Abstract
Background:
The phase III S0819 trial investigated addition of cetuximab to first-line chemotherapy (CT) in NSCLC. Subgroup analyses suggested an OS benefit among patients with EGFR copy number gain in squamous cell carcinomas (SCC), (HR=0.58 [0.39–0.86], p=0.0071). A more detailed model based on EGFR FISH, EGFR IHC and KRAS mutation status was evaluated to yield a more precise predictive paradigm of cetuximab-based therapy in advanced NSCLC.
Methods:
FISH was performed using the Colorado Scoring Criteria; H-Score was used to quantify EGFR IHC expression (cut-off ≥ 200). A Cox model was used to assess treatment effects for OS and PFS within biomarker and clinical subgroups. KRAS mutation was analyzed using Therascreen. The false discovery rate controlled for multiple comparisons. S0819 ClinicalTrials.gov Identifier: NCT00946712.
Results:
Of 1,313 eligible patients, assay results were obtained for FISH on 976 patients (41% positive), for IHC on 945 patients (31% positive), and KRAS mutation status on 627 patients (26% positive). In SCC patients, OS was significantly improved with addition of cetuximab when both EGFR FISH and EGFR IHC were positive (N= 58), (OS HR: 0.32 [95% CI 0.18–0.59]; p=0.0002, q=0.08), median 12.6 vs 4.6 months. The results were independent of KRAS mutation status. In Non-SCC, no predictive value of EGFR IHC, EGFR FISH status and/or KRAS status was seen.
Conclusions:
In NSCLC SCC, a combination index of EGFR FISH plus EGFR IHC results was associated with improved OS when cetuximab was added to CT, representing a potential predictive molecular paradigm for patients suitable for EGFR-antibody therapy.
Keywords: NSCLC, EGFR antibodies, EGFR FISH, EGFR IHC, KRAS, Survival Analysis
MicroAbstract
A molecular model was investigated to predict outcome of patients enrolled on the S0819 clinical trial which investigated the addition of cetuximab to first-line chemotherapy (with or without bevacizumab) in 1,313 NSCLC patients. In squamous cell histology with evaluable markers (n = 321), increased EGFR copy number by FISH plus elevated EGFR protein expression by IHC was associated with significantly improved OS when cetuximab was added to CT.
Introduction
The SWOG S0819 trial (1) was an open-label, phase III study, designed to evaluate the efficacy of cetuximab, a highly specific chimeric monoclonal IgG1 antibody targeting EGFR, in addition to carboplatin-paclitaxel or carboplatin-paclitaxel-bevacizumab as first-line treatment for advanced NSCLC. The design of this study has been previously described (2) and the primary results have been published (1). Prior to S0819, cetuximab had been investigated in combination with platinum-based chemotherapies for the treatment of CT-naïve patients with advanced NSCLC in unselected patient populations and among patients with higher expression levels by immunohistochemistry (3–8). Accordingly, the design of S0819 included an evaluation of EGFR by fluorescence in situ hybridization (FISH) as a potential predictive biomarker for cetuximab in this population. To investigate this directly, S0819 included two co-primary objectives: a comparison of OS in the entire study population and PFS in patients with an increased gene copy number of EGFR, assessed by FISH. While there were no significant differences in the primary endpoints, secondary analyses suggested that in the sub-population of SCC patients with EGFR FISH-positive (FISH+), OS was significantly improved in the cetuximab-containing arm compared with the control arm, with an HR of 0.58 (95% CI 0.39–0.86; p = 0.007), median OS of 6.1 months (95% CI 4.2–8.7) and 11.8 months (95% CI 8.6–13.5) in the control and cetuximab-containing arms, respectively. These data suggested that further biomarker analysis was warranted to determine which patients derive benefit from the addition of cetuximab to platinum chemotherapy.
The S0819 study also included a prospective evaluation of KRAS mutation status, a factor highly relevant to the success of EGFR-directed antibodies in colorectal cancer(9). KRAS mutations are rare in lung SCC histology, but frequent in lung non-SCC (>25%) (10). Evaluation of KRAS status was incorporated into S0819 to determine whether it played a similar predictive role in lung non-SCC cases.
Here, we evaluated the impact of EGFR protein expression and copy number, along with KRAS mutation status, on patient outcome in the S0819 clinical trial, the largest randomized study of cetuximab plus chemotherapy conducted to date.
Methods
Study design
The S0819 study design, characteristics, arm treatments, and eligibility criteria have been previously reported (1–2). This was a randomized, multicenter, phase III trial of carboplatin + paclitaxel or carboplatin + paclitaxel + bevacizumab with or without concurrent cetuximab in patients with histologically or cytologically proven stage IV primary NSCLC that was newly diagnosed or recurrent after previous surgery and/or radiation. A co-primary objective of the study was to evaluate clinical outcomes within patients with EGFR FISH-positive (FISH+) NSCLC. Secondary objectives included an evaluation of outcomes by EGFR by IHC. An additional objective was to evaluate the role of KRAS; specifically to evaluate if there was evidence that KRAS mutations were associated with reduced efficacy of cetuximab treatment, as has been seen in colorectal cancer (11–12). Written informed consent for the use of tumor tissue for molecular analysis was obtained from each patient before enrollment into the study, as previously reported (1,).
Procedures
Formalin-fixed, paraffin-embedded (FFPE) tumor specimens or needle aspirates on charged glass slides were required to be submitted prior to the start of therapy. As assessment of PFS in the FISH+ group was a co-primary objective, FISH testing was given priority. If adequate tissue remained, IHC testing was then performed, and subsequently any remaining tissue was evaluated for KRAS mutation status.
EGFR FISH analysis was performed at the University of Colorado using the “Colorado EGFR Scoring System” (13). Tumors were considered to be EGFR FISH+ if they harbored 4 or more copies of EGFR in ≥ 40% of cells, or if they showed EGFR amplification (defined as gene-to-chromosome ratio ≥ 2 or presence of gene cluster or ≥ 15 gene copies in ≥ 10% of cells), all other tumors were classified as EGFR FISH-negative (FISH-).
IHC was also performed at the University of Colorado with the Dako EGFR pharmDX kit (Carpinteria, CA). Staining was performed according to the protocol and membrane staining was scored in four different categories, including no staining (0), weak staining (1+, light brown membrane staining, visible only with high magnification), intermediate staining (2+, between 1+ and 3+) and strong staining (3+, dark-brown linear membrane staining, visible just with low magnification). The H-Score system was used to generate a semi-quantitative score, ranging from 0 to 300 and was calculated with the following formula: 1 x (percentage of 1+ cells) + 2 x (percentage of 2+ cells) + 3 x (percentage of 3+ cells); the percentage of tumor cells showing the different staining intensities were assessed visually by the pathologist. A cut-off of ≥ 200 was considered as positive based on the above H-Score calculations.
KRAS mutation status was determined using the Therascreen KRAS test (QIAGEN Sciences, MD. USA.), conducted in a CLIA-certified diagnostic laboratory at the UC Davis Comprehensive Cancer Center.
Statistical analysis
Baseline characteristics were compared between patients with tumors that were FISH+, H-Score ≥ 200, KRAS mutant, using an analysis of variance F-Score for age, and a Cochrane-Mantel-Haenszel test of general association for categorical variables. To evaluate potential subgroup treatment effects, a stratified Cox proportional-hazards model was used for both overall survival and progression-free survival, with the randomized group as a covariate and including only patients within the subgroup. Forest plots were constructed to visually display hazard ratios and associated 95% confidence intervals; the column headings describe the major subgroup evaluated and row labels within these panels describe the combination subgroup evaluated (e.g. within the IHC+ panel, the F+ row denotes the subgroup with both FISH+ and IHC+ disease). To account for multiple comparisons, we used a false discovery rate (FDR) as measured by the q-value with significance set at 10% (14). In addition, SAS (version 9.4) and R (version 3.6.1) were used for all statistical analyses.
Results
From August 13, 2009 to May 30, 2014, 1,333 patients were enrolled in the study; 20 were deemed ineligible, resulting in 1,313 patients in the intention-to-treat (ITT) population: CT plus cetuximab arm (N = 656) and CT without cetuximab arm (N = 657) (Figure 1). A description of the full patient population and the population characteristics within the biomarker-defined subgroups is presented in Table 1. Of the 1,313 eligible patients, EGFR FISH was available for 976 patients (74%), EGFR IHC was available for 945 (72%) and KRAS molecular assessment was available for 627 (48%). Availability of biomarker data did not vary by patient characteristics. Of the patients with available biomarker data, 400 of 976 (41%) were EGFR FISH positive (355 (88.8%) with high polysomy and 45 (11.2%) with gene amplification), 295 of 945 (31%) had an H-score ≥200 (IHC+), and 166 of 627 (26%) had KRAS mutations detected. The histology distribution among the patient subgroups are shown in Table 1. Consistent with the FLEX trial[15], patients with EGFR IHC+ disease relative to all NSCLC (and compared to FISH+/KRAS MT) were more likely to have SCC versus adenocarcinoma (ADC) (45% ADC in IHC+ versus 63% ADC in FISH+ and 81% ADC in KRAS MT).
