Abstract
The invasive plant pathogen Xylella fastidiosa currently threatens European flora through the loss of economically and culturally important host plants. This emerging vector-borne bacterium, native to the Americas, causes several important diseases in a wide range of plants including crops, ornamentals, and trees. Previously absent from Europe, and considered a quarantine pathogen, X. fastidiosa was first detected in Apulia, Italy in 2013 associated with a devastating disease of olive trees (Olive Quick Decline Syndrome, OQDS). OQDS has led to significant economic, environmental, cultural, as well as political crises. Although the biology of X. fastidiosa diseases have been studied for over a century, there is still no information on the determinants of specificity between bacterial genotypes and host plant species, which is particularly relevant today as X. fastidiosa is expanding in the naive European landscape. We analysed the genomes of 79 X . fastidiosa samples from diseased olive trees across the affected area in Italy as well as genomes of the most genetically closely related strains from Central America. We provided insights into the ecological and evolutionary emergence of this pathogen in Italy. We first showed that the outbreak in Apulia is due to a single introduction from Central America that we estimated to have occurred in 2008 [95 % HPD: 1930–2016]. By using a combination of population genomic approaches and evolutionary genomics methods, we further identified a short list of genes that could play a major role in the adaptation of X. fastidiosa to this new environment. We finally provided experimental evidence for the adaptation of the strain to this new environment.
Keywords: outbreak, adaptation, Olive Quick Decline Syndrome (OQDS), emerging pathogen
Data Summary
All the sequencing data have been deposited in the NCBI SRA database under the BioProject PRJNA505446 (Submission ID: SUB4579352). All the supporting data have been provided through supplementary data files.
Impact Statement.
Xylella fastidiosa is an emerging plant pathogenic bacterium causing several diseases of economic importance worldwide. First detected in Europe in 2013, it has already caused the death of millions of Italian olive trees and is continuing to spread in the affected region. By analysing a population of strains from Italian olive trees along with closely related genomes from other plant species and locations, we have shown that the outbreak is due to a single introduction from Central America which would have occurred in 2008 [95 % HPD: 1930–2016]. We have further identified a few genes likely to have played a role in the adaptation of the introduced strain in this naive environment. This is the first study tackling the enduring question of the genetic determinants of X. fastidiosa adaptation to a new environment and sheds light on processes linked to the emergence of devastating plant diseases.
Introduction
The introduction of a pathogen in a new environment can have highly detrimental impacts on human health or agriculture. While several factors underlying the emergence of pathogens have been identified, the expansion of the human population and globalization of trade are considered as the two most important ones [1]. A number of human disease outbreaks are of zoonotic origin while many agricultural plant pathogens result from the spillover of wild reservoirs into cultivated areas [2]. Several recent outbreaks emphasize the rapidity with which an emerging pathogen can then spread across the world thanks to human activities when the environmental conditions are met. However, only a few microorganisms introduced in a naive environment will lead to an outbreak. The ability of the introduced microorganism to withstand the biotic and abiotic pressures that it will encounter in this new habitat is indeed key to its establishment and growth [3]. The adaptation of the invader is a crucial factor contributing to invasion success.
In the case of pathogenic microorganisms, interactions with their hosts are crucial to colonization and establishment [4], as hosts represent their primary, and sometimes only, habitat. The composition and genetic diversity of the host population and of its resident microbiota present in the new environment play a major role in the establishment, growth and spread of the introduced microorganism. For instance, in agricultural landscapes, the high density and uniformity of crop plants that microorganisms encountered in monocultural systems are highly conducive to the emergence and spread of pathogens [5]. Although the main factors leading to the emergence of pathogens and the stepwise process leading to it have been deciphered [3, 6, 7], the origins of the emerging pathogens as well as the genetic mechanisms underlying host adaptation are usually difficult to assess [2]. Understanding the ecological and evolutionary history of disease outbreaks is however crucial to developing sustainable plant protection strategies [5].
The plant-associated bacterium Xylella fastidiosa is known for causing devastating epidemics in several crop species. Indeed, while it does not produce disease symptoms on most of its host plants, X. fastidiosa has been reported to cause several diseases in crops (e.g. grapevines, almonds, coffee, sweet orange, peach), ornamentals (e.g. oleander, hibiscus), and trees (e.g. oak, sycamore, elm) [8]. Despite having long been thought to be a generalist because of its broad host range of at least 595 plant species [8, 9], the host range of a given X. fastidiosa strain is much narrower, infecting a limited number of plant species, and causing disease in a small number of plant species [10, 11]. Three main X. fastidiosa ’s subspecies, namely fastidiosa, multiplex, and pauca, have so far been described, each of them displaying a different host range [12, 13]. These subspecies have been further divided into sequence types (ST) based on their allelic profile of seven housekeeping genes (MLST genotyping) [14]. Besides this wide host range, X. fastidiosa is transmitted by all xylem-sap feeding hemipterans without apparent specificity [8], implying that native insect vector species are readily available for pathogen spread virtually anywhere in the world [8, 13].
Previously thought to be restricted to the Americas, the recent detection of X. fastidiosa in Europe has raised new concerns worldwide due to its highly noxious potential. Reported for the first time in Italy in 2013 [15], its subsequent spread across Southern Apulia has led to the death of several millions of olive trees, with significant environmental, social, political and economic consequences. This first confirmed report in Europe was soon followed by its progressive detection in several other E.U. countries, with X. fastidiosa reportedly infecting over one hundred plant species [9]. A major concern regarding this pathogen gradually expanding its geographical range worldwide is its ability to form novel associations with host plants [8]. However, the genetic mechanisms driving X. fastidiosa adaptation to novel plants or environments remain unknown. The emergence of X. fastidiosa in Italy represents an opportunity to study the ecological and evolutionary processes driving and unfolding with an emerging bacterial plant disease epidemic. Within this framework, here we used genomic tools to analyse a population of X. fastidiosa isolates obtained from olive trees across the affected area in Southern Italy.
Methods
Samples collection
Twig samples from 75 olive trees (Olea europaea cv. ‘Cellina di Nardò’ or ‘Ogliarola Salentina’) showing typical OQDS symptoms were collected between 2013 and 2017 in Apulia, Italy, from different foci emerging as the epidemic continued spreading in the region. The data can be visualized here: https://microreact.org/project/9Ywdh72Fhz7gihnvuuJya2. X. fastidiosa was successfully cultured from 65 of these samples onto BCYE medium [16] as previously described [17]. After triple cloning, X. fastidiosa clones preserved in a −80 °C freezer had their DNA extracted for sequencing purposes using the DNeasy Plant Mini kit (Qiagen, Valencia, CA, USA). For the ten samples for which colonies could not be obtained, DNA was directly extracted from twigs using the CTAB-based protocol as described in [18]. Samples were sent for Illumina sequencing at the QB3 Vincent J. Coates Genomics Sequencing Laboratory (Berkeley, CA, U.SA.). Each sample was sequenced twice (40 samples) or three times independently (35 samples - including the ten total DNA samples) using the Illumina HiSeq 4000. Raw reads of individual isolates were combined for further analyses. One isolate (APL16) was removed from the analyses because of low-quality sequencing. All raw reads and metadata have been submitted to the NCBI SRA database (Submission ID: SUB4579352, BioProject PRJNA505446).
