Abstract
Laboratory studies of pathogens aim to limit complexity in order to disentangle the important parameters contributing to an infection. However, pathogens rarely exist in isolation, and hosts may sustain co-infections with multiple disease agents. These interact with each other and with the host immune system dynamically, with disease outcomes affected by the composition of the community of infecting pathogens, their order of colonization, competition for niches and nutrients, and immune modulation. While pathogen-immune interactions have been detailed elsewhere, here we examine the use of ecological and experimental studies of trypanosome and malaria infections to discuss the interactions between pathogens in mammal hosts and arthropod vectors, including recently developed laboratory models for co-infection. The implications of pathogen co-infection for disease therapy are also discussed.
Keywords: parasite, co-infection, trypanosoma, plasmodium
1. Introduction
The aim of scientific exploration in the laboratory is to control as many factors as possible, allowing the understanding of the impact of only one, or a few, variables. This has underpinned almost all pathogen research and generated impressive insight into the molecular mechanisms of pathogen invasion, establishment and immune evasion, as well as the response of the host defences. However, in almost all cases, such studies cannot represent the context and setting of pathogen infections in the real world. In particular, pathogens rarely exist in isolation but rather enter hosts already harbouring a diversity of pre-existing commensal or pathogenic organisms, each imposing their own effects on the within-host environment and immune system [1–3]. Furthermore, the community of organisms within a host may be unstable or destabilized by the new ingression. As a consequence, a host represents a complex and dynamic environment in which pathogens must establish and optimize their survival in competition with co-infecting organisms while also avoiding the defences of the host [4]—which themselves may be modulated by the pre-existing congregation. This introduces considerable complexity at any given time and also dynamically over time. This complexity is compounded where pathogens are transmitted by disease vectors, where the contribution of the vector's biology and epidemiology, as well as its own community of microbial passengers, generates additional interactions, potentially further influenced by the impact of environmental change [5]. These combined interactions, which are so important in the real world, cannot be fully disentangled using laboratory studies alone. In this review, we discuss the different approaches that can be used to understand the impact of co-infections, with a focus on two microbial eukaryotic pathogens, causing malaria (Plasmodium spp.; transmitted by mosquitoes) [6] and African trypanosomiasis (Trypanosoma brucei, Trypanosoma congolense and Trypanosoma vivax; transmitted by tsetse flies) [7,8]. Focusing on these parasites, we review diverse studies on the epidemiology of co-infection, and the factors that are important to consider when using experimental systems to understand how these organisms sustain themselves in their mammalian host in the context of co-infection, extending beyond the elegant immunological and molecular studies focused on immune evasion by individual infections in host model systems. The mechanisms through which trypanosomes and Plasmodium spp. detect and respond to co-infecting parasites are then discussed. Finally, we highlight how these interactions can be best understood through an interdisciplinary approach involving a combination of infection biology, epidemiology, mathematical modelling and evolutionary theory (figure 1).
Figure 1.
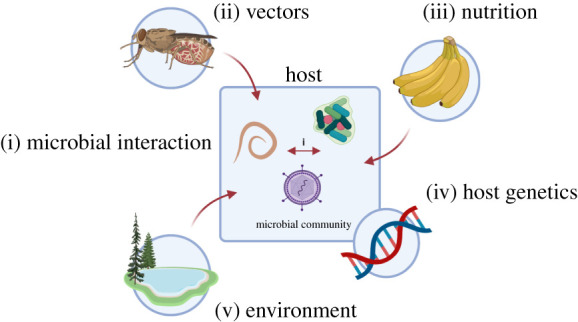
A hypothetical scenario which illustrates some of the factors that shape the composition of a microbial community. These include (i) direct and indirect interactions between co-infecting microorganisms; (ii) the availability of vectors and their capacity to transmit different parasite species or strains; (iii) nutrition status and (iv) host genetics which impact upon host immune responses and susceptibility to infection; (v) environmental factors such as soil conditions and climate. Created with BioRender.com. (Online version in colour.)
2. Epidemiology of co-infection
Most studies of co-infection have focused on the analysis of hosts and pathogens at the epidemiological level. Hosts in natural systems are frequently infected with a diverse community of microorganisms composed of different taxa, different species or even strains of the same organism. This can be illustrated by a recent longitudinal study from western Kenya, where a cohort of 548 zebu calves were found to be infected with over 50 different pathogens, including many trypanosome and apicomplexan parasites [9]. With respect to trypanosomes, the potential for infection with multiple species is particularly high because these parasites have an unusually broad host range, allowing frequent interactions between species in diverse hosts. Reflecting this, a large number of studies in different regions have reported the co-circulation of T. brucei, T. congolense and T. vivax, and these have been detected both in surveyed livestock and game animals [10], as well as in trapped tsetse flies (e.g. [11,12]). The major human infective Plasmodium species, Plasmodium falciparum, in contrast shows strict host specificity although other human infective malaria parasites, such as Plasmodium knowlesi, are more promiscuous and zoonotic.
Alongside mammalian hosts that are multiply infected with different parasite species, vectors can also sustain and transmit multiple parasites. In some cases, the same vector can transmit different pathogens, as is the case for mosquito transmission of Plasmodium spp. and Wuchereria bancrofti [13]. The same mosquito species can also be infected by different species of Plasmodium [14], although host preference might impose some limits on the prevalence of co-infecting parasites in the vector. Similarly, multiple trypanosome species, and strains of the same species, can simultaneously infect their tsetse vector [15,16] although different subspecies of tsetse vary in their transmission of distinct trypanosome species, contributed to by their geographical distribution and anthropophilic or zoophilic preferences [17]. Climate factors can also determine the distribution of vector species [18,19] and may alter the infection dynamics of the parasites that they harbour, influencing their potential to sustain or establish co-infections. For example, tsetse flies are particularly sensitive to changes in temperature and humidity, as illustrated by the shifts in fly distribution observed in northern Zimbabwe [20,21]. Interestingly, vector availability itself does not necessarily mean high disease incidence. For example, a longitudinal cohort study of calves in western Kenya found a low incidence of clinical trypanosomiasis in an area with high tsetse challenge [9].
While epidemiological studies continue to provide insight into the complicated drivers of co-infections, important knowledge gaps remain, sometimes driven by the tractability of the pathogen or the research priorities and focus of the research community exploring the pathogen. This was highlighted in a recent meta-analysis which found that only 0.05% of co-infection studies in humans focused on helminth parasites, despite their profound global burden [22]. A consequence of this bias can be a focus in laboratory studies on species such as T. brucei or P. falciparum, at the expense of other species that are not, or less established, human pathogens (T. vivax and T. congolense; P. knowlesi). This has the potential to limit surveillance studies in the field to the better-studied parasites, although the advent of metagenomic rather that species-targeted analyses will alleviate this issue. It has also been highlighted that abundance studies do not consider the within-host dynamics of co-infecting organisms, reflected by the variation seen in such data between individuals in the same geographical location [23–25]. Combined, this highlights the need for more experimental and field studies to tease apart the delicate interactions between different species or strains co-infecting the same host and the value of longitudinal surveillance and chronic infection studies to monitor dynamic temporal changes in co-infection prevalence. Only with data from comprehensive and unbiased epidemiological studies can researchers in the laboratory understand what are the important interactions happening in the real world, and as a result focus their research on co-infections that have the potential to impact human and animal health most severely.
