Abstract
Advances in sequencing technologies have revealed the complex and diverse microbial communities present in ticks (Ixodida). As obligate blood-feeding arthropods, ticks are responsible for a number of infectious diseases that can affect humans, livestock, domestic animals and wildlife. While cases of human tick-borne diseases continue to increase in the northern hemisphere, there has been relatively little recognition of zoonotic tick-borne pathogens in Australia. Over the past 5 years, studies using high-throughput sequencing technologies have shown that Australian ticks harbour unique and diverse bacterial communities. In the present study, free-ranging wildlife (n=203), representing ten mammal species, were sampled from urban and peri-urban areas in New South Wales (NSW), Queensland (QLD) and Western Australia (WA). Bacterial metabarcoding targeting the 16S rRNA locus was used to characterize the microbiomes of three sample types collected from wildlife: blood, ticks and tissue samples. Further sequence information was obtained for selected taxa of interest. Six tick species were identified from wildlife: Amblyomma triguttatum, Ixodes antechini, Ixodes australiensis, Ixodes holocyclus, Ixodes tasmani and Ixodes trichosuri. Bacterial 16S rRNA metabarcoding was performed on 536 samples and 65 controls, generating over 100 million sequences. Alpha diversity was significantly different between the three sample types, with tissue samples displaying the highest alpha diversity (P<0.001). Proteobacteria was the most abundant taxon identified across all sample types (37.3 %). Beta diversity analysis and ordination revealed little overlap between the three sample types (P<0.001). Taxa of interest included Anaplasmataceae , Bartonella , Borrelia , Coxiellaceae , Francisella , Midichloria , Mycoplasma and Rickettsia . Anaplasmataceae bacteria were detected in 17.7% (95/536) of samples and included Anaplasma , Ehrlichia and Neoehrlichia species. In samples from NSW, ‘Ca. Neoehrlichia australis’, ‘Ca. Neoehrlichia arcana’, Neoehrlichia sp. and Ehrlichia sp. were identified. A putative novel Ehrlichia sp. was identified from WA and Anaplasma platys was identified from QLD. Nine rodent tissue samples were positive for a novel Borrelia sp. that formed a phylogenetically distinct clade separate from the Lyme Borrelia and relapsing fever groups. This novel clade included recently identified rodent-associated Borrelia genotypes, which were described from Spain and North America. Bartonella was identified in 12.9% (69/536) of samples. Over half of these positive samples were obtained from black rats (Rattus rattus), and the dominant bacterial species identified were Bartonella coopersplainsensis and Bartonella queenslandensis . The results from the present study show the value of using unbiased high-throughput sequencing applied to samples collected from wildlife. In addition to understanding the sylvatic cycle of known vector-associated pathogens, surveillance work is important to ensure preparedness for potential zoonotic spillover events.
Keywords: tick, microbiome, wildlife, sylvatic cycle, tick-borne diseases
Data Summary
Sequence data have been deposited in relevant data repositories (see above). Additional information is available within supplementary files (available in the online version of this article) including links to electronic files which have been deposited on FigShare: https://doi.org/10.6084/m9.figshare.14363627.v1. Code used for analysis and RData files used for bioinformatics and statistical analysis are available at https://github.com/siobhon-egan/wildlife-bacteria and https://githubcom/siobhon-egan/wildlife-ticks.
The authors confirm all supporting data, code and protocols have been provided within the article or through supplementary data files.
Impact Statement.
Globally, the incidence and distribution of tick-borne diseases is increasing. In contrast, there is currently limited data available regarding tick-borne pathogens in Australia. This research employed bacterial 16S metabarcoding on wildlife blood and tissue samples and their ticks. It aimed to understand the sylvatic cycle of microbes and identify potential pathogens. This study identified several recently described and novel microbes from both wildlife and ticks. It identified Neoehrlichia and Ehrlichia species from native wildlife and the introduced black rat. It describes the first identification of the rodent-associated Borrelia clade in Australia; this unique clade is distinct from both the Lyme Borrelia and relapsing fever groups. Black rats were identified as an important reservoir of both endemic and novel microbes. Importantly, these findings provide support for the absence of northern hemisphere tick-borne pathogens. In conclusion, it is likely that any potential human tick-borne pathogen(s) in Australia are probably endemic and distinct from currently described pathogens from the northern hemisphere.
Introduction
Ticks carry a diverse range of infectious microbes such as viruses, piroplasms, spirochaetes and Rickettsiales [1]. Globally recognized infectious human tick-borne diseases include Lyme borreliosis, ehrlichiosis, babesiosis, tick-borne encephalitis and Powassan viral disease. Additionally, a tick bite can cause reactions such as paralysis and anaphylaxis [2]. The incidence of tick-borne diseases is rapidly increasing in both prevalence and geographical distribution [3]. The sylvatic cycles of tick-borne pathogens in the northern hemisphere are generally well understood with respect to competent tick vectors and the reservoir hosts (e.g. Babesia microti [4] and Borrelia burgdorferi sensu lato [5]). The value of wildlife health surveillance as a tool for the detection of emerging zoonotic infectious disease has been demonstrated in outbreaks historically, such as malaria [6], and more recently with SARS-CoV-2 [7].
In Australia, three human tick-borne diseases are recognized: Queensland tick typhus ( Rickettsia australis ), spotted fever ( Rickettsia honei subspp.) and Q fever ( Coxiella burnetii ) [8]. Over the past decade there have been increasing concerns about unidentified tick-borne illnesses (syn. Lyme-like disease) in Australia [9], recently designated ‘Debilitating Symptom Complexes Attributed To Ticks’ (DSCATT) by the Australian Government Department of Health [10]. Presently, 74 tick species have been described in Australia [11, 12], including five introduced species, of which 22 species have been identified biting humans [13]. However, data show that Ixodes holocyclus and Amblyomma triguttatum account for >90 % of human tick bites, with the remainder of reports largely attributed to Ixodes tasmani, Haemaphysalis bancrofti, Haemaphysalis longicornis and Bothriocroton hydrosauri [13]. Small to medium-sized mammals are important in maintaining these tick populations, in particular during the immature life stages [14–16]. While data on the incidence of human tick bites in Australia are limited, evidence suggests that tick bites are common in peri-urban areas, including residential yards (C. Taylor, pers. comm.).
Over the past decade, due to new technologies there is mounting evidence that Australian ticks harbour a unique array of microbes [17, 18]. In addition to these molecular studies, more targeted screening of potential reservoirs such as dogs [19] and sea bird ticks [20] have added to the growing body of evidence suggesting that human tick-borne pathogens described in the northern hemisphere are not endemic in Australia. Some of the microbes that have recently been identified from Australian ticks, however, include novel species of Borrelia [21, 22], Ehrlichia [23], Neoehrlichia [24], Babesia and Theileria [25, 26] and several viruses [27, 28]. Little is known about the sylvatic life cycle of these microbes or their wildlife species reservoir hosts. Even for the few tick-borne pathogens recognized in Australia (e.g. Rickettsia spp.), much of this information remains elusive.
For a tick-borne disease to become established it requires a temporal and spatial overlap between the microbe (pathogens), competent vector tick(s), and one or more vertebrate reservoir hosts. In the northern hemisphere studies have shown that there are important differences in the role of host species in maintaining the sylvatic cycle of tick-borne pathogens [29–31]. Broadly, reservoir hosts can be classified as amplification and dilution hosts. Amplification hosts, in most cases, are small mammals that host immature ticks (larvae or nymph stages) and increase the environmental density of infected nymphs [29]. In contrast, dilution hosts are inefficient reservoirs of the microbial agent(s) [32, 33]. An additional factor is the role of hosts throughout the tick life cycle. Larger animals are considered important in maintaining adult stages of the tick: white-tailed deer (Odocoileus virginianus) and Ixodes scapularis [34] for example. In the Australian context it is generally assumed that small mammals such as bandicoots, possums and rodents are likely to be reservoirs for tick-associated microbes. In turn, larger mammals such as kangaroos and wallabies may then play an important role in maintaining tick populations, filling a similar ecological niche as deer in the northern hemisphere.
While studies have used molecular tools to investigate microbes present within ticks in Australia [20, 24, 25], very little research has been conducted at the vertebrate host level to further elucidate the sylvatic cycle of tick-transmitted bacteria. Bacterial amplicon sequencing is valuable in the setting of wildlife epidemiology surveys where investigations are not confined to existing presumptions of microbes [35], but such studies of the ticks alone, without concurrent analysis of vertebrate host tissues, cannot distinguish infectious components of the tick’s total microbiome from those that are non-transmissible. Therefore, the search for tick-associated microbes in vertebrate host(s) is the next logical piece of information required to untangle the complex dynamics of tick-borne pathogens. In addition to novel pathogen discovery, high-throughput sequencing technologies applied concomitantly to tick and vertebrate host tissues can provide insights into the dynamics of microbial epidemiology and interactions within landscapes [36]. Identification of microbes may inform our understanding of the tick-borne disease risk and potential differences among temporal and spatial gradients.
The present study used metabarcoding to explore the bacterial communities present in Australian wildlife and their ticks, with a focus on bacteria related to known (vector-borne) pathogens, to provide insights into the sylvatic life cycle of tick-associated organisms. This research was conducted in an ecological zone referred to as the urban–wildland interface, where there is a transition between wilderness and land developed by human activity – an area where a built environment meets or intermingles with a natural environment.
Methods
Study sites and sample collection
Small mammal trapping
Small mammal trapping was performed at various sites in Perth, Western Australia, and Sydney, New South Wales (Fig. 1). Each site was targeted in urban and peri-urban areas to reflect the urban–wildland interface. Elliot and cage traps were set and baited with universal bait (rolled oats, peanut butter and sardines). Traps were set at dusk and cleared at sunrise over three or four consecutive nights. To ensure a comprehensive assessment and sampling, target mammals were briefly anaesthetized in the field using isoflurane (I.S.O., 1 ml ml−1) vaporized in medical oxygen. Animals were weighed and examined systematically for ectoparasites. Up to 1 ml of blood was collected from either the caudal (tail) vein, femoral vein or ear capillary and stored in Mini Collect EDTA tubes. If practical, a 2 mm punch biopsy was taken at the tick bite site; where possible, a biopsy was taken from the ear and stored in RNAlater or 80% ethanol. Animals were systematically examined for ectoparasites, which were removed and stored in 80 % ethanol. Animals were recovered from anaesthesia by providing medical oxygen, and once fully alert were released at the trap point. An individual mark was applied to identify animals by using either a microchip administered subcutaneously or a unique patch of hair was removed. The number of animals and samples analysed, and locations from the present study are available in Table 1 and Fig. 1 respectively.
Fig. 1.
Map of study sites for collection of wildlife samples used in bacterial profiling. Sampling sites are denoted by blue triangles and capital cities by pink circles (for geographical reference). Insert maps of sites in (1) Perth, Western Australia, and (2) Sydney, New South Wales.
Table 1.
Animal sampling numbers for bacterial microbiome sequencing: number of individual animals (n) by host species with number of genomic DNA samples analysed in parentheses
|
Common name |
Scientific name |
n |
Blood |
Tick |
Tissue |
|---|---|---|---|---|---|
|
Black rat |
Rattus rattus |
88 |
68 (84) |
54 (79) |
71 (80) |
|
Brown antechinus |
Antechinus stuartii |
5 |
0 (0) |
4 (6) |
2 (2) |
|
Brush-tailed possum |
Trichosurus vulpecula |
27 |
18 (21) |
15 (33) |
16 (20) |
|
Bush rat |
Rattus fuscipes |
3 |
2 (2) |
3 (4) |
3 (3) |
|
Chuditch |
Dasyurus geoffroii |
22 |
22 (24) |
1 (1) |
9 (9) |
|
Deer |
Rusa timorensis |
3 |
0 (0) |
0 (0) |
3 (6) |
|
Long-nosed bandicoot |
Perameles nasuta |
44 |
20 (25) |
31 (60) |
40 (44) |
|
Quenda |
Isoodon fusciventer |
2 |
1 (1) |
2 (2) |
0 (0) |
|
Rabbit |
Oryctolagus cuniculus |
7 |
0 (0) |
7 (20) |
7 (7) |
|
Swamp rat |
Rattus lutreolus |
2 |
1 (1) |
0 (0) |
1 (1) |
|
Total |
|
203 |
132 (159) |
117 (205) |
152 (172) |
Opportunistic collection
In a small number of cases, animal carcasses were also obtained opportunistically. These specimens were collected through incidental findings during fieldwork or from situations where animals were humanely euthanized in accordance with animal ethics and state and federal guidelines.
Tick identification
Ticks were collected and stored in tubes containing 70–90% ethanol. Samples were visualized using an Olympus SZ61 stereomicroscope (Olympus) with an external Schott KL 1500 LED (Schott) light source. Photographs of tick specimens were taken using an Olympus SC30 digital camera (Olympus) and analysis getIT software (Olympus). Instar, sex and species were identified using a combination of available morphological keys and species descriptions [37–41].
DNA extractions
Total genomic DNA (gDNA) was extracted from 200 μl of blood using a MasterPure DNA purification kit (EpicentreBiotechnologies) following the manufacturer’s recommendations. Where 200 μl of blood was not available, sterile DNA-free PBS was used to make samples up to 200 μl. Genomic DNA was eluted in 30–40 μl of TE buffer and stored at −20 °C until further processing.
Tissue samples (skin and spleen) were first rinsed in sterile, DNA-free PBS and cut into small pieces (<1–2 mm) using a sterile scalpel blade. Samples were homogenized in 180 μl of buffer ATL and 20 μl of proteinase K was added and incubated at 56 °C for ~16 h. gDNA was extracted using the QIAamp DNA Mini Kit (Qiagen) following the manufacturer’s protocols with the exception that the final elution volume was decreased to 40–50 μl to increase gDNA yield.
Once ticks were identified, they were surface-sterilized by washing in 10% hypochlorite solution, rinsed in 70% ethanol and DNA-free PBS, and then air-dried. Genomic DNA was extracted using the DNeasy Blood and Tissue kit (Qiagen) for adults, or the QIAamp DNA Mini Kit (Qiagen) for nymphs and larvae. Due to the large number of immature tick stages collected from some animals and the expected low DNA yield, up to five specimens were pooled for extraction based on host, instar, engorgement status and species identification as determined by morphological methods. Ticks were placed in a 2 ml safe lock Eppendorf tube with a 5 mm steel bead, frozen in liquid nitrogen for 1 min and homogenized by shaking at 40 Hz in a Tissue Lyser LT (Qiagen). Final elution of DNA was adjusted according to tick size, and 30–150 μl of AE buffer was added to the silicon membrane.
Extraction controls (EXBs) consisting of 200 μl sterile DNA-free PBS were included randomly in each extraction batch (total EXBs=34). Blood and tissue samples were extracted in a separate laboratory, away from where ticks were processed, to avoid cross-contamination between sample types. Sterile procedures were followed throughout the laboratory process.
Metabarcoding sequencing
A high-throughput metabarcoding approach was used to sequence samples using the Illumina MiSeq platform. Libraries were built following the 16S Metagenomic Sequencing Library Preparation (Illumina Part no. 15044223 Rev. B), with amplicon PCR primers containing Illumina MiSeq adapter sequences (Table 2). A total of 536 samples from 203 individuals (159 blood, 205 tick pools and 172 tissue samples) underwent bacterial profiling.
Table 2.
