC or EpiC differentiation, respectively. We used fluorescence activated cell sorting (FACS) to isolate live cells via DAPI exclusion (flow gating strategy shown in Figure S4) and created two libraries, monoculture and coculture, for Illumina 10X sequencing. Note that the monoculture library contained both monoculture CPCs and monocultured EpiCs, distinguished by expression of GFP and RFP. In total, we identified 5,470 monoculture cells and 7,138 coculture cells with over 64,000 reads per cell mapping to approximately 5,000 genes per cell.
Figure 4: Single cell sequencing of EpiC and CM coculture.
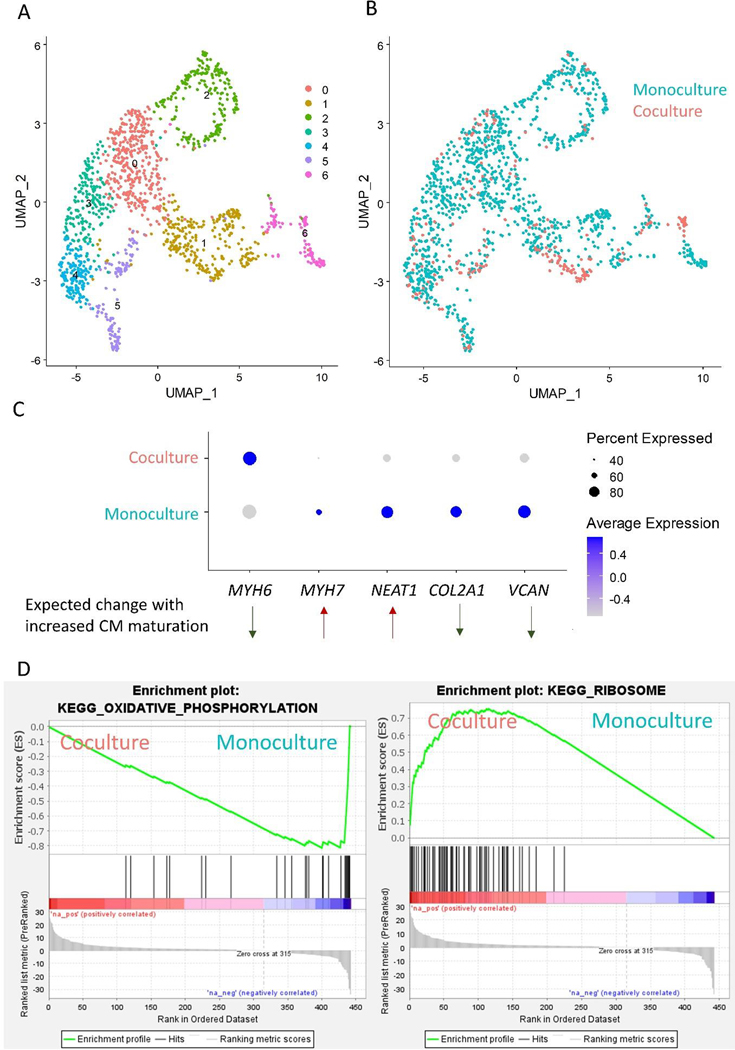
(A) Schematic of single cell RNA sequencing experiment. hPSC-CPCs were differentiated in the WTC-CAAX-RFP cell line and hPSC-EpiCs were differentiated in the WTC-LMNB1-eGFP line. Cells were cultured for two weeks, then monoculture and coculture (1:1 EpiCs:CPCs) samples were collected for sequencing. (B) UMAP projections of single cell clusters. (C) Bar chart and feature plots highlighting abundance of cells from monoculture/coculture conditions in each cluster. (D) Feature plots of monoculture/coculture conditions, RFP indicating cells arising from CPCs and MYH6 indicating CMs. Slingshot lineage trajectory denoting potential differentiation pathways between clusters.
Non-biased clustering identified 18 different populations which we categorized into cell types based on expression of known markers (Data File S1). Figure 4B shows a UMAP projection of these clusters and Data File S1 shows differentially enriched genes in each cluster. We identified two EpiC populations (Clusters 6 and 9) enriched for ITLN1, EFEMP1, and UPK3B, and four different CM populations (Clusters 5, 12, 16, 18) enriched for sarcomere transcripts including TNNT2, MYL6, and MYL7, and twelve stromal cell populations (Clusters 0–4, 7–8, 10–11, 13, 15, 17) (Figure S8). Additionally, we identified one cluster (Cluster 14) enriched in cell-cycle associated genes which likely represents proliferative stromal cells. We then identified which clusters were present in the monoculture and coculture conditions (Figure 4C–D). RFP+ cells were identified in the CM clusters and Clusters 2, 11, 13, and 15, which we can further classify as FB and stromal cell side populations of the CM differentiation. Unfortunately, we were unable to map a significant portion of GFP+ cells because we performed 3’ library prep and the heterozygous GFP fusion was located at the N-terminus of the LMNB1 protein. However, it is likely that the vast majority of cells in Clusters 0–1, 3–4, 6, 8–9, and 14 arose from the EpiC differentiation since they did not express RFP.
We validated identification of cell types by combining our dataset with a publicly available hPSC-CM differentiation single cell sequencing dataset and two single cell sequencing datasets of the developing human heart (Figure S9, Dataset S2). Interestingly, the main side populations in the hPSC-CM differentiations aligned with FBs, smooth muscle cells, EpiCs, and a population of cells (Cluster 6) that does not correlate with a population in the developing human heart. Additionally, the hPSC datasets did not contain many endothelial cells, macrophages, or other immune cells (Clusters 14 and 18).
We then analyzed differences in EpiC and CPC lineage potential in the monoculture and coculture conditions. Slingshot trajectories suggest a fate map from epicardial cluster 6 through EMT to cluster 4 (monoculture) and from epicardial cluster 9 through EMT to cluster 0 (coculture) (Figure 4C). Additionally, we observed high expression of WT1 in Cluster 0 and 6, confirming epicardial clusters, expression of TBX18 primarily in monoculture clusters 6 and 9, and expression of TCF21, a marker of EPDCs, in Cluster 3 indicating distinct monoculture and coculture epicardial lineages (Figure S10A).
