Abstract
A rapid and accurate diagnostic assay represents an important means to detect Mycobacterium tuberculosis , identify drug-resistant strains and ensure treatment success. Currently employed techniques to diagnose drug-resistant tuberculosis include slow phenotypic tests or more rapid molecular assays that evaluate a limited range of drugs. Whole-genome-sequencing-based approaches can detect known drug-resistance-conferring mutations and novel variations; however, the dependence on growing samples in culture, and the associated delays in achieving results, represents a significant limitation. As an alternative, targeted sequencing strategies can be directly performed on clinical samples at high throughput. This study proposes a targeted sequencing assay to rapidly detect drug-resistant strains of M. tuberculosis using the Nanopore MinION sequencing platform. We designed a single-tube assay that targets nine genes associated with drug resistance to seven drugs and two phylogenetic-determining regions to determine strain lineage and tested it in nine clinical isolates and six sputa. The study’s main aim is to calibrate MinNION variant calling to detect drug-resistance-associated mutations with different frequencies to match the accuracy of Illumina (the current gold-standard sequencing technology) from both culture and sputum samples. After calibrating Nanopore MinION variant calling, we demonstrated 100% agreement between Illumina WGS and our MinION set up to detect known drug resistance and phylogenetic variants in our dataset. Importantly, other variants in the amplicons are also detected, decreasing the recall. We identify minority variants and insertions/deletions as crucial bioinformatics challenges to fully reproduce Illumina WGS results.
Keywords: Mycobacterium tuberculosis, amplicon sequencing, Nanopore, MinION, genomics, drug-resistance, genotyping
Data Summary
Table 1 describes the accession numbers of fastq files from Illumina whole-genome sequencing.
Table 1.
Accession numbers of samples included in this study
|
Illumina |
MinION |
|||
|---|---|---|---|---|
|
Sample |
Study accession |
RUN accession |
Study accession |
RUN accession |
|
G841 |
PRJEB22237 |
ERR2099789 |
PRJEB44496 |
ERS6293816 |
|
N0067 |
PRJEB3223 |
ERR233356 |
PRJEB44496 |
ERS6293819 |
|
G1335_W27 |
PRJEB37609 |
ERR3994128 |
PRJEB44496 |
ERS6293807 |
|
G1646_W19 |
PRJEB37609 |
ERR3994105 |
PRJEB44496 |
ERS6293808 |
|
G2103_W23 |
PRJEB37609 |
ERR3994108 |
PRJEB44496 |
ERS6293812 |
|
G2267_W20 |
PRJEB37609 |
ERR3994106 |
PRJEB44496 |
ERS6293813 |
|
G2284_W26 |
PRJEB37609 |
ERR3994111 |
PRJEB44496 |
ERS6293814 |
|
G107 |
PRJEB38719 |
ERR4195665 |
PRJEB44496 |
ERS6293806 |
|
G1800 |
PRJEB38719 |
ERR4195305 |
PRJEB44496 |
ERS6293809 |
|
G1961 |
PRJEB38719 |
ERR4195372 |
PRJEB44496 |
ERS6293811 |
|
G770 |
PRJEB38719 |
ERR4195357 |
PRJEB44496 |
ERS6293815 |
|
G870 |
PRJEB38719 |
ERR4195619 |
PRJEB44496 |
ERS6293817 |
|
G981 |
PRJEB38719 |
ERR4195376 |
PRJEB44496 |
ERS6293818 |
|
182320_M20 |
PRJEB44496 |
ERS6337728 |
PRJEB44496 |
ERS6293805 |
|
G1952 |
PRJEB44496 |
ERS6337729 |
PRJEB44496 |
ERS6293810 |
Impact Statement.
Nanopore long-amplicon sequencing represents an under-used technology for diagnosing tuberculosis despite its potential to predict drug resistance and genotype at almost real-time. This work describes a proof-of-concept study in which a single-tube PCR assay to identify drug resistance and phylogenetic-determining mutations has been designed and evaluated with clinical cultures and diagnostic sputa. In addition, this study demonstrates the viability of the long-amplicon sequencing strategy regardless of the sample type. Our bioinformatics analyses establish a high degree of congruence to Illumina short-reads (as the gold-standard technique); however, we note the challenges associated with interpreting heteroresistance from Nanopore low-frequency variants.
Introduction
Tuberculosis (TB) remains a significant health problem worldwide and represents the principal cause of death by a single infectious agent, Mycobacterium tuberculosis complex (MTBC). Approximately 10 million new cases and 1.5 million deaths were reported in 2019 [1]. TB has been successfully treated since the late 1940s thanks to the development of the first anti-tubercular drugs. However, the number of drug-resistant cases has increased 13-fold in the last decade, complicating the global control of TB and delaying its eradication [1–3].
Standard TB treatment consists of a combination of four different drugs administered daily for up to 4 months. Treatment of drug-resistant TB (DR-TB) represents a more complex task, comprising a combination of second-line antibiotics, which requires more extended treatment times and is more expensive [4, 5]. Multidrug-resistant TB (MDR-TB) strains are resistant to both rifampicin (RIF) and isoniazid (INH), two key first-line drugs. MDR-TB is treatable by second-line drugs including oral and injectable drugs. Since 2019, injectable drugs are being replaced by all-oral regimens following World Health Organization (WHO) guidelines [6]. Consequently MDR- and XDR-TB definitions are being revisited [7]. However, injectable drugs are still being used in many regimes and countries including those that have not yet approved bedaquiline, an oral drug, to treat MDR-TB cases [8].
The current reference technique for DR-TB diagnosis is based on phenotypic tests that evaluate the susceptibility of MTBC strains to different antibiotics (drug susceptibility tests, DSTs). DSTs assess the ability of mycobacteria to grow in a solid or liquid medium containing antimicrobial drugs [9]. Despite being a commonly used technique, phenotypic diagnosis methods are laborious, time-consuming due to the slow growth rate of mycobacteria and require dedicated facilities [10]; furthermore, the results are not entirely reproducible in some cases [11]. Such limitations can be overcome with rapid molecular tests; however, these approaches target only a reduced number of genomic positions located in hot-spot regions for the most frequent drug-resistance-associated mutations [12–16]. This limitation increases false-negative results due to silent mutations and other mutations located outside the hot-spot regions [17, 18]. As an alternative, whole-genome sequencing (WGS) provides a maximum level of detail regarding the complete genomic sequence. Published catalogues of mutations robustly associated with drug-resistant phenotypes can be used to design targeted approaches against a wide number of regions involved in drug resistance. However, obtaining the whole-genome sequence from sputum samples is challenging and expensive (see, for example, Goig et al. [19]).
Compared to WGS, targeted sequencing of drug-resistance-associated genes represents a cheaper approach that can be routinely applied to sputum samples. Two assays based on amplicon sequencing have been developed by next generation sequencing (NGS) technologies – the Deeplex Myc-TB assay (GenoScreen, Lille, France) [20] and the Next Gen-RDST assay (Translational Genomics Research Institute, Phoenix, AZ, USA). Both assays target specific genomic regions known to be hot-spots for drug-resistance-conferring variants. Due to its flexibility, custom panels can incorporate other target regions, such as those that allow M. tuberculosis genotyping. Importantly, strain type plays an important role in disease outcome, transmission, drug-resistance emergence, host responses to the pathogen, disease epidemiology and virulence [21, 22]. Amplicon sequencing can be directly performed on patient samples, forgoing the need for long in vitro culture times, and obtaining high depth of coverage [23]. In addition, an increasing body of evidence has suggested the role of heteroresistance to some drugs, in which drug-resistance subpopulation undetected during in vitro culture may impact treatment outcomes [24].
Nanopore (Oxford Technologies) released MinION, a portable device that allows real-time sequencing and analysis. MinION long-read sequencing is based on nanopores that act as biosensors, measuring the changes in voltage produced when a nucleotide passes through them [25]. This sequencing technology has been recently applied to the point-of-care diagnosis of viral and bacterial infections [26, 27]. In MTBC research, MinION technology has been employed to assemble M. tuberculosis Beijing XDR strains and detect DR-TB in Madagascar [28, 29]. The Nanopore MinION platform presents certain limitations, including the generation of noisy reads and a high error rate [30]. However, these limitations are balanced by the higher depth of coverage obtained compared to Illumina sequencing technology, which makes free-culture sequencing and shorter diagnostic turnaround times possible [31].