Figure 1:
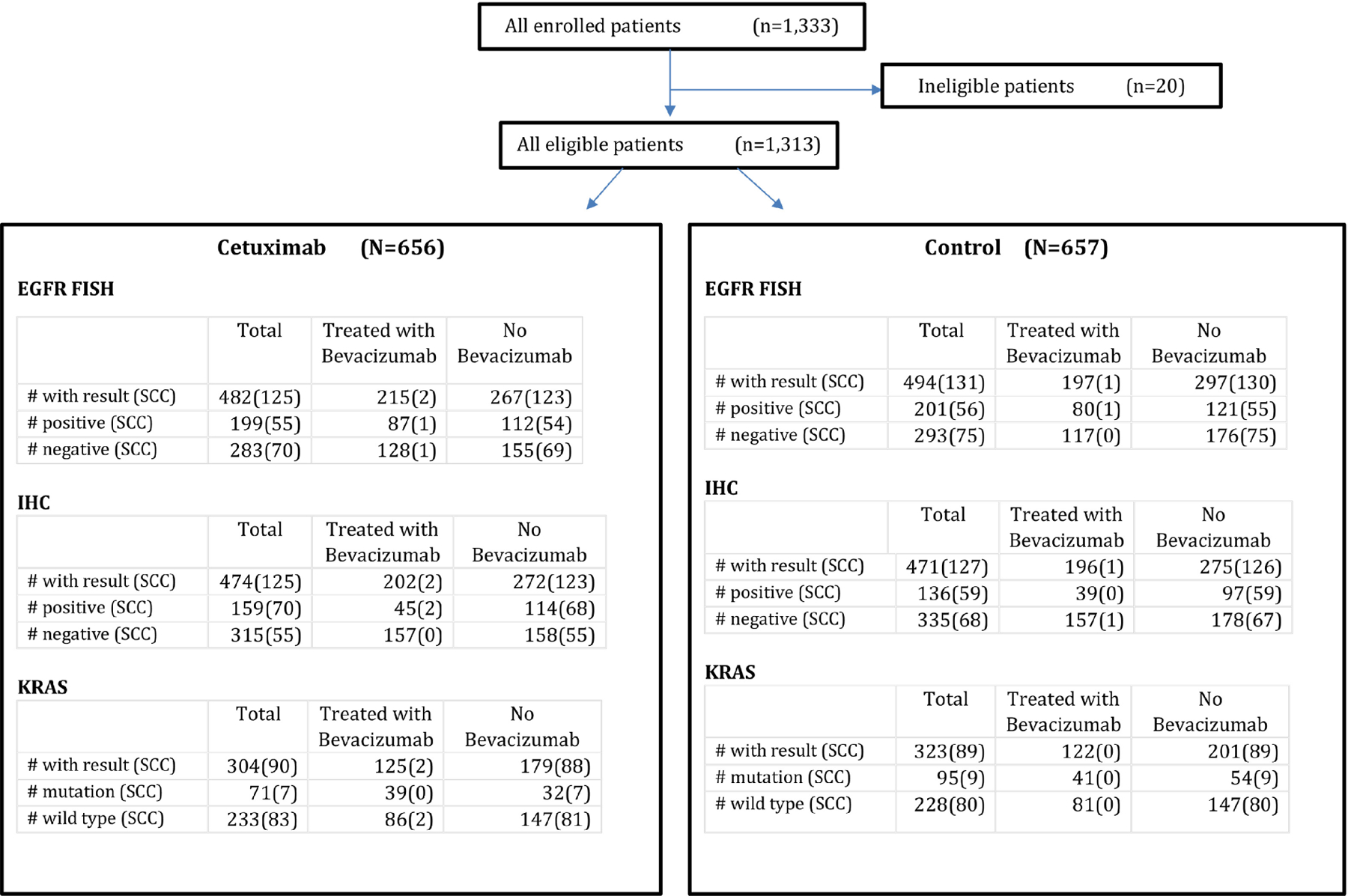
CONSORT S0819 trial profile. Eligible patients are dichotomized by treatment arm and further subdivided by assay conducted. Parentheses indicate the subset that is SCC within each group. (n: number of patients; EGFR: epidermal growth factor receptor; FISH: fluorescence in situ hybridization; H-Score: immunohistochemistry score; SCC: squamous cell carcinoma)
Table 1.
Baseline characteristics and demographics by biomarker (n: number of patients; FISH: fluorescence in situ hybridization; H-Score: immunohistochemistry score)
| All patients (N = 1,313) |
FISH+ (N = 400) |
H-score ≥ 200 (N = 295) |
KRAS mutant (N = 166) |
p-value | |
|---|---|---|---|---|---|
| Age median (range) | 63 (20–86) | 63 (35–84) | 63 (20–83) | 62 (36–82) | 0.93 |
|
| |||||
| Sex | |||||
| • Female | 569 (43%) | 160 (40%) | 116 (39%) | 84 (51%) | |
| • Male | 744 (57%) | 240 (60%) | 179 (61%) | 82 (49%) | 0.04 |
|
| |||||
| Race | |||||
| • Caucasian | 1,133 (86%) | 341 (85 %) | 254 (86%) | 147 (89%) | |
| • African-American | 116 (9%) | 36 (9%) | 29 (10%) | 12 (7%) | |
| • Other/Unknown | 64 (5%) | 23 (6%) | 12 (4%) | 7 (4%) | 0.58 |
|
| |||||
| Treated with Bevacizumab | |||||
| • Yes | 560 (43%) | 167 (42%) | 84 (28%) | 80 (48%) | |
| • No | 753 (57 %) | 233 (58 %) | 211 (72%) | 86 (52%) | 0.00003 |
|
| |||||
| Smoking status | |||||
| • Current/former smoker | 1,197 (91%) | 364 (91%) | 267 (91%) | 161 (97%) | |
| • Never smoker | 36 (9%) | 28 (9%) | 5 (3%) | 0.02 | |
|
| |||||
| Stage | |||||
| • M1a | 298 (23%) | 97 (24%) | 85 (29%) | 41 (25%) | |
| • M1b | 1,015 (77%) | 303 (76%) | 210 (71%) | 125 (75%) | 0.37 |
|
| |||||
| Histology | |||||
| • Adenocarcinoma | 819 (62%) | 250 (63%) | 133 (45%) | 135 (81%) | |
| • Squamous cell | 321 (24%) | 111 (28%) | 129 (44%) | 16 (10%) | |
| • Other | 172 (13%) | 39 (10%) | 33 (11%) | 15 (9%) | |
| • Not reported | 1 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | 8 x 10−14 |
|
| |||||
| Performance status | |||||
| • 0 | 485 (37%) | 145 (36%) | 108 ((37%) | 73 (44%) | |
| • 1 | 827 (63%) | 255 (64%) | 187 (63%) | 93 (56%) | |
| • Not reported | 1 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | 0.19 |
|
| |||||
| FISH | |||||
| Has result | 976 (74%) | 271 (92%) | 158 (95%) | ||
| Positive | 400 (41%) | 142 (52%) | 59 (37%) | ||
|
| |||||
| H-score | |||||
| Has result | 945 (72%) | 360 (90%) | 154 (93%) | ||
| >200 | 295 (31%) | 142 (39%) | 34 (22%) | ||
|
| |||||
| KRAS | |||||
| Has result | 627 (48%) | 243 (61%) | 200 (68%) | ||
| Mutation positive | 166 (26%) | 59 (24%) | 34 (17%) | ||
Among those with known KRAS status, KRAS mutations were observed in 9% SCC, 36% with ADC, and 21% in other histologies. As is typical in NSCLC, the majority of cases were smoking-associated G12C (53%), with G12V at 17% and G12D at 16%.
Forest plots for OS and PFS hazard ratios are provided in Figure 2 and Supplementary Figure S1, respectively for subgroups defined by biomarker, histology, and treatment including bevacizumab. Supplementary Tables S1 and S2 include the sample size, number of events, median times, and summary statistics for the comparisons presented in the forest plots for OS and PFS, respectively. Details of the subgroup analyses follow.
Figure 2.