To further investigate these epicardial lineage differences, we performed subclustering of all epicardial lineages (Clusters 0–1, 3–4, 6, 8–9, and 14), defined by lack of sarcomeric genes and RFP expression, and identified 10 subclusters (Figure S10B). Interestingly, both EpiCs (ITLN1+) and stromal cells (ITLN1−) clustered by monoculture and coculture conditions (Figure S10B–C). Monoculture epicardial lineages were enriched for SFRP2, a WNT signaling ligand. Coculture epicardial lineages were enriched for two lineages primarily driven by expression of DLK1, a non-canonical ligand in the Notch family, and C3, an activator of the complement system. Interestingly, a recent single cell sequencing study identified two distinct mouse epicardial cell-derived populations, one enriched for Sfrp2 in epicardial-derived mesenchymal cells and the other enriched for Dlk1 in epicardial-derived mesothelial cells [29]. The authors of this paper suggested that these two populations are nodes of two distinct lineages and that epicardial EMT is required for proper endothelial cell localization and specification. Together, this suggests that EpiC coculture with developing CMs alters EpiC transcriptional profiles.
We next performed subclustering of CMs (Clusters 5, 12, 16, and 18), defined by enrichment of transcripts encoding sarcomeric proteins and RFP expression, and identified 7 subclusters (Figure 5A, Data File S3). We did not observe enrichment of monoculture or coculture cells in any of the subclusters which may be due to the fact that many sarcomeric genes are regulated at the protein level rather than the transcriptomic level (Figure 5B).
Figure 5: Sub-clustering of hPSC-CMs in scRNA-seq dataset.
(A) Subclustering and UMAP projections of CMs from data in Figure 4 (Clusters 5, 12, 16, and 18, determined by high expression of sarcomere protein encoding genes). (B) Feature plot of cells from monoculture and coculture conditions. (C) Dot plot highlighting cells from monoculture and coculture conditions. Genes selected were differentially expressed in this dataset and two single cell sequencing datasets of hPSC-CM differentiation and maturation during extended culture (E-MTAB-6268)[27,37]. Direction of differential expression was further validated in three bulk sequencing datasets of hPSC-CM maturation (GSE154294, GSE64189, and GSE81585) [30–32] and a single cell sequencing dataset of human heart CM development (GSE106118) [33]. (D) GSEA Enrichment plots of KEGG pathways enriched in monoculture and cocultured CMs.
To investigate hPSC-CM maturation, we analyzed two publicly available single cell sequencing datasets to identify transcripts associated with CM maturation over extended culture. hPSC-CMs were identified by clusters enriched in sarcomeric genes, and differential expression analysis was performed comparing D15 vs. D30 hPSC-CMs (Friedman et al., GSE106118) and D12 vs. D24 hPSC-CMs (Gerbin et al.) These timepoints span our D20 hPSC-CMs coculture samples and are representative of an expected level of maturation in our study. Cross referencing differential expression across these datasets, we identified increased expression of 13 genes, and 7 of these genes (VCAN, MASP1, COL2A1, DOK4, NEAT1, MYH6, and MYH7) were differentially regulated between monoculture and coculture hPSC-CMs (Data File S3). We then cross referenced this list with three publicly available bulk sequencing datasets of hPSC-CM differentiation around similar timepoints (GSE154294, GSE64189, and GSE81585) [30–32] and verified a decrease in VCAN, COL2A1, and MYH6 expression and increase in NEAT1 and MYH7 with increasing hPSC-CM maturation. Lastly, we validated differential expression and directionality of these transcripts using CMs from a single cell sequencing dataset of human heart development (Figure S11) (GSE106118) [33]. We identified similar patterns of gene expression of MYH6 and COL2A1 which were inversely correlated to expression of MYH7 in the developing human heart dataset, confirming that several of the identified genes in our hPSC-CM maturation analysis are spatiotemporally regulated in human heart development.
Using the set of genes that correlate with hPSC-CM maturation over extended culture, we benchmarked maturation of our hPSC-CM cocultures. We observed decreased structural maturation in coculture CMs (increased expression of MYH6 and decreased expression of MYH7) (Figure 5C). A similar trend was observed by qPCR in RFP+ FACS populations of three independent differentiations and was validated by Western blotting of total coculture lysates in four independent differentiations (Figure S12). Additionally, we observed decreased expression of NEAT1, a long non-coding RNA shown to regulate processing of pro-apoptotic microRNA-22 in hPSC-CMs [34]. ECM related signaling has been associated with cardiomyogenesis and development [35]. In our cocultures, we observed changes in ECM transcripts in cocultured CMs demonstrated by decreased expression of COL2A1 and VCAN. This analysis suggests that coculture of EpiCs with hPSC-CMs leads to complex changes in CM maturation.
We then performed Gene Ontology analysis on a pre-ranked list of genes generated from our differential expression analysis (Figure 5D). We identified the Kegg pathway terms “oxidative phosphorylation enriched” in the monoculture CMs and “ribosome” in the EpiC coculture CMs. During development, CMs transition from primarily glycolytic metabolism to oxidative phosphorylation and this transition has been used as a metric to assess hPSC-CM maturation [14]. Ribosome biogenesis has been associated with cell growth and proliferation, and correlated with CM hypertrophy [36].
Overall, the single cell transcriptomic analysis suggests that coculture of EpiCs and differentiating CMs imparts bi-directional effects on differentiation potential and the resulting cell populations. EpiCs cocultured with differentiating CMs are enriched for DLK1 and C3 and monoculture EpiCs are enriched for SFRP2. Additionally, we identified 5 differentially regulated genes (MYL6, MYL7, NEAT1, COL2A1, and VCAN) between monoculture and coculture CMs suggesting decreased MHCβ/α ratios in cocultured CMs. Taken together, this suggests the importance of crosstalk between CM and EpiCs during early developmental/differentiation stages.
3. Discussion
EpiCs are required for proper heart development and play an important role in cardiac repair and regeneration. Thus, we hypothesized that coculture of hPSC-derived EpiCs or EPDCs with differentiating hPSC-CMs would influence CM maturation.