This study aims to develop a single-tube PCR amplification protocol to sequence a panel of genes of interest using Nanopore technology and calibrate MinION variant-calling parameters for prospective sequencing by comparing them to paired Illumina samples. We analyse recall and true negative rate to identify known drug-resistance-associated variants in culture and sputum samples. We also analyse accuracy in identifying new variants and the source of errors associated with Nanopore MinION amplicon sequencing by comparing results with Illumina MiSeq WGS technology, the current gold-standard technique.
Methods
Samples and DNA extraction
Fifteen samples were barcoded and sequenced in two MinION runs. In the first run, nine samples extracted from solid pure cultures were sequenced, including lineage control strains (L1-L6) and one sample extracted from smear-positive sputum (no smear grading was provided for sample 182320_M20). In the second run, five smear-positive sputa extracted directly from respiratory specimens of TB culture-positive patients were sequenced. Acid-fast bacilli (AFB) smear results were four 3+ sputa (G2267_W20, G2103_W23, G2284_W26, G1335_W27), and one 1+ sputum (G1646_W19). Cultures from all samples were previously whole-genome sequenced in our laboratory using the Illumina MiSeq platform (Table 2).
Table 2.
Description of the dataset included in this study
|
Sample |
Sample type |
Run MinION |
Resistance profile |
Resistance agreement |
Lineage |
Lineage agreement |
|---|---|---|---|---|---|---|
|
182320_M20 |
Sputum |
Run1 |
Susceptible |
100 |
Lineage4 |
100 |
|
G107 |
Culture |
Run1 |
Susceptible |
100 |
Lineage3 |
100 |
|
G1800 |
Culture |
Run1 |
FQ, RMP, INH, KAN, EMB |
100 |
Lineage2 |
100 |
|
G1952 |
Culture |
Run1 |
Susceptible |
100 |
Lineage6 |
100 |
|
G1961 |
Culture |
Run1 |
RMP, EMB |
100 |
Lineage5 |
100 |
|
G770 |
Culture |
Run1 |
Susceptible |
100 |
Lineage4 |
100 |
|
G841 |
Culture |
Run1 |
RMP |
100 |
Lineage4 |
100 |
|
G870 |
Culture |
Run1 |
FQ, RMP, INH, KAN, EMB |
100 |
Lineage2 |
100 |
|
G981 |
Culture |
Run1 |
Susceptible |
100 |
Lineage2 |
100 |
|
N0067 |
Culture |
Run1 |
Susceptible |
100 |
Lineage1 |
100 |
|
G1335_W27 |
Sputum |
Run2 |
Susceptible |
100 |
Lineage2 |
100 |
|
G1646_W19 |
Sputum |
Run2 |
Susceptible |
100 |
Lineage4 |
100 |
|
G2103_W23 |
Sputum |
Run2 |
Susceptible |
100 |
Lineage4 |
100 |
|
G2267_W20 |
Sputum |
Run2 |
Susceptible |
100 |
Lineage4 |
100 |
|
G2284_W26 |
Sputum |
Run2 |
Susceptible |
100 |
Lineage4 |
100 |
TP, true positive variants; FN, false negative variants; FP, false positive variants; TN, true negative variants; TPR, true positive rate; TNR, true negative rate.
DNA from culture samples was extracted following the standard CTAB protocol [32], while DNA from sputum samples was extracted following a differential cell lysis-based protocol to remove, as much as possible, all the contaminant non-MTBC DNA, according to Goig et al. [19]. All steps performed before library preparation were carried out in a BSL-3 laboratory.
Culture and sputum samples included in this study were derived from an ongoing surveillance programme of communicable diseases by the General Directorate of Public Health of the Comunitat Valenciana (Spain); therefore, the samples fall outside the mandate of the corresponding Ethics Committee for Biomedical Research. All personal information was anonymized, and no data allowing individual identification were retained.
Primer design
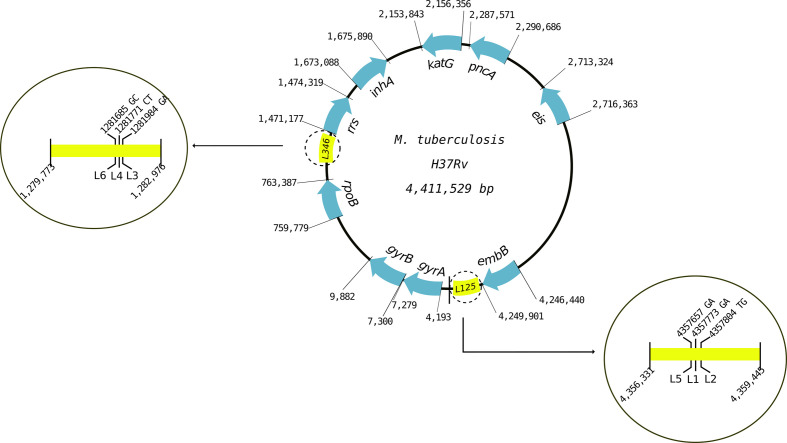
The gene panel consisted of nine resistance genes and two phylogenetic-determining regions. We selected nine genes known to be associated with first- and second-line drug resistance in MTBC (rpoB, inhA, katG, pncA, embB, rrs, gyrA, gyrB and eis) and two regions containing phylogenetic-determining SNPs (one region for lineages 1, 2 and 5 and another region for lineages 3, 4 and 6) (Table 3, Fig. 1) [33]. Primers were designed to amplify the entire genomic regions, obtaining products between 2500–3600 bp. Their specificity for MTBC was verified with Primer-blast (NCBI). The primer concentration for PCR reactions was optimized for an equimolar amplification of all genes (Methods S1, available in the online version of this article). Table S1 contains all primer concentrations and sequences.
Table 3.
Genes and phylogenetic regions included in the gene panel, their corresponding positions in the MTBC genome and the antibiotic they confer resistance to
|
Amplicon MTBC genomic positions |
Gene |
Prediction |
|
|---|---|---|---|
|
Start |
End |
||
|
4193 |
7279 |
gyrB |
Fluoroquinolones |
|
7300 |
9882 |
gyrA |
Fluoroquinolones |
|
759779 |
763387 |
rpoB |
Rifampicin |
|
1471177 |
1474319 |
rrs |
Streptomycin |
|
1673088 |
1675890 |
inhA |
Isoniazid |
|
2153843 |
2156356 |
katG |
Isoniazid |
|
2287571 |
2290686 |
pncA |
Pirazinamid |
|
2713324 |
2716363 |
eis |
Kanamycin |
|
4246440 |
4249901 |
embB |
Ethambutol |
|
4356331 |
4359445 |
Rv3878/Rv3879c |
L1, L2, L5 |
|
1279773 |
1282976 |
Rv1155/intergenic region Rv1156-Rv1157c |
L3, L4, L6 |
Fig. 1.
Diagram of the target regions of the gene panel. The lengths of regions are not scaled to genome size.
Single-tube multiplex PCR
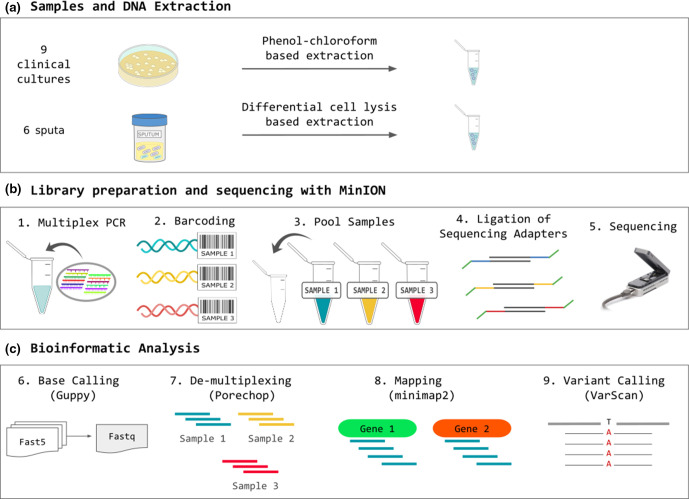
A single-tube multiplex PCR reaction was performed for each sample using the Kapa HiFi HS polymerase kit (Kapa Biosystems). The final reaction volume was adjusted depending on the DNA concentration of the samples to optimize yield (50 µl for samples above 10−3 ng μl−1, and 100 µl for samples below 10−3 ng μl−1). The optimum concentration of primers was previously set-up (see Methods S1 for the evaluation of optimal PCR conditions). PCR conditions were 3 min of DNA denaturation at 95 °C followed by 30 cycles of amplification as follows: 20 s at 98 °C for denaturing, 15 s at 65 °C for primer annealing, 2 min at 72 °C for extension, followed by 5 min at 72 °C for a final extension. The PCR product was purified with AMPure XP magnetic beads at 0.6X volume (following the Agencourt AMPure XP purification protocol). The pure PCR product was eluted with 10 mM Tris (pH=8.5) and quantified with Qubit to evaluate reaction yield. Fig. 2 summarizes the steps of the workflow.
