Abstract
Genomic surveillance can inform effective public health responses to pathogen outbreaks. However, integration of non-local data is rarely done. We investigate two large hospital outbreaks of a carbapenemase-carrying Klebsiella pneumoniae strain in Germany and show the value of contextual data. By screening about 10 000 genomes, over 400 000 metagenomes and two culture collections using in silico and in vitro methods, we identify a total of 415 closely related genomes reported in 28 studies. We identify the relationship between the two outbreaks through time-dated phylogeny, including their respective origin. One of the outbreaks presents extensive hidden transmission, with descendant isolates only identified in other studies. We then leverage the genome collection from this meta-analysis to identify genes under positive selection. We thereby identify an inner membrane transporter (ynjC) with a putative role in colistin resistance. Contextual data from other sources can thus enhance local genomic surveillance at multiple levels and should be integrated by default when available.
Keywords: genomic surveillance, meta-analysis, antimicrobial resistance, KPC, plasmids, colistin
Data Summary
All data and metadata used in the analyses have been deposited with the Open Science Foundation (OSF) under project ID n78q3. Extensive metadata on all samples used in the analyses is available there and in the Supplementary Material (Table S1, available in the online version of this article), including curated phenotype data on colistin resistance (Table S2). In addition, all genomes collected specifically for this study have been deposited with NCBI GenBank under project ID PRJNA742413. For all contextual genome data, please refer to the corresponding studies (Table S1). Table S3 presents all genomes excluded manually from the analysis (see Methods). Table S4 holds isolate feature annotations from kleborate such as sequence type and a selection of resistance determinants. Table S5 provides metadata for all genomes that did not pass the tiered quality protocol (see Methods) and were thus not included in the analyses.
Impact Statement.
Usually, hospital outbreaks are analysed in isolation. However, here we demonstrate how to screen publicly available genomes, metagenomes and culture isolates. We place a local multidrug-resistant bla KPC-2 -carrying strain of Klebsiella pneumoniae in a larger genomic context. This context provides important details about the origin and distribution of the outbreak-causing strain and reveals hidden transmission far from where the outbreak occurred. Contextual genomes can also facilitate inquiries into genomic determinants of resistance. We demonstrate this by identifying a new putative target for colistin resistance, an antibiotic of last resort, thereby using the collected data to move beyond the local outbreak in a step-function improvement.
Introduction
Multiresistant strains of Klebsiella pneumoniae (Kp) are a global health threat [1]. Among all known resistance mechanisms, carbapenemases are one of the most concerning, as carbapenem antibiotics are considered a last-line drug. These enzymes are typically encoded on mobile genetic elements such as the Tn4401 transposon [2], which mediates transfer between plasmids [3] and bacterial species [4]. Furthermore, the prevalence of carbapenemase-producing Kp has increased in recent years [5], leaving only few antibiotics such as colistin to treat those cases. Such pathogen spread can be prevented by molecular surveillance and derived public health measures: isolate genomes reveal transmission routes by accumulating characteristic mutations, from which ancestry can be inferred through time-dated phylogeny [6].
While it has become standard practice to reconstruct such phylogenies of within-hospital outbreaks [5, 7], few studies assess ‘contextual’ information, i.e. genome sequences from isolates that were not part of the local outbreak but closely related. From a public health perspective, this is suboptimal. While many larger hospitals run screening programmes to detect the carriage of resistant strains on admission [8, 9], peripheral institutions rarely do. However, there is a significant transfer of patients, e.g. from operation theatre to rehabilitation centre or from one country to another. For an outbreak investigation with only local scope, these boundary-crossing transmission events remain hidden.
Here we reanalyze a large outbreak at the University Hospital Leipzig (UHL) from 2010 to 2013 [10] in light of new data from a nearby institution, which experienced an outbreak with a closely related, albeit non-descending, strain. We performed a genomic meta-analysis to link both outbreaks, discovering hundreds of related genomes distributed across 28 different studies. We identify the likely sources of both outbreaks and illustrate hidden transmission across study boundaries. Only the integration of data from several sources provided a ‘complete picture’. However, we highlight several obstacles that need to be addressed before cross-boundary genomic surveillance can work in practice.
Beyond epidemiology, we show how outbreak meta-analyses can generate new hypotheses about host adaptation and antimicrobial resistance: the genomes under study underly similar selective pressures, such as treatment with colistin, an antibiotic of last resort. Thus, recurring mutations in the same gene(s) but across different genomes can signal putative causes for an observed phenotype, such as colistin resistance [11]. For colistin, several such inducible genomic changes have been described that mediate resistance [12]. Nevertheless, the exact mechanisms remain incompletely understood and seem to be multifactorial [13]. We show how contextual data can be leveraged to generate hypotheses about putative factors contributing to colistin resistance.
Methods
Culture and sequencing
All samples were streaked on CHROMagar KPC chromogenic agar plates (CHROMagar, Paris, France). KPC carriage of isolates was confirmed using PCR. In total, 142 Kp-1 isolates had been collected from 105 patients in a previous investigation [10] and were complemented in the present study with an additional ten isolates discovered using PCR screening of two culture collections (see below) and 13 samples collected from Kp-2. All of the isolates were sequenced using short reads (Illumina). Short-read sequencing for isolates from other studies is described in the respective publications (Table S1). For genomes sequenced for the current study, a read length of 150 bases (paired-end) was used on an Illumina MiSeq sequencer. The libraries were constructed using a previously established protocol [14]. For the current study, 28 Kp-1 samples were additionally sequenced using long reads (Nanopore) to enable hybrid assembly (see below). DNA extraction for Nanopore sequencing and quality control was done as reported elsewhere [15]. Care must be taken, especially for Nanopore sequencing, not to damage the extracted DNA to achieve a sizeable median fragment length (target 8 kb) for sequencing to be effective. Nanopore sequencing was performed using the MinION sequencer and the 1D ligation library kit (LSK109) on an R9.4 flow cell (all Oxford Nanopore Technologies, ONT).
In silico screening of isolate and metagenomes
Through screening, our aim was to collect as many genomes as possible with a putative relation to the outbreak clone Kp-1, yielding a total of 9409 genomes. From NCBI RefSeq, we retrieved all 9163 genomes that were labelled as Klebsiella pneumoniae (Taxonomy ID: 573, last accessed 1 August 2020) [16]. In a comprehensive literature search using the search terms ‘KPC, Klebsiella pneumoniae , outbreak’ we identified 80 genomes from various studies that had only deposited reads with NCBI SRA, and which we reassembled for this study (see below).
For metagenomic search, we screened about 400 000 metagenomic read sets in a reduced representation known as MinHash signature [17] using wort (no version, unpublished, github.com/dib-lab/wort). These read sets represent a random sample from the entire NCBI SRA corpus, which we were limited to when this analysis took place (last accessed 1 June 2020). Hashing was performed using sourmash (v3.5) [18]. As the query we used the Kp-1 index genome (k=51, sampling rate 0.001) and manually reviewed all 15 hits reported with a threshold 0.01 Jaccard similarity, a measure that approximates average nucleotide identity (ANI) [17]. Most hits were synthetic consortia of microbes, assembled for an undetermined experimental purpose. Three hits were repeat stool samples from a single person, and we continued our analysis with the earliest sample (see Results).
Strain-specific screening PCR
We then screened two culture collections (National Reference Centre for multidrug-resistant Gram-negative bacteria, Bochum, and Department of Medical Microbiology and Virology, Leipzig) for related isolates using a strain-specific marker PCR, designed using a proprietary, pangenome-based algorithm (nanozoo GmbH). Each 50 µl PCR reaction contained 10 µl template DNA, 2 µl 10 nM primer mix for each primer (primer 1: ATGCGTCCACGAAGAATTAT; primer 2: CATCGCCAAGATACTGTACA), 25 µl 2× polymerase master mix (Superfi II, Invitrogen) and 11 µl ultra-pure water. Thermal cycling consisted of initial denaturation at 98 C for 1 min followed by 35 cycles of denaturation at 98 C for 20 s, annealing at 55 C for 20 s, extension at 72 C for 1 min, followed by final extension at 72 C for 5 min.
Data processing
Unless otherwise stated, default parameters were used. Of the 9409 collected genome assemblies, 1461 passed a minimum Jaccard similarity of 0.97 compared to the Kp-1 index genome (15.5 %, parameters: k-mer size 51 nt, scale 0.001). Jaccard similarity was computed using sourmash (see above). In a subsequent filtering step, 415 (4.4 %) were included for tree construction based on a minimum in silico DNA–DNA-hybridization threshold of 99.98 % computed using FastANI (v1.32) [19] as well as a minimum genome length of 5 Mb and an alignment of 90 % of the query genome to the Kp-1 index isolate (completed, circular), excluding all extra-chromosomal sequences. This alignment was performed using minimap2 (v2.17-r941) [20] with the asm5 option for an expected sequence divergence of 0.1 %. This sequential approach allows for laxer but computationally efficient methods with fewer constraints to screen many genomes in the beginning. Subsequently, the selection is refined using more computationally expensive methods. We conservatively removed 16 samples from the timetree because they did not fit the estimated molecular clock model, likely due to unidentified recombination (Table S3).