We observed increased proliferation in CMs cocultured with EpiCs only when EpiC fate was maintained by A83–01. This was accompanied with a decrease in sarcomere organization and cell size. CM proliferation was affected by the ratio of EpiCs to CPCs, and we observed changes in proliferation up to a ratio of EpiCs:CPCs of 0.33:1. We further demonstrated that EpiC conditioned medium and indirect cocultures did not replicate the effects of direct coculture, suggesting that direct contact is required which is consistent with a previous study investigating coculture effects of quail EPDCs and mouse CMs [20].
hPSC-CM maturation has also been benchmarked by other metrics including MLC2a/v isoform shift, MHCα/β isoform shift, and cTnI expression [2,8,9]. We detected similar expression levels of MLC2v [38], cTnI [39], and Ki67 [40,41] in our D20 CMs compared to previous reports of hPSC-CMs at a similar maturation timepoint. Surprisingly, we observed increased MLC2v protein expression in the LAEC coculture condition, no increase in cTnI expression, and a decrease in sarcomere organization. We hypothesize that the EpiC coculture accelerates the isoform switch from MLC2a to MLC2v, but that the cells lack a mechanism to organize sarcomeres.
In an attempt to classify hPSC-CM maturation induced by our EpiC cocultures, we compared our single cell sequencing data with other publicly available sequencing datasets of hPSC-maturation maturation. We identified 3 novel markers of hPSC-CM maturation (NEAT1, COL2A1, and VCAN). In our LAEC cocultures, we observed changes in ECM transcripts (decreased expression of COL2A1 and VCAN) and decreased MYH7/6 ratios which we confirmed by Western blotting. This suggests that MLC and MHC isoform shifts may not be directly related in maturing hPSC-CMs as previously demonstrated by Lee et al. after treatment of hPSC-CMs with thyroid hormone T3 [42] and emphasizes the idea that coculture may improve hPSC-CM maturation by some benchmarks but not others. Taken together, direct coculture of hPSC-CM with hPSC-EpiCs increased MLC2v expression while decreasing sarcomere organization, cell size, and MHCβ/α ratios.
Furthermore, we observed bidirectional effects of EpiC/CM coculture suggesting that CM coculture affects EpiC lineage bias. First, we identified two EpiC clusters, one from the monoculture and one from the coculture conditions. Using slingshot analysis, we determined that these clusters were terminal nodes of two different lineages. Subclustering of the EpiC lineages identified that monoculture clusters are enriched for SFRP2 and coculture clusters are enriched for DLK1 or C3 suggesting that CM coculture alters EpiC lineage bias and cell signaling.
While our study provides mechanistic insight into CPC-EpiC interactions in a 2D platform, we have not yet explored how EpiC migration and localization in a 3D-tissue like structure would affect these interactions. Self-assembling human cardiac organoids, such as those developed by Lewis-Israeli et al., could be the next platform to study the increasingly complex differentiation of hPSCs into cTnT+ CMs surrounded by WT1+ EpiCs [43]. We hypothesize that these organoids or spheroids of pre-differentiated cells will provide further insight into complex spatiotemporal crosstalk mechanisms.
4. Conclusions
In conclusion, direct coculture of hPSC-EpiCs with differentiating CMs induces CM proliferation, decreases CM sarcomere structural organization, decreases CM cell size, and directs EpiC lineage bias from a SFRP2 enriched population to a DLK1 or C3 enriched population. This work suggests important crosstalk between EpiCs and CMs during differentiation can be used to influence cell fate and improve the ability to generate cardiac cells and tissues for in vitro models and development of cardiac cellular therapies.
5. Materials and Methods
5.1. hPSC culture with CPC and EpiC differentiations
For this study, hESC line H9 (WiCell) and hiPSC lines WTC-CAAX-RFP (Allen Institute for Cell Science), WTC-LMNB1-eGFP (Allen Institute for Cell Science), and 19–9-11 (WiCell) were used. Human pluripotent stem cells (hPSCs) were maintained on Matrigel (Corning)-coated plates in mTeSR1 (STEMCELL Technologies) according to previously published methods [2]. At 80–90% confluency hPSCs were passaged with Versene to maintain colonies. As previously described and detailed below, hPSCs were differentiated into CPCs and EpiCs [2,44].
5.2. Cardiac progenitor cell differentiation via modulation of canonical Wnt signaling
As previously published in the GiWi protocol to derive cardiac progenitors, hPSCs were singularized with Accutase at 37°C for 5 minutes, quenched in mTeSR1, and centrifuged at 200 g for 5 minutes [2]. hPSCs were seeded at 100,000–600,000 cells/cm2 in mTeSR1 supplemented with 5 μM ROCK inhibitor Y-27632 (Selleckchem) (day −2) for 24 hours. The following day (day −1), cells were treated with fresh mTeSR1. At day 0, cells were treated with 6–15 μM CHIR99021 (Selleckchem) in RPMI1640 supplemented with B27 minus insulin (RPMI/B27−) media. Exactly 23–24 hours later, media was changed to fresh RPMI/B27− (day 1). At day 3, 5 μM IWP2 (Tocris) was added to 1:1 conditioned media to fresh RPMI/B27− media. At day 5, cells were treated with RPMI/B27− media. At day 6, cardiac progenitors were either frozen in cryomedia (60% DMEM/F12, 30% FBS, 10% DMSO) or singularized for further differentiation.
5.3. EpiC differentiation via activation of canonical Wnt signaling
Following our previously published protocol for epicardial differentiation, day 6 CPCs were either singularized in Accutase at 37°C for 10 minutes or thawed from cryo and seeded onto a gelatin (Sigma-Aldrich) or Matrigel-coated cell culture plate at 20,000–80,000 cells/cm2 (approximately a 1:3 or 1:12 split) in LaSR basal media (advanced DMEM/F12 (Life Technologies) with 0.06g/L L-ascorbic acid 2-phosphate sesquimagnesium salt hydrate (Sigma-Aldrich) and 12.5 mL GlutaMAX) supplemented with 5 μM ROCK inhibitor Y-27632[44]. At day 7 and 8, cells were treated with fresh LaSR basal media supplemented with 3 μM CHIR99021. At days 9, 10, and 11, cells were treated with fresh LaSR basal media. At day 12, EpiCs were singularized with Accutase for 5 minutes at 37°C and either cryopreserved for later use or replated in LaSR basal media supplemented with 0.5μM A83–01 (R&D Systems) and 5μM ROCK inhibitor Y-27632. Subsequently, EpiCs were treated with LaSR supplemented with A83–01 daily until 90–100% confluent. EpiCs were then passaged using Versene into fresh LaSR basal media supplemented with 0.5μM A83–01 without ROCK inhibitor Y-27632 to maintain colonies, prevent further differentiation, and improve attachment for up to five passages. Alternatively, cells were frozen in cryomedia (60% DMEM/F21, 30% FBS, 10% DMSO). Differentiations were validated to have at least 90% WT1 positive cells by flow cytometry.
