Abstract
Despite evidence of a positive effect of functional diversity on ecosystem productivity, the importance of functionally distinct species (i.e. species that display an original combination of traits) is poorly understood. To investigate how distinct species affect ecosystem productivity, we used a forest-gap model to simulate realistic temperate forest successions along an environmental gradient and measured ecosystem productivity at the end of the successional trajectories. We performed 10 560 simulations with different sets and numbers of species, bearing either distinct or indistinct functional traits, and compared them to random assemblages, to mimic the consequences of a regional loss of species. Long-term ecosystem productivity dropped when distinct species were lost first from the regional pool of species, under the harshest environmental conditions. On the contrary, productivity was more dependent on ordinary species in milder environments. Our findings show that species functional distinctiveness, integrating multiple trait dimensions, can capture species-specific effects on ecosystem productivity. In a context of an environmentally changing world, they highlight the need to investigate the role of distinct species in sustaining ecosystem processes, particularly in extreme environmental conditions.
Keywords: functional rarity, functional distinctiveness, biodiversity and ecosystem functioning, productivity, virtual ecology, forest-gap model
1. Introduction
Two decades of research have shown that ecosystem processes—such as productivity, nutrient cycling or temporal stability—depend on emergent properties of ecological communities, species number and functional diversity in particular [1–3]. This focus on community-aggregated properties tends to view the roles of individual species as idiosyncratic [4], or, when particular species are considered, the focus is put on dominant species only [5,6], which are seen as optimal phenotypes in a given environment [7]. As a consequence, there has been a blind spot regarding the contribution of species that have an original combination of functional traits, namely functionally distinct species [8].
Recent studies suggest that functionally distinct species can play important roles in the functioning of ecosystems, mediated by various mechanisms. First, they are likely to sustain functions that are not performed by other species [9], thus increasing the whole ecosystem functionality through complementarity mechanisms [1,10]. Second, they can increase the resistance of communities in response to environmental change by being adapted to a broader range of environmental conditions [8]. Third, they can contribute to lowering community-wide competition through greater trait dispersion [11]. However, empirical evidence supporting the importance of distinct species in regulating ecosystem functioning remains scarce [12,13], and the lack of experimental manipulation of the number and identity of functionally distinct species impedes a thorough exploration of their role in driving ecosystem dynamics and functioning.
The effects of biodiversity on ecosystem functioning depend on environmental conditions such as climate or soil [14–16], which have both direct impacts on plant physiology and indirect influence on community composition [17,18]. Changes in assembly rules and community composition along environmental gradients can impact ecosystem properties in various ways, including by affecting species interactions [19], or by modifying the identity of dominant species (‘mass-ratio effect’ [5,6,20]), which could be either functionally ordinary or distinct depending on the environment [8,21]. Whether and how much environmental conditions modulate the effects of distinct species on ecosystem productivity remains unexplored, partly because experiments manipulating the composition of communities on gradients at large scales can be difficult to perform. One way of overcoming this problem is to use simulation experiments [22], which can be performed over large spatial and temporal scales, and at the same time manipulate various parameters of interest.
Here, we used ForCEEPS (Forest Community Ecology and Ecosystem ProcesseS [23,24]), a process-based forest succession model that explicitly involves ecological processes, mainly succession in small patches (up to 1000 m2) and competition for light between trees. This model has several features that make it a useful simulation tool to test the effects of functionally distinct species on ecosystem functioning. First, the species in the model have functional trade-offs (e.g. between growth and tolerance to competition) calibrated to existing tree species [24–26], making it a realistic tool to compute distinctiveness in a multidimensional trait space. Second, this model has originally been developed to study successions independently from ecosystem functioning, and thus ecosystem properties are emergent properties that arise from the modelled forest dynamics and are not directly controlled in the simulations. ForCEEPS has successfully been applied to study biodiversity–ecosystem functioning theory [26,27] and to implement biodiversity loss experiments [28]. Third, its calibration and validation rely on predictions of both annual productivity (which reflects ecosystem energy and matter dynamics and is one of the most commonly measured ecosystem processes) and community composition (including species relative abundance in the long term) [24]. Fourth, it has been calibrated for a wide range of environmental conditions [24,29], which provides an excellent opportunity to investigate how the effects of distinct species will change along environmental gradients.
We used the ForCEEPS forest-gap model [24] to simulate communities undergoing 2000 years of succession from bare ground along an environmental gradient, and to measure ecosystem annual productivity at equilibrium. We initiated independent successions with varying species richness—from 30 to 1 tree species—to mimic the consequences of regional species loss. At each regional richness, we compared ecosystem productivity, measured in three scenarios (including only the most distinct species, the least distinct species or random assemblages), to test the following predictions:
-
(1)
The loss of functionally distinct species reduces ecosystem functioning in the long term. We expect ecosystem productivity to decrease faster when distinct species are lost from the regional pool first than in any other configuration (figure 1).
-
(2)
Environmental conditions modulate the effects of distinct species on ecosystem functioning. If so, support for prediction (1) depends on the environmental conditions across the 11 sites.
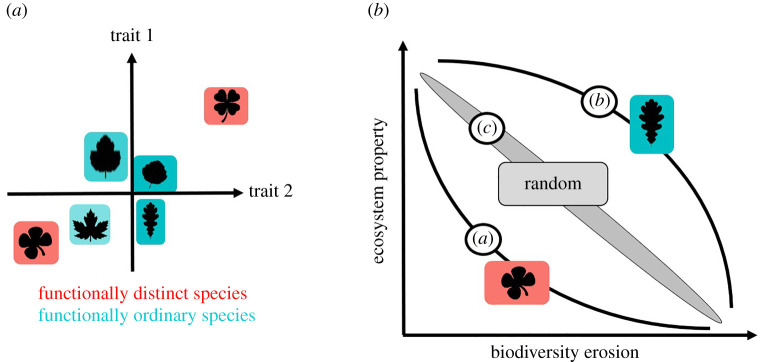
Figure 1.
Conceptual framework of the study (adapted from [8]). A species is schematically represented by a leaf. (a) Six species are located in a two-dimensional functional trait space. Ordinary species (blue background) are those located in the centre of the distribution in that space, whereas distinct species (red background, clover shape) are away from that centre. (b) Diagram showing the expected level of ecosystem property (in this study, productivity) as biodiversity declines, in the hypothesis that distinct phenotypes support important functions in the ecosystem. Orders of species loss are distinct first (a), ordinary first (b) or random (c). (Online version in colour.)