List of sequences used for metabarcoding and target PCRs to screen blood, tissue and tick samples for bacteria
References: 1, Gofton et al. [17]; 2, Turner et al. [144]; 3, Lopez et al. [145]; 4, Caporaso et al. [146]; 5, Beard et al. [147]; 6, Anderson et al. [148]; 7, Paddock et al. [149]; 8, Kawahara et al. [82]; 9, Beati and Keirans 2001 [42].
|
Primer |
Sequence (5′−3′) |
Reference |
|---|---|---|
|
Bacteria 16S rRNA |
||
|
27F-Y |
AGAGTTTGATCCTGGCTYAG |
[1] |
|
338R |
TGCTGCCTCCCGTAGGAGT |
[2] |
|
338F |
ACTCCTACGGGAGGCAGCAG |
[3] |
|
806R |
GGACTACHVGGGTWTCTAAT |
[4] |
|
Bartonella 16S rRNA/ITS |
||
|
438s |
GGTTTTCCGGTTTATCCCGGAGGGC |
[5] |
|
1100as |
GAACCGACGACCCCCTGCTTGCAAAGC |
[5] |
|
Anaplasmataceae 16S rRNA |
||
|
EC9 |
TACCTTGTTACGACTT |
[6] |
|
EC12 |
TGATCCTGGCTCAGAACGAACG |
[7] |
|
A171a |
GCGGCAAGCCTCCCACAT |
[8] |
|
IS58-1345r |
CACCAGCTTCGAGTTAAACC |
[8] |
|
Ixodida 12S rRNA |
||
|
T1B |
AAACTAGGATTAGATACCCT |
[9] |
|
T2A |
AATGAGAGCGACGGGCGATGT |
[9] |
Bacterial 16S rRNA libraries were built targeting the 16S hypervariable region 1–2 using primers 27F-Y and 338R [17]. Reactions were carried out in 25 μl volumes each containing: 1× buffer (KAPA Biosystems), 1.5 mM MgCl2, 0.4 mg ml−1 BSA (Fisher Biotech), 0.4 μM of each primer, 0.25 mM of each dNTP, 0.5 U of Taq (KAPA Biosystems) and 2 μl of gDNA. Thermal cycling conditions were as follows: 95 °C for 5 min, followed by 35 cycles of 95 °C for 30 s, 55 °C for 30 s, 72 °C for 45 s; and a final extension of 72 °C for 5 min. For a subset of samples where microbes of interest were identified after initial screening at the v1–2 hypervariable region, additional bacterial 16S rRNA libraries targeting the v3–4 hypervariable region were also prepared. Primers 338F and 806R were used to target an ~450 bp product. Reactions and thermal cycling conditions were carried out as per the 27F-Y/338R assay, except for an increased concentration of MgCl2 to 2.0 mM.
To confirm morphological identification, tick gDNA underwent an amplicon metabarcoding approach targeting the 12S rDNA gene. Pan-Ixodida primers T1B/T2A [42] were used to amplify an ~370 bp product of the 12S rRNA locus. Reactions were carried out as per the bacteria 27F-Y/338R assay, except only 1 μM of tick gDNA template was added. Thermal cycling conditions were as follows: 95 °C for 5 min, followed by five cycles of 95 °C for 15 s, 51 °C for 30 s, 68 °C for 30 s; 25 cycles of 95 °C for 30 s, 53 °C for 30 s, 72 °C for 1 min; and a final extension of 72 °C for 5 min.
Amplicon PCR products were then indexed using the Nextera XT DNA library preparation kit in 25 μl volumes following the manufacturer’s recommendations. All PCRs included no-template controls (NTC; total=31) during each reaction set up and PCRs were performed under strict laboratory conditions. Amplicons were then dual-indexed using the Nextera XT index kit. Reactions were performed in 25 μl volumes following the manufacturer’s recommendations. Libraries were purified with Axygen PCR clean up beads and quantified using a Qubit High Sensitivity dsDNA assay kit (Thermo Fisher Scientific) and pooled in equimolar amounts. Libraries were shipped to the Australian Genomic Research Facility (Melbourne, Australia) for final QC and sequenced on an Illumina MiSeq using v2 chemistry (2×250 paired-end).
Bioinformatics
Metabarcoding 16S rRNA sequence data were analysed using Quantitative Insights into Microbial Ecology (QIIME 2 2020.11) [43]. Briefly, raw sequences were demultiplexed and quality filtered using the q2-demux plugin, followed by denoising using DADA2 (via q2-dada2) [44] resulting in amplicon sequence variations (ASVs) (i.e. 100% identical operational taxonomic units or OTUs [45]). Bacterial data taxonomy was assigned to ASVs using the q2‐feature‐classifier [46] classify‐sklearn naive Bayes taxonomy classifier against the silva database [47] (release 132). Taxonomic assignments were confirmed using blast analysis (blastn 2.11.0+ [48, 49]) against the NCBI nucleotide collection (nt) database (accessed January 2021). Taxonomic lineage was then retrieved from the NCBI taxonomy database using TaxonKit [50] and adjusted to the lowest common ancestor based on percentage identity and e-value score.
Tick 12S metabarcoding sequences generated were analysed via the USEARCH v11 [51] pipeline. Briefly paired-end reads were merged and sequences matching forward and reverse primers were retrieved (maximum number of mismatches=2). Sequences were then quality filtered and singletons were removed. The unoise3 [52] algorithm was used to perform denoising (error-correction) and generate zero-radius taxonomic units (zOTUs) equivalent to ASVs. Taxonomy was assigned using blast analysis and lineage was retrieved as outlined above for 16S metabarcoding.
Data visualization and statistical analysis was performed in RStudio (v1.4) with R version 4.0.2. The main R packages used were ampvis2 v2.6.7, phyloseq v1.34.0, microbiomeutilities v1.00.12 and vegan v2.5–7. The R package decontam v1.10.0 [53] was used to inspect data for cross-contamination and cross-talk between samples and to establish read cut-off thresholds (prevalence threshold=0.05). Identified contaminant ASVs (n=104), control samples (n=65) and sequence values <100 were removed. Rarefaction curves were generated using a step size of 100. Alpha diversity was measured using four indexes (number of observed ASVs, Chao1, Shannon and invSimpson) and statistical analysis was calculated using a Wilcoxon pairwise test (non-parametric) between sample types (blood, tick and tissue). Heatmaps were generated using ampvis2, where data were first aggregated to the family level and transformed to relative abundance.
Constrained ordination analysis was performed using two methods, canonical correspondence analysis and redundancy analysis (constrained principal component analysis), to investigate the relationships between sample types. Prior to the analysis, ASVs of <0.1 relative abundance and samples with <1000 sequences were removed. Data were first transformed using the Hellinger transformation and distance was measured using the Bray–Curtis method. Beta diversity statistical analysis was performed using ANOVA and permutational multivariate analysis of variance (PERMANOVA) (999 permutations) via the adonis function in the vegan package [54] (Hellinger transformation and Bray–Curtis distance measure); the F statistic and P value are presented (full statistical output available in Supplementary file 1). Hierarchical cluster analysis was performed using the euclidean distance measure (average), where data were first transformed using Hellinger transformation.
The composition for taxa of interest was aggregated to the family level and compared between the three sample types. Taxa of interest were defined as bacteria related to known pathogens associated with ticks and other vectors described globally, as outlined in Egan et al. [18]. Statistical analysis was performed using the Wilcoxon pairwise test (non-parametric). To identify shared ASVs (i.e. core microbiome) between sample types and species, data were transformed to relative abundance and assigned to the best taxonomic hit using the microbiomeutilities R package. Similarities of the core microbiome between the three sample types were investigated using thresholds of abundance and prevalence of 0.0001−0.001 and 0.001–0.05 respectively. For subsequent comparisons of the core microbiome between host and tick species, abundance and prevalence levels were set at 0.001 and 0.05, respectively. Prevalence data for taxa of interest are presented for samples based on 16S rRNA metabarcoding. Where ticks have been pooled, prevalence is reported based on minimum infection rates (i.e. assuming only one positive tick in each pool) [55].
Scripts for data analysis are available at https://github.com/siobhon-egan/wildlife-bacteria and https://github.com/siobhon-egan/wildlife-ticks, and raw Illumina MiSeq data have been deposited in the European Nucleotide Archive under project accession numbers PRJEB46056 (16S rRNA bacteria) and PRJEB46056 (12S rRNA ticks).
Target PCRs
A nested PCR was used to amplify an ~1.4 kb fragment of 16S rRNA locus for Neoehrlichia and Ehrlichia . Amplicon PCRs were carried out in 25 μl reactions each containing: 1× buffer (KAPA Biosystems), 2.5 mM MgCl2, 0.4 μM of each primer, 0.25 mM of each dNTP, 0.5 U of Taq (KAPA Biosystems) and 2 μl of gDNA or 1 µl of primary product. Thermal cycling conditions were as follows: 95 °C for 3 min, followed by 40 cycles of 95 °C for 30 s, 48 °C (primary) or 54 °C (secondary) for 1 min, 72 °C for 2 min; and a final extension of 72 °C for 5 min.
For Bartonella , a PCR was used to amplify an ~370–450 bp fragment of the 16S rRNA – 23S internal transcribed spacer (ITS) region. Amplicon PCRs were carried out in 25 μl reactions each containing: 1× buffer (KAPA Biosystems), 2.0 mM MgCl2, 0.4 μM of each primer, 0.25 mM of each dNTP, 0.5 U of Taq (KAPA Biosystems) and 2 μl of gDNA. Thermal cycling conditions were as follows: 95 °C for 3 min, followed by 40 cycles of 95 °C for 15 s, 66 °C 15 s, 72 °C for 18 s; and a final extension of 72 °C for 5 min.
Amplicons were visualized on agarose gel and products of the expected size were excised with a sterile scalpel blade and purified using an in-house filtered pipette tip method [56]. Sanger sequencing was performed at the Australian Genome Research Facility (Perth, Western Australia) on an Applied Biosystems 3730xl DNA Analyzer using a BigDye Terminator v3.1 Cycle Sequencing Kit.
Phylogeny
Nucleotide sequences were inspected and quality filtered using Geneious 10.2.6 (https://www.geneious.com). Identity was confirmed using blast analysis (blastn 2.11.0+ [48, 49]) against the NCBI nucleotide collection (nt) database. Sequences generated were aligned with references retrieved from GenBank [57] using muscle [58] (Anaplasmataceae and Borrelia ) or clustal w [59] ( Bartonella ). The clustal w aligment method was identified as most suitable for Bartonella analysis due to the variable length of the 16S rRNA and ITS regions targeted. Phylogenies were inferred using the maximum-likelihood (ML) method. The optimal evolutionary model was selected using ModelFinder [60] based on the Bayesian information criterion. Phylogenetic analysis was performed in IQ-TREE v1.6.11 [61] and bootstrap support was calculated using the ultrafast (UFBoot2) method with 10000 replicates [62].
Results
Next-generation sequencing
The hosts species sampled in the present study were the black rat (Rattus rattus), brown antechinus (Antechinus stuartii), brush-tailed possum (Trichosurus vulpecula), bush rat (Rattus fuscipes), chuditch (Dasyurus geoffroii), Rusa deer (Rusa timorensis), long-nosed bandicoot (Perameles nasuta), quenda (Isoodon fusciventer), rabbit (Oryctolagus cuniculus) and swamp rat (Rattus lutreolus) (Table 1). Ticks identified from wildlife hosts were Am. triguttatum, Ix. antechini, Ix. australiensis, Ix. holocyclus, Ix. tasmani and Ix. trichosuri. Molecular screening was performed on 257 ticks pooled into 205 gDNA extracts (Table 3). Metabarcoding of ticks at the 12S rRNA locus confirmed morphological analysis and accurate differentiation of morphologically similar species. These results identified that mixed tick species were detected in five gDNA pools and, in all cases, these were larvae of Ix. holocyclus and Ix. trichosuri. A subset of representative 12S rRNA tick zOTU sequences were deposited under accession numbers MW665133–MW665150.
Table 3.
Summary of tick species collected from wildlife; sample numbers in the table refer to the number of gDNA extracts with total number of tick specimens included in parentheses
|
Host |
Tick species |
Larvae |
Nymph |
Male |
Female |
|---|---|---|---|---|---|
|
Black rat |
Ix. holocyclus |
6 (12) |
5 (5) |
0 |
0 |
|
|
Ix. tasmani |
30 (38) |
31 (34) |
0 |
0 |
|
|
Ix. trichosuri |
4 (9) |
2 (3) |
0 |
0 |
|
|
Ix. holocyclus; Ix. trichosuri |
1 (3) |
0 |
0 |
0 |
|
Brown antechinus |
Ix. antechini |
0 |
0 |
0 |
1 (1) |
|
|
Ix. holocyclus |
1 (1) |
0 |
0 |
0 |
|
|
Ix. tasmani |
2 (2) |
2 (2) |
0 |
0 |
|
Brush-tailed possum |
Am. triguttatum |
2 (3) |
0 |
0 |
0 |
|
|
Ix. holocyclus |
0 |
3 (3) |
0 |
5 (5) |
|
|
Ix. tasmani |
0 |
0 |
0 |
2 (2) |
|
|
Ix. trichosuri |
1 (1) |
2 (2) |
0 |
18 (18) |
|
Bush rat |
Ix. tasmani |
1 (1) |
3 (3) |
0 |
0 |
|
Chuditch |
Am. triguttatum |
1 (1) |
0 |
0 |
0 |
|
Long-nosed bandicoot |
Ix. holocyclus |
8 (9) |
5 (5) |
5 (5) |
20 (20) |
|
|
Ix. tasmani |
5 (7) |
12 (12) |
0 |
0 |
|
|
Ix. trichosuri |
3 (3) |
2 (2) |
0 |
0 |
|
Quenda |
Am. triguttatum |
0 |
1 (1) |
0 |
0 |
|
|
Ix. australiensis |
0 |
1 (1) |
0 |
0 |
|
Rabbit |
Ix. holocyclus |
3 (6) |
6 (10) |
0 |
0 |
|
|
Ix. tasmani |
0 |
2 (2) |
0 |
0 |
|
|
Ix. trichosuri |
1 (4) |
4 (6) |
0 |
0 |
|
|
Ix. holocyclus; Ix. trichosuri |
3 (12) |
1 (3) |
0 |
0 |
Bacteria 16S metabarcoding
A total of 536 gDNA extracts from 203 individual animals (159 blood, 205 tick pools, 172 tissue samples) underwent bacterial profiling. An additional 65 control samples (EXB and NTC) were also sequenced to obtain background laboratory and reagent profiles. Bacterial 16S metabarcoding produced 102,503, 643 raw sequences (Table S1, Fig. S1; samples=96,969,355, controls=5,534,288). Blood samples produced 35,160,160 sequences (mean=185 053), tick samples produced 24,844, 688 sequences (mean=95,556) and tissue samples produced 36,964, 507 sequences (mean=197, 671). Following quality filtering, denoising and merging, a total of 64, 186,307 sequences (66.2 %) were obtained from the samples; the mean for blood samples was 121 376 (65.6 %), mean for tick samples 70, 650 (73.9 %) and mean for tissue samples 121,689 (61.6 %).
After contaminating taxa and controls were removed, a total of 31,194 bacterial ASVs were identified and 5243 ASVs had an abundance of over 1000 sequences. Rarefaction curves (Fig. S2) showed the number of ASVs generally plateaued at a depth of 60, 000, 40,000 and 50,000 sequences for blood, tick and tissue samples, respectively. Alpha diversity analysis (Fig. 2) identified significant variation between sample types (Wilcoxon pairwise test, P<0.001). The highest number of observed ASVs was in tissue samples (21, 811), followed by blood (9,401) and ticks (3,568). Tissue samples showed the highest level of alpha diversity across all measures of alpha diversity. Ixodes australiensis and Ix. trichosuri had the highest alpha diversity of the tick species sampled (Fig. S3). Once identified, contaminant taxa and controls were removed and the dominant phyla identified were Proteobacteria (37.3 %), Firmicutes (27.8 %), Actinobacteria (14.7 %) and Bacteroidetes (10.7 %) (Figs S4 and S5).