De novo assembly and phylogenetic analyses
The De Donno genome was reassembled with Unicycler v0.4.4 [19] using a combination of the Illumina and Pacbio raw reads previously published [20] and the default parameters. The assembly was then circularized using Circlator v1.5.5 [21]. As the De Donno plasmid could not be closed and circularized using a combination of Unicycler and Circlator, the Recycler v0.7 algorithm dedicated to plasmid assembly [22] and Circlator were used instead. An assembly generated by SPAdes v3.9.0 using the careful parameter with -k of 21, 33, 55, 77 and 99 [23] was used as an input of Recycler. The combined De Donno assembly thus generated was of similar length (2492475 bp for the chromosome and 35441 bp for the plasmid) as the one previously obtained [20] but of a higher quality; notably the sequencing errors from PacBio reads were corrected by the Illumina reads. The assembly was eventually manually curated to correct for the few remaining sequencing errors detected. All other isolates – including the three Costa Rican isolates previously sequenced and for which raw reads are publicly available on NCBI - were de novo assembled using SPAdes v3.9.0 with the careful parameter and -k of 21, 33, 55, 77 and 99. The contigs were then reordered using progressiveMauve v2.4.0 [24] before being annotated with Prokka v1.13.3 [25]. A ST53 pangenome was obtained using Roary v1.14.6 [26] and the 64 Apulian isolates sequenced for this project, the five isolates from this region previously sequenced and published (CoDiRO, De Donno, SQ here named APL87, Salento-1 and Salento-2) [12, 20, 27, 28] as well as the three ST53 Costa Rican isolates (COF0407: XFAS006-SEQ-1-ASM-1, OLS0478: XFAS005-SEQ-1-ASM-1 and OLS0479: XFAS004-SEQ-2-ASM-1). The core genome alignment was used to build a ST53 Maximum Likelihood tree using RaxML v8.2.12 [29]. The best-scoring tree was generated using the GTRGAMMA substitution model and 1000 bootstrap replicates. In a similar manner, a ML tree of subsp. pauca was built using all the published pauca genomes as well as the ones generated in this study. The Apulian core genome alignment was used to construct a haplotype network. The network was built using the HaploNet function in the R package ‘pegas’ v1.0 [30], which calculates the number of mutations within the dataset.
Whole genome comparisons
Two different approaches were used to compare the whole genomic sequences of the X. fastidiosa isolates infecting olive trees (Olea europaea) in Italy and the isolates infecting coffee (Coffea sp.) and oleander (Nerium oleander) in Costa Rica. First, the Harvest suite designed to obtain a rapid core-genome alignment for bacterial genomes [31] was applied to all ST53 isolates. Secondly, we used the breseq pipeline developed by Deatherage and coauthors to detect mutations in microbial re-sequenced samples [32]. In both cases, the Costa Rican coffee strain COF0407 was used as a reference. SNPs and indels were independently plotted using the circlize R package v0.4.12 [33]. Ancestral (lineage-specific) and recent (strain-specific) introgression events in the ST53 data set were determined using FastGEAR and the default parameters [34]. Genomic region copy number variation was calculated with the CNOGpro R package v1.1 [35]. Briefly, FASTQ files for each Apulian field collected isolated were mapped against genome assembly Pr8x (ASM145629v1) using Bowtie2 v2.3.4.1 [36]. Samtools v1.8 [37] was then used to convert SAM files into BAM and subsequently sort them. Finally, individual hit files were constructed following the instructions listed in the CNOGpro manual. Copy number variation (CNV) was calculated through a Hidden Markov Model and using 1000 bootstrap replicates to estimate confidence intervals. CNV was calculated over a window size of 100 bp after correcting for GC bias. The Pr8x genome assembly was again used as a reference. A flow chart summarizing the different tools used in this study is provided (Fig. S1, available in the online version of this article).
Detection of regions under positive selection and selective sweeps
First, Tajima’s D test were performed on the Apulian core genome alignment using the PopGenome R package v2.7.5 [38] and a sliding window of 500 bp. Second, in order to assess evidence of selection at the gene level, a gene-based approach using a combination of maximum-likelihood and counting-based approaches to infer the ratio of substitution rates at non-synonymous and synonymous sites (dN/dS) was used. This test, called SLAC (Hyphy’s v2.5.14), enables to test for pervasive selection within a population [39, 40]. We found homologous sequences for each functionally characterized gene within the Apulian population. We focused on genes with defined functions since a significant proportion of the hypothetical genes in the data set were partial sequences or singletons. Homologous sequences were defined and aligned using GET_HOMOLOGUES v2 [41] and Macse v2.03 [42]; individual ML gene trees were constructed with RAxML v8.2.12. The gene alignments and ML trees were then used as input for SLAC.
Molecular dating
To perform inference of divergence times, we performed a core genome alignment using the 72 ST53 isolates as well as 48 previously published X. fastidiosa genomes [12]. Recombining regions were identified using ClonalFrameML [43] and subsequently removed. Due to the limited number of SNPs found within our Italian isolates, we only applied molecular rate dating to extract node ages for branching events; Bayesian inferences were performed using BEAST v1.8.4 [44] using the evolutionary rate previously found for X. fastidiosa (7.62×10−7 substitutions per site per year [12]) and the same parameters as in that work. Five independent chains in which samples were drawn every 10 000 MCMC steps from a total of 100 000 000 steps were run, after a discarded burn-in of 10 000 000 steps. Convergence to the stationary distribution and sufficient sampling and mixing were checked by inspection of posterior samples (effective sample size >200). Parameter estimation was based on the samples combined from the different chains. The best-supported tree was estimated from the combined samples using the maximum clade credibility method implemented in TreeAnnotator as previously described [12].
Read mapping, SNP calling and Mantel test
Read mapping and SNP calling were performed as in [12], using as reference the newly assembled De Donno genome. Reads statistics are summarized in Table S1. Correlation between genetic and geographic distance among Apulian isolates was assessed using a Mantel test using the ecodist R package and 10 000 permutations [45]. Genetic distance was calculated as the Bray-Curtis dissimilarity between isolates while geographic distances were computed as Euclidean distances.
Plant inoculations, X. fastidiosa pathogenicity and transmissibility assessment
Ten Olea europeae and three Coffea sp. plants were needle-inoculated in January 2018 using a turbid suspension of the strain De Donno (~109 cells ml−1) as previously described [17]. In parallel, three coffee and three olive plants were mock-inoculated (i.e. inoculated with resuspension buffer only). The small number of coffee plants used was due to the limited availability of this plant species for experimentation when the work was performed. Bacterial colonization of the inoculated plants was monitored by real time PCR [46] on leaf petioles sampled at the inoculation points and at the distal points (up to 20 cm far from the inoculation point) [17]. From the plants that yielded positive qPCR-results at the distal points, culturing was then performed to confirm bacterial multiplication and systemic infections. In the case of olives, isolation was performed from twigs as previously described. For coffee, isolation was made from four leaf petioles and midribs homogenized in Phosphate-Buffered Saline – PBS (1 : 10 W:v); ten-fold serial dilutions of the extracts were plated onto Periwinkle Wilt Gelrite (PWG) medium and incubated at 28 °C for 4–8 weeks. The tissues used for bacterial isolation from olive and coffee plants were chosen according to previous publications to yield the highest X. fastidiosa populations while limiting contamination and inhibitors [17, 47]. Symptoms were visually assessed every month for a period of almost 2 years.