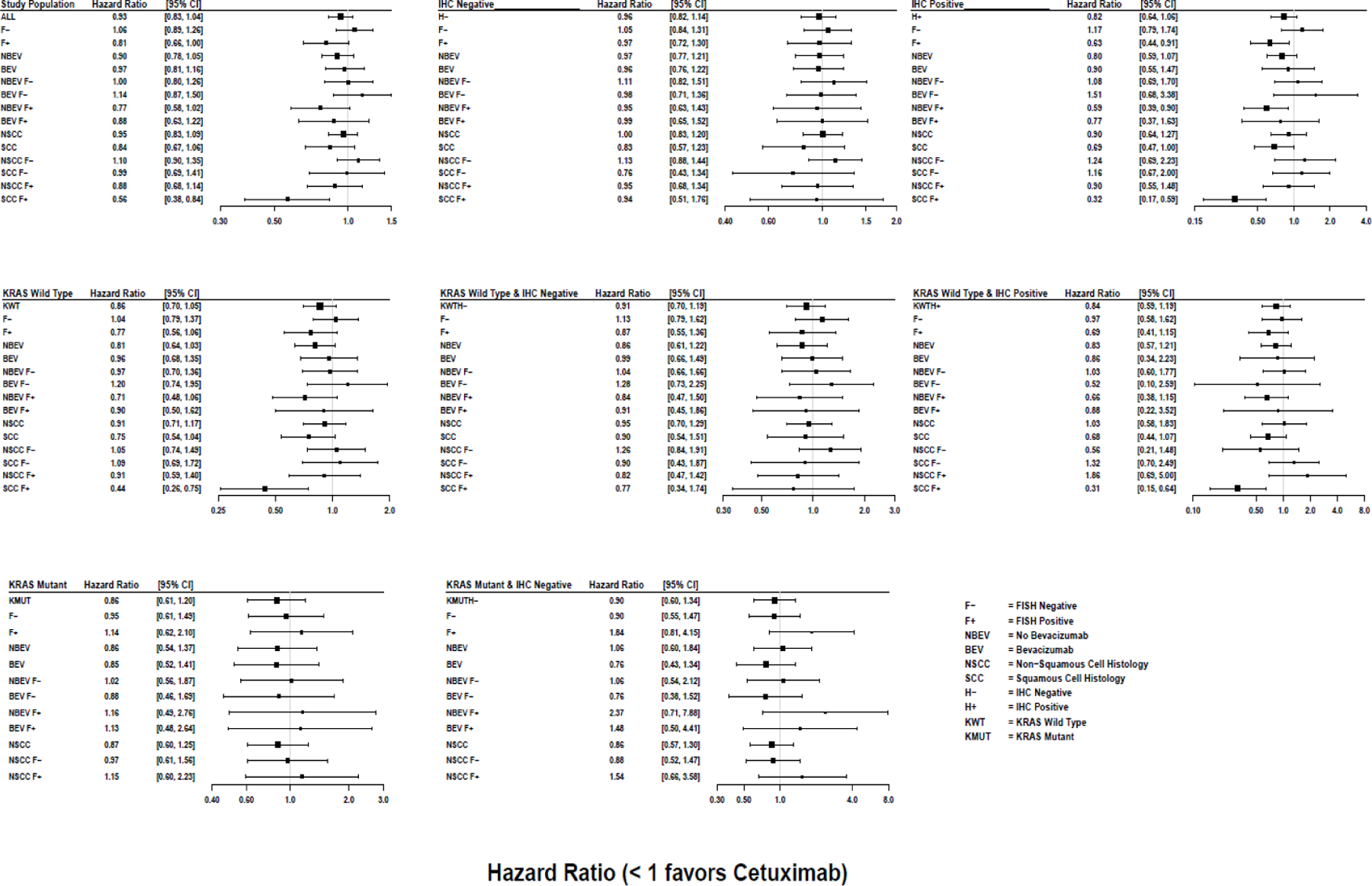
Forest plot of OS hazard ratios along with their 95% confidence limits, comparing the Cetuximab arm to the control arm in various sub-populations
Treatment Effects for patients with IHC+ disease
Evaluation of IHC+ disease on its own did not identify a subgroup that benefited from the addition of cetuximab (OS HR (95% CI) 0.82 (0.64 – 1.06), p = 0.13, q = 0.98)). However, for those with dual positive IHC+ and FISH+ disease, the HR for OS was 0.63 (95% CI 0.44–0.91, p = 0.01, q =0.49). Among those who did not receive bevacizumab, the HR was 0.59 (95% CI 0.39–0.90, p = 0.02, q = 0.49). For either bevacizumab group, this did not meet the multiplicity-adjusted threshold for significance.
Treatment Effects for patients with IHC+ plus FISH+ combination index
Treatment effects for the subgroup of patients with a positive combination index are summarized in Figure 2. When considering all histologic subtypes, treatment outcomes were not significantly different by the combination index. However, by histology, OS was significantly improved with cetuximab among patients with SCC histology and a positive combination index (FISH+/IHC+); HR OS = 0.32 (95% CI 0.18–0.59, p = 0.0002, q = 0.08); this is the only comparison that met the multiplicity-adjusted threshold for statistical significance (q-values < 0.10). The median OS was 12.6 months (95% CI 7.9–15.9) for the cetuximab arm and 4.6 months (95% CI 3.4–7.3) for the control arm.
Figure 3 displays the Kaplan-Meier curves for OS among patients with SCC histology by the combination index status. Figure 4 displays the data among non-SCC histology.
Figure 3: Comparison of Overall Survival by Treatment Arm among Patients with Squamous Cell Lung Cancer.
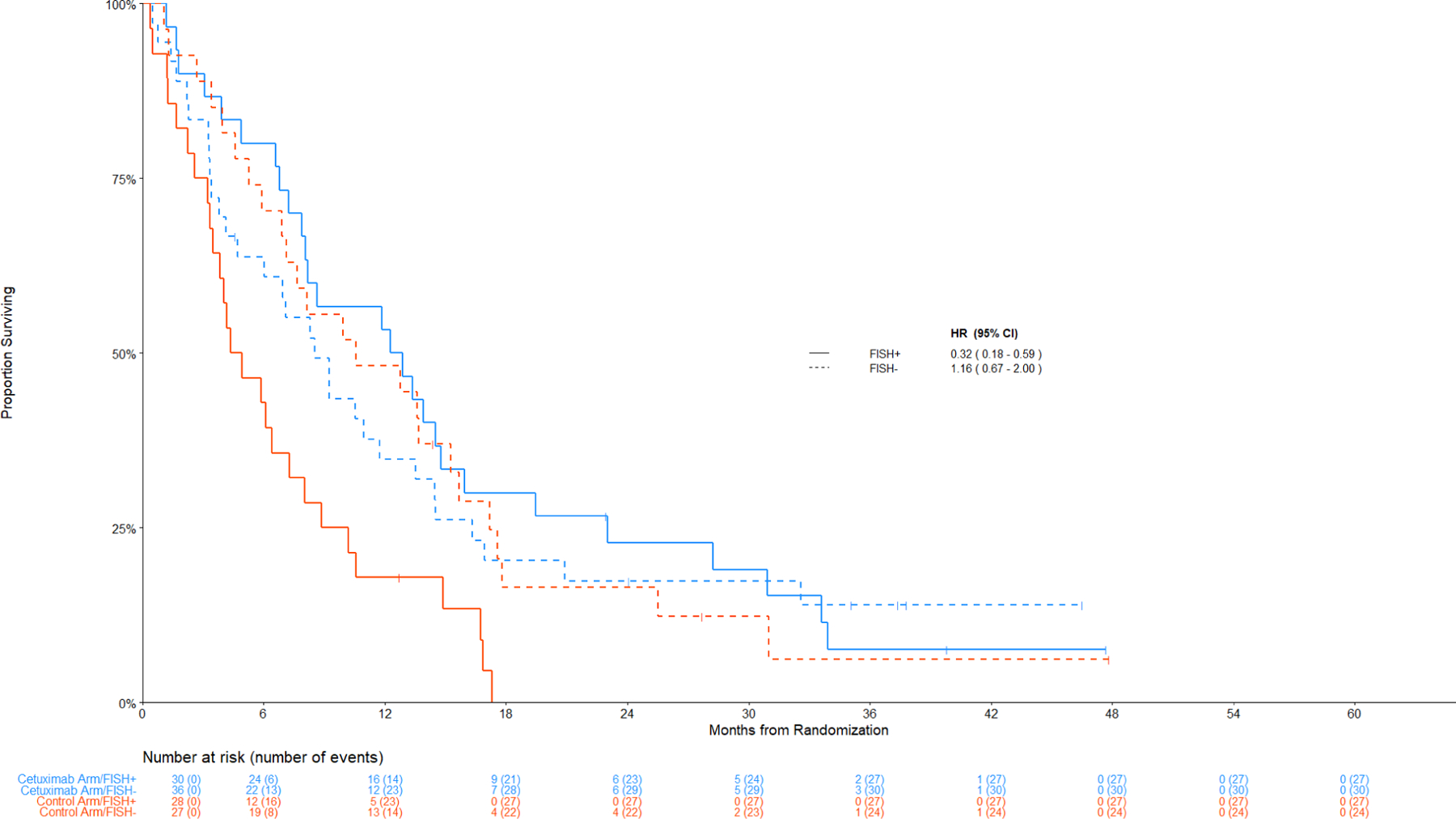
Panel A: Among IHC high, FISH+ v FISH –
Figure 4: Comparison of Overall Survival By Treatment Arm among Patients with Non-Squamous Cell Lung Cancer.