Fig. 2.
Boxplot of Alpha-diversity indices. Diversity indexes: (a) observed number of ASVs, (b) Chao1 index, (c) Shannon index and (d) inverse Simpson index. Boxplots and violin plots represent the distribution of diversity among samples within their category: blood (159), tick (205 pools) and tissue (172). Statistical analysis between sample types was done using Wilcoxon pairwise (non-parametric) test with significance values indicated as follows: NS for P>0.05; * for P≤0.05; ** for P≤0.01; *** for P≤0.001; **** for P≤0.0001.
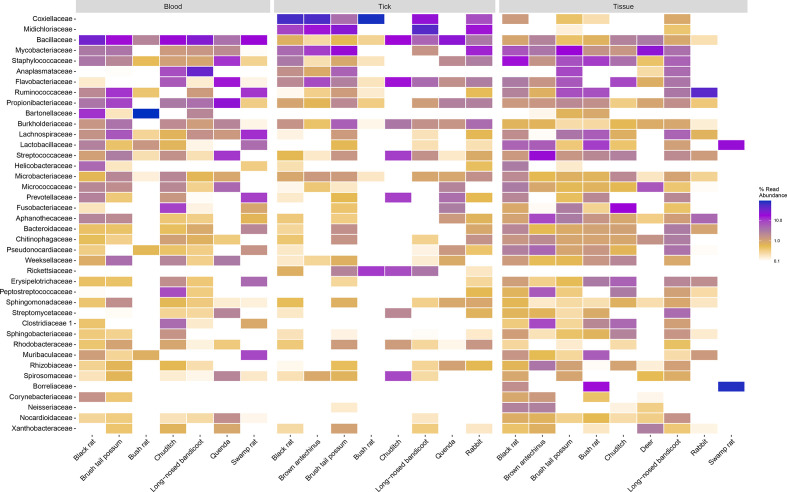
In blood samples, the most abundant families were Bacillaceae (22.9 %), Bartonellaceae (11.3 %), Anaplasmataceae (7.3 %) and Mycobacteriaceae (2.6 %) (Fig. S6). In tick samples, the most abundant families were Coxiellaceae (33.8 %), Midichloriaceae (31.0 %), Mycobacteriaceae (6.0 %), Flavobacteriaceae (2.6 %) and Anaplasmataceae (1.8 %) (Fig. S7). In tissue samples, the most abundant families were Staphylococcaceae (8.2 %), Mycobacteriaceae (6.5 %), Ruminococcaceae (5.5 %), Flavobacteriaceae (4.7 %) and Anaplasmataceae (2.7 %) (Fig. S8). Comparison of family taxa showed differences in abundance among sample types and species (and tick instar) (Figs. 3 and 4).
Fig. 3.
Heatmap of the top 40 most prevalent bacterial family taxa identified in wildlife blood, tick and tissue samples. Data were first transformed to relative sequence abundance.
Fig. 4.
Heatmap of the top 15 most prevalent bacterial family taxa identified in tick samples, showing abundance in species and instar. Data were first transformed to relative sequence abundance. Tick samples consisting of mixed species pools were excluded.
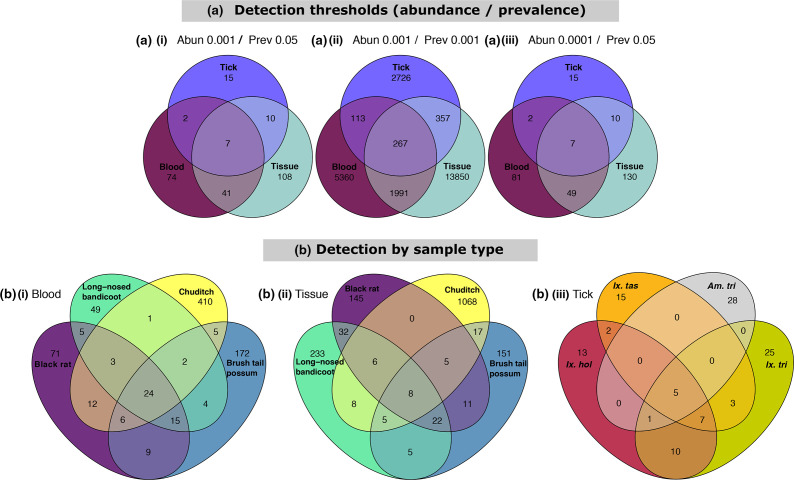
The overlap of samples at the ASV level was investigated using various thresholds to define the ‘core microbiome’, with two out of three models showing only seven shared ASVs between all three sample types (Fig. 5). Shared core microbiome ASVs of blood and tissue samples were compared between the four most abundant host samples and tick species (Fig. 5).
Fig. 5.
Venn diagram of core taxa present within samples presented as detection of ASVs using abundance and prevalence. Data were first transformed to relative abundance and ASVs assigned to the best hit. (a) Impact of similarity based on detection thresholds of abundance and prevalence as (i) 0.001 and 0.05, (ii) 0.001 and 0.001, and (iii) 0.0001 and 0.05. (b) Core taxa present within sample types using detection levels set at 0.001 and 0.05 for abundance and prevalence, respectively, for (i) blood, (ii) tissue and (iii) ticks. For within-sample comparisons, only the four most abundant vertebrate host/tick species were selected. Tick species abbreviations: Amblyomma triguttatum (Am. tri), Ixodes holocyclus (Ix. hol), Ixodes tasmani (Ix. tas), Ixodes trichosuri (Ix. tri). Tick samples consisting of mixed species pools were excluded.
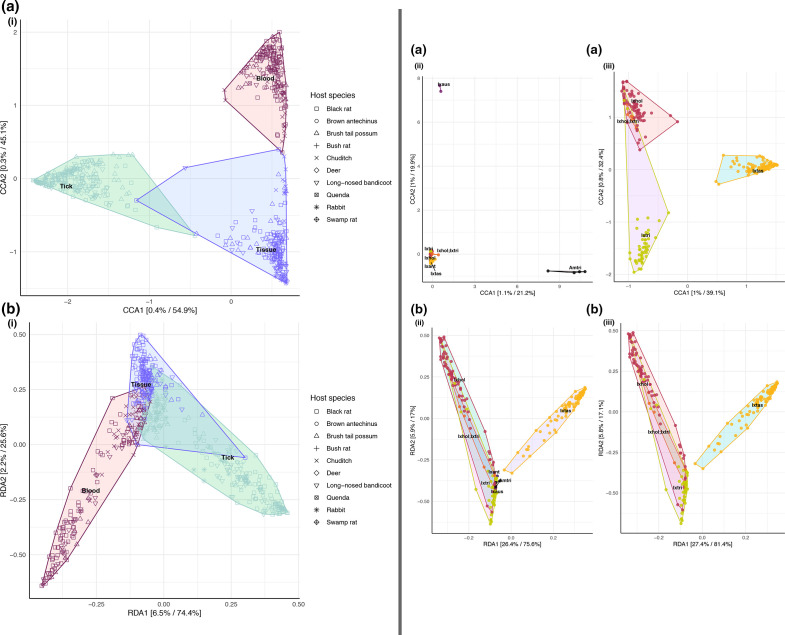
Ordination analysis revealed a distinct difference between the bacterial composition of blood, tick and tissue samples (Fig. 6). Hierarchical cluster analysis (Fig. S9) and statistical analysis (PERMANOVA F=36.209, P=0.001, see Supplementary file S1) also supported this finding showing that there was a statistical difference in sample types, and there was little overlap in the microbiome composition of blood and tick samples. Tissue samples were identified as an intermediate sample type and showed some (albeit low) similarities to blood and tick samples. Ordination analysis of tick samples showed distinction between tick species (Fig. 6). The tick species Ix. holocyclus and Ix. trichosuri showed the most similar bacterial composition with ordination analysis demonstrating overlap between these two species (PERMANOVA, F=14.01, P=0.001, see Supplementary file S1). Although sampling numbers were uneven among host species, which limited statistical inference, hierarchical cluster analysis did show that samples grouped were based on host (and tick) species (Fig. S10).
Fig. 6.
Constrained ordination plot of microbiome composition from using constrained methods: (a) canonical correspondence analysis and (b) redundancy analysis (constrained principal component analysis). (i) Ordination analysis of all samples, N=536 (159 blood, 205 tick pools and 172 tissue) by sample type. (ii) Ordination of all tick samples by tick species. (iii) Ordination of three co-habiting tick species in Sydney Northern Beaches area. Prior to the analysis, ASVs with <0.1 relative abundance were removed. Analysis was done using the Hellinger transformation and Bray–Curtis distance measure. The relative contribution (eigen value) of each axis to the total inertia in the data as well as to the constrained space only, respectively, are indicated in percent on the axis titles. Tick species abbreviations: Amblyomma triguttatum (Am. tri), Ixodes antechinus (Ix. ant), Ixodes australiensis (Ix. aus), Ixodes holocyclus (Ix. hol), Ixodes tasmani (Ix. tas), Ixodes trichosuri (Ix. tri).
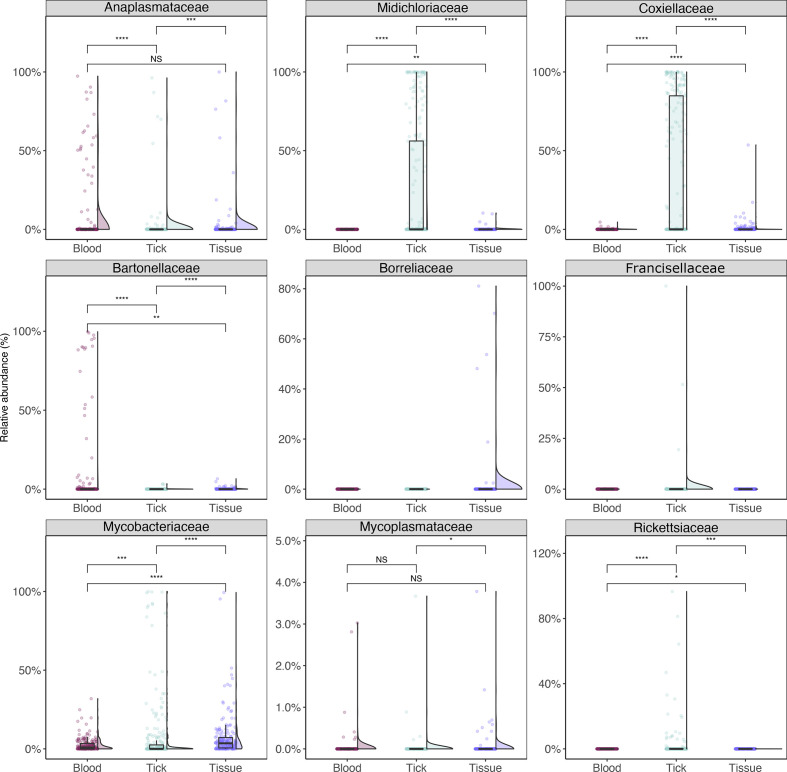
Taxa of interest
Nine family taxa were chosen for comparison among sample types (Fig. 7). Anaplasmataceae bacteria were most prevalent in blood samples (both by relative abundance and prevalence). Blood and tissue samples showed no statistical difference in relative abundance (Wilcoxon pairwise test, P=0.26), with significantly lower abundance in tick samples (Wilcoxon pairwise test, P<0.001). There was weak support for differences in Mycoplasmataceae between sample types (Wilcoxon pairwise test, P=0.023–0.43). Midichloriaceae , Coxielleaceae and Rickettsiaceae were all significantly more abundant in ticks than in blood or tissue samples. Statistical comparisons of Borreliaceae and Francisellaceae were not made due to the small number of positive samples.
Fig. 7.
Relative abundance for select bacterial taxa of interest (aggregated to the family level) from wildlife samples. Statistical analysis between sample types was done using a Wilcoxon pairwise (non-parametric) test with significance values indicated as follows: NS for P>0.05; * for P≤0.05; ** for P≤0.01; *** for P≤0.001; **** for P≤0.0001. Taxa of interest are defined as bacteria related to known pathogens associated with ticks and other vectors described globally, as outlined in Egan et al. [18].
For subsequent prevalence analysis only taxa of interest that could be accurately defined to the genus level were retained (as per blast results). This identified 235 ASVs from 364 samples and 159 individuals (see Table 4 and full sequence data and blast results in Supplementary file S2).
Table 4.
Presence of taxa of interest* by bacteria metabarcoding
Prevalence is reported as the proportion of individual hosts positive with the proportion of samples positive in parentheses (accounting for instances where more than one sample type was collected from an individual).