5.4. Coculture of CPCs and EpiCs
CPCs were thawed and plated in monoculture or combined with EpiCs on gelatin or Matrigel-coated plates at a density of 571k cells/cm2 in LaSR medium supplemented with 5 μM Y-27632 with or without 0.5μM A83–01. We chose this medium as it is conducive for EpiC maintenance and efficient differentiation of CPCs into CMs [45]. Medium was changed every two days for 14 days without Y-27632. For conditioned media studies, CMs were treated every two days with fresh LA medium or 1:1 fresh LA medium to medium that was conditioned by EpiCs for two days. For indirect coculture studies, CPCs were seeded in adjacent chambers to CPCs or EpiCs with an empty cell-free well between them on ibidi chamber slides and treated every two days with LA medium supplemented with Antibiotic-Antimycotic. Then the cells were singularized with ACCUTASE™ for at least 30 minutes and quenched with DMEM/F12. The cells were then counted before being replated for immunostaining in LaSR with 5 μM Y-27632 with or without 0.5μM A83–01 or collected for flow cytometry.
5.5. Flow cytometry
As previously described, cells were singularized with Accutase then fixed with 1% paraformaldehyde for 20 minutes at room temperature and stained with primary antibodies overnight at 4°C in BSA buffer (PBS plus 0.1% Triton X-100 and 0.5% BSA) [2]. The following day, cells were washed and stained with secondary antibodies in BSA buffer at room temperature for one hour. Antibody dilutions and product information are in Data File S3. At least 10,000 events/sample were collected on a BD Accuri C6 flow cytometer and analyzed using FlowJo. Flow cytometry gating was based on a no-primary control and negative cell type control.
5.6. Immunochemistry
As explained previously, cells were fixed with 4% paraformaldehyde for 10 minutes or ice cold methanol for 5 minutes at room temperature and then blocked in milk buffer (PBS plus 0.4% Triton X-100 and 5% non-fat dry milk or BSA buffer (PBS plus 0.1% Triton X-100 and 0.5% BSA) for one hour at room temperature [2]. Then, primary antibodies were added, and samples were incubated overnight at 4°C on a shaker. The following day, cells were washed with PBS and stained with secondary antibodies at room temperature for one hour or overnight at 4°C on a shaker. Antibody dilutions and information are provided Data File S3. Hoechst counterstain was added at 5 μg/mL in PBS for five minutes. For image analysis, an epifluorescence microscope Olympus IX70 or Nikon Ti2 was used or cells were plated on slides and imaged using a Nikon A1R-SI+ or Nikon A1RS HD confocal microscope.
5.7. Automated Sarcomere Analysis
We performed automated sarcomere analysis using SarcTrack2 available at https://github.com/HMS-IDAC/SarcTrack2 on sarcomeres stained for α-actinin [28]. The code was modified to process images instead of real-time videos and the following input parameters were used: ds= 10:0.3:19, stretch=1, scale=1.5, and nangs=8.
5.8. FACS Sorting for mRNA extractions
CPCs were differentiated in the WTC-CAAX-RFP constitutive reporter hiPSC line and EpiCs were differentiated in the WTC-LMNB1-eGFP constitutive reporter hiPSC line. Cells were cocultured for 2 weeks, lifted for flow cytometry with Accutase, and resuspended in PBS (Ca2+/Mg2+ free) supplemented with 0.5% BSA, 5 μM Rock Inhibitor Y-27632 and 5μM DAPI. Dead cells were gated out based on DAPI staining. FACS was performed on a BD FACSAria in a BioBubble. For single cell sequencing, two samples were prepared of live cells from CMs and EpiCs monocultured or cocultured. For further analysis, RFP+ monoculture, GFP+ monoculture, RFP+ coculture, and GFP+ coculture samples were collected from three differentiations with three well replicates per differentiation and stored at −80°C until further extraction. All samples were collected in PBS (Ca2+/Mg2+ free) supplemented with 0.5% BSA, 5 μM Rock Inhibitor Y-27632 and 5 μM DAPI.
5.9. Single Cell Sequencing and Analysis
Two cellular suspension (monoculture and coculture) were loaded into two separate wells (estimated loading of 6,000 cells/well) in a 10X Chromium instrument using the Single Cell 3’ reagent kit according to the manufacturer’s instructions. MiSeq analysis confirmed greater than 90% of droplets contained cells compared to empty droplets, so we continued with paired-end sequencing performed on Illumina NovaSeq6000. Each fraction generated approximately 390 million raw reads which corresponded to approximately 71,000 reads per cell.
Single cell sequencing data was processed using the CellRanger Pipeline (4.0.0-released July 7, 2020). Reads were mapped against the human genome (genome assembly version 2020-A) and annotated with the ENCODE gene annotations for the GRCh38 genome assembly (GENCODE v32). GFP and RFP construct sequences, as provided by the Allen Institute for Cell Science, were added to the genome. Normalization, dimensionality reduction, scaling of data, and clustering of the single cell data were performed using the Seurat package (version 4.0) [46]. Principle component analysis was performed on the 20 most significant components (or 15 for CM and EpiC sub-clustering) as determined by an ElbowPlot showing the standard deviation of principle components. Clusters were identified using the FindNeighbors function (settings: reduction=”pca” dims=1:20 or 1:15) and FindClusters function (settings: resolution=0.5). We used UMAP to visualize the single cell clusters in a 2D space. To identify differentially expressed genes in an individual cluster compared to all other clusters we used the FindAllMarkers function (settings: only.pos=TRUE, min.pct=0.25, logfc.threshold=0.25) in the Seurat package. To compare two clusters or groups directly, we used FindMarkers function and sorted by the largest fold change. Lineage trajectories were determined using Bioconductor (version 3.12) and Slingshot (version 1.8.0) (settings: stretch=0 embeddings= “umap”)[47]. GSEA analysis was performed on preranked list of differentially expressed genes between monocultured CMs and cocultured CMs based on log2(FC)*log10(p) using publicly downloadable software from https://www.gsea-msigdb.org/gsea/index.jsp [48,49].