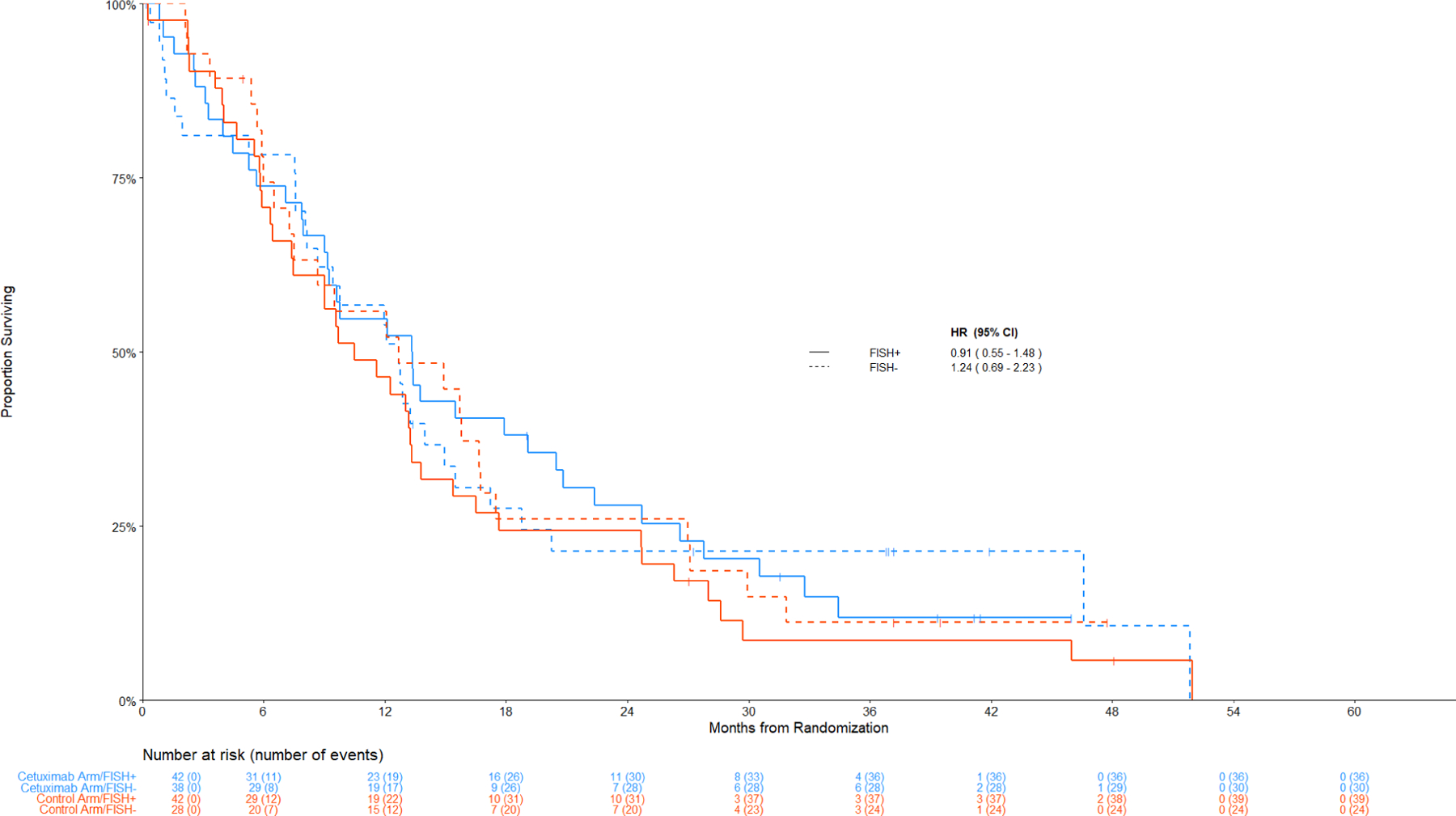
Panel A: Among IHC high, FISH+ v FISH –
The hazard ratios for PFS tracked similarly in the subset with a positive combination index (HR PFS = 0.49 [95% CI 0.28 – 0.88], p = 0.02, q=0.49); however, this result did not meet the multiplicity threshold. Kaplan-Meier curves for PFS by IHC/FISH and histology are shown in Figure S2 and S3). In contrast to SCC, no significant differences between treatment arms were observed in nonSCC (Figure 2 and Figure 4).
Treatment Effects for patients with an H-score <= 200 (IHC-)
OS, PFS, and response did not differ between the treatment arms in any subgroup of patients with IHC-low/negative disease (p ≥ 0.20 and q ≥ 0.99 for all comparisons) (Figures 3B and 4B).
Figure 3:
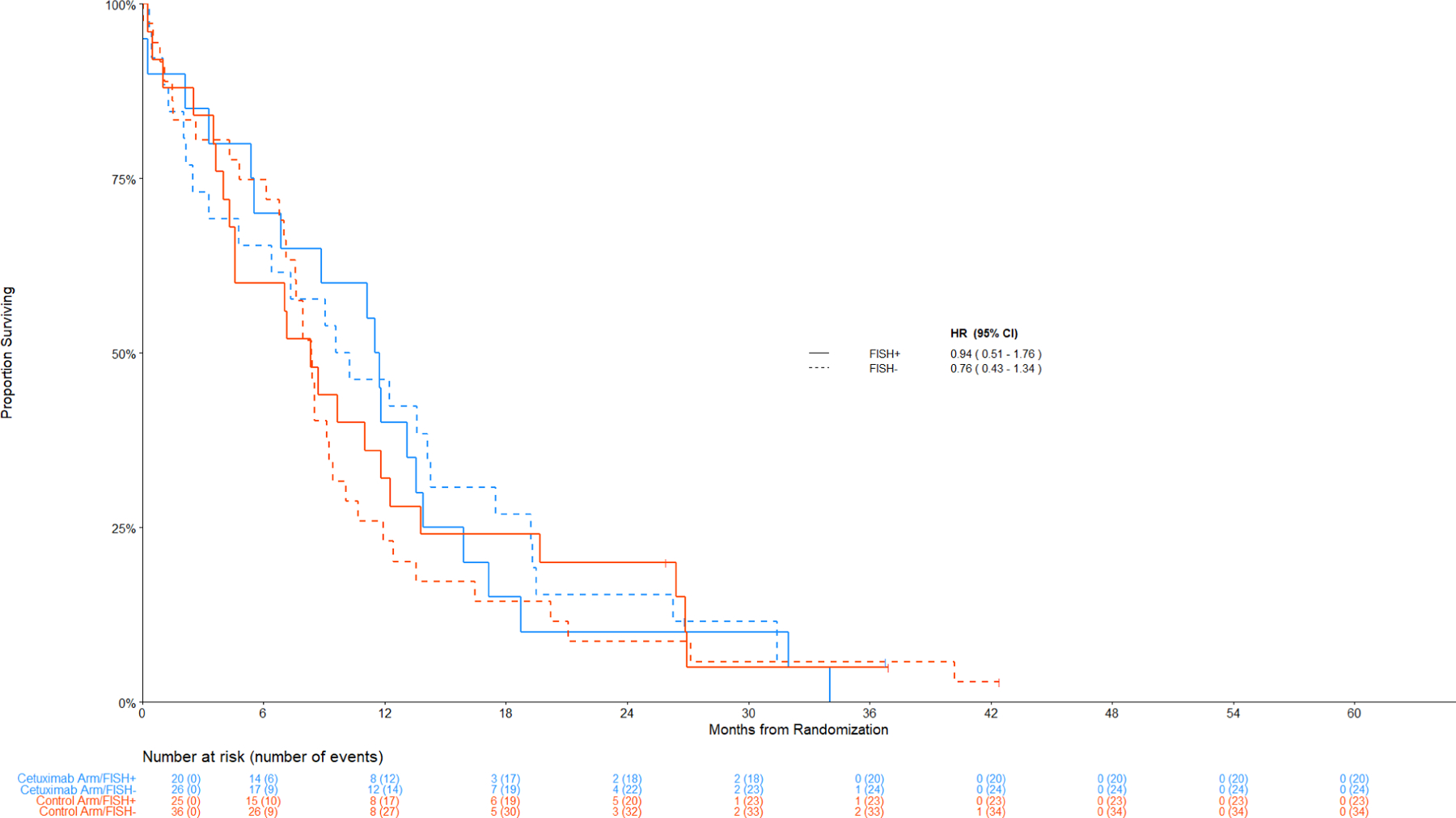
Panel B: Among IHC low/negative, FISH+ v FISH -
Figure 4:
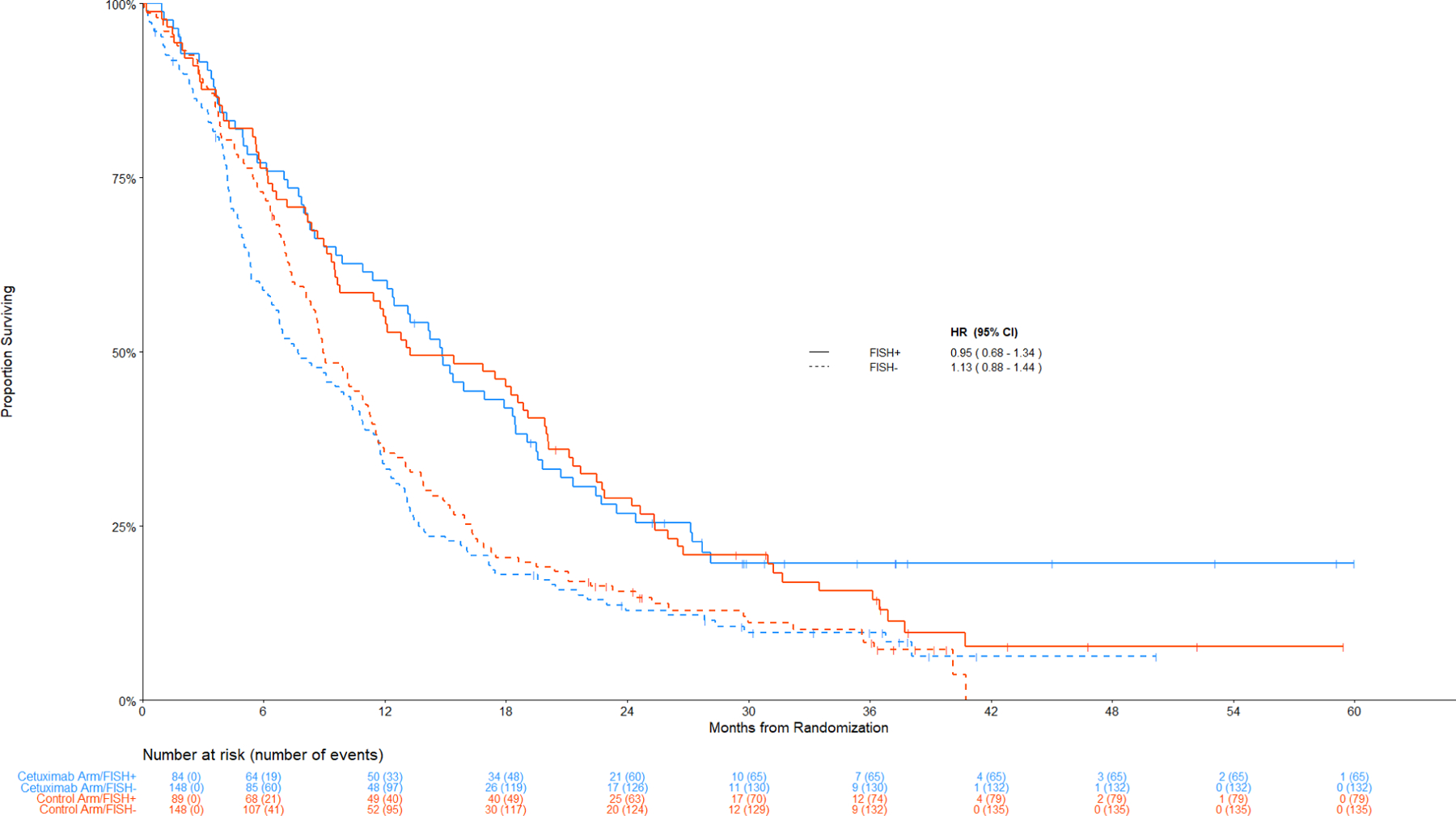
Panel B: Among IHC low/negative, FISH+ v FISH -
Treatment effects by KRAS mutation status
KRAS mutation status was not associated with a treatment benefit. The HR (95% CI) for OS was 0.86 (0.61 – 1.20) for KRAS mutant and 0.86 (0.70 – 1.05) for wild-type KRAS. Similarly, for PFS the HR (95% CI) values were 0.99 (0.72 – 1.37) for KRAS mutant and 0.94 (0.78 – 1.14) for KRAS wild type. Additionally, there was no evidence of a differential effect by KRAS status among patients with non-SCC disease as depicted in Figure 2. Figure 5 shows the Kaplan-Meier curves of the 166 patients with a known KRAS mutation: 16 had SCC, 7 with an H-score ≥ 200, and 7 with FISH+ disease. Since KRAS mutations predominantly occur in non-squamous, Figure 5 depicts the distribution of OS and PFS by arms and by KRAS MT versus WT patients within the non-SCC population. The distribution of specific KRAS amino acid substitutions is shown in Table S3.