2. Methods
(a) . Forest succession model
We used the ForCEEPS forest-gap model (http://capsis.cirad.fr/capsis/help_en/forceeps) [24], which was developed on the Capsis modelling platform [30]. A forest-gap model simulates forest successions in small, independent patches of forest, by explicitly modelling the establishment, growth and mortality of tree individuals. This model relies on the same basic assumptions as the historical ForClim model [31,32]. First, the establishment and growth of individuals depend on the environment: bioclimatic conditions (temperature and water availability), soil nutrient content and browsing intensity [24]. Second, they are affected by biotic interactions that are implemented through competition for light. Finally, individual tree mortality is stochastic, with an increasing probability with age. A thorough description of the model is provided in electronic supplementary material, appendix S1, and more details on the model calibration and equations can be found in [23,24,26].
(b) . Species pool and species traits
We considered 30 forest tree species occurring in European mountains, whose behaviour is simulated by the model. The parameters describing species properties were calibrated on traits from forest inventories and from measures available from the literature, and take into account critical trade-offs in species biology (e.g. growth in full light/survival under shade) [24].
A set of 14 parameters was used to characterize each species (the meaning and values of parameters are given in the electronic supplementary material, table S1). Response-to-driver parameters mechanistically drive species establishment and survival (through response to water and nitrogen availability, browsing tolerance and temperature requirements), and intrinsic parameters determine species growth, competition for light, and succession dynamics (see electronic supplementary material, table S1). The congruence of these parameters with classical functional traits extracted from the literature was assessed in a previous study [24]. Because they are involved in mechanisms that functional traits approximate, and because they correlate with usual functional traits, these 14 parameters will be referred to as ‘traits’ hereafter.
(c) . Functional distinctiveness computation
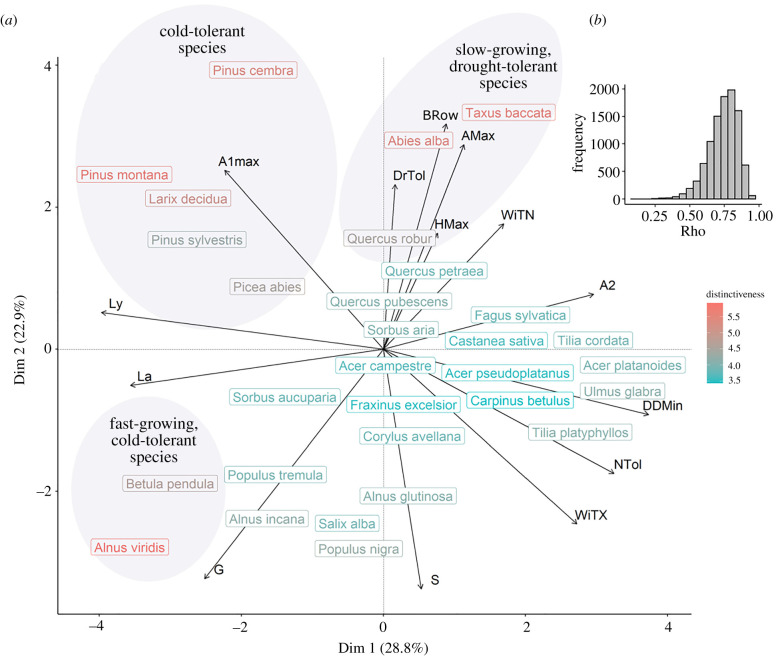
We ranked species according to their functional distinctiveness computed within the 30-species-rich regional pool of species. Functional distinctiveness is a measure of species originality in a multidimensional trait space, which can be performed irrespective of species abundance [33] (figure 1a). To avoid over-emphasizing a particular set of correlated traits (electronic supplementary material, figure S1), we first performed a principal component analysis on the fourteen traits and retained the first four axes that explained 72.2% of the total variance (figure 3a; electronic supplementary material, figure S2). We then computed Euclidean distances between species using their scores on these axes with the compute_dist_matrix() function from the funrar package v. 1.4.0 [33]. We used these distances to compute each species's functional distinctiveness, which is the average functional distance of a given species to all the other species, using the distinctiveness_com() function from the same package. We finally ranked species according to their distinctiveness value.
Figure 3.
Position of the species in the trait space and distinctiveness computation. (a): Position of the species on the two first axes of a PCA computed on ForCEEPS traits. Species are labelled. Their distinctiveness is coded by a gradient of colour, from blue (functionally ordinary species) to red (functionally distinct species). The 30% most distinct species are evidenced by three grey circles, and the name of strategies describing their trait combinations is given. (b): Sensitivity of distinctiveness ranking to the traits used. Traits were bootstrapped 10 000 times, and for each bootstrap, the new distinctiveness ranking was correlated with the one computed on all the traits, using Spearman's rank correlation coefficient. The distribution of rho is given (mean = 0.739, median = 0.747, s.d. = 0.096). Parameters are, in alphabetic order: Amax: maximum age (years); A1max and A2: crown size allometry parameters; Brown: Browsing susceptibility of seedlings (from 1, least susceptible, to 5, most susceptible); DDMin: minimal required annual degree-days sum (°C); DrTol: drought tolerance index (unitless, continuous from 0, sensitive, to 1, tolerant); G: optimal growth (unitless); HMax: maximum height (m); La: shade tolerance of adults (from 0, tolerant, to 1, sensitive); Ly: shade tolerance of seedlings (from 0, tolerant, to 1, sensitive); NTol: soil nitrogen requirements (from 1, weak requirements, to 5, strong requirements); S: allometry between diameter and height (unitless); WiTN: monthly minimum winter temperature tolerated for regeneration (°C); WiTX: monthly maximum winter temperature tolerated for regeneration (°C). (Online version in colour.)
We investigated the sensitivity of this ranking to the traits used through a bootstrap procedure. We did so by subsampling species trait values with replacement, recomputing the functional distinctiveness index for all species with the same procedure and correlating the new ranking of the species with the one computed on the 14 traits, using Spearman's rank correlation coefficient (figure 3b). The procedure was repeated 10 000 times.