For comparison to hPSC-CM datasets, we identified CM clusters in of hPSC-CM differentiations at D15 and D30 samples of E-MTAB-6268 by expression of sarcomeric genes. We then performed differential expression comparing D15 and D30 hPSC-CMs as described previously. Differential expression analysis of hPSC-CMs at D12 and D24 from Gerbin et al. was obtained in their supplementary information as the raw single cell sequencing data was not yet publicly available.
To evaluate temporal expression of hPSC-CM maturation genes in human single cell sequencing datasets, we first downloaded publicly available ArrayExpress E-MTAB-6268. Normalization, dimensionality reduction, scaling of data, and clustering of the single cell data were performed using the Seurat package (version 4.0) [46]. Principle component analysis was performed on the 15 most significant components as determined by an ElbowPlot showing the standard deviation of principle components. Clusters were identified using the FindNeighbors function (settings: reduction=”pca” dims=1:15) and FindClusters function (settings: resolution=0.5). We used UMAP to visualize the single cell clusters in a 2D space. We then identified pseudotime trajectory using Slingshot (version 1.8.0) (settings: stretch=0 embeddings= “umap”)[47] and plotted pseudotime versus gene expression.
For cell type comparison with publicly available datasets, we first downloaded developing human heart single cell sequencing datasets EGAS00001003996 and GSE106118 and hPSC-CM differentiation single cell sequencing dataset ArrayExpress E-MTAB-6268. Data was normalized and variable features were identified using the Seurat package (version 4.0). We then integrated the datasets by using the FindIntegrationAnchors() and IntegrateData() functions. Principle component analysis was performed on the 30 most significant components. Clusters were identified using the FindNeighbors function (settings: reduction=”pca” dims=1:30) and FindClusters function (settings: resolution=0.5). We used UMAP to visualize the single cell clusters in a 2D space. To identify differentially expressed genes in an individual cluster compared to all other clusters we used the FindAllMarkers function (settings: only.pos=TRUE, min.pct=0.25, logfc.threshold=0.25) in the Seurat package.
5.10. Bulk Sequencing Analysis
We downloaded publicly available GSE154294 (D15, D21, D35), GSE64189 (D14, D21, D28), and GSE81585 (D14, D30, and D90) processed data (TPM or FPKM) of hPSC-CM differentiation and validated the direction of differential expression of MYH6, MYH7, NEAT1, COL2A1, and VCAN.
5.11. Total mRNA Extractions and qPCR Analysis
Total RNA was isolated using the RNeasy mini kit (Qiagen) and treated with DNase (Qiagen). Extracted mRNA was stored in nuclease-free water at −20°C and 1 μg RNA was reverse transcribed into cDNA using the RT SuperScript III First-Strand kit (Invitrogen). Real-time quantitative PCR in 25 uL reactions using PowerUP Syber Master Mix (Applied Biosystems) on an AriaMx Real-Time PCR System at 60°C (Agilent Technologies). GAPDH was used as housekeepers and analysis was performed using the 2^−ΔΔCt method. Primer sequences are as follows: GAPDH_FW 5’-GAAGGTGAAGGTCGGAGTCAACG, GAPDH_RV 5’-TCCTGGAAGATGGTGATGGGAT, MYH6_FW 5’AGCTCACCTACCAGACAGAGG, MYH6_RV 5’-TTGCTTGGCACCAATGTCAC, MYH7_FW 5’-GAGGAGCAAGCCAACACCAA, and MYH7_RV 5’-CTCATTCAAGCCCTTCGTGC.
5.12. Western Blot Analysis
Cocultures were lysed in RIPA buffer in the presence of Halt Protease Inhibitor Cocktail (ThermoFisher). BCA assay was used to determine protein concentration. Equal amounts of lysates were loaded on 6% tris-glycine gels and transferred to nitrocellulose membranes. Membranes were blocked in tris-buffered saline + 0.1% Tween20 (TBST) + 5% dry milk for 1 hour and incubated with primary antibodies (MHCα and MHCβ) overnight at 4°C on a shaker. The following day, membranes were washed with TBST and incubated with secondary antibodies at 1:5000 in 15 mL antibody solution/blot for 1 hr on a shaker. Blots were washed again. Then, blots were imaged on a LICOR Odyssey and bands were quantified using Image Studio 5.2.
5.13. Statistics
All experiments (aside from sequencing) were conducted using at least three technical replicates (e.g., three 12-wells) from the same differentiation and replicated (independent differentiations) at least three times with one replicate in the 19–9-11 hiPSC line and one replicate in the H9 hESC line. Indirect cocultures were performed using at least three technical replicates from the same differentiation and replicated in one or two differentiations in either the 19–9-11 hiPSC or H9 hESC line. Statistical analysis was performed using JMP PRO 15 software and statistical tests are reported in figure captions.
5.14. Data Availability
Sequencing data is publicly available at GSE168956.
Supplementary Material
Investigate important cardiac crosstalk during development and disease
Coculture of human pluripotent stem cell cardiomyocytes and epicardial cells
Crosstalk causes cocultured cardiomyocytes to be more proliferative
Cocultured epicardial cells undergo lineage bias
Implications in tissue engineering and development of cardiac therapies
6. Acknowledgements
The authors would like to acknowledge Benjamin Gastfriend and Koji Foreman for their assistance with data analysis and figure design. We would like to thank the University of Wisconsin Optical Imaging Core at the Wisconsin Institute for Medical Research and at Biochemistry for providing imaging facilities and services, University of Wisconsin Carbone Cancer Center Flow Cytometry Core for FACS services, and the University of Wisconsin Biotechnology Center DNA Sequencing Facility for providing RNA sequencing facilities and services. This work was funded by NIH grants R01HL148058 and R01EB007534, and NSF grants EEC-1648035 and CBET-1743346. We acknowledge a seed grant from 10X Genomics which defrayed costs of scRNAseq library preparation.