Figure 5. Comparison of Overall and Progression-Free for KRAS mutant versus wild type in non-squamous.
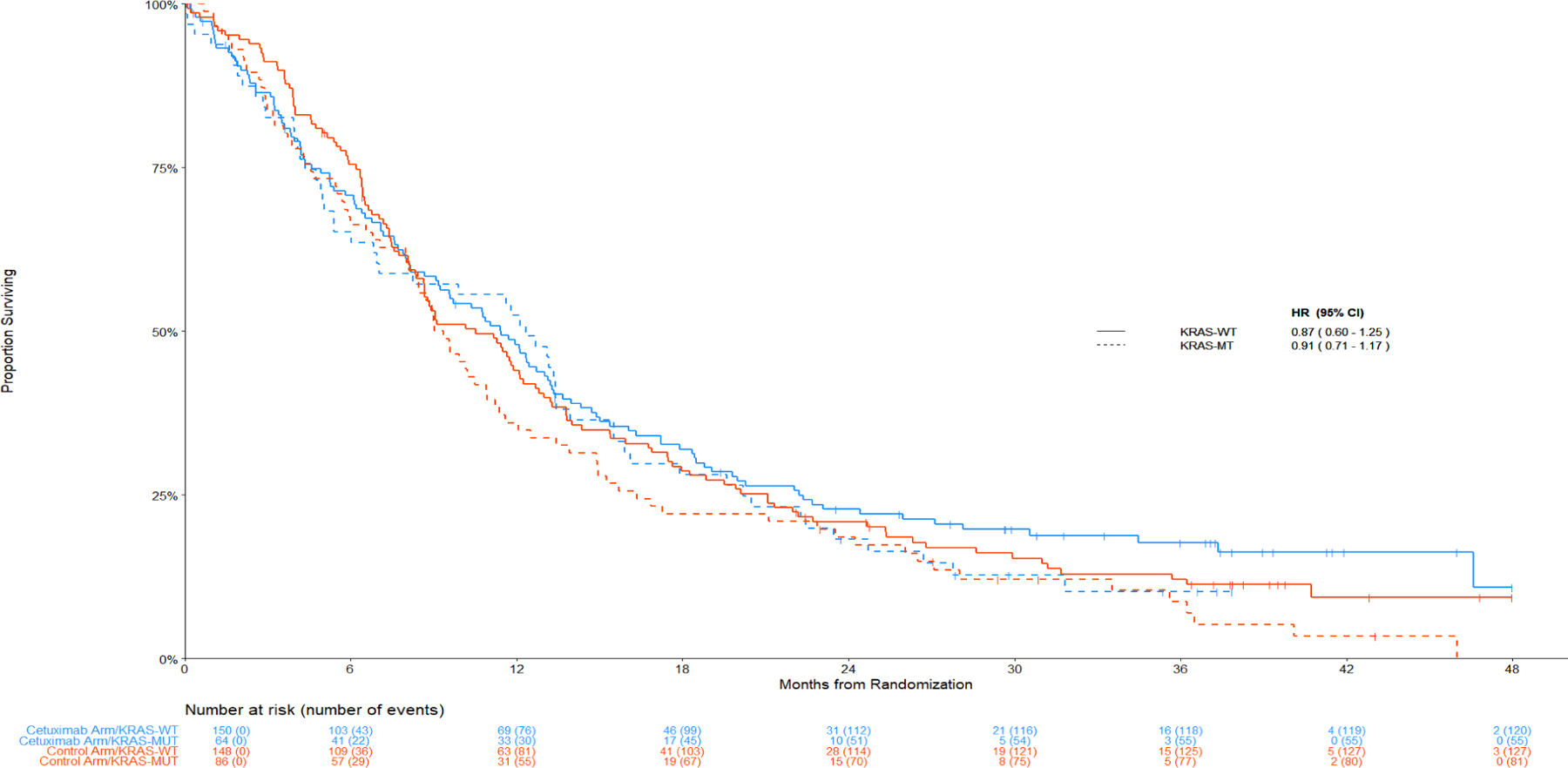
Panel A: Overall Survival
Discussion
The S0819 trial assessed the efficacy and safety of carboplatin/paclitaxel, with bevacizumab in non-SCC histology, with or without cetuximab in the first-line treatment of patients with advanced NSCLC. While the addition of cetuximab did not result in a statistically significant difference in OS in the entire study population, or in PFS in the EGFR FISH+ patients (primary endpoints), analyses presented here indicate a significant benefit with cetuximab treatment in SCC patients who had a positive combination index of EGFR IHC+ plus EGFR FISH+ cancers (23% of SCC population), while no impact was seen in patients with non-SCC NSCLC. In our first report of the S0819 study, there was a suggestion that patients with FISH+ SCC showed benefit from the addition of cetuximab to chemotherapy (1,). The difference in the predictive role of EGFR IHC and FISH in patients with SCC versus Non-SCC histologies is likely to reflect differences in EGFR biology between the NSCLC subtypes (15–16).
In contrast to colorectal cancer (CRC), we did not observe a predictive role for KRAS mutation status in this NSCLC population. In CRC, the presence of a KRAS mutation is a strong predictor for the lack of efficacy of EGFR-targeted monoclonal Abs (12). One opportunity in this large, randomized study was to determine whether a similar effect could be observed in NSCLC since previous investigations of EGFR monoclonal antibodies lacked the power to make this distinction (5,8). In the current analysis, the apparent benefit from the addition of cetuximab to chemotherapy was limited to the biomarker-marker defined subgroup of the SCC sub-population. However, KRAS mutations are rare in SCC precluding a realistic evaluation of KRAS status in this histology. In non-SCC, KRAS mutations were observed in 33% of patients. However, in this largely ADC subpopulation, KRAS wild-type status did not predict benefit from the addition of cetuximab, in contrast to the CRC (12) .
Biomarker analysis in advanced stage NSCLC requires consideration of factors related to the heterogeneity of this tumor type, limited tissue availability in many cases and now eight different actionable oncogenes for upfront testing (17–18). Furthermore, variability in technology and assessment methods adds to the complexity. While these considerations today therapeutically are most applicable to lung ADC, it is becoming increasingly recognized that SCC is non-homogeneous as well. High expression levels of WT EGFR, in particular, are most common in lung SCC. Because anti-EGFR mAbs activity is mechanistically linked to direct interaction with EGFR, and high levels of EGFR protein expression correlate with sensitivity to anti-EGFR mAbs in vitro(9), there is a strong rationale to consider the level of tumor protein expression of EGFR, a significant potential predictive biomarker in this setting. EGFR pathway analysis of NSCLC cell lines and patient tumor tissues has described the predictive value of several potential biomarkers for EGFR-inhibitor activity, including EGFR protein expression by immunohistochemistry (IHC) and EGFR gene copy number by FISH or chromogenic in situ hybridization (CISH)(19). While a retrospective analysis(20) of phase II S0342 study(3), in which 76 patients with NSCLC were treated with CT (carboplatin + paclitaxel) plus cetuximab administered concurrently or sequentially, suggested that increased EGFR gene copy number might be associated with improved clinical outcome.