Isolates where only short reads could be obtained were assembled using shovill (v1.1.0, unpublished, github.com/tseemann/shovill). Metagenomic reads were preprocessed using fastp (v0.20.1) [21] and assembled using megahit (v1.2.9) [22]. All contigs with a minimum length of 2 kb were then mapped to the reference genome (Kp-1 index patient, VA13414, Table S1) using minimap2 (see above). The Nanopore sequencing data were basecalled using Albacore (v2.3.2, available from Oxford Nanopore Technologies) and adapters removed using Porechop (v0.2.3, unpublished, github.com/rrwick/Porechop). Genome hybrid assembly using long and short reads was performed using Unicycler (v0.4.6) [23].
Genome annotation was performed using prokka (v1.14.6) [24]. Annotation of Klebsiella -specific features was done using kleborate (v0.4.0-beta) [25]. Plasmids were annotated using abricate (v1.0.1, unpublished, github.com/tseemann/abricate) using the plasmidfinder database (version 2021-01-13) [26]. Antimicrobial resistance genes were annotated using the same programme with the Comprehensive Antibiotic Resistance Database (CARD, v3.1.2) [27]. Genes from toxin-antitoxin systems were searched using mmseqs2 (v12-113e3) [28] against the Toxin-Antitoxin Database (TADB) (v2) [29]. Phages were annotated using uv (v0.1, unpublished, github.com/phiweger/uv). Recombinant regions were annotated using gubbins (v2.4.1) [30]. Single nucleotide variant (SNV) calling was performed using the snippy workflow (v4.6.0, unpublished, github.com/tseemann/snippy), which proved the most accurate programme in a recent benchmark study [7]. Note that SNV calling was based on the assemblies of all samples in our collection; where only reads could be obtained from the literature, we assembled the data first (see above). In short, snippy simulates reads from input genomes and maps them to the provided reference using bwa (v0.7.17-r1188) [31], before calling variants with freebayes (v1.3.2, unpublished, github.com/freebayes/freebayes). Putative recombinant, repetitive and prophage regions were masked before SNV calling. Sites with SNVs were extacted using snp-sites (v2.5.1) [32].
Reconstruction of time-dated phylogeny
A time-dated phylogeny was calculated using timetree (v0.7.6) [33], a maximum-likelihood-based approach starting from a core-genome SNV alignment. The derived mutation rate was scaled by the total genome size. Homoplasy was assessed using the treetime homoplasy function. The final tree and associated metadata were visualized using the microreact webservice [34]. Bootstrap support values were extracted from the guide tree, a prerequisite of the timetree, and calculated with raxml-ng (v0.9.0) [35].
Analysis of genomic variants
A genome-wide association study was performed using pyseer (v1.3.7) [36] and included the aggregation of mutations across genes in a burden test [37]. Gene-set enrichment was performed using the Gene Ontology webservice (last accessed 1 April 2021) [38, 39]. Positive selection was assessed by first aligning all sequences for a particular gene using nextalign (no version, unpublished, github.com/nextstrain/nextclade). The multiple sequence alignment was then analysed using the BUSTED algorithm [40] as part of the HyPhy suite (v2.5.31) [41]. In silico folding of proteins was done using the trRosetta model (no version) [42].
Plasmid containment and plasmid gene content
For plasmid containment, we first calculated one MinHash signature (target, T) for each of the four plasmids found in the Kp-1 index isolate, again using sourmash (k=21, scaled=100). Next, we sketched all isolate genomes (query, Q) with the same parameters. MinHash containment can then be calculated as the size of the union over the size of the target . In other words, how much of each Kp-1 plasmid is contained in a query isolate genome, ranging from 0 (no containment) to 1 (all sampled k-mers are contained, i.e. target and query likely contain the same plasmid).
To explain the choice of k-mer size and sampling rate in MinHash-based genome similarity and containment calculations further: Generally, the larger the k-mer size, the more stringent the comparison, because as variation increases between two sequences, fewer k-mers are shared when fixing k to any particular value. For the genome filtering (see above), we thus chose a community-standard value for k to compare closely related sequences on the taxonomic scale of ‘strain’ (k=51). Species-level comparisons are usually conducted with k set to 31. Note that such values derive from experiments [17, 43], but there are no canonical values, and one has to experiment given the individual use case. For the plasmid containment analysis, we relaxed the k-mer size to 21 bases, which is commonly used to compare sequences where one expects more genetic variation, roughly at the level of ‘genera’ [43]. We justify this setting because compared to bacterial chromosomes, plasmids can present more genetic variation [44]. More genetic diversity requires a smaller k-mer size to approximate ANI accurately [17].
The sampling rate affects how many k-mers are sampled. The higher the rate, the more sensitive similarity and containment calculations are, but the less computationally efficient. Based on our experience and in line with values commonly used by the community, we typically use sampling rates of 1 in 10 000 for large metagenomes, 1 in 1000 for bacterial genomes, and 1 in 100 for small genomes and genomic elements like viruses and plasmids. We roughly target to sample at least 400 k-mers per sequence as this has been shown to suffice for average nucleotide identity (ANI) estimation [17].
Pangenome reconstruction for plasmids from hybrid assemblies (‘plasmidome’, n=28) was done using panaroo (v1.2.8) [45]. This resulted in a set of genes, each one representing an orthologous gene cluster. After translation into protein sequence, we searched for these genes in all outbreak genomes using mmseqs2 (v12-113e3) [28], resulting in a presence–absence matrix (Fig. 2b).
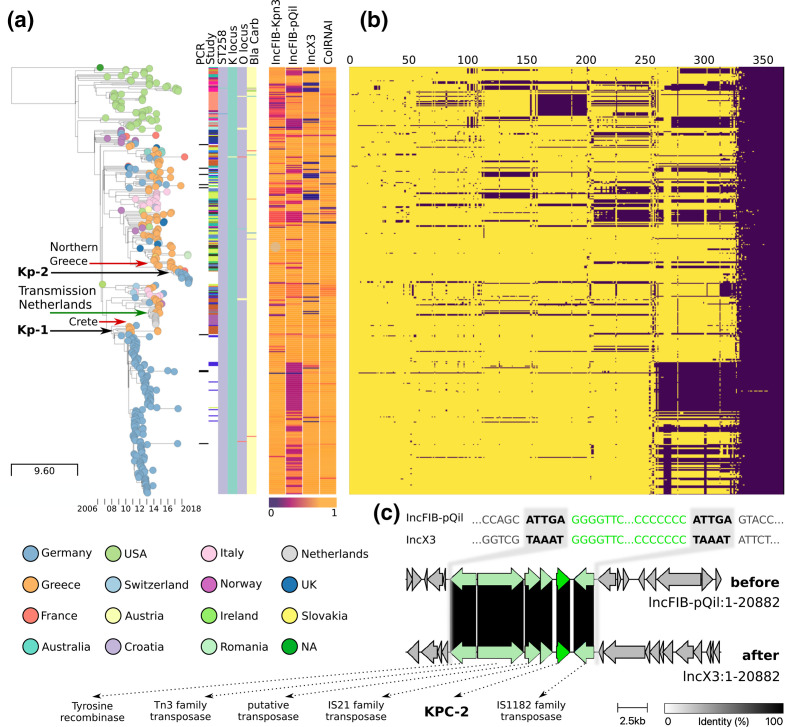
Fig. 2.
Time-dated phylogeny of 415 bla KPC-2 carrying Kp genomes and associated metadata blocks. For interactive exploration see visualization in microreact [34] (https://microreact.org/project/6bBfAYXswvY691LfbVytLT). (a) A timetree reveals how both outbreak strains, Kp-1 and Kp-2 (black arrows), most likely originated from southern and northern Greece (red arrows), respectively. Two scales are provided: SNV distance (left) and a timeline depicting year of isolation (right). In the leftmost metadata block columns, read from left to right, genomes are marked that have been identified using our strain-specific screening PCR. The second column indicates which study they were recruited from (white is our study, all of the other 28 colours are study-specific, see Table S1). The next four columns show the sequence type (purple is ST258), capsule type (turquoise is KL106), O antigen (LPS) type (purple is O2v2), and carbapenemase variant (‘Bla Carb’, yellow is bla KPC-2, other colours indicate either combinations with other carbapenemases like OXA or other KPC variants, see interactive plot). Note how the majority of genomes that pass our tiered filtering approach are homogenous in these features (see also Table S4). The following metadata block shows what fraction of each of the four plasmids in the Kp-1 index genome is contained in each isolate genome from our collection. Containment values range from 0 (blue, no containment) to 1 (orange, Kp-1 plasmid is wholly contained in the query isolate). (b) Matrix indicates presence (yellow) or absence (purple) of plasmid genes (columns) for each genome (row) in the phylogenetic tree. (c) Alignment of genes around the bla KPC-2 locus between the two plasmids IncFIB(pQil) (top) and IncX3 (bottom) shows a transposition event that allows shedding of IncFIB(pQil) while maintaining bla KPC-2 on the IncX3 plasmid, likely increasing host fitness. Transposed sequence in light green, bla KPC-2 in dark green. The 5 bp target site duplications (bold sequence) indicate that Tn4401 (green sequence) moved through transposition rather than recombination [58].