3. Considerations in experimental studies of co-infection
Given the challenges of experimentally manipulating co-infection in the field, experimental systems in the laboratory are particularly valuable. In recent years, both trypanosome and Plasmodium systems have become excellent experimental models to explore parasite co-infections in the mammalian host, providing a bridge between the complexity of field-based studies and the accessibility of laboratory studies that have traditionally focused on monoinfection. This is because, for each parasite, there are available tools to mark or identify distinct parasite strains or species and the ability to discriminate proliferative and transmission-adapted forms. There are also relevant culture systems and rodent infection models. This has permitted experimental studies of co-infection, the design of which can substantially alter the outcome. Considerations include the timing and order of infection, as well as how pathogen virulence and transmission potential are modulated in multiply infected hosts. The impact of acute and chronic co-infection profiles is also relevant. In this section, we discuss the factors that can influence the outcome of experimental studies of co-infection.
(a) . Time and space
An important consideration for co-infection studies relates to when the respective parasites are introduced experimentally. In one scenario, there is infection with both pathogens at the same time, for example by a single insect bite from a co-infected vector, or through simultaneous inoculation of more than one parasite strain or species via syringe. The second scenario involves temporally offset superinfection of an already infected host by a second pathogen strain or species. Given the chronic nature of many infections, superinfection is likely to be the more common route to establishing a co-infection in the real world and evidence suggests that there is often a competitive advantage to being first on the scene. For example, goats already infected with T. congolense showed delayed appearance of T. brucei (compared to uninfected goats) after superinfection by tsetse fly bite [26]. Experimental sequential infection with T. congolense isolates in cattle also suppressed the prevalence of the incoming parasites [27]. On the other hand, there was no delay in superinfection by another trypanosome species, T. vivax, in goats already infected with T. congolense [26].
The importance of timing has also been shown by different studies analysing the consequence of co-infection between T. brucei and Plasmodium berghei in mice. One study revealed a heightened number of both parasites when co-infected, with more severe anaemia, hypoglycaemia and lower survival [28]. More recently, however, an initial infection with T. brucei was found to limit subsequent P. berghei establishment in the liver, and protect mice from cerebral malaria and prolong survival [29]. In the first study, parasites were inoculated simultaneously generating enhanced Plasmodium virulence, whereas in the latter study T. brucei was established first, generating reduced virulence. Similarly, a recent study of mixed-species malaria co-infection in mice found increased virulence when Plasmodium yoelli was inoculated at the same time as Plasmodium vinckei or Plasmodium chabaudi [30]. The authors highlight differences between their findings and others, acknowledging that timing, route of inoculation and host strain or species can all alter the co-infection outcome. Such studies indicate that interactions which modulate virulence may occur in human malaria co-infections, but their nature remains unpredictable. Thus, it is important when designing co-infection studies to consider how different experimental designs can deliver contrasting outcomes. It may be necessary to carry out multiple permutations of any one experiment to better understand how two pathogens could influence each other in the field. Epidemiological information, for example whether two parasites commonly share a vector, can help to predict whether simultaneous or sequential inoculation better represents the field setting.
In addition to the relative timing of infection, the spatial coincidence of parasites can be important in co-infection studies where the proximal interaction of parasites might be relevant. For example, two parasites which inhabit macrophages, Leishmania infantum and Toxoplasma gondii, can reside within the same cell when inoculated on the same day [31]. However, when T. gondii was inoculated five days later than L. infantum, both parasites established infection but not within the same macrophage. Interestingly, mice infected with L. infantum were also protected from the usual virulence of T. gondii. For Plasmodium, the simultaneous infection of a single red blood cell can occur frequently in culture and in vivo using parasites of the same species [32]. However, there is complexity in the potential for the co-occupation of erythrocytes between co-infecting species, since P. vivax and P. ovale favour young erythrocytes, P. malariae favours older red blood cells, whereas P. falciparum is quite promiscuous with respect to red blood cell age. These preferences are not absolute and the relative proportion of red blood cell types available to co-infecting parasites might be dynamic within the host. The relative success of each parasite strain or species in a Plasmodium co-infection could therefore vary substantially depending on the host's red blood cell landscape. This points to the value of incorporating studies focused on host biology in affected populations to build a more complete picture of co-infections. Experimental studies could benefit from testing co-infection interactions in diverse host environments to see whether observations are widespread or restricted to a given scenario.
(b) . Transmission and virulence
Monitoring the modulation of virulence is an important consideration in co-infection studies, whether virulence is defined as overall parasite number, or parasite-induced host pathology which is the focus of most of the following discussion [33,34]. A meta-analysis of co-infection data found that most studies reported negative impacts for human health [22] and co-infection can exacerbate detrimental effects caused by individual pathogens. For example, P. falciparum and hookworm contribute to host anaemia by distinct mechanisms and in co-infection these effects could be additive [35].
Alternatively, co-infection can result in reduced virulence where parasites with different levels of relatedness directly interfere with each other. This interference could have an associated parasite fitness cost, reducing the extent to which co-infecting parasite species are able to exploit the host [36]. For example, a survey of African trypanosomiasis in the Gambia revealed that co-infection with T. vivax and T. congolense was associated with reduced pathology compared to T. congolense infection alone [37], suggesting these different species could interfere to reduce virulence. However, a mixed infection comprising an avirulent strain and a virulent strain of the same species, T. brucei, also reduced the deleterious effects on the host relative to an infection with the virulent strain alone [38]. This might reflect that overall relatedness between co-infecting parasites is not important in determining virulence outcomes, but instead the extent of diversity or relatedness at key virulence gene loci.
As well as virulence, transmissibility can be affected by co-infections, with virulence and transmissibility often interacting [39]. Various studies have focused on the impact of co-infection on malaria transmission potential through monitoring gametocyte investment. One study featuring two P. chabaudi clones of differing virulence did not reveal increased investment in transmission stages by either clone, despite competitive suppression of asexual parasite density in the co-infection [40]. However, the investment in transmission in the host may change dynamically in response to parasite numbers, allowing a short-term or long-term transmission strategy in the context of signals from co-infecting Plasmodium strains [41]. Likewise, in studies of human malaria, co-infection with P. malariae was associated with increased gametocyte production by P. falciparum in three out of four study sites in malaria-endemic regions [42]. On the other hand, co-infection with P. vivax was associated with lower P. falciparum gametocyte density in patients in Thailand, although it was unclear whether this was influenced by patients seeking medical attention earlier in cases of co-infection or whether there was an inhibition of P. falciparum gametocytogenesis [43]. Co-infection with other parasites can also affect malaria transmission. Humans simultaneously infected with helminths and P. falciparum presented a higher malaria transmission potential through increased gametocyte production [44]. Interactions between parasites within the vector may also influence transmission success of individual strains. For example, mosquitoes infected with one strain of P. chabaudi are more susceptible to infection by further strains during subsequent blood meals [45]. All of these examples highlight that the balance between within-host replication and the preparation for onward transmission can be sensitive to the presence of co-infecting parasites, of different strains or species, but that the outcome can be difficult to predict and may show plasticity in different experimental or clinical conditions. This emphasizes the need for wide-ranging and collaborative research, as transmission and virulence impacts observed in one study are unlikely to be applicable universally. With more data, the impact of co-infection and the potential outcomes of intervention can be better predicted at a local level.