Fig. 2.
Workflow diagram summarizing the methodology.
MinION library preparation and sequencing
Samples were barcoded by ligation using customized barcodes (see Methods S2) and pooled in an equimolar manner. An additional PCR enrichment step after barcoding was performed in samples that failed to reach 500 ng of DNA after the first PCR (see Methods S3).
The library was prepared using the 1D SQK-LSK108 Ligation Sequencing Kit (Oxford Nanopore Technologies) following the recommended protocol with modifications to increase process efficiency (see Methods S4).
Analysis of nanopore reads
After sequencing, Fast5 files were base-called with Guppy version 2.3.5 (Oxford Nanopore Technologies) using the ‘flip-flop’ model described by Wick et al. [34], which is slow but provides high read accuracy (see Methods S5 for the code). The resulting fastq file was demultiplexed with Porechop version 0.2.3 (https://github.com/rrwick/Porechop), generating a fastq file for each sample containing its corresponding reads (Methods S6). The source code of Porechop was modified by substituting the default sequences of Oxford Nanopore barcodes with the sequences of our custom barcodes (Table S2). Reads were mapped to the MTBC reference sequences of target genes included in the gene panel using minimap2 version 2.5-r607-dirty [35] with default parameters (Methods S7) [reference sequences were obtained from an inferred MTBC ancestral genome (NC_000962.3) [36]]. Aligned reads with a map quality under 60 (MAPQ60) were discarded. The remaining BAM file was sorted using samtools [37] (see Methods S7 for the mapping code). Variant calling and indel (insertion/deletion) calling were performed by running VarScan v2.3.7 [38] (https://github.com/dkoboldt/varscan) (see Methods S8 for the variant-calling parameters).
Illumina sequencing and analysis
DNA libraries for WGS were prepared according to the standardized NexteraXT protocol and loaded into the Illumina MiSeq sequencer. Reads were analysed following our tested and publicly available analysis pipeline (https://gitlab.com/tbgenomicsunit/ThePipeline) for mapping and variant calling (http://tgu.ibv.csic.es/?page_id=1794). This pipeline includes a first step to filter out low-quality and non-MTBC reads with fastp (https://github.com/OpenGene/fastp) and Kraken [39]. Filtered MTBC reads were aligned to an inferred MTBC ancestral genome [40] with BWA-MEM version 0.7.10 [41] and applied the MAPQ60 filter. Next, SNP calling and indel calling were performed with VarScan v2.3.7 [42] considering only positions covered at least by six reads, a minimum depth of coverage of 10, a minimum base quality of 25, and that present a minimum frequency of the variant allele above 0.1 (for variant positions, vSNPs) and 0.9 (for fixed variants, fSNPs). Additional information and details of the pipeline can be found at http://tgu.ibv.csic.es/wp-content/uploads/2020/03/ThePipeline_flowchart.pdf.
Resistance profile and phylogenetic classification
The resistance profile of each strain was obtained using a customized script (Python 2.7.5) that searched for drug-conferring variants in the target genes of the panel using a catalogue of well-known variants obtained from two databases [PhyResSe (http://www.phyresse.org/) and ReSeqTB (https://platform.reseqtb.org/), 9 July 2019]. In addition, lineage-defining variants (shown in Table S3 and described in a previously published study [33]) were searched to determine the major MTBC lineage of each strain. The customized script for resistance prediction and genotyping is available at GitLab https://gitlabcom/tbgenomicsunit/paneltb_scripts/.
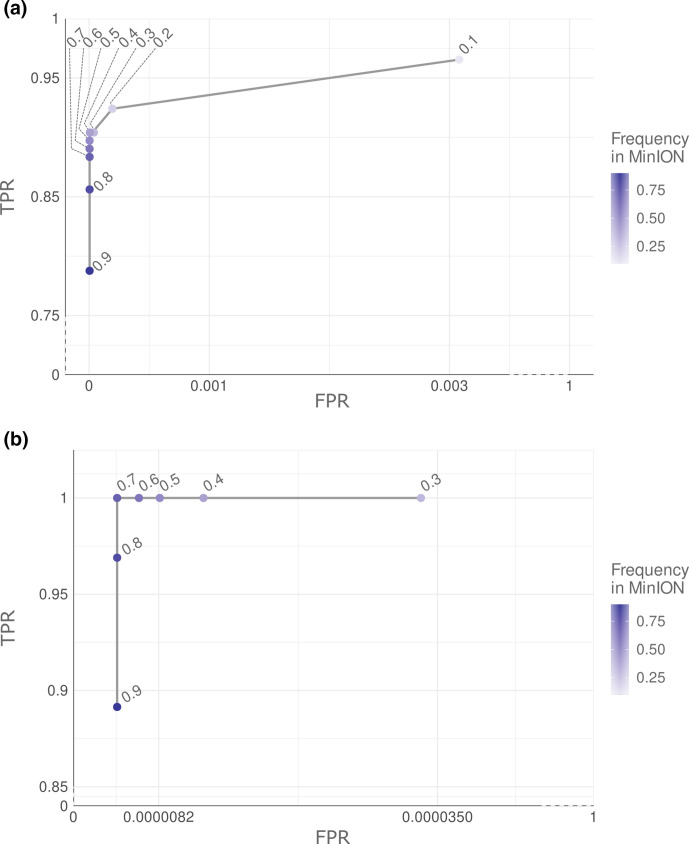
Calibration of variant calling in MinION
A receiver operating characteristic (ROC) curve analysis was performed to determine the optimum variant allele frequency to reduce false-positive calls by calling variants at different frequencies (from 0.1 to 0.9 using increments of 0.1) taking Illumina MiSeq variants (Illumina variants from now to the end) at 0.1 frequency as the reference. These analyses focused on positions that pass all variant-calling thresholds, as described above in the case of Illumina. For Nanopore MinION variants (MinION variants from now), stricter filtering was applied to reduce false-positive calls that increase background noise and decrease the recall. Positions appearing in all 15 samples sequenced by MinION were considered systematic errors and discarded. Triallelic positions with more than one nucleotide change were also discounted from downstream analysis. In addition, only positions that obtained a good depth value in both sequencing platforms were compared (positions above 10× depth in Illumina and 50× depth in MinION). Illumina was selected as the gold standard. Thus, true-positive variants (TP) were defined as variants detected in both sequencing technologies, false-positive variants (FP) as variants detected in MinION but not in Illumina, false negative (FN) as missing variants in MinION, and true negatives (TN) as sites that contained the wild-type allele in both technologies. According to these definitions, true-positive rate (TPR) or recall, true-negative rate (TNR), precision, agreement, error rate and F1-score were obtained (see Methods S9)
Insertion/deletion analysis
We performed an indel calling in MinION at different frequencies (from 0.1 to 1 with 0.1 increment) and validated with indels called by Illumina at 0.1 frequency. Two different comparisons were performed, one considering all indels regardless of the length and the other one filtering out indels longer than 50 bp.
Results
Selection of genes, samples and qPCR assay set-up
We selected nine culture samples from an ongoing TB population study of the Comunitat Valenciana (Spain), while the Hospital Clínico Universitario de Valencia (Spain) provided six sputum samples. Samples were selected to maximize the number of MTBC lineages and antibiotic resistance profiles for this proof-of-concept study. All the 15 samples were barcoded and then sequenced in two MinION runs. In the first run, we sequenced nine samples extracted from solid pure cultures, including lineage control strains (L1–L6) and one sample extracted from smear-positive sputum. In the second run, we sequenced five samples extracted directly from respiratory specimens of TB culture-positive patients. All 15 samples were previously WGS by Illumina MiSeq.