Abbreviations
- ANOVA
analysis of variance
- CPC
cardiac progenitor cell
- CM
cardiomyocyte
- EPDC
epicardial derived cell
- EpiC
epicardial cell
- EMT
epithelial-to-mesenchymal transition
- FACS
fluorescence activated cell sorting
- FB
fibroblast
- hPSC
human pluripotent stem cells
- LAC
cardiac progenitor cells cultured in LaSR + A83–01
- LAEC
cardiac progenitor cells and epicardial cells cultured in LaSR + A83–01 media
- LC
cardiac progenitor cells cultured in LaSR media
- LEC
cardiac progenitor cells and epicardial cells cultured in LaSR media
- MFI
median fluorescence intensity
- SMC
smooth muscle cell
Footnotes
Declaration of Interests
S.P.P. holds patent US9663764B2 and US10131878B2 on the cardiomyocyte and epicardial differentiation protocols.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
9 References
- [1].Burridge PW, Matsa E, Shukla P, Lin ZC, Churko JM, Ebert AD, Lan F, Diecke S, Huber B, Mordwinkin NM, Plews JR, Abilez OJ, Cui B, Gold JD, Wu JC, Chemically defined generation of human cardiomyocytes, Nat. Methods 11 (2014) 855–860. doi: 10.1038/nmeth.2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Lian X, Zhang J, Azarin SM, Zhu K, Hazeltine LB, Bao X, Hsiao C, Kamp TJ, Palecek SP, Directed cardiomyocyte differentiation from human pluripotent stem cells by modulating Wnt/β-catenin signaling under fully defined conditions, Nat. Protoc 8 (2013) 162–175. doi: 10.1038/nprot.2012.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Mummery CL, Perspectives on the Use of Human Induced Pluripotent Stem Cell-Derived Cardiomyocytes in Biomedical Research, Stem Cell Rep. 11 (2018) 1306–1311. doi: 10.1016/j.stemcr.2018.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Jarrell DK, Vanderslice EJ, VeDepo MC, Jacot JG, Engineering Myocardium for Heart Regeneration-Advancements, Considerations, and Future Directions, Front. Cardiovasc. Med 7 (2020) 586261. doi: 10.3389/fcvm.2020.586261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Gao L, Gregorich ZR, Zhu W, Mattapally S, Oduk Y, Lou X, Kannappan R, Borovjagin AV, Walcott GP, Pollard AE, Fast VG, Hu X, Lloyd SG, Ge Y, Zhang J, Large Cardiac Muscle Patches Engineered From Human Induced-Pluripotent Stem Cell-Derived Cardiac Cells Improve Recovery From Myocardial Infarction in Swine, Circulation. 137 (2018) 1712–1730. doi: 10.1161/CIRCULATIONAHA.117.030785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Liu Y-W, Chen B, Yang X, Fugate JA, Kalucki FA, Futakuchi-Tsuchida A, Couture L, Vogel KW, Astley CA, Baldessari A, Ogle J, Don CW, Steinberg ZL, Seslar SP, Tuck SA, Tsuchida H, Naumova AV, Dupras SK, Lyu MS, Lee J, Hailey DW, Reinecke H, Pabon L, Fryer BH, MacLellan WR, Thies RS, Murry CE, Human embryonic stem cell-derived cardiomyocytes restore function in infarcted hearts of non-human primates, Nat. Biotechnol 36 (2018) 597–605. doi: 10.1038/nbt.4162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Xu C, Police S, Rao N, Carpenter MK, Characterization and enrichment of cardiomyocytes derived from human embryonic stem cells, Circ. Res 91 (2002) 501–508. doi: 10.1161/01.res.0000035254.80718.91. [DOI] [PubMed] [Google Scholar]
- [8].Yang X, Pabon L, Murry CE, Engineering adolescence: maturation of human pluripotent stem cell-derived cardiomyocytes, Circ. Res 114 (2014) 511–523. doi: 10.1161/CIRCRESAHA.114.300558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Yu T, Miyagawa S, Miki K, Saito A, Fukushima S, Higuchi T, Kawamura M, Kawamura T, Ito E, Kawaguchi N, Sawa Y, Matsuura N, In vivo differentiation of induced pluripotent stem cell-derived cardiomyocytes, Circ. J. Off. J. Jpn. Circ. Soc 77 (2013) 1297–1306. doi: 10.1253/circj.cj-12-0977. [DOI] [PubMed] [Google Scholar]
- [10].Saggin L, Gorza L, Ausoni S, Schiaffino S, Troponin I switching in the developing heart, J. Biol. Chem 264 (1989) 16299–16302. [PubMed] [Google Scholar]
- [11].Bedada FB, Chan SS-K, Metzger SK, Zhang L, Zhang J, Garry DJ, Kamp TJ, Kyba M, Metzger JM, Acquisition of a quantitative, stoichiometrically conserved ratiometric marker of maturation status in stem cell-derived cardiac myocytes, Stem Cell Rep. 3 (2014) 594–605. doi: 10.1016/j.stemcr.2014.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Feric NT, Radisic M, Maturing human pluripotent stem cell-derived cardiomyocytes in human engineered cardiac tissues, Adv. Drug Deliv. Rev 96 (2016) 110–134. doi: 10.1016/j.addr.2015.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Lundy SD, Zhu W-Z, Regnier M, Laflamme MA, Structural and Functional Maturation of Cardiomyocytes Derived from Human Pluripotent Stem Cells, Stem Cells Dev. 22 (2013) 1991–2002. doi: 10.1089/scd.2012.0490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Dunn KK, Palecek SP, Engineering Scalable Manufacturing of High-Quality Stem Cell-Derived Cardiomyocytes for Cardiac Tissue Repair, Front. Med 5 (2018) 110. doi: 10.3389/fmed.2018.00110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Giacomelli E, Meraviglia V, Campostrini G, Cochrane A, Cao X, van Helden RWJ, Krotenberg Garcia A, Mircea M, Kostidis S, Davis RP, van Meer BJ, Jost CR, Koster AJ, Mei H, Míguez DG, Mulder AA, Ledesma-Terrón M, Pompilio G, Sala L, Salvatori DCF, Slieker RC, Sommariva E, de Vries AAF, Giera M, Semrau S, Tertoolen LGJ, Orlova VV, Bellin M, Mummery CL, Human-iPSC-Derived Cardiac Stromal Cells Enhance Maturation in 3D Cardiac Microtissues and Reveal Non-cardiomyocyte Contributions to Heart Disease, Cell Stem Cell. 26 (2020) 862–879.e11. doi: 10.1016/j.stem.2020.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Branco MA, Cabral JMS, Diogo MM, From Human Pluripotent Stem Cells to 3D Cardiac Microtissues: Progress, Applications and Challenges, Bioengineering. 7 (2020) 92. doi: 10.3390/bioengineering7030092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Ali SR, Ranjbarvaziri S, Talkhabi M, Zhao P, Subat A, Hojjat A, Kamran P, Müller AMS, Volz KS, Tang Z, Red-Horse K, Ardehali R, Developmental heterogeneity of cardiac fibroblasts does not predict pathological proliferation and activation, Circ. Res 115 (2014) 625–635. doi: 10.1161/CIRCRESAHA.115.303794. [DOI] [PubMed] [Google Scholar]
- [18].Lie-Venema H, van den Akker NMS, Bax NAM, Winter EM, Maas S, Kekarainen T, Hoeben RC, deRuiter MC, Poelmann RE, Gittenberger-de Groot AC, Origin, fate, and function of epicardium-derived cells (EPDCs) in normal and abnormal cardiac development, ScientificWorldJournal. 7 (2007) 1777–1798. doi: 10.1100/tsw.2007.294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Cao J, Poss KD, The epicardium as a hub for heart regeneration, Nat. Rev. Cardiol 15 (2018) 631–647. doi: 10.1038/s41569-018-0046-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Weeke-Klimp A, Bax NAM, Bellu AR, Winter EM, Vrolijk J, Plantinga J, Maas S, Brinker M, Mahtab EAF, Gittenberger-de Groot AC, van Luyn MJA, Harmsen MC, Lie-Venema H, Epicardium-derived cells enhance proliferation, cellular maturation and alignment of cardiomyocytes, J. Mol. Cell. Cardiol 49 (2010) 606–616. doi: 10.1016/j.yjmcc.2010.07.007. [DOI] [PubMed] [Google Scholar]
- [21].Zhou B, Honor LB, He H, Ma Q, Oh J-H, Butterfield C, Lin R-Z, Melero-Martin JM, Dolmatova E, Duffy HS, von Gise A, Zhou P, Hu YW, Wang G, Zhang B, Wang L, Hall JL, Moses MA, McGowan FX, Pu WT, Adult mouse epicardium modulates myocardial injury by secreting paracrine factors, J. Clin. Invest 121 (2011) 1894–1904. doi: 10.1172/JCI45529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Wei K, Serpooshan V, Hurtado C, Diez-Cuñado M, Zhao M, Maruyama S, Zhu W, Fajardo G, Noseda M, Nakamura K, Tian X, Liu Q, Wang A, Matsuura Y, Bushway P, Cai W, Savchenko A, Mahmoudi M, Schneider MD, van den Hoff MJB, Butte MJ, Yang PC, Walsh K, Zhou B, Bernstein D, Mercola M, Ruiz-Lozano P, Epicardial FSTL1 reconstitution regenerates the adult mammalian heart, Nature. 525 (2015) 479–485. doi: 10.1038/nature15372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Bargehr J, Ong LP, Colzani M, Davaapil H, Hofsteen P, Bhandari S, Gambardella L, Le Novère N, Iyer D, Sampaziotis F, Weinberger F, Bertero A, Leonard A, Bernard WG, Martinson A, Figg N, Regnier M, Bennett MR, Murry CE, Sinha S, Epicardial cells derived from human embryonic stem cells augment cardiomyocyte-driven heart regeneration, Nat. Biotechnol 37 (2019) 895–906. doi: 10.1038/s41587-019-0197-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Niderla-BieliŃska J, Jankowska-Steifer E, Flaht-Zabost A, Gula G, Czarnowska E, Ratajska A, Proepicardium: Current Understanding of its Structure, Induction, and Fate, Anat. Rec. Hoboken NJ 2007. (2018). doi: 10.1002/ar.24028. [DOI] [PubMed] [Google Scholar]
- [25].Ronaldson-Bouchard K, Ma SP, Yeager K, Chen T, Song L, Sirabella D, Morikawa K, Teles D, Yazawa M, Vunjak-Novakovic G, Advanced maturation of human cardiac tissue grown from pluripotent stem cells, Nature. 556 (2018) 239–243. doi: 10.1038/s41586-018-0016-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Dunn KK, Reichardt IM, Simmons AD, Jin G, Floy ME, Hoon KM, Palecek SP, Coculture of Endothelial Cells with Human Pluripotent Stem Cell-Derived Cardiac Progenitors Reveals a Differentiation Stage-Specific Enhancement of Cardiomyocyte Maturation, Biotechnol. J (2019) e1800725. doi: 10.1002/biot.201800725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Friedman CE, Nguyen Q, Lukowski SW, Helfer A, Chiu HS, Miklas J, Levy S, Suo S, Han J-DJ, Osteil P, Peng G, Jing N, Baillie GJ, Senabouth A, Christ AN, Bruxner TJ, Murry CE, Wong ES, Ding J, Wang Y, Hudson J, Ruohola-Baker H, Bar-Joseph Z, Tam PPL, Powell JE, Palpant NJ, Single-Cell Transcriptomic Analysis of Cardiac Differentiation from Human PSCs Reveals HOPX-Dependent Cardiomyocyte Maturation, Cell Stem Cell. 23 (2018) 586–598.e8. doi: 10.1016/j.stem.2018.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Toepfer CN, Sharma A, Cicconet M, Garfinkel AC, Mücke M, Neyazi M, Willcox JAL, Agarwal R, Schmid M, Rao J, Ewoldt J, Pourquié O, Chopra A, Chen CS, Seidman JG, Seidman CE, SarcTrack, Circ. Res 124 (2019) 1172–1183. doi: 10.1161/CIRCRESAHA.118.314505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Quijada P, Trembley MA, Misra A, Myers JA, Baker CD, Pérez-Hernández M, Myers JR, Dirkx RA, Cohen ED, Delmar M, Ashton JM, Small EM, Coordination of endothelial cell positioning and fate specification by the epicardium, Nat. Commun 12 (2021) 4155. doi: 10.1038/s41467-021-24414-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Churko JM, Garg P, Treutlein B, Venkatasubramanian M, Wu H, Lee J, Wessells QN, Chen S-Y, Chen W-Y, Chetal K, Mantalas G, Neff N, Jabart E, Sharma A, Nolan GP, Salomonis N, Wu JC, Defining human cardiac transcription factor hierarchies using integrated single-cell heterogeneity analysis, Nat. Commun 9 (2018). doi: 10.1038/s41467-018-07333-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Piccini I, Rao J, Seebohm G, Greber B, Human pluripotent stem cell-derived cardiomyocytes: Genome-wide expression profiling of long-term in vitro maturation in comparison to human heart tissue, Genomics Data. 4 (2015) 69–72. doi: 10.1016/j.gdata.2015.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Zhang F, Qiu H, Dong X, Wang C, Na J, Zhou J, Wang C, Transferrin improved the generation of cardiomyocyte from human pluripotent stem cells for myocardial infarction repair, J. Mol. Histol 52 (2021) 87–99. doi: 10.1007/s10735-020-09926-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Cui Y, Zheng Y, Liu X, Yan L, Fan X, Yong J, Hu Y, Dong J, Li Q, Wu X, Gao S, Li J, Wen L, Qiao J, Tang F, Single-Cell Transcriptome Analysis Maps the Developmental Track of the Human Heart, Cell Rep. 26 (2019) 1934–1950.e5. doi: 10.1016/j.celrep.2019.01.079. [DOI] [PubMed] [Google Scholar]
- [34].Gidlöf O, Bader K, Celik S, Grossi M, Nakagawa S, Hirose T, Metzler B, Olde B, Erlinge D, Inhibition of the long non-coding RNA NEAT1 protects cardiomyocytes from hypoxia in vitro via decreased pri-miRNA processing, Cell Death Dis. 11 (2020) 1–15. doi: 10.1038/s41419-020-02854-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Happe CL, Engler AJ, Mechanical Forces Reshape Differentiation Cues That Guide Cardiomyogenesis, Circ. Res. 118 (2016) 296–310. doi: 10.1161/CIRCRESAHA.115.305139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Hannan RD, Luyken J, Rothblum LI, Regulation of ribosomal DNA transcription during contraction-induced hypertrophy of neonatal cardiomyocytes, J. Biol. Chem 271 (1996) 3213–3220. doi: 10.1074/jbc.271.6.3213. [DOI] [PubMed] [Google Scholar]
- [37].Gerbin K et al. , Cell states beyond transcriptomics: integrating structural organization and gene expression in hiPSC-derived cardiomyocytes, bioRxiv. (2020). [DOI] [PubMed] [Google Scholar]
- [38].Pei F, Jiang J, Bai S, Cao H, Tian L, Zhao Y, Yang C, Dong H, Ma Y, Chemical-defined and albumin-free generation of human atrial and ventricular myocytes from human pluripotent stem cells, Stem Cell Res. 19 (2017) 94–103. doi: 10.1016/j.scr.2017.01.006. [DOI] [PubMed] [Google Scholar]
- [39].Veevers J, Farah EN, Corselli M, Witty AD, Palomares K, Vidal JG, Emre N, Carson CT, Ouyang K, Liu C, van Vliet P, Zhu M, Hegarty JM, Deacon DC, Grinstein JD, Dirschinger RJ, Frazer KA, Adler ED, Knowlton KU, Chi NC, Martin JC, Chen J, Evans SM, Cell-Surface Marker Signature for Enrichment of Ventricular Cardiomyocytes Derived from Human Embryonic Stem Cells, Stem Cell Rep. 11 (2018) 828–841. doi: 10.1016/j.stemcr.2018.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Titmarsh DM, Glass NR, Mills RJ, Hidalgo A, Wolvetang EJ, Porrello ER, Hudson JE, Cooper-White JJ, Induction of Human iPSC-Derived Cardiomyocyte Proliferation Revealed by Combinatorial Screening in High Density Microbioreactor Arrays, Sci. Rep 6 (2016) 24637. doi: 10.1038/srep24637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Funakoshi S, Fernandes I, Mastikhina O, Wilkinson D, Tran T, Dhahri W, Mazine A, Yang D, Burnett B, Lee J, Protze S, Bader GD, Nunes SS, Laflamme M, Keller G, Generation of mature compact ventricular cardiomyocytes from human pluripotent stem cells, Nat. Commun 12 (2021) 3155. doi: 10.1038/s41467-021-23329-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Lee Y-K, Ng K-M, Chan Y-C, Lai W-H, Au K-W, Ho C-YJ, Wong L-Y, Lau C-P, Tse H-F, Siu C-W, Triiodothyronine Promotes Cardiac Differentiation and Maturation of Embryonic Stem Cells via the Classical Genomic Pathway, Mol. Endocrinol 24 (2010) 1728–1736. doi: 10.1210/me.2010-0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Lewis-Israeli YR, Wasserman AH, Gabalski MA, Volmert BD, Ming Y, Ball KA, Yang W, Zou J, Ni G, Pajares N, Chatzistavrou X, Li W, Zhou C, Aguirre A, Self-assembling human heart organoids for the modeling of cardiac development and congenital heart disease, Nat. Commun 12 (2021) 5142. doi: 10.1038/s41467-021-25329-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Bao X, Lian X, Qian T, Bhute VJ, Han T, Palecek SP, Directed differentiation and long-term maintenance of epicardial cells derived from human pluripotent stem cells under fully defined conditions, Nat. Protoc 12 (2017) 1890–1900. doi: 10.1038/nprot.2017.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Bao X, Lian X, Hacker TA, Schmuck EG, Qian T, Bhute VJ, Han T, Shi M, Drowley L, Plowright A, Wang Q-D, Goumans M-J, Palecek SP, Long-term self-renewing human epicardial cells generated from pluripotent stem cells under defined xeno-free conditions, Nat. Biomed. Eng 1 (2016). doi: 10.1038/s41551-016-0003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM, Hao Y, Stoeckius M, Smibert P, Satija R, Comprehensive Integration of Single-Cell Data, Cell. 177 (2019) 1888–1902.e21. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Street K, Risso D, Fletcher RB, Das D, Ngai J, Yosef N, Purdom E, Dudoit S, Slingshot: cell lineage and pseudotime inference for single-cell transcriptomics, BMC Genomics. 19 (2018) 477. doi: 10.1186/s12864-018-4772-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Mootha VK, Lindgren CM, Eriksson K-F, Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E, Ridderstråle M, Laurila E, Houstis N, Daly MJ, Patterson N, Mesirov JP, Golub TR, Tamayo P, Spiegelman B, Lander ES, Hirschhorn JN, Altshuler D, Groop LC, PGC-1α-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes, Nat. Genet 34 (2003) 267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- [49].Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP, Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles, Proc. Natl. Acad. Sci. U. S. A 102 (2005) 15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequencing data is publicly available at GSE168956.