(d) . Environmental gradient
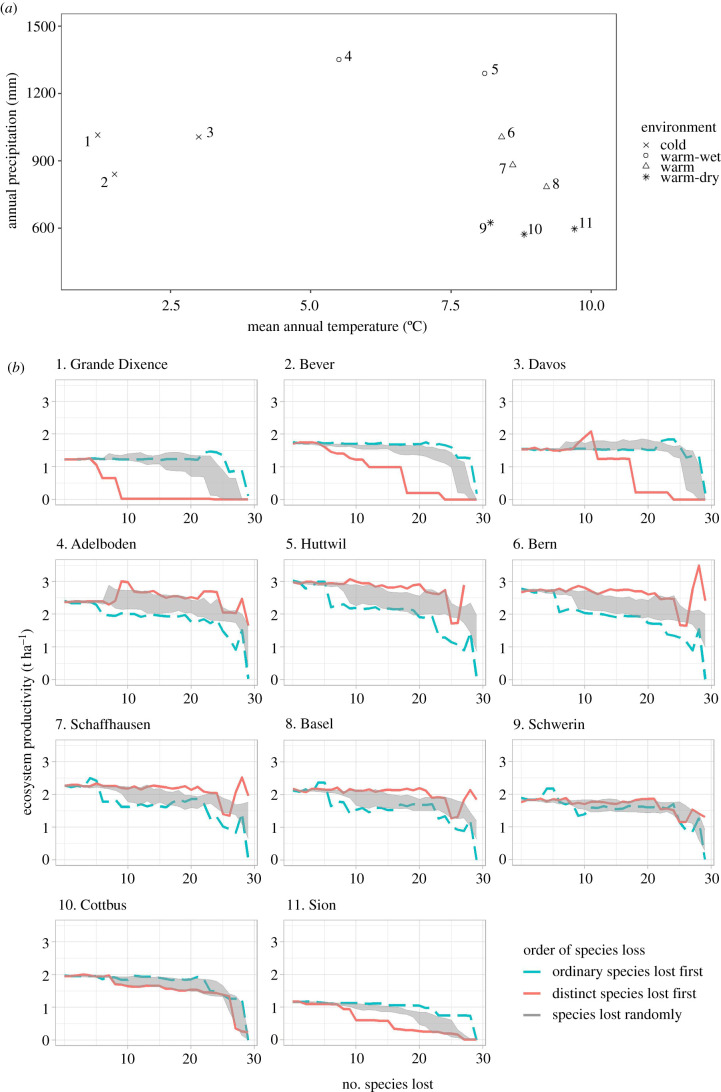
Relying on the ForClim model [26,32], from which ForCEEPS has been derived, the model was parameterized for 11 sites in Switzerland and Germany, distributed with a broad gradient of temperature and water availability (figure 4a). Each site was defined by its geographic position (latitude, longitude and elevation), temperature and annual precipitation, and was divided into 50 patches of 800 m2 each (4 ha per site in total). For each site, 2000-year-long fluctuations of climatic parameters were implemented based on historical records [25], providing climate data with inter-annual variability but with no general trend across the 2000 years (i.e. no climate change effect).
Figure 4.
Changes in productivity of the simulated forests caused by species loss in different environmental conditions. (a) The 11 sites are numbered and located by dots in a temperature/precipitation graph, and classified into four categories. (b) The consequences on ecosystem productivity of the loss of functionally distinct species (red continuous line), ordinary species (blue dashed line), or of random species losses (grey surface), are shown for each site, and the correspondence with site number in (a) is given. (Online version in colour.)
(e) . Effects of species distinctiveness on ecosystem productivity
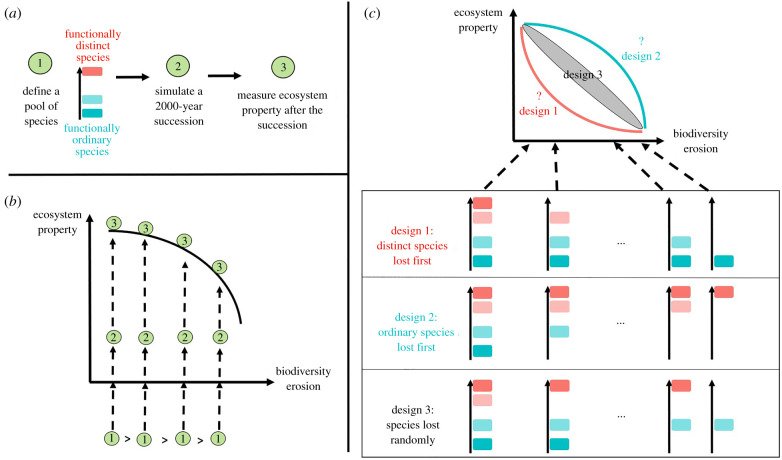
Across the environmental gradient, we generated scenarios in which the regional pool, composed of 30 European tree species, lost sequentially one species, then two, three, etc. These scenarios correspond to richness gradients (from 30 to one species), in which the most distinct or the most ordinary species were lost between each richness level of the gradient (figure 2). In each site, and for each richness gradient, we thus simulated 30 independent forest communities (one at each richness level), undergoing 2000 years of succession starting from bare ground, and measured ecosystem productivity in the last 1000 years (figure 2a,b). For the sake of clarity, the richness gradient scenario in which species were ranked from the most distinct to the most ordinary will be referred to as ‘distinct species lost first’ (and conversely, the other scenario will be referred to as ‘ordinary species lost first’, figure 2c). To compare the results to random expectations, we generated 30 random species richness gradients and implemented the same procedure, i.e. for each of the 30 random rankings, species were sequentially lost from the regional pool, and we simulated a community undergoing succession at each richness level. A total of 10 560 simulations were run for the 11 sites.
Figure 2.
Experimental design. (a) A simulation followed three steps. Species were ranked according to their distinctiveness, which is represented by a gradient of colours, from blue (ordinary species, bottom of the arrow) to red (distinct species, top of the arrow). (b) To implement biodiversity loss scenarios, simulations were made using several pools of species. Each pool on the x-axis is a subset of the pool located at its left (which is represented by the sign >). For each pool of species, a simulation was made and the ecosystem productivity was measured and represented on the y-axis. (c) The process was repeated for three designs. Design 3, in which species were lost randomly, was repeated 30 times to give a null distribution against which the results of designs 1 and 2 could be plotted. (Online version in colour.)