Results
In silico and PCR-based screening identifies hundreds of outbreak-related, contextual genomes
From 2010 to 2013, UHL experienced a large outbreak of a multiresistant, bla KPC-2-carrying Kp strain (hereafter referred to as ‘Kp-1’) of sequence type ST258, characterized by capsule type KL106 and O antigen (lipopolysaccharide, LPS) serotype O2v2. Overall, 105 patients were affected, and it took a multidisciplinary team many months to contain it [10] (Fig. S1). In 2018, we received 13 isolates from a bla KPC-2 Kp outbreak from the University Hospital Jena, located about 80 km from UHL (‘Kp-2’). We hypothesized that these were related to the Kp-1 outbreak at UHL due to the spatiotemporal proximity. A comparison of two genomes from Kp-1 and Kp-2, isolated from the respective index cases, showed that they were closely related. Kp-2 had the same sequence type (ST258), capsule type (KL106) and serotype (O2v2) as Kp-1. The genomes differed in only 69 SNVs, without larger rearrangements or differences in gene content. While within-hospital Kp outbreaks have been estimated to differ by fewer than 21 SNVs [5], we are unaware of recommendations for isolates further apart in space and time. Therefore, more ‘contextual’ genomes were needed to populate the genomic distance between Kp-1 and 2 and to fill the genomic ‘gap’. We, therefore, performed a comprehensive, multimodal screening, consisting of (1) a comprehensive literature search including manual extraction of genomes and metadata, (2) an in vitro screening of two German culture collections and (3) an in silico screening of publicly available genomic and metagenomic datasets.
In total, we obtained 9409 Kp genomes. Of those, 142 were collected during the Kp-1 outbreak from 105 patients [10], and 28 isolates were randomly selected for additional long-read sequencing (Nanopore) to obtain accurate plasmid reconstructions. The sampling aimed to uniformly represent all branches of the Kp-1 outbreak phylogeny. No long-read sequencing was performed on Kp-2 isolates, as we only obtained them later on in the analysis. A further ten isolates were identified in two culture collections through PCR-based screening using strain-specific primers (see Methods). The local UHL collection holds several hundred multiresistant Gram-negative bacteria, all recruited from the hospital over the last 20 years. The second collection is the German National Reference Center for multidrug-resistant Gram-negative bacteria in Bochum and holds an order of magnitude more isolates, which are subject to a standardized characterization. Sequencing of the ten isolates confirmed that all were closely related to Kp-1. The primers were designed using a proprietary algorithm (nanozoo GmbH) to recognize Kp-1 and close relatives but not other Kp strain genomes, e.g. different sequence types. Interestingly, the algorithm selected a putative intact prophage region as the most specific PCR template, and >99 % of genomes closely related to Kp-1 and selected for further analyses (see below) contained the target. Thus, even though phages are mobile, they can be remarkably stable across decades and still be used as effective markers [46].
The remaining 9257 genomes were collected from public sources. The majority was retrieved from NCBI RefSeq [16]. However, 80 isolate read datasets were only identified through a literature survey, as they did not have an associated genome assembly deposited. In addition, extensive metadata were extracted where available. Furthermore, we searched the index Kp-1 isolate in a k-mer database of over 400 000 metagenomic read datasets (SRA, NCBI, see Methods). We identified a single sample from an unpublished study of ICU patient colonization (NCBI, project ID PRJNA561398) where we could recover a closely related, metagenome-assembled Kp genome.
From the collected 9409 genomes, we wanted to select the ones most similar to the Kp-1 index genome. Altogether, 415 genomes (4.4 %) passed a tiered quality-control protocol (see Methods), resulting in a collection of high-quality genomes (ANI >99.98 %, alignment to Kp-1 index isolate >90 %) for further analyses. The 415 accepted genomes represent isolate and metagenomic read data, from a total of 28 studies and 16 countries (Fig. 1a, b, Table S1). Sequence type, capsule type and serotype of most selected isolates closely match Kp-1 (ST258, KL106, O2v2). Isolation dates span 15 years, from 2006 to 2018. Most isolates were recruited from European countries, particularly Germany and Greece. All but one isolate were clinical; that one was isolated from wastewater (NCBI, project ID PRJNA579879) [47]. Unfortunately, the metadata on isolation sources was sparse, and we did not collect this information for further analyses. Our impression, however, was that roughly half of the isolates were obtained through some kind of screening effort. Additional metadata, including data sources and associated publications, where available, can be found in Table S1.
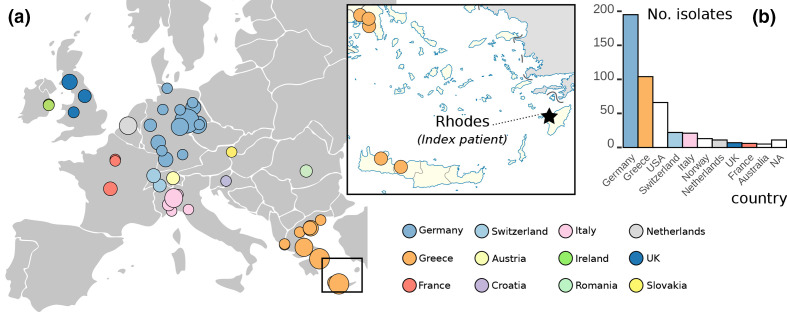
Fig. 1.
Geographic distribution of the genomes under study. (a) Location of isolation sources in Europe. Circle size is proportional to the number of genomes collected from this location. Leaves were coloured by country code. In the detailed map to the right, genomes found in Crete are shown (bottom). Our index patient was hospitalized in nearby Rhodes (star), and endemic transmission across these islands, which are connected by boat, is plausible. (b) Distribution of countries from which isolates were collected. Only countries with more than five isolates are depicted.
Time-dated phylogeny resolves outbreak origin and reveals hidden transmission
We observed 69 SNVs between the genomes of Kp-1 and Kp-2. However, relating isolates based on mutation rates alone can be misleading because rates of Kp are variable and thus difficult to estimate [48–50]. Therefore, to see whether Kp-2 descended from Kp-1, we constructed a time-dated phylogeny based on an alignment of 3720 core SNV sites (total alignment length 5 384 856 sites) from the 415 genomes in our filtered collection (Fig. 2). This phylogeny clearly shows that Kp-2 is not a direct descendant of Kp-1. The tree topology did not change when we fixed it to mutation rates reported in the literature instead of estimating the mutation rate from the data.
The index patient’s travel history and symptom onset led to the hypothesis that the origin of the Kp-1 outbreak was a Kp strain imported from Rhodes, an island in southern Greece and a popular tourist location for German travellers. After being acutely hospitalized there, the patient was transferred to UHL, where bla KPC-2 was detected for the first time in the patient’s medical history. A Greek origin seemed plausible because of the travel and the high prevalence of carbapenemase-carrying strains in Greece [51]. However, this hypothesis could not be substantiated with this data alone, and no supporting isolate genomes from Greece were available [10]. We identified several closely related genomes from Crete [52], a neighbouring island of Rhodes (see detailed map in Fig. 1a), which populate the timetree around the time of the start of the Kp-1 outbreak (Fig. 2a, black arrow ‘Kp-1’). With frequent travel by boat between these islands, it is plausible that an ancestor of Kp-1 was circulating in this region. Interestingly, the originating strain for the Kp-2 outbreak also seems to have come from Greece, albeit from northern provinces. Here, we could identify closely related genomes from two studies [5, 53] (Fig. 2a, black arrow ‘Kp-2’). We even identified a third transmission from Greece to mainland Europe, with a strain from northern Greece causing an outbreak in the Netherlands [54] (Fig. 2a, green arrow). The authors of the corresponding study did not identify this origin because they limited their investigation to local cases, supporting our argument for an integrative approach across study borders. All nodes in the tree where these transmissions out of Greece appeared had over 95 % bootstrap support. However, it is important to consider potential sampling bias when inferring origins. While we identified many samples from Greece (Fig. 1b), the screening methods were blind towards genome origin and considered an exhaustive set of Kp genomes. Furthermore, several studies have described the high prevalence of carbapenemase-carrying Kp in southern Europe [51]. Therefore, we argue that the large number of Greek samples likely represents the true distribution of bla KPC-2 Kp and is not an artefact of sampling bias, though it cannot be ruled out completely.