To assess the bacterial transmissibility from the two host plant species, two olive and two coffee plants testing positive by qPCR, and from which colonies were successfully isolated, were used for transmission tests using X. fastidiosa -free adults of Philaenus spumarius (the vector species of importance locally in Italy). Between 20 and 25 adult specimens were caged on each infected plant for 3 days of acquisition access period (AAP). Following the AAP, insects in groups of five individuals were transferred to periwinkle or young olive plants (cultivar Cellina di Nardò), for a 3 day inoculation access period (IAP), as previously done [48, 49]. Insects recovered after the IAP were individually tested by qPCR for X. fastidiosa , and transmission was then assessed by testing the recipient plants for the presence of X. fastidiosa ; periwinkle recipient plants were tested 3 months after the IAP and olive trees 6 months after the IAP. A higher number of olive trees than periwinkle plants were used as recipient plants since (i) periwinkle plants after the IAP often suffer from fungal root attack compromising the experiments and (ii) olive trees correspond to the plant species of interest in Apulia, Italy. Three independent transmission experiments were performed in late summer 2019 (approx. 20 months after the inoculation of the source plants).
Results
The outbreak in Apulia is the result of a recent single pathogen introduction
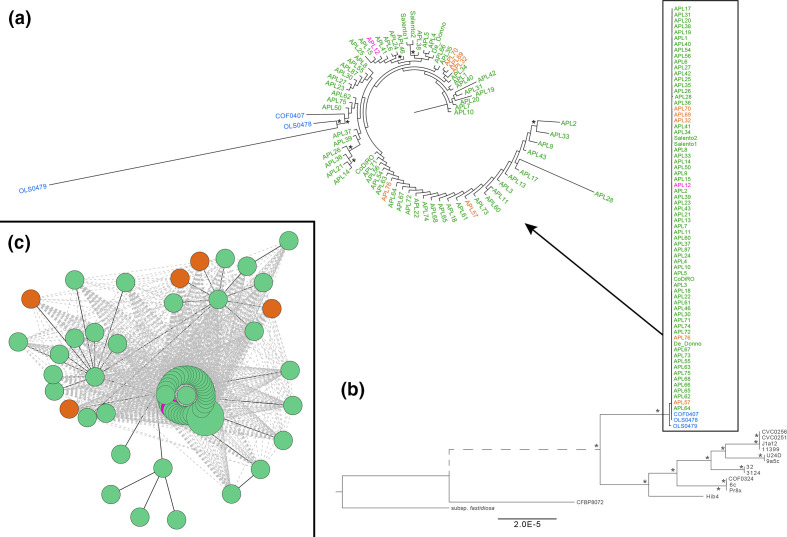
The genome sequences of 75 X . fastidiosa samples collected from olive trees between 2013 and 2017 were generated. The average depth of coverage for most samples was 144-fold, while ten samples were sequenced from total plant DNA with average 11-fold depth of coverage (Table S1; one isolate was dropped due to poor quality). Because the latter ten samples had lower depth of coverage they led to incomplete de novo assemblies; as such those samples were only used for genomic comparisons described below and are summarized in Fig. S1. Five previously published isolates from olive trees in Apulia were used, as well as three Costa Rican strains that served as the most closely related outgroups available [12], and confirmed with a phylogenetic approach (Fig. 1b). The tree showed a monophyletic group including all Apulian isolates along with three closely related isolates from Costa Rica (this clade/population with the Costa Rican and Apulian isolates is commonly known as ST53). The Costa Rican strain from coffee (COF0407) is a sister taxon to the Apulian isolates while the two other Costa Rican strains isolated from oleander (OLS0478 and OLS0479) share an older ancestor on the phylogeny (Fig. 1a). A total number of 1355 SNPs were detected in the dataset including the Costa Rican isolates (ST53 clade) when mapped to the reference strain De Donno, while 455 SNPs were observed within the olive population alone.
Fig. 1.
The X. fastidiosa outbreak in Apulia is due to a single introduction from Costa Rica (a, b) followed by its establishment and spread (c). X. fastidiosa subsp. pauca (b) and ST53 (a) maximum likelihood phylogenetic trees including all the sequenced Apulian isolates. A X. fastidiosa subsp. fastidiosa strain was used as an outgroup in (b). The Apulian isolates appear in green (region of origin: Lecce), orange (Brindisi) and pink (Taranto) based on their region of origin while the Costa Rican strains appear in blue. Bootstrapping values over 80 % corresponding to 1000 replicates are indicated by an asterisk (a, b). C shows the haplotype network linking X. fastidiosa isolates from Apulia.
To estimate the introduction date of X. fastidiosa in Apulia a maximum likelihood SNP-based phylogeny of the ST53 population was constructed using additional previously published genomic data [12], resulting in 120 genomes and a time span of 33 years. Applying this previously obtained substitution rate, an introduction date of 9.4 years [95 % highest posterior density (HPD): 1.4–87.4 years] before our latest sample of 2017 was obtained, that is to say 2008 [95 % HPD: 1930–2016] (Fig. S2). The credible interval obtained here is similar as in a prior study that included X. fastidiosa genomes from various subspecies and regions (95 % HPD: 1952–2015) [12]. In order to test whether we could use a phylogeography approach to reconstruct the spread of X. fastidiosa in Apulia, we performed a Mantel test. The test indicated no association between genetic and geographic distances among isolates (r=5.35×10−3, P-value=0.37). This lack of spatial correlation is illustrated by a star-like haplotype network of the OQDS isolates (Fig. 1c), and can be further visualized here: https://microreact.org/project/9Ywdh72Fhz7gihnvuuJya2.
Recombination events within the ST53 population
Two different lineages were detected within the ST53 population. The first one includes all Apulian isolates and the Costa Rican coffee strain (APL-COF) while the second one includes the two Costa Rican strains coming from oleander (OLS, Fig. S3). No ancestral recombination event (between lineages) was detected, while four recent (between isolates) events were identified. Two introgression events of 19.2 kb and 23.7 kb of the APL-COF lineage into Costa Rican oleander isolate OLS0478 were observed while the two other events consisted of the introgression of segments from an ‘unknown’ lineage into COF0407 and APL38, of 1.9 kb and of 14.8 kb respectively (Fig. S3). The removal of the recombinant segments did not change the ST53 phylogeny.
Whole genome comparison between Costa Rican and Apulian isolates
An analysis of the pangenome of the ST53 clade revealed that its core genome was composed of 1557 genes, which represents 65 % of the De Donno genes (Fig. S4). The ST53 core genome was primarily composed of genes involved in essential physiological functions (Fig. S4). An additional 518 genes were present in the soft-core genome – corresponding to the genes present in ≥95 % of the genomes (Fig. S4). The core and soft-core genomes together are more likely to represent the core genome due to assembly and annotation limitations [50–52].
To identify potential loci linked to X. fastidiosa adaptation to olive trees and Apulia biotic and abiotic stresses, we hypothesized that the genes should either be present in all isolates from olive trees and absent in the ST53 strains from Costa Rica (i.e. gene acquisition), country where olive trees are rare [53], or contain non-synonymous mutations in all Apulian isolates in comparison to the Costa Rica strains [54, 55]. As loss of functional mutations (including null mutations, i.e. complete gene deletion) have been shown to play crucial roles in the adaptation of several pathogens to new hosts, a third non-exclusive scenario that could lead to host adaptation would be deletion of certain genes within the olive-infecting population that were present in the Costa Rican strains [56, 57]. We thus looked for (i) insertion or duplication events, (ii) deletions and (iii) non-synonymous mutations shared by the Apulian population. We found five genes present in all Apulian isolates and absent in those from Costa Rica: a frameshifted alkaline phosphatase (PhoA, B9J09_02565), an autotransporter translocation and assembly factor annotated as a pathogenicity protein (TamB, B9J09_08570), a zonula occludens toxin (P-loop_NTPase, B9J09_08545), a protein of unknown function (B9J09_05845) and a predicted transcriptional regulator (B9J09_01520).