Since we started all simulations from bare ground, we measured ecosystem productivity after the community reached a pseudo-equilibrium for biomass and species composition (from year 1000 on), to avoid any effects due to transient dynamics [25] (electronic supplementary material, figure S3). After this 1000-year-long transient period, mean productivity was computed by averaging the productivity (accumulated biomass during a year) of 10 years sampled every 100 years (i.e. at year 1100, 1200, … , 2000), to minimize temporal autocorrelation [26,27]. A confidence interval of the productivity of the community at each species richness level was computed on the 30 random removal rankings. Since data were not normally distributed, we used a non-parametric approach to build a confidence interval of the median at each species richness level. This interval is not biased by the sample size: it covers the true median of a population using a subsample that depends on the population size (30 here) and on the confidence chosen (95% here) [26,34].
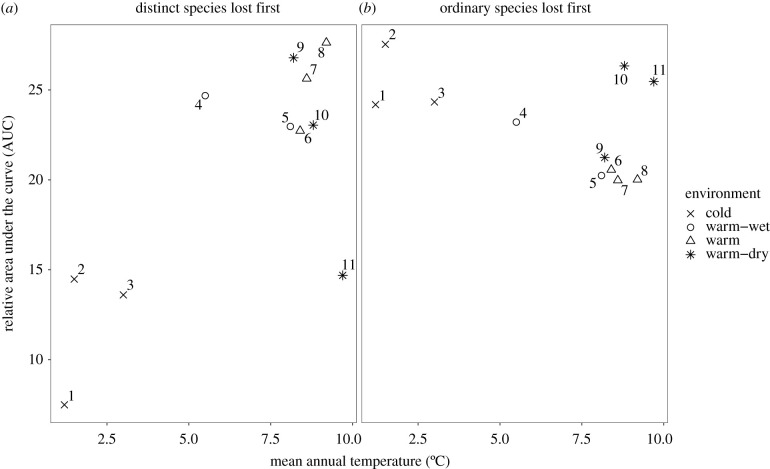
To summarize the effects of distinct and ordinary species in each site, we computed the area under the red curves (figure 4b) for scenario 1 (distinct species lost first) and under the blue curves for scenario 2 (ordinary species lost first). To allow intersite comparison, we then divided this area under the curve (AUC) by the productivity of the site computed when all the 30 species were present in the regional pool. We finally plotted the resulting relative area under the curve against site mean annual temperature to compare the sites.
Finally, to study the behaviour of species in the absence of interspecific interactions, we measured the biomass and productivity of each species in monoculture. We then tested potential links between species biomass and productivity in monoculture in each site and their distinctiveness using Spearman's rank correlation coefficient.
All analyses were performed using R v. 4.0.3 [35].
3. Results
The first two PCA axes represented 52% of total trait variance and were used to map the position of the 30 species in trait space (figure 3a). On the first axis, strategies ranged from shade-intolerant species (high values of shade-sensitivity traits Ly and La—see figure 3 for descriptions of traits) that were adapted to cold (low annual required degree-day sum DDMin; e.g. Pinus montana and Alnus viridis, figure 3a), to shade-tolerant, cold-sensitive, nutrient-demanding species (e.g. Ulmus glabra, Tilia platyphyllos). On the second axis, positive values corresponded to species with long lifespan and tall stature (high maximum age AMax and height HMax), tolerant to drought (DrTol trait), but sensitive to herbivory at a juvenile stage (Browsing trait; e.g. Abies alba, Taxus baccata). Species with negative values on the second axis included those sensitive to drought but not susceptible to herbivory (e.g. Populus nigra and Salix alba), and with a high growth speed (trait G), but a low maximum age and height (AMax and HMax). Many functionally ordinary species showed, as expected, intermediate values for most of the traits (e.g. Sorbus aria and Acer campestre). A portion of ordinary species was located towards positive values on the first dimension (e.g. Acer platanoides and Ulmus glabra) and the others towards negative values on the second dimension (e.g. Populus nigra and Salix alba). On the contrary, the 30% most distinct species were located in three peripheral zones of the trait space (figure 3a, grey circles). Species distinctiveness ranking computed after bootstrapping the traits correlated with that computed on all the traits, with an average Spearman's rho of 0.739 (figure 3b). Rho followed an unimodal distribution centred on rho = 0.747, with a standard deviation of 0.096.
Ecosystem productivity generally decreased with the number of species lost and varied with the environment (figure 4). Warm and wet sites (e.g. Adelboden, Huttwil) had the highest productivities, with values up to 3 t ha−1, whereas it did not exceed 1.5 to 2 t ha−1 in the cold sites (e.g. Grande Dixence), or warm and dry sites (e.g. Sion). When species were removed in random order (grey surface, figure 4), the productivity of the ecosystem either decreased steeply (in warm-wet sites, e.g. at Adelboden or Bern), or first remained constant and then decreased (e.g. in cold sites such as Bever or Cottbus). The effects of distinctiveness-driven species removal on ecosystem productivity varied across the environmental gradient. In the three coldest sites (Grande Dixence, Bever and Davos), productivity decreased more rapidly when distinct species were lost first (red curves, figure 4), than when species loss was random (grey surface) or when ordinary species were lost first (blue line). This was also the case for one warm-dry site, Sion. On the contrary, in three warm-wet sites (Adelboden, Huttwil, Bern), productivity decreased more rapidly when ordinary species were lost first (blue curves, figure 4). This trend was also apparent, but less significant, in warm sites with intermediate levels of rainfall (Schaffhausen, Basel). Finally, in the two remaining sites (Schwerin and Cottbus), there was no significant difference in productivity between species richness gradients. These results are summed up in figure 5: when distinct species were lost first, the cumulative productivity at all richness levels (standardized by site productivity) was indeed lower in the extreme sites (sites 1, 2, 3 and 11; figure 5a) than in the other sites. When ordinary species were lost first, on the contrary, cumulative productivity was slightly higher in these harsh sites than in other sites. Productivity was thus dependent on the presence of functionally distinct species in harsh (either cold or warm and dry) environments, whereas it depended more on ordinary species in milder conditions.