Shortly after the outbreak onset, a systematic screening of patients on hospital admission was established. From then on, we did not detect any new Kp-1 isolates on admission but only after, which argues against a re-importation from Greece. In many cases, a causal transmission chain could be reconstructed [10]. As the Kp-1 outbreak unfolded, local health authorities assumed that the outbreak was likely not limited to one hospital. They based their assessment on the long duration and the large number of patients involved in the outbreak, with frequent transfers to and from the hospital as a tertiary care centre. While these factors make non-local transmission more likely, no evidence was available to support this hypothesis. Surprisingly, we identified 13 isolates that were collected outside of UHL, but are part of the Kp-1 outbreak (Fig. 2a, Table S2). Most of them come from the same federal state that UHL is in, but several were isolated in other states hundreds of kilometres away. We did not detect descendant isolates of the Kp-1 outbreak in other countries. The Kp-2 outbreak seems to have been contained within the affected hospital, as no published genomes were found in other places. On an international level, the data supports repeated introduction of KPC-carrying Kp strains from Greece, likely due to it being a popular travel site. In fact, travel-related carbapenemase-producing Enterobacterales have been recognized as an important source of resistance transmission [55]. The above described hidden transmission events would not have been observed without integration of data across study borders, and illustrate the value of our approach.
Carbapenemase preservation in spite of frequent plasmid changes
Plasmids serve many functions, but a central one is as a gene-delivery platform [44]. Their payload is manifold, and here includes the bla KPC-2 carbapenemase. However, to the host genome, plasmids can come at a considerable fitness cost, which creates pressure to remove them unless they provide a selection advantage [44]. At the same time, plasmids resist removal through, e.g. toxin-antitoxin systems and compete with rivalling plasmids [44]. In search of persistence in the host, frequent changes to the genetic material of plasmids can be observed [56]. In the Kp-1 index genome, we found four types of circular plasmid using Nanopore-based hybrid assembly: IncFIB(Kpn3), IncX3, ColRNAl and IncFIB(pQil), the latter carrying bla KPC-2.
Plasmid reconstruction is often difficult for short-read-based assemblies due to assembly fragmentation. Thus, comparing plasmids between isolates usually means comparing sets of putative plasmid contigs. However, if any set is incomplete, such an analysis can be misleading. We, therefore, decided to use the newly sequenced, hybrid-assembled, circular plasmid reconstructions from the Kp-1 index genome as a reference and determined what fraction of these plasmids was contained in any of the 142 Kp-1 isolates that Kaiser et al. sequenced in 2017 (Fig. 2a) [10]. Containment here refers to MinHash containment (see Methods). To complement this data, we aggregated the plasmid-encoded gene content (‘plasmid pangenome’) across 28 Nanopore-sequenced isolates, leveraging the superior ability of this technology to reconstruct plasmids. We then determined whether these genes were present in all members of our isolate collection (Fig. 2b). The contextual genomes show repeat loss of IncFIB(pQil) (109 Kb) in phylogenetically distinct isolates, which likely constitute independent events (Fig. 2b). Interestingly, besides bla KPC-2 and the extended-spectrum beta-lactamase (ESBL) TEM, we also found a toxin-antitoxin system on IncFIB(pQil) (vagC and vagD [57]), which can prevent the host from shedding the plasmid. Nevertheless, we observed the complete loss of IncFIB(pQil) during the Kp-1 outbreak. About one-third of the Kp-1 outbreak genomes were affected, initially confusingly so because this plasmid carried the bla KPC-2 carbapenemase. However, we found one isolate with two bla KPC-2 copies, one on IncFIB(pQil) and IncX3 (43 Kb), respectively. The only other resistance gene on IncX3 was the SHV ESBL. As bla KPC-2 is carried on the Tn4401 transposon, it could be copied from IncFIB(pQil) to IncX3 (Fig. 2c). Tn4401 moving between plasmids was the result of transposition and not recombination, which can be inferred from 5 bp target site duplications (Fig. 2c) [58]. After this transposition event, the host could discard the IncFIB(pQil) plasmid, which likely confers a fitness advantage, but retain bla KPC-2 simultaneously.
Contextual genomes reveal positive selection of virulence and resistance genes
Modified lipopolysaccharides often cause colistin and, more generally, polymyxin resistance (PR). They result in a positive charge to the bacterial membrane that repels polymyxins [12]. Several proteins are involved, though ‘the exact mode of action of polymyxins still remains unclear’ [12]. For 171 of the 415 genomes in our collection (41.2 %), we were able to assess from the original records and publications whether the isolate was colistin-resistant or not (Table S2). Where minimum inhibitory concentration (MIC) measurements were available, breakpoints by the EUCAST committee (v11) were used to classify isolates into colistin sensitive and resistant.
We found several known genes involved in PR to be mutated. Overall, 60 of the 171 isolates (35.1 %) with phenotype data were colistin-resistant. In 39 out of these 60 cases (65 %) we found a truncated or missing mgrB gene product, a negative regulator of the PhoPQ signalling system [59]. Interestingly, we detected an incomplete mgrB gene product in eight isolates reported as colistin-sensitive. mgrB-mediated resistance can occur rapidly: in the Kp-1 outbreak, we identified three different mgrB loss-of-function mutations (Fig. S2). Furthermore, we found frequent truncations in pmrB and non-synonymous but not truncating or missense mutations in phoQ (17 UM) and phoP (9 UM), all regulatory proteins involved in LPS modification [60]. These ‘canonical genes’ [59] cause PR by acting on the outer membrane. We did not detect the plasmid-encoded mcr-1 gene, which encodes a transferase that modifies lipid A and thereby causes PR [12].
To identify other genomic regions associated with PR, we first performed a genome-wide association study (GWAS) based on SNVs, small insertions and deletions. This analysis did not return a significant result after correcting for population structure (P>0.05), neither when considering each SNV individually nor when aggregating SNVs over genes in a so-called burden test [37]. This failure might be due to technical limitations of GWAS, especially in light of few genomes [61] or strong population structure [62, 63]. Furthermore, while single SNVs can induce colistin resistance [64], PR is generally assumed to be a polygenic phenomenon [12]. To test which genes were mutated more than expected, we first aggregated unique haplotypes for each gene across all genomes, similar to a burden test [37]. To conservatively correct for population structure, we counted mutations only once per position in the reference genome. Recombinant sites, putative phages and sites within repetitive sequences were excluded. This procedure would not detect convergent evolution where mutations arise in the same position in two different clades. However, we did not detect any homoplastic mutations outside of recombinant regions. At a mutation rate of 0.68 per Mb per year and 5.5 million bases' genome size, we expect about four mutations per year in the Kp genomes under study. Consequently, most of the roughly 4000 genes in the genome remain unaltered, even in the ten years our genome collection is distributed across (Fig. 3b). Conversely, if we observe more mutations than expected in a gene, this might indicate evolutionary selection.
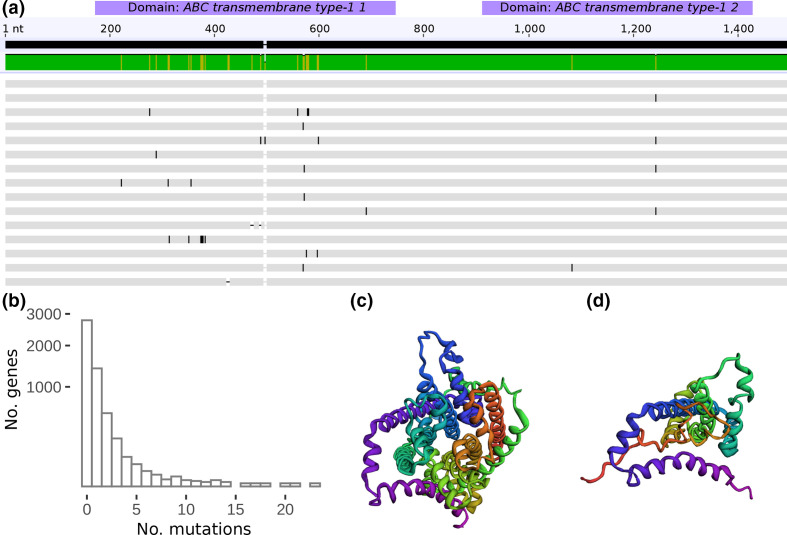
Fig. 3.