|
Taxa of interest |
Sample type |
Black rat |
Brown antechinus |
Brush tail possum |
Bush rat |
Chuditch |
Deer |
Long-nosed bandicoot |
Quenda |
Rabbit |
Swamp rat |
|---|---|---|---|---|---|---|---|---|---|---|---|
|
Blood |
4/68 (4/84) |
0/0 (0/0) |
5/18 (6/21) |
0/2 (0/2) |
7/22 (9/24) |
0/0 (0/0) |
18/20 (22/25) |
0/1 (0/2) |
0/0 (0/0) |
0/1 (0/1) |
|
|
|
Tick |
3/54 (3/79) |
1/4 (1/6) |
4/15 (5/33) |
0/3 (0/4) |
0/1 (0/1) |
0/0 (0/0) |
8/31 (9/60) |
0/2 (0/2) |
0/7 (0/20) |
0/0 (0/0) |
|
|
Tissue |
6/71 (8/80) |
0/2 (0/2) |
4/16 (5/20) |
0/3 (0/3) |
1/9 (1/9) |
3/3 (3/6) |
18/40 (19/44) |
0/0 (0/0) |
0/7 (0/7) |
0/1 (0/1) |
|
Blood |
22/68 (27/84) |
0/0 (0/0) |
7/18 (8/21) |
2/2 (2/2) |
0/22 (0/24) |
0/0 (0/0) |
4/20 (4/25) |
0/1 (0/2) |
0/0 (0/0) |
0/1 (0/1) |
|
|
|
Tick |
3/54 (4/79) |
0/4 (0/6) |
0/15 (0/33) |
1/3 (1/4) |
0/1 (0/1) |
0/0 (0/0) |
1/31 (1/60) |
0/2 (0/2) |
0/7 (0/20) |
0/0 (0/0) |
|
|
Tissue |
4/71 (5/80) |
1/2 (1/2) |
9/16 (11/20) |
2/3 (2/3) |
0/9(0/9) |
0/3 (0/6) |
3/40 (3/44) |
0/0 (0/0) |
0/7 (0/7) |
0/1 (0/1) |
|
Blood |
0/68 (0/84) |
0/0 (0/0) |
0/18 (0/21) |
0/2 (0/2) |
0/22 (0/24) |
0/0 (0/0) |
0/20 (0/25) |
0/1 (0/2) |
0/0 (0/0) |
0/1 (0/1) |
|
|
|
Tick |
0/54 (0/79) |
0/4 (0/6) |
0/15 (0/33) |
0/3 (0/4) |
0/1 (0/1) |
0/0 (0/0) |
0/31 (0/60) |
0/2 (0/2) |
0/7 (0/20) |
0/0 (0/0) |
|
|
Tissue |
7/71 (7/80) |
0/2 (0/2) |
0/16 (0/20) |
1/3 (1/3) |
0/9 (0/9) |
0/3 (0/6) |
0/40 (0/44) |
0/0 (0/0) |
0/7 (0/7) |
1/1 (1/1) |
|
Blood |
7/68 (7/84) |
0/0 (0/0) |
1/18 (1/21) |
0/2 (0/2) |
0/22 (0/24) |
0/0 (0/0) |
3/20 (5/25) |
0/1 (0/2) |
0/0 (0/0) |
0/1 (0/1) |
|
|
|
Tick |
50/54 (63/79) |
3/4 (4/6) |
8/15 (9/33) |
3/3 (4/4) |
0/1 (0/1) |
0/0 (0/0) |
22/31 (24/60) |
0/2 (0/2) |
2/7 (2/20) |
0/0 (0/0) |
|
|
Tissue |
33/71 (38/80) |
0/2 (0/2) |
2/16 (2/20) |
2/3 (2/3) |
0/9 (0/9) |
0/3 (0/6) |
20/40 (22/44) |
0/0 (0/0) |
0/7 (0/7) |
0/1 (0/1) |
|
Blood |
0/68 (0/84) |
0/0 (0/0) |
0/18 (0/21) |
0/2 (0/2) |
0/22 (0/24) |
0/0 (0/0) |
0/20 (0/25) |
0/1 (0/2) |
0/0 (0/0) |
0/1 (0/1) |
|
|
|
Tick |
0/54 (0/79) |
0/4 (0/6) |
2/15 (2/33) |
0/3 (0/4) |
0/1 (0/1) |
0/0 (0/0) |
0/31 (0/60) |
2/2 (2/2) |
0/7 (0/20) |
0/0 (0/0) |
|
|
Tissue |
0/71(0/80) |
0/2(0/2) |
0/16 (0/20) |
0/3 (0/3) |
0/9 (0/9) |
0/3 (0/6) |
0/40 (0/44) |
0/0 (0/0) |
0/7 (0/7) |
0/1 (0/1) |
|
Blood |
0/68 (0/84) |
0/0 (0/0) |
0/18 (0/21) |
0/2 (0/2) |
0/22 (0/24) |
0/0 (0/0) |
0/20 (0/25) |
0/1 (0/2) |
0/0 (0/0) |
0/1 (0/1) |
|
|
|
Tick |
17/54 (21/79) |
2/4 (2/6) |
12/15 (20/33) |
0/3 (0/4) |
0/1 (0/1) |
0/0 (0/0) |
19/31 (41/60) |
0/2 (0/2) |
5/7 (10/20) |
0/0 (0/0) |
|
|
Tissue |
5/71 (5/80) |
0/2 (0/2) |
1/16 (1/20) |
0/3(0/3) |
0/9 (0/9) |
0/3 (0/6) |
3/40 (3/44) |
0/0 (0/0) |
0/7 (0/7) |
0/1 (0/1) |
|
Blood |
1/68 (1/84) |
0/0 (0/0) |
0/18 (0/21) |
0/2(0/2) |
0/22 (0/24) |
0/0 (0/0) |
0/20 (0/25) |
0/1 (0/2) |
0/0 (0/0) |
1/1 (1/1) |
|
|
|
Tick |
0/54 (0/79) |
0/4 (0/6) |
0/15 (0/33) |
0/3 (0/4) |
0/1 (0/1) |
0/0 (0/0) |
0/31 (0/60) |
0/2 (0/2) |
0/7 (0/20) |
0/0 (0/0) |
|
|
Tissue |
1/71 (1/80) |
0/2 (0/2) |
0/16 (0/20) |
0/3 (0/3) |
0/9 (0/9) |
0/3 (0/6) |
0/40 (0/44) |
0/0 (0/0) |
0/7 (0/7) |
0/1 (0/1) |
|
Blood |
0/68 (0/84) |
0/0 (0/0) |
0/18 (0/21) |
0/2 (0/2) |
0/22 (0/24) |
0/0 (0/0) |
0/20 (0/25) |
0/1 (0/2) |
0/0 (0/0) |
0/1 (0/1) |
|
|
|
Tick |
7/54 (7/79) |
0/4 (0/6) |
2/15 (2/33) |
2/3 (3/4) |
1/1 (1/1) |
0/0 (0/0) |
9/31 (10/60) |
0/2 (0/2) |
1/7 (1/20) |
0/0 (0/0) |
|
|
Tissue |
0/71 (0/80) |
0/2 (0/2) |
0/16 (0/20) |
0/3 (0/3) |
0/9 (0/9) |
0/3 (0/6) |
0/40 (0/44) |
0/0 (0/0) |
0/7 (0/7) |
0/1 (0/1) |
*Taxa of interest are defined as bacteria related to known pathogens associated with ticks and other vectors described globally, as outlined in Egan et al. [18].
Anaplasmataceae
The family Anaplasmataceae was identified in 95 samples from 59 individuals. The most prevalent genera were Neoehrlichia (43 individuals), followed by Ehrlichia (eight individuals) and Anaplasma (three individuals).
‘Candidatus Neoehrlichia arcana’ was identified in 52 samples: 29 blood, seven ticks (Ix. holocyclus) and 16 tissue. ‘Candidatus Neoehrlichia australis’ was identified in 33 samples: eight blood, six ticks (Ix. holocyclus, Ix. trichosuri) and 19 tissue. Sixteen samples had mixed infections of ‘Ca. N. arcana’ and ‘Ca. N. australis’ (six blood and ten tissue). No ticks had mixed Neoehrlichia infections. Three putative novel Anaplasmataceae species were identified from the present study, two Ehrlichia spp. and a single Neoehrlichia sp. The novel Neoehrlichia sp. was identified in three samples: a tissue sample from a black rat and a corresponding tick from the same individual (Ix. tasmani); and a tick from a brown antechinus (Ix. antechini). One novel Ehrlichia sp. was identified in five samples from two brush-tailed possums: two blood, one tick (Ix. holocyclus) and two tissue. A second novel Ehrlichia sp. was identified in seven samples from six chuditch (six blood and one tissue). Only one chuditch was positive in corresponding tissue and blood samples. Three deer tissue samples were infected with Anaplasma platys (Supplementary file S2). Detection of Anaplasmataceae bacteria differed between tick species and instars. Ixodes holocyclus females had the highest prevalence of this taxon, followed by nymph and larval ticks, while no males were positive. Only Ix. tasmani nymph samples were positive and in the case of Ix. trichosuri, females and nymphs were identified with Anaplasmataceae and all larvae were negative.
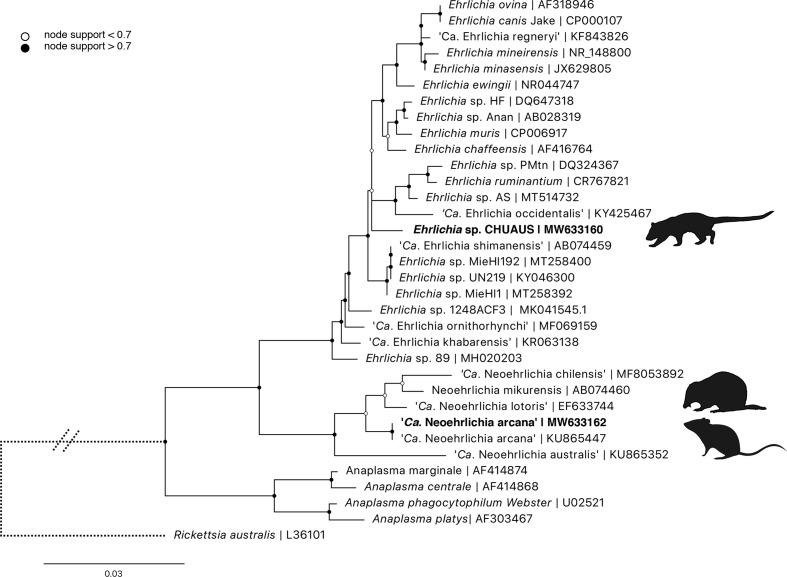
Phylogenetic analysis was performed using a 1,244 bp fragment of the 16S rRNA locus (Fig. 8). All ‘Ca. N. arcana’ amplified were identical to each other and showed 100% similarity to ‘Ca. Neoehrlichia arcana’ isolate HT94 from Ix. holocyclus, Australia (KU865447). Ehrlichia sequences from chuditch blood samples were 100% identical to each other and most similar to Ehrlichia sp. Anan from Ix. ovatus, Japan (AB028319, 98.94% similarity). The nearest named species was Ehrlichia muris (CP006917, 98.86% similarity) isolated from a wild mouse (Eothenomys kageus) in Japan (genetic distances in Supplementary file S3).
Fig. 8.
ML phylogenetic reconstruction of Anaplasmataceae based on a 1244 bp alignment of the 16S rRNA locus. Substitution model: K80+F with 10000 replicates. Node values correspond to bootstrap support where values >0.7 are indicated by shaded circles. Bar, number of substitutions per nucleotide position. Sequences generated in the present study are in bold type.
Midichloria
A total of 103 samples were positive for Midichloria , which were identified in 94 tick samples from 62 individual animals. Positive tick species included Ix. holocyclus, Ix. tasmani and Ix. trichosuri from black rats, brown antechinuses, brush-tailed possums, long-nosed bandicoots and rabbits from New South Wales. While Midichloria was identified in 13 Ix. tasmani, the relative abundance was low (108–363 sequences; 0.1–0.4% relative abundance). Nine tissue samples (nine individuals) were positive for Midichloria , but the number of sequences was generally low (111–14,002) when compared to tick samples; hosts included black rats (five), a brush-tailed possum (one) and long-nosed bandicoots (two). Only two individuals had positive tissue and tick samples, one brush-tailed possum and one long-nosed bandicoot. Midichloria was identified in all instars of Ix. holocyclus but only female and nymph stages of Ix. trichosuri (Fig. 4). No blood samples were positive for Midichloria (Fig. 3).
Coxiellaceae
The family Coxiellaceae was identified in 183 samples from 113 individuals. Eleven individuals had positive blood samples, although there was a relatively low number of sequences (100–2819), from hosts including the black rat (seven), brush-tailed possum (one) and long-nosed bandicoot (three). In total, 106 ticks from 88 individuals were positive: Am triguttatum (one), Ix. tasmani (90) Ix. holocyclus (nine), and Ix. trichosuri. Fifty-seven individuals were positive for Coxiellaceae in tissue samples, including the black rat (33), brush-tailed possum (two), bush rat (two) and long-nosed bandicoot (20). Forty-five individuals had at least two sample types positive for Coxiellaceae , and only two black rats had positive blood, tick and tissue samples. All instars of Ix. tasmani sampled (female, nymph, larva) were positive for Coxiellaceae , but only female Ix. holocyclus were positive.
Rickettsiaceae
Rickettsia was identified exclusively in 24 tick samples from 22 individuals. Positive samples from Western Australia were all identified as originating in Am. triguttatum (three) from brush-tailed possums and quenda. Ticks identified as positive from New South Wales were: Ix. holocyclus (nine) from black rats and long-nosed bandicoots, Ix. tasmani (15) from black rats, bush rats, long-nosed bandicoots and rabbits, and Ix. trichosuri (three) from long-nosed bandicoots. No tissue or blood samples were positive for Rickettsia .
Borrelia
A novel Borrelia sp. was identified in nine tissue samples: seven black rats, one bush rat and one swamp rat. The identity of these Borrelia sequences showed they were most similar to Borrelia R57 (AY626138, 95.68–96.70% similarity, Supplementary file S2). Corresponding blood and ticks from these individuals were negative and no other Borrelia sequences were identified from any other samples.
Attempts to amplify and perform Sanger sequencing on the flaB gene using pan- Borrelia primers [63] were unsuccessful. Given these failed attempts and the small volume of gDNA from samples (tissue biopsy punch only 2 mm), an alternative approach was employed to amplify additional fragments of the 16S rRNA v3–4 hypervariable region on the Illumina MiSeq.
After bioinformatic analysis outlined above, a single ASV was generated from all nine samples. Phylogeny was reconstructed using the ML method (GTR+R model) based on a 426 bp alignment of the 16S rRNA locus (hypervariable region v3–4) (Fig. 9). The novel Borrelia sequence showed 98.4% similarity to Borrelia sp. ALEPB216 (KF957671) from R. rattus in California, USA (genetic distances in Supplementary file S4). The next most similar sequences were Borrelia sp. CA682 (KF957670, similarity 98.1%) and Borrelia sp. R57 (AY626138, similarity 97.6%). The topology showed that these sequences formed a distinct ‘rodent’ clade, basal to the three major groups currently described (Fig. 9). The nearest named species was Borrelia theileri (U38375, similarity 94.50%).
Fig. 9.
ML phylogenetic reconstruction of Borrelia based on a 431 bp alignment of the 16S rRNA locus (hypervariable region 3–4). Substitution model: K2P+G4 with 10000 replicates. Node values correspond to bootstrap support where values >0.7 are indicated by shaded circles. Bar, number of substitutions per nucleotide position. The sequence generated in the present study is shown in bold type.
Bartonella
Bartonella sp. was identified in 69 samples from 47 individuals. Blood samples represented the majority of Bartonella positives with 35 individuals, which included black rats (22), brush-tailed possums (seven), bush rats (two) and long-nosed bandicoots (four). Nineteen individuals had positive tissue samples: black rats (four), brown antechinus (one), brush-tailed possums (nine), bush rats (two) and long-nosed bandicoots (three); seven individuals had positive tissue and blood samples. Six tick samples were positive for Bartonella (from five individuals) with three tick species (Ix. holocyclus, Ix. tasmani and Ix. trichosuri) from black rats and long-nosed bandicoots. All tick samples that were positive also had corresponding positive blood or tissue samples.
Targeted sequencing on a subset of positive samples showed two genotypes of Bartonella queenslandensis that were 97.2% similar to each other with ten SNPs (genetic distances in Supplementary file S5). The next most similar sequence to genotype BR025 and BR048 was Ba. queenslandensis strain AUST/NH8 (EU111767) at 96.1 and 94.8% similarity, respectively. Comparison of Ba. queenslandensis type sequences showed divergence of up to 7.2% (between EU111765 and EU111766) within the species. Five genotypes of Ba. coopersplainsensis were identified with 94.4–99.2% similarity to each other, and 96.7–98.7% similarity to the Ba. coopersplainsensis type sequence (EU111770) (Fig. 10).
Fig. 10.
ML phylogenetic reconstruction of Bartonella based on a 533 bp alignment of the 16S–23S ITS region. Substitution model: HKY+F with 10000 replicates. Node values correspond to bootstrap support, where values >0.7 are indicated by shaded circles. Bar, number of substitutions per nucleotide position. Sequences generated in the present study are in bold type. Taxa of interest are defined as bacteria related to known pathogens associated with ticks and other vectors described globally, as outlined in Egan et al. [18].
Francisella and Mycoplasma
Francisella was identified in four ticks, Am. triguttatum (three) and Ix. australiensis (one) from brush-tailed possums and quenda in Western Australia (Table 4, Supplementary file S2). Mycoplasma was present in three samples, from three individuals: two blood samples from a black rat and a swamp rat and one tissue sample from a black rat (Table 4, Supplementary file S2).
Discussion
This body of work represents the first large-scale investigation into tick (and vector)-associated bacteria concurrently with their vertebrate hosts in urban and peri-urban locations, and identified novel tick-associated bacteria in Australian wildlife. Bacterial amplicon sequencing showed that there were limited taxa shared between blood, tick and tissue samples. The identification of bacterial taxa of interest was generally confined to certain hosts and/or sample types. To the best of the authors’ knowledge, this study represents the first report of Neoehrlichia and Ehrlichia from Australian wildlife (blood and/or tissue samples). The first identification of the rodent-associated Borrelia clade in Australia is also described. These findings show that geographical boundaries and host associations influence the prevalence of tick-associated bacteria. Importantly, these findings also provide support for the absence of northern hemisphere tick-borne pathogens, adding to the overwhelming body of evidence that any potential human tick-borne pathogens in Australia are probably endemic and genetically distinct from currently described pathogens in parts of North America and Europe.