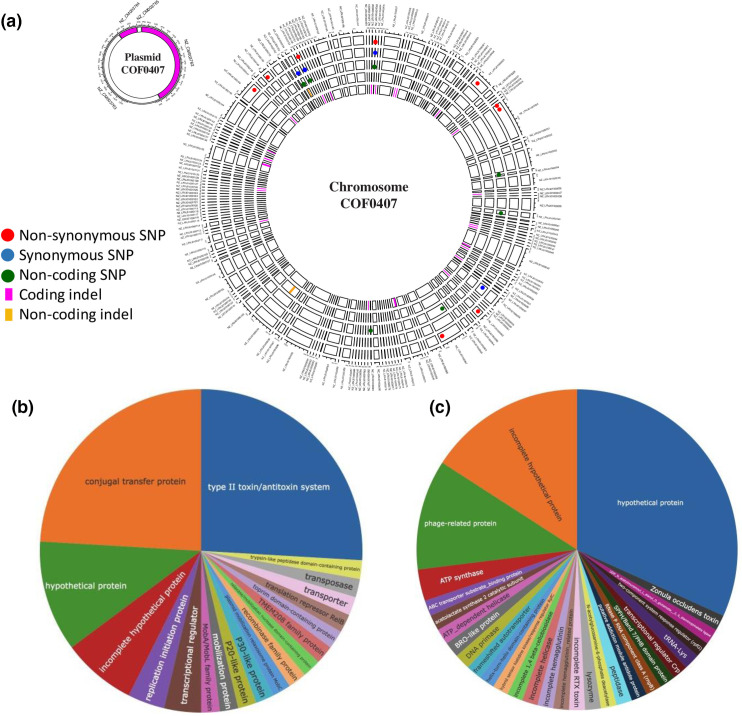
We then compared the Apulian and Costa Rican strains using breseq [32] using the coffee strain (COF0407) as reference. Sixty-three genes in the chromosome and fifty-four in the plasmid evidenced mutations in all Apulian isolates when compared to the Costa Rican ones (Fig. 2). All shared mutations present in the plasmid corresponded to complete gene deletions (Fig. 2a, Table S2), primarily genes involved in plasmid maintenance, conjugation and replication (Fig. 2b, Table S2). Apart from phage-related genes, those in the chromosome were involved in metabolism (acetolactate synthase, ABC transporter substrate-binding protein), DNA replication (DNA primase and ATP-dependent helicase), signalling (hybrid sensor histidine kinase (rpfC), two-component system response regulator (rpfG) [58] and SPFH/Band 7/PHB domain protein [59, 60]), gene expression (helix-turn-helix domain containing protein, transcriptional regulator Crp), cell wall growth/lysis [61] (lysozyme and UDP-N-acetylmuramoyl-l-alanyl-d-glutamate-−2, 6-diaminopimelate ligase), peptidase, ATP synthase and pathogenesis (zonula occludens toxin, Fig. 2c, Table S2). We note some of the genes also encoded for proteins of unknown function that were either complete or incomplete (i.e. pseudogenes, Fig. 2, Table S2).
Fig. 2.
Mutations shared by the entire Apulian population. The types (one circle corresponding to one mutation type) and positions of the shared mutations are shown on the Costa Rican coffee strain genome, COF0407 (a). The names appearing outside the circle correspond to the names of COF0407 contigs (a). Functions of the proteins with mutations (SNPs or indels) encoded in the plasmid (b, n=54) and chromosome (c, n=63).
Using a second comparative approach, the rapid core-genome alignment software Parsnp, we found shared synonymous and non-synonymous mutations among the Apulian population. These mutations were also found using the previous approach confirming our results (Table S2). Finally, analyses of the copy number variations of protein-coding and intergenic regions of the ST53 genomes did not reveal conserved duplication events in the Apulian population (Fig. S5).
Detection of selection in the Apulian population
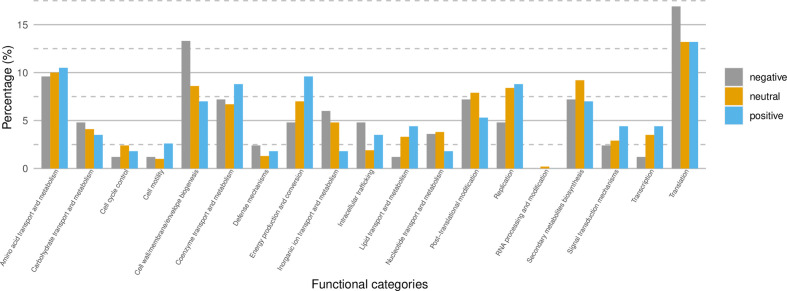
As positive selection is considered to be an important mechanism of adaptation, two different statistical methods were undertaken to investigate signs of adaptation in the Apulian population: a frequency-based method (Tajima’s D) and a gene-based method (SLAC) [39]. A negative value of −2.9 was obtained for the Tajima’s D test indicating a surplus of rare alleles [62]. In order to determine whether we could detect genes under positive selection, a test combining maximum-likelihood and counting-based approaches to measure the dN/dS ratio was used (SLAC). This approach tested for pervasive selection at individual sites [39]. As many of the genes encoding for proteins of unknown functions were either incomplete or shared only by a few isolates, they were not included in the analyses (see, Methods). Most of the genes that were tested showed no variation - i.e. genes were found to be under neutral selection, dN/dS ≈ 1 (Table S3). However, dN/dS>1 were obtained for 115 genes, indicating positive selective pressure on those genes. Eighty-four genes were also found to have dN/dS<1, indicating the presence of genes being under purifying selection in the data set. The following categories showed at least a 25 % enrichment among the genes under positive selection in comparison to the genes under neutral evolution: cell motility, intracellular trafficking, signal transduction, energy production and conversion, defence mechanisms, coenzyme transport and metabolism, lipid transport and metabolism with cell motility displaying the highest enrichment (Fig. 3).
Fig. 3.
Functional categories of the genes found to be under negative selection (Ω<1, n=84), neutral (Ω=1, n=628) and under positive selection (Ω>1, n=115) within the Apulian data set.
Coffee plants are poor sources of the Apulian strain for vector spread
In order to test whether the Apulian strain De Donno could infect coffee, we mechanically inoculated this strain into three coffee and ten olive plants in a greenhouse. While mock-inoculated plants remained negative for the length of the experiment, 1 month post-inoculation, all plants were positive for X. fastidiosa at the inoculation point (qPCR-based detection). At 10 months post-inoculations, distal leaves (~10–15 cm above the point of inoculation) were tested; seven of ten olive plants tested positive at these distal points, but all coffee plants were negative. The test was repeated at 18 months post-inoculation with two out of three coffee and seven out of ten olive trees testing positive at distal points; X. fastidiosa colonies were recovered from these positive plants. At 10 months post-inoculation, initial shoot dieback was first observed in three out of the seven infected olives. Leaf scorching was observed in the coffee plants but symptoms could not be associated with X. fastidiosa infections as they were also present in the mock-inoculated plants. By the end of the second year post-inoculation, all seven infected olive plants showed typical OQDS symptoms, whereas no clear symptom could be observed in the two X. fastidiosa infected coffee plants. A transmission experiment using the insect species driving the OQDS epidemic in Italy, namely the spittlebug Philaenus spumarius, was performed to test for pathogen spread from olive and coffee plants (Table S4). Results showed that the acquisition and the overall transmission rate when coffee plants were used as a source for the spittlebugs were lower than when using olive plants. Overall, these experiments showed that (i) the olive infecting strain can colonize and multiply in coffee plants and that (ii) coffee plants may represent a latent carrier of the bacterium; (iii) coffee plants are a poor acquisition source for De Donno transmission by spittlebugs when compared to olive trees.
Discussion
While Xylella fastidiosa causes severe disease symptoms in some crop plant species, most strains-plant species interactions result in asymptomatic infections, which unfortunately facilitate the global trade of X. fastidiosa infected plants. After the detection of X. fastidiosa in Apulia in 2013, several confirmed reports have now been made elsewhere in Italy, as well as in France, Spain, Portugal [63]. We focused on the first X. fastidiosa outbreak in Europe and confirmed that the epidemic devastating olive trees in Apulia was caused by the introduction and subsequent establishment of a single X. fastidiosa subsp. pauca strain from Central America, likely Costa Rica – as previously suggested by studies using MLST data [64, 65]. By using a tip-dating approach, we estimated that this introduction would have occurred in ~2008 [95 % HPD: 1930–2016]. We note that this estimate is similar to the previous one (i.e. 2008 [95 % HPD: 1952–2015]), the latter estimation having been previously made using a similar approach with only a subset of the strains used here [12]. Besides the higher number of strains used in our study, the reassembly of the De Donno strain generated here, correcting for sequencing errors generated by PacBio reads, did not impact this estimation. One important component of such dating efforts is that we cannot ascertain if the proposed introduction date of 2008 represents an inference on the time when the pathogen was first introduced into Apulia, when it first adapted to olive trees as a host plant, or when the epidemic effectively initiated. Lastly, while the pathways for the different X. fastidiosa introduction events that have taken place in Europe might differ, similar introduction dates were obtained regarding the introduction of X. fastidiosa subsp. multiplex strains in Corsica, France using a meta-population approach (estimated to have occurred between 1985 and 2000 [66]); and the introduction of X. fastidiosa subsp. multiplex and fastidiosa strains in Mallorca, Spain, estimated to have occurred around 1993 using a range of approaches including tree ring data and a molecular clock as used here [67]. Combined, these studies indicate that X. fastidiosa infected plant material was introduced multiple times into Europe, predominantly from the USA, when trade regulations were less strict.
The genetic relatedness among the ST53 coffee isolate from Costa Rica and the Apulian isolates observed in this study (Figs 1 and S2) along with the numerous reports of coffee plants infected with X. fastidiosa imported into Europe from Central America [68–70] point towards an introduction from a X. fastidiosa infected coffee plant - probably as an ornamental plant - into Italy. This introduction was then followed by subsequent establishment and adaptation to this new environment as suggested by the evidence of positive selection. First, while the ST53 Costa Rican coffee strain, COF0407, was isolated from coffee plants displaying disease symptoms in 2009 [71], no symptom associated to X. fastidiosa infection was observed for coffee plants inoculated with an Apulian isolate in our study. In addition, our results indicate a slower colonization process of coffee plants by the Apulian strain, which could be the result of the adaptation of this strain to its new environment (olive trees), decreasing its ability to infect coffee plants. However, as no direct comparison could be made between the colonization and virulence of a ST53-coffee strain and the Apulian strain in coffee and olive plants, the conclusions based on a loss of pathogenicity in coffee plants require further testing. We also observed a higher number of X. fastidiosa infected insects and higher transmission rates when olive trees were used as a source of the Italian De Donno strain compared to coffee plants. These higher transmission rates observed when using olive trees as source plants could be due to higher bacterial population sizes within these plants, transmission efficiency being positively correlated with bacterial population in plants [72, 73]. Altogether, these data support a scenario in which the selection of a strain for faster movement and multiplication within the main host plant species found in this new environment - that is olive trees - may have occurred, thus increasing both its transmission efficiency and its virulence. The uniform physical and genetic environments found in monocultures has indeed been hypothesized to favour the emergence of host-specialized pathogens with increased virulence; the pathogen being under homogeneous selective pressure [2, 5, 13] and dispersal being a key step in the invasion process [3].
Another scenario, not contradictory with the previous one, explaining the high virulence of the Apulian strain on olive trees is that the interactions between the bacterium and this new host have not yet been finely regulated by selection leading to highly virulent infections [74]. The higher number of complete zonula occludens toxins (zot), a known virulence factor [75–77], within the De Donno strain (five against two complete zot in COF0407 and OLS0478 and one complete zot in OLS0479) may contribute to the pathogenicity of this strain in olive trees, as previously reported in grapevines [77]. In addition to these differences in zot copy numbers, four other genes - including another pathogenicity factor and a transcriptional regulator - were found to be present in the Apulian population while absent in the Costa Rican strains. While zot copy number could be the result of duplication followed by diversification events, the four other genes would need to have been horizontally acquired. In addition, 78 genes mainly present within the plasmid (70 % of the deleted genes) were found to be absent in the Apulian isolates while present in the Costa Rican ones. These gene losses could either be adaptive, contributing to X. fastidiosa strain propagation success in Apulia or due to genetic drift [2, 78]. While gene deletion events, frequently found during bacterial adaptation, have been hypothesized to be the first phases that generally occur in the adaptation of bacterial populations to a new environment [57], deletion of genes under relaxed selection are also often observed during bacterial evolution undergoing population bottlenecks [78]. Interestingly, further comparison between the Costa Rican coffee strain and the Apulian strain genomes revealed the presence of non-synonymous mutations affecting 11 different genes (Table S2). Most of these mutations were found using two different comparative approaches. Six of these genes were also found to be under positive selection, making them good candidates for X. fastidiosa ’s adaptation to this new environment. This included the transcriptional regulator crp, the genes encoding for the two proteins of X. fastidiosa ’s quorum sensing system (rpfC, DSF membrane sensor kinase and rpfG, the response regulator), an ATP-dependent helicase, an ABC transporter substrate-binding protein and an enzyme involved in the biosynthesis of bacterial cell-wall peptidoglycan (murE). Interestingly, a study by Killiny and Almeida [79] showed that disruption of quorum sensing signalling by knocking out rpfF (which is required to produce the DSF signal) increased the host range of X. fastidiosa . Moreover, among studies that have identified the molecular basis of some bacterial species host specificity, a structural protein from the cell envelope was shown to mediate the specificity between Xhenorhabdus nematophila and its worm host [80] while a two-component sensor kinase was shown to modify the host range of Vibrio fischeri [81]. Another ~100 genes were under positive selection within the Apulian population. It is noteworthy that these genes were enriched in categories corresponding to pathogen virulence (cell motility, energy production, defence mechanisms). Moreover, shared indels or 2 bp-substitutions were detected in three different genes. In order to determine whether these mutated genes and/or genes under positive selection are indeed involved in the adaptation of X. fastidiosa to this new environment and, to distinguish between mutations linked to the founder event and those involved in adaptation, functional genomic studies would be required. While essential, such studies might be difficult to carry out with the Apulian isolates, which remain difficult to genetically manipulate [82].
We were able to use X. fastidiosa genomic data to accurately reconstruct the emergence of OQDS in Italy, including highlighting the similarity in disease emergence data to other recent reports of X. fastidiosa introductions in Europe [66, 67]. This is the first study aiming at addressing the long-standing question of the genetic determinants of X. fastidiosa adaptation to a new environment. While we could not provide a decisive answer, we have identified a short number of candidate genes that could be involved in the adaptation of X. fastidiosa to olive trees in Italy. Finally, although homologous recombination has so far been considered as the main driver of X. fastidiosa evolution and adaptation, our work suggests that mutations also play an important role in X. fastidiosa evolution and may lead to new outbreaks.
Supplementary Data
Funding information
This work received funding from the European Union’s Horizon 2020 research and innovation programme under grant agreement N. 727987 ‘ Xylella fastidiosa Active Containment Through a Multidisciplinary-Oriented Research Strategy’ XF-ACTORS, and the PD/GWSS Research Programme from the California Department of Food and Agriculture. AS was supported by a Marie Skłodowska-Curie Fellowship (European Union’s Horizon 2020 Research and Innovation Programme, grant agreement 707013). The Vincent J. Coates Genomics Sequencing Laboratory at the University of California–Berkeley was supported by a National Institutes of Health S10 OD018174 Instrumentation Grant.
Acknowledgements
We wish to thank Giuseppe Altamura (CNR), Francesco Specchia (CNR) and Fabio Lazzari for technical assistance in collecting the isolates and for the vector-mediated transmission experiments and Adrien Rieux for helpful discussions on tip dating. We thank the Vincent J. Coates Genomics Sequencing Laboratory at the University of California–Berkeley for sequencing our samples.
Author contributions
A.S. and R.P.P.A., originally conceived the study. Apulian isolates were provided by M.S., A.G., G.L. and P.S. A.S,. M.V. and A.I.C., contributed to the data curation, formal analysis and interpretation of the data. Inoculation and transmission experiments were carried out by M.S. D.B., C.N. and R.P.P.A., supervised the project and contributed to funding acquisition. A.S., wrote the original draft of the manuscript. All authors contributed to the manuscript revision and read and approved the submitted version.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Footnotes
Abbreviations: APL, Apulia; HPD, highest posterior density; OQDS, olive quick decline syndrome; ST53, sequence type 53; X. fastidiosa, Xylella fastidiosa.
All supporting data, code and protocols have been provided within the article or through supplementary data files. Four supplementary tables and five supplementary figures are available with the online version of this article.
References
- 1.Brown C. Emerging zoonoses and pathogens of public health significance – an overview. Rev Sci Tech OIE. 2004;23:435–442. doi: 10.20506/rst.23.2.1495. [DOI] [PubMed] [Google Scholar]
- 2.McCann HC. Skirmish or war: the emergence of agricultural plant pathogens. Curr Opin Plant Biol. 2020;56:147–152. doi: 10.1016/j.pbi.2020.06.003. [DOI] [PubMed] [Google Scholar]
- 3.Mallon CA, Elsas J van, Salles JF. Microbial invasions: The process, patterns, and mechanisms. Trends Microbiol. 2015;23:719–729. doi: 10.1016/j.tim.2015.07.013. [DOI] [PubMed] [Google Scholar]
- 4.Braga RM, Dourado MN, Araújo WL. Microbial interactions: ecology in a molecular perspective. Braz J Microbiol. 2016;47 Suppl 1:86–98. doi: 10.1016/j.bjm.2016.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McDonald BA, Stukenbrock EH. Rapid emergence of pathogens in agro-ecosystems: global threats to agricultural sustainability and food security. Philos Trans R Soc Lond B Biol Sci. 2016;371:20160026. doi: 10.1098/rstb.2016.0026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morse SS. Factors in the emergence of infectious diseases. Emerg Infect Dis. 1995;1:7–15. doi: 10.3201/eid0101.950102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hajek AE, Júnior ID, McManus ML. In: Field Manual of Techniques in Invertebrate Pathology. Lacey LA, Kaya HK, editors. Dordrecht: Springer Netherlands; Introduction of exotic pathogens and documentation of their establishment and impact; pp. 299–325. [Google Scholar]
- 8.Almeida RPP, Nunney L. How do plant diseases caused by Xylella fastidiosa emerge? Plant Dis. 2015;99:1457–1467. doi: 10.1094/PDIS-02-15-0159-FE. [DOI] [PubMed] [Google Scholar]
- 9.EFSA (European Food Safety Authority) Scientific report on the update of the Xylella spp. host plant database – systematic literature search up to 30 June 2019. EFSA Journal. 2020;18:6114. doi: 10.2903/j.efsa.2020.6114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nunney L, Vickerman DB, Bromley RE, Russell SA, Hartman JR, et al. Recent evolutionary radiation and host plant specialization in the Xylella fastidiosa subspecies native to the United States. Appl Environ Microbiol. 2013;79:2189–2200. doi: 10.1128/AEM.03208-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nunney L, Azad H, Stouthamer R. An experimental test of the host-plant range of nonrecombinant strains of North American Xylella fastidiosa subsp. multiplex. Phytopathology®. 2019;109:294–300. doi: 10.1094/PHYTO-07-18-0252-FI. [DOI] [PubMed] [Google Scholar]
- 12.Vanhove M, Retchless AC, Sicard A, Rieux A, Coletta-Filho HD, et al. Genomic diversity and recombination among Xylella fastidiosa subspecies. Appl Environ Microbiol. 2019;85:aem. doi: 10.1128/AEM.02972-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sicard A, Zeilinger AR, Vanhove M, Schartel TE, Beal DJ, et al. Xylella fastidiosa: Insights into an emerging plant pathogen. Annu Rev Phytopathol. 2018;56:181–202. doi: 10.1146/annurev-phyto-080417-045849. [DOI] [PubMed] [Google Scholar]
- 14.Scally M, Schuenzel EL, Stouthamer R, Nunney L. Multilocus sequence type system for the plant pathogen Xylella fastidiosa and relative contributions of recombination and point mutation to clonal diversity. Appl Environ Microbiol. 2005;71:8491–8499. doi: 10.1128/AEM.71.12.8491-8499.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Saponari M, Boscia D, Nigro F, Martelli GP. Identification of DNA sequences related to Xylella fastidiosa in oleander, almond and olive trees exhibiting leaf scorch symptoms in Apulia (Southern Italy) J Plant Pathol. 2013;95:668. [Google Scholar]
- 16.Wells JM, Raju BC, Nyland G, Lowe SK. Medium for isolation and growth of bacteria associated with plum leaf scald and phony peach diseases. Appl Environ Microbiol. 1981;42:357–363. doi: 10.1128/aem.42.2.357-363.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saponari M, Boscia D, Altamura G, Loconsole G, Zicca S, et al. Isolation and pathogenicity of Xylella fastidiosa associated to the olive quick decline syndrome in southern Italy. Sci Rep. 2017;7:17723. doi: 10.1038/s41598-017-17957-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Loconsole G, Potere O, Boscia D, Altamura G, Djelouah K, et al. Detection of Xylella fastidiosa in olive trees by molecular and serological methods. J Plant Pathol. 2014;96:1–8. [Google Scholar]
- 19.Wick RR, Judd LM, Gorrie CL, Holt KE. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol. 2017;13:e1005595. doi: 10.1371/journal.pcbi.1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Giampetruzzi A, Saponari M, Almeida RPP, Essakhi S, Boscia D, et al. Complete genome sequence of the olive-infecting strain Xylella fastidiosa subsp. pauca De Donno. Genome Announc. 2017;5:e00569-17. doi: 10.1128/genomeA.00569-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hunt M, Silva ND, Otto TD, Parkhill J, Keane JA, et al. Circlator: automated circularization of genome assemblies using long sequencing reads. Genome Biol. 2015;16:294. doi: 10.1186/s13059-015-0849-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rozov R, Brown Kav A, Bogumil D, Shterzer N, Halperin E, et al. Recycler: an algorithm for detecting plasmids from de novo assembly graphs. Bioinformatics. 2016:btw651. doi: 10.1093/bioinformatics/btw651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Darling AE, Mau B, Perna NT. progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PloS one. 2010;5:e11147. doi: 10.1371/journal.pone.0011147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 26.Page AJ, Cummins CA, Hunt M, Wong VK, Reuter S, et al. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics. 2015;31:3691–3693. doi: 10.1093/bioinformatics/btv421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Giampetruzzi A, Chiumenti M, Saponari M, Donvito G, Italiano A, et al. Draft Genome Sequence of the Xylella fastidiosa CoDiRO Strain. Genome Announc. 2015;3:e01538-14. doi: 10.1128/genomeA.01538-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ramazzotti M, CimagliaI F, Gallo A, Ranaldi F, Surico G, et al. Insights on a founder effect: the case of Xylella fastidiosa in the Salento area of Apulia, Italy. Phytopathol Mediterr. 2018;57:8–25. doi: 10.14601/Phytopathol_Mediterr-22263. [DOI] [Google Scholar]
- 29.Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Paradis E. pegas: an R package for population genetics with an integrated-modular approach. Bioinformatics. 2010;26:419–420. doi: 10.1093/bioinformatics/btp696. [DOI] [PubMed] [Google Scholar]
- 31.Treangen TJ, Ondov BD, Koren S, Phillippy AM. The Harvest suite for rapid core-genome alignment and visualization of thousands of intraspecific microbial genomes. Genome Biol. 2014;15:524. doi: 10.1186/s13059-014-0524-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Deatherage DE, Barrick JE. In: Engineering and Analyzing Multicellular Systems. Sun L, Shou W, editors. New York, NY: Springer New York; Identification of mutations in laboratory-evolved microbes from next-generation sequencing data using breseq; pp. 165–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gu Z, Gu L, Eils R, Schlesner M, Brors B. Circlize implements and enhances circular visualization in R. Bioinformatics. 2014;30:2811–2812. doi: 10.1093/bioinformatics/btu393. [DOI] [PubMed] [Google Scholar]
- 34.Mostowy R, Croucher NJ, Andam CP, Corander J, Hanage WP, et al. Efficient inference of recent and ancestral recombination within bacterial populations. Mol Biol Evol. 2017;34:1167–1182. doi: 10.1093/molbev/msx066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brynildsrud O, Snipen L-G, Bohlin J. CNOGpro: detection and quantification of CNVs in prokaryotic whole-genome sequencing data. Bioinformatics. 2015;31:1708–1715. doi: 10.1093/bioinformatics/btv070. [DOI] [PubMed] [Google Scholar]
- 36.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, et al. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pfeifer B, Wittelsbürger U, Ramos-Onsins SE, Lercher MJ. PopGenome: an efficient swiss army knife for population genomic analyses in R. Mol Biol Evol. 2014;31:1929–1936. doi: 10.1093/molbev/msu136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kosakovsky Pond SL, Frost SDW. Not so different after all: a comparison of methods for detecting amino acid sites under selection. Mol Biol Evol. 2005;22:1208–1222. doi: 10.1093/molbev/msi105. [DOI] [PubMed] [Google Scholar]
- 40.Pond SLK, Frost SDW, Muse SV. HyPhy: hypothesis testing using phylogenies. Bioinformatics. 2005;21:676–679. doi: 10.1093/bioinformatics/bti079. [DOI] [PubMed] [Google Scholar]
- 41.Contreras-Moreira B, Vinuesa P. GET_HOMOLOGUES, a versatile software package for scalable and robust microbial pangenome analysis. Appl Environ Microbiol. 2013;79:7696–7701. doi: 10.1128/AEM.02411-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ranwez V, Harispe S, Delsuc F, Douzery EJP. MACSE: Multiple Alignment of Coding SEquences accounting for frameshifts and stop codons. PLoS ONE. 2011;6:e22594. doi: 10.1371/journal.pone.0022594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Didelot X, Wilson DJ. ClonalFrameML: efficient inference of recombination in whole bacterial genomes. PLoS Comput Biol. 2015;11:e1004041. doi: 10.1371/journal.pcbi.1004041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Goslee SC, Urban DL. The ecodist package for dissimilarity-based analysis of ecological data. J Stat Soft. 2007;22:1–19. doi: 10.18637/jss.v022.i07. [DOI] [Google Scholar]
- 46.Harper SJ, Ward LI, Clover GRG. Development of LAMP and real-time PCR methods for the rapid detection of Xylella fastidiosa for quarantine and field applications. Phytopathology. 2010;100:1282–1288. doi: 10.1094/PHYTO-06-10-0168. [DOI] [PubMed] [Google Scholar]
- 47.Montero-Astúa M, Chacón-Díaz C, Aguilar E, Rodríguez CM, Garita L, et al. Isolation and molecular characterization of Xylella fastidiosa from coffee plants in Costa Rica. J Microbiol. 2008;46:482–490. doi: 10.1007/s12275-008-0072-8. [DOI] [PubMed] [Google Scholar]
- 48.Cornara D, Cavalieri V, Dongiovanni C, Altamura G, Palmisano F, et al. Transmission of Xylella fastidiosa by naturally infected Philaenus spumarius (Hemiptera, Aphrophoridae) to different host plants. J Appl Entomol. 2017;141:80–87. doi: 10.1111/jen.12365. [DOI] [Google Scholar]
- 49.Cavalieri V, Altamura G, Fumarola G, di Carolo M, Saponari M, et al. Transmission of Xylella fastidiosa subspecies pauca sequence type 53 by different insect species. Insects. 2019;10:E324. doi: 10.3390/insects10100324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schnoes AM, Brown SD, Dodevski I, Babbitt PC. Annotation error in public databases: misannotation of molecular function in enzyme superfamilies. PLoS Comput Biol. 2009;5:e1000605. doi: 10.1371/journal.pcbi.1000605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Griesemer M, Kimbrel JA, Zhou CE, Navid A, D’haeseleer P. Combining multiple functional annotation tools increases coverage of metabolic annotation. BMC Genomics. 2018;19:948. doi: 10.1186/s12864-018-5221-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tonkin-Hill G, MacAlasdair N, Ruis C, Weimann A, Horesh G, et al. Producing polished prokaryotic pangenomes with the Panaroo pipeline. Genome Biol. 2020;21:180. doi: 10.1186/s13059-020-02090-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ali N, Chapuis E, Tavoillot J, Mateille T. Plant-parasitic nematodes associated with olive tree (Olea europaea L.) with a focus on the Mediterranean Basin: A review. Comptes Rendus Biologies. 2014;337:423–442. doi: 10.1016/j.crvi.2014.05.006. [DOI] [PubMed] [Google Scholar]
- 54.Viana D, Comos M, McAdam PR, Ward MJ, Selva L, et al. A single natural nucleotide mutation alters bacterial pathogen host tropism. Nat Genet. 2015;47:361–366. doi: 10.1038/ng.3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sheppard SK, Guttman DS, Fitzgerald JR. Population genomics of bacterial host adaptation. Nat Rev Genet. 2018;19:549–565. doi: 10.1038/s41576-018-0032-z. [DOI] [PubMed] [Google Scholar]