Co-infection might also contribute to the trypanosome transmission potential of the tsetse for example through influencing the coordinated ‘social motility’ of the parasites in tsetse fly stages of the life cycle. Experimental studies of this phenomenon in vitro demonstrate avoidance behaviours of parasites of the same species and strain, monitored via the trajectory of growth spurs on culture plates [46]; whether a similar phenomenon occurs between species or strains has not been tested but has the potential to alter the migration of the parasites during maturation in the vector if operational in the fly. Also, the distinct swimming behaviours of different trypanosome species [47] could generate the potential for interference if different parasite species occupy the same vector simultaneously, since coordinated swimming may contribute to the parasites' journey through the fly [48]. Interestingly, colonization of tsetse fly salivary glands by T. brucei has also been shown to alter the anti-haemostatic activity of the saliva, impairing blood-feeding ability [49]—this promoting multiple feeding cycles and potentially increasing co-infection likelihood.
(c) . Acute versus chronic infections
Another aspect to consider when investigating experimental co-infections is dynamic interactions in acute versus chronic stages of infection, reflecting the mismatch that can be seen in longitudinal versus cross-sectional field studies. Co-infection may be missed in cross-sectional studies when one pathogen is competitively suppressing another at one point in time, but with fluctuations in their relative dominance over time. This is illustrated in a study of children with asymptomatic Plasmodium infection in Papua New Guinea, which monitored the dynamics of multiple Plasmodium species and genotypes over a 60-day period. This revealed that overall parasitaemia remained relatively stable but that there were dramatic shifts in the representation of different parasite genotypes over time [50]. These complex co-infection dynamics would have been missed in a cross-sectional study. The authors suggest that the observed dynamics may be explained by a combination of density-dependent regulation between species, antigenic variation and immune clearance. Once a species-specific response clears the majority species the density would fall below the threshold allowing other co-infecting species to expand, leading to the sequential pattern of infection observed.
These examples all highlight that tractable experimental studies can provide important information on the outcome of co-infections. However, they also highlight that the observed outcomes can be significantly affected by the experimental design and that this should be informed by knowledge of the biological interactions between co-infecting parasites in the field, where possible.
4. Mechanisms of interaction that operate in co-infections
Once experimental conditions are established for monitoring co-infections, a mechanistic understanding of interactions becomes feasible. Clearly, modulation of the host immune response by pathogens has important consequences for co-infection opportunity and outcome but this topic has been thoroughly explored in other reviews [51–54]. Below we focus on the parasite-intrinsic molecular processes that parasites in co-infections use to interact, directly or indirectly, specifically through the exchange of genes and protein factors, whether as part of sexual processes, compartmentalized in vesicles or as soluble signals (figure 2).
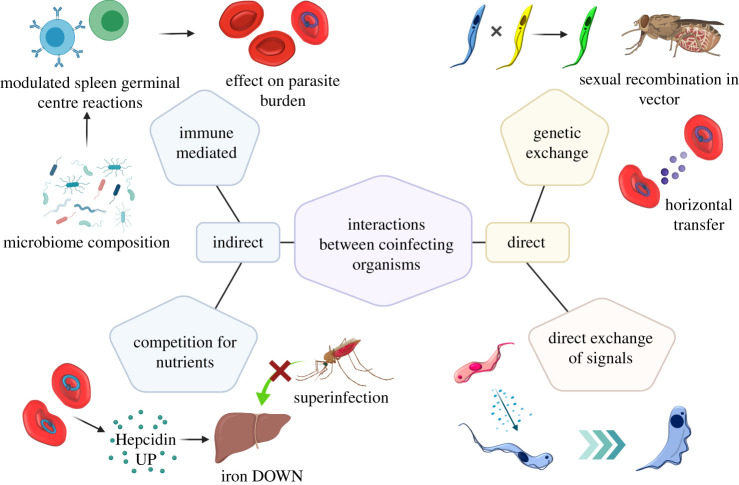
Figure 2.
Mechanisms that can mediate the interactions between co-infecting organisms. Examples of direct and indirect mechanisms of interaction are illustrated. Genetic exchange: sexual exchange between African trypanosomes in a co-infected tsetse fly, or horizontal transfer of DNA between Plasmodium infected red blood cells via extracellular vesicles. Direct exchange of signals: interspecies quorum sensing between African trypanosomes altering transmission potential. Competition for nutrients: an established Plasmodium infection increases host production of hepcidin diverting iron away from the liver, inhibiting invasion by superinfecting parasites. Immune mediated: the microbiota can influence the immune environment of a host to affect malaria infection. Created with BioRender.com. (Online version in colour.)
(a) . Sexual exchange
Eukaryotic parasites typically have a capacity for sexual exchange. In T. brucei meiosis is not an obligatory part of the life cycle but can take place within the tsetse fly. This permits new variants of human infective trypanosome to be generated through sexual exchange between T. b. rhodesiense and T. b. brucei in a co-infected tsetse [55], providing the opportunity for the acquisition of the SRA gene, important for human infectivity, but also other alleles. This is possible because T. brucei and T. b. rhodesiense cohabit the tsetse salivary glands, where meiosis occurs. T. b. gambiense type 1 also occupies this niche but appears asexual, without evidence of sexual exchange within or between species; there is evidence for T. b. gambiense type 2 sexual exchange, but these parasites may now have become extinct [56]. Beyond the T. brucei group of trypanosomes, opportunities for cross species sexual exchanges are absent because each species matures at a different site in the fly: T. brucei spp. (salivary glands), T. congolense (proboscis) and T. vivax (mouthparts). However, there is evidence for within-species sexual exchange for T. congolense [57] although there is some controversy around this, with others supporting a clonal population structure [58]. By contrast to T. brucei and T. congolense, T. vivax does not appear to undergo mating [59].
Unlike trypanosomes, sex is essential in the life cycle of Plasmodium resulting from the fusion of male and female gametocytes produced in the bloodstream to form a zygote in the mosquito gut. Where mosquito infections are initiated with gametocytes of different genetic backgrounds, there are significant opportunities for parasite co-infection to have important consequences for the epidemiology of the parasite in the field, for example in the exchange and spread of drug resistance between parasite strains [60]. Interestingly, infections with a single genotype can enhance mosquito infectivity compared with mixed genotype infections [61]. Consequently, where the component parasites differ in drug sensitivity, there is a risk that therapeutic intervention could generate enhanced disease transmission in a geographical location through the selection for monoinfections after competitive release [61–63].
(b) . Extracellular vesicles
A mechanism where co-infecting pathogens can interact directly throughout the life cycle is through the exchange of virulence factors, via either gene or protein transfer. In bacterial systems, horizontal gene transfer can lead to the transfer of antimicrobial resistance genes between strains and species, and potentially from commensals to pathogens [64]. Horizontal gene transfer may occur through direct cell-to-cell contact by bacterial conjugation or via the exchange of extracellular vesicles [65]. Extracellular vesicles have also been proposed to exchange the SRA virulence factor between subspecies of T. brucei, transferring resistance to human serum. In co-culture experiments, SRA was transferred to T. brucei via extracellular vesicles generated from shed nanotubes [66], rendering the recipient cells resistant to trypanolysis in human serum. In tsetse fly stages, extracellular vesicles can also affect social motility that may influence the coordinated migration of the parasites through the insect gut [67]. Extracellular vesicles can also mediate transfer of DNA between infected red blood cells in P. falciparum co-cultures resulting in the transfer of drug-resistance genes [68]. Additionally, signalling through extracellular vesicles can promote differentiation of Plasmodium parasites to transmissible gametocyte stages in culture [69] such that, in combination, virulence or drug-resistance genes can be exchanged between strains in a co-infection, helping new variants to be generated and transmitted. More recently extracellular vesicles containing lactate dehydrogenase derived from density-stressed P. falciparum cultures were found to limit the growth of low-density parasite populations in vitro by triggering apoptotic events, suggesting an intercellular signalling mechanism that could regulate parasite population density [70].