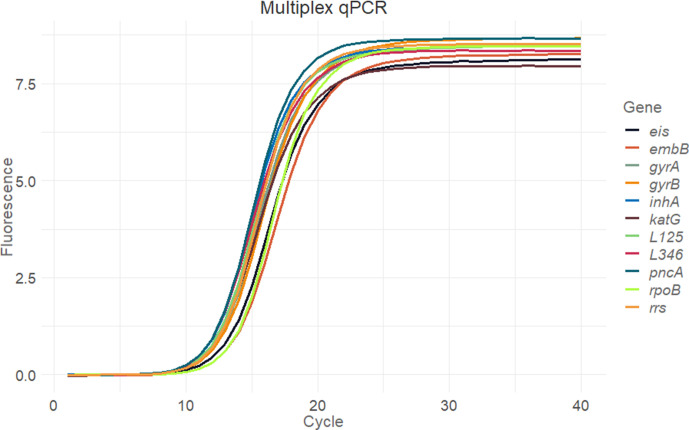
The gene panel consisted of nine genes (gyrA, gyrB, rpoB, rrs, inhA, embB, eis, pncA and katG) associated with resistance to first- and second-line drugs such as isoniazid, rifampicin, pyrazinamide, ethambutol, streptomycin, kanamycin and fluoroquinolones; and two phylogenetic regions containing lineage determining SNPs (L1, L2 and L5 in the first region and L3, L4 and L6 in the second region) (Fig. 1). We designed this panel to cover the whole region of interest and as a one-tube test. We carried out qPCR of the multiplex PCR product as a quality-control step to assay the proportion of each amplicon before sequencing. Observed Cq values from 9.81 (L346 region) to 11.42 (rpoB) suggested that all target regions were amplified at the same ratio (Fig. 3).
Fig. 3.
qPCR of the multiplex PCR product. Each colour represents a different gene. Two replicates per gene and negative controls were included.
Nanopore MinION sequencing
Samples were loaded in two Nanopore MinION R9 flow cells with 764 (first run) and 1375 (second run) active pores. In the first run, 848583 reads passed base calling with an N50 read length of 3123 bp, median read length of 2941 bp, median read quality of 11.4, and median depth of 4194.5× per gene. In the second run, a total number of 623989 reads passed base calling with an N50 read length of 3153 bp, median read length of 3102 bp, median read quality of 10.6, and median depth 1846× per gene. We hypothesize that the quick depletion of the library prompted the lower yield in the second run.
Porechop demultiplexed 65.9 and 61.6% of reads in the first and second runs, respectively; however, we failed to detect barcodes for around 22% of reads in both runs due to problems in the barcode ligation step. In addition, 11.4 and 16.2% of reads in the first and second runs, respectively, could not be demultiplexed as they did not pass the default threshold used in Porechop (see demultiplexing parameters in Methods S6).
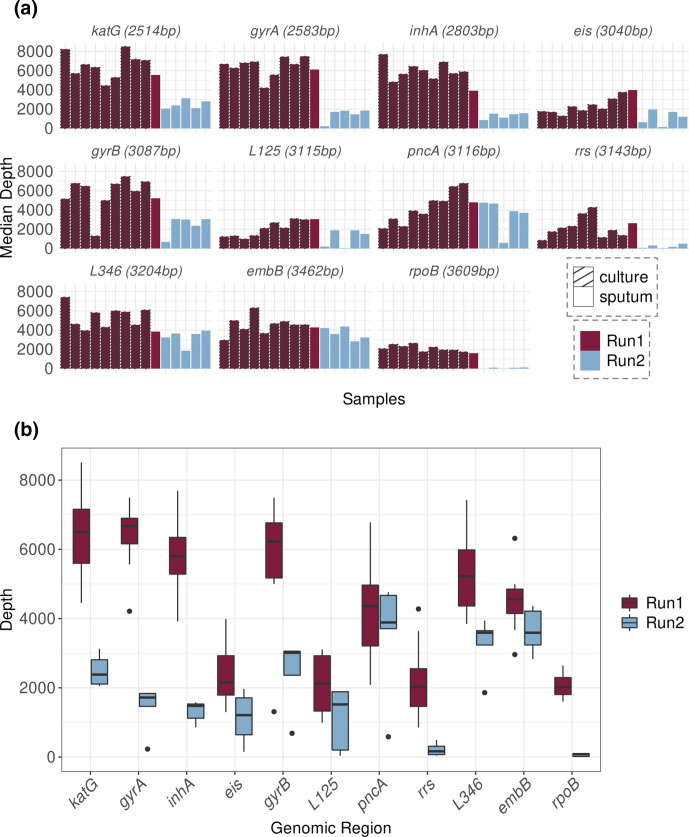
Regarding the depth of coverage, it was higher in the first run, perhaps due to the sample type, the library or flow-cell differences. The first run included DNA extracted from cultures (only one sputum sample), while the second run included only sputum samples. The greater heterogeneity of sputum samples generally associates with lower DNA quality and DNA fragmentation. Fig. 4 depicts median depth-coverage values for each gene per sample; we found that the rpoB and rrs genes had the lowest depth of coverage in both runs (Table 4).
Fig. 4.
Depth of coverage by gene in each MinION sequencing run. Genes are ordered by length (from the shortest to the longest). (a) Bar plot representing the median depth by sample and by region of the gene panel. Colours represent the sequencing run and the pattern of the bars represent sample type. (b) Box plot representing the distribution of sequencing depths of each genomic region in the 15 samples. Plots are coloured according to the MinION sequencing run (purple for the first run, blue for the second run).
Table 4.
Median depth of coverage for each gene per MinION run
|
Region |
Run 1 |
Run 2 |
||||
|---|---|---|---|---|---|---|
|
Median depth |
IQR |
Min–max |
Median depth |
IQR |
Min–max |
|
|
gyrA |
6677 |
737.25 |
4213–7493 |
1719 |
373 |
231–1856 |
|
katG |
6505 |
1561 |
4453–8510 |
2383 |
702 |
2056–3129 |
|
inhA |
5802 |
1059.25 |
3916–7695 |
1479 |
410 |
863–1580 |
|
gyrB |
6222 |
1590 |
1310–7489 |
3011 |
695 |
686–3058 |
|
embB |
4564 |
801 |
2962–6318 |
3592 |
976.5 |
2833–4370 |
|
pncA |
4358 |
1754.75 |
2086–6780 |
3888 |
960 |
587–4763 |
|
eis |
2156 |
1135.75 |
1299–3985 |
1211 |
1067 |
152–1967 |
|
rpoB |
2016 |
2011.75 |
1605–2643 |
84 |
78 |
3–118 |
|
rrs |
2022 |
1088 |
852–4278 |
166 |
241 |
40–495 |
|
L125 |
2124 |
1597.5 |
994–3109 |
1520 |
1684 |
37–1896 |
|
L346 |
5222 |
1618 |
3844–7428 |
3593 |
3930.625 |
1860–3943 |
IQR, interquartile range.