- 56.Sun Y-C, Jarrett CO, Bosio CF, Hinnebusch BJ. Retracing the evolutionary path that led to flea-borne transmission of Yersinia pestis . Cell Host and Microbe. 2014;15:578–586. doi: 10.1016/j.chom.2014.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hottes AK, Freddolino PL, Khare A, Donnell ZN, Liu JC, et al. Bacterial adaptation through loss of function. PLoS Genet. 2013;9:e1003617. doi: 10.1371/journal.pgen.1003617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chatterjee S, Wistrom C, Lindow SE. A cell-cell signaling sensor is required for virulence and insect transmission of Xylella fastidiosa . Proceedings of the National Academy of Sciences. 2008;105:2670–2675. doi: 10.1073/pnas.0712236105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Padilla-Vaca F, Vargas-Maya NI, Elizarrarás-Vargas NU, Rangel-Serrano Á, Cardoso-Reyes LR, et al. Flotillin homologue is involved in the swimming behavior of Escherichia coli. Arch Microbiol. 2019;201:999–1008. doi: 10.1007/s00203-019-01670-8. [DOI] [PubMed] [Google Scholar]
- 60.Guzmán-Flores JE, Steinemann-Hernández L, González de la Vara LE, Gavilanes-Ruiz M, Romeo T, et al. Proteomic analysis of Escherichia coli detergent-resistant membranes (DRM) PLoS ONE. 2019;14:e0223794. doi: 10.1371/journal.pone.0223794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vollmer W, Joris B, Charlier P, Foster S. Bacterial peptidoglycan (murein) hydrolases. FEMS Microbiol Rev. 2008;32:259–286. doi: 10.1111/j.1574-6976.2007.00099.x. [DOI] [PubMed] [Google Scholar]
- 62.Tajima F. Simple methods for testing the molecular evolutionary clock hypothesis. Genetics. 1993;135:599–607. doi: 10.1093/genetics/135.2.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.EFSA Panel on Plant Health (PLH) Bragard C, Dehnen‐Schmutz K, Di Serio F, Gonthier P, et al. Update of the Scientific Opinion on the risks to plant health posed by Xylella fastidiosa in the EU territory. EFS2. 2019;17 doi: 10.2903/j.efsa.2019.5665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Giampetruzzi A, Saponari M, Loconsole G, Boscia D, Savino VN, et al. Genome-wide analysis provides evidence on the genetic relatedness of the emergent Xylella fastidiosa genotype in Italy to isolates from Central America. Phytopathology. 2017;107:816–827. doi: 10.1094/PHYTO-12-16-0420-R. [DOI] [PubMed] [Google Scholar]
- 65.Marcelletti S, Scortichini M. Xylella fastidiosa CoDiRO strain associated with the olive quick decline syndrome in southern Italy belongs to a clonal complex of the subspecies pauca that evolved in Central America. Microbiology (Reading) 2016;162:2087–2098. doi: 10.1099/mic.0.000388. [DOI] [PubMed] [Google Scholar]
- 66.Soubeyrand S, de Jerphanion P, Martin O, Saussac M, Manceau C, et al. Inferring pathogen dynamics from temporal count data: the emergence of Xylella fastidiosa in France is probably not recent. New Phytol. 2018;219:824–836. doi: 10.1111/nph.15177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Moralejo E, Gomila M, Montesinos M, Borràs D, Pascual A, et al. Phylogenetic inference enables reconstruction of a long-overlooked outbreak of almond leaf scorch disease (Xylella fastidiosa) in Europe. Commun Biol. 2020;3:560. doi: 10.1038/s42003-020-01284-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jacques M-A, Denancé N, Legendre B, Morel E, Briand M, et al. New coffee plant-infecting Xylella fastidiosa variants derived via homologous recombination. Appl Environ Microbiol. 2016;82:1556–1568. doi: 10.1128/AEM.03299-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bergsma-Vlami M, van de Bilt JLJ, Tjou-Tam-Sin NNA, Helderman CM, Gorkink-Smits PPMA, et al. Assessment of the genetic diversity of Xylella fastidiosa in imported ornamental Coffea arabica plants. Plant Pathol. 2017;66:1065–1074. doi: 10.1111/ppa.12696. [DOI] [Google Scholar]
- 70.Bergsma-Vlami M, Van de Bilt JLJ, Tjou-Tam-Sin NNA, BTLH V de V, Westenberg M. Xylella fastidiosa in Coffea arabica ornamental plants imported from Costa Rica and Honduras in the Netherlands. J Plant Pathol. 2015;1 doi: 10.4454/JPP.V97I2.032. [DOI] [Google Scholar]
- 71.Nunney L, Ortiz B, Russell SA, Ruiz Sánchez R, Stouthamer R. The complex biogeography of the plant pathogen Xylella fastidiosa: genetic evidence of introductions and subspecific introgression in Central America. PLoS ONE. 2014;9:e112463. doi: 10.1371/journal.pone.0112463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hill BL, Purcell AH. Populations of Xylella fastidiosa in plants required for transmission by an efficient vector. Phytopathology. 1997;87:1197–1201. doi: 10.1094/PHYTO.1997.87.12.1197. [DOI] [PubMed] [Google Scholar]
- 73.Cornara D, Sicard A, Zeilinger AR, Porcelli F, Purcell AH, et al. Transmission of Xylella fastidiosa to grapevine by the meadow spittlebug. Phytopathology. 2016;106:1285–1290. doi: 10.1094/PHYTO-05-16-0202-R. [DOI] [PubMed] [Google Scholar]
- 74.Toft C, Andersson SGE. Evolutionary microbial genomics: insights into bacterial host adaptation. Nat Rev Genet. 2010;11:465–475. doi: 10.1038/nrg2798. [DOI] [PubMed] [Google Scholar]
- 75.Waldor MK, Mekalanos JJ. Lysogenic conversion by a filamentous phage encoding cholera toxin. Science. 1996;272:1910–1914. doi: 10.1126/science.272.5270.1910. [DOI] [PubMed] [Google Scholar]
- 76.Liu F, Lee H, Lan R, Zhang L. Zonula occludens toxins and their prophages in Campylobacter species. Gut Pathog. 2016;8:43. doi: 10.1186/s13099-016-0125-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhang S, Chakrabarty PK, Fleites LA, Rayside PA, Hopkins DL, et al. Three new pierce’s disease pathogenicity effectors identified using Xylella fastidiosa biocontrol strain EB92-1. PLoS ONE. 2015;10:e0133796. doi: 10.1371/journal.pone.0133796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Albalat R, Cañestro C. Evolution by gene loss. Nat Rev Genet. 2016;17:379–391. doi: 10.1038/nrg.2016.39. [DOI] [PubMed] [Google Scholar]
- 79.Killiny N, Almeida RPP. Gene regulation mediates host specificity of a bacterial pathogen. Environ Microbiol Rep. 2011;3:791–797. doi: 10.1111/j.1758-2229.2011.00288.x. [DOI] [PubMed] [Google Scholar]
- 80.Cowles CE, Goodrich-Blair H. The Xenorhabdus nematophila nilABC genes confer the ability of Xenorhabdus spp. to colonize Steinernema carpocapsae nematodes. J Bacteriol. 2008;190:4121–4128. doi: 10.1128/JB.00123-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mandel MJ, Wollenberg MS, Stabb EV, Visick KL, Ruby EG. A single regulatory gene is sufficient to alter bacterial host range. Nature. 2009;458:215–218. doi: 10.1038/nature07660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.D’Attoma G, Morelli M, De La Fuente L, Cobine PA, Saponari M, et al. Phenotypic characterization and transformation attempts reveal peculiar traits of Xylella fastidiosa subspecies pauca strain De Donno. Microorganisms. 2020;8:1832. doi: 10.3390/microorganisms8111832. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.