(c) . Soluble signalling factors
Single-celled organisms have developed soluble communication systems that enable composite members to coordinate behaviours as a community. One example is ‘quorum sensing', which involves regulation of gene expression through the production and detection of signalling molecules whose concentration increases with cell density. Quorum sensing is best understood in bacteria where a variety of signalling mechanisms are known, but also plays a role in the community behaviour of diverse eukaryotic pathogens. For example, the development of T. brucei from a proliferative slender form to a transmission-adapted stumpy form in its mammal host is regulated by quorum sensing via ‘stumpy induction factor' activity [71,72]. This has been found to comprise oligopeptide signals generated by parasite-released peptidases [73], with intracellular signalling pathway components required for quorum sensing also identified [74]. Where both the signal generation and transduction pathways are shared between co-infecting strains or species there is the potential to influence the infection outcome for the host and the pathogen. This has been observed experimentally, with T. congolense able to promote accelerated T. brucei differentiation to stumpy forms in a co-infection through shared quorum-sensing signals [75]. Trypanosoma congolense also exhibits density-dependent arrest and has the machinery for quorum sensing, which can functionally complement the equivalent molecules in T. brucei.
(d) . Indirect interactions
Infectious organisms may also interfere with each other's growth and establishment through indirect interactions involving modification of the host environment and competition for nutrients. Iron is a valuable commodity to compete for in co-infections and microbes have developed mechanisms to increase their share, for example, the deployment of iron-scavenging siderophores by bacterial and fungal pathogens. For Plasmodium, modified host iron responses can allow parasites that have established a blood-stage infection to prevent superinfection by a competitor species or strain [76]. This is possible because the blood-stage Plasmodium infection increases host production of the iron-regulatory hormone hepcidin in a density-dependent manner. This diverts iron away from the liver, impairing the liver-stage development of newly inoculated parasites, and making the host refractory to superinfection. Environmental factors can also modulate asexual growth and the generation of transmission stages in Plasmodium. In particular, P. falciparum (but not all Plasmodium species) monitor the presence of Lyso-phosphatidyl choline (LyosPC) within the infected mammalian host, with LysoPC repressing gametocyte formation. Upon LyosPC depletion with elevated infection levels, gametocyte formation is promoted [77]. Consequently, differing sensitivity or competition for LysoPC could potentially generate distinct probabilities of sexual maturation between strains, favouring relative virulence or transmission potential for competing genotypes in a co-infection.
Finally, pathogens can modify the host environment in diverse ways with consequences for neighbours locally and in distant niches within the host. For example, experimental murine infection revealed that P. chabaudi may diminish the barrier function of the intestinal wall resulting in enhanced translocation and dissemination of non-typhoidal Salmonella from the intestine. Correspondingly, Salmonella co-infection resulted in alterations in the immune response to P. chabaudi [78]. This is consistent with a pathological association between malaria and gastrointestinal disturbance involving non-typhoidal salmonella [79].
5. Evolutionary implications and areas that need development
In addition to field evaluations and experimental laboratory studies, insight into the dynamics of parasite co-infection benefit from modelling and theoretical approaches. Modelling can assist with the interpretation of spatial/geographical mapping to identify areas of high co-infection risk, as well as determining the strength of interactions between co-infecting parasites in vivo [80,81] and therapeutic impact [82]. Such data will become increasingly important in future, as climate change and human activities alter parasite and vector distributions, which may increase or reduce opportunities for species to interact. Single- and co-infection data from the field and multi-omic analyses in the laboratory can also be extended through mathematical models to predict the effects of co-infections.
More analyses and theoretical input are needed on the evolutionary consequences of co-infection. Simplistically, theory predicts that co-infection will favour the selection of parasites with increased virulence or heightened transmission potential [83]. There are data, however, which suggests that intermediate virulence levels may be optimal, as a trade-off between transmission and persistence [84]. Experimental evidence shows that virulent strains can have a competitive advantage, which can alter the distribution of less competitive strains. For example, a study of mixed strain P. chabaudi infections revealed that more virulent parasite strains had a competitive advantage, so that mixed infections could favour selection of yet more virulent parasites [85,86]. However, in the case of another rodent malaria parasite, P. yoelii, virulence was not linked to increased competitive success [87], indicating that within-host competition does not always select for more virulent parasites, as trade-offs come in to play [88]. Ultimately, the infection success of even the most virulent strains and species is governed by the composition of the infecting community within a host. Statistical models have illustrated positive and negative effects of different species combinations, while other experiments, using trematode parasites of amphibian hosts, showed that increased species richness diminished the infection success of the most virulent parasite species [80,89]. Which traits are selected for during within-host competition will vary in different contexts, depending upon the diverse biology of the hosts and infectious organisms involved in the interaction. Further, short-term experimental studies may not fully reflect the selective consequences of long-term coexistence between pathogens in the field.
It is clear that species composition can alter the infection success within a host and at the population level. Hence, more careful analyses are required before public health-and veterinary-interventions are undertaken in the field, given that a targeted approach against a particular parasite may alter the distribution and infection success of another species in co-infection scenarios. For example, malaria and lymphatic filariasis (LF) are co-endemic and transmitted by the same mosquito vector. Incorporating data into a susceptible-infected model indicated that the introduction of LF into a population reduced the prevalence of malaria [13]. Given these data, the authors caution against targeted interventions against LF through mass-drug administration, which may have unintended perverse effects such as an increase in the R0 of malaria. The predicted effects of different strategies to control animal African trypanosomiasis (AAT) were also recently explored using modelling approaches. These data indicated that insecticidal treatment of cattle alone could eliminate T. brucei from the local population, but that it would have little effect on two other species, namely T. congolense and T. vivax, which are maintained in reservoirs such as wildlife and small ruminants [90].
Simplistically, theory predicts that two parasites, which compete directly or indirectly, are incapable of occupying the same niche indefinitely [91]. Hence, niche adaptation and speciation as a result of co-infection and the resultant effects on parasite interactions warrants exploration. One species may actively exclude another from a particular host niche, or a species may exhibit avoidance behaviours to promote their fitness. This may underlie the tissue compartmentalization of different African trypanosome species, with T. brucei exhibiting tropism for the adipose tissue [92] and skin, whereas other African trypanosomes preferentially occupy other niches. The differential preference for red blood cell types may reflect a similar phenomenon in malaria species.
Finally, we have focused primarily on interactions between parasites during co-infection but the ability of a pathogen to colonize a host can be strongly influenced by the pre-existing community of non-pathogenic organisms within that host, adding complexity to the network of potential interactions. Although beyond what can be discussed here, the impact of the microbiome, for example, on the immune system of the hosts has clear relevance in the context of parasite infections in their hosts. This is true of malaria, where the gut microbiota influences disease severity caused by, and the immune response to, the parasite (e.g. [93–95]). A further particularly relevant example concerns trypanosome infections in the tsetse fly, where tsetse endosymbionts can have significant impact on the vectorial capacity of the arthropod [96]. The ability to manipulate both the microbiome in humans via probiotics [95] or tsetse endosymbionts through gene drive [97] or paratransgenesis [98] offers exciting possibility to control the pathology and transmission of each parasite.