Calibrating variant-calling parameters in MinION
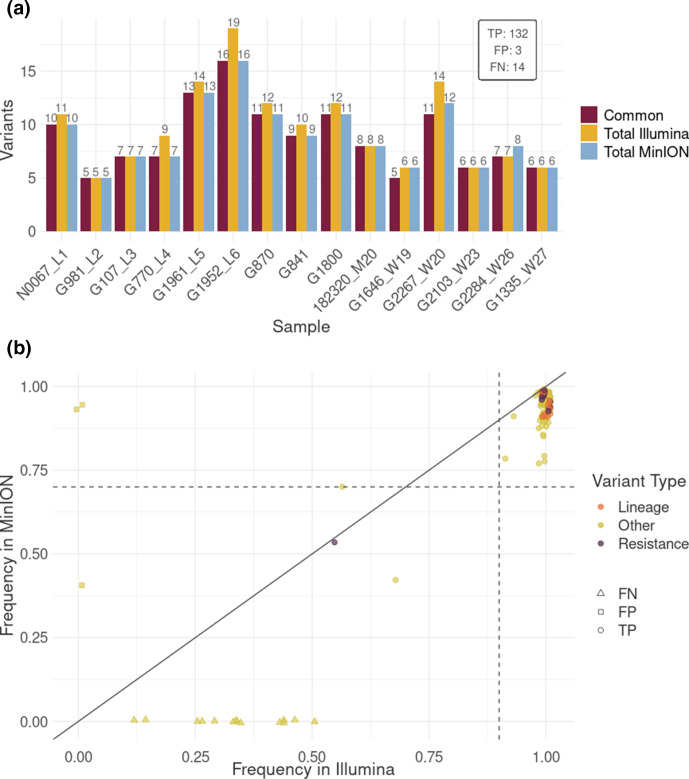
A ROC analysis was performed to determine the frequency threshold for MinION Nanopore variant-calling optimization, taking Illumina variants as a reference (those passing the filters and at least 0.1 frequency) to set the parameters for future applications. The nature of the vSNPs obtained in MinION and Illumina in all 15 samples was compared after filtering (i) variants detected in all MinION samples (systematic errors), (ii) triallelic positions (more than one nucleotide change), and (iii) by depth threshold under 50×. While whole-genome sequences were available in the case of Illumina, only positions belonging to the gene panel regions were included in the comparison study. This analysis highlighted 0.4 as the optimum frequency threshold to obtain the highest TPR and TNR to call variants with >0.1 frequency in Illumina (Fig. 5a, Table S4). Based on these cut-offs, calls from MinION were classified as TP if they were detected in both sequencing platforms, FP if they were detected only in MinION reads, and FN if they were detected in Illumina reads and not in MinION. A total of 132 TP, 3 FP and 14 FN were obtained, which resulted in a 90.41% TPR, 99.99% TNR, 88.59% agreement, 97.77% precision, 99.99% accuracy, 12.59% error rate and a 0.94 F1-score. All FN positions were Illumina low-frequency variants missed in MinION. Globally, we found a lower variant frequency in MinION than Illumina (Wilcoxon test, P-value<0.001) with a median of 4.44 [IQR=4.51] change in frequency (Fig. 6b). This difference explains the loss of low-frequency variants in MinION.
Fig. 5.
ROC curve used to set the frequency threshold employed to call variants in MinION. Points represent the values for recall and false positive rate obtained when applying different frequency values in MinION variant calling. Both axes are truncated. (a) ROC curve used to set the frequency threshold to call variable SNPs in MinION. Recall and false positive rate value obtained using different variant calling frequency cut-offs for MinION (from 0.1 to 0.9 using increments of 0.1) and comparing with Illumina variant calls at a 0.1 fixed threshold. (b) ROC curve used to determine the frequency threshold to call fixed SNPs in MinION. Both axes are truncated. Recall and false positive rate value obtained using different variant calling frequency cut-offs for MinION (from 0.3 to 0.9 using increments of 0.1) and comparing with Illumina variant calls at a 0.9 fixed threshold. For a detailed result see Fig. S1.
Fig. 6.
Comparison of variants found in Illumina and MinION. (a) Number of variants by sample obtained in MinION and Illumina that passed all quality filters for credible variants defined in Methods. Blue bars represent all variants detected in MinION reads, yellow bars represent variants obtained in Illumina reads, and purple bars represent common variants detected by both sequencing platforms. TP, FP and FN values in the inset box were obtained considering 15 samples. (b) Variant frequency correlation between Illumina and MinION reads. Only SNPs that passed all filters are represented. Points represent variants that appear in both platforms, squares represent discrepant variants only present in MinION reads, and triangles represent variants only present in Illumina. Orange represents phylogenetic variants, purple represents variants associated with antibiotic resistance, and yellow represents other variants. Dashed lines represent the threshold values to define fixed variants for MinION (horizontal) and Illumina (vertical).
Regarding the identification of variants fixed in Illumina (>0.9 frequency) that are typically used for phylogenetic and epidemiological investigations, ROC analysis identified a cut-off value of 0.7 in MinION to maximize TPR and TNR (Fig. 5b). A strong concordance between both platforms was obtained with an F1-score of 0.99, 98.47% agreement, 99.99% accuracy, 100% TPR, 99.99% TNR, and 1.53% error rate (Table 5). Of note, the 0.7 threshold in MinION involved the identification of two FPs.
Table 5.
Comparison of common and discrepant fSNPs
|
Frequency in MinION |
TP |
FP |
FN |
TN |
TPR |
TNR |
|---|---|---|---|---|---|---|
|
0.1 |
129 |
1489 |
0 |
478248 |
1 |
0.9968962 |
|
0.2 |
129 |
98 |
0 |
479592 |
1 |
0.9997957 |
|
0.5 |
129 |
4 |
0 |
479678 |
1 |
0.9999917 |
|
0.4 |
129 |
6 |
0 |
479676 |
1 |
0.9999875 |
|
0.3 |
129 |
16 |
0 |
479666 |
1 |
0.9999666 |
|
0.6 |
129 |
3 |
0 |
479679 |
1 |
0.9999937 |
|
0.7 |
129 |
2 |
0 |
479680 |
1 |
0.9999958 |
|
0.8 |
125 |
2 |
4 |
479680 |
0.9689922 |
0.9999958 |
|
0.9 |
115 |
2 |
14 |
479679 |
0.8914729 |
0.9999958 |
TP, true positive; FN, false negative; FP, false positive; TN, true negative; TPR, true-positive rate; TNR, true-negative rate.
Same ROC analysis was performed classifying SNPs by sample type for vSNPs and fSNPs obtaining the same cut-off frequencies mentioned above (see Tables S7 and S8, Fig. S1).
Indels' analysis
We tried to assess the capability of MinION to call insertions and deletions by calling indels at different frequencies (0.1 to 1 with 0.1 increments) and comparing the results with indels called by Illumina at 0.1. The total amount of indels detected by each sequencing platform is shown in Table 6. TPs are considered as indels detected by both sequencing platforms. We observed a very low agreement in indel calling between both sequencing platforms suggesting that most of them could be due to sequencing or mapping errors. So that, indel calling still remains a limitation for drug-resistance prediction from MinION data.
Table 6.
Indels' comparison
|
Frequency in MinION |
Indels MinION |
Indels Illumina |
TP |
|---|---|---|---|
|
Freq 10 |
14413 |
16 |
6 |
|
Freq 20 |
3735 |
16 |
6 |
|
Freq 30 |
1159 |
16 |
5 |
|
Freq 40 |
356 |
16 |
5 |
|
Freq 50 |
81 |
16 |
5 |
|
Freq 60 |
49 |
16 |
5 |
|
Freq 70 |
19 |
16 |
5 |
|
Freq 80 |
17 |
16 |
4 |
|
Freq 90 |
1 |
16 |
1 |
|
Freq 100 |
0 |
16 |
0 |
TP, true positive.
Drug resistance and lineage prediction
Regarding phylogenetic and drug-resistance-associated variants, 100% of the variants found in Illumina reads were also called by MinION when applying the cut-off frequency obtained in the ROC analysis (0.4). All samples (lineage control samples, sputa and cultures) could be classified correctly in their correspondent lineage by evaluating lineage-specific variants (Table S3). From 15 samples, we obtained one lineage 1, four lineage 2, one lineage 3, seven lineage 4, one lineage 5, and one lineage 6 strains (Table S5). We determined resistance profiles by evaluating known variants in panel genes associated with drug-resistant phenotypes [43, 44] resulting in one sample resistant to RMP (G841), one MDR strain with heteroresistance to INH (G1961), two pre-XDR-TB strains (G1800, G870) and 11 drug-susceptible strains (Table S5). Phylogenetic and drug-resistance SNPs detected are shown in Table 7. Both lineage classifications and antibiotic resistance predictions by MinION agree with the results obtained with Illumina WGS, when identifying known DR associated SNPs (0.4 frequency), giving a 100% agreement between both sequencing technologies (see Table S9). However, the agreement decreased for some regions when examining all variants. For RMP, the agreement was 50 % in sputa and 90% in cultures; for SM it was 60% in cultures; for KAN it was 75% in sputa; for L125 the agreement was 54 and 60% in sputa and culture, respectively; and for L346 it was 60% in sputa (see Table S9). These findings provide robust evidence that the gene panel sequenced using MinION can predict resistance profiles and strain lineages associated with known variants, but if novel variants are found additional validation will be required.
Table 7.
Phylogenetic and drug-resistance-associated variants detected by the gene panel
|
Sample |
Variant |
Gene |
Antibiotic or lineage |
Frequency MinION |
Frequency llumina |
Resistance profile |
|---|---|---|---|---|---|---|
|
G1961_L5 |
761155CT |
rpoB |
RMP |
96.33 |
100 |
MDR |
|
G1961_L5 |
1673425CT |
inhA |
INH |
52.94 |
55.67 |
|
|
G1961_L5 |
4247431GA |
embB |
EMB |
89.12 |
100 |
|
|
G1961_L5 |
4357657GA |
L125 |
Lineage 5 |
95.37 |
100 |
|
|
G1800 |
7570CT |
gyrA |
FQ |
87.44 |
100 |
pre-XDR |
|
G1800 |
761095TC |
rpoB |
RMP |
92.72 |
100 |
|
|
G1800 |
2155168CG |
katG |
INH |
98.06 |
100 |
|
|
G1800 |
2715344GA |
eis |
KAN |
96.54 |
100 |
|
|
G1800 |
4247429AG |
embB |
EMB |
98.49 |
100 |
|
|
G1800 |
4357804TG |
L125 |
Lineage 2 |
98.17 |
100 |
|
|
G870 |
6750CT |
gyrB |
FQ |
95.26 |
100 |
pre-XDR |
|
G870 |
761155CT |
rpoB |
RMP |
95.76 |
99.17 |
|
|
G870 |
2155168CG |
katG |
INH |
98.11 |
100 |
|
|
G870 |
2715346GA |
eis |
KAN |
92.68 |
100 |
|
|
G870 |
4247429AG |
embB |
EMB |
98.28 |
100 |
|
|
G870 |
4357804TG |
L125 |
Lineage 2 |
98.49 |
100 |
|
|
G841 |
761277AT |
rpoB |
RMP |
98.67 |
100 |
RIF-R |
|
G841 |
1281771CT |
L346 |
Lineage 4 |
93.07 |
100 |
|
|
N0067_L1 |
4357773GA |
L125 |
Lineage 1 |
94.77 |
100 |
Susceptible |
|
G1335_W27 |
4357804TG |
L125 |
Lineage 2 |
97.43 |
100 |
Susceptible |
|
G981_L2 |
4357804TG |
L125 |
Lineage 2 |
98.4 |
100 |
Susceptible |
|
G107_L3 |
1281984GA |
L346 |
Lineage 3 |
97.62 |
100 |
Susceptible |
|
182320_M20 |
1281771CT |
L346 |
Lineage 4 |
92.89 |
100 |
Susceptible |
|
G1646_W19 |
1281771CT |
L346 |
Lineage 4 |
90.75 |
100 |
Susceptible |
|
G2103_W23 |
1281771CT |
L346 |
Lineage 4 |
89.69 |
100 |
Susceptible |
|
G2267_W20 |
1281771CT |
L346 |
Lineage 4 |
90.18 |
100 |
Susceptible |
|
G2284_W26 |
1281771CT |
L346 |
Lineage 4 |
90.96 |
100 |
Susceptible |
|
G770_L4 |
1281771CT |
L346 |
Lineage 4 |
90.9 |
100 |
Susceptible |
|
G1952_L6 |
1281685CG |
L346 |
Lineage 6 |
84.47 |
100 |
Susceptible |
FQ, fluoroquinolones; RMP, rifampicin; SM, streptomicin; INH, isoniazid; PZA, pyrazinamide; KAN, kanamycin; EMB, ethambutol.
Discussion
The rapid and accurate detection of drug-resistant mycobacterial strains represents a crucial means to ensure treatment effectiveness; however, conventional diagnostic techniques are often time-intensive due to the requirement for in vitro culture steps. Amplicon-based approaches coupled with MinION sequencing are a promising strategy towards culture-free characterization of TB bacilli. Amplicons can detect resistant strains in culture-free samples with very low limit-of-detection meaning that drug-resistance results can be obtained in around a week, which is faster than culture-dependent strategies that can last up to a month. In addition, Illumina MiSeq takes around 72 h to produce 20 GB of output while MinION takes between 24 and 48 h and the main advantage is that the user can stop it when the output is enough for downstream analysis, as it allows real-time base calling. Here we design an amplicon-based approach aimed to take advantage of MinION long reads. Current targeted amplicon-sequencing techniques to identify DR-TB are based on short-read NGS technologies (i.e. Deeplex Myc-TB), which mainly focus on small amplicons (around 300 bp) within regions containing known drug-resistance-associated mutations [20]. Low DNA concentrations and high levels of fragmentation drive the need for small amplicons, particularly in complex samples, but this precludes certain advantages associated with long-read sequencing technologies like Nanopore. MinION long-read technology can sequence entire genes for variant phasing when genomic DNA has high integrity. Notably, the high error rate (as the main associated limitation) has decreased with time [45–47] through improvements in base-calling and variant-calling steps [34].
We present a long-amplicon alternative for research applications and demonstrate its utility using diagnostic samples. The strategy proposed in this study involves sequencing entire genes (around 2000–3000 bp), which is an advantage to identify all variants in each genomic region but it could compromise the efficiency of the multiplex PCR for some amplicons. We observed differences in depth of coverage between the genes included in our panel being rpoB and rrs the ones with the lowest depth value. Our data suggests that the length of the amplicon could represent an important bias during multiplex PCR amplification as shorter fragments are amplified preferently (as seen in Fig. 4 and reported by Tafess et al. [48]). In addition, we can not discard that sample type and between-run differences could also be responsible for the depth inconsistency. In any case, our results showed that the sequencing depth obtained was enough to detect drug-resistance-associated variants.
For clinical diagnosis, calling susceptibility is as important as calling resistance. This study shows the potential of a long-amplicon panel in predicting susceptibility and drug-resistance from both culture and sputum samples when scanning known markers. We designed a gene panel targeting nine genes (gyrA, gyrB, rpoB, rrs, inhA, embB, eis, pncA and katG) associated with resistance to commonly used first-line drugs (isoniazid, rifampicin, pyrazinamide and ethambutol) and injectable anti-TB drugs (streptomycin, kanamycin and fluoroquinolones). Since 2019, the WHO has updated the treatment for MDR-TB patients by recommending all-oral regimens including new drugs like bedaquiline over injectable drugs and also, some antibiotics, such as streptomycin, fell into disuse in some countries. In this assay, Streptomycin was included for historical reasons, but we have to emphasize the flexibility of the amplicon approaches to include or drop amplicons for different regions both for legacy drugs and new drugs by re-setting up the conditions of the multiplex PCR. The major obstacle however, is that many clinically relevant regions for new drugs remain unknown [49].
Beyond the limit of detection of our design, which would require a large number of samples, in this proof-of-concept study we focused on the bioinformatic challenges to identify variation, especially SNPs, associated with drug resistance, when using the Nanopore MinION sequencing technology. Reducing the high number of FP variant calls required a high cut-off frequency (0.4) compared with Illumina (0.1), which compromised recall in our study. Other studies have also shown 100% of agreement in the prediction of drug-resistance-associated variants between Illumina and MinION Nanopore when variants detected by MinION at an allele frequency below 0.4 are excluded from the analysis [50, 51]. Similarly, two recent studies showed complete concordance between both sequencing platforms when applied a threshold 0.8 to call fixed variants [52, 53]. Accurate identification of low-frequency variants is relevant for clinical management [11, 54] especially for drugs such as fluoroquinolones, for which frequencies ranging from 14–38 % have been reported [55].
Thus, the identification of heteroresistance below 0.4 remains challenging for MinION. MinION is also currently limited to call insertions/deletions due to the high error rate. Insertions/deletions is a type of variation that has been associated with resistance to diferencrent drugs. Our analysis shows the limited correlation between indel calls in MinION compared to WGS. Fortunately, Nanopore has recently released new flow cells with an improved chemistry that increases the variant-calling accuracy and, therefore, the resolution in low-frequency variant studies [56]. Our proof-of-concept study demonstrates the power of single-molecule sequencing to determine the resistance profile and lineage regardless of sample type and points out the need to overcome variant calling challenges to fully take advantage of its potential for the point-of-care prediction of drug resistance (especially heteroresistance and insertions/deletions).
Supplementary Data
Funding information
This project has been funded by the European Research Council (638553-TB-ACCELERATE, 101001038-TB-RECONNECT), the Ministerio de Economía, Indústria y Competividad (SAF2016-77346-R, PID2019-104477RB-I00) and the Conselleria d'Educació, Investigació, Cultura i Esport (PROMETEO 2020/012).