Although a comparatively deep level of sequencing was achieved compared to similar studies [64, 65], rarefaction plots showed that blood samples did not reach the same asymptote as tick and tissue, indicating that deeper sequencing of these samples is needed to ensure complete coverage of bacterial diversity. Across all four alpha diversity measures, tissue samples showed the highest diversity and variance. In comparison, tick samples showed a less diverse bacterial composition. This is likely to be due to the dominance of endosymbiont bacteria in the tick microbiome, which has been demonstrated in previous studies of the same tick species as identified in the present study [17, 18], and in tick species globally [66, 67]. The overall lack of similarity in bacterial communities between sample types observed in the present work has been reported in previous studies on small mammals and their vectors (ticks and fleas), indicating the complexity of microbial assemblages inhabiting these biological niches [65, 68, 69]. The diagnostic value of 16S rRNA amplicon sequencing methods has not been thoroughly validated for tick-borne pathogens. One study, however, showed that ticks which tested positive for Borrelia using quantitative PCR methods also had a high abundance of Borrelia using amplicon bacterial metabarcoding [70]. Therefore, even though Ix. holocyclus and Ix. tasmani had abundant endosymbionts, and the presence of bacterial species in lower abundance cannot be ruled out completely, it is likely that our finding of the absence of some bacterial species here reflects the true situation.
The microbiome of Ix. holocyclus and Ix. tasmani was dominated by a limited number of highly abundant ASVs. Other tick species, such as Ix. trichosuri and Ix. australiensis, showed a more diverse bacterial composition. The interaction between bacterial endosymbionts and pathogens in ticks is not yet fully understood. The presence of endosymbiont bacteria is thought to inhibit the colonization by other potentially pathogenic bacterial species. For example, colonization by the rickettsial endosymbiont ( Rickettsia peacockii ) in Dermacentor andersoni tick ovaries is thought to render the ticks more resistant to co-infection by a second rickettsial species ( Rickettsia rickettsii ), which is the main cause of Rocky Mountain Spotted Fever [71]. Likewise, Dermacentor variabilis ticks infected with one strain of Rickettsia were shown to be less susceptible to secondary rickettsial infections [72]. However, this theory is not uniformly accepted, and there is some rejection of Burgdorfer’s [71] initial findings. Instead, it is hypothesized that differences in R. peacockii prevalence in De. andersoni are due to microclimatic factors [73]. Additionally, in recent years microbiome profiling has led to conflicting conclusions about the interaction between endosymbiont and pathogens [74]. Based on what is known from the northern hemisphere examples, tick species without a dominant endosymbiont may pose a greater risk of harbouring potentially pathogenic microbes [74].
Equivalent studies from the northern hemisphere using 16S rRNA metabarcoding methods and wildlife surveillance to identify tick-borne pathogens are limited. Instead, pathogen surveillance studies are generally targeted to well-known pathogens. For example, studies of B. burgdorferi sensu lato have shown infection is not detrimental to the health of natural rodent reservoirs [75], which have been shown to harbour a diverse range of Borrelia species [76]. In the present study, chuditch and long-nosed bandicoots had a high abundance of Anaplasmataceae in blood samples, while brush-tailed possums had the highest abundance of this taxon in tissue samples. In addition, Anaplasmataceae bacteria were mostly absent from larval ticks. This suggests that the Anaplasmataceae bacteria identified are probably maintained via horizontal transmission (i.e. between host and tick) and transstadial routes, which has been observed with related bacteria in companion animals [77, 78]. In contrast, the identification of Midichloria across all life stages of Ix. holocyclus shows evidence of vertical (transovarial) transmission. Both horizontal and vertical transmission have been identified previously in other tick species with Midichloria [79, 80]. The absence of Midichloria in vertebrate hosts (blood and skin) in the present study suggests the horizontal infection route is not a major contributor to the presence of this endosymbiont in Ix. holocyclus and is consistent with previous studies [81].
Taxa of interest
Anaplasmataceae
The present study has provided the first identification of Neoehrlichia spp. from Australian wildlife. Two Neoehrlichia species, ‘Ca. N. arcana’ and ‘Ca. N. australis’, were recently characterized from Ix. holocyclus along the east coast of Australia at a prevalence of 11.3% (44/391) [24]. This finding is consistent with other species of Neoehrlichia globally, which have been described circulating in a range of mammals and associated ticks [82, 83]. In contrast to Gofton et al. [24], the results presented here show that ‘Ca. N. arcana’ is more prevalent than ‘Ca. N. australis’ in small mammals, which is consistent with a recent study of Australian wildlife ticks [18]. The main reservoir host for ‘Ca. N australis’ is likely to be medium to large macropods [e.g. eastern grey kangaroo (Macropus giganteus) or swamp wallaby (Wallabia bicolor)], which were not sampled in the present study.
The identification of three putative novel Anaplasmataceae species further demonstrates the high diversity of Anaplasmataceae from Australian ticks and wildlife. This finding adds to recent discoveries identifying new species from Am. triguttatum [23], Bothriocroton concolor [84], Ix. holocyclus [24], and from the platypus (Ornithorhynchus anatinus) and its associated ticks Ix. ornithorhynchi [85]. The novel Anaplasmataceae species were less readily identified in tick vectors compared to ‘Ca. N. arcana’ and ‘Ca. N. australis’; however, more targeted studies are needed to understand their prevalence and distribution.
The first identification of Anaplasma platys (formerly Ehrlichia platys) in Australia was made in 2001 [86], and has since been described from dogs throughout the country [87, 88]. To date it has not been identified in any native Australian wildlife, although A. platys has been reported from deer overseas [89]. To the best of our knowledge, this is the first report of A. platys from wild deer in Australia. The vector of A. platys overseas, Rhipicephalus sanguineus [90], is probably also the vector in Australia. The brown dog tick, syn. Rh. sanguineus, tropical lineage was recently named Rhipicephalus linnaei [91]. Importantly, Ehrlichia canis , which was first identified in dogs and brown dog ticks in northern Australia during 2020, was not identified in ticks, tissue or blood from wildlife in this study.
Midichloria
While ‘Ca. M. mitochondrii’ (CMm) is known to be highly prevalent and abundant in ticks identified in the present study, the present study did not employ a blocking primer as used in previous studies [17, 18]. The justification for this was a deliberate choice to include CMm in the ‘taxa of interest’ investigation, i.e. provide insight into the ecology of this microbe. The absence of CMm in both blood and tissue was surprising, especially given the high proportion and abundance of the bacteria in Ix. holocyclus in the present study. Studies in France have identified CMm from roe deer (Capreolus capreolus) blood samples using molecular and serology techniques [80]; while that study reported a higher sensitivity for serology, molecular methods were also successful at identifying infection. In vitro studies of Ix. ricinus and rabbits suggest that the endosymbiont replicates within the vertebrate host [92]. The bacterial microbiome of many tick species is generally dominated by the presence of one (or a few) highly abundant endosymbiont microbes [17, 93, 94]. This suggests that rare, less abundant bacteria are not easily identified using standard 16S rRNA bacterial metabarcoding methods. While the possibility cannot be ruled out, given the high abundance of CMm in tick samples, it is likely that the methods employed here would have identified it in wildlife hosts (either blood or tissue) if present. Therefore, given the absence of CMm in the hosts sampled in the present study, vertical transmission may be the primary source of Midichloria infection for Ix. holocyclus. Alternatively, the reservoir host for CMm may not have been sampled.
Coxiellaceae
All Coxiellaceae ASVs identified were most similar to members of the genus Rickettsiella and dominated the microbiome of Ix. tasmani. Coxiella was not detected in the present study. The absence of Coxiella from host blood and tissue samples probably reflects a low abundance of this bacteria in the small mammal hosts sampled. Serological methods are more commonly used to detect exposure to C. burnetii [95] but lack the ability to detect acute infections. A recent study using molecular techniques detected C. burnetii in kangaroo meat samples [96]. This suggests that animals sampled in the present study were negative for an active infection with Coxiella .
Rickettsiaceae
While several forms of rickettsiosis are amongst the few recognized human tick-borne diseases in Australia, remarkably little is known about the sylvatic life cycle of these intracellular bacteria. At least five agents of human rickettsial disease have been described in Australia [97], together with several additional species and genotypes with unknown pathogenicity [97–99]. Bandicoots and other small mammals have previously been described as reservoirs for Queensland tick typhus ( R. australis ), and Ix. holocyclus and Ix. tasmani are known vectors [100, 101]. Although the relationship between bandicoots and rickettsial infections is largely accepted, there has been little validation of this association since the early publications.
In the present study, the genus Rickettsia was predominately identified from ticks, with a low prevalence and abundance in four tissue samples and was absent from all blood samples. Areas sampled in Sydney’s northern beaches are endemic for Queensland tick typhus and spotted fever. Therefore, the absence of spotted fever or typhus group Rickettsia from mammal hosts in the present study was unexpected. Recent studies utilizing qPCR have reported Rickettsia in 15.4% (23/149) Ix. holocyclus from NSW [102] and 6.4% (13/203) ticks collected from wildlife in north-east Queensland [103]. Additional reports also recognize the presence of spotted fever group Rickettsia in Australian ticks [100, 104, 105].
The lack of Rickettsia identified in ticks may be due to the highly abundant endosymbionts Midichloria or Coxiellaceae in Ix. holocyclus and Ix. tasmani respectively. While there are limited comparable studies that have used bacterial metabarcoding on wildlife blood samples, previous research has shown that molecular methods successfully identify Rickettsia in reservoir hosts [106, 107, 108]. Flinders Island Spotted Fever, caused by R. honei , is associated with reptiles and the southern reptile tick Bo. hydrosauri [109]. A closely related strain, named R. honei marmionii, has been identified from various regions in Australia and is associated with Haemaphysalis novaeguineae [110, 111]. Interestingly no Haemaphysalis ticks were identified from hosts in the present study. Further investigation into the sylvatic cycle of Rickettsia in Australia would benefit from the inclusion of samples from reptile hosts and vertebrate hosts of Haemaphysalis ticks.
Borrelia
The novel Borrelia identified here came exclusively from black rats and was found at two sites in Sydney’s Northern Beaches area. All samples that tested positive were tissue, and no Borrelia sequences were identified from corresponding blood or tick samples.
The Borrelia species identified from black rats formed a clade with other described Borrelia species identified from rodents. Borrelia sp. R57 was identified from bank voles (Myodes glareolus; formerly Clethrionomys glareolus) and wood mice (Apodemus sylvaticus) in Spain at a prevalence rate of 8.5–12.0% using PCR methods [112]. That study identified serological cross-reactivity of this species with B. burgdorferi sensu lato using western blot testing. A follow-up study (in Spain) further identified the genotype from 24.5% (62/253) of rodent tissue samples [113]. Identification of similar sequences was subsequently made from black rats (R. rattus) in California, USA, with a prevalence of 43.5% (10/23) in tissue (skin) biopsy samples [114]. The absence of this Borrelia species from our tick samples is interesting and mirrors the previous studies in Europe and the USA [112, 114]. Despite multiple attempts during this study, amplification at the flaB locus was unsuccessful, consistent with previous research of members within the clade [112].
The extensive geographical range of this clade, yet tight host associations identified to date (i.e. confined to the order Rodentia), raises questions regarding its origin. Particularly interesting is the paucity of reports globally, despite small mammals being the most widely studied reservoir host of tick-borne diseases [115]. Our phylogenetic analysis shows strong support for these Borrelia sequences forming a distinct clade and is consistent with other studies [116]. Its basal phylogenetic position may provide important insights into the evolutionary history of the genus Borrelia . The lack of its identification in tick samples means that the vector for this Borrelia sp. remains unknown, and the transmissibility and pathogenicity of this microbe requires further research. Importantly, no identifications were made of the recently characterized Borrelia species from Australia, ‘Ca. B. tachyglossi’ [21] or reptile Borrelia sp. [22], nor any sequences from the B. burgdorferi sensu lato group.
Currently there is no evidence that the novel rodent-associated Borrelia genotype identified in the present study is transmissible to humans or capable of causing disease. Phylogenetic reconstruction indicates that it is distinct from the B. burgdorferi sensu lato and relapsing fever groups [116]. The absence of detection from tick samples and its identification only in rodents more probably implies another vector that presumably has a tight host relationship with Rattus species.
Bartonella
Bartonella spp. were identified in a relatively high proportion of wildlife blood samples, and most prevalent in blood samples from black rats (31.0%). Bartonella coopersplainsensis and Ba. queenslandensis were described from native Australian rodents (Melomys spp. and Rattus spp.) in Queensland [117]. Since then, a number of novel Bartonella strains have been identified from Australia, including novel species described from marsupials and their ectoparasites (ticks and fleas) [118–120]. While generally considered to be transmitted by fleas, reports of Bartonella species isolated from ticks are increasing, as studies take a broader approach to surveillance of vector-associated microbes [65, 121, 122]. Identification of Bartonella in fed ticks does not mean they are a viable vector, but it does highlight that future studies should include ticks when exploring candidate vectors. The high prevalence of identification of Bartonella in rodents has been reported in other metabarcoding studies from blood samples [69].
Francisella and Mycoplasma
Francisella was identified exclusively from ticks in Western Australia, Am. triguttatum and Ix. australiensis. Previous bacterial profiling of Am. triguttatum has shown a high proportion of Francisella species [18, 123]. Given its high abundance in tick samples, the absence of Francisella in a corresponding host may indicate a lack of transmission. Tularemia (caused by Francisella tularensis ) can cause disease in humans and animals. While the infection is more commonly acquired via handling infected animal material or aerosol inhalation, F. tularensis has been reported in ticks [124]. Francisella has been reported in several tick species [125]. While the species identified in the present study is not closely related to F. tularensis , considering the significance of this genus [124], further characterization should be of high importance.
The present study did not identify any haemoprotic mycoplasma species. The Mycoplasma sequences identified were most closely related to Mycoplasma arthritidis , which is recognized as a common pathogen in rats [126]. It was identified in two black rats and a single identification in the native swamp rat.
Limitations and recommendations
While the cost of next-generation or second-generation sequencing platforms continues to decline, analysis of large numbers of samples is still constrained by resources and funds. Amplicon high-throughput sequencing remains a cost-effective approach for characterizing bacterial microbiomes. With the advent of long-read sequencing technologies (e.g. PacBio), more studies are moving towards obtaining full-length 16S rRNA sequences which allow for more robust phylogenetic reconstruction and in many cases identification of bacteria to strain/genotype level [127, 128]. In addition to describing the bacteria present, the idea of the functional tick microbiome has been suggested [129]. While adaptation of these methods can seem promising, their use outside of human microbiome studies is questionable due to the reliability of databases [74, 130]. An alternative approach such as the ‘common core’ or ecological ‘core microbiome’ may be more suitable in the context of tick microbiome research [131]. Studies incorporating transcriptomics will probably also prove useful for understanding the functional components of the microbiome.
While the present study reported on the relative abundance of microbes between ticks sampled, it is well known that differences in the 16S copy number limit inferences [132, 133]. In the context of the present study, relative abundance of taxa was compared among samples, as it was considered a suitable measure given samples were processed in the same manner. Calculations of absolute abundance using 16S rRNA metabarcoding are challenging. While copy number is one factor that can be modelled, it relies on information that in many cases is not accurately known. Using mock communities can help assess bias without baseline data on copy number, but these analysis methods are still limited.
The sensitivity of molecular diagnostic testing for tick-borne pathogens has been shown to vary greatly. Factors that can influence molecular detection of pathogens include level of parasitaemia/bacteraemia, sample type, time of sample collection from initial infection, and pre-analytical preparation techniques; additionally, this is compounded by differences between taxa due to host–pathogen interactions [134–136]. A strong advantage of using generic metabarcoding assays is their unbiased approach; in the context of the present study, the most important factor was to achieve unbiased characterization of bacterial communities from Australian wildlife. However, it is acknowledged that the use of bacterial metabarcoding for the identification of low-abundance and rare organisms is limited. The lack of recognized endemic bacterial pathogens such as Coxiella burnetii and Rickettsia spp. was surprising. Future studies should use serological and culture methods in addition to these highly specific and sensitive molecular tools (e.g. qPCR).
Another limitation of the present study was the bias in animals sampled. Small mammals were chosen as it was hypothesized they would have the highest prevalence of potential tick-borne pathogens based on what is known from the northern hemisphere [137–139]. Further sampling with expanded host and spatial aspects are vital to understand the full epidemiology of tick- (and vector) associated microbes. Likewise, alternative trapping methods (e.g. differing trap type, bait) may expand the suite of hosts captured, importantly those with territorial overlap (e.g. semi-arboreal marsupials).
In the present study, most taxa of interest were confined to specific sample types. For example, Borrelia was identified from tissue samples of rats but absent from any blood or tick samples. This finding has also been reported in white-footed mice (Peromyscus leucopus) [140]. While in many cases disease surveillance is targeted to known pathogens where sampling protocols and diagnostics are well defined, in scoping studies such as the present one, a range of sample types are essential. This is evident in the different ways pathogens act within their host. As the first line of defence, the innate immune system has a key role in protecting against cutaneous and systemic infection [141]. Alternatively pathogens can quickly evade host innate immune responses by rapidly disseminating into the bloodstream after invasion [142, 143]. In the context of Australia, however, many wildlife species are endangered and therefore their capture and sampling in the wild is subject to strict ethical conditions and government regulations. As such, collection of multiple sample types is restricted to those requiring only a minimally invasive method and is also dependent on the availability of trained personnel and resources. In many instances it is standard procedure to collect a tissue biopsy for animal genetic (population) purposes, and this presents a simple alternative to other procedures which may require field anaesthesia. The results here highlight the value of this sample type for use in pathogen surveillance.
Conclusions
Increased land-use and climate change will continue to affect the spread of vectors and associated diseases globally. There is an urgent need for expanded disease surveillance, particularly at the urban interface with wildlife. Through collaborative approaches, discoveries of novel microbes and important epidemiological links will be critical to ensure adequate and timely responses to human and animal diseases. Investigations into taxa of interest in the present study showed Anaplasmataceae bacteria had a relatively high prevalence in the vertebrate hosts examined here and a wide geographical distribution. This group of bacteria is therefore a viable candidate for continued research into Australian zoonotic tick-borne diseases.
Supplementary Data
Funding information
This study was part-funded by the Australian Research Council (LP160100200), Bayer HealthCare (Germany) and Bayer Australia. S.L.E. was supported by an Australian Government Research Training Program (RTP) Scholarship. C.L.T. was supported by a scholarship from the Northern Beaches Council. This project was also part supported by The Holsworth Wildlife Research Endowment from The Ecological Society of Australia (awarded to S.L.E.) and the Paddy Pallin Science Grant from The Royal Zoological Society (awarded to C.L.T.).
Acknowledgements
This work was supported by resources provided by the Pawsey Supercomputing Centre with funding from the Australian Government and the Government of Western Australia. We thank Dr David Chandler, Dr Christopher Noune and Dr Matthew Stevens at the Australian Genomics Research Facility for their assistance. We thank the Department of Biodiversity, Conservation and Attractions staff for their invaluable assistance in Western Australian fieldwork, in particular Rebecca Kay, Douglas Giles and Hannah Kilian. We also thank Jade Kelly and volunteers for assistance in fieldwork and sample collection.
Author contributions
Conceptualization: S.L.E., U.M.R., P.J.I., C.L.O. Data curation: S.L.E., C.L.T. Formal Analysis: S.L.E. Funding acquisition: S.L.E., C.L.T., P.B.B., L.A.A., U.M.R., P.J.I., C.L.O. Investigation: S.L.E., C.L.T. Methodology: S.L.E., C.L.T., A.S.N. Project administration: P.J.I., C.L.O. Resources: A.S.N., P.B.B., P.J.I., C.L.O. Software: S.L.E. Supervision: P.B.B., U.M.R., P.J.I., C.L.O. Visualization: S.L.E. Writing – original draft: S.L.E. Writing – review and editing: S.L.E., C.L.T., P.B.B., A.S.N., L.A.A., U.M.R., P.J.I., C.L.O.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Ethical statement
Murdoch University Animal Ethics Committee permit number R3026/18. Department of Biodiversity Conservation and Attractions permit numbers 2018/54B, 2018/55B, 2018/57D and 2017/30. University of Sydney Animal Ethics Committee Permit number 2018/1429.
Footnotes
Abbreviations: ASV, amplicon sequence variation; gDNA, genomic DNA; ML, maximum-likelihood; OTU, operational taxonomic unit.
All supporting data, code and protocols have been provided within the article or through supplementary data files. Five supplementary files are available with the online version of this article.
References
- 1.Pfäffle M, Littwin N, Muders SV, Petney TN. The ecology of tick-borne diseases. Int J Parasitol. 2013;43:1059–1077. doi: 10.1016/j.ijpara.2013.06.009. [DOI] [PubMed] [Google Scholar]
- 2.Van Nunen SA, O’Connor KS, Clarke LR, Boyle RX, Fernando SL. An association between tick bite reactions and red meat allergy in humans. Med J Aust. 2009;190:510–511. doi: 10.5694/j.1326-5377.2009.tb02533.x. [DOI] [PubMed] [Google Scholar]
- 3.Madison-Antenucci S, Kramer LD, Gebhardt LL, Kauffman E. Emerging Tick-Borne Diseases. Clin Microbiol Rev. 2020;33:e00083-18. doi: 10.1128/CMR.00083-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Westblade LF, Simon MS, Mathison BA, Kirkman LA. Babesia microti: from mice to ticks to an increasing number of highly susceptible humans. J Clin Microbiol. 2017;55:2903–2912. doi: 10.1128/JCM.00504-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Radolf JD, Caimano MJ, Stevenson B, Hu LT. Of ticks, mice and men: Understanding the dual-host lifestyle of Lyme disease spirochaetes. Nat Rev Microbiol. 2012;10:87–99. doi: 10.1038/nrmicro2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rondón S, León C, Link A, González C. Prevalence of Plasmodium parasites in non-human primates and mosquitoes in areas with different degrees of fragmentation in Colombia. Malar J. 2019;18:276. doi: 10.1186/s12936-019-2910-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Plowright RK, Reaser JK, Locke H, Woodley SJ, Patz JA, et al. Land use-induced spillover: A call to action to safeguard environmental, animal, and human health. Lancet Planet Health. 2021;5:e237–e245. doi: 10.1016/S2542-5196(21)00031-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Graves SR, Stenos J. Tick-borne infectious diseases in Australia. Med J Aust. 2017;206:320–324. doi: 10.5694/mja17.00090. [DOI] [PubMed] [Google Scholar]
- 9.Chalada MJ, Stenos J, Bradbury RS. Is there a Lyme-like disease in Australia? Summary of the findings to date. One Health. 2016;2:42–54. doi: 10.1016/j.onehlt.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Radcliffe J, Rusjakovski N, Forkert J, Jones M, Stewart C. Canberra, ACT: Community Affairs References Committee; 2016. Growing evidence of an emerging tick-borne disease that causes a Lyme-like illness for many Australian patients. [Google Scholar]
- 11.Evans ML, Egan S, Irwin PJ, Oskam CL. Automatic Barcode Gap Discovery reveals large COI intraspecific divergence in Australian Ixodidae. Zootaxa. 2018;4656:393–396. doi: 10.11646/zootaxa.4656.2.13. [DOI] [PubMed] [Google Scholar]
- 12.Barker D. Ixodes barkeri n. sp. (Acari: Ixodidae) from the short-beaked echidna, Tachyglossus aculeatus, with a revised key to the male Ixodes of Australia, and list of the subgenera and species of Ixodes known to occur in Australia. Zootaxa. 2019;4658:331–342. doi: 10.11646/zootaxa.4658.2.7. [DOI] [PubMed] [Google Scholar]
- 13.Kwak ML. The first records of human infestation by the hard tick Ixodes (Endopalpiger) australiensis (Acari: Ixodidae), with a review of human infestation by ticks in Australia. Exp Appl Acarol. 2018;74:185–190. doi: 10.1007/s10493-018-0217-3. [DOI] [PubMed] [Google Scholar]
- 14.Lydecker HW, Hochuli DF, Banks PB. Peri-urban black rats host a rich assembly of ticks and healthier rats have more ticks. Ticks Tick Borne Dis. 2019;10:749–753. doi: 10.1016/j.ttbdis.2019.03.007. [DOI] [PubMed] [Google Scholar]
- 15.Murdoch FA, Spratt DM. Ecology of the common marsupial tick (Ixodes tasmani Neumann) (Acarina : Ixodidae), in eastern Australia. Aust J Zool. 2005;53:383. doi: 10.1071/ZO05032. [DOI] [Google Scholar]
- 16.Taylor CL, Lydecker HW, Lo N, Hochuli DF, Banks PB. Invasive rabbits host immature Ixodes ticks at the urban-forest interface. Ticks and Tick-borne Diseases. 2020;11:101439. doi: 10.1016/j.ttbdis.2020.101439. [DOI] [PubMed] [Google Scholar]
- 17.Gofton AW, Oskam CL, Lo N, Beninati T, Wei H, et al. Inhibition of the endosymbiont “Candidatus Midichloria mitochondrii” during 16S rRNA gene profiling reveals potential pathogens in Ixodes ticks from Australia. Parasites Vectors. 2015;8 doi: 10.1186/s13071-015-0958-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Egan SL, Loh S-M, Banks PB, Gillett A, Ahlstrom L, et al. Bacterial community profiling highlights complex diversity and novel organisms in wildlife ticks. Ticks Tick Borne Dis. 2020;11:101407. doi: 10.1016/j.ttbdis.2020.101407. [DOI] [PubMed] [Google Scholar]
- 19.Irwin PJ, Robertson ID, Westman ME, Perkins M, Straubinger RK. Searching for Lyme borreliosis in Australia: Results of a canine sentinel study. Parasites Vectors. 2017;10 doi: 10.1186/s13071-017-2058-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moon KL, Chown SL, Loh SM, Oskam CL, Fraser CI. Australian penguin ticks screened for novel Borrelia species. Ticks Tick Borne Dis. 2018;9:410–414. doi: 10.1016/j.ttbdis.2017.12.007. [DOI] [PubMed] [Google Scholar]
- 21.Loh SM, Gillett A, Ryan U, Irwin P, Oskam C. Molecular characterization of “Candidatus Borrelia tachyglossi” (family Spirochaetaceae) in echidna ticks, Bothriocroton concolor . Int J Syst Evol Microbiol. 2017;67:1075–1080. doi: 10.1099/ijsem.0.001929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Panetta JL, Šíma R, Calvani NED, Hajdušek O, Chandra S, et al. Reptile-associated Borrelia species in the goanna tick (Bothriocroton undatum) from Sydney, Australia. Parasit Vectors. 2017;10:616. doi: 10.1186/s13071-017-2579-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gofton AW, Waudby HP, Petit S, Greay TL, Ryan UM, et al. Detection and phylogenetic characterisation of novel Anaplasma and Ehrlichia species in Amblyomma triguttatum subsp. From four allopatric populations in Australia. Ticks Tick Borne Dis. 2017;8:749–756. doi: 10.1016/j.ttbdis.2017.05.009. [DOI] [PubMed] [Google Scholar]
- 24.Gofton AW, Doggett S, Ratchford A, Ryan UM, Irwin PJ. Phylogenetic characterisation of two novel Anaplasmataceae from Australian Ixodes holocyclus ticks: “Candidatus Neoehrlichia australis” and “Candidatus Neoehrlichia arcana”. Int J Syst Evol Microbiol. 2016;66:4256–4261. doi: 10.1099/ijsem.0.001344. [DOI] [PubMed] [Google Scholar]
- 25.Loh S-M, Egan S, Gillett A, Banks PB, Ryan UM, et al. Molecular surveillance of piroplasms in ticks from small and medium-sized urban and peri-urban mammals in Australia. Int J Parasitol Parasites Wildl. 2018;7:197–203. doi: 10.1016/j.ijppaw.2018.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Greay TL, Zahedi A, Krige A-S, Owens JM, Rees RL, et al. Endemic, exotic and novel apicomplexan parasites detected during a national study of ticks from companion animals in Australia. Parasit Vectors. 2018;11:197. doi: 10.1186/s13071-018-2775-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harvey E, Rose K, Eden J-S, Lo N, Abeyasuriya T, et al. Extensive diversity of RNA viruses in Australian ticks. J Virol. 2019;9:e01358–18. doi: 10.1128/JVI.01358-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.O’Brien CA, Hall-Mendelin S, Hobson-Peters J, Deliyannis G, Allen A, et al. Discovery of a novel iflavirus sequence in the eastern paralysis tick Ixodes holocyclus. Arch Virol. 2018;163:2451–2457. doi: 10.1007/s00705-018-3868-9. [DOI] [PubMed] [Google Scholar]
- 29.Levi T, Keesing F, Holt RD, Barfield M, Ostfeld RS. Quantifying dilution and amplification in a community of hosts for tick-borne pathogens. Ecol Appl. 2016;26:484–498. doi: 10.1890/15-0122. [DOI] [PubMed] [Google Scholar]
- 30.Stewart Merrill TE, Johnson PTJ. Towards a mechanistic understanding of competence: A missing link in diversity–disease research. Parasitology. 2020;147:1159–1170. doi: 10.1017/S0031182020000943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang C-I, Kay SC, Davis S, Tufts DM, Gaffett K, et al. High burdens of Ixodes scapularis larval ticks on white-tailed deer may limit Lyme disease risk in a low biodiversity setting. Ticks Tick Borne Dis. 2019;10:258–268. doi: 10.1016/j.ttbdis.2018.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ginsberg HS, Hickling GJ, Burke RL, Ogden NH, Beati L, et al. Why Lyme disease is common in the northern US, but rare in the south: The roles of host choice, host-seeking behavior, and tick density. PLoS Biol. 2021;19:e3001066. doi: 10.1371/journal.pbio.3001066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Takumi K, Sprong H, Hofmeester TR. Impact of vertebrate communities on Ixodes ricinus-borne disease risk in forest areas. Parasit Vectors. 2019;12:434. doi: 10.1186/s13071-019-3700-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ostfeld RS, Levi T, Keesing F, Oggenfuss K, Canham CD. Tick-borne disease risk in a forest food web. Ecology. 2018;99:1562–1573. doi: 10.1002/ecy.2386. [DOI] [PubMed] [Google Scholar]
- 35.Galan M, Razzauti M, Bard E, Bernard M, Brouat C, et al. 16S rRNA Amplicon Sequencing for Epidemiological Surveys of Bacteria in Wildlife. mSystems. 2016;1:e00032-16. doi: 10.1128/mSystems.00032-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Titcomb GC, Jerde CL, Young HS. High-throughput sequencing for understanding the ecology of emerging infectious diseases at the wildlife-human interface. Front Ecol Evol. 2019;7:126. doi: 10.3389/fevo.2019.00126. [DOI] [Google Scholar]
- 37.Roberts FSH. Australian Ticks. Melbourne, Vic: CSIRO; 1970. [Google Scholar]
- 38.Barker SC, Walker AR. Ticks of Australia. The species that infest domestic animals and humans. Zootaxa. 2014;3816:1. doi: 10.11646/zootaxa.3816.1.1. [DOI] [PubMed] [Google Scholar]
- 39.Jackson J, Beveridge I, Chilton NB, Andrews RH, Dixon B. Morphological comparison of the adult and larval stages of the Australian ticks Ixodes holocyclus Neumann, 1899 and I. cornuatus Roberts, 1960 (Acari: Ixodoidea) Systematic and Applied Acarology. 2002;7:91. doi: 10.11158/saa.7.1.10. [DOI] [Google Scholar]
- 40.Laan B, Handasyde K, Beveridge I. Observations of the biology and distribution of the tick Ixodes hirsti Hassall. Proceedings of the Royal Society of Victoria. 2011;123:200–216. [Google Scholar]
- 41.Kwak ML, Beveridge I, Koehler AV, Malipatil M, Gasser RB, et al. Phylogenetic analysis of the Australasian paralysis ticks and their relatives (Ixodidae: Ixodes: Sternalixodes) Parasit Vectors. 2017;10:122. doi: 10.1186/s13071-017-2045-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Beati L, Keirans JE. Analysis of the Systematic Relationships among Ticks of the Genera Rhipicephalus and Boophilus (Acari: Ixodidae) Based on Mitochondrial 12S Ribosomal DNA Gene Sequences and Morphological Characters. J Parasitol. 2001;87:32–48. doi: 10.1645/0022-3395(2001)087[0032:AOTSRA]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 43.Bolyen E, Rideout JR, Dillon MR, Bokulich NA, Abnet CC, et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol. 2019;37:852–857. doi: 10.1038/s41587-019-0209-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, et al. DADA2: High-resolution sample inference from Illumina amplicon data. Nat Methods. 2016;13:581–583. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Callahan BJ, McMurdie PJ, Holmes SP. Exact sequence variants should replace operational taxonomic units in marker-gene data analysis. ISME J. 2017;11:2639–2643. doi: 10.1038/ismej.2017.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bokulich NA, Kaehler BD, Rideout JR, Dillon M, Bolyen E, et al. Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2’s q2-feature-classifier plugin. Microbiome. 2018;6:90. doi: 10.1186/s40168-018-0470-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, et al. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2012;41:D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang Z, Schwartz S, Wagner L, Miller W. A greedy algorithm for aligning DNA sequences. J Comput Biol. 2000;7:203–214. doi: 10.1089/10665270050081478. [DOI] [PubMed] [Google Scholar]
- 49.Morgulis A, Coulouris G, Raytselis Y, Madden TL, Agarwala R, et al. Database indexing for production MegaBLAST searches. Bioinformatics. 2008;24:1757–1764. doi: 10.1093/bioinformatics/btn322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Weiss S, Taggart D, Smith I, Helgen KM, Eisenhofer R. Host reproductive cycle influences the pouch microbiota of wild southern hairy-nosed wombats (Lasiorhinus latifrons) Anim Microbiome. 2021;3:13. doi: 10.1186/s42523-021-00074-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 52.Edgar RC. UNOISE2: Improved error-correction for Illumina 16S and ITS amplicon sequencing. biorxviv. Epub ahead of print. 2016 doi: 10.1101/081257. [DOI] [Google Scholar]
- 53.Davis NM, Proctor DM, Holmes SP, Relman DA, Callahan BJ. Simple statistical identification and removal of contaminant sequences in marker-gene and metagenomics data. Microbiome. 2018;6:226. doi: 10.1186/s40168-018-0605-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Oksanen J, Blanchet FG, Friendly M, Kindt R, Legendre P, et al. Vegan: Community ecology package. 2020. https://CRAN.R-project.org/package=vegan
- 55.Estrada-Peña A, Cevidanes A, Sprong H, Millán J. Pitfalls in tick and tick-borne pathogens research, some recommendations and a call for data sharing. Pathogens. 2021;10:712. doi: 10.3390/pathogens10060712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yang R, Murphy C, Song Y, Ng-Hublin J, Estcourt A, et al. Specific and quantitative detection and identification of Cryptosporidium hominis and C. parvum in clinical and environmental samples. Exp Parasitol. 2013;135:142–147. doi: 10.1016/j.exppara.2013.06.014. [DOI] [PubMed] [Google Scholar]
- 57.Benson DA, Cavanaugh M, Clark K, Karsch-Mizrachi I, Lipman DJ, et al. GenBank. Nucleic Acids Res. 2017;45:D37–D42. doi: 10.1093/nar/gkw1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Edgar RC. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 60.Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat Methods. 2017;14:587–589. doi: 10.1038/nmeth.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 2015;32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hoang DT, Chernomor O, von Haeseler A, Minh BQ, Vinh LS. UFBoot2: improving the ultrafast bootstrap approximation. Mol Biol Evol. 2018;35:518–522. doi: 10.1093/molbev/msx281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Loh S-M, Gofton AW, Lo N, Gillett A, Ryan UM, et al. Novel Borrelia species detected in echidna ticks, Bothriocroton concolor, in Australia. Parasites Vectors. 2016;9:1–7. doi: 10.1186/s13071-016-1627-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ge Y, Guo G, Ge B, Yin H, Yin H. The spleen microbiota of small wild mammals reveals distinct patterns with tick-borne bacteria. PLoS Negl Trop Dis. 2018;12:e0006499. doi: 10.1371/journal.pntd.0006499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rynkiewicz EC, Hemmerich C, Rusch DB, Fuqua C, Clay K. Concordance of bacterial communities of two tick species and blood of their shared rodent host. Mol Ecol. 2015;24:2566–2579. doi: 10.1111/mec.13187. [DOI] [PubMed] [Google Scholar]
- 66.Gil JC, Helal ZH, Risatti G, Hird SM. Ixodes scapularis microbiome correlates with life stage, not the presence of human pathogens, in ticks submitted for diagnostic testing. PeerJ. 2020;8:e10424. doi: 10.7717/peerj.10424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lalzar I, Harrus S, Mumcuoglu KY, Gottlieb Y. Composition and seasonal variation of Rhipicephalus turanicus and Rhipicephalus sanguineus bacterial communities. Appl Environ Microbiol. 2012;78:4110–4116. doi: 10.1128/AEM.00323-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Li H, Li T, Qu J. Stochastic processes govern bacterial communities from the blood of pikas and from their arthropod vectors. FEMS Microbiol Ecol. 2018;94 doi: 10.1093/femsec/fiy082. [DOI] [PubMed] [Google Scholar]
- 69.Cohen C, Toh E, Munro D, Dong Q, Hawlena H. Similarities and seasonal variations in bacterial communities from the blood of rodents and from their flea vectors. ISME J. 2015;9:1662–1676. doi: 10.1038/ismej.2014.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sperling JLH, Fitzgerald D, Sperling FAH, Magor KE. Microbiome composition and borrelia detection in Ixodes scapularis ticks at the northwestern edge of their range. TropicalMed. 2020;5:173. doi: 10.3390/tropicalmed5040173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Burgdorfer W, Hayes SF, Mavros AJ. In: Rickettsiae and Rickettsial Diseases. Burgdorfer W, Anacker RL, editors. New York: Academic Press Inc; 1981. Non-pathogenic rickettsiae in Dermacentor andersoni: A limiting factor for the distribution of Rickettsia rickettsii ; pp. 585–594. [Google Scholar]
- 72.Macaluso KR, Sonenshine DE, Ceraul SM, Azad AF. Rickettsial Infection in Dermacentor variabilis (Acari: Ixodidae) Inhibits Transovarial Transmission of a Second Rickettsia . J Med Entomol. 2002;39:809–813. doi: 10.1603/0022-2585-39.6.809. [DOI] [PubMed] [Google Scholar]
- 73.Telford SR. Status of the “East Side Hypothesis” (Transovarial Interference) 25 Years Later. Ann N Y Acad Sci. 2009;1166:144–150. doi: 10.1111/j.1749-6632.2009.04522.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bonnet SI, Pollet T. Update on the intricate tango between tick microbiomes and tick-borne pathogens. Parasite Immunol. 2021;43:e12813. doi: 10.1111/pim.12813. [DOI] [PubMed] [Google Scholar]
- 75.Voordouw MJ, Lachish S, Dolan MC. The lyme disease pathogen has no effect on the survival of its rodent reservoir host. PLOS ONE. 2015;10:e0118265. doi: 10.1371/journal.pone.0118265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bunikis J, Barbour AG. Third Borrelia species in white-footed mice. Emerg Infect Dis. 2005;11:1150–1151. doi: 10.3201/eid1107.041355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fourie JJ, Stanneck D, Luus HG, Beugnet F, Wijnveld M, et al. Transmission of Ehrlichia canis by Rhipicephalus sanguineus ticks feeding on dogs and on artificial membranes. Vet Parasitol. 2013;197:595–603. doi: 10.1016/j.vetpar.2013.07.026. [DOI] [PubMed] [Google Scholar]
- 78.Almazán C, Fourniol L, Rouxel C, Alberdi P, Gandoin C, et al. Experimental Ixodes ricinus-sheep cycle of Anaplasma phagocytophilum NV2Os propagated in tick cell cultures. Front Vet Sci. 2020;7:40. doi: 10.3389/fvets.2020.00040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mariconti M, Epis S, Gaibani P, Dalla Valle C, Sassera D, et al. Humans parasitized by the hard tick Ixodes ricinus are seropositive to Midichloria mitochondrii: Is Midichloria a novel pathogen, or just a marker of tick bite? Pathog Glob Health. 2012;106:391–396. doi: 10.1179/2047773212Y.0000000050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Serra V, Cafiso A, Formenti N, Verheyden H, Plantard O, et al. Molecular and serological evidence of the presence of Midichloria mitochondrii in roe deer (Capreolus capreolus) in France. J Wildl Dis. 2018;54:597–600. doi: 10.7589/2017-09-241. [DOI] [PubMed] [Google Scholar]
- 81.Mukhacheva TA, Kovalev SY. Bacteria of the Family “Candidatus Midichloriaceae” in sympatric zones of Ixodes ticks: genetic evidence for vertical transmission. Microb Ecol. 2017;74:185–193. doi: 10.1007/s00248-017-0932-z. [DOI] [PubMed] [Google Scholar]
- 82.Kawahara M, Rikihisa Y, Isogai E, Takahashi M, Misumi H, et al. Ultrastructure and phylogenetic analysis of “Candidatus Neoehrlichia mikurensis” in the family Anaplasmataceae, isolated from wild rats and found in Ixodes ovatus ticks. Int J Syst Evol Microbiol. 2004;54:1837–1843. doi: 10.1099/ijs.0.63260-0. [DOI] [PubMed] [Google Scholar]
- 83.Müller A, Monti G, Otth C, Sepúlveda P, Bittencourt P, et al. “Candidatus Neoehrlichia chilensis” sp. nov.: Molecular detection and characterization of a novel Anaplasmataceae in wild rodents from Valdivia, southern Chile. Transbound Emerg Dis. 2018;65:357–362. doi: 10.1111/tbed.12815. [DOI] [PubMed] [Google Scholar]
- 84.Loh S-M. Identification and Characterisation of Microorganisms in Australian wildlife Tick. PhD Thesis. Murdoch University; 2018. [Google Scholar]
- 85.Gofton AW, Loh S-M, Barbosa AD, Paparini A, Gillett A, et al. A novel Ehrlichia species in blood and Ixodes ornithorhynchi ticks from platypuses (Ornithorhynchus anatinus) in Queensland and Tasmania, Australia. Ticks Tick Borne Dis. 2018;9:435–442. doi: 10.1016/j.ttbdis.2017.12.011. [DOI] [PubMed] [Google Scholar]
- 86.Brown GK, Martin AR, Roberts TK, Aitken RJ. Detection of Ehrlichia platys in dogs in Australia. Aust Vet J. 2001;79:554–558. doi: 10.1111/j.1751-0813.2001.tb10747.x. [DOI] [PubMed] [Google Scholar]
- 87.Brown GK, Canfield PJ, Dunstan RH, Roberts TK, Martin AR, et al. Detection of Anaplasma platys and Babesia canis vogeli and their impact on platelet numbers in free-roaming dogs associated with remote Aboriginal communities in Australia. Aust Vet J. 2006;84:321–325. doi: 10.1111/j.1751-0813.2006.00029.x. [DOI] [PubMed] [Google Scholar]
- 88.Hii SF, Kopp SR, Thompson MF, O’Leary CA, Rees RL, et al. Canine vector-borne disease pathogens in dogs from south-east Queensland and north-east Northern Territory. Aust Vet J. 2012;90:130–135. doi: 10.1111/j.1751-0813.2012.00898.x. [DOI] [PubMed] [Google Scholar]
- 89.Yu S, Modarelli J, Tomeček JM, French JT, Hilton C, et al. Prevalence of common tick-borne pathogens in white-tailed deer and coyotes in south Texas. Int J Parasitol Parasites Wildl. 2020;11:129–135. doi: 10.1016/j.ijppaw.2020.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Snellgrove AN, Krapiunaya I, Ford SL, Stanley HM, Wickson AG, et al. Vector competence of Rhipicephalus sanguineus sensu stricto for Anaplasma platys . Ticks Tick Borne Dis. 2020;11:101517. doi: 10.1016/j.ttbdis.2020.101517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Šlapeta J, Chandra S, Halliday B. The “tropical lineage” of the brown dog tick Rhipicephalus sanguineus sensu lato identified as Rhipicephalus linnaei (Audouin, 1826) Int J Parasitol. 2021;51:431–436. doi: 10.1016/j.ijpara.2021.02.001. [DOI] [PubMed] [Google Scholar]
- 92.Cafiso A, Sassera D, Romeo C, Serra V, Hervet C, et al. Midichloria mitochondrii, endosymbiont of Ixodes ricinus: Evidence for the transmission to the vertebrate host during the tick blood meal. Ticks and Tick-borne Diseases. 2019;10:5–12. doi: 10.1016/j.ttbdis.2018.08.008. [DOI] [PubMed] [Google Scholar]
- 93.Varela-Stokes AS, Park SH, Kim SA, Ricke SC. Microbial communities in North American ixodid ticks of veterinary and medical importance. Front Vet Sci. 2017;4:179. doi: 10.3389/fvets.2017.00179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Clayton KA, Gall CA, Mason KL, Scoles GA, Brayton KA. The characterization and manipulation of the bacterial microbiome of the Rocky Mountain wood tick, Dermacentor andersoni . Parasites Vectors. 2015;8:632. doi: 10.1186/s13071-015-1245-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Cooper A, Goullet M, Mitchell J, Ketheesan N, Govan B. Serological evidence of Coxiella burnetii exposure in native marsupials and introduced animals in Queensland, Australia. Epidemiol Infect. 2011;140:1304–1308. doi: 10.1017/S0950268811001828. [DOI] [PubMed] [Google Scholar]