6. Conclusion
In this review we have discussed, using trypanosome and Plasmodium parasites as exemplars, how an understanding of pathogen biology requires a broad analysis of the context of infection—particularly relating to competition and cooperation with other organisms. Traditionally, these interactions have been difficult to study comprehensively and pathogen research has focused on simple but unrepresentative one-host one-pathogen models. While informative and tractable, this inevitably omits the contribution of multiple other factors that can substantially impact the virulence and transmission of pathogens, and on the ability to control them immunologically or therapeutically. With the advent of high-resolution and high-throughput genomic and metagenomic surveillance and big data approaches to epidemiological, molecular and immunological study, multi-species analysis is now possible as is a dynamic study of interactions over time. Nonetheless, these methodological developments do not remove the value of well-controlled laboratory studies where complex interactions can be studied with limited variables. Fortunately, such experimental systems are now tractable for trypanosomes and Plasmodium parasites in particular, with the availability of molecular reporters for different parasite stages of development, antigenic expression or the dynamic monitoring of niche occupation. This represents a particularly exciting platform where the rigour of laboratory study can be combined with a wide bandwidth of information input, helping to deconvolve the contributions and interactions of many different components in a pathogen infection with closer proximity to the real world. Inevitably such studies will remain incomplete and oversimplified but they will provide the structural framework on which relevant parameters can be identified or tested for impact. The field of co-infection biology is one that particularly lends itself to an interdisciplinary approach. By working together epidemiologists, molecular biologists, modellers and evolutionary biologists can build a picture of where and how co-infections are negatively impacting human and animal health. Most importantly, through the combination of quantitative information and evolutionary theory, the consequences of the deployment of therapies can be better understood to avoid the perverse impact of controlling one pathogen while increasing the virulence, transmission or pathology of others. Indeed, drug development often focuses on a target organism without consideration of the wider impact on human and animal health over time, where vacant niches become occupied by new threats, or there is selection for enhanced transmission or virulence in related and unrelated pathogens. Understanding the interactions between the community of organisms that contribute to pathogen impact in the field is necessary if control approaches and therapies are to be sustained, economical and safe.
Supplementary Material
Acknowledgements
We thank Dr Petra Schneider, University of Edinburgh, UK, for comments on the manuscript.
Data accessibility
This article has no additional data.
Authors' contributions
F.V.: writing—original draft, writing—review and editing; K.R.M.: funding acquisition, writing—original draft, writing—review and editing; E.S.: writing—original draft, writing—review and editing. All authors gave final approval for publication and agreed to be held accountable for the work performed therein.
Competing interests
We declare we have no competing interests.
Funding
Work in K.R.M.'s laboratory is funded by the Wellcome Trust (grant nos. 221 717/Z/20/Z, 206815/Z/17/Z, 103740/Z/14/Z). F.V. is funded by a Wellcome Trust PhD studentship on the University of Edinburgh Wellcome Trust ‘Hosts, Pathogens and Global Health’ Programme (grant no. 108905/Z/15/Z).
References
- 1.Devi P, Khan A, Chattopadhyay P, Mehta P, Sahni S, Sharma S, Pandey R. 2021. Co-infections as Modulators of disease outcome: minor players or major players? Front. Microbiol. 12, 664386. ( 10.3389/fmicb.2021.664386) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rynkiewicz EC, Pedersen AB, Fenton A. 2015. An ecosystem approach to understanding and managing within-host parasite community dynamics. Trends Parasitol. 31, 212-221. ( 10.1016/j.pt.2015.02.005) [DOI] [PubMed] [Google Scholar]
- 3.Balmer O, Tanner M. 2011. Prevalence and implications of multiple-strain infections. Lancet Infect. Dis. 11, 868-878. ( 10.1016/S1473-3099(11)70241-9) [DOI] [PubMed] [Google Scholar]
- 4.Matthews KR. 2011. Controlling and coordinating development in vector-transmitted parasites. Science 331, 1149-1153. ( 10.1126/science.1198077) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rossati A, Bargiacchi O, Kroumova V, Zaramella M, Caputo A, Garavelli PL. 2016. Climate, environment and transmission of malaria. InfezMed 24, 93-104. [PubMed] [Google Scholar]
- 6.Cowman AF, Healer J, Marapana D, Marsh K. 2016. Malaria: biology and disease. Cell 167, 610-624. ( 10.1016/j.cell.2016.07.055) [DOI] [PubMed] [Google Scholar]
- 7.Giordani F, Morrison LJ, Rowan TG, De Koning HP, Barrett MP. 2016. The animal trypanosomiases and their chemotherapy: a review. Parasitology 143, 1862-1889. ( 10.1017/S0031182016001268) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kennedy PG. 2013. Clinical features, diagnosis, and treatment of human African trypanosomiasis (sleeping sickness). Lancet Neurol. 12, 186-194. ( 10.1016/S1474-4422(12)70296-X) [DOI] [PubMed] [Google Scholar]
- 9.De Clare Bronsvoort BM, et al. 2013. Design and descriptive epidemiology of the Infectious Diseases of East African Livestock (IDEAL) project, a longitudinal calf cohort study in western Kenya. BMC Vet. Res. 9, 171. ( 10.1186/1746-6148-9-171) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kasozi KI, et al. 2021. Epidemiology of Trypanosomiasis in wildlife-implications for humans at the wildlife interface in Africa. Front. Vet. Sci. 8, 621699. ( 10.3389/fvets.2021.621699) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Signaboubo D, Payne VK, Moussa IMA, Hassane HM, Berger P, Kelm S, Simo G. 2021. Diversity of tsetse flies and trypanosome species circulating in the area of Lake Iro in southeastern Chad. Parasites Vectors 14, 293. ( 10.1186/s13071-021-04782-7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kamdem CN, Tiofack AAZ, Mewamba EM, Ofon EA, Gomseu EBD, Simo G. 2020. Molecular identification of different trypanosome species in tsetse flies caught in the wildlife reserve of Santchou in the western region of Cameroon. Parasitol. Res. 119, 805-813. ( 10.1007/s00436-020-06606-6) [DOI] [PubMed] [Google Scholar]
- 13.Slater HC, Gambhir M, Parham PE, Michael E. 2013. Modelling co-infection with malaria and lymphatic filariasis. PLoS Comput. Biol. 9, e1003096. ( 10.1371/journal.pcbi.1003096) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Imwong M, Nakeesathit S, Day NP, White NJ. 2011. A review of mixed malaria species infections in anopheline mosquitoes. Malar. J. 10, 253. ( 10.1186/1475-2875-10-253) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Masiga DK, McNamara JJ, Laveissiere C, Truc P, Gibson WC. 1996. A high prevalence of mixed trypanosome infections in tsetse flies in Sinfra, Cote d'Ivoire, detected by DNA amplification. Parasitology 112, 75-80. ( 10.1017/S0031182000065094) [DOI] [PubMed] [Google Scholar]