Positive selection of the inner membrane ABC transporter permease ynjC. (a) Multiple sequence alignment of representative haplotypes of the nucleotide sequence of ynjC. Most mutations occur between positions 75 to 730, which includes both transmembrane and topological domains. Three of those haplotypes lead to premature stop codons. (b) Distribution of unique mutations observed in all genes. As expected by the estimated mutation rate of 0.68 mutations per Mb per year, most genes remain unchanged over the ten years that our study covers. Several genes, however, accrue over 20 unique mutations across 415 genomes. (c) Three-dimensional (3D) protein structure of the ynjC permease. In the centre is the pore through which small molecules are shuttled. (d) 3D protein structure of a truncated form of the protein [same orientation as (c)], created through a premature stop codon. Clearly, the channel structure is lost, and the protein is likely dysfunctional.
We, therefore, ranked genes by the number of unique mutations (UM) per gene. We define unique mutations as specific to a single gene and position. We used the number of UMs per gene as a heuristic to rank and prioritize genes for further analyses. Note that if an isolate presents a mutation at a particular position in some gene like gyrA, all descendants will likely share this mutation. However, we only count this mutation as a single UM for gyrA; only mutations in other positions in the gene would increase the UM count after that. A gene-set enrichment of all genes with 10 UMs (n=15) showed two overrepresented biological processes. For one, the phosphorelay signal transduction system was enriched (19.8-fold, P=0.001) [65], which is known to be implicated in PR [12]. Furthermore, genes associated with nitrate assimilation were enriched (41.8-fold, P=0.001), which to our knowledge has neither been described nor could we assess the biological significance of this finding.
We tested the top-ranking genes with the highest number of unique mutations for gene-wide evidence of episodic positive selection. For each candidate gene, we used a random-effects framework to pool evidence across multiple sites and thereby increase statistical power [40]. All genes discussed hereafter exhibited significant positive selection (dN/dS>1, likelihood-ratio test, P=0.05). We found two positively selected genes that affect virulence: the transcriptional activator cadC (18 UM) has been linked to increased Kp colonization [66] and fimH (16 UM) is a critical virulence factor in urinary tract infection, a common complication of Kp colonization [67]. Note that fimH has a direct role in epithelial binding, and allele switching via homologous recombination is common [68].
Recently, colistin has also been found to target the inner cytoplasmic membrane [69]. Interestingly, we identified a highly mutated inner-membrane ABC transporter permease [70] under strong positive selection (all detected mutations non-synonymous), named ynjC (21 UM, Uniprot, P76224). Proteins of this group utilize ATP to import many small molecules such as nutrients and antibiotics [71–73]. Mutations in permeases have been shown to ‘lock’ the transporter in one of its two states [74, 75], such as inward-facing [76], disrupting the shuttle function [77]. Additionally, we found three mutations that caused premature stop codons and subsequent dysfunctional proteins (Fig. 3c, d). Most mutations accumulate in a region between residues 75 to 730, spanning both transmembrane and topological domains (Fig. 3a). In 12 isolates with ynjC mutations, 7 (58.3 %) were resistant to colistin; however, for none of the haplotypes with premature stop codons, phenotype data could be obtained, and future functional validation is needed. Nevertheless, ABC family transporters have been proposed to transport nascent core-lipid A molecules across the inner membrane [78], with a putative effect on colistin resistance. They have also been proposed as an antibiotic target [79]. We thus argue that the ynjC permease could have a role in PR.
Discussion
Genomic surveillance is a powerful public health tool to reduce the spread of resistant bacteria. We show that genomic meta-analysis of outbreak genomes can provide important contextual information when interpreting local outbreaks. To construct the context, we employed both in vitro and in silico search methods to aggregate more than 400 genomes to supplement the local outbreak under investigation, screening more than 10 000 genomes and nearly half a million metagenomes in the process. As a result, we discovered critical epidemiologic details that would have been missed in a traditional outbreak study focusing on local data only. For example, we determined the likely source of the Kp-1 outbreak, its relation to an outbreak at a nearby institution, and it being an instance of the repeated introduction of bla KPC-2 Kp isolates into mainland Europe from Greece. We also identified isolates from other studies that are direct descendants of Kp-1.
We then illustrated the plasmid dynamics across our genome collection. We found frequent loss of genetic material associated with IncFIB(pQil)-type plasmids, even though they often carry the bla KPC-2 gene. We resolved this paradox by showing how bla KPC-2 can still be preserved in the host: the carrier transposon is first transferred to another plasmid before IncFIB(pQil) removal from the host. Such spread of KPC genes between plasmids over short time scales has been documented previously [80, 81].
Besides phylogenomic insights, our context-enriched genome collection informs about adaptation to selective pressure. For one, we found several positively selected genes that are known to mediate, e.g. colistin resistance. We also discovered positive selection of the inner-membrane transporter ynjC together with an overrepresentation of mutated gene copies in colistin-resistant isolates. However, future experiments will have to validate if an effect on colistin resistance can indeed be shown, e.g. by introducing loss-of-function mutations using CRISPR [59].
The pathogens in our filtered dataset can be assumed to be under similar selective pressures, as all but one were isolated from hospital patients. All isolates carry a carbapenemase, where few antibiotics such as colistin remain as a rational treatment option, sometimes combined with rifampicin for synergy [82]. Based on standard clinical practice, we argue that many patients will have received colistin to either decolonize them or treat an infection. Kp colonization has been described as a risk factor for subsequent infection [83], and colonization can persist for years [84], increasing the chance of infection. Indeed, it is likely that Kp isolates will evolve to facilitate long-term carriage. In the Kp-1 outbreak, about half the patients with bla KPC-2 detection presented with clinical disease, mainly pneumonia, and sepsis, and were subsequently treated with colistin either alone or in combination [8]. We assume that these numbers apply to other hospitals as well. In the absence of infection, decolonization is sometimes attempted using colistin. While we observed low efficacy and rapid resistance induction [85], colistin continues to be used for decolonization [86]. In the metadata collected in this study, we did not find any treatment descriptions other than anecdotal ones.
Many genomes in our collection were assembled using different and often insufficiently documented methods. It is thus possible that observed mutations are technical artefacts. However, most genomes were reconstructed from accurate short reads. Therefore, we think the effect of spurious SNVs (in contrast to structural variants, which are a problem with this method) is negligible. However, it must be taken into account as a potential source of error. We also chose a conservative approach to control for unobserved effects other than technical error, such as geography, or sampling site, limiting our analyses to unique mutations and only counting mutations once per gene and locus across the entire dataset.
Colistin resistance can be unstable and transient [87]. Also, colistin resistance can be observed in the absence of explanatory genetic changes [88]. A genome-wide association study (GWAS) of this trait is thus challenging, and in fact, our analysis did not yield any significant target that could predict colistin resistance. The heterogeneous genome collection also complicates the GWAS. Because we often start from pre-assembled genomes, we have no control over the sequencing and assembly process. Similarly, we have no way to check the accuracy of the sparse metadata. Also, we completely lack data on which drugs or other selective pressures most isolates have undergone and can only infer them. Thus, our GWAS using contextual genomes was limited in its explanatory power.
Several components are still missing until we can analyse putative outbreak genomes in a real-time, integrated surveillance system. The main bottleneck, counter-intuitively, is not sequencing but data management and bioinformatics [89]. For example, there is no common repository for bacterial outbreak metadata in active use by the community. We manually aggregated metadata from 28 studies, which frequently involved squinting at low-resolution images to extract, e.g. data on colistin resistance. For most genomes, important information besides the year and country of isolation was missing. Without this metadata, the sequenced genomes cannot easily be integrated into any analysis other than the one they were originally sequenced for. This could be aided in the short term if authors published Supplementary Material giving genome accessions alongside all relevant isolate data, genotypes and phenotypes explored in the study.
Also, more sophisticated tools for outbreak genome sharing are needed [90]. Most outbreak studies appear 1 to 2 years after the outbreak took place (personal observation). However, by then, the value of the results is primarily academic. Only prospective data analysis [91] in real-time would enable a practical outbreak response. A recent example of this is nextstrain, where the virus genomics community converged on a set of protocols and databases [92], which allowed a data-driven public health response. When combined with real-time sequencing of bacterial genomes [93], this set of technologies could substantially improve outbreak response.
Our metagenomic screening returned an isolate closely related to Kp-1, sampled on a different continent. Generally, we think that metagenomic screening for isolates of interest holds great promise. The construction of the required index for an entire read collection such as the NCBI SRA is challenging, as is the design of an easy search interface. However, once operational, such a system allows resistance and virulence tracking in, e.g. mobile genetic elements across species and habitats, which is especially relevant for pathogen outbreak investigations.
Supplementary Data
Funding information
This work is funded in part by the Gordon and Betty Moore Foundation’s Data-Driven Discovery Initiative through Grants GBMF4551 to C. Titus Brown.
Acknowledgements
The authors would like to thank Professor A.C. Rodloff for extensive discussion of the manuscript. The authors would also like to thank T. Kaiser (Institute of Laboratory Medicine, Clinical Chemistry und Molecular Diagnostics, University Hospital Leipzig) and K. Finstermeier (Max Planck Unit for the Science of Pathogens, Berlin) for access to and guidance through the collected Kp-1 outbreak metadata.
Author contributions
A.V. designed the study. A.V., C.Bl. and N.L. performed all laboratory work. A.V. and J.B.H. screened isolates using PCR, A.V. and C.Bl. implemented Nanopore sequencing. A.V., C.Bl. and N.L. collected metadata. A.V., C.Bl., N.L., K.L.W., C.Br., L.I. and M.H. conducted data analysis. B.K. supervised the work. All authors interpreted the results, wrote the text, created the figures and approved the submitted paper.
Conflicts of interest
A.V. has received travel expenses to speak at Oxford Nanopore meetings. A.V., C.Br. and M.H. are co-founders of nanozoo GmbH and hold shares in the company.
Ethical statement
This retrospective study was performed in accordance with the ethical guidelines of the 1964 Declaration of Helsinki and its later amendments and was approved by the local ethics committee (University of Leipzig, register no. 411-1211032013). The need for informed consent was waived according to the ethics approval.
Footnotes
Abbreviations: ANI, average nucleotide identity; ESBL, extended-spectrum beta-lactamase; GWAS, genome-wide association study; ICU, intensive care unit; Kp, Klebsiella pneumoniae; MIC, minimum inhibitory concentration; PR, polymyxin resistance; UHJ, University Hospital Jena; UHL, University Hospital Leipzig; UM, unique mutations.
All supporting data, code and protocols have been provided within the article or through supplementary data files. Five supplementary tables and two supplementary figures are available with the online version of this article.
References
- 1.Holt KE, Wertheim H, Zadoks RN, Baker S, Whitehouse CA, et al. Genomic analysis of diversity, population structure, virulence, and antimicrobial resistance in Klebsiella pneumoniae, an urgent threat to public health. Proc Natl Acad Sci U S A. 2015;112:E3574–81. doi: 10.1073/pnas.1501049112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Munoz-Price LS, Quinn JP. The spread of Klebsiella pneumoniae carbapenemases: a tale of strains, plasmids, and transposons. Clin Infect Dis. 2009;49:1739–1741. doi: 10.1086/648078. [DOI] [PubMed] [Google Scholar]
- 3.Brandt C, Viehweger A, Singh A, Pletz MW, Wibberg D, et al. Assessing genetic diversity and similarity of 435 KPC-carrying plasmids. Sci Rep. 2019;9:11223. doi: 10.1038/s41598-019-47758-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sidjabat HE, Silveira FP, Potoski BA, Abu-Elmagd KM, Adams-Haduch JM, et al. Interspecies spread of Klebsiella pneumoniae carbapenemase gene in a single patient. Clin Infect Dis. 2009;49:1736–1738. doi: 10.1086/648077. [DOI] [PubMed] [Google Scholar]
- 5.David S, Reuter S, Harris SR, Glasner C, Feltwell T, et al. Epidemic of carbapenem-resistant Klebsiella pneumoniae in Europe is driven by nosocomial spread. Nat Microbiol. 2019;4:1919–1929. doi: 10.1038/s41564-019-0492-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lam MMC, Wick RR, Watts SC, Cerdeira LT, Wyres KL, et al. A genomic surveillance framework and genotyping tool for Klebsiella pneumoniae and its related species complex. Nat Commun. 2021;12 doi: 10.1038/s41467-021-24448-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bush SJ, Foster D, Eyre DW, Clark EL, De Maio N, et al. Genomic diversity affects the accuracy of bacterial single-nucleotide polymorphism-calling pipelines. Gigascience. 2020;9:giaa007. doi: 10.1093/gigascience/giaa007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ducomble T, Faucheux S, Helbig U, Kaisers UX, König B, et al. Large hospital outbreak of KPC-2-producing Klebsiella pneumoniae: investigating mortality and the impact of screening for KPC-2 with polymerase chain reaction. J Hosp Infect. 2015;89:179–185. doi: 10.1016/j.jhin.2014.11.012. [DOI] [PubMed] [Google Scholar]
- 9.Ambretti S, Bassetti M, Clerici P, Petrosillo N, Tumietto F, et al. Screening for carriage of carbapenem-resistant enterobacteriaceae in settings of high endemicity: a position paper from an italian working group on CRE infections. Antimicrob Resist Infect Control. 2019;8:136. doi: 10.1186/s13756-019-0591-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kaiser T, Finstermeier K, Häntzsch M, Faucheux S, Kaase M, et al. Stalking a lethal superbug by whole-genome sequencing and phylogenetics: Influence on unraveling a major hospital outbreak of carbapenem-resistant Klebsiella pneumoniae . Am J Infect Control. 2018;46:54–59. doi: 10.1016/j.ajic.2017.07.022. [DOI] [PubMed] [Google Scholar]
- 11.Lübbert C, Faucheux S, Becker-Rux D, Laudi S, Dürrbeck A, et al. Rapid emergence of secondary resistance to gentamicin and colistin following selective digestive decontamination in patients with KPC-2-producing Klebsiella pneumoniae: a single-centre experience. Int J Antimicrob Agents. 2013;42:565–570. doi: 10.1016/j.ijantimicag.2013.08.008. [DOI] [PubMed] [Google Scholar]
- 12.Poirel L, Jayol A, Nordmann P. Polymyxins: Antibacterial activity, susceptibility testing, and resistance mechanisms encoded by plasmids or chromosomes. Clin Microbiol Rev. 2017;30:557–596. doi: 10.1128/CMR.00064-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pitt ME, Elliott AG, Cao MD, Ganesamoorthy D, Karaiskos I, et al. Multifactorial chromosomal variants regulate polymyxin resistance in extensively drug-resistant Klebsiella pneumoniae . Microb Genom. 2018;4 doi: 10.1099/mgen.0.000158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meyer M, Kircher M. Illumina sequencing library preparation for highly multiplexed target capture and sequencing. Cold Spring Harb Protoc. 2010;2010:pdb.prot5448. doi: 10.1101/pdb.prot5448. [DOI] [PubMed] [Google Scholar]
- 15.Wick RR, Judd LM, Gorrie CL, Holt KE. Completing bacterial genome assemblies with multiplex MinION sequencing. Microb Genom. 2017;3:e000132. doi: 10.1099/mgen.0.000132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li W, O’Neill KR, Haft DH, DiCuccio M, Chetvernin V, et al. RefSeq: expanding the prokaryotic genome annotation pipeline reach with protein family model curation. Nucleic Acids Res. 2021;49:D1020–D1028. doi: 10.1093/nar/gkaa1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ondov BD, Treangen TJ, Melsted P, Mallonee AB, Bergman NH, et al. Mash: fast genome and metagenome distance estimation using MinHash. Genome Biol. 2016;17:132. doi: 10.1186/s13059-016-0997-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pierce NT, Irber L, Reiter T, Brooks P, Brown CT. Large-scale sequence comparisons with sourmash. F1000Res. 2019;8:1006. doi: 10.12688/f1000research.19675.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jain C, Rodriguez-R LM, Phillippy AM, Konstantinidis KT, Aluru S. High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat Commun. 2018;9:5114. doi: 10.1038/s41467-018-07641-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li H. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics. 2018;34:3094–3100. doi: 10.1093/bioinformatics/bty191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen S, Zhou Y, Chen Y, Gu J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34:i884–i890. doi: 10.1093/bioinformatics/bty560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li D, Liu CM, Luo R, Sadakane K, Lam TW. MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de bruijn graph. Bioinformatics. 2015;31:1674–1676. doi: 10.1093/bioinformatics/btv033. [DOI] [PubMed] [Google Scholar]
- 23.Wick RR, Judd LM, Gorrie CL, Holt KE. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol. 2017;13:e1005595. doi: 10.1371/journal.pcbi.1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 25.Lam MMC, Wick RR, Wyres KL, Holt KE. Genomic surveillance framework and global population structure for Klebsiella pneumoniae . bioRxiv. 2020 [Google Scholar]
- 26.Carattoli A, Zankari E, García-Fernández A, Voldby Larsen M, Lund O, et al. In silico detection and typing of plasmids using plasmid finder and plasmid multilocus sequence typing. Antimicrob Agents Chemother. 2014;58:3895–3903. doi: 10.1128/AAC.02412-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jia B, Raphenya AR, Alcock B, Waglechner N, Guo P, et al. CARD 2017: expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res. 2017;45:D566–D573. doi: 10.1093/nar/gkw1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Steinegger M, Söding J. MMseqs2 enables sensitive protein sequence searching for the analysis of massive data sets. Nat Biotechnol. 2017;35:1026–1028. doi: 10.1038/nbt.3988. [DOI] [PubMed] [Google Scholar]
- 29.Xie Y, Wei Y, Shen Y, Li X, Zhou H, et al. TADB 2.0: an updated database of bacterial type II toxin-antitoxin loci. Nucleic Acids Res. 2018;46:D749–D753. doi: 10.1093/nar/gkx1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Croucher NJ, Page AJ, Connor TR, Delaney AJ, Keane JA, et al. Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using gubbins. Nucleic Acids Res. 2015;43:e15. doi: 10.1093/nar/gku1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li H, Durbin R. Fast and accurate short read alignment with burrows-wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Page AJ, Taylor B, Delaney AJ, Soares J, Seemann T, et al. SNP-sites: rapid efficient extraction of SNPs from multi-FASTA alignments. Microb Genom. 2016;2:e000056. doi: 10.1099/mgen.0.000056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sagulenko P, Puller V, Neher RA. Tree time: Maximum-likelihood phylodynamic analysis. Virus Evol. 2018;4:vex042. doi: 10.1093/ve/vex042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Argimón S, Abudahab K, Goater RJE, Fedosejev A, Bhai J, et al. Microreact: visualizing and sharing data for genomic epidemiology and phylogeography. Microb Genom. 2016;2:e000093. doi: 10.1099/mgen.0.000093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kozlov AM, Darriba D, Flouri T, Morel B, Stamatakis A. RAxML-NG: a fast, scalable and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics. 2019;35:4453–4455. doi: 10.1093/bioinformatics/btz305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lees JA, Galardini M, Bentley SD, Weiser JN, Corander J. pyseer: a comprehensive tool for microbial pangenome-wide association studies. Bioinformatics. 2018;34:4310–4312. doi: 10.1093/bioinformatics/bty539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee S, Abecasis GR, Boehnke M, Lin X. Rare-variant association analysis: study designs and statistical tests. Am J Hum Genet. 2014;95:5–23. doi: 10.1016/j.ajhg.2014.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Consortium GO. The gene ontology resource: enriching a GOld mine. Nucleic Acids Res. 2021;49:D325–D334. doi: 10.1093/nar/gkaa1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mi H, Muruganujan A, Ebert D, Huang X, Thomas PD. PANTHER version 14: more genomes, a new PANTHER GO-slim and improvements in enrichment analysis tools. Nucleic Acids Res. 2019;47:D419–D426. doi: 10.1093/nar/gky1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Murrell B, Weaver S, Smith MD, Wertheim JO, Murrell S, et al. Gene-wide identification of episodic selection. Mol Biol Evol. 2015;32:1365–1371. doi: 10.1093/molbev/msv035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Spielman SJ, Weaver S, Shank SD, Magalis BF, Li M, et al. Evolution of viral genomes: Interplay between selection, recombination, and other forces. Methods Mol Biol. 2019;1910:427–468. doi: 10.1007/978-1-4939-9074-0_14. [DOI] [PubMed] [Google Scholar]
- 42.Yang J, Anishchenko I, Park H, Peng Z, Ovchinnikov S, et al. Improved protein structure prediction using predicted interresidue orientations. Proc Natl Acad Sci U S A. 2020;117:1496–1503. doi: 10.1073/pnas.1914677117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Koslicki D, Falush D, Jansson JK. MetaPalette: a k-mer painting approach for metagenomic taxonomic profiling and quantification of novel strain variation. mSystems. 2016;1:e00020-16. doi: 10.1128/mSystems.00020-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rodríguez-Beltrán J, DelaFuente J, León-Sampedro R, MacLean RC, San Millán Á. Beyond horizontal gene transfer: the role of plasmids in bacterial evolution. Nat Rev Microbiol. 2021;19:347–359. doi: 10.1038/s41579-020-00497-1. [DOI] [PubMed] [Google Scholar]
- 45.Tonkin-Hill G, MacAlasdair N, Ruis C, Weimann A, Horesh G, et al. Producing polished prokaryotic pangenomes with the panaroo pipeline. Genome Biol. 2020;21:180. doi: 10.1186/s13059-020-02090-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rezaei Javan R, Ramos-Sevillano E, Akter A, Brown J, Brueggemann AB. Prophages and satellite prophages are widespread in streptococcus and may play a role in pneumococcal pathogenesis. Nat Commun. 2019;10:4852. doi: 10.1038/s41467-019-12825-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Surleac M, Czobor Barbu I, Paraschiv S, Popa LI, Gheorghe I, et al. Whole genome sequencing snapshot of multi-drug resistant Klebsiella pneumoniae strains from hospitals and receiving wastewater treatment plants in southern romania. PLoS One. 2020;15:e0228079. doi: 10.1371/journal.pone.0228079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Duchêne S, Holt KE, Weill FX, Le Hello S, Hawkey J, et al. Genome-scale rates of evolutionary change in bacteria. Microb Genom. 2016;2:e000094. doi: 10.1099/mgen.0.000094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jousset AB, Bonnin RA, Rosinski-Chupin I, Girlich D, Cuzon G, et al. A 4.5-year within-patient evolution of a colistin-resistant Klebsiella pneumoniae carbapenemase-producing K. pneumoniae sequence type 258. Clin Infect Dis. 2018;67:1388–1394. doi: 10.1093/cid/ciy293. [DOI] [PubMed] [Google Scholar]
- 50.Gibson B, Eyre-Walker A. Investigating evolutionary rate variation in bacteria. J Mol Evol. 2019;87:317–326. doi: 10.1007/s00239-019-09912-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Munoz-Price LS, Poirel L, Bonomo RA, Schwaber MJ, Daikos GL, et al. Clinical epidemiology of the global expansion of Klebsiella pneumoniae carbapenemases. Lancet Infect Dis. 2013;13:785–796. doi: 10.1016/S1473-3099(13)70190-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bathoorn E, Tsioutis C, da Silva Voorham JM, Scoulica EV, Ioannidou E, et al. Emergence of pan-resistance in KPC-2 carbapenemase-producing Kklebsiella pneumoniae in Crete, Greece: a close call. J Antimicrob Chemother. 2016;71:1207–1212. doi: 10.1093/jac/dkv467. [DOI] [PubMed] [Google Scholar]
- 53.Meletis G, Chatzopoulou F, Fragkouli A, Alexandridou M, Mavrovouniotis I, et al. Whole-genome sequencing study of KPC-encoding Kklebsiella pneumoniae isolated in Greek private laboratories from non-hospitalised patients. J Glob Antimicrob Resist. 2020;20:78–81. doi: 10.1016/j.jgar.2019.07.027. [DOI] [PubMed] [Google Scholar]
- 54.Zhou K, Lokate M, Deurenberg RH, Tepper M, Arends JP, et al. Use of whole-genome sequencing to trace, control and characterize the regional expansion of extended-spectrum β-lactamase producing ST15 Klebsiella pneumoniae . Sci Rep. 2016;6:20840. doi: 10.1038/srep20840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lübbert C, Straube L, Stein C, Makarewicz O, Schubert S, et al. Colonization with extended-spectrum beta-lactamase-producing and carbapenemase-producing enterobacteriaceae in international travelers returning to Germany. Int J Med Microbiol. 2015;305:148–156. doi: 10.1016/j.ijmm.2014.12.001. [DOI] [PubMed] [Google Scholar]
- 56.Stohr J, Verweij JJ, Buiting AGM, Rossen JWA, Kluytmans J, et al. Within-patient plasmid dynamics in Klebsiella pneumoniae during an outbreak of a carbapenemase-producing Klebsiella pneumoniae. PLoS One. 2020;15:e0233313. doi: 10.1371/journal.pone.0233313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pullinger GD, Lax AJ. A salmonella dublin virulence plasmid locus that affects bacterial growth under nutrient-limited conditions. Mol Microbiol. 1992;6:1631–1643. doi: 10.1111/j.1365-2958.1992.tb00888.x. [DOI] [PubMed] [Google Scholar]
- 58.Cuzon G, Naas T, Nordmann P. Functional characterization of tn4401, a tn3-based transposon involved in blaKPC gene mobilization. Antimicrob Agents Chemother. 2011;55:5370–5373. doi: 10.1128/AAC.05202-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.McConville TH, Annavajhala MK, Giddins MJ, Macesic N, Herrera CM, et al. CrrB positively regulates High-Level polymyxin resistance and virulence in Klebsiella pneumoniae . Cell Rep. 2020;33:108313. doi: 10.1016/j.celrep.2020.108313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jayol A, Nordmann P, Brink A, Poirel L. Heteroresistance to colistin in Klebsiella pneumoniae associated with alterations in the PhoPQ regulatory system. Antimicrob Agents Chemother. 2015;59:2780–2784. doi: 10.1128/AAC.05055-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Saber MM, Shapiro BJ. Benchmarking bacterial genome-wide association study methods using simulated genomes and phenotypes. Microb Genom. 2020;6 doi: 10.1099/mgen.0.000337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Barton N, Hermisson J, Nordborg M. Why structure matters. Elife. 2019;8:e45380. doi: 10.7554/eLife.45380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sul JH, Martin LS, Eskin E. Population structure in genetic studies: Confounding factors and mixed models. PLoS Genet. 2018;14:e1007309. doi: 10.1371/journal.pgen.1007309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jayol A, Poirel L, Brink A, Villegas MV, Yilmaz M, et al. Resistance to colistin associated with a single amino acid change in protein PmrB among Klebsiella pneumoniae isolates of worldwide origin. Antimicrob Agents Chemother. 2014;58:4762–4766. doi: 10.1128/AAC.00084-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hoch JA, Varughese KI. Keeping signals straight in phosphorelay signal transduction. J Bacteriol. 2001;183:4941–4949. doi: 10.1128/JB.183.17.4941-4949.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hsieh PF, Lin HH, Lin TL, Wang JT. CadC regulates cad and tdc operons in response to gastrointestinal stresses and enhances intestinal colonization of Klebsiella pneumoniae . J Infect Dis. 2010;202:52–64. doi: 10.1086/653079. [DOI] [PubMed] [Google Scholar]
- 67.Stahlhut SG, Chattopadhyay S, Struve C, Weissman SJ, Aprikian P, et al. Population variability of the FimH type 1 fimbrial adhesin in Klebsiella pneumoniae . J Bacteriol. 2009;191:1941–1950. doi: 10.1128/JB.00601-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Paul S, Linardopoulou EV, Billig M, Tchesnokova V, Price LB, et al. Role of homologous recombination in adaptive diversification of extraintestinal Escherichia coli . J Bacteriol. 2013;195:231–242. doi: 10.1128/JB.01524-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sabnis A, Hagart KL, Klöckner A, Becce M, Evans LE, et al. Colistin kills bacteria by targeting lipopolysaccharide in the cytoplasmic membrane. Elife. 2021;10:e65836. doi: 10.7554/eLife.65836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schneider E, Hunke S. ATP-binding-cassette (ABC) transport systems: functional and structural aspects of the ATP-hydrolyzing subunits/domains. FEMS Microbiol Rev. 1998;22:1–20. doi: 10.1111/j.1574-6976.1998.tb00358.x. [DOI] [PubMed] [Google Scholar]
- 71.McDaniel C, Su S, Panmanee W, Lau GW, Browne T, et al. A putative ABC transporter permease is necessary for resistance to acidified nitrite and EDTA in pseudomonas aeruginosa under aerobic and anaerobic planktonic and biofilm conditions. Front Microbiol. 2016;7:291. doi: 10.3389/fmicb.2016.00291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Greene NP, Kaplan E, Crow A, Koronakis V. Antibiotic resistance mediated by the MacB ABC transporter family: A structural and functional perspective. Front Microbiol. 2018;9:950. doi: 10.3389/fmicb.2018.00950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Karalewitz AA, Miller SI. Multidrug-resistant Acinetobacter baumannii chloramphenicol resistance requires an inner membrane permease. Antimicrob Agents Chemother. 2018;62:e00513-18. doi: 10.1128/AAC.00513-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Abramson J, Smirnova I, Kasho V, Verner G, Iwata S, et al. The lactose permease of Escherichia coli: overall structure, the sugar-binding site and the alternating access model for transport. FEBS Lett. 2003;555:96–101. doi: 10.1016/s0014-5793(03)01087-1. [DOI] [PubMed] [Google Scholar]
- 75.Guan L, Mirza O, Verner G, Iwata S, Kaback HR. Structural determination of wild-type lactose permease. Proc Natl Acad Sci U S A. 2007;104:15294–15298. doi: 10.1073/pnas.0707688104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhan HQ, Najmi M, Lin K, Aluri S, Fiser A, et al. A proton-coupled folate transporter mutation causing hereditary folate malabsorption locks the protein in an inward-open conformation. J Biol Chem. 2020;295:15650–15661. doi: 10.1074/jbc.RA120.014757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sahin-Tóth M, Frillingos S, Lawrence MC, Kaback HR. The sucrose permease of Escherichia coli: functional significance of cysteine residues and properties of a cysteine-less transporter. Biochemistry. 2000;39:6164–6169. doi: 10.1021/bi000124o. [DOI] [PubMed] [Google Scholar]
- 78.Zhou Z, White KA, Polissi A, Georgopoulos C, Raetz CR. Function of Escherichia coli MsbA, an essential ABC family transporter, in lipid a and phospholipid biosynthesis. J Biol Chem. 1998;273:12466–12475. doi: 10.1074/jbc.273.20.12466. [DOI] [PubMed] [Google Scholar]
- 79.Garmory HS, Titball RW. ATP-binding cassette transporters are targets for the development of antibacterial vaccines and therapies. Infect Immun. 2004;72:6757–6763. doi: 10.1128/IAI.72.12.6757-6763.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Stoesser N, Phan HTT, Seale AC, Aiken Z, Thomas S, et al. Genomic epidemiology of complex, multispecies, plasmid-borne bla KPC carbapenemase in enterobacterales in the united kingdom from 2009 to 2014. Antimicrob Agents Chemother. 2020;64:e02244-19. doi: 10.1128/AAC.02244-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.David S, Cohen V, Reuter S, Sheppard AE, Giani T, et al. Integrated chromosomal and plasmid sequence analyses reveal diverse modes of carbapenemase gene spread among Klebsiella pneumoniae . Proc Natl Acad Sci U S A. 2020;117:25043–25054. doi: 10.1073/pnas.2003407117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tascini C, Tagliaferri E, Giani T, Leonildi A, Flammini S, et al. Synergistic activity of colistin plus rifampin against colistin-resistant KPC-producing Klebsiella pneumoniae . Antimicrob Agents Chemother. 2013;57:3990–3993. doi: 10.1128/AAC.00179-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gorrie CL, Mirceta M, Wick RR, Edwards DJ, Thomson NR, et al. Gastrointestinal carriage is a major reservoir of Klebsiella pneumoniae infection in intensive care patients. Clin Infect Dis. 2017;65:208–215. doi: 10.1093/cid/cix270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lübbert C, Lippmann N, Busch T, Kaisers UX, Ducomble T, et al. Long-term carriage of Klebsiella pneumoniae carbapenemase-2-producing K pneumoniae after a large single-center outbreak in germany. Am J Infect Control. 2014;42:376–380. doi: 10.1016/j.ajic.2013.12.001. [DOI] [PubMed] [Google Scholar]
- 85.Lübbert C, Faucheux S, Becker-Rux D, Laudi S, Dürrbeck A, et al. Rapid emergence of secondary resistance to gentamicin and colistin following selective digestive decontamination in patients with KPC-2-producing Klebsiella pneumoniae: a single-centre experience. Int J Antimicrob Agents. 2013;42:565–570. doi: 10.1016/j.ijantimicag.2013.08.008. [DOI] [PubMed] [Google Scholar]
- 86.Janssen AB, van Hout D, Bonten MJM, Willems RJL, van Schaik W. Microevolution of acquired colistin resistance in enterobacteriaceae from ICU patients receiving selective decontamination of the digestive tract. J Antimicrob Chemother. 2020;75:3135–3143. doi: 10.1093/jac/dkaa305. [DOI] [PubMed] [Google Scholar]
- 87.Lee JY, Park YK, Chung ES, Na IY, Ko KS. Evolved resistance to colistin and its loss due to genetic reversion in pseudomonas aeruginosa. Sci Rep. 2016;6:25543. doi: 10.1038/srep25543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kapel N, Caballero JD, Craig MacLean R. Localized hypermutation drives the evolution of unstable colistin resistance in pseudomonas aeruginosa. bioRxiv. 2021 doi: 10.1101/2021.08.10.455869. [DOI] [Google Scholar]
- 89.Hodcroft EB, De Maio N, Lanfear R, MacCannell DR, Minh BQ, et al. Want to track pandemic variants faster? Fix the bioinformatics bottleneck. Nature. 2021;591:30–33. doi: 10.1038/d41586-021-00525-x. [DOI] [PubMed] [Google Scholar]
- 90.Viehweger A, Brandt C, Hölzer M. DarkQ: Continuous genomic monitoring using message queues. F1000Res. 2021;10:998 doi: 10.12688/f1000research.54255.1. [DOI] [Google Scholar]
- 91.Kwong JC, Mercoulia K, Tomita T, Easton M, Li HY, et al. Prospective whole-genome sequencing enhances national surveillance of listeria monocytogenes. J Clin Microbiol. 2016;54:333–342. doi: 10.1128/JCM.02344-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hadfield J, Megill C, Bell SM, Huddleston J, Potter B, et al. Nextstrain: real-time tracking of pathogen evolution. Bioinformatics. 2018;34:4121–4123. doi: 10.1093/bioinformatics/bty407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Steinig E, Duchêne S, Aglua I, Greenhill A, Ford R, et al. Phylodynamic modelling of bacterial outbreaks using nanopore sequencing. bioRxiv. 2021 doi: 10.1101/2021.04.30.442218. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.