In regard to EGFR protein expression, a retrospective analysis(21) of data from the phase III FLEX study(14) suggested that high EGFR expression, as determined by IHC using the same assay and threshold as in the current study, was associated with survival benefit from the addition of cetuximab to first-line platinum chemotherapy in patients with advanced NSCLC. In particular, for patients in the high EGFR expression group, OS was longer in the chemotherapy plus cetuximab arm than in the chemotherapy-only arm (HR: 0.73 [95% CI 0.58–0.93]; p = 0.011, median 12.0 vs 9.6 months). No corresponding survival benefit was observed for patients in the low EGFR expression group (HR: 0.99 [95% CI 0.84–1.16]; p = 0.88, median 9.8 vs 10.3 months). In the retrospective analysis(21) of the FLEX trial’s results, authors investigated the predictive and prognostic use of four tumor-associated molecular characteristics (KRAS mutation, increased EGFR copy number, EGFR mutation, and PTEN expression status) linked to the EGFR signaling pathway. EGFR copy number assessed by FISH (positive vs negative) was not predictive for the efficacy of CT plus cetuximab in relation to OS [HR = 0.85 (0.56–1.29), p = 0.44], PFS [HR = 0.80 (0.51–1.25), p = 0.33], or RR [HR = 1.62 (0.70–3.76), p = 0.26]. However, the assessment method for EGFR FISH was not done by the same laboratory as in the current study, and there might be technical- or interpretative differences in the EGFR FISH assessment between the FLEX study and the current study. Of interest, EGFR mutation status (positive vs negative) was not predictive for the efficacy of CT plus cetuximab in relation to OS [HR = 1.48 (0.77–2.85), p = 0.24], PFS [HR = 0.92 (0.53–1.60), p = 0.76] and RR [HR = 1.36 (0.50–3.70), p = 0.55].
Correlation of EGFR-expression with outcomes in the SQUIRE trial(22) showed similar results as in the current study in patients with SCC treated with another EGFR monoclonal antibody (necitumumab). In the SQUIRE trial, the large majority of patients (95%) had tumor samples expressing EGFR protein, as determined by IHC (EGFR > 0, when at least one positive cell was identified); OS for the overall population in this study was significantly longer in the necitumumab plus gemcitabine-cisplatin group than in the gemcitabine-cisplatin group (HR = 0.79 [95% CI 0.69–0.92]; p = 0.002, median 11.7 vs 10.0 months), which led to US FDA approval of the drug. In the same study, while patients with high EGFR expression (H-Score ≥ 200) had a more favorable HR for OS (HR = 0.75; [0.60–0.94]) compared to the patients with low expression of EGFR protein (HR = 0.90; 0.75–1.07), no conclusion could be drawn regarding the predictive value of EGFR protein expression(23). Additionally in the SQUIRE study, EGFR amplification was associated with imnproved OS and EGFR high polysomy was associated with increased PFS and “EGFR FISH-positivity” (e.g. high polysomy plus amplification) had a non-significant trend towards improved outcome (HR=0.70) (24). In a meta-analysis based on results from several studies of EGFR antibody therapies, a predictive role of EGFR IHC and EGFR FISH for EGFR antibody therapy was observed in advanced NSCLC, including SCC histology patients(25).
In our analysis of the S0819 trial, we applied a biomarker-enriched model including EGFR FISH, EGFR IHC, and KRAS; to our knowledge, none of the previous secondary analyses of the above-mentioned trials, have evaluated FISH and IHC together to create a combination index. Based on the previous findings with cetuximab or necitumumab related to EGFR protein expression and/or EGFR gene copy gain, the chance of our results being a random finding seems less likely. Our data suggest that this IHC+/FISH+ combination index is worthy of further study and could potentially be used as a selection factor for treatment with EGFR antibody-based therapy in advanced SCC lung cancer. In this regard, studies combining EGFR monoclonal antibodies with checkpoint immunotherapy might consider incorporation of these markers in SCC patient population. The techniques for both FISH and IHC are well-established and widely used in routine clinical practice, with relatively low costs.
Conclusion
Despite recent advances in personalized therapy for lung cancer, lung SCC remains largely excluded from targeted therapy approaches, with the rare exception of those who are never-smokers with oncogene-driven disease. Our results suggest that leveraging the recognized high expression of WT EGFR by a FISH/IHC combination index could provide a step forward toward personalizing therapy with EGFR-directed monoclonal antibody therapy. Further prospective validation studies of this concept are warranted.
Supplementary Material
Figure 3:
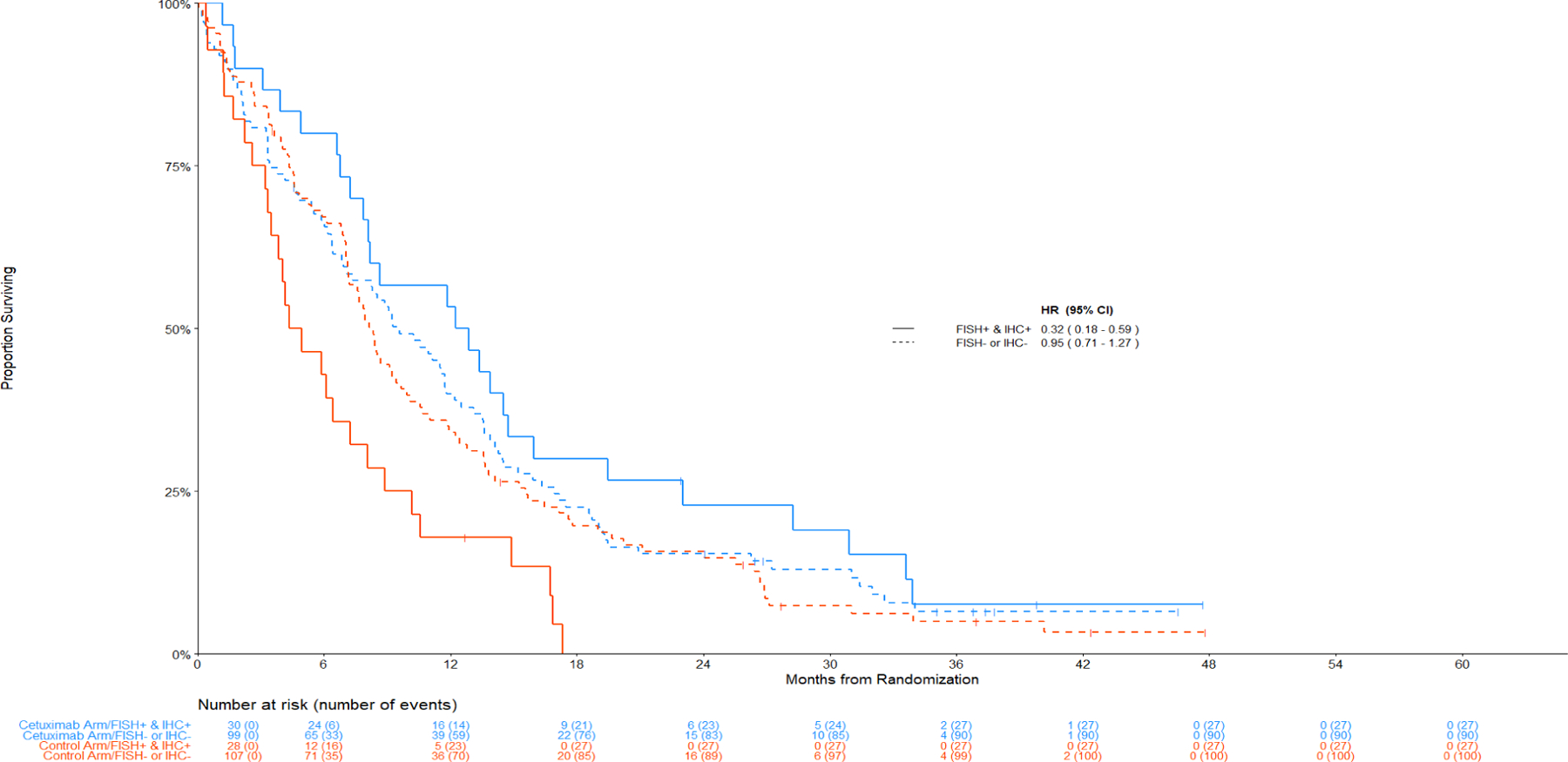
Panel C: (IHC+ and FISH+) versus (IHC- or FISH-)
Figure 4:
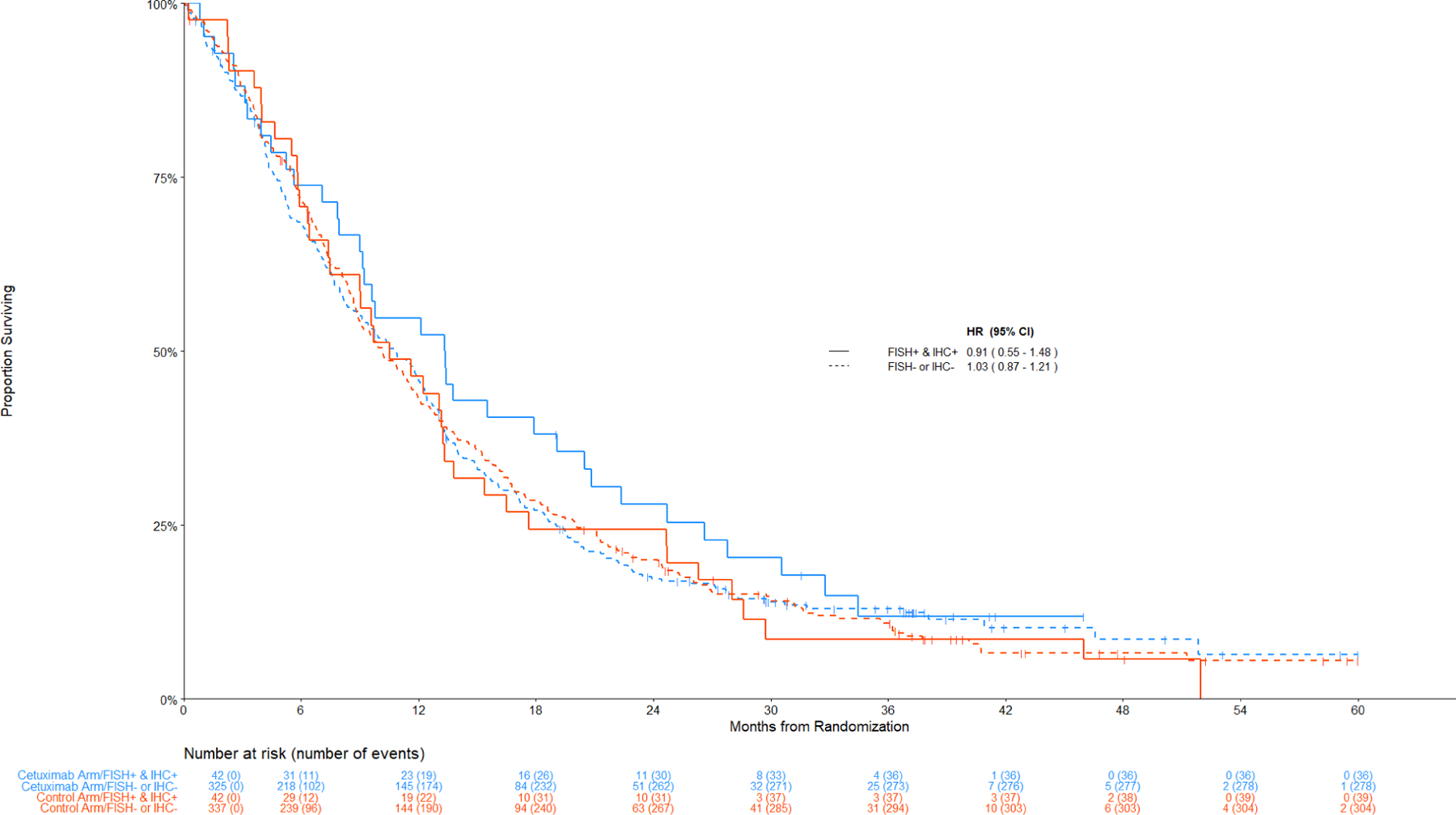
Panel C: (IHC+ and FISH+) versus (IHC- or FISH-)
Figure 5:
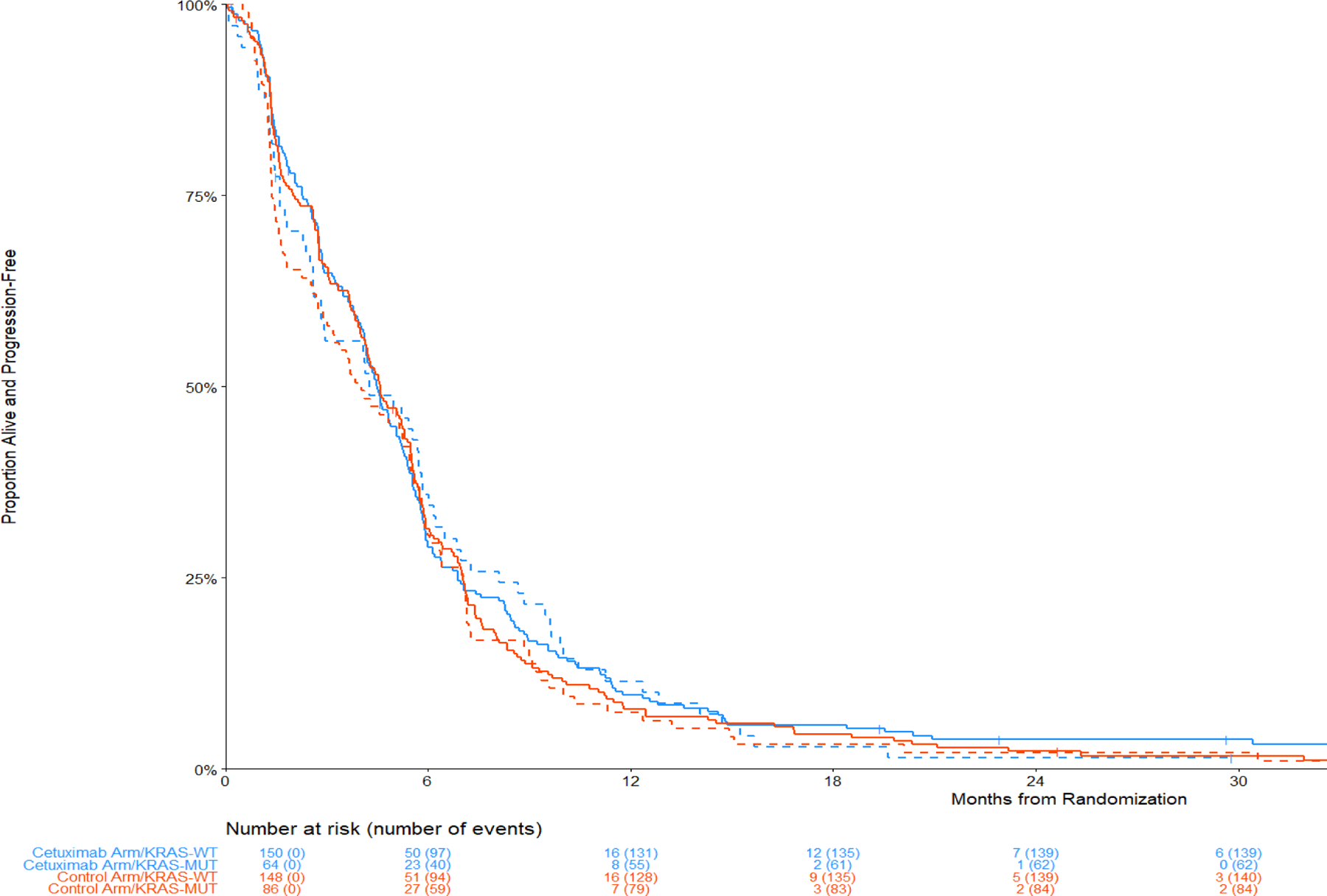
Panel B: Progression-Free Survival
Clinical Pratice Points:
Clinical and preclinical investigations of EGFR monoclonal antibodies such as cetuximab have shown limited benefit in combination with chemotherapy in NSCLC. S0819, a large, randomized phase III trial of chemotherapy with or without cetuximab in NSCLC was designed to prospectively identify molecular subsets of patients with enhanced benefit from the addition of cetuximab. An analysis of EGFR gene copy number and protein expression demonstrated that a combination index of EGFR FISH and IHC positivity was associated with improved overall survival in patients with squamous cell carcinoma histology (n = 321). EGFR monoclonal antibodies may be of utility in the treatment in this NSCLC subset, with further focused investigations warranted.