- 96.Shapiro A, Bosward K, Mathews K, Vincent G, Stenos J, et al. Molecular detection of Coxiella burnetii in raw meat intended for pet consumption. Zoonoses Public Health. 2020;67:443–452. doi: 10.1111/zph.12707. [DOI] [PubMed] [Google Scholar]
- 97.Tadepalli M, Vincent G, Hii SF, Watharow S, Graves S, et al. Molecular Evidence of Novel Spotted Fever Group Rickettsia Species in Amblyomma albolimbatum Ticks from the Shingleback Skink (Tiliqua rugosa) in Southern Western Australia. Pathogens. 2021;10:35. doi: 10.3390/pathogens10010035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Vilcins I-M, Old JM, Deane EM. Detection of a spotted fever group Rickettsia in the tick Ixodes tasmani collected from koalas in Port Macquarie, Australia. J Med Entomol. 2008;45:745–750. doi: 10.1603/0022-2585(2008)45[745:doasfg]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 99.Vilcins I-ME, Fournier P-E, Old JM, Deane E. Evidence for the presence of Francisella and spotted fever group rickettsia DNA in the tick Amblyomma fimbriatum (Acari: Ixodidae), Northern Territory, Australia. J Med Entomol. 2009;46:926–933. doi: 10.1603/033.046.0427. [DOI] [PubMed] [Google Scholar]
- 100.Campbell RW, Domrow R. Rickettsioses in Australia: Isolation of Rickettsia tsutsugamushi and R. australis from naturally infected arthropods. Transactions of the Royal Society of Tropical Medicine and Hygiene. 1974;68:397–402. doi: 10.1016/0035-9203(74)90156-4. [DOI] [PubMed] [Google Scholar]
- 101.Sexton DJ, Dwyer BW, Kemp R, Graves S. Spotted fever group rickettsial infections in Australia. Rev Infect Dis. 1991;13:876–886. doi: 10.1093/clinids/13.5.876. [DOI] [PubMed] [Google Scholar]
- 102.Graves SR, Jackson C, Hussain-Yusuf H, Vincent G, Nguyen C, et al. Ixodes holocyclus tick-transmitted human pathogens in north-eastern New South Wales, Australia. Trop Med Infect Dis. 2016;1:E4. doi: 10.3390/tropicalmed1010004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Hussain-Yusuf H, Stenos J, Vincent G, Shima A, Abell S, et al. Screening for Rickettsia, Coxiella and Borrelia species in ticks from Queensland, Australia. Pathogens. 2020;9:E1016. doi: 10.3390/pathogens9121016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Graves S, Unsworth N, Stenos J. Rickettsioses in Australia. Ann N Y Acad Sci. 2006;1078:74–79. doi: 10.1196/annals.1374.008. [DOI] [PubMed] [Google Scholar]
- 105.Cook I, Scott W, Campbell RW. Scrub typhus and other infections in North Queensland animals. Transactions of the Royal Society of Tropical Medicine and Hygiene. 1967;61:343–350. doi: 10.1016/0035-9203(67)90007-7. [DOI] [Google Scholar]
- 106.Martello E, Mannelli A, Grego E, Ceballos LA, Ragagli C, et al. Borrelia burgdorferi sensu lato and spotted fever group rickettsiae in small rodents and attached ticks in the Northern Apennines, Italy. Ticks Tick Borne Dis. 2019;10:862–867. doi: 10.1016/j.ttbdis.2019.04.005. [DOI] [PubMed] [Google Scholar]
- 107.Chaorattanakawee S, Korkusol A, Tippayachai B, Promsathaporn S, Poole-Smith BK, et al. Amplicon-based next generation sequencing for rapid identification of Rickettsia and ectoparasite species from entomological surveillance in Thailand. Pathogens. 2021;10:215. doi: 10.3390/pathogens10020215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ndeereh D, Thaiyah A, Muchemi G, Miyunga AA. Molecular surveillance of spotted fever group rickettsioses in wildlife and detection of Rickettsia sibirica in a Topi (Damaliscus lunatus ssp. Jimela) in Kenya. Onderstepoort J Vet Res. 2017;84:e1–e7. doi: 10.4102/ojvr.v84i1.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Stenos J, Graves S, Popov VL, Walker DH. Aponomma hydrosauri, the reptile-associated tick reservoir of Rickettsia honei on Flinders Island, Australia. Am J Trop Med Hyg. 2003;69:314–317. [PubMed] [Google Scholar]
- 110.Unsworth NB, Stenos J, Graves SR, Faa AG, Cox GE, et al. Flinders island spotted fever rickettsioses caused by “marmionii” strain of Rickettsia honei, Eastern Australia. Emerg Infect Dis. 2007;13:566–573. doi: 10.3201/eid1304.050087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Lane AM, Shaw MD, McGraw EA, O’Neill SL. Evidence of a spotted fever-like Rickettsia and a potential new vector from northeastern Australia. J Med Entomol. 2005;42:918–921. doi: 10.1093/jmedent/42.5.918. [DOI] [PubMed] [Google Scholar]
- 112.Gil H, Barral M, Escudero R, García-Pérez AL, Anda P. Identification of a new Borrelia species among small mammals in areas of Northern Spain where lyme disease is endemic. Appl Environ Microbiol. 2005;71:1336–1345. doi: 10.1128/AEM.71.3.1336-1345.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Barandika JF, Hurtado A, García-Esteban C, Gil H, Escudero R, et al. Tick-borne zoonotic bacteria in wild and domestic small mammals in Northern Spain. Appl Environ Microbiol. 2007;73:6166–6171. doi: 10.1128/AEM.00590-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Fedorova N, Kleinjan JE, James D, Hui LT, Peeters H, et al. Remarkable diversity of tick or mammalian-associated Borreliae in the metropolitan San Francisco Bay Area, California. Ticks Tick Borne Dis. 2014;5:951–961. doi: 10.1016/j.ttbdis.2014.07.015. [DOI] [PubMed] [Google Scholar]
- 115.Zikeli S, Zohdy S. Why research on lyme disease and Ixodes scapularis needs wildlife ecology without taxonomic bias: a review. Southeastern Naturalist. 2020;19:1. doi: 10.1656/058.019.0101. [DOI] [Google Scholar]
- 116.Margos G, Fingerle V, Cutler S, Gofton A, Stevenson B, et al. Controversies in bacterial taxonomy: The example of the genus Borrelia . Ticks and Tick-borne Diseases. 2020;11:101335. doi: 10.1016/j.ttbdis.2019.101335. [DOI] [PubMed] [Google Scholar]
- 117.Gundi VAKB, Taylor C, Raoult D, La Scola B. Bartonella rattaustraliani sp. nov., Bartonella queenslandensis sp. nov. and Bartonella coopersplainsensis sp. nov., identified in Australian rats. Int J Syst Evol Microbiol. 2009;59:2956–2961. doi: 10.1099/ijs.0.002865-0. [DOI] [PubMed] [Google Scholar]
- 118.Kaewmongkol G, Kaewmongkol S, Owen H, Fleming PA, Adams PJ, et al. Candidatus Bartonella antechini: A novel Bartonella species detected in fleas and ticks from the yellow-footed antechinus (Antechinus flavipes), an Australian marsupial. Vet Microbiol. 2011;149:517–521. doi: 10.1016/j.vetmic.2010.12.003. [DOI] [PubMed] [Google Scholar]
- 119.Vilcins I-M, Kosoy M, Old JM, Deane EM. Bartonella-like DNA detected in Ixodes tasmani ticks (Acari: Ixodida) infesting Koalas (Phascolarctos cinereus) in Victoria, Australia. Vector Borne Zoonotic Dis. 2009;9:499–503. doi: 10.1089/vbz.2008.0132. [DOI] [PubMed] [Google Scholar]
- 120.Kaewmongkol G, Kaewmongkol S, McInnes LM, Burmej H, Bennett MD, et al. Genetic characterization of flea-derived Bartonella species from native animals in Australia suggests host-parasite co-evolution. Infect Genet Evol. 2011;11:1868–1872. doi: 10.1016/j.meegid.2011.07.021. [DOI] [PubMed] [Google Scholar]
- 121.Asyikha R, Sulaiman N, Mohd-Taib FS. Detection of Bartonella sp. In ticks and their small mammal hosts in mangrove forests of Peninsular Malaysia. Trop Biomed. 2020;37:919–931. doi: 10.47665/tb.37.4.919. [DOI] [PubMed] [Google Scholar]
- 122.Król N, Militzer N, Stöbe E, Nijhof AM, Pfeffer M, et al. Evaluating transmission paths for three different Bartonella spp. in Ixodes ricinus ticks using artificial feeding. Microorganisms. 2021;9:901. doi: 10.3390/microorganisms9050901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Gofton AW, Doggett S, Ratchford A, Oskam CL, Paparini A, et al. Bacterial profiling reveals novel “Ca. Neoehrlichia”, Ehrlichia, and Anaplasma species in australian human-biting ticks. PLoS One. 2015;10:e0145449. doi: 10.1371/journal.pone.0145449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Regier Y, Komma K, Weigel M, Kraiczy P, Laisi A, et al. Combination of microbiome analysis and serodiagnostics to assess the risk of pathogen transmission by ticks to humans and animals in central Germany. Parasit Vectors. 2019;12:11. doi: 10.1186/s13071-018-3240-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Garcia-Vozmediano A, Giglio G, Ramassa E, Nobili F, Rossi L, et al. Dermacentor marginatus and Dermacentor reticulatus, and their infection by SFG rickettsiae and Francisella-like endosymbionts, in mountain and periurban habitats of Northwestern Italy. Veterinary Sciences. 2020;7:157. doi: 10.3390/vetsci7040157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Dybvig K, Zuhua C, Lao P, Jordan DS, French CT, et al. Genome of Mycoplasma arthritidis . Infect Immun. 2008;76:4000–4008. doi: 10.1128/IAI.00516-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Callahan BJ, Wong J, Heiner C, Oh S, Theriot CM, et al. High-throughput amplicon sequencing of the full-length 16S rRNA gene with single-nucleotide resolution. Nucleic Acids Res. 2019;47:e103. doi: 10.1093/nar/gkz569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Earl JP, Adappa ND, Krol J, Bhat AS, Balashov S, et al. Species-level bacterial community profiling of the healthy sinonasal microbiome using Pacific Biosciences sequencing of full-length 16S rRNA genes. Microbiome. 2018;6:190. doi: 10.1186/s40168-018-0569-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Estrada-Peña A, Cabezas-Cruz A, Obregón D. Behind taxonomic variability: the functional redundancy in the tick microbiome. Microorganisms. 2020;8:1829. doi: 10.3390/microorganisms8111829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Sun S, Jones RB, Fodor AA. Inference-based accuracy of metagenome prediction tools varies across sample types and functional categories. Microbiome. 2020;8:46. doi: 10.1186/s40168-020-00815-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Risely A. Applying the core microbiome to understand host-microbe systems. J Anim Ecol. 2020;89:1549–1558. doi: 10.1111/1365-2656.13229. [DOI] [PubMed] [Google Scholar]
- 132.Brooks JP, Edwards DJ, Harwich MD, Jr, Rivera MC, Fettweis JM, et al. The truth about metagenomics: Quantifying and counteracting bias in 16S rRNA studies. BMC Microbiol. 2015;15:66. doi: 10.1186/s12866-015-0351-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Louca S, Doebeli M, Parfrey LW. Correcting for 16S rRNA gene copy numbers in microbiome surveys remains an unsolved problem. Microbiome. 2018;6:41. doi: 10.1186/s40168-018-0420-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Bonnet SI, Nijhof AM, de la Fuente J. Editorial: tick-host-pathogen interactions. Front Cell Infect Microbiol. 2018;8:194. doi: 10.3389/fcimb.2018.00194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Estrada-Peña A, de la Fuente J, Cabezas-Cruz A. Functional redundancy and ecological innovation shape the circulation of tick-transmitted pathogens. Front Cell Infect Microbiol. 2017;7:234. doi: 10.3389/fcimb.2017.00234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.de la Fuente J, Antunes S, Bonnet S, Cabezas-Cruz A, Domingos AG, et al. Tick-pathogen interactions and vector competence: identification of molecular drivers for tick-borne diseases. Front Cell Infect Microbiol. 2017;7:114. doi: 10.3389/fcimb.2017.00114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Swei A, O’Connor KE, Couper LI, Thekkiniath J, Conrad PA, et al. Evidence for transmission of the zoonotic apicomplexan parasite Babesia duncani by the tick Dermacentor albipictus . Int J Parasitol. 2019;49:95–103. doi: 10.1016/j.ijpara.2018.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Bastian S, Jouglin M, Brisseau N, Malandrin L, Klegou G, et al. Antibody prevalence and molecular identification of Babesia spp. In roe deer in France. J Wildl Dis. 2012;48:416–424. doi: 10.7589/0090-3558-48.2.416. [DOI] [PubMed] [Google Scholar]
- 139.Bouchard C, Leighton PA, Beauchamp G, Nguon S, Trudel L, et al. Harvested white-tailed deer as sentinel hosts for early establishing Ixodes scapularis populations and risk from vector-borne zoonoses in southeastern Canada. J Med Entomol. 2013;50:384–393. doi: 10.1603/me12093. [DOI] [PubMed] [Google Scholar]
- 140.Zawada SG, von Fricken ME, Weppelmann TA, Sikaroodi M, Gillevet PM. Optimization of tissue sampling for Borrelia burgdorferi in white-footed mice (Peromyscus leucopus) PLOS ONE. 2020;15:e0226798. doi: 10.1371/journal.pone.0226798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Coates M, Blanchard S, MacLeod AS. Innate antimicrobial immunity in the skin: A protective barrier against bacteria, viruses, and fungi. PLoS Pathog. 2018;14:e1007353. doi: 10.1371/journal.ppat.1007353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Hyde JA. Borrelia burgdorferi keeps moving and carries on: A review of borrelial dissemination and invasion. Front Immunol. 2017;8:114. doi: 10.3389/fimmu.2017.00114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Kumar D, Ristow LC, Shi M, Mukherjee P, Caine JA, et al. Intravital imaging of vascular transmigration by the lyme spirochete: Requirement for the integrin binding residues of the B. burgdorferi P66 protein. PLoS Pathog. 2015;11:e1005333. doi: 10.1371/journal.ppat.1005333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Turner S, Pryer KM, Miao VP, Palmer JD. Investigating deep phylogenetic relationships among cyanobacteria and plastids by small subunit rRNA sequence analysis. J Eukaryot Microbiol. 1999;46:327–338. doi: 10.1111/j.1550-7408.1999.tb04612.x. [DOI] [PubMed] [Google Scholar]
- 145.Lopez I, Ruiz-Larrea F, Cocolin L, Orr E, Phister T, et al. Design and evaluation of PCR primers for analysis of bacterial populations in wine by denaturing gradient gel electrophoresis. Appl Environ Microbiol. 2003;69:6801–6807. doi: 10.1128/AEM.69.11.6801-6807.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, et al. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci U S A. 2011;108 Suppl 1:4516–4522. doi: 10.1073/pnas.1000080107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Beard AW, Maggi RG, Kennedy-Stoskopf S, Cherry NA, Sandfoss MR, et al. Bartonella spp. in feral pigs, Southeastern United States. Emerg Infect Dis. 2011;17:893–895. doi: 10.3201/eid1705.100141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Anderson BE, Dawson JE, Jones DC, Wilson KH. Ehrlichia chaffeensis, a new species associated with human ehrlichiosis. J Clin Microbiol. 1991;29:2838–2842. doi: 10.1128/jcm.29.12.2838-2842.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Paddock CD, Sumner JW, Shore GM, Bartley DC, Elie RC, et al. Isolation and characterization of Ehrlichia chaffeensis strains from patients with fatal ehrlichiosis. J Clin Microbiol. 1997;35:2496–2502. doi: 10.1128/jcm.35.10.2496-2502.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.