- 16.Kubi C, Van den Abbeele J, Dorny P, Coosemans M, Marcotty T, Van den Bossche P. 2005. Ability of trypanosome-infected tsetse flies (Diptera: Glossinidae) to acquire an infection with a second trypanosome species. J. Med. Entomol. 42, 1035-1038. ( 10.1093/jmedent/42.6.1035) [DOI] [PubMed] [Google Scholar]
- 17.Aksoy S, Weiss BL, Attardo GM. 2014. Trypanosome transmission dynamics in Tsetse. Curr. Opin. Insect Sci. 3, 43-49. ( 10.1016/j.cois.2014.07.003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lord JS, Hargrove JW, Torr SJ, Vale GA. 2018. Climate change and African trypanosomiasis vector populations in Zimbabwe's Zambezi Valley: a mathematical modelling study. PLoS Med. 15, e1002675. ( 10.1371/journal.pmed.1002675) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Couper LI, et al. 2021. How will mosquitoes adapt to climate warming? Elife 10, e69630. ( 10.7554/eLife.69630) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pagabeleguem S, et al. 2016. Influence of temperature and relative humidity on survival and fecundity of three tsetse strains. Parasites Vectors 9, 520. ( 10.1186/s13071-016-1805-x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Longbottom J, Caminade C, Gibson HS, Weiss DJ, Torr S, Lord JS. 2020. Modelling the impact of climate change on the distribution and abundance of tsetse in Northern Zimbabwe. Parasites Vectors 13, 526. ( 10.1186/s13071-020-04398-3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Griffiths EC, Pedersen AB, Fenton A, Petchey OL. 2011. The nature and consequences of coinfection in humans. J. Infect. 63, 200-206. ( 10.1016/j.jinf.2011.06.005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Buck AA, Anderson RI, MacRae AA. 1978. Epidemiology of poly-parasitism. I. Occurrence, frequency and distribution of multiple infections in rural communities in Chad, Peru, Afghanistan, and Zaire. Tropenmed Parasitol 29, 61-70. [PubMed] [Google Scholar]
- 24.Pullan RL, Bethony JM, Geiger SM, Cundill B, Correa-Oliveira R, Quinnell RJ, Brooker S. 2008. Human Helminth co-infection: analysis of spatial patterns and risk factors in a Brazilian community. PLOS Negl. Trop. Dis. 2, e352. ( 10.1371/journal.pntd.0000352) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Viney ME, Graham AL. 2013. Patterns and processes in parasite co-infection. In Advances in parasitology, vol. 82 (ed. Rollinson D), pp. 321-369. San Diego, CA: Academic Press. [DOI] [PubMed] [Google Scholar]
- 26.Dwinger RH, Murray M, Luckins AG, Rae PF, Moloo SK. 1989. Interference in the establishment of tsetse-transmitted Trypanosoma congolense, T. brucei or T. vivax superinfections in goats already infected with T. congolense or T. vivax. Vet. Parasitol. 30, 177-189. ( 10.1016/0304-4017(89)90013-7) [DOI] [PubMed] [Google Scholar]
- 27.Morrison WI, Wells PW, Moloo SK, Paris J, Murray M. 1982. Interference in the establishment of superinfections with Trypanosoma congolense in cattle. J. Parasitol. 68, 755-764. ( 10.2307/3280980) [DOI] [PubMed] [Google Scholar]
- 28.Ademola IO, Odeniran PO. 2016. Co-infection with Plasmodium berghei and Trypanosoma brucei increases severity of malaria and trypanosomiasis in mice. Acta Trop. 159, 29-35. ( 10.1016/j.actatropica.2016.03.030) [DOI] [PubMed] [Google Scholar]
- 29.Sanches-Vaz M, Temporao A, Luis R, Nunes-Cabaco H, Mendes AM, Goellner S, Carvalho T, Figueiredo LM, Prudêncio M. 2019. Trypanosoma brucei infection protects mice against malaria. PLoS Pathog. 15, e1008145. ( 10.1371/journal.ppat.1008145) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tang J, Templeton TJ, Cao J, Culleton R. 2019. The consequences of mixed-species malaria parasite co-infections in mice and mosquitoes for disease severity, parasite fitness, and transmission success. Front. Immunol. 10, 3072. ( 10.3389/fimmu.2019.03072) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Christodoulou V, Messaritakis I, Svirinaki E, Tsatsanis C, Antoniou M. 2011. Leishmania infantum and Toxoplasma gondii: mixed infection of macrophages in vitro and in vivo. Exp. Parasitol. 128, 279-284. ( 10.1016/j.exppara.2011.02.022) [DOI] [PubMed] [Google Scholar]
- 32.Simpson JA, Silamut K, Chotivanich K, Pukrittayakamee S, White NJ. 1999. Red cell selectivity in malaria: a study of multiple-infected erythrocytes. Trans. R. Soc. Trop. Med. Hyg. 93, 165-168. ( 10.1016/S0035-9203(99)90295-X) [DOI] [PubMed] [Google Scholar]
- 33.Cressler CE, McLeod DV, Rozins C, Van Den Hoogen J, Day T. 2016. The adaptive evolution of virulence: a review of theoretical predictions and empirical tests. Parasitology 143, 915-930. ( 10.1017/S003118201500092X) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Frank SA. 1996. Models of parasite virulence. Q. Rev. Biol. 71, 37-78. ( 10.1086/419267) [DOI] [PubMed] [Google Scholar]
- 35.Brooker S, Akhwale W, Pullan R, Estambale B, Clarke SE, Snow RW, Clarke S. 2007. Epidemiology of plasmodium-helminth co-infection in Africa: populations at risk, potential impact on anemia, and prospects for combining control. Am. J. Trop. Med. Hyg. 77(Suppl. 6), 88-98. ( 10.4269/ajtmh.2007.77.88) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chao L, Hanley KA, Burch CL, Dahlberg C, Turner PE. 2000. Kin selection and parasite evolution: higher and lower virulence with hard and soft selection. Q. Rev. Biol. 75, 261-275. ( 10.1086/393499) [DOI] [PubMed] [Google Scholar]
- 37.Pinchbeck GL, Morrison LJ, Tait A, Langford J, Meehan L, Jallow S, Jallow A, Christley RM. 2008. Trypanosomosis in The Gambia: prevalence in working horses and donkeys detected by whole genome amplification and PCR, and evidence for interactions between trypanosome species. BMC Vet. Res. 4, 7. ( 10.1186/1746-6148-4-7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Balmer O, Stearns SC, Schotzau A, Brun R. 2009. Intraspecific competition between co-infecting parasite strains enhances host survival in African trypanosomes. Ecology 90, 3367-3378. ( 10.1890/08-2291.1) [DOI] [PubMed] [Google Scholar]
- 39.Mackinnon MJ, Marsh K. 2010. The selection landscape of malaria parasites. Science 328, 866-871. ( 10.1126/science.1185410) [DOI] [PubMed] [Google Scholar]
- 40.Wargo AR, de Roode JC, Huijben S, Drew DR, Read AF. 2007. Transmission stage investment of malaria parasites in response to in-host competition. Proc. R. Soc. B 274, 2629-2638. ( 10.1098/rspb.2007.0873) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schneider P, Greischar MA, Birget PLG, Repton C, Mideo N, Reece SE. 2018. Adaptive plasticity in the gametocyte conversion rate of malaria parasites. PLoS Pathog. 14, e1007371. ( 10.1371/journal.ppat.1007371) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bousema JT, Drakeley CJ, Mens PF, Arens T, Houben R, Omar SA, Gouagna LC, Schallig H, Sauerwein RW. 2008. Increased Plasmodium falciparum gametocyte production in mixed infections with P. malariae. Am. J. Trop. Med. Hyg. 78, 442-448. ( 10.4269/ajtmh.2008.78.442) [DOI] [PubMed] [Google Scholar]
- 43.Price R, Nosten F, Simpson JA, Luxemburger C, Phaipun L, ter Kuile F, Phaipun L, Luxemburger C, Simpson JA. 1999. Risk factors for gametocyte carriage in uncomplicated falciparum malaria. Am. J. Trop. Med. Hyg. 60, 1019-1023. ( 10.4269/ajtmh.1999.60.1019) [DOI] [PubMed] [Google Scholar]
- 44.Nacher M, Singhasivanon P, Silachamroon U, Treeprasertsu S, Krudsood S, Gay F, Mazier D, Looareesuwan S. 2001. Association of helminth infections with increased gametocyte carriage during mild falciparum malaria in Thailand. Am. J. Trop. Med. Hyg. 65, 644-647. ( 10.4269/ajtmh.2001.65.644) [DOI] [PubMed] [Google Scholar]
- 45.Pollitt LC, Bram JT, Blanford S, Jones MJ, Read AF. 2015. Existing infection facilitates establishment and density of malaria parasites in their mosquito vector. PLoS Pathog. 11, e1005003. ( 10.1371/journal.ppat.1005003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Saada EA, DeMarco SF, Shimogawa MM, Hill KL. 2015. ‘With a little help from my friends’: social motility in Trypanosoma brucei. PLoS Pathog. 11, e1005272. ( 10.1371/journal.ppat.1005272) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bargul JL, Jung J, McOdimba FA, Omogo CO, Adung'a VO, Kruger T, Masiga DK, Engstler M. 2016. Species-specific adaptations of trypanosome morphology and motility to the mammalian host. PLoS Pathog. 12, e1005448. ( 10.1371/journal.ppat.1005448) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schuster S, Kruger T, Subota I, Thusek S, Rotureau B, Beilhack A, Engstler M. 2017. Developmental adaptations of trypanosome motility to the tsetse fly host environments unravel a multifaceted in vivo microswimmer system. Elife 6, e27656. ( 10.7554/eLife.27656) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Van Den Abbeele J, Caljon G, De Ridder K, De Baetselier P, Coosemans M. 2010. Trypanosoma brucei modifies the tsetse salivary composition, altering the fly feeding behavior that favors parasite transmission. PLoS Pathog. 6, e1000926. ( 10.1371/journal.ppat.1000926) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bruce MC, Donnelly CA, Alpers MP, Galinski MR, Barnwell JW, Walliker D, Day KP. 2000. Cross-species interactions between malaria parasites in humans. Science 287, 845-848. ( 10.1126/science.287.5454.845) [DOI] [PubMed] [Google Scholar]
- 51.Mabbott NA. 2018. The influence of parasite infections on host immunity to co-infection with other pathogens. Front. Immunol. 9, 2579. ( 10.3389/fimmu.2018.02579) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wiedemann M, Voehringer D. 2020. Immunomodulation and immune escape strategies of gastrointestinal helminths and schistosomes. Front. Immunol. 11, 572865. ( 10.3389/fimmu.2020.572865) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Maizels RM, Smits HH, McSorley HJ. 2018. Modulation of host immunity by helminths: the expanding repertoire of parasite effector molecules. Immunity 49, 801-818. ( 10.1016/j.immuni.2018.10.016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Khan N, Gowthaman U, Pahari S, Agrewala JN. 2012. Manipulation of costimulatory molecules by intracellular pathogens: veni, vidi, vici!! PLoS Pathog. 8, e1002676. ( 10.1371/journal.ppat.1002676) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gibson W, Peacock L, Ferris V, Fischer K, Livingstone J, Thomas J, Bailey M. 2015. Genetic recombination between human and animal parasites creates novel strains of human pathogen. PLOS Negl. Trop. Dis. 9, e0003665. ( 10.1371/journal.pntd.0003665) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Weir W, et al. 2016. Population genomics reveals the origin and asexual evolution of human infective trypanosomes. Elife 5, e11473. ( 10.7554/eLife.11473) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tihon E, Imamura H, Dujardin JC, Van Den Abbeele J, Van den Broeck F. 2017. Discovery and genomic analyses of hybridization between divergent lineages of Trypanosoma congolense, causative agent of animal African trypanosomiasis. Mol. Ecol. 26, 6524-6538. ( 10.1111/mec.14271) [DOI] [PubMed] [Google Scholar]
- 58.Tibayrenc M, Ayala F. 2018. Hybridization in Trypanosoma congolense does not challenge the predominant clonal evolution model: a comment on Tihon et al., 2017. Mol. Ecol. 27, 3421-3424. ( 10.1111/mec.14714) [DOI] [PubMed] [Google Scholar]
- 59.Duffy CW, Morrison LJ, Black A, Pinchbeck GL, Christley RM, Schoenefeld A, Tait A, Turner CMR, Macleod A. 2009. Trypanosoma vivax displays a clonal population structure. Int. J. Parasitol. 39, 1475-1483. ( 10.1016/j.ijpara.2009.05.012) [DOI] [PubMed] [Google Scholar]
- 60.Gibson W. 2021. The sexual side of parasitic protists. Mol. Biochem. Parasitol. 243, 111371. ( 10.1016/j.molbiopara.2021.111371) [DOI] [PubMed] [Google Scholar]
- 61.Morlais I, et al. 2015. Plasmodium falciparum mating patterns and mosquito infectivity of natural isolates of gametocytes. PLoS ONE 10, e0123777. ( 10.1371/journal.pone.0123777) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Huijben S, Chan BHK, Nelson WA, Read AF. 2018. The impact of within-host ecology on the fitness of a drug-resistant parasite. Evol. Med. Public Health 2018, 127-137. ( 10.1093/emph/eoy016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wargo AR, Huijben S, de Roode JC, Shepherd J, Read AF. 2007. Competitive release and facilitation of drug-resistant parasites after therapeutic chemotherapy in a rodent malaria model. Proc. Natl Acad. Sci. USA 104, 19 914-19 919. ( 10.1073/pnas.0707766104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.von Wintersdorff CJ, Penders J, van Niekerk JM, Mills ND, Majumder S, van Alphen LB, Savelkoul PHM, Wolffs PFG. 2016. Dissemination of antimicrobial resistance in microbial ecosystems through horizontal gene transfer. Front. Microbiol. 7, 173. ( 10.3389/fmicb.2016.00173) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yaron S, Kolling GL, Simon L, Matthews KR. 2000. Vesicle-mediated transfer of virulence genes from Escherichia coli O157:H7 to other enteric bacteria. Appl. Environ. Microbiol. 66, 4414-4420. ( 10.1128/AEM.66.10.4414-4420.2000) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Szempruch AJ, et al. 2016. Extracellular vesicles from Trypanosoma brucei mediate virulence factor transfer and cause host anemia. Cell 164, 246-257. ( 10.1016/j.cell.2015.11.051) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Eliaz D, et al. 2017. Exosome secretion affects social motility in Trypanosoma brucei. PLoS Pathog. 13, e1006245. ( 10.1371/journal.ppat.1006245) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Regev-Rudzki N, et al. 2013. Cell-cell communication between malaria-infected red blood cells via exosome-like vesicles. Cell 153, 1120-1133. ( 10.1016/j.cell.2013.04.029) [DOI] [PubMed] [Google Scholar]
- 69.Mantel PY, et al. 2013. Malaria-infected erythrocyte-derived microvesicles mediate cellular communication within the parasite population and with the host immune system. Cell Host Microbe 13, 521-534. ( 10.1016/j.chom.2013.04.009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Correa R, Coronado L, Caballero Z, Faral P, Robello C, Spadafora C. 2019. Extracellular vesicles carrying lactate dehydrogenase induce suicide in increased population density of Plasmodium falciparum in vitro. Sci. Rep. 9, 5042. ( 10.1038/s41598-019-41697-x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Reuner B, Vassella E, Yutzy B, Boshart M. 1997. Cell density triggers slender to stumpy differentiation of Trypanosoma brucei bloodstream forms in culture. Mol. Biochem. Parasitol. 90, 269-280. ( 10.1016/S0166-6851(97)00160-6) [DOI] [PubMed] [Google Scholar]
- 72.Vassella E, Reuner B, Yutzy B, Boshart M. 1997. Differentiation of African trypanosomes is controlled by a density sensing mechanism which signals cell cycle arrest via the cAMP pathway. J. Cell Sci. 110, 2661-2671. ( 10.1242/jcs.110.21.2661) [DOI] [PubMed] [Google Scholar]
- 73.Rojas F, et al. 2019. Oligopeptide signaling through TbGPR89 drives trypanosome quorum sensing. Cell 176, 306-317; e16. ( 10.1016/j.cell.2018.10.041) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mony BM, MacGregor P, Ivens A, Rojas F, Cowton A, Young J, Horn D, Matthews K. 2014. Genome-wide dissection of the quorum sensing signalling pathway in Trypanosoma brucei. Nature 505, 681-685. ( 10.1038/nature12864) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Silvester E, Young J, Ivens A, Matthews KR. 2017. Interspecies quorum sensing in co-infections can manipulate trypanosome transmission potential. Nat. Microbiol. 2, 1471-1479. ( 10.1038/s41564-017-0014-5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Portugal S, et al. 2011. Host-mediated regulation of superinfection in malaria. Nat. Med. 17, 732-737. ( 10.1038/nm.2368) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Brancucci NMB, et al. 2017. Lysophosphatidylcholine regulates sexual stage differentiation in the human malaria parasite Plasmodium falciparum. Cell 171, 1532-1544; e15. ( 10.1016/j.cell.2017.10.020) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Alamer E, et al. 2019. Dissemination of non-typhoidal salmonella during Plasmodium chabaudi infection affects anti-malarial immunity. Parasitol. Res. 118, 2277-2285. ( 10.1007/s00436-019-06349-z) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sey ICM, Ehimiyein AM, Bottomley C, Riley EM, Mooney JP. 2020. Does malaria cause diarrhoea? A systematic review. Front. Med. (Lausanne). 7, 589379. ( 10.3389/fmed.2020.589379) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Telfer S, Lambin X, Birtles R, Beldomenico P, Burthe S, Paterson S, Begon M. 2010. Species interactions in a parasite community drive infection risk in a wildlife population. Science 330, 243-246. ( 10.1126/science.1190333) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Clark NJ, et al. 2020. Parasite associations predict infection risk: incorporating co-infections in predictive models for neglected tropical diseases. Parasites Vectors 13, 138. ( 10.1186/s13071-020-04016-2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Orwa TO, Mbogo RW, Luboobi LS. 2019. Multiple-strain malaria infection and its impacts on Plasmodium falciparum resistance to antimalarial therapy: a mathematical modelling perspective. Comput. Math. Methods Med. 2019, 9783986. ( 10.1155/2019/9783986) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bremermann HJ, Pickering J. 1983. A game-theoretical model of parasite virulence. J. Theor. Biol. 100, 411-426. ( 10.1016/0022-5193(83)90438-1) [DOI] [PubMed] [Google Scholar]
- 84.Levin S, Pimentel D. 1981. Selection of intermediate rates of increase in parasite-host systems. Am. Nat. 117, 308-315. ( 10.1086/283708) [DOI] [Google Scholar]
- 85.de Roode JC, et al. 2005. Virulence and competitive ability in genetically diverse malaria infections. Proc. Natl Acad. Sci. USA 102, 7624-7628. ( 10.1073/pnas.0500078102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bell AS, de Roode JC, Sim D, Read AF. 2006. Within-host competition in genetically diverse malaria infections: parasite virulence and competitive success. Evolution 60, 1358-1371. ( 10.1111/j.0014-3820.2006.tb01215.x) [DOI] [PubMed] [Google Scholar]
- 87.Abkallo HM, Tangena J-A, Tang J, Kobayashi N, Inoue M, Zoungrana A, Colegrave N, Culleton R. 2015. Within-host competition does not select for virulence in malaria parasites; studies with Plasmodium yoelii. PLoS Pathog. 11, e1004628. ( 10.1371/journal.ppat.1004628) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Schneider P, Reece SE. 2021. The private life of malaria parasites: strategies for sexual reproduction. Mol. Biochem. Parasitol. 244, 111375. ( 10.1016/j.molbiopara.2021.111375) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Johnson PT, Hoverman JT. 2012. Parasite diversity and coinfection determine pathogen infection success and host fitness. Proc. Natl Acad. Sci. USA 109, 9006-9011. ( 10.1073/pnas.1201790109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Odeniran PO, Onifade AA, MacLeod ET, Ademola IO, Alderton S, Welburn SC. 2020. Mathematical modelling and control of African animal trypanosomosis with interacting populations in West Africa-Could biting flies be important in maintaining the disease endemicity? PLoS ONE 15, e0242435. ( 10.1371/journal.pone.0242435) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hardin G. 1960. The competitive exclusion principle. Science 131, 1292-1297. ( 10.1126/science.131.3409.1292) [DOI] [PubMed] [Google Scholar]
- 92.Trindade S, et al. 2016. Trypanosoma brucei parasites occupy and functionally adapt to the adipose tissue in mice. Cell Host Microbe 19, 837-848. ( 10.1016/j.chom.2016.05.002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Villarino NF, et al. 2016. Composition of the gut microbiota modulates the severity of malaria. Proc. Natl Acad. Sci. USA 113, 2235-2240. ( 10.1073/pnas.1504887113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Mandal RK, Denny JE, Namazzi R, Opoka RO, Datta D, John CC, Schmidt NW. 2021. Dynamic modulation of spleen germinal center reactions by gut bacteria during Plasmodium infection. Cell Rep. 35, 109094. ( 10.1016/j.celrep.2021.109094) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Romoli O, Mancio-Silva L, Gendrin M. 2021. Linking microbiota composition with antimalarial antibody response. Trends Parasitol. 37, 853-855. ( 10.1016/j.pt.2021.07.007) [DOI] [PubMed] [Google Scholar]
- 96.Weiss BL, Wang J, Maltz MA, Wu Y, Aksoy S. 2013. Trypanosome infection establishment in the tsetse fly gut is influenced by microbiome-regulated host immune barriers. PLoS Pathog. 9, e1003318. ( 10.1371/journal.ppat.1003318) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Alam U, et al. 2011. Wolbachia symbiont infections induce strong cytoplasmic incompatibility in the tsetse fly Glossina morsitans. PLoS Pathog. 7, e1002415. ( 10.1371/journal.ppat.1002415) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Yang L, et al. 2021. Paratransgenic manipulation of a tsetse microRNA alters the physiological homeostasis of the fly's midgut environment. PLoS Pathog. 17, e1009475. ( 10.1371/journal.ppat.1009475) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This article has no additional data.