Figure 5.
Relative AUC of each scenario of species loss (distinct species lost first (a), or ordinary species lost first (b); cf. figure 2). For each site, in each of the two scenarios, relative AUC corresponds to the sum of the productivity of all the 30 simulations, divided by the productivity of the site when all the 30 species were present in the regional pool, to allow intersite comparison. Sites are numbered from 1 to 11 and classified into four categories, following figure 4.
Trends of decreasing productivity with biodiversity erosion were not monotonous. In some cases, the loss of one single species led to a strong decrease in ecosystem productivity—for instance when the fifth and sixth species (Pinus cembra and Larix decidua) were removed from the coldest site (Grande Dixence), the productivity dropped from about 1.2 t ha−1 year−1 to almost 0.6 (figure 4; see electronic supplementary material, table S1 for species distinctiveness ranking). In other cases, species removal led to an increase in ecosystem productivity, indicating the suppression of a negative interspecific interaction (e.g. when the ninth most distinct species, Picea abies, was lost from the wettest site, Adelboden, productivity increased from about 2.4 to 3 t ha−1 year−1, figure 4; electronic supplementary material, table S1). To study the behaviour of species across the environmental gradient in the absence of these interspecific interactions, we measured the productivity of each species grown in monoculture in each site. The number of species persisting in monoculture was smaller in the cold and warm-dry environments than in milder conditions (electronic supplementary material, figure S4), indicating a stronger abiotic filtering in harsh environmental conditions. Species that persisted in these sites belonged mostly to the 30% most distinct species. In addition, the correlation between species productivity in monoculture and their distinctiveness was significantly positive in the three coldest sites (Bever, Grande Dixence and Davos—table 1). In all the other sites, there was no correlation between species functional distinctiveness and either biomass or productivity.
Table 1.
Correlation between species distinctiveness and species biomass and productivity in monoculture for each site. Spearman's rank correlation coefficient, and the p-value of the corresponding test, are given for both biomass and productivity. Significant values are highlighted in italics. Sites are ordered as in figure 4.
| site | biomass |
productivity |
||
|---|---|---|---|---|
| correlation | p-value | correlation | p-value | |
| Grande Dixence | 0.62 | <0.01 | 0.6 | <0.01 |
| Bever | 0.52 | <0.01 | 0.61 | <0.01 |
| Davos | 0.49 | 0.01 | 0.47 | 0.01 |
| Adelboden | 0.29 | 0.12 | 0.13 | 0.5 |
| Huttwil | 0.07 | 0.71 | −0.14 | 0.47 |
| Bern | 0.12 | 0.52 | −0.15 | 0.42 |
| Schaffhausen | 0.01 | 0.97 | −0.15 | 0.44 |
| Basel | −0.02 | 0.93 | −0.19 | 0.31 |
| Schwerin | 0 | 0.99 | −0.07 | 0.7 |
| Cottbus | −0.11 | 0.56 | −0.15 | 0.42 |
| Sion | −0.11 | 0.55 | −0.13 | 0.51 |
4. Discussion
Although rarity is a long-studied attribute of species, the rarity of functions has received little attention [36] until recently [8], and no test of the effects of functionally distinct species on ecosystem properties has been performed so far. Our simulations of temperate forest ecosystems dynamics aimed at investigating distinct species' effects on ecosystem productivity along an environmental gradient. The loss of functionally distinct species from the regional pool strongly reduced ecosystem-level productivity in sites at the extremes of the environmental conditions.
The reasons for the strong effects of distinct species loss on ecosystem productivity in harsh (cold and warm-dry) environmental conditions could be that (i) they dominated the community by being the most abundant and productive species in such conditions [20], (ii) they provided a larger breadth of options to maintain productivity under annually fluctuating conditions [8] or (iii) they could maintain ecosystem productivity through interspecific interactions (i.e. via niche complementarity and/or facilitation) [4,37,38]. The latter, especially niche complementarity, could be expected since complex interspecific interactions have been shown to be central to the forest ecosystem dynamics simulated by ForCEEPS [26] and resulted in nonlinear, non-monotonous decrease of ecosystem productivity along the species richness gradient. When simulating monoculture experiments along the environmental gradient, we showed that distinct species remained the most productive in cold sites: their overall effect on ecosystem productivity could still be seen without biotic interactions. In cold sites, distinct species were the most abundant and productive of the community because of their response to the abiotic environment. On the contrary, at the other extreme of the environmental gradient (i.e. in warm-dry sites), although distinct species sustained productivity too, they were neither more abundant nor more productive than ordinary ones. We further performed a partitioning analysis of biodiversity effects (electronic supplementary material, appendix S3), which tends to show that distinct species might have sustained productivity in warm-dry sites through complementarity with the other species and not by being the dominant ones in such environments. Even if the relatively low number of sites along our environmental gradient does not allow for generalization, this result opens an interesting direction for future research. Species distinctiveness, measured on a multidimensional trait space involving traits linked to species, species growth, size and age, their ability to compete for light and tolerate shade, and their response to the abiotic environment, thus emerged as an integrated index indicating the ability of species to sustain ecosystem productivity in environments at the extremes of the gradient, although the mechanisms involved might vary depending on the environment.
Interestingly, our analysis revealed that, in the coldest sites, the system experienced sudden drops in productivity when few of the most distinct species went extinct. Few species were indeed both cold-adapted and productive, leading to low functional redundancy in the community. As functional redundancy can act as an insurance against the consequences of species losses [39–41], the loss of the few species displaying the adequate traits was not compensated for by the remaining species, which was sufficient to trigger abrupt changes and lose most of the productivity of the ecosystem. Even though functional redundancy is often measured on effect traits [41,42], and contrary to the tendency of biodiversity and ecosystem functioning studies to put the emphasis on grouping species according to their functional effect traits [43,44], our results also evidenced the primary importance of species response to the abiotic environment. Likely explanations for this bias of emphasize on effect traits can be that (1) measures in field experiments are made on plants that have already passed the abiotic filter, in which case effect traits are a more relevant grouping criteria than response traits, or (2) manipulative experiments are often made in homogeneous abiotic environments, which explains the emphasis put on effect traits (but see [45]). Prior to considering effect traits, assessing the diversity of response to the abiotic environment appears to be a necessary step [42]. The importance of environmental gradients and their interactions with species response traits in studying the effects of biodiversity on ecosystems cannot be overstated. Yet, since response and effect traits are difficult to disentangle in practice [46], how much the response of particular (here, functionally distinct) species can drive ecosystem properties can be fruitfully investigated along environmental gradients, as shown in this study.
The measure of distinctiveness is, by construction, relative to the traits included in the calculation [8,39]. Interestingly, distinctiveness rankings computed after bootstraps significantly correlated with the distinctiveness ranking computed on all the traits. The scores on the main PCA axes used to compute distinctiveness were thus robust to bootstrapping because several traits contributed to each of the main axes, indicating that distinctiveness informed on phenotypes integrated in a multi-trait space. When using traits from the TRY database [47], the distinctiveness ranking computed on traits linked to growth and leaf economics strategy (specific leaf area, nitrogen content and plant height) correlated with the ranking computed on the 14 parameters of the model used as traits, giving confidence in the robustness of this metrics (see electronic supplementary material, analysis). Yet, the correlation was not significant when distinctiveness was computed on the six traits used by Díaz et al. [48] (electronic supplementary material, appendix S4 and figure S6). This is not surprising, since these six traits were chosen by the authors to maximize the dispersion of species in a multivariate analysis and are thus likely not to reflect with accuracy the ecological processes, such as growth or competition for light, that are modelled in ForCEEPS. More broadly, our results should, of course, be taken with care, since the present study relies on simulations from a model, which cannot consider all the processes operating in nature. First, many mechanisms not included in the model could mediate an effect of distinct species on ecosystem properties. For instance, in addition to complementarity in the access to light, distinct species can be involved in other mechanisms (e.g. plant-soil feedbacks), which are not included in the model, but which may have an effect on ecosystem processes such as nutrient cycling [16,49] (e.g. nitrogen-fixing plants should be distinct and should affect nitrogen cycle more than other species via their interactions with soil bacteria). Second, the distinctiveness index was computed at a regional scale, with a limited set of species implemented in the model (e.g. many shrubs are absent from ForCEEPS). Although this enabled us to evidence that functionally distinct species were driving ecosystem productivity in the extremes of a regional climatic gradient, and that this effect was independent from biotic interactions in cold sites (where the effect was the strongest), computing distinctiveness at a local scale (which should be done only when realized species richness is high enough for this index to be meaningful, electronic supplementary material, figure S4) may be a way to explore potential roles of distinct species mediated by complementarity in resource use [1,10], or by a reduction of competition through trait dispersion [11]. At a local scale (i.e. that of realized community), distinctiveness ranking might or might not be correlated to that computed at the regional scale, depending on the number of species persisting and the functional diversity of the realized community [50]. Overall, depending on the question and the mechanisms implemented in the models, simulation experiments can be powerful tools to generate predictions and hypotheses based on a mechanistic examination of ecological systems [22,51] and can pave the way for subsequent hypothesis-driven empirical tests.
5. Conclusion
Relying on a forest-gap model, we found that functional distinctiveness, a measure of the originality of a phenotype, can be linked to its role in ecosystem functioning. In particular, at the regional scale, we showed that functionally distinct species’ response to the abiotic environment enabled them to sustain ecosystem productivity in harsh conditions, whereas productivity depended more on functionally ordinary species in milder conditions. If distinct species appear to be vulnerable to extinction, they should be considered in conservation plans aiming at maintaining ecosystem functioning and services in an environmentally changing world.
Supplementary Material
Acknowledgement
We thank Nicolas Beudez for his help with the Capsis simulation platform.
Data accessibility
The data are provided in the electronic supplementary material [52].
Authors' contributions
L.D.: conceptualization, data curation, formal analysis, investigation, methodology, validation, visualization, writing—original draft and writing—review and editing; P.G.: conceptualization, formal analysis, methodology, project administration, resources, supervision, validation, visualization and writing—review and editing; W.T.: conceptualization, formal analysis, methodology, project administration, validation, visualization and writing—review and editing; N.M.: formal analysis, methodology, validation, visualization and writing—review and editing; D.M.: formal analysis, validation, visualization and writing—review and editing; M.C.: formal analysis, validation and writing—review and editing; F.M.: conceptualization, validation and writing—review and editing; P.D.: validation and writing—review and editing; N.L.: validation and writing—review and editing; X.M.: conceptualization, formal analysis, methodology, project administration, resources, software, supervision, validation, visualization and writing—review and editing; C.V.: conceptualization, formal analysis, funding acquisition, methodology, project administration, supervision, visualization and writing—review and editing. All authors gave final approval for publication and agreed to be held accountable for the work performed therein.
Competing interests
We declare we have no competing interests.
Funding
This research is supported by the Fondation pour la Recherche sur la Biodiversité (FRB) and Electricité de France (EDF) in the context of the CESAB project ‘Causes and consequences of functional rarity from local to global scales' (FREE). C.V. was supported by the European Research Council (ERC) Starting Grant Project ‘Ecophysiological and biophysical constraints on domestication in crop plants’ (grant ERC-StG-2014-639706-CONSTRAINTS). The TRY database is hosted at the Max Planck Institute for Biogeochemistry (Jena, Germany) and supported by DIVERSITAS/Future Earth, the German Centre for Integrative Biodiversity Research (iDiv) Halle-Jena-Leipzig and BACI (grant ID 640176).
References
- 1.Hooper DU, et al. 2005. Effects of biodiversity on ecosystem functioning: a consensus of current knowledge. Ecol. Monogr. 75, 3-35. ( 10.1890/04-0922) [DOI] [Google Scholar]
- 2.Loreau M. 2010. Linking biodiversity and ecosystems: towards a unifying ecological theory. Phil. Trans. R. Soc. B 365, 49-60. ( 10.1098/rstb.2009.0155) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mouillot D, Villéger S, Scherer-Lorenzen M, Mason NWH. 2011. Functional structure of biological communities predicts ecosystem multifunctionality. PLoS ONE 6, e17476. ( 10.1371/journal.pone.0017476) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Díaz S, Lavorel S, deBello F, Quétier F, Grigulis K, Robson TM. 2007. Incorporating plant functional diversity effects in ecosystem service assessments. Proc. Natl Acad Sci. USA 104, 20 684-20 689. ( 10.1073/pnas.0704716104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Garnier E, et al. 2004. Plant functional markers capture ecosystem properties during secondary succession. Ecology 85, 2630-2637. ( 10.1890/03-0799) [DOI] [Google Scholar]
- 6.Grime JP. 1998. Benefits of plant diversity to ecosystems: immediate, filter and founder effects. J. Ecol. 86, 902-910. ( 10.1046/j.1365-2745.1998.00306.x) [DOI] [Google Scholar]
- 7.Denelle P, Violle C, Munoz F. 2019. Distinguishing the signatures of local environmental filtering and regional trait range limits in the study of trait–environment relationships. Oikos 128, 960-971. ( 10.1111/oik.05851) [DOI] [Google Scholar]
- 8.Violle C, Thuiller W, Mouquet N, Munoz F, Kraft NJB, Cadotte MW, Livingstone SW, Mouillot D. 2017. Functional rarity: the ecology of outliers. Trends Ecol. Evol. 32, 356-367. ( 10.1016/j.tree.2017.02.002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jaillard B, Deleporte P, Isbell F, Loreau M, Violle C. 2021. Consistent functional clusters explain the effects of biodiversity on ecosystem productivity in a long-term experiment. Ecology 102, e03441. [DOI] [PubMed] [Google Scholar]
- 10.Loreau M. 1998. Biodiversity and ecosystem functioning: a mechanistic model. Proc. Natl Acad. Sci. USA 95, 5632-5636. ( 10.1073/pnas.95.10.5632) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mahaut L, Fort F, Violle C, Freschet GT. 2020. Multiple facets of diversity effects on plant productivity: species richness, functional diversity, species identity and intraspecific competition. Funct. Ecol. 34, 287-298. ( 10.1111/1365-2435.13473) [DOI] [Google Scholar]
- 12.Le Bagousse-Pinguet Y, et al. 2021. Functional rarity and evenness are key facets of biodiversity to boost multifunctionality. Proc. Natl Acad. Sci. USA 118, e2019355118. ( 10.1073/pnas.2019355118) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maire E, et al. 2018. Community-wide scan identifies fish species associated with coral reef services across the Indo-Pacific. Proc. R. Soc. B 285, 20181167. ( 10.1098/rspb.2018.1167) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cardinale BJ, Nelson K, Palmer MA. 2000. Linking species diversity to the functioning of ecosystems: on the importance of environmental context. Oikos 91, 175-183. ( 10.1034/j.1600-0706.2000.910117.x) [DOI] [Google Scholar]
- 15.Jing X, et al. 2015. The links between ecosystem multifunctionality and above- and belowground biodiversity are mediated by climate. Nat. Commun. 6, 1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martinez-Almoyna C, Thuiller W, Chalmandrier L, Ohlmann M, Foulquier A, Clément JC, Zinger L, Münkemüller T. 2019. Multi-trophic β-diversity mediates the effect of environmental gradients on the turnover of multiple ecosystem functions. Funct. Ecol. 33, 2053-2064. ( 10.1111/1365-2435.13393) [DOI] [Google Scholar]
- 17.Hillebrand H, Matthiessen B. 2009. Biodiversity in a complex world: consolidation and progress in functional biodiversity research. Ecol. Lett. 12, 1405-1419. ( 10.1111/j.1461-0248.2009.01388.x) [DOI] [PubMed] [Google Scholar]
- 18.Morin X, Fahse L, Jactel H, Scherer-Lorenzen M, García-Valdés R, Bugmann H. 2018. Long-term response of forest productivity to climate change is mostly driven by change in tree species composition. Sci. Rep. 8, 5627. ( 10.1038/s41598-018-23763-y) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fugère V, Andino P, Espinosa R, Anthelme F, Jacobsen D, Dangles O. 2012. Testing the stress-gradient hypothesis with aquatic detritivorous invertebrates: insights for biodiversity-ecosystem functioning research. J. Anim. Ecol. 81, 1259-1267. ( 10.1111/j.1365-2656.2012.01994.x) [DOI] [PubMed] [Google Scholar]
- 20.Vile D, Shipley B, Garnier E. 2006. Ecosystem productivity can be predicted from potential relative growth rate and species abundance. Ecol. Lett. 9, 1061-1067. ( 10.1111/j.1461-0248.2006.00958.x) [DOI] [PubMed] [Google Scholar]
- 21.Jain M, et al. 2014. The importance of rare species: a trait-based assessment of rare species contributions to functional diversity and possible ecosystem function in tall-grass prairies. Ecol. Evol. 4, 104-112. ( 10.1002/ece3.915) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zurell D, et al. 2010. The virtual ecologist approach: simulating data and observers. Oikos 119, 622-635. ( 10.1111/j.1600-0706.2009.18284.x) [DOI] [Google Scholar]
- 23.Morin X, Damestoy T, Toigo M, Castagneyrol B, Jactel H, de Coligny F, Meredieu C. 2020. Using forest gap models and experimental data to explore long-term effects of tree diversity on the productivity of mixed planted forests. Ann. Forest Sci. 77, 1-9. ( 10.1007/s13595-020-00954-0) [DOI] [Google Scholar]
- 24.Morin X, et al. 2021. Beyond forest succession: a gap model to study ecosystem functioning and tree community composition under climate change. Func. Ecol. 35, 955-975. ( 10.1111/1365-2435.13760) [DOI] [Google Scholar]
- 25.Chauvet M, Kunstler G, Roy J, Morin X. 2017. Using a forest dynamics model to link community assembly processes and traits structure. Funct. Ecol. 31, 1452-1461. ( 10.1111/1365-2435.12847) [DOI] [Google Scholar]
- 26.Morin X, Fahse L, Scherer-Lorenzen M, Bugmann H. 2011. Tree species richness promotes productivity in temperate forests through strong complementarity between species. Ecol. Lett. 14, 1211-1219. ( 10.1111/j.1461-0248.2011.01691.x) [DOI] [PubMed] [Google Scholar]
- 27.Morin X, Fahse L, de Mazancourt C, Scherer-Lorenzen M, Bugmann H. 2014. Temporal stability in forest productivity increases with tree diversity due to asynchrony in species dynamics. Ecol. Lett. 17, 1526-1535. ( 10.1111/ele.12357) [DOI] [PubMed] [Google Scholar]
- 28.García-Valdés R, Bugmann H, Morin X. 2018. Climate change-driven extinctions of tree species affect forest functioning more than random extinctions. Divers. Distrib. 24, 906-918. ( 10.1111/ddi.12744) [DOI] [Google Scholar]
- 29.Gauzere P, Morin X, Violle C, Caspeta I, Ray C, Blonder B. 2020. Vacant yet invasible niches in forest community assembly. Funct. Ecol. 34, 1945-1955. ( 10.1111/1365-2435.13614) [DOI] [Google Scholar]
- 30.Dufour-Kowalski S, Courbaud B, Dreyfus P, Meredieu C, de Coligny F. 2012. Capsis: an open software framework and community for forest growth modelling. Ann. Forest Sci. 69, 221-233. ( 10.1007/s13595-011-0140-9) [DOI] [Google Scholar]
- 31.Bugmann HKM. 1996. A simplified forest model to study species composition along climate gradients. Ecology 77, 2055-2074. ( 10.2307/2265700) [DOI] [Google Scholar]
- 32.Didion M, Kupferschmid AD, Zingg A, Fahse L, Bugmann H. 2009. Gaining local accuracy while not losing generality—extending the range of gap model applications. Can. J. For. Res. 39, 1092-1107. ( 10.1139/X09-041) [DOI] [Google Scholar]
- 33.Grenié M, Denelle P, Tucker CM, Munoz F, Violle C. 2017. funrar: An R package to characterize functional rarity. Divers. Distrib. 23, 1365-1371. ( 10.1111/ddi.12629) [DOI] [Google Scholar]
- 34.Rice JA. 2006. Mathematical statistics and data analysis. Belmont, CA: Cengage Learning. [Google Scholar]
- 35.R Core Team. 2020. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing. [Google Scholar]
- 36.Pavoine S, Ollier S, Dufour AB. 2005. Is the originality of a species measurable? Ecol. Lett. 8, 579-586. ( 10.1111/j.1461-0248.2005.00752.x) [DOI] [Google Scholar]
- 37.Barry KE, et al. 2019. The future of complementarity: disentangling causes from consequences. Trends Ecol. Evol. 34, 167-180. ( 10.1016/j.tree.2018.10.013) [DOI] [PubMed] [Google Scholar]
- 38.Turnbull LA, Isbell F, Purves DW, Loreau M, Hector A. 2016. Understanding the value of plant diversity for ecosystem functioning through niche theory. Proc. R. Soc. B 283, 20160536. ( 10.1098/rspb.2016.0536) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Grenié M, Mouillot D, Villéger S, Denelle P, Tucker CM, Munoz F, Violle C. 2018. Functional rarity of coral reef fishes at the global scale: hotspots and challenges for conservation. Biol. Conserv. 226, 288-299. ( 10.1016/j.biocon.2018.08.011) [DOI] [Google Scholar]
- 40.McLean M, Auber A, Graham NAJ, Houk P, Villéger S, Violle C, Thuiller W, Wilson SK, Mouillot D. 2019. Trait structure and redundancy determine sensitivity to disturbance in marine fish communities. Glob. Change Biol. 25, 3424-3437. ( 10.1111/gcb.14662) [DOI] [PubMed] [Google Scholar]
- 41.Mori AS, et al. 2015. Functional redundancy of multiple forest taxa along an elevational gradient: predicting the consequences of non-random species loss. J. Biogeogr. 42, 1383-1396. ( 10.1111/jbi.12514) [DOI] [Google Scholar]
- 42.Laliberté E, et al. 2010. Land-use intensification reduces functional redundancy and response diversity in plant communities. Ecol. Lett. 13, 76-86. ( 10.1111/j.1461-0248.2009.01403.x) [DOI] [PubMed] [Google Scholar]
- 43.Díaz S, Cabido M. 2001. Vive la différence: plant functional diversity matters to ecosystem processes. Trends Ecol. Evol. 16, 646-655. ( 10.1016/S0169-5347(01)02283-2) [DOI] [Google Scholar]
- 44.Lavorel S, Garnier E. 2002. Predicting changes in community composition and ecosystem functioning from plant traits: revisiting the Holy Grail. Funct. Ecol. 16, 545-556. ( 10.1046/j.1365-2435.2002.00664.x) [DOI] [Google Scholar]
- 45.Hector A, et al. 1999. Plant diversity and productivity experiments in European grasslands. Science 286, 1123-1127. ( 10.1126/science.286.5442.1123) [DOI] [PubMed] [Google Scholar]
- 46.Garnier E, Navas ML, Grigulis K. 2016. Plant functional diversity: organism traits, community structure, and ecosystem properties. Oxford, UK: Oxford University Press. [Google Scholar]
- 47.Kattge J, et al. 2020. TRY plant trait database–enhanced coverage and open access. Glob. Change Biol. 26, 119-188. ( 10.1111/gcb.14904) [DOI] [PubMed] [Google Scholar]
- 48.Díaz S, et al. 2016. The global spectrum of plant form and function. Nature 529, 167-171. ( 10.1038/nature16489) [DOI] [PubMed] [Google Scholar]
- 49.Cheeke TE, Phillips RP, Brzostek ER, Rosling A, Bever JD, Fransson P. 2017. Dominant mycorrhizal association of trees alters carbon and nutrient cycling by selecting for microbial groups with distinct enzyme function. New Phytol. 214, 432-442. ( 10.1111/nph.14343) [DOI] [PubMed] [Google Scholar]
- 50.Gaüzère P, et al. Submitted. The functional trait distinctiveness of plant species is scale dependent.
- 51.Maréchaux I, et al. 2021. Tackling unresolved questions in forest ecology: The past and future role of simulation models. Ecol. Evol. 11, 3746-3770. ( 10.1002/ece3.7391) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Delalandre L, et al. 2022. Functionally distinct tree species support long-term productivity in extreme environments. FigShare . [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Delalandre L, et al. 2022. Functionally distinct tree species support long-term productivity in extreme environments. FigShare . [DOI] [PMC free article] [PubMed]
Supplementary Materials
Data Availability Statement
The data are provided in the electronic supplementary material [52].