Abstract
Shigella and enteroinvasive Escherichia coli (EIEC) cause human bacillary dysentery with similar invasion mechanisms and share similar physiological, biochemical and genetic characteristics. Differentiation of Shigella from EIEC is important for clinical diagnostic and epidemiological investigations. However, phylogenetically, Shigella and EIEC strains are composed of multiple clusters and are different forms of E. coli , making it difficult to find genetic markers to discriminate between Shigella and EIEC. In this study, we identified 10 Shigella clusters, seven EIEC clusters and 53 sporadic types of EIEC by examining over 17000 publicly available Shigella and EIEC genomes. We compared Shigella and EIEC accessory genomes to identify cluster-specific gene markers for the 17 clusters and 53 sporadic types. The cluster-specific gene markers showed 99.64% accuracy and more than 97.02% specificity. In addition, we developed a freely available in silico serotyping pipeline named Shigella EIEC Cluster Enhanced Serotype Finder (ShigEiFinder) by incorporating the cluster-specific gene markers and established Shigella and EIEC serotype-specific O antigen genes and modification genes into typing. ShigEiFinder can process either paired-end Illumina sequencing reads or assembled genomes and almost perfectly differentiated Shigella from EIEC with 99.70 and 99.74% cluster assignment accuracy for the assembled genomes and read mapping respectively. ShigEiFinder was able to serotype over 59 Shigella serotypes and 22 EIEC serotypes and provided a high specificity of 99.40% for assembled genomes and 99.38% for read mapping for serotyping. The cluster-specific gene markers and our new serotyping tool, ShigEiFinder (installable package: https://github.com/LanLab/ShigEiFinder, online tool: https://mgtdb.unsw.edu.au/ShigEiFinder/), will be useful for epidemiological and diagnostic investigations.
Keywords: phylogenetic clusters, cluster-specific gene markers, Shigella, enteroinvasive E. coli , serotyping
Data Summary
Sequencing data have been deposited at the National Center for Biotechnology Information under BioProject number PRJNA692536.
Impact Statement.
The differentiation of Shigella strains from enteroinvasive Escherichia coli (EIEC) is important for clinical diagnosis and public health epidemiological investigations. The similarities between Shigella and EIEC strains make this differentiation very difficult as both share common ancestries within E. coli . However, Shigella and EIEC are phylogenetically separated into multiple clusters, making high-resolution separation using cluster-specific genomic markers possible. In this study, we identified 17 Shigella or EIEC clusters including five that were newly identified through examination of over 17000 publicly available Shigella and EIEC genomes. We further identified cluster-specific gene marker sets for each cluster using comparative genomic analysis. These markers were used to classify isolates into clusters and then to develop an in silico pipeline, ShigEiFinder (https://github.com/LanLab/ShigEiFinder), for accurate differentiation, cluster typing and serotyping of Shigella and EIEC from Illumina sequencing reads or assembled genomes. This study will have broad application ranging from understanding the evolution of Shigella and EIEC to diagnosis and epidemiology.
Introduction
Shigella is a leading cause of diarrhoea with a very low infective dose [1, 2]. The infections can vary from mild diarrhoea to severe bloody diarrhoea referred to as bacillary dysentery. The estimated cases of Shigella infections are 190 million with >210000 deaths annually, predominantly in children younger than 5 years old in developing countries [3–7]. Shigella infections also have a significant impact on public health in developed countries, although most cases are travel-associated [8].
The genus Shigella consists of four species, Shigella sonnei , Shigella flexneri , Shigella boydii and Shigella dysenteriae [9]. Serological testing has further classified Shigella species into more than 55 serotypes through the agglutination reaction of antisera to Shigella serotype-specific O-antigens [10, 11]. Up to 89.6% of Shigella infections were caused by S. flexneri (65.9%) and S. sonnei (23.7%) globally [12, 13]. The predominant serotype reported in Shigella infections has been S. flexneri serotype 2a while S. dysenteriae serotype 1 has caused the most severe disease [10, 14]. Note that for brevity, in all references to Shigella serotypes below, S. sonnei , S. flexneri , S. boydii and S. dysenteriae are abbreviated as SS, SF, SB and SD respectively and a serotype is designated with an abbreviated ‘species’ name plus the serotype number (e.g. S. dysenteriae serotype 1 is abbreviated as SD1).
Enteroinvasive Escherichia coli (EIEC) is a pathovar of E. coli that causes diarrhoea with less severe symptoms than Shigella infections in humans worldwide, particularly in developing countries [8, 13, 15–18]. EIEC infections in developed countries are mainly imported [19]. EIEC has more than 18 specific E. coli O-serotypes [19, 20]. Although the incidence of EIEC is low [17], EIEC serotypes have been associated with outbreaks and sporadic cases of infections [20–22]. In contrast to Shigella , EIEC infections are not notifiable in many countries [23, 24].
Shigella and EIEC have always been considered very closely related and share several characteristics [25–28]. Shigella and EIEC are both non-motile and lack the ability to ferment lactose [24]. Some EIEC O antigens are identical or similar to Shigella O antigens (O112ac, O124, O136, O143, O152 and O164) [26, 29–31]. Furthermore, Shigella and EIEC both carry the virulence plasmid pINV, which encodes virulence genes required for invasion [32, 33] and contain ipaH (invasion plasmid antigen H) genes with the exception of some SB13 isolates [11, 23, 24, 34, 35]. Shigella and EIEC have arisen from E. coli in multiple independent events and should be regarded as a single pathovar of E. coli [25, 26, 28, 36–38]. Previous phylogenetic studies suggested that Shigella isolates were divided into three clusters (C1, C2 and C3) with five outliers (SS, SB13, SD1, SD8 and SD10) [25, 28] whereas EIEC isolates were grouped into four clusters (C4, C5, C6 and C7) [26]. The seven Shigella and EIEC clusters and five outliers of Shigella are within the broader E. coli species except for SB13 some of which are in fact Escherichia albertii [39, 40]. Whole genome sequencing (WGS)-based phylogenomic studies have also defined multiple alternative clusters of Shigella and EIEC [23, 28, 41].
The traditional biochemical test for motility and lysine decarboxylase (LDC) activity [42] and molecular test for the presence of the ipaH gene have been used to differentiate Shigella and EIEC from non-EIEC [24, 43–45]. Agglutination with Shigella- and EIEC-associated antiserum further classifies Shigella and EIEC to the serotype level. However, cross-reactivity, strains not producing O antigens and newly emerged Shigella serotypes may all prevent accurate serotyping [11, 46]. Serotyping by antigenic agglutination is being replaced by molecular serotyping [46–48], which can be achieved through examination of the sequences of O antigen biosynthesis and modification genes [8, 24, 49–52].
Recently, PCR-based molecular detection methods targeting the gene lacY were developed to distinguish Shigella from EIEC [53, 54]. However, the ability of the primers described in these methods to accurately differentiate between Shigella and EIEC was later questioned [23, 28]. With the uptake of WGS technology, several studies have identified phylogenetic clade-specific markers, species-specific markers and EIEC lineage-specific genes for discrimination between Shigella and EIEC and between Shigella species [23, 27, 28, 41, 55, 56]. More recently, genetic markers, lacY, cadA and SS_methylase gene, were used for identification of Shigella and EIEC [11]. However, these markers failed to discriminate between Shigella and EIEC when a larger genetic diversity is considered [23, 28, 55]. A Kmer-based approach can identify Shigella isolates to the species level but misidentification was also observed [56].
In this study, we aimed to (i) identify phylogenetic clusters of Shigella and EIEC through large-scale examination of publicly available genomes; (ii) identify cluster-specific gene markers using comparative genomic analysis of Shigella and EIEC accessory genomes for differentiation of Shigella and EIEC; and (iii) develop a pipeline for Shigella and EIEC in silico serotyping based on the cluster-specific gene markers combined with Shigella and EIEC serotype-specific O antigen and H antigen genes. We demonstrate that these cluster-specific gene markers enhance in silico serotyping using genomic data. We also developed an automated pipeline for cluster typing and serotyping of Shigella and EIEC from WGS data.
Methods
Identification of Shigella and EIEC isolates from the NCBI database
E. coli and Shigella isolates from the NCBI SRA (National Center for Biotechnology Information Sequence Read Archive) in May 2019 were queried. The keywords ‘ Escherichia coli ’ and ‘ Shigella ’ were used to retrieve SRA accession numbers of E. coli and Shigella isolates. Raw reads were retrieved from the ENA (European Nucleotide Archive). The ipaH gene (GenBank accession number M32063.1) was used to screen E. coli and Shigella reads using Salmon v0.13.0 [57]. Taxonomic classification for E. coli and Shigella was confirmed by Kraken v1.1.1 [58]. Molecular serotype prediction of ipaH-negative Shigella isolates was performed using ShigaTyper v1.0.6 [11]. Isolates that were ipaH positive and isolates with designation of SB13 by ShigaTyper were selected to form the Shigella and EIEC database.
The sequence types (STs) and ribosomal STs (rSTs) of ipaH-negative E. coli (non-enteroinvasive E. coli ) isolates were examined. STs and rSTs for these isolates were obtained from the E. coli and Shigella database in Enterobase [59] in May 2019. For STs and rSTs with only one isolate, the isolate was selected. For STs and rSTs with more than one isolate, one representative isolate for each ST and rST was randomly selected to cover the diversity. In total, 12743 ipaH-negative E. coli isolates representing 3800 STs and 11463 rSTs were selected as a non-EIEC control database.
Genome sequencing
Genome sequencing of 31 EIEC strains used in a previous study [26] was performed by Illumina NextSeq (Illumina). DNA libraries were constructed using Nextera XT Sample preparation kits (Illumina) and sequenced using the NextSeq sequencer (Illumina). FASTQ sequences of the strains sequenced in this study were deposited in the NCBI under the BioProject PRJNA692536.
Genome assembly and data processing
Raw reads were de novo assembled using SPADES v3.14.0 with default settings [http://bioinf.spbau.ru/spades] [60]. The metrics of assembled genomes were obtained with QUAST v5.0.0 [61]. Three standard deviations (sd) from the mean for contig number, largest contig, total length, GC, N50 and genes were used as quality filters for the assembled genomes.
The STs for isolates in the Shigella and EIEC database were checked by using mlst (https://github.com/tseemann/mlst) with the E. coli scheme from PubMLST [62]. rSTs were extracted from the E. coli and Shigella rMLST database in Enterobase [59] in May 2019. Serotype prediction for isolates in the Shigella and EIEC database was performed using ShigaTyper v1.0.6 [11]. Serotyping of E. coli O and H antigens was predicted using SerotypeFinder v2.0.1 [63].
Selection of isolates for Shigella and EIEC identification dataset
The selection of isolates for the identification dataset was based on the representative isolates for each ST, rST, and serotype of Shigella and EIEC in the Shigella and EIEC database. For STs, rSTs and serotypes with only one isolate, the isolate was selected. For STs, rSTs and serotypes with more than one isolate, one representative isolate for each ST, rST and serotype was randomly selected. The 72 ECOR isolates [64] and 18 E. albertii isolates downloaded from Enterobase [59] were used as controls for the identification dataset. Details of the identification dataset are given in Table S1, Supplementary Material file 1 (available in the online version of this article). The remaining isolates in the Shigella and EIEC database were referred to as the validation dataset (Table S2).
The identification dataset was used to characterize the phylogenetic relationships of Shigella and EIEC. The identification dataset was also used to identify cluster-specific gene markers. The validation dataset was used to evaluate the performance of cluster-specific gene markers using the in silico serotyping pipeline.
Phylogeny of Shigella and EIEC based on WGS
Nine phylogenetic trees including an identification tree, a confirmation tree and seven validation trees were constructed using Quicktree v1.3 [65] with the default parameters to identify and confirm the phylogenetic clustering of Shigella and EIEC isolates. The phylogenetic trees were visualized using Grapetree and ITOL v5 [66, 67].
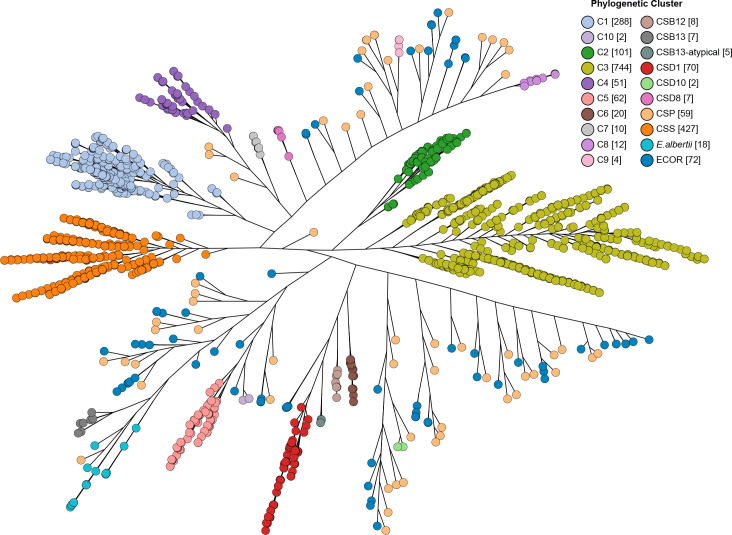
The identification phylogenetic tree was generated based on isolates in the identification dataset for characterization of clusters of Shigella and EIEC isolates (Fig. 1). A subset of 485 Shigella and EIEC isolates known to represent each identified cluster from the identification dataset in the present study was then selected. These 485 isolates consisted of all 74 isolates from the sporadic EIEC lineages (59 isolates) and four clusters (four C9 isolates, two C10 isolates, two CSD10 isolates and seven CSD8 isolates), and 411 isolates representing the remaining 13 clusters with number of isolates per cluster ranging from three to 168 isolates. These 411 isolates were selected manually to represent each independent clade from each cluster based on the identification phylogenetic tree of Shigella and EIEC isolates (Fig. 1).
Fig. 1.
Shigella and EIEC cluster identification phylogenetic tree. Representative isolates from the identification dataset were used to construct the phylogenetic tree using Quicktree v1.3 [65] to identify Shigella and EIEC (enteroinvasive E. coli ) clusters and visualized using Grapetree. The dendrogram shows the phylogenetic relationships of 1879 Shigella and EIEC isolates represented in the identification dataset. Branch lengths are on a log scale for clarity. Bar, 0.2 substitutions per site. Shigella and EIEC clusters are coloured. Numbers in square brackets after the cluster name are the number of isolates for each identified cluster. CSP indicates sporadic EIEC lineages. ECOR is the Escherichia coli reference collection. E. albertii is Escherichia albertii which was included to show the location of ‘typical’ S. boydii serotype 13 strains. CSS, CSB12, CSB13, CSD1, CSD8 and CSD10 are the clusters of S. sonnei , S. boydii serotype 12, S. boydii serotype 13, S. dysenteriae serotype 1, S. dysenteriae serotype 8 and S. dysenteriae serotype 10 respectively.
The confirmation tree was constructed based on the subset of 485 isolates from the identification dataset and 1872 non-EIEC isolates from the non-EIEC control dataset (2357 isolates in total). This tree was used for confirmation of the phylogenetic relationships between identified Shigella and EIEC clusters in the identification dataset and non-EIEC isolates. The validation trees were generated based on Shigella and EIEC isolates from the validation dataset and a subset of 575 isolates (485 Shigella and EIEC isolates representing each cluster and lineage, 72 ECOR isolates and 18 E. albertii isolates) from the identification dataset to assign validation dataset isolates to the clusters defined.
Investigation of Shigella virulence plasmid pINV
The presence of the Shigella virulence plasmid pINV in isolates was investigated by using BWA-MEM v0.7.17 (Burrows-Wheeler Aligner) [68] to align the raw reads of an isolate onto the reference sequence of pINV [69] (NC_024996.1). Mapped reads were sorted and indexed using Samtools v1.9 [70]. The individual gene coverage from mapping was obtained using Bedtools coverage v2.27.1 [71].
Identification of cluster-specific gene markers
Cluster-specific gene markers were identified from Shigella and EIEC accessory genomes. The genomes from the identification dataset were annotated using PROKKA v1.13.3 [72]. Pan- and core-genomes were analysed using roary v3.12.0 [73] using an 80% sequence identity threshold. An in-house python script was used to generate the candidate-specific gene markers for each cluster from the profile of gene presence or absence in each genome, which was produced by roary. The script is available on https://github.com/LanLab/ShigEiFinder/tree/main/scripts and the process to identify potential candidates is described in Dataset S1, Supplementary Material file 2. The best performing cluster-specific gene marker set was selected from the candidates by using blastn to search against the identification dataset.
In this study, the genomes from a given cluster containing all specific gene markers for that cluster were termed true positives (TP), and the genomes from the same cluster lacking any of those same gene markers were termed false negatives (FN). The genomes from other clusters containing all of those same gene markers were termed false positives (FP), the genomes from other clusters lacking any of those same gene markers were termed true negatives (TN). Relaxed cut-offs (40% FP) were used in initial screening to ensure that all clusters had candidate-specific gene markers which could be further investigated.
The sensitivity (true positive rate, TPR) of each cluster-specific gene marker was defined as TP/(TP +FN). The specificity (true negative rate, TNR) was defined as TN/(TN +FP).
Validation of the cluster-specific gene markers
The ability of cluster-specific gene markers to assign Shigella and EIEC isolates was examined by using blastn to search against the validation dataset (Table S2) and non-EIEC control database for the presence of any of the cluster-specific gene marker sets. The blastn thresholds were defined as 80% sequence identity and 50% gene length coverage.
Development of ShigEiFinder, an automated pipeline for molecular serotyping of Shigella and EIEC
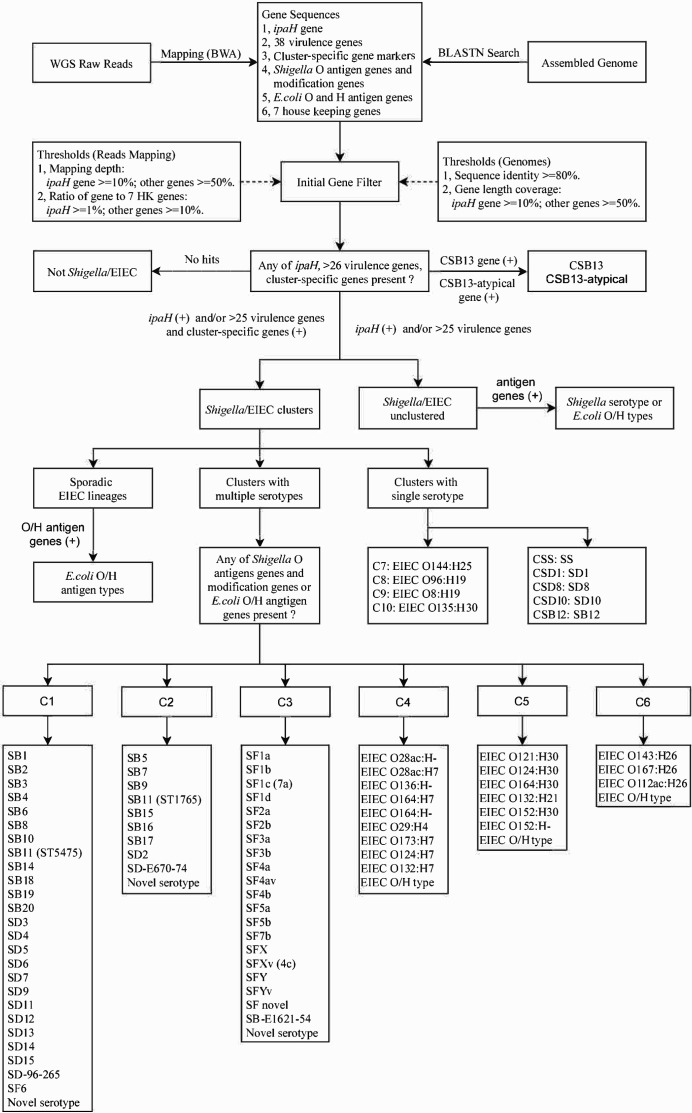
ShigEiFinder was developed using paired-end illumina genome sequencing reads or assembled genomes to type Shigella and EIEC isolates to the serotype level using cluster-specific gene markers combined with Shigella and EIEC serotype-specific O antigen genes (wzx and wzy) and modification genes (Fig. 2). Further details of the algorithms used are presented in Dataset S2. We used the same signature O and H sequences from ShigaTyper and SerotypeFinder (Dataset S3) [11, 63]. These include Shigella serotype-specific wzx/wzy genes and modification genes from ShigaTyper and E. coli O antigen and fliC (H antigen) genes from SerotypeFinder. The ipaH gene and 38 virulence genes used in analysis of virulence of 59 sporadic EIEC isolates were also included in the typing reference sequences database. Seven housekeeping (HK) genes, recA, purA, mdh, icd, gyrB, fumC and adk downloaded from NCBI, were used for contamination checking.
Fig. 2.
In silico serotyping pipeline workflow. Schematic of in silico serotyping of Shigella and EIEC (enteroinvasive E. coli ) by cluster-specific genes combined with the ipaH gene, O antigen and modification genes and H antigen genes, implemented in ShigEiFinder. Both assembled genomes and raw reads are accepted as data input. The dotted arrows show the cutoff value applied for initial gene filtering. WGS, whole-genome sequencing; HK, housekeeping; SS, SF, SB and SD, S. sonnei , S. flexneri , S. boydii and S. dysenteriae respectively. The abbreviated ‘species’ name plus the serotype number is the designation of a Shigella serotype (e.g. S. dysenteriae serotype 1 is abbreviated as SD1). For SB11, there were two sequence types (STs) with ST5475 and ST1765 located within clusters C1 and C2 respectively.
For raw reads input, raw reads were aligned to the typing reference sequences by using BWA-MEM v0.7.17 [68]. The mapping length percentage and the mean mapping depth for all genes were calculated using Samtools coverage v1.10 [70]. To determine whether the genes were present or absent, 50% of mapping length for all cluster-specific gene markers, virulence genes and O antigen genes, and 10% for the ipaH gene were used as cutoff values. The ratio of mean mapping depth to the mean mapping depth of the seven HK genes was used to determine a contamination threshold with ratios less than 1% for the ipaH gene and less than 10% for other genes assigned as contamination. Read coverage mapped to particular regions of genes was checked by using samtools mpileup v1.10 [70].
For assembled genome input, assembled genomes were searched against the typing reference sequences using blastn v2.9.0 [74] with 80% sequence identity and 50% gene length coverage for all genes, with the exception of the ipaH gene which was defined as 10% gene length coverage.
ShigEiFinder was tested with the identification dataset and validated with the Shigella and EIEC validation dataset and the non-EIEC control database. The specificity defined as (1 − the number of non-EIEC isolates being detected/the total number of non-EIEC isolates) × 100.
Results
Screening sequenced genomes for Shigella and EIEC isolates
We first screened available E. coli and Shigella genomes based on the presence of the ipaH gene. We examined 122361 isolates with the species annotation of E. coli (104256) or Shigella (18105) with paired-end Illumina sequencing reads available in the NCBI SRA database. Of 122361 isolates, 17989 were positive for the ipaH gene, including 455 out of 104256 E. coli isolates and 17434 out of 18105 Shigella isolates. The 17989 ipaH-positive E. coli and Shigella isolates and 571 ipaH-negative ‘Shigella’ isolates were checked for taxonomic classification and genome assembly quality . In total, 17320 ipaH-positive E. coli and Shigella genomes and 246 ipaH-negative ‘Shigella’ genomes passed quality filters. Among 246 ipaH-negative ‘Shigella’ isolates, 11 were predicted as SB13 by using ShigaTyper [11] while the remaining 235 were classified with a taxonomic identifier of E. coli by Kraken v1.1.1 [58] and their E. coli O/H antigen types predicted using SerotypeFinder were not classic EIEC serotypes or their O antigen was untypable. These 235 isolates were removed from analysis. A total of 17331 isolates including 17320 ipaH positives and 11 SB13 isolates were selected to form the Shigella and EIEC database. The Shigella and EIEC database contained 429 isolates with species identifier of E. coli and 16902 isolates with species identifier of Shigella .
Isolates in the Shigella and EIEC database were typed using MLST, ShigaTyper and SerotypeFinder. MLST and rMLST divided the 17331 Shigella and EIEC isolates into 252 STs (73 isolates were untypeable by MLST) and 1128 rSTs (3513 isolates were untypeable by rMLST). Of 16902 isolates with a species identifier of Shigella , 8313 and 8189 were typed as Shigella and EIEC respectively by ShigaTyper while 400 isolates were untypeable. ShigaTyper typed the majority of the 8313 isolates as SF (66.82%) including 25.43% SF2a isolates, followed by SS (19.69%), SB (7.22%) and SD (6.27%).
SerotypeFinder typed 293 of the 429 E. coli isolates into 71 E. coli O/H antigen types. Among these 293 isolates with typable O/H antigen types, 190 belonged to 22 known EIEC serotypes (O28ac:H-, O28ac:H7, O29:H4, O112ac:H26, O121:H30, O124:H30, O124:H24, O124:H7, O132:H7, O132:H21, O135:H30, O136:H7, O143:H26, O144:H25, O152:H-, O152:H30, O164:H-, O164:H30, O167:H26, O173:H7 and two newly emerged EIEC serotypes, O96:H19 and O8:H19) [20–22]. The remaining 136 of the 429 isolates were O antigen untypable and typed to 15 H antigen types by SerotypeFinder, of which H16 was the predominant type.
Identification of Shigella and EIEC clusters
Shigella and EIEC are known to have been derived from E. coli independently. To identify previously defined clusters [25, 26] and any new clusters from the 17331 Shigella and EIEC isolates, we selected representative isolates to perform phylogenetic analysis as it was impractical to construct a tree with all isolates. The selection was based on ST, rST and serotype of the 17331 Shigella and EIEC isolates. One isolate was selected to represent each ST, rST and serotype for a total of 1830 isolates. Note that in the case that STs or rSTs overlapped with serotype, an isolate would have only been selected once to avoid duplicates of the same isolate. The selection included 252 STs, 1128 rSTs, 59 Shigella serotypes (21 SB serotypes, 20 SF serotypes, 17 SD serotypes and one SS serotype), 22 EIEC known serotypes, and 31 other or partial antigen types. A further 31 in-house sequenced EIEC isolates, 18 EIEC isolates used in a previous typing study [41], 72 ECOR isolates [64] and 18 E. albertii isolates were also included to form the identification dataset of 1969 isolates. Details are listed in Table S1. A phylogenetic tree was constructed based on the identification dataset to identify the clusters (Fig. 1).
All known clusters were identified (Fig. 1) including three Shigella clusters (C1, C2, C3) and five outliers (SD1, SD8, SD10, SB13 and SS) as defined by Pupo et al. [25] and four EIEC clusters (C4, C5, C6 and C7) as defined by Lan et al. [26]. Each of these clusters was supported by a bootstrap value of 80% or greater (Fig. S1, Supplementary Material file 3). In total, 1789 of the 1879 Shigella and EIEC isolates (1830 isolates from the Shigella and EIEC database, 31 in-house sequenced EIEC isolates and 18 EIEC isolates from Hazen et al. [41]) fell within these clusters.
Of the remaining 90 Shigella and EIEC unclustered isolates, 31 belonged to typical or known Shigella or EIEC serotypes, including five SB13 isolates, eight SB12 isolates, two EIEC O135:H30 isolates, 12 EIEC O96:H19 isolates and four EIEC O8:H19 isolates, while 59 isolates were separated from the identified clusters by non- Shigella /EIEC isolates and interspersed among non- Shigella /EIEC isolates. Of these 59 isolates, 34 were singletons with a single member in the group, while the remaining 25 isolates formed 12 groups of two or more isolates. Furthermore their E. coli O/H antigen types were not classic EIEC serotypes or their O antigen was untypable. These 59 isolates were named as sporadic EIEC isolates which are described in detail in the separate section below.
The five SB13 isolates were grouped into one lineage within E. coli and close to known Shigella and EIEC clusters rather than the established SB13 cluster outside E. coli which was within the species E. albertii . The former was previously named as atypical SB13 while the latter was previously named as typical SB13 [39]. The eight SB12 isolates formed one single cluster close to SD1 and atypical SB13 clusters. SB12 was previously grouped into C3 based on HK gene trees [25, 28] but was seen as outliers in two other studies [28, 56]. Two EIEC O135:H30 isolates were grouped as a separate cluster close to C5. Twelve isolates belonging to EIEC serotype O96:H19 and four isolates typed as O8:H19 were clustered into two separate clusters, both of which were more closely related to SD8 than other Shigella and EIEC clusters. Each of these five groups was phylogenetically distinct and represented the classic Shigella or EIEC serotypes. Furthermore, each of the five groups was supported by a bootstrap value of 80% or greater (Fig. S1). Therefore, atypical SB13 and SB12 were defined as new clusters of Shigella while EIEC O96:H19, EIEC O8:H19 and EIEC O135:H30 were defined as C8, C9 and C10 respectively. In total there were 10 Shigella clusters and seven EIEC clusters (Table 1).
Table 1.
Summary of identified Shigella and EIEC clusters and outliers in the identification dataset
|
Clusters (no. of serotypes)* |
No. of isolates |
No. of STs |
No. of rSTs |
Serotypes |
|---|---|---|---|---|
|
C1 (25) |
288 |
36 |
166 |
SB1–4, SB6, SB8, SB10, SB14, SB18, SB11†, SB19–20†; SD3–7, SD9, SD11–13, SD14–15†, SD96-26b†; SF6 |
|
C2 (9) |
101 |
19 |
56 |
SB5, SB7, SB9, SB11, SB15, SB16, SB17; SD2, SD-E670-74†; SD2 |
|
C3 (20) |
744 |
81 |
437 |
SF1a, SF1b, SF1c (7 a), SF2a, SF2b, SF3a, SF3b, SF4a, SF4av, SF4b, SF4bv, SF5a, SF5b, SF7b, SFX, SFXv (4 c), SFY, SFYv, SF novel serotype; SB-E1621-54† |
|
C4 (9) |
51 |
6 |
21 |
O28ac:H-, O28ac:H7, O136:H7, O164:H-, O164:H7, O29:H4, O173:H7, O124:H7, O132:H7† |
|
C5 (6) |
62 |
4 |
15 |
O121:H30, O124:H30, O164:H30, O132:H21, O152:H30, O152:H- |
|
C6 (3) |
20 |
2 |
6 |
O143:H26, O167:H26, O112ac:H26† |
|
C7 |
10 |
1 |
3 |
O144:H25 |
|
C8‡ |
12 |
2 |
1 |
O96:H19 |
|
C9‡ |
4 |
1 |
2 |
O8:H19 |
|
C10‡ |
2 |
1 |
1 |
O135:H30 |
|
CSS |
427 |
39 |
294 |
|
|
CSD1 |
70 |
8 |
56 |
SD1 |
|
CSD8 |
7 |
3 |
3 |
SD8 |
|
CSD10 |
2 |
2 |
1 |
SD10 |
|
CSB12‡ |
8 |
2 |
6 |
SB12 |
|
CSB13 |
7 |
3 |
3 |
SB13 |
|
CSB13-atypical‡ |
5 |
3 |
3 |
SB13 |
|
Sporadic EIEC lineages‡ [53] |
59 |
49 |
53 |
53 antigen types |
*Numbers in parentheses are the number of serotypes within that cluster.
†Serotypes were inconsistent with previous analyses.
‡Clusters identified as new clusters in this study.
Analysis of the 59 sporadic EIEC isolates
To determine the phylogenetic relationships of the above defined clusters and the remaining 59 sporadic EIEC isolates within the larger non-EIEC population, a confirmation tree was generated using 485 isolates representing the known clusters and 1872 representative non- Shigella /EIEC isolates (Fig. S2). The 59 sporadic EIEC isolates were interspersed among non- Shigella /EIEC isolates and did not form large clusters. Groups of these isolates that were not previously identified were named as sporadic EIEC lineages followed by their serotype. For example, isolate M2330 (O152:H51), which was sequenced in this study, was named ‘sporadic EIEC lineage O152:H51’. There were 53 sporadic EIEC lineages including five lineages with two or more isolates and 48 lineages with only one isolate. The STs, rSTs and antigen types of these 59 isolates are listed in Table S1.
Some of the sporadic EIEC isolates fell into STs containing ipaH-negative isolates. We therefore examined the presence of the pINV virulence plasmid in the sporadic EIEC isolates. We selected 38 genes that are essential for virulence, including 35 genes (12 mxi genes, nine spa genes, five ipaA-J genes, six ipgA-F genes as well as acp, virB and icsB) in the conserved entry region encoding the Mxi-Spa-Ipa type III secretion system and its effectors and three regulator genes (virF, virA and icsA/virG) [24, 32, 69], and determined the presence of pINV in the 59 sporadic EIEC isolates by mapping the sequence reads onto a pINV reference sequence [69]. Reads from 18 non- Shigella /EIEC isolates that shared the same ST as one of 59 sporadic isolates were also mapped onto a pINV reference sequence [69].
The distribution of essential virulence genes with mapped reads in the 59 sporadic EIEC isolates was analysed (Fig. S3). Based on the distribution of the number of virulence genes present among the 59 isolates, 26 was selected as the minimum for the pINV to be considered present. Thus, this number was used as the cutoff to call pINV presence/absence. Those isolates containing more than 25 of the 38 essential virulence genes were defined as virulence plasmid positive, while isolates containing between 13 and 25 were defined as indeterminate and fewer than 13 were defined as virulence plasmid negative.
The two newly sequenced sporadic EIEC isolates (M2330 and M2339) were positive for the virulence plasmid, and of the other 57 sporadic EIEC isolates, 39 were positive, nine were negative and another nine were indeterminate (Table S1). The results were compared with 18 non- Shigella /EIEC isolates mentioned above. The virulence plasmid was absent in all non- Shigella /EIEC isolates while all sporadic EIEC isolates in these STs were either positive or indeterminate. Therefore, this analysis confirmed the sporadic isolates belonged to EIEC and the STs contained both EIEC and non-EIEC isolates.
Identification of cluster-specific gene markers
In this study, a cluster-specific gene marker set (single gene or two or more genes) was present in all isolates of a cluster and absent in all isolates of other cluters. For the marker sets with two or more genes, a subset of cluster-specific genes for a given cluster could be found in other clusters but the entire set was only found in the target cluster.
Comparative genomic analysis on 1969 accessory genomes from the identification dataset was used to identify the potential cluster-specific gene marker sets. Multiple candidate cluster-specific gene marker sets for each of the 17 Shigella and EIEC clusters and 53 sporadic EIEC lineages were identified through initial screening of the accessory genes from the 1969 genomes. Genes associated with Shigella and EIEC O antigen clusters were excluded from the analysis. The candidate cluster-specific gene marker sets were 100% sensitive to clusters but with varying specificity. The cluster-specific gene marker sets with the lowest FP rates were then selected from candidate cluster-specific gene marker sets by blastn searches against genomes in the identification dataset using 80% sequence identity and 50% gene length threshold.
The final cluster-specific gene marker sets were all 100% sensitive and 100% specific, with the exception of those for C1 (99.94% specificity), C3 (99.91% specificity) and SS (99.8% specificity). The sensitivity and specificity for each cluster-specific gene marker or marker set for the identification dataset are listed in Table 2. A single specific gene for each of the 53 sporadic EIEC lineages was also selected with the exception of sporadic EIEC lineage 27 which had a set of two genes. These genes were all 100% sensitive and specific for a given sporadic EIEC lineage.
Table 2.
The sensitivity and specificity of cluster-specific genes
|
Cluster |
Cluster-specific genes (single/sets) |
Identification dataset (1969 isolates) |
||
|---|---|---|---|---|
|
No. of isolates |
Sensitivity |
Specificity |
||
|
C1 |
Set of 4 genes |
288 |
100 |
99.94* |
|
C2 |
Set of 3 genes |
101 |
100 |
100 |
|
C3 |
Set of 3 genes |
744 |
100 |
99.59* |
|
C4 |
Set of 2 genes |
51 |
100 |
100 |
|
C5 |
Set of 3 genes |
62 |
100 |
100 |
|
C6 |
Set of 2 genes |
20 |
100 |
100 |
|
C7 |
Single gene |
10 |
100 |
100 |
|
C8 |
Set of 2 genes |
12 |
100 |
100 |
|
C9 |
Set of 2 genes |
4 |
100 |
100 |
|
C10 |
Single gene |
2 |
100 |
100 |
|
CSS |
Set of 5 genes |
427 |
100 |
99.87* |
|
CSD1 |
Set of 2 genes |
70 |
100 |
100 |
|
CSD8 |
Single gene |
7 |
100 |
100 |
|
CSD10 |
Single gene |
2 |
100 |
100 |
|
CSB12 |
Single gene |
8 |
100 |
100 |
|
CSB13 |
Single gene |
7 |
100 |
100 |
|
CSB13-atypical |
Single gene |
5 |
100 |
100 |
|
53 Sporadic EIEC lineages |
Single gene/lineage |
59 |
100 |
100 |
*A cluster-specific gene set specificity of less than 100% was due to at least one FP found in that set.
All 37 cluster-specific gene markers and 54 sporadic EIEC lineage-specific gene markers were located on the chromosome except for one of the C4 gene markers and five sporadic EIEC lineage-specific genes which were located on plasmids by NCBI blast searches. None of the cluster-specific gene markers was contiguous in the genomes. The location of these cluster-specific gene markers was determined by blastn against representative complete genomes of Shigella and EIEC containing gene features downloaded from GenBank (accession numbers listed in Table S3). In those cluster or sporadic lineages with no representative complete genome, specific gene markers were named using their cluster or sporadic EIEC lineage followed by the cluster or lineage number. For example, the C7 specific gene marker was named ‘C7 specific gene’.
Validation of cluster-specific gene markers
The ability of cluster-specific gene markers to correctly assign Shigella and EIEC isolates to a cluster was evaluated with 15501 Shigella and EIEC isolates in the validation dataset and 12743 isolates from the non-EIEC control database. Using cluster-specific gene markers, 15442 of the 15501 (99.63%) Shigella and EIEC isolates were assigned to a single cluster, which included 15336 Shigella isolates, 102 EIEC isolates and four sporadic EIEC isolates. However, 38 (0.24%) isolates were assigned to more than one cluster and 21 isolates were not assigned to any of the identified clusters.
To confirm the cluster assignment by cluster-specific gene markers, we divided the 15501 validation isolates into seven groups as it was impractical to construct a tree with all 15501 genomes. We then constructed seven ‘validation’ phylogenetic trees (Fig. S4) using each of the seven groups and a subset of 575 isolates from the identification dataset consisting of 485 isolates representing each cluster, 72 ECOR isolates and 18 E. albertii strains. The cluster identity of a ‘validation’ isolate was confirmed if the isolate was found manually within a branch that exclusively contained identification dataset isolates from that cluster or lineage and that the branch had a bootstrap support value of 80 % or greater (Fig. S4). The seven phylogenies of 15501 validation isolates showed that all 15501 isolates were assigned to expected clusters with the exception of four isolates which were not grouped with any of the identified clusters or sporadic EIEC lineages (Table S2, column E).
Compared to cluster assignment by phylogenetic trees as the ground truth, cluster-specific gene markers assigned 15442 of the 15501 (99.63%) Shigella and EIEC isolates correctly to clusters and correctly identified three of the 21 isolates without cluster assignments. The accuracy of cluster assignments by cluster-specific gene markers was 99.64%. The sensitivity and specificity for each cluster-specific gene marker set for the validation dataset are listed in Table S4.
We tested cluster-specific gene markers with the 12743 non-EIEC isolates. The Shigella and EIEC cluster-specific gene markers were highly specific with specificity varying from 98.8 to 100% for cluster-specific gene markers and from 97.02 to 100% for sporadic EIEC-specific gene markers. Details are listed in Table S4.
Development of an automated pipeline for molecular serotyping of Shigella and EIEC
The above results showed that cluster-specific gene markers were sensitive and specific and can distinguish Shigella and EIEC isolates. Therefore, we used these gene markers combined with established Shigella and EIEC serotype-specific O and H antigen genes to develop an automated pipeline for in silico serotyping of Shigella and EIEC (Fig. 2). The pipeline is named Shigella EIEC Cluster Enhanced Serotype Finder (ShigEiFinder). ShigEiFinder can process either paired-end Illumina sequencing reads or assembled genomes (installable package: https://github.com/LanLab/ShigEiFinder, online tool: https://mgtdb.unsw.edu.au/ShigEiFinder/). Details of the performance and algorithms incorporated into ShigEiFinder are documented in Dataset S2.
ShigEiFinder classifies isolates into three categories: Not Shigella /EIEC, Shigella or EIEC clusters, and Shigella or EIEC unclustered, based on the presence of the ipaH gene, the number of virulence genes and the entire cluster-specific gene marker set. The ‘Not Shigella /EIEC’ assignment was determined by the absence of the ipaH gene, fewer than 26 pINV encoded virulence genes and the absence of the entire cluster-specific gene marker set. The cutoff for the number of virulence genes to call the presence or absence of pINV was determined above. The ‘ Shigella or EIEC clusters’ assignment was made based on the presence of the ipaH gene, and/or more than 25 pINV encoded virulence genes together with the presence of any of the entire cluster-specific gene marker set. The presence of the ipaH gene and/or more than 25 pINV encoded virulence genes with the absence of the entire cluster-specific gene marker set was assigned as ‘ Shigella or EIEC unclustered’.
Shigella and EIEC isolates were differentiated and serotypes were assigned after cluster assignment. ShigEiFinder predicts a serotype through examining the presence of any established Shigella serotype-specific O antigen and modification genes and E. coli O and H antigen genes that differentiate the serotypes as used by ShigaTyper and SerotypeFinder [11, 63]. A ‘novel serotype’ is assigned if there is no match to known serotypes.
Two pairs of Shigella serotypes, SB1/SB20 and SB6/SB10, are known to be difficult to differentiate as they share identical O antigen genes [11, 46, 75]. ShigaTyper used a heparinase gene for the differentiation of SB20 from SB1 and the wbaM gene for the separation of SB6 from SB10. We found that fragments of the heparinase and wbaM genes may be present in other serotypes and cannot accurately differentiate SB1/SB20 and SB6/SB10. We identified a SB20-specific gene which encoded a hypothetical protein with unknown function and was located on a plasmid by comparative genomic analysis of all isolates in C1 accessory genomes. The SB20-specific gene can reliably differentiate SB20 from SB1 and also one SNP each in wzx and wzy genes can differentiate SB6 from SB10. We used these differences (Dataset S2)in ShigEiFinder for the prediction of these serotypes.
The accuracy and specificity of ShigEiFinder in cluster typing
The accuracy of ShigEiFinder was tested with 1969 isolates [1969 assembled genomes and 1951 Illumina reads (note no reads available for 18 EIEC isolates from NCBI)] from the identification dataset and 15501 isolates (15501 assembled genomes and 15501 Illumina reads) from the validation dataset. The results are listed in Table 3.
Table 3.
The accuracy of ShigEiFinder with the identification dataset and validation dataset
|
ShigEiFinder assignments |
Identification dataset (n=1969)* |
Validation dataset (n=15501) |
||
|---|---|---|---|---|
|
Assembled genome |
Read mapping |
Assembled genome |
Read mapping |
|
|
Shigella or EIEC clusters |
1871 |
1848 |
15455 |
15471 |
|
Multiple Shigella or EIEC clusters |
9 |
6 |
33 |
7 |
|
Shigella or EIEC unclustered |
0 |
8 |
13 |
23 |
|
Not Shigella /EIEC |
89 |
89 |
0 |
0 |
|
Accuracy† |
99.54% |
99.28% |
99.70% |
99.81% |
*Reads were not available for 18 EIEC isolates downloaded from NCBI in the identification dataset. The identification dataset has 90 non-Shigella/EIEC isolates including 72 ECOR isolates and 18 E.albertii isolates. One E. albertii isolate was assigned to SB13 by ShigaTyper which was grouped into cluster SB13 on the phylogenetic tree.
†Accuracy was defined as the number of Shigella and EIEC isolates being correctly assigned to a cluster over the total number tested.
ShigEiFinder was able to assign 99.54 and 99.28% of the isolates in the identification dataset to clusters for assembled genomes and read mapping respectively. Accuracy was 99.70 and 99.81% for assembled genomes and read mapping respectively when applied to the validation dataset. Discrepancies were observed between assembled genomes and read mapping (Table 3). There were more isolates assigned to ‘ Shigella or EIEC unclustered’ in read mapping, whereas there were more isolates assigned to multiple clusters in genome assemblies. The specificity of ShigEiFinder was 99.40% for assembled genomes and 99.38% for read mapping when evaluated with 12743 non- Shigella /non-EIEC isolates. An additional two isolates were detected as sporadic EIEC lineages by read mapping.
Comparison of ShigEiFinder and ShigaTyper
To demonstrate the use of ShigEiFinder for differentiation of Shigella from EIEC and enhancement of cluster-based serotyping, the comparison of read mapping results between ShigEiFinder and the existing in silico Shigella identification pipeline ShigaTyper [11] was performed. Since ShigaTyper recommends the use of read mapping, we compared ShigEiFinder read mapping results with ShigaTyper read mapping results.
The 488 isolates used in Wu et al. [11] were tested using ShigEiFinder. These 488 isolates consisted of 25 EIEC isolates, 420 Shigella isolates and 45 non- Shigella /non-EIEC isolates. The assignment of 477 of 488 isolates by ShigEiFinder agreed with that by ShigaTyper. Of the remaining 11 isolates (one EIEC isolate and 10 Shigella isolates), two Shigella isolates were assigned to EIEC and eight Shigella isolates and one EIEC isolate were untypeable (either multiple wzx or no wzx genes found) by ShigaTyper, whereas one EIEC isolate was assigned to EIEC (C4) and 10 Shigella isolates were assigned to Shigella clusters by ShigEiFinder.
The read mapping results for 15501 Shigella and EIEC isolates from the validation dataset were then compared. ShigEiFinder assigned 15460 of 15501 Shigella and EIEC isolates to Shigella or EIEC clusters and then to a serotype. By contrast, ShigaTyper assigned 7277 isolates to Shigella , 7976 isolates to EIEC and 177 isolates to multiple wzx genes, and failed to type 71 isolates. A total of 7353 isolates predicted as Shigella (7252) or EIEC (101) by ShigaTyper agreed with the results of ShigEiFinder (Table 4). For the 8148 isolates typed as EIEC or untypable by ShigaTyper, 8107 were assigned to Shigella or EIEC clusters by ShigEiFinder (Table 4). Of these isolates, the majority belonged to SS, SD1 and SF, which were erroneously predicted as EIEC by ShigaTyper.
Table 4.
The assignments of 15501 validation isolates by ShigEiFinder and Shigatyper
|
ShigEiFinder assignment |
ShigaTyper assignment |
Total |
|||
|---|---|---|---|---|---|
|
Agreement with ShigEiFinder |
Discrepant with ShigEiFinder |
||||
|
EIEC |
Non-assignment* |
||||
|
SS |
1515 |
0 |
7465 |
19 |
8999 |
|
SF |
4644 |
0 |
117 |
71 |
4832 |
|
C1 and C2 (SB and SD) |
1004 |
0 |
17 |
151 |
1172 |
|
SB12 |
4 |
0 |
0 |
2 |
6 |
|
SB13 |
1 |
0 |
0 |
0 |
1 |
|
SB13-atypical |
2 |
0 |
0 |
0 |
2 |
|
SD1 |
80 |
0 |
244 |
2 |
326 |
|
SD8 |
2 |
0 |
1 |
0 |
3 |
|
SD10 |
0 |
0 |
0 |
1 |
1 |
|
EIEC |
101 |
1 |
0 |
0 |
102 |
|
Sporadic EIEC lineages |
0 |
1 |
15 |
0 |
16 |
|
Multiple clusters |
0 |
0 |
5 |
2 |
7 |
|
Shigella or EIEC unclustered |
0 |
23 |
11 |
0 |
34 |
|
Total |
7353 |
25 |
7875 |
248 |
15501 |
*Non-assignment: multiple wzx genes and non-prediction.
Compared to the phylogenetic analysis results of cluster identity of the isolates as ground truth, ShigEiFinder had 99.74% (15460/15501) accuracy to differentiate Shigella isolates from EIEC, while ShigaTyper assigned only 47.6% isolates correctly in the same dataset we tested.
Discussion
Determining phylogenetic clusters for better separation of Shigella isolates from EIEC
From a phylogenetic perspective, Shigella and EIEC strains consisted of multiple phylogenetic lineages derived from commensal E. coli , which do not reflect the taxonomic classification of Shigella as a genus [23, 25, 26, 28, 38, 41]. In the present study, we identified all phylogenetic clusters of Shigella and EIEC through large-scale examination of publicly available genomes. The phylogenetic results demonstrated that Shigella isolates had at least 10 clusters while EIEC isolates had at least seven clusters. The 10 Shigella clusters included the eight previously defined lineages including three major clusters (C1, C2 and C3) and five outliers (SD1, SD8, SD10, SB13 and SS) [25] and two newly identified clusters (SB12 and SB13-atypical). The seven EIEC clusters consisted of four previously defined EIEC clusters (C4, C5, C6 and C7) [26] and three newly identified EIEC clusters (C8, C9 and C10).
Our WGS-based phylogeny provided high resolution for assigning Shigella and EIEC isolates to clusters. Several serotypes that are currently increasing in frequency (SB19, SB20, SD14, SD15, SD provisional serotype 96-626) [76–79] were assigned to clusters and five new clusters/outliers were identified. Newly identified clusters C8 (EIEC O96:H19) and C9 (EIEC O8:H19) represented the emergence of novel EIEC serotypes. A recent study revealed that EIEC serotype O96:H19 (C8) could be the result of a recent acquisition of the invasion plasmid by commensal E. coli [80]. The EIEC serotype O8:H19 (C9) had not been reported previously.
Apart from the 17 major clusters of Shigella and EIEC, the presence of 53 sporadic EIEC lineages indicated greater genetic diversity than has been observed previously. Isolates belonging to these sporadic EIEC lineages were more closely related to non-EIEC isolates than to major Shigella and EIEC lineages. However, 41 of these isolates, representing 38 sporadic EIEC lineages, carried pINV. Shigella and EIEC both carry the Shigella virulence plasmid pINV which is vital for virulence and distinguishes Shigella and EIEC from other E. coli [24, 32, 69]. Therefore, these isolates may represent recently formed EIEC lineages through acquisition of the pINV. The remaining 18 isolates contained the ipaH gene but may or may not carry pINV. It is possible that these strains carried a very low copy number of the pINV or the pINV plasmid was lost during isolation or culture.
Highly sensitive and cluster-specific gene markers for differentiation of Shigella and EIEC isolates
The cluster-specific gene marker sets can be used to differentiate Shigella and EIEC from non-EIEC independent of the presence of the ipaH gene. The ipaH gene as a molecular target has been used to differentiate Shigella and EIEC from non-EIEC [24, 43–45]. In our study, the cluster-specific gene markers were specific to Shigella and EIEC with 98.8–100% specificity when evaluated on a non-EIEC control dataset, providing confidence that the cluster-specific genes or sets are robust markers for the identification of Shigella and EIEC.
Several studies have identified phylogenetically related genomic markers for discrimination of Shigella and EIEC [23, 27, 28, 41, 55, 56]. However, these phylogenetic analyses were performed only with a small number of genomes [23, 28, 55]. In addition, non-EIEC isolates were included in some of the phylogenetic clusters identified [28], which led to non-EIEC isolates being identified by the markers. We identified cluster-specific gene markers for each respective cluster which were exclusively composed of Shigella or EIEC isolates. A previous study identified six loci to distinguish EIEC from Shigella [23]. We searched the six loci against our Shigella and EIEC database and found that some Shigella isolates were misidentified as EIEC; for example, SD8 isolates were incorrectly identified as EIEC subtype 13. Our cluster-specific genes can differentiate SD8 from EIEC with 100% accuracy. Overall, the cluster-specific gene marker sets described here provided nearly perfect differentiation of Shigella from EIEC.
The cluster-specific gene marker sets can differentiate SS and SF (with the exception of SF6) from SB and SD. SF and SS are the major cause of Shigella infections, accounting for up to 89.6% of annual cases [10, 12, 13]. Differentiation of SS and SF isolates from SB and SD is also beneficial for diagnosis and surveillance. A recent study identified ‘species’-specific markers for the detection of each of the four Shigella ‘species’ and validated with only one isolate per species [55]. By contrast, a set of SF-specific genes and SS-specific genes in our study can correctly identify SF isolates and SS isolates with 99.64% accuracy when applied to 15501 Shigella and EIEC isolates.
It should be noted that we were unable to validate cluster-specific gene markers of C6, C7, C10 and CSD10. These clusters are rare and once isolates were included in the identification dataset, none remained for validation. Therefore, the markers for the C6, C7, C10 and SD10 clusters are tentative and require future validation when more genomes become available. Genes specific to each of the 53 sporadic EIEC lineages were also based on a very small number of genomes and should be used with caution. However, since these sporadic lineages are very low in frequency, they may be rarely encountered in practice and thus have relatively little effect on the overall applicability of the lineage-specific markers to Shigella and EIEC typing.
ShigEiFinder can accurately type Shigella and EIEC
ShigEiFinder can accurately differentiate Shigella from EIEC whereas there were a large proportion of isolates incorrectly assigned by ShigaTyper. The majority of the isolates predicted as EIEC by ShigaTyper were SS or SD1 as they belonged to SS- and SD1-specific STs and were positive to a set of SS- or SD1-specific gene markers and grouped into SS or SD1 clusters on our phylogenetic trees. The genes used in ShigaTyper were the SS-specific marker Ss_methylase gene [81, 82] together with SS O antigen wzx gene. However, the SS-specific marker Ss_methylase gene was found in other Shigella serotypes and EIEC [11] and SS O antigen wzx gene was located on a plasmid which is frequently lost [83]. Similarly, the SD1 O antigen genes used in ShigaTyper were plasmid-borne, which may also lead to inconsistent detection [84, 85]. By contrast, the cluster-specific gene markers used in ShigEiFinder for identification of Shigella and EIEC and nearly all chromosomal and provided higher discriminatory power than ShigaTyper.
ShigEiFinder was able to serotype over 59 Shigella serotypes and 22 EIEC serotypes. ShigEiFinder can assign Shigella and EIEC isolates to the serotype level using cluster-specific markers to enhance its accuracy. For clusters containing more than one serotype, including the major Shigella and EIEC clusters C1–C6, once an isolate is assigned to a cluster, only serotype-associated O antigen and modification genes found in that cluster need be examined. This allows the elimination of ambiguous or incorrect serotype assignments that may otherwise occur, increasing the overall accuracy of the method. For the clusters that contain only one serotype such as SD1, SD8, SD10, SB13, SB12 and EIEC C7–C10, cluster-specific markers can also be used a proxy for serotyping but with increased robustness when the combination of cluster-specific gene marker and O antigen and modification genes was used.
ShigEiFinder will be useful for clinical, epidemiological and diagnostic investigations, and the cluster-specific gene markers identified could be adapted for metagenomics or culture-independent typing.
Conclusion
This study analysed over 17000 publicly available Shigella and EIEC isolates and identified 10 clusters of Shigella , seven clusters of EIEC and 53 sporadic types of EIEC. Cluster-specific gene marker sets for the 17 major clusters and 53 sporadic types were identified and found to be valuable for in silico typing. We additionally developed ShigEiFinder, a freely available in silico serotyping pipeline, incorporating the cluster-specific gene markers to facilitate serotyping of Shigella and EIEC isolates using genome sequences with very high specificity and sensitivity.
Supplementary Data
Funding information
This work was funded in part by a National Health and Medical Research Council project grant (grant number 1129713) and an Australian Research Council Discovery Grant (DP170101917).
Acknowledgements
The authors thank Duncan Smith and Robin Heron from UNSW Research Technology Services for high performance computing assistance.
Author contributions
Conceptualization: R.L., M.P.; Investigation: X.Z., M.P., T.N., S.K.; Methodology: M.P., R.L., X.Z.; Writing – original draft: X.Z.; Writing – review and editing: M.P., R.L.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Footnotes
Abbreviations: ECOR, Escherichia coli reference collection; EIEC, enteroinvasive Escherichia coli; FN, false negative; FP, false positive; HK, housekeeping; MLST, multilocus sequence typing; NCBI SRA, National Center for Biotechnology Information Sequence Read Archive; rMLST, ribosomal MLST; rST, ribosomal ST; SB, Shigella boydii; SD, Shigella dysenteriae; SF, Shigella flexneri; SS, Shigella sonnei; ST, sequence type; TP, true positive; WGS, whole-genome sequencing.
All supporting data, code and protocols have been provided within the article or through supplementary data files. Three supplementary data files, five supplementary tables and four supplementary figures are available with the online version of this article.
References
- 1.DuPont HL, Levine MM, Hornick RB, Formal SB. oculum size in shigellosis and implications for expected mode of transmission. J Infect Dis. 1989;159:1126–1128. doi: 10.1093/infdis/159.6.1126. [DOI] [PubMed] [Google Scholar]
- 2.GBD Diarrhoeal Diseases Collaborators Estimates of global, regional, and national morbidity, mortality, and aetiologies of diarrhoeal diseases: a systematic analysis for the Global Burden of Disease Study 2015. Lancet Infect Dis. 2017;17:909–948.:S1473-3099(17)30276-1. doi: 10.1016/S1473-3099(17)30276-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kirk MD, Pires SM, Black RE, Caipo M, Crump JA, et al. Correction: World Health Organization Estimates of the Global and Regional Disease Burden of 22 Foodborne Bacterial, Protozoal, and Viral Diseases, 2010: A Data Synthesis. PLoS Med. 2015;12:e1001940. doi: 10.1371/journal.pmed.1001940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.World HO. Guidelines for the Control of Shigellosis, Including Epidemics Due to Shigella dysenteriae Type 1. World Health Organization; 2005. [Google Scholar]
- 5.Brengi SP, Sun Q, Bolaños H, Duarte F, Jenkins C, et al. PCR-based method for Shigella flexneri serotyping: international multicenter validation. J Clin Microbiol. 2019;57:e01592-18. doi: 10.1128/JCM.01592-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Khalil IA, Troeger C, Blacker BF, Rao PC, Brown A, et al. Morbidity and mortality due to Shigella and enterotoxigenic Escherichia coli diarrhoea: the Global Burden of Disease Study 1990-2016. Lancet Infect Dis. 2018;18:1229–1240.:S1473-3099(18)30475-4. doi: 10.1016/S1473-3099(18)30475-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kotloff KL, Nataro JP, Blackwelder WC, Nasrin D, Farag TH, et al. Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): a prospective, case-control study. Lancet. 2013;382:209–222.:S0140-6736(13)60844-2. doi: 10.1016/S0140-6736(13)60844-2. [DOI] [PubMed] [Google Scholar]
- 8.van den Beld MJC, Warmelink E, Friedrich AW, Reubsaet FAG, Schipper M, et al. Incidence, clinical implications and impact on public health of infections with Shigella spp. and entero-invasive Escherichia coli (EIEC): results of a multicenter cross-sectional study in the Netherlands during 2016-2017. BMC Infect Dis. 2019;19:1037. doi: 10.1186/s12879-019-4659-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Edwards PR, Ewing WH. Identification of Enterobacteriaceae. Identification of Enterobacteriaceae. 3rd ed. 1972. [Google Scholar]
- 10.The HC. Thanh DP, Holt KE, Thomson NR, Baker S. The genomic signatures of Shigella evolution, adaptation and geographical spread. Nat Rev Microbiol. 2016;14:235–250. doi: 10.1038/nrmicro.2016.10. [DOI] [PubMed] [Google Scholar]
- 11.Wu Y, Lau HK, Lee T, Lau DK, Payne J, et al. In silico serotyping based on whole-genome sequencing improves the accuracy of Shigella identification. Appl Environ Microbiol. 2019;85 doi: 10.1128/AEM.00165-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Livio S, Strockbine NA, Panchalingam S, Tennant SM, Barry EM, et al. Shigella isolates from the global enteric multicenter study inform vaccine development. Clin Infect Dis. 2014;59:933–941. doi: 10.1093/cid/ciu468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Group OW. Monitoring the incidence and causes of diseases potentially transmitted by food in Australia: Annual report of the OzFoodNet network, 2011. Commun Dis Intell Q Rep. 2015;39:E236. [PubMed] [Google Scholar]
- 14.Connor TR, Barker CR, Baker KS, Weill F-X, Talukder KA, et al. Species-wide whole genome sequencing reveals historical global spread and recent local persistence in Shigella flexneri. Elife. 2015;4:e07335. doi: 10.7554/eLife.07335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gomes TAT, Elias WP, Scaletsky ICA, Guth BEC, Rodrigues JF, et al. Diarrheagenic Escherichia coli . Braz J Microbiol. 2016;47 Suppl 1:3–30.:S1517-8382(16)31091-7. doi: 10.1016/j.bjm.2016.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Levine MM. Escherichia coli that cause diarrhea: enterotoxigenic, enteropathogenic, enteroinvasive, enterohemorrhagic, and enteroadherent. J Infect Dis. 1987;155:377–389. doi: 10.1093/infdis/155.3.377. [DOI] [PubMed] [Google Scholar]
- 17.Tai AYC, Easton M, Encena J, Rotty J, Valcanis M, et al. A review of the public health management of shigellosis in Australia in the era of culture-independent diagnostic testing. Aust N Z J Public Health. 2016;40:588–591. doi: 10.1111/1753-6405.12590. [DOI] [PubMed] [Google Scholar]
- 18.Taylor DN, Echeverria P, Sethabutr O, Pitarangsi C, Leksomboon U, et al. Clinical and microbiologic features of Shigella and enteroinvasive Escherichia coli infections detected by DNA hybridization. J Clin Microbiol. 1988;26:1362–1366. doi: 10.1128/jcm.26.7.1362-1366.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pasqua M, Michelacci V, Di Martino ML, Tozzoli R, Grossi M, et al. The Intriguing Evolutionary Journey of Enteroinvasive E. coli (EIEC) toward Pathogenicity. Front Microbiol. 2017;8:2390. doi: 10.3389/fmicb.2017.02390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Herzig CTA, Fleischauer AT, Lackey B, Lee N, Lawson T, et al. Notes from the Field: Enteroinvasive Escherichia coli Outbreak Associated with a Potluck Party - North Carolina, June-July 2018. MMWR Morb Mortal Wkly Rep. 2019;68:183–184. doi: 10.15585/mmwr.mm6807a5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pettengill EA, Hoffmann M, Binet R, Roberts RJ, Payne J, et al. Complete genome sequence of enteroinvasive Escherichia coli O96:H19 associated with a severe foodborne outbreak. Genome Announc. 2015;3:e00883-15. doi: 10.1128/genomeA.00883-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Escher M, Scavia G, Morabito S, Tozzoli R, Maugliani A, et al. A severe foodborne outbreak of diarrhoea linked to a canteen in Italy caused by enteroinvasive Escherichia coli, an uncommon agent. Epidemiol Infect. 2014;142:2559–2566. doi: 10.1017/S0950268814000181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dhakal R, Wang Q, Lan R, Howard P, Sintchenko V. Novel multiplex PCR assay for identification and subtyping of enteroinvasive Escherichia coli and differentiation from Shigella based on target genes selected by comparative genomics. J Med Microbiol. 2018;67:1257–1264. doi: 10.1099/jmm.0.000784. [DOI] [PubMed] [Google Scholar]
- 24.van den Beld MJ, Reubsaet FA. Differentiation between Shigella, enteroinvasive Escherichia coli (EIEC) and noninvasive Escherichia coli . European journal of clinical microbiology & infectious diseases: official publication of the European Society of Clinical Microbiology. 2012;31:899–904. doi: 10.1007/s10096-011-1395-7. [DOI] [PubMed] [Google Scholar]
- 25.Pupo GM, Lan R, Reeves PR. Multiple independent origins of Shigella clones of Escherichia coli and convergent evolution of many of their characteristics. Proc Natl Acad Sci U S A. 2000;97:10567–10572. doi: 10.1073/pnas.180094797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lan R, Alles MC, Donohoe K, Martinez MB, Reeves PR. Molecular evolutionary relationships of enteroinvasive Escherichia coli and Shigella spp. Infect Immun. 2004;72:5080–5088. doi: 10.1128/IAI.72.9.5080-5088.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sahl JW, Morris CR, Emberger J, Fraser CM, Ochieng JB, et al. Defining the phylogenomics of Shigella species: a pathway to diagnostics. J Clin Microbiol. 2015;53:951–960. doi: 10.1128/JCM.03527-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pettengill EA, Pettengill JB, Binet R. Phylogenetic analyses of Shigella and enteroinvasive Escherichia coli for the identification of molecular epidemiological markers: whole-genome comparative analysis does not support distinct genera designation. Front Microbiol. 2015;6:1573. doi: 10.3389/fmicb.2015.01573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cheasty T, Rowe B. Antigenic relationships between the enteroinvasive Escherichia coli O antigens O28ac, O112ac, O124, O136, O143, O144, O152, and O164 and Shigella O antigens. J Clin Microbiol. 1983;17:681–684. doi: 10.1128/jcm.17.4.681-684.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Landersjö C, Weintraub A, Widmalm G. Structure determination of the O-antigen polysaccharide from the enteroinvasive Escherichia coli (EIEC) O143 by component analysis and NMR spectroscopy. Carbohydr Res. 1996;291:209–216. doi: 10.1016/S0008-6215(96)00168-1. [DOI] [PubMed] [Google Scholar]
- 31.Linnerborg M, Weintraub A, Widmalm G. Structural studies of the O-antigen polysaccharide from the enteroinvasive Escherichia coli O164 cross-reacting with Shigella dysenteriae type 3. Eur J Biochem. 1999;266:460–466. doi: 10.1046/j.1432-1327.1999.00878.x. [DOI] [PubMed] [Google Scholar]
- 32.Lan R, Lumb B, Ryan D, Reeves PR. Molecular evolution of large virulence plasmid in Shigella clones and enteroinvasive Escherichia coli . Infect Immun. 2001;69:6303–6309. doi: 10.1128/IAI.69.10.6303-6309.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sansonetti PJ, d’Hauteville H, Ecobichon C, Pourcel C. Molecular comparison of virulence plasmids in Shigella and enteroinvasive Escherichia coli . Ann Microbiol (Paris) 1983;134a:295–318. [PubMed] [Google Scholar]
- 34.Hale TL. Genetic basis of virulence in Shigella species. Microbiol Rev. 1991;55:206–224. doi: 10.1128/mr.55.2.206-224.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Venkatesan MM, Buysse JM, Kopecko DJ. Use of Shigella flexneri ipaC and ipaH gene sequences for the general identification of Shigella spp. and enteroinvasive Escherichia coli . J Clin Microbiol. 1989;27:2687–2691. doi: 10.1128/jcm.27.12.2687-2691.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jin Q, Yuan Z, Xu J, Wang Y, Shen Y, et al. Genome sequence of Shigella flexneri 2a: insights into pathogenicity through comparison with genomes of Escherichia coli K12 and O157. Nucleic Acids Res. 2002;30:4432–4441. doi: 10.1093/nar/gkf566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang F, Yang J, Zhang X, Chen L, Jiang Y, et al. Genome dynamics and diversity of Shigella species, the etiologic agents of bacillary dysentery. Nucleic Acids Res. 2005;33:6445–6458. doi: 10.1093/nar/gki954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang J, Nie H, Chen L, Zhang X, Yang F, et al. Revisiting the molecular evolutionary history of Shigella spp. J Mol Evol. 2007;64:71–79. doi: 10.1007/s00239-006-0052-8. [DOI] [PubMed] [Google Scholar]
- 39.Hyma KE, Lacher DW, Nelson AM, Bumbaugh AC, Janda JM, et al. Evolutionary genetics of a new pathogenic Escherichia species: Escherichia albertii and related Shigella boydii strains. J Bacteriol. 2005;187:619–628. doi: 10.1128/JB.187.2.619-628.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Walters LL, Raterman EL, Grys TE, Welch RA. Atypical Shigella boydii 13 encodes virulence factors seen in attaching and effacing Escherichia coli. FEMS Microbiol Lett. 2012;328:20–25. doi: 10.1111/j.1574-6968.2011.02469.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hazen TH, Leonard SR, Lampel KA, Lacher DW, Maurelli AT, et al. Investigating the relatedness of enteroinvasive Escherichia coli to Other E. coli and Shigella isolates by using comparative genomics. Infect Immun. 2016;84:2362–2371. doi: 10.1128/IAI.00350-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Silva RM, Toledo MR, Trabulsi LR. Biochemical and cultural characteristics of invasive Escherichia coli . J Clin Microbiol. 1980;11:441–444. doi: 10.1128/jcm.11.5.441-444.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.de Boer RF, Ott A, Kesztyüs B, Kooistra-Smid AMD. Improved detection of five major gastrointestinal pathogens by use of a molecular screening approach. J Clin Microbiol. 2010;48:4140–4146. doi: 10.1128/JCM.01124-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van den Beld MJC, Friedrich AW, van Zanten E, Reubsaet FAG, Kooistra-Smid MAMD, et al. Multicenter evaluation of molecular and culture-dependent diagnostics for Shigella species and Entero-invasive Escherichia coli in the Netherlands. J Microbiol Methods. 2016;131:10–15.:S0167-7012(16)30273-1. doi: 10.1016/j.mimet.2016.09.023. [DOI] [PubMed] [Google Scholar]
- 45.Van Lint P, De Witte E, Ursi JP, Van Herendael B, Van Schaeren J. A screening algorithm for diagnosing bacterial gastroenteritis by real-time PCR in combination with guided culture. Diagnostic Microbiology and Infectious Disease. 2016;85:255–259. doi: 10.1016/j.diagmicrobio.2016.03.017. [DOI] [PubMed] [Google Scholar]
- 46.Liu B, Knirel YA, Feng L, Perepelov AV, Senchenkova SN, et al. Structure and genetics of Shigella O antigens. FEMS Microbiol Rev. 2008;32:627–653. doi: 10.1111/j.1574-6976.2008.00114.x. [DOI] [PubMed] [Google Scholar]
- 47.Cai HY, Lu L, Muckle CA, Prescott JF, Chen S. Development of a novel protein microarray method for serotyping Salmonella enterica strains. J Clin Microbiol. 2005;43:3427–3430. doi: 10.1128/JCM.43.7.3427-3430.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wattiau P, Boland C, Bertrand S. Methodologies for Salmonella enterica subsp. enterica subtyping: gold standards and alternatives. Appl Environ Microbiol. 2011;77:7877–7885. doi: 10.1128/AEM.05527-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li Y, Cao B, Liu B, Liu D, Gao Q, et al. Molecular detection of all 34 distinct O-antigen forms of Shigella. J Med Microbiol. 2009;58:69–81. doi: 10.1099/jmm.0.000794-0. [DOI] [PubMed] [Google Scholar]
- 50.Sun Q, Lan R, Wang Y, Zhao A, Zhang S, et al. Development of a multiplex PCR assay targeting O-antigen modification genes for molecular serotyping of Shigella flexneri . J Clin Microbiol. 2011;49:3766–3770. doi: 10.1128/JCM.01259-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.van der Ploeg CA, Rogé AD, Bordagorría XL, de Urquiza MT, Castillo ABC, et al. Design of two multiplex PCR assays for serotyping Shigella flexneri . Foodborne Pathog Dis. 2018;15:33–38. doi: 10.1089/fpd.2017.2328. [DOI] [PubMed] [Google Scholar]
- 52.van den Beld MJC, de Boer RF, Reubsaet FAG, Rossen JWA, Zhou K, et al. Evaluation of a culture-dependent algorithm and a molecular algorithm for identification of Shigella spp., Escherichia coli, and enteroinvasive E. coli . J Clin Microbiol. 2018;56:e00510-18. doi: 10.1128/JCM.00510-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Løbersli I, Wester AL, Kristiansen Å, Brandal LT. Molecular differentiation of Shigella spp. from enteroinvasive E. coli . European Journal of Microbiology and Immunology. 2016;6:197–205. doi: 10.1556/1886.2016.00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pavlovic M, Luze A, Konrad R, Berger A, Sing A, et al. Development of a duplex real-time PCR for differentiation between E. coli and Shigella spp. J Appl Microbiol. 2011;110:1245–1251. doi: 10.1111/j.1365-2672.2011.04973.x. [DOI] [PubMed] [Google Scholar]
- 55.Kim HJ, Ryu JO, Song JY, Kim HY. Multiplex polymerase chain reaction for identification of Shigellae and four Shigella species using novel genetic markers screened by comparative genomics. Foodborne Pathog Dis. 2017;14:400–406. doi: 10.1089/fpd.2016.2221. [DOI] [PubMed] [Google Scholar]
- 56.Chattaway MA, Schaefer U, Tewolde R, Dallman TJ, Jenkins C. Identification of Escherichia coli and Shigella species from whole-genome sequences. J Clin Microbiol. 2017;55:616–623. doi: 10.1128/JCM.01790-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C. Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods. 2017;14:417–419. doi: 10.1038/nmeth.4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wood DE, Salzberg SL. Kraken: ultrafast metagenomic sequence classification using exact alignments. Genome Biol. 2014;15:R46. doi: 10.1186/gb-2014-15-3-r46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Alikhan NF, Zhou Z, Sergeant MJ, Achtman M. A genomic overview of the population structure of Salmonella . PLoS Genet. 2018;14:e1007261. doi: 10.1371/journal.pgen.1007261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gurevich A, Saveliev V, Vyahhi N, Tesler G. QUAST: quality assessment tool for genome assemblies. Bioinformatics. 2013;29:1072–1075. doi: 10.1093/bioinformatics/btt086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jolley KA, Maiden MCJ. BIGSdb: Scalable analysis of bacterial genome variation at the population level. BMC Bioinformatics. 2010;11:595. doi: 10.1186/1471-2105-11-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Joensen KG, Tetzschner AMM, Iguchi A, Aarestrup FM, Scheutz F. Rapid and easy in silico serotyping of Escherichia coli isolates by use of whole-genome sequencing data. J Clin Microbiol. 2015;53:2410–2426. doi: 10.1128/JCM.00008-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hartl DL, Dykhuizen DE. The population genetics of Escherichia coli . Annu Rev Genet. 1984;18:31–68. doi: 10.1146/annurev.ge.18.120184.000335. [DOI] [PubMed] [Google Scholar]
- 65.Hu D, Liu B, Wang L, Reeves PR. Living trees: high-quality reproducible and reusable construction of bacterial phylogenetic trees. Mol Biol Evol. 2020;37:563–575. doi: 10.1093/molbev/msz241. [DOI] [PubMed] [Google Scholar]
- 66.Letunic I, Bork P. teractive Tree Of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 2019;47:W256–W259. doi: 10.1093/nar/gkz239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhou Z, Alikhan N-F, Sergeant MJ, Luhmann N, Vaz C, et al. GrapeTree: visualization of core genomic relationships among 100,000 bacterial pathogens. Genome Res. 2018;28:1395–1404. doi: 10.1101/gr.232397.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Li H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv preprint arXiv [Google Scholar]
- 69.Buchrieser C, Glaser P, Rusniok C, Nedjari H, D’Hauteville H, et al. The virulence plasmid pWR100 and the repertoire of proteins secreted by the type III secretion apparatus of Shigella flexneri . Mol Microbiol. 2000;38:760–771. doi: 10.1046/j.1365-2958.2000.02179.x. [DOI] [PubMed] [Google Scholar]
- 70.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 73.Page AJ, Cummins CA, Hunt M, Wong VK, Reuter S, et al. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics. 2015;31:3691–3693. doi: 10.1093/bioinformatics/btv421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, et al. BLAST+: architecture and applications. BMC Bioinformatics. 2009;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Senchenkova SN, Feng L, Yang J, Shashkov AS, Cheng J, et al. Structural and genetic characterization of the Shigella boydii type 10 and type 6 O antigens. J Bacteriol. 2005;187:2551–2554. doi: 10.1128/JB.187.7.2551-2554.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ansaruzzaman M, Kibriya AK, Rahman A, Neogi PK, Faruque AS, et al. Detection of provisional serovars of Shigella dysenteriae and designation as S. dysenteriae serotypes 14 and 15. J Clin Microbiol. 1995;33:1423–1425. doi: 10.1128/jcm.33.5.1423-1425.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Balows A. In: Diagnostic Microbiology. 8th. Murray PR, Baron EJ, Jorgenson JH, Pfaller MA, Yolken RH, editors. ASM Press; 2003. Manual of clinical microbiology; p. 2113. edn. p. [Google Scholar]
- 78.Woodward DL, Clark CG, Caldeira RA, Ahmed R, Soule G, et al. Identification and characterization of Shigella boydii 20 serovar nov., a new and emerging Shigella serotype. J Med Microbiol. 2005;54:741–748. doi: 10.1099/jmm.0.46095-0. [DOI] [PubMed] [Google Scholar]
- 79.Kim J, Lindsey RL, Garcia-Toledo L, Loparev VN, Rowe LA, et al. High-quality whole-genome sequences for 59 historical Shigella strains generated with PacBio sequencing. Genome Announc. 2018;6:15. doi: 10.1128/genomeA.00282-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Michelacci V, Prosseda G, Maugliani A, Tozzoli R, Sanchez S, et al. Characterization of an emergent clone of enteroinvasive Escherichia coli circulating in Europe. Clin Microbiol Infect. 2016;22:287. doi: 10.1016/j.cmi.2015.10.025. [DOI] [PubMed] [Google Scholar]
- 81.Liu J, Platts-Mills JA, Juma J, Kabir F, Nkeze J, et al. Use of quantitative molecular diagnostic methods to identify causