Abstract
Isolates of enterohemorrhagic Escherichia coli (EHEC) of serotype O104:H21 implicated in a 1994 outbreak of hemorrhagic colitis in Montana were analyzed for the presence of trait EHEC virulence markers. By using a multiplex PCR that specifically amplifies several genes, the O104:H21 strains were found to carry only the Shiga toxin 2 gene (stx2) and to express Stx2. They did not have the eaeA gene for γ-intimin, which is typically found in O157:H7, or the α- or β-intimin derivatives, which are common in other EHEC and enteropathogenic E. coli serotypes. Results of the multiplex PCR also indicated that the ehxA gene for enterohemolysin was absent from O104:H21. This, however, was not consistent with the results of a phenotypic assay that showed them to be hemolytic or a PCR analysis with another set of ehxA-specific primers, which indicated the presence of ehxA. To resolve this discrepancy, the ehxA region in O104:H21 and O157:H7 strains, to which the multiplex PCR primers anneal, was cloned and sequenced. Comparison of the sequences showed that the upstream primer binding site in the ehxA gene of O104:H21 was not identical to that of O157:H7. Specifically, there were several base mutations, including an A-to-G substitution at the 3′ end of the primer binding site. These base mutations are presumably not unique to O104:H21, since other enterohemolytic serotypes were also not detected with the ehxA primers used in the multiplex PCR. Comparison of the ehxA sequences of O104:H21 strains with those of other Stx-producing E. coli strains showed that they more closely resembled those of O8:H19 strains, which have cluster II ehxA genes, than those of O157:H7 strains, which have cluster I ehxA sequences. By modifying the upstream ehxA primer, the multiplex PCR was able to detect ehxA genes in both O157:H7 and O104:H21 strains.
Symptoms of food-borne infections with enterohemorrhagic Escherichia coli (EHEC) can range from mild diarrhea to bloody diarrhea or hemorrhagic colitis (HC), which may progress to the life-threatening hemolytic-uremic syndrome (HUS). EHEC strains are distinguished from the other pathogenic E. coli groups by their unique virulence factors. These include the production of Shiga toxins (Stxs); the presence of the locus for the enterocyte effacement island, which carries the eaeA gene (which encodes the intimin protein essential for cellular attachment), and the presence of a 60-MDa plasmid (pO157) that carries the ehxA (EHEC hlyA) gene that encodes EHEC hemolysin or enterohemolysin. Although Stx is considered the primary virulence factor, in the more severe forms of EHEC infections, it appears that combinations of these factors are required for full virulence (2, 19).
EHEC serotype O157:H7 strains account for most of the food-borne outbreaks worldwide; however, non-O157:H7 serotypes are also implicated in illness (17, 19). In the United States, food-borne outbreaks caused by non-O157:H7 strains are infrequent. However, this may in part be attributed to the fact that most detection methods used are specific for the O157:H7 serotype. Still, few outbreaks caused by non-O157:H7 strains have occurred, such as the 1994 outbreak in Montana, in which serotype O104:H21 was implicated. In that incident, there were 11 confirmed cases and 7 suspected cases of infection, with 16 of the patients having hemorrhagic colitis. It was speculated that none of the cases progressed to HUS because those infected were 8 years of age or older and therefore were not as susceptible as the younger populations (18). Consumption of contaminated pasteurized milk was suspected but not confirmed, as O104:H21 was isolated only from the stool samples of infected patients and not from the product (10). Characterization of the O104:H21 isolates showed that all had identical patterns on pulsed-field gel electrophoresis, produced only Stx2, and did not ferment sorbitol. The latter phenotype actually enabled their isolation, since sorbitol-containing medium was used in the analysis. Previously, an O104:H2 serotype that produced only Stx2 and that did not have ehxA or eaeA genes was implicated in a HUS infection in the United Kingdom (28). In Germany, an O104:H− strain isolated from a patient with HUS was also found to produced only Stx2, but the strain was not tested for other virulence markers (6). Since Stx2 was also the only virulence factor reported for the O104:H21 isolates from the Montana outbreak, we examined these strains for the presence of other EHEC virulence factors.
MATERIALS AND METHODS
Bacterial strains.
Isolates of the O104:H21 serotype, strains G5506, G5507, and G5508, were obtained from the Centers for Disease Control and Prevention, Atlanta, Ga. Strain 35150, an O157:H7 serotype strain that produces both Stx1 and Stx2, was obtained from the American Type Culture Collection, Manassas, Va. Strain E2348/69, an enteropathogenic E. coli (EPEC) strain of the O127:H6 serotype, was obtained from T. Whittam, Pennsylvania State University. Other strains were from the culture collection at the Food and Drug Administration and included 13C60, a patient isolate of the O26:H11 serotype that produces Stx1, and 13C09, an O48:H21 serotype strain that produces Stx2 that was isolated from a 1.5-year-old patient with bloody diarrhea.
Multiplex PCR analysis.
Isolates were analyzed by PCR for the presence of virulence genes. The multiplex assay used was a combination and modification of two PCR assays that allowed amplification of five genes in a single reaction. Three primer pairs, indicated in parentheses, were derived from the mismatch amplification mutation assay that amplifies 348 bp from the stx1 allele (primers LP30 and LP31), 584 bp from the stx2 allele (primers LP43 and LP44), and 252 bp from the uidA (also known as gusA) allele (primers PT-2 and PT-3) that is specific for the O157:H7 serotype (9). These were combined with primers that amplify a 397-bp region from the eaeA gene (primers AE20-2 and AE22) and a 166-bp fragment from the ehxA gene (primers MFS-1F and MFS-1R) (16). Each 100 μl of the PCR mixture contained each deoxynucleotide triphosphate at a concentration of 200 μM, each primer at a concentration of 300 nM, 1× PCR buffer (50 mM KCl, 2.5 mM MgCl2, 10 mM Tris-HCl [pH 8.3]), and 0.5 μl (2.5 U) of AmpliTaq Gold polymerase (PE Applied Biosystems, Foster City, Calif.). The template DNA was prepared by suspending a single colony in 100 μl of TE (Tris-EDTA) buffer; the suspension was then heated for 5 min in a boiling water bath, chilled on ice for 5 min, and centrifuged to remove debris. Prior to amplification, the reagents were mixed and heated at 95°C for 7 min to activate AmpliTaq Gold, and then 5 μl of template was added and the mixture was heated for an additional 5 min. Samples were amplified for 25 cycles of 1 min at 94°C, 1 min at 56°C, and 1 min at 72°C, followed by a final extension of 7 min at 72°C. The PCR products were examined by agarose gel (1%) electrophoresis in TBE (Tris-borate-EDTA) buffer (pH 8.2).
Analysis for intimin derivatives.
Pathogenic E. coli produces at least five distinct intimin derivatives (1), which are designated α, β, γ, δ, and NT (nontypeable). The eaeA primers used in our multiplex PCR are specific for the 3′ end of the eaeA gene of serotype O157:H7, which produces γ-intimin (22). However, other EHEC serotypes are reported to carry other intimin derivatives; hence, O104:H21 strains were also tested separately by PCR for the presence of genes for these derivatives. The primer sequences and procedure for amplifying α-eaeA and β-eaeA have been described by Reid et al. (22). Strains E2348/69 (EPEC O127:H6) and 13C60 (EHEC O26:H11) were used as positive controls for α- and β-eaeA, respectively.
Analysis for enterohemolysin.
Phenotypic assay for enterohemolysin activity was done on washed sheep blood agar plates containing calcium (3). The medium was prepared by supplementing tryptic soy agar with 10 mM CaCl2 (pH 7.3) and 5% defibrinated sheep blood that was washed three times in phosphate-buffered saline. Isolates were also tested for the presence of the ehxA gene with another set of PCR primers (hlyA1 and hlyA4) as described by Schmidt et al. (25).
To examine the genetic sequence of the MFS-1F and MFS-1R primer binding region, the ehxA gene of O157:H7 and O104:H21 was amplified, cloned, and sequenced as follows. A small amount of bacterial colony was directly added to 44 μl of an amplification mixture containing 1× Thermopol buffer (New England Biolabs, Beverly, Mass.), 2 mM MgSO4, each deoxynucleoside triphosphate at a concentration of 200 μM, and the 5′ primer hlyAalt1 (5′-CCA GGA GAA GAA GTT AGA G-3′) and the 3′ primer MFS-1R each at a concentration of 200 nM. This primer pair amplifies a 368-bp fragment (nucleotides [nt] 1612 to 1980) of the ehxA gene that includes the MFS-1F binding site. The PCR mixtures were heated at 95°C for 5 min, at which time 0.5 U of Vent DNA polymerase (New England Biolabs) in 5 μl of 1× Thermopol buffer was added to yield a final reaction volume of 50 μl. After heating for another 5 min at 95°C, the samples were amplified for 32 cycles, with each cycle consisting of 95°C for 1 min, 51°C for 45 s, and 75°C for 1 min, followed by a final 10-min extension at 75°C. The PCR products were examined on a 1% agarose gel in 0.5× TBE buffer. By using the described procedure (23), the desired fragments were excised, recovered from the gel, ligated into EcoRV-digested pBluescript SK(−), and electroporated into E. coli DH5α. Recombinants were examined for plasmids containing fragments of the expected size by screening by PCR with the vector-specific T3 and T7 promoter primers. Plasmids prepared from these clones were sequenced by Amplicon Express (Pullman, Wash.) by using the Automated Applied Biosystems Sequencing system.
RESULTS
PCR analysis for virulence genes.
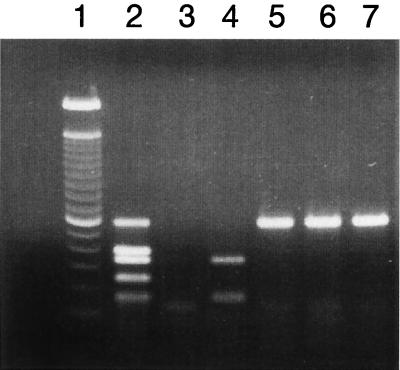
Multiplex PCR amplification showed that the O157:H7-positive control strain carried all four trait EHEC virulence markers as well as the +93 uidA mutation that is characteristic for this serotype (9) (Fig. 1, lane 2). The O104:H21 strains examined did not have the uidA mutation, eaeA, or ehxA and carried only the stx2 gene (Fig. 1, lanes 5, 6, and 7). Serological testing with Verotox-F (Denka Seiken, Tokyo, Japan), a reverse passive latex agglutination test, confirmed that Stx2 was produced by O104:H21. The absence of both eaeA and ehxA genes in O104:H21 seemed unusual. Many non-O157:H7 EHEC serotypes that have been implicated in illness usually carried combinations of the stx genes and the eaeA or ehxA gene, or both (2, 8, 20); hence, the apparent absence of the last two virulence markers in O104:H21 strains was investigated further.
FIG. 1.
Agarose gel electrophoresis of DNA fragments amplified by multiplex PCR specific for EHEC virulence genes and other trait markers of O157:H7 serotype. Lane 1, 100-bp DNA ladder (Life Technologies, Rockville, Md.); lane 2, 35150 (EHEC O157:H7), positive control; lane 3, E2348/69 (EPEC O127:H6), negative control; lane 4, 13C60 (EHEC O26:H11); lanes 5 (G5507), 6 (G5506), and 7 (G5508), strains of the O104:H21 serotype. The amplified products (sizes are in base pairs) from the O157:H7-positive control shown in lane 2 are, from top to bottom, stx2 (584 bp), eaeA (397 bp), stx1 (348 bp), uidA (252 bp), and ehxA (166 bp).
Analysis for intimin derivatives.
Multiplex PCR assay showed that O104:H21 strains did not carry the eaeA gene for the γ-intimin derivative that is typical of the O157:H7 serotype. PCR analysis with primers specific for α- and β-intimin derivatives showed that amplicons of the expected sizes for these primers for α-intimin (1,648 bp) and β-intimin (1,926 bp) were obtained from the respective control strains. However, none of the O104:H21 strains carried the eaeA genes for either the α-intimin or the β-intimin derivative (data not shown).
Analysis for enterohemolysin.
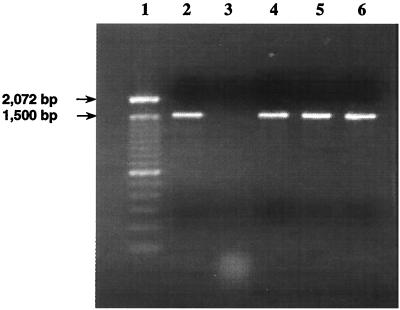
Multiplex PCR also showed that O104:H21 strains did not carry the ehxA gene that encodes enterohemolysin. However, phenotypic assays showed that these strains exhibited enterohemolysin activity on enhanced blood agar plates (data not shown). To resolve this discrepancy, we tested O104:H21 isolates using another set of primers, hlyA1 and hlyA4 (25), which amplify a 1,551-bp fragment (nt 238 to 1789) that is 25 bp upstream from the 166-bp fragment (nt 1814 to 1900) amplified by MFS-1F and MFS-1R primers used in the multiplex PCR. Agarose gel electrophoresis showed that all isolates of O104:H21 had the expected 1,551-bp fragment from the ehxA gene (Fig. 2, lanes 4, 5, and 6), confirming the phenotypic data that indicated that these strains are hemolytic.
FIG. 2.
Agarose gel electrophoresis of DNA fragments amplified from the ehxA gene by PCR with primers hlyA1 and hlyA4. Lane 1, 100-bp DNA ladder (Life Technologies); lane 2, 35150 (O157:H7); positive control; lane 3, DH5α, negative control; lanes 4 to 6, O104:H21 strains G5508, G5507, and G5506, respectively. The top two bands of the size ladder are 2,072 and 1,500 bp, respectively.
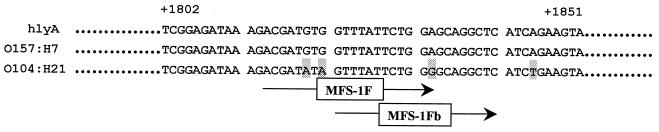
Since O104:H21 strains carry the ehxA gene, the absence of the ehxA-specific product from these strains in PCRs with our multiplex primers (primers MFS-1F and MFS-1R) was puzzling, especially since good amplification of ehxA was obtained from O157:H7 with these primers (Fig. 1, lane 2), as well as with primers hlyA1 and hlyA4 (Fig. 2, lane 2). The presence of base mutations in either the MFS-1F or MFS-1R binding sites may cause the absence of the ehxA-specific PCR product from O104:H21. To determine if MFS-1R was functional, a PCR combining upstream primer hlyA1 and downstream primer MFS-1R was used to test O104:H21 isolates. The expected size of the product would be 1,742 bp, consisting of the 1,551 bp from hlyA1 to hlyA4, the 166 bp from MFS-1F to MFS-1R, and the 25 bp between the two fragments. PCR analysis showed that amplicons approximating the expected size of 1,742 bp were obtained from the O157:H7-positive control, as well as from the O104:H21 strains examined (data not shown). This finding showed that the downstream MFS-1R binding region in O104:H21 is functional and that genetic variation most likely resides in the MFS-1F binding site. Sequence analysis of a 368-bp fragment that contained this region showed that the ehxA sequence of O157:H7 was identical to the published sequence (25). However, the ehxA gene of O104:H21 contained 9 base mutations, of which 3 were in the MFS-1F region, including an A-to-G substitution at the 3′ end of the primer binding site (Fig. 3).
FIG. 3.
Partial nucleotide sequence of ehxA gene showing the MFS-1F primer binding site. Sequences shown are hlyA (O157:H7), published by Schmidt et al. (25); O157:H7 (strain 35150); and O104:H21 (strain G5507). The mismatched bases are shaded. The region indicated by MFS-1Fb shows the new primer sequence, which, with MFS-1R, amplifies the ehxA genes from strains of both serotypes O157:H7 and O104:H21.
DISCUSSION
Evolutionary analyses have subdivided the EHEC and EPEC pathogenic groups into four distinct clones. EHEC 1 is composed mostly of the O157:H7 serotype, but includes the O55:H7 serotype; EHEC 2 includes Stx-producing O26 and O111 serotypes; EPEC 1 is represented by strain E2348/69, an O127:H6 serotype strain; and EPEC 2 includes Stx-nonproducing O26 and O111 serotype strains (26, 27). A genetic relatedness analysis of an O104:H21 isolate (isolate G5506) from the Montana outbreak showed that this strain was not an EHEC 1 or EHEC 2 clone; rather, it is closer in genetic distance to EHEC 2 (26). Consistent with those findings, our results showed that O104:H21 strains did not have the +93 uidA mutation, which is found only in O157:H7 serotype strains of the EHEC 1 clone. The production of Stx varies among the various EHEC serotypes and clonal groups, as some produce Stx1, or Stx2, or both. The O104:H21 strains examined here produced only Stx2.
Among EHEC virulence factors, intimin and Stx2 seem to be strongly associated with severe illness in humans (8). The eaeA gene, which encodes intimin, is found in both EHEC and EPEC strains. There are at least five distinct intimin derivatives (1), and of these, α-, β-, and γ-intimins are characteristic of the EPEC 1, EPEC 2, and EHEC 1 groups, respectively (22). The eaeA-specific primers used in our multiplex PCR are designed to detect the 3′ end of the eaeA gene of O157:H7 and, therefore, are specific only for γ-intimin (15). These primers have been shown to react only with isolates of the O157:H7 and EPEC O55:H7 serotypes (15), which are very closely related genetically and which are in the same clonal group (14). The O104:H21 isolates did not react with these eaeA-specific primers and therefore did not carry the gene encoding the γ-intimin derivative. Strains in the EHEC 2 clonal group are related genetically to strains in the EPEC 2 clonal group, as both groups include strains of the O26 and O111 serotypes and produce β-intimin (26). Since O104:H21 is closer in genetic distance to EHEC 2 (26), it was possible that it also produced β-intimin. However, PCR analysis showed that O104:H21 strains carried neither the α-intimin nor the β-intimin gene. We did not look for δ-intimin or NT intimin, but these derivatives are rare and are found in only a few serotypes (1); therefore, they are unlikely to be present in the O104:H21 isolates. Although O104:H21 strains do not appear to carry intimin, it does not rule out the presence of other adherence factors. An O113:H21 strain, isolated from a HUS patient, also produced Stx2 and did not carry eaeA, but it exhibited an adhesion pattern that was distinct from the typical attachment-and-effacing lesion of intimin (11).
The enterohemolytic phenotype is present in most O157:H7 isolates and is regarded as a trait characteristic of this serotype; however, it is also closely associated with other Stx-producing E. coli strains, as most O111 isolates from HUS patients were enterohemolytic (24). The enterohemolysin or EHEC hemolysin is encoded by the ehxA gene, which resides on a large plasmid (25), and the toxin is related to but is not identical to the α-hemolysin of E. coli in that they share about 60% sequence homology (24). Since enterohemolysin production seems to be closely associated with Stx production (5, 18), it was suggested that it may be a reliable epidemiological marker when testing for Stx-producing E. coli in clinical samples (4). The MFS-1F and MFS-1R primers used in our multiplex PCR are designed to be specific for the ehxA gene of O157:H7; but they also detected ehxA genes in strains of serotypes O26:H11, O111:NM (NM indicates nonmotile), O165:H25, O145:NM, O103:H2, O22:H8, O121:H19, O125:NM, and O45:H2 and strains of other serotypes (15; unpublished data). Although the ehxA genes of most serotypes have not been sequenced, that of O111 strains has been determined and found to be 99.4% identical to that of O157:H7 strains (24); therefore, it supports the observations that primers MFS-1F and MFS-1R will recognize ehxA genes of other serotypes. Results of our analyses, however, showed that these same primers are not effective in detecting the ehxA gene of O104:H21 strains from the Montana outbreak. A partial sequence analysis showed that the ehxA gene of O104:H21 was not identical to that of O157:H7, as it contained several base mutations, some of which are in the MFS-1F binding site. These mutations do not appear to be unique to O104:H21, as the ehxA genes of enterohemolytic O48:H21 (strain 13C09) and ON:HN (nontypeable) strains were also not amplified by MFS-1F and MFS-1R but were amplified by hlyA1 and hlyA4 (data not shown). Boerlin et al. (7) showed that the ehxA genes of Stx-producing E. coli strains cluster in two groups. The ehxA genes of strains in cluster I, which include strains of serotypes O157:H7, O26, and O111 and strains of other serotypes, share 98 and 97.3% homologies at the DNA and amino acid levels, respectively, with strains in cluster II, such as strains of serotypes O113:H21 and O8:H19 and strains of other serotypes. Specifically, the regions between amino acids 312 and 660 and amino acids 835 and 948 showed the highest degree of variation, with rates of substitution ranging from 5.7 to 7% (7). The ehxA gene region of the O104:H21 strains that we examined lies between amino acids 538 and 661; hence, the genetic variation that we noted is consistent with the findings of Boerlin et al. (7). Moreover, the sequence of the 368-bp fragment cloned from the ehxA gene of O104:H21 closely resembled that of the fragment cloned from the ehxA gene of O8:H19, which consists of cluster II strains (7), suggesting that O104:H21 also belongs in the cluster II group.
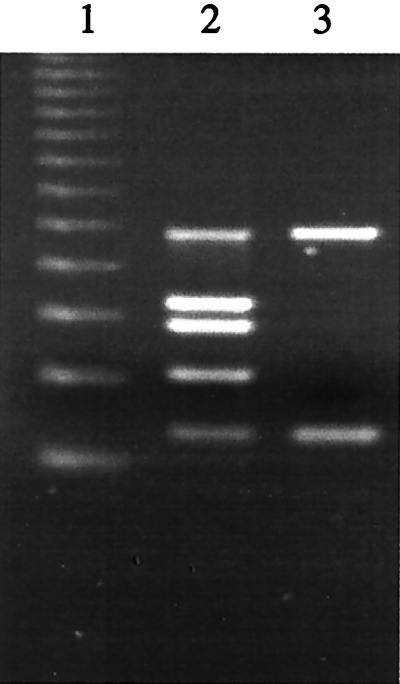
Primers MFS-1F and MFS-1R have been used to test for ehxA genes in E. coli isolates (15) and in other multiplex PCRs to characterize the virulence factors of Stx-producing E. coli strains isolated from animal feces (12). However, since these primers did not amplify the ehxA genes of some EHEC strains, presumably due to the A-to-G substitution at the 3′ end of the MFS-1F binding site, we modified this primer by shifting it 8 bases downstream and renaming it MFS-1Fb (Fig. 3). We compared the MFS-1Fb and MFS-1R sequences with that of hlyA, which encodes α-hemolysin (13), and also tested them by PCR with α-hemolytic E. coli strains to ensure that these primers did not bind to hlyA. Substituting MFS-1Fb for MFS-1F in the multiplex PCR cocktail and using identical PCR conditions, we found that we were able to effectively amplify the ehxA genes of EHEC strains from cluster I (O157:H7), cluster II (O113:H21) (not shown), and O104:H21 (Fig. 4).
FIG. 4.
Agarose gel electrophoresis of DNA fragments amplified by multiplex PCR with primer MFS-1Fb instead of primer MFS-1F for amplification of ehxA. Lane 1, 123-bp DNA ladder (Sigma, St. Louis, Mo.); lane 2, 35150 (O157:H7), positive control; lane 3, G5507 (O104:H21). The amplified products (sizes are in base pairs) from O157:H7 shown in lane 2 are, from top to bottom, stx2 (584 bp), eaeA (397 bp), stx1 (348 bp), uidA (252 bp), and ehxA (158 bp).
Results from these studies show that the O104:H21 strain, implicated in the HC outbreak in Montana, does not have eaeA but produces Stx2 and enterohemolysin. Other EHEC serotypes with similar combinations of virulence factors have also been implicated in HUS (21). Genetic analysis showed that the ehxA genes of O104:H21 strains contained base mutations that are not found in O157:H7 strains and that the ehxA sequences of O104:H21 strains resembled those of cluster II Stx-producing strains rather than those of cluster I Stx-producing strains (O157:H7). By modifying one of the ehxA primers, the multiplex PCR assay is able to detect ehxA genes from both hemolysin gene clusters, including O104:H21, and may be useful in the analysis of virulence factors in other EHEC isolates.
ACKNOWLEDGMENTS
We thank N. Strockbine and S. Abbott for providing cultures and T. Whittam for helpful discussions and for critical reading of the manuscript.
REFERENCES
- 1.Adu-Robie J, Frankel G, Bain C, Goncalves A G, Trabulsi L R, Douce G, Knutton S, Dougan G F. Detection of intimins α, β, γ, and δ, four intimin derivatives expressed by attachment and effacing microbial pathogens. J Clin Microbiol. 1998;36:662–668. doi: 10.1128/jcm.36.3.662-668.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barrettt T J, Kaper J B, Jerse A E, Wachsmuth I K. Virulence factors in Shiga-like toxin-producing Escherichia coli isolated from humans and cattle. J Infect Dis. 1992;165:979–980. doi: 10.1093/infdis/165.5.979. [DOI] [PubMed] [Google Scholar]
- 3.Bettelheim K A. Identification of enterohaemorrhagic Escherichia coli by means of their production of enterohaemolysin. J Appl Bacteriol. 1995;79:178–180. doi: 10.1111/j.1365-2672.1995.tb00932.x. [DOI] [PubMed] [Google Scholar]
- 4.Beutin L, Aleksic S, Zimmermann S, Gleier K. Virulence factors and phenotypic traits of verotoxigenic strains of Escherichia coli isolated from human patients in Germany. Med Microbiol Immunol. 1994;183:13–21. doi: 10.1007/BF00193627. [DOI] [PubMed] [Google Scholar]
- 5.Beutin L, Montenegro M, Orskov I, Orskov F, Prada J, Zimmermann S, Stephan R. Close association of verotoxin (Shiga-like toxin) production with enterohemolysin production in strains of Escherichia coli. J Clin Microbiol. 1989;27:2559–2564. doi: 10.1128/jcm.27.11.2559-2564.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bockemuhl J, Aleksic S, Karch H. Serological and biochemical properties of Shiga-like toxin (verocytotoxin)-producing strains of Escherichia coli, other than O-group 157, from patients in Germany. Zentbl Bakteriol Parasitenkd Infektcankh Nyg Abt 1 Orig. 1992;276:189–195. doi: 10.1016/s0934-8840(11)80005-8. [DOI] [PubMed] [Google Scholar]
- 7.Boerlin P, Chen S, Colbourne J K, Johnson R, DeGrandis S, Gyles C. Evolution of enterohemorrhagic Escherichia coli hemolysin plasmid and the locus for enterocyte effacement in Shiga toxin-producing E. coli. Infect Immun. 1998;66:2553–2561. doi: 10.1128/iai.66.6.2553-2561.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boerlin P, McEwen S A, Boerlin-Petzold F, Wilson J B, Johnson R, Gyles C. Association between virulence factors of Shiga toxin-producing Escherichia coli and disease in human. J Clin Microbiol. 1999;37:497–503. doi: 10.1128/jcm.37.3.497-503.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cebula T A, Payne W L, Feng P. Simultaneous identification of strains of Escherichia coli serotype O157:H7 and their Shiga-like toxin type by mismatch amplification mutation assay-multiplex PCR. J Clin Microbiol. 1995;33:248–250. doi: 10.1128/jcm.33.1.248-250.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Centers for Disease Control and Prevention. Outbreak of acute gastroenteritis attributed to Escherichia coli serotype O104:H21—Helena, Montana, 1994. JAMA. 1995;274:529–530. [PubMed] [Google Scholar]
- 11.Dytoc M T, Ismaili A, Philpott D J, Soni R, Brunton J L, Sherman P M. Distinct binding properties of eaeA-negative verocytotoxin-producing Escherichia coli of serotype O113:H21. Infect Immun. 1994;62:3494–3505. doi: 10.1128/iai.62.8.3494-3505.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fagan P K, Hornitzky M A, Bettelheim K A, Djordjevic S P. Detection of Shiga-like toxin (stx1 and stx2), intimin (eaeA), and enterohemorrhagic Escherichia coli (EHEC) hemolysin (EHEC hlyA) genes in animal feces by multiplex PCR. Appl Environ Microbiol. 1999;65:868–872. doi: 10.1128/aem.65.2.868-872.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Felmlee T, Pellett S, Welch R A. Nucleotide sequence of an Escherichia coli chromosomal hemolysin. J Bacteriol. 1985;163:94–105. doi: 10.1128/jb.163.1.94-105.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Feng P, Lampel K A, Karch H, Whittam T S. Sequential genotypic and phenotypic changes in the emergence of Escherichia coli O157:H7. J Infect Dis. 1998;177:1750–1753. doi: 10.1086/517438. [DOI] [PubMed] [Google Scholar]
- 15.Fratamico P M, Sackitey S K, Wiedmann M, Deng M Y. Detection of Escherichia coli O157:H7 by multiplex PCR. J Clin Microbiol. 1995;33:2188–2191. doi: 10.1128/jcm.33.8.2188-2191.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fratamico P M, Strobaugh T P. Simultaneous detection of Salmonella spp and Escherichia coli O157:H7 by multiplex PCR. J Ind Microbiol Biotechnol. 1998;21:92–98. [Google Scholar]
- 17.Goldwater P N, Bettelheim K A. New perspectives on the role of Escherichia coli O157:H7 and other enterohemorrhagic E. coli serotypes in human disease. J Med Microbiol. 1998;47:1039–1045. doi: 10.1099/00222615-47-12-1039. [DOI] [PubMed] [Google Scholar]
- 18.Gyles C, Johnson R, Gao A, Ziebell K, Pierard D, Aleksic S, Boerlin P. Association of enterohemorrhagic Escherichia coli hemolysin with serotypes of Shiga-like-toxin-producing Escherichia coli of human and bovine origin. Appl Environ Microbiol. 1998;64:4131–4141. doi: 10.1128/aem.64.11.4134-4141.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johnson R P, Clarke R C, Wilson J B, Read S C, Rahn K, Renwick S A, Sandhu K A, Alves D, Karmali M A, Lior H, Mcewen S A, Spika J S, Gyles C L. Growing concerns and recent outbreak involving non-O157:H7 serotypes of verotoxigenic Escherichia coli. J Food Prot. 1996;59:1112–1122. doi: 10.4315/0362-028X-59.10.1112. [DOI] [PubMed] [Google Scholar]
- 20.Levine M M, Xu J G, Kaper J B, Lior H, Prado V, Tall B, Nataro J, Karch H, Wachsmuth I K. A DNA probe to identify enterohemorrhagic Escherichia coli of O157:H7 serotype and other serotypes that cause hemorrhagic colitis and hemolytic uremic syndrome. J Infect Dis. 1987;156:175–182. doi: 10.1093/infdis/156.1.175. [DOI] [PubMed] [Google Scholar]
- 21.Paton A W, Woodrow M C, Doyle R M, Lanser J A, Paton J C. Molecular characterization of a Shiga toxigenic Escherichia coli O113:H21 strain lacking eae responsible for a cluster of cases of hemolytic-uremic syndrome. J Clin Microbiol. 1999;37:3357–3361. doi: 10.1128/jcm.37.10.3357-3361.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reid S D, Betting D J, Whittam T S. Molecular identification of intimin alleles in pathogenic Escherichia coli by multiplex PCR. J Clin Microbiol. 1999;37:2719–2722. doi: 10.1128/jcm.37.8.2719-2722.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 24.Schmidt H, Karch H. Enterohemolytic phenotypes and genotypes of Shiga toxin-producing Escherichia coli O111 strains from patients with diarrhea and hemolytic-uremic syndrome. J Clin Microbiol. 1996;34:2364–2367. doi: 10.1128/jcm.34.10.2364-2367.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schmidt H, Beutin L, Karch H. Molecular analysis of the plasmid-encoded hemolysin of Escherichia coli O157:H7 strain EDL 933. Infect Immun. 1995;63:1055–1061. doi: 10.1128/iai.63.3.1055-1061.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Whittam T S. Evolution of Escherichia coli O157:H7 and other Shiga-toxin producing E. coli strains. In: Kaper J B, O'Brien A D, editors. Escherichia coli O157:H7 and other Shiga toxin-producing E. coli strains. Washington, D.C.: American Society for Microbiology; 1998. pp. 195–209. [Google Scholar]
- 27.Whittam T S, McGraw E A. Clonal analysis of EPEC serogroups. Rev Microbiol Sao Paulo. 1996;27:7–16. [Google Scholar]
- 28.Willshaw G A, Scotland S M, Smith H R, Rowe B. Properties of Verocytotoxin-producing Escherichia coli of human origin of O sergroups other than O157. J Infect Dis. 1992;166:797–802. doi: 10.1093/infdis/166.4.797. [DOI] [PubMed] [Google Scholar]