Acknowledgments
Funding
This work was supported by the National Cancer Institute of the National Institutes of Health [U10CA180888, U10CA180819, U10CA180846]; and in part by Eli Lilly and Company. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health or Eli Lily and Company. SWOG Statistics and Data Management Center is funded by the National Cancer Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosures
Fred R. Hirsch has participated in scientific advisory boards (compensated) for Bristol-Myers Squibb, Amgen, Merck, Novartis, Genentech/Roche, Sanofi, Genzyme, AstraZeneca/Daiichi, Lilly, Abbvie, and OncoCyte. Francesco Agustoni received honoraria from Roche, Bristol-Myers Squibb, Merck Sharp & Dohme, and Astra Zeneca, travel/meeting support from Incyte, and has participated in scientific advisory boards for Boehringer-Ingelheim, Merck Sharp & Dohme, and Bristol-Myers Squibb. Roy S. Herbst received research grants from AstraZeneca, Eli Lilly and Company, Genentech/Roche, and Merck and Company, compensation for consultation from Abbvie Pharmaceuticals, ARMO Biosciences, AstraZeneca, Bayer HealthCare Pharmaceuticals, Inc., Bolt Biotherapeutics, Bristol‐Myers Squibb, Candel Therapeutics, Inc., Checkpoint Therapeutics, Cybrexa Therapeutics, DynamiCure Biotechnology, LLC., eFFECTOR Therapeutics, Inc., Eli Lilly and Company, EMD Serono, Foundation Medicine, Inc., Genentech/Roche, Genmab, Gilead, Halozyme Therapeutics, Heat Biologics, HiberCell, Inc., I‐Mab Biopharma, Immune‐Onc Therapeutics, Inc., Immunocore, Infinity Pharmaceuticals, Johnson and Johnson, Loxo Oncology, Merck and Company, Mirati Therapeutics, Nektar, Neon Therapeutics, NextCure, Novartis, Ocean Biomedical, Inc., Oncocyte Corp, Oncternal Therapeutics, Pfizer, Refactor Health, Inc., Ribbon Therapeutics, Sanofi, Seattle Genetics, Shire PLC, Spectrum Pharmaceuticals, STCube Pharmaceuticals, Inc., Symphogen, Takeda, Tesaro, Tocagen, Ventana Medical Systems, Inc., WindMIL Therapeutics, and Xencor, Inc, travel/meeting support from AstraZeneca, Genentech/Roche, Merck and Company, and Ventana Medical Systems, Inc., has participated in scientific advisory boards for AstraZeneca, Bolt Biotherapeutics, Candel Therapeutics, Inc., Checkpoint Therapeutics, Cybrexa Therapeutics, EMD Serono, Halozyme Therapeutics, Heat Biologics, I‐Mab Biopharma, Immune‐Onc Therapeutics, Inc., Immunocore, Infinity Pharmaceuticals, Neon Therapeutics, Novartis, Ocean Biomedical, Inc. , Ribbon Therapeutics, STCube Pharmaceuticals, Inc., and Xencor, Inc., has served as a fiduciary role in American Association for Cancer Research, International Association for the Study of Lung Cancer, Society for Immunotherapy of Cancer, and Southwest Oncology Group, owns stock in Bolt Biotherapeutics, Checkpoint Therapeutics, and Immunocore Holdings Limited, and has been a non-executive/independent board member in Immunocore Holdings Limited and Junshi Pharmaceuticals. Karen Kelly received compensation for consultation from Eli Lilly Advisory Board. David R. Gandara received research grants from Amgen, AstraZeneca, Genentech, and Merck, compensation for consultation from Lilly, Merck, and Novartis, and has participated in scientific advisory boards for AstraZeneca, Roche-Genentech, Guardant Health, IO Biotech, and Oncocyte. Philip C. Mack has participated in scientific advisory boards and speaking engagements for Amgen and Guardant Health. Mary W. Redman, James Moon, Thomas J. Semrad, Marileila Varella-Garcia, and Chris J. Rivard declare that they have no conflict of interest.
References
- 1.Herbst RS, Redman MW, Kim ES et al. A randomized, phase III study of carboplatin / paclitaxel/bevacizumab with or without concurrent cetuximab in patients with advanced non-small-cell lung cancer: SWOG S0819. Lancet 2018; 19(1): 101–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Redman MW, Crowley JJ, Herbst RS et al. Design of a phase III clinical trial with prospective biomarker validation: SWOG S0819. Clin Cancer Res 2012; 18: 4004–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Herbst RS, Kelly K, Chansky K et al. Phase II selection design trial of concurrent chemotherapy and cetuximab versus chemotherapy followed by cetuximab in advanced non-small-cell lung cancer: Southwest Oncology Group Study S0342. J ClinOncol 2010; 28: 4747–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim ES, Moon J, Herbst RS et al. Phase II trial of carboplatin, paclitaxel, cetuximab and bevacizumab followed by cetuximab and bevacizumab in advanced non-squamous non-small-cell lung cancer SWOG S0536. J ThoracOncol 2013; 8: 1519–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lynch TJ, Patel T, Dreisbach L et al. Cetuximab and first-line taxane/carboplatin chemotherapy in advanced non-small-cell lung cancer: results of the randomized multicenter phase III trial BMS099. J ClinOncol 2010; 28: 911–17. [DOI] [PubMed] [Google Scholar]
- 6.Rosell R, Robinet G, Szczesna A et al. Randomized phase II study of cetuximab plus cisplatin/vinorelbine compared with cisplatin/vinorelbine alone as first-line therapy in EGFR-expressing advanced non-small-cell lung cancer. Ann Oncol 2008; 19: 362–69. [DOI] [PubMed] [Google Scholar]
- 7.Butts CA, Bodkin D, Middleman EL et al. Randomized phase II study of gemcitabine plus cisplatin or carboplatin, with or without cetuximab, as first-line therapy for patients with advanced or metastatic non-small-cell lung cancer. J ClinOncol 2007; 25: 5777–84. [DOI] [PubMed] [Google Scholar]
- 8.Pirker R, Pereira JR, Szczesna A et al. Cetuximab plus chemotherapy in patients with advanced non-small-cell lung cancer (FLEX): an open-label randomized phase III trial. Lancet 2009; 373: 1525–31. [DOI] [PubMed] [Google Scholar]
- 9.Kimura H, Sakai K, Arao T et al. Antibody-dependent cellular cytotoxicity of cetuximab against tumor cells with wild-type or mutant epidermal growth factor receptor. Cancer Sci 2007; 98: 1275–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dearden S, Stevens J, Wu Y-L, Blowers D. Mutation incidence and coincidence in non small-cell lung cancer: meta-analyses by ethnicity and histology (mutMap). Ann Oncol 24(9);2371–2376, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cancer Genome Atlas N. Comprehensive molecular characterization of human colon and rectal cancer. Nature 487:330–337, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Karapetis CS, Khambata-Ford S, Jonker DJ et al. K-ras mutations and benefit from cetuximab in advanced colorectal cancer. N Engl J Med 2008; 359(17):1757–1765. [DOI] [PubMed] [Google Scholar]
- 13.Varella-Garcia M, Diebold J, Eberhard DA et al. EGFR fluorescence in situ hybridization assay: guidelines for application to non-small-cell lung cancer. J ClinPathol 2009; 62: 970–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Storey JD, Tibshirani R. Statistical significance for genomewide studies. ProcNatlAcadSci USA 2003; 100(16): 9440–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pirker R, Pereira JR, von Pawel J et al. EGFR expression as a predictor of survival for first-line chemotherapy plus cetuximab in patients with advanced non-small-cell lung cancer: analysis of data from the phase 3 FLEX study. Lancet Oncol 2012; 13: 33–42. [DOI] [PubMed] [Google Scholar]
- 16.Hirsch FR, Varella-Garcia M, Bunn PA Jr et al. Epidermal growth factor receptor in non-small-cell lung carcinomas: correlation between gene copy number and protein expression and impact on prognosis. J Clin Oncol 2003;21(20):3798–3807. [DOI] [PubMed] [Google Scholar]
- 17.Hirsch FR, Suda K, Wiens J et al. New and emerging targeted treatments in advanced non-small-cell lung cancer. Lancet 2016;388(10048):1012–1024. [DOI] [PubMed] [Google Scholar]
- 18.Mosele F. et al. ; Recommendation for the use of Next-Generation Sequencing (NGS) for patients with metastatic cancer: a report from the ESMO Precision Medicine Working Group. Ann Oncol 2020. (August 24 online) [DOI] [PubMed] [Google Scholar]
- 19.Bunn PA Jr, Dziadziuszko R, Varella-Garcia M et al. Biological markers for non-small cell lung cancer patient selection for epidermal growth factor receptor tyrosine kinase inhibitor therapy. Clin Cancer Res 2006; 12: 3652–56. [DOI] [PubMed] [Google Scholar]
- 20.Hirsch FR, Herbst RS, Olsen C et al. Increased EGFR gene copy number detected by fluorescent in situ hybridization predicts outcome in non-small-cell cancer patients treated with cetuximab and chemotherapy. J ClinOncol 2008; 26: 3351–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.O’Byrne KJ, Gatzemeier U, Bondarenko I et al. Molecular biomarkers in non-small-cell lung cancer: a retrospective analysis of data from the phase 3 FLEX study. Lancet Oncol 2011; 12: 795–805. [DOI] [PubMed] [Google Scholar]
- 22.Thatcher N, Hirsch FR, Luft AV et al. Necitumumab plus gemcitabine and cisplatin versus gemcitabine and cisplatin alone as first-line therapy in patients with stage IV squamous non-small-cell lung cancer (SQUIRE): an open-label, randomized, controlled phase 3 trial. Lancet Oncol 2015; 16: 763–74. [DOI] [PubMed] [Google Scholar]
- 23.Paz-Ares L, Socinski MA, Shahidi J et al. Correlation of EGFR-expression with safety and efficacy outcomes in SQUIRE: a randomized, multicenter, open-label, phase III study of gemcitabine-cisplatin plus necitumumab versus gemcitabine-cisplatin alone in the first-line treatment of patients with stage IV squamous non-small-cell lung cancer. Ann Oncol 2016; 27: 1573–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Genova C, Socinski MA, Hozak RR, Mi G, Kurek R, Shahidi J, Paz-Ares L, Thatcher N, Rivard CJ, Varella-Garcia M, Hirsch FR. EGFR Gene Copy Number by FISH May Predict Outcome of Necitumumab in Squamous Lung Carcinomas: Analysis from the SQUIRE Study. J Thorac Oncol 2018. Feb;13(2):228–236. doi: 10.1016/j.jtho.2017.11.109. Epub 2017 Nov 20. PMID: 29158193; PMCID: PMC6233716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bonomi PD, Gandara D, Hirsch FR et al. Predictive biomarkers for response to EGFR-directed monoclonal antibodies for advanced squamous cell lung cancer. Ann Oncol (29); 1701–1709, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