Author contributions
Author contributions are described with CRedit taxonomy. Conceptualization: I.C. Investigation: M.T.P., G.A.G., L.M.V., L.Z.I., C.M.L. Methodology: M.T.P., G.A.G., L.M.V. Resources: M.T.P., L.M.V., R.B., D.N., I.C. Software: G.A.G., C.M.L. Formal analysis: L.Z.I., C.M.L. Data curation: C.M.L. Supervision: A.C.O., I.C. Funding acquisition: I.C. Validation: I.C., M.T.P. Visualization: C.M.L., A.C.O., I.C. Writing original draft: C.M.L., A.C.O., I.C. Writing - reviewing and editing: M.T.P., A.C.O., I.C. Project administration: I.C.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Ethical statement
Culture and sputum samples included in this study came from an ongoing surveillance programme of communicable diseases by the General Directorate of Public Health of the Comunitat Valenciana (Spain), and therefore fall outside the mandate of the corresponding Ethics Committee for Biomedical Research. All personal information was anonymized, and no data allowing individual identification was retained.
Footnotes
Abbreviations: DR-TB, drug resistant tuberculosis; DST, drug susceptibility test; EMB, ethambutol; FN, false negative; FP, false positive; FPR, false positive rate; FQ, fluoroquinolones; Indel, insertion or deletion; INH, isoniazid; IQR, interquartile range; KAN, kanamycin; MDR-TB, multidrug resistant tuberculosis; MTBC, Mycobacterium tuberculosis complex; NGS, next generation seuencing; PCR, polymerase chain reaction; PZA, pyrazinamide; RMP, rifampicin; ROC, receiver operating characteristic curve; SM, streptomycin; SNP, single-nucleotide polymorphism; TB, tuberculosis; TN, true negative; TP, true positive; TPR, true positive rate; WGS, whole genome sequencing; XDR-TB, extensively drug-resistant tuberculosis.
All supporting data, code and protocols have been provided within the article or through supplementary data files. Supplementary material is available with the online version of this article.
References
- 1.World Health Organization Global Tuberculosis Report 2019. 2019.
- 2.Hoang TTT, Nguyen NV, Dinh SN, Nguyen HB, Cobelens F, et al. Challenges in detection and treatment of multidrug resistant tuberculosis patients in Vietnam. BMC Public Health. 2015;15:1–10. doi: 10.1186/s12889-015-2338-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.World Health Organization Global tuberculosis control: a short update to the 2009 report. World Health Organization; 2009. Report No.: WHO/HTM/TB/2009.426. 2009.
- 4.Lee A, Xie YL, Barry CE, Chen RY. Current and future treatments for tuberculosis. BMJ. 2020:m216. doi: 10.1136/bmj.m216. [DOI] [PubMed] [Google Scholar]
- 5.Marks SM, Hirsch-Moverman Y, Salcedo K, Graviss EA, Oh P, et al. Characteristics and costs of multidrug-resistant tuberculosis in-patient care in the United States, 2005–2007. Int J Tuberc Lung Dis. 2016;20:435–441. doi: 10.5588/ijtld.15.0575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.World Health Organization Rapid сommunication: key changes to treatment of multidrug-and rifampicin-resistant tuberculosis (MDR/RR-TB) [Internet]. World Health Organization. 2019. https://www.who.int/tb/publications/2019/WHO_RapidCommunicationMDR_TB2019.pdf
- 7.Roelens M, Battista Migliori G, Rozanova L, Estill J, Campbell JR, et al. Evidence-based definition for extensively drug-resistant tuberculosis. Am J Respir Crit Care Med. 2021;204:713–722. doi: 10.1164/rccm.202009-3527OC. [DOI] [PubMed] [Google Scholar]
- 8.Mirzayev F, Viney K, Linh NN, Gonzalez-Angulo L, Gegia M, et al. World Health Organization recommendations on the treatment of drug-resistant tuberculosis, 2020 update. Eur Respir J. 2021;57:2003300. doi: 10.1183/13993003.03300-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim SJ. Drug-susceptibility testing in tuberculosis: methods and reliability of results. European Respiratory Journal. 2005;25:564–569. doi: 10.1183/09031936.05.00111304. [DOI] [PubMed] [Google Scholar]
- 10.Falkinham JO. Molecular Medical Microbiology. Academic Press; 2015. The Mycobacterium avium complex and slowly growing mycobacteria; pp. 1669–1678. [DOI] [Google Scholar]
- 11.Cancino-Muñoz I, Moreno-Molina M, Furió V, Goig GA, Torres-Puente M, et al. Cryptic resistance mutations associated with misdiagnoses of multidrug-resistant tuberculosis. J Infect Dis. 2019;220:316–320. doi: 10.1093/infdis/jiz104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.World Health Organization The use of molecular line probe assay for the detection of resistance to isoniazid and rifampicin: policy update. World Health Organization. 2016 [Google Scholar]
- 13.Mäkinen J, Marttila HJ, Marjamäki M, Viljanen MK, Soini H. Comparison of two commercially available DNA line probe assays for detection of multidrug-resistant Mycobacterium tuberculosis . J Clin Microbiol. 2006;44:350–352. doi: 10.1128/JCM.44.2.350-352.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bai Y, Wang Y, Shao C, Hao Y, Jin Y, et al. GenoType MTBDRplus assay for rapid detection of multidrug resistance in Mycobacterium tuberculosis: a meta-analysis. PLoS ONE. 2016;11:e0150321. doi: 10.1371/journal.pone.0150321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ninan MM, Gowri M, Christopher DJ, Rupali P, Michael JS. The diagnostic utility of line probe assays for multidrug-resistant tuberculosis. Pathog Glob Health. 2016;110:194–199. doi: 10.1080/20477724.2016.1214350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Osei Sekyere J, Maphalala N, Malinga LA, Mbelle NM, Maningi NE. A comparative evaluation of the new genexpert MTB/RIF ultra and other rapid diagnostic assays for detecting tuberculosis in pulmonary and extra pulmonary specimens. Sci Rep. 2019;9:16587. doi: 10.1038/s41598-019-53086-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bunsow E, Ruiz-Serrano MJ, López Roa P, Kestler M, Viedma DG, et al. Evaluation of GeneXpert MTB/RIF for the detection of Mycobacterium tuberculosis and resistance to rifampin in clinical specimens. J Infect. 2014;68:338–343. doi: 10.1016/j.jinf.2013.11.012. [DOI] [PubMed] [Google Scholar]
- 18.Sanchez-Padilla E, Merker M, Beckert P, Jochims F, Dlamini T, et al. Detection of drug-resistant tuberculosis by Xpert MTB/RIF in Swaziland. N Engl J Med. 2015;372:1181–1182. doi: 10.1056/NEJMc1413930. [DOI] [PubMed] [Google Scholar]
- 19.Goig GA, Cancino-Muñoz I, Torres-Puente M, Villamayor LM, Navarro D, et al. Whole-genome sequencing of Mycobacterium tuberculosis directly from clinical samples for high-resolution genomic epidemiology and drug resistance surveillance: an observational study. The Lancet Microbe. 2020;1:e175–e183. doi: 10.1016/S2666-5247(20)30060-4. [DOI] [PubMed] [Google Scholar]
- 20.Jouet A, Gaudin C, Badalato N, Allix-Béguec C, Duthoy S, et al. Deep amplicon sequencing for culture-free prediction of susceptibility or resistance to 13 anti-tuberculous drugs. Eur Respir J. 2021;57:2002338. doi: 10.1183/13993003.02338-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.López B, Aguilar D, Orozco H, Burger M, Espitia C, et al. A marked difference in pathogenesis and immune response induced by different Mycobacterium tuberculosis genotypes. Clin Exp Immunol. 2003;133:30–37. doi: 10.1046/j.1365-2249.2003.02171.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ford CB, Shah RR, Maeda MK, Gagneux S, Murray MB, et al. Mycobacterium tuberculosis mutation rate estimates from different lineages predict substantial differences in the emergence of drug-resistant tuberculosis. Nat Genet. 2013;45:784–790. doi: 10.1038/ng.2656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Colman RE, Schupp JM, Hicks ND, Smith DE, Buchhagen JL, et al. Detection of low-level mixed-population drug resistance in Mycobacterium tuberculosis using high fidelity amplicon sequencing. PLoS One. 2015;10:e0126626. doi: 10.1371/journal.pone.0126626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rinder H, Mieskes KT, Löscher T. Heteroresistance in Mycobacterium tuberculosis . Int J Tuberc Lung Dis. 2001;5:339–345. [PubMed] [Google Scholar]
- 25.Magi A, Semeraro R, Mingrino A, Giusti B, D’Aurizio R. Nanopore sequencing data analysis: state of the art, applications and challenges. Brief Bioinform. 2018;19:1256–1272. doi: 10.1093/bib/bbx062. [DOI] [PubMed] [Google Scholar]
- 26.Xu Y, Lewandowski K, Lumley S, Pullan S, Vipond R, et al. Detection of viral pathogens with multiplex nanopore MinION sequencing: be careful with cross-talk. Front Microbiol. 2018;9:2225. doi: 10.3389/fmicb.2018.02225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schmidt K, Mwaigwisya S, Crossman LC, Doumith M, Munroe D, et al. Identification of bacterial pathogens and antimicrobial resistance directly from clinical urines by nanopore-based metagenomic sequencing. J Antimicrob Chemother. 2017;72:104–114. doi: 10.1093/jac/dkw397. [DOI] [PubMed] [Google Scholar]
- 28.Bainomugisa A, Duarte T, Lavu E, Pandey S, Coulter C, et al. A complete high-quality MinION nanopore assembly of an extensively drug-resistant Mycobacterium tuberculosis Beijing lineage strain identifies novel variation in repetitive PE/PPE gene regions. Microbial Genomics. 2018;4 doi: 10.1099/mgen.0.000188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Public health teams in Madagascar pioneer use of portable, real time DNA sequencing to fight drug-resistant tuberculosis (TB) [Internet] 2018. http://nanoporetech.com/about-us/news/public-health-teams-madagascar-pioneer-use-portable-real-time-dna-sequencing-fight
- 30.Tyler AD, Mataseje L, Urfano CJ, Schmidt L, Antonation KS, et al. Evaluation of Oxford Nanopore’s MinION sequencing device for microbial whole genome sequencing applications. Sci Rep. 2018;8:10931. doi: 10.1038/s41598-018-29334-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Votintseva AA, Bradley P, Pankhurst L, Del Ojo Elias C, Loose M, et al. Same-day diagnostic and surveillance data for tuberculosis via whole-genome sequencing of direct respiratory samples. J Clin Microbiol. 2017;55:1285–1298. doi: 10.1128/JCM.02483-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Somerville W, Thibert L, Schwartzman K, Behr MA. Extraction of Mycobacterium tuberculosis DNA: a question of containment. J Clin Microbiol. 2005;43:2996–2997. doi: 10.1128/JCM.43.6.2996-2997.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cancino-Muñoz I, Gil-Brusola A, Torres-Puente M, Mariner-Llicer C, Dogba J, et al. Development and application of affordable SNP typing approaches to genotype Mycobacterium tuberculosis complex strains in low and high burden countries. Sci Rep. 2019;9:15343. doi: 10.1038/s41598-019-51326-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wick RR, Judd LM, Holt KE. Performance of neural network basecalling tools for Oxford Nanopore sequencing. Genome Biol. 2019;20:129. doi: 10.1186/s13059-019-1727-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li H. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics. 2018;34:3094–3100. doi: 10.1093/bioinformatics/bty191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Comas I, Chakravartti J, Small PM, Galagan J, Niemann S, et al. Human T cell epitopes of Mycobacterium tuberculosis are evolutionarily hyperconserved. Nat Genet. 2010;42:498–503. doi: 10.1038/ng.590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Koboldt DC, Zhang Q, Larson DE, Shen D, McLellan MD, et al. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 2012;22:568–576. doi: 10.1101/gr.129684.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wood DE, Salzberg SL. Kraken: ultrafast metagenomic sequence classification using exact alignments. Genome Biol. 2014;15:R46. doi: 10.1186/gb-2014-15-3-r46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Comas I, Chakravartti J, Small PM, Galagan J, Niemann S, et al. Human T cell epitopes of Mycobacterium tuberculosis are evolutionarily hyperconserved. Nat Genet. 2010;42:498–503. doi: 10.1038/ng.590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Koboldt DC, Zhang Q, Larson DE, Shen D, McLellan MD, et al. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 2012;22:568–576. doi: 10.1101/gr.129684.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Feuerriegel S, Schleusener V, Beckert P, Kohl TA, Miotto P, et al. PhyResSE: a web tool delineating Mycobacterium tuberculosis antibiotic resistance and lineage from whole-genome sequencing data. J Clin Microbiol. 2015;53:1908–1914. doi: 10.1128/JCM.00025-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.ReSeqTB [Internet] 2021. https://platform.reseqtb.org/main/home.html
- 45.Minervini CF, Cumbo C, Orsini P, Anelli L, Zagaria A, et al. Nanopore sequencing in blood diseases: a wide range of opportunities. Front Genet. 2020;11 doi: 10.3389/fgene.2020.00076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Orsini P, Minervini CF, Cumbo C, Anelli L, Zagaria A, et al. Design and MinION testing of a nanopore targeted gene sequencing panel for chronic lymphocytic leukemia. Sci Rep. 2018;8:11798. doi: 10.1038/s41598-018-30330-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ammar R, Paton TA, Torti D, Shlien A, Bader GD. Long read nanopore sequencing for detection of HLA and CYP2D6 variants and haplotypes. F1000Res. 2015;4:17. doi: 10.12688/f1000research.6037.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tafess K, Ng TTL, Lao HY, Leung KSS, KKG T, et al. Targeted sequencing workflows for comprehensive drug resistance profiling of Mycobacterium tuberculosis cultures using Illumina MiSeq and Nanopore MinION: Comparison of analytical and diagnostic performance, turnaround time and cost [Internet] bioRxiv. 2019:760462. doi: 10.1093/clinchem/hvaa092. [DOI] [PubMed] [Google Scholar]
- 49.World Health Organization Meeting report of the WHO expert consultation on the definition of extensively drug-resistant tuberculosis, 27-29 October 2020, Geneva. 2021. https://apps.who.int/iris/bitstream/handle/10665/338776/9789240018662-eng.pdf
- 50.Greig DR, Jenkins C, Gharbia S, Dallman TJ. Comparison of single-nucleotide variants identified by Illumina and Oxford Nanopore technologies in the context of a potential outbreak of Shiga toxin–producing Escherichia coli . GigaScience. 2019;8 doi: 10.1093/gigascience/giz104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tafess K, Ng TTL, Lao HY, Leung KSS, KKG T, et al. Targeted-sequencing workflows for comprehensive drug resistance profiling of Mycobacterium tuberculosis cultures using two commercial sequencing platforms: comparison of analytical and diagnostic performance, turnaround time, and cost. Clin Chem. 2020;66:809–820. doi: 10.1093/clinchem/hvaa092. [DOI] [PubMed] [Google Scholar]
- 52.Cabibbe AM, Spitaleri A, Battaglia S, Colman RE, Suresh A, et al. Application of targeted next-generation sequencing assay on a portable sequencing platform for culture-free detection of drug-resistant tuberculosis from clinical samples. J Clin Microbiol. 2020;58 doi: 10.1128/JCM.00632-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chan WS, Au CH, Chung Y, Leung HCM, Ho DN, et al. Rapid and economical drug resistance profiling with Nanopore MinION for clinical specimens with low bacillary burden of Mycobacterium tuberculosis . BMC Res Notes. 2020;13:444. doi: 10.1186/s13104-020-05287-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vargas R, Freschi L, Marin M, Epperson LE, Smith M, et al. In-host population dynamics of complex during active disease. Elife [Internet] 2021;10 doi: 10.7554/eLife.61805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rigouts L, Miotto P, Schats M, Lempens P, Cabibbe AM, et al. Fluoroquinolone heteroresistance in Mycobacterium tuberculosis: detection by genotypic and phenotypic assays in experimentally mixed populations. Sci Rep. 2019;9:11760. doi: 10.1038/s41598-019-48289-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Karst SM, Ziels RM, Kirkegaard RH, Sørensen EA, McDonald D, et al. High-accuracy long-read amplicon sequences using unique molecular identifiers with Nanopore or PacBio sequencing. Nat Methods. 2021;18:165–169. doi: 10.1038/s41592-020-01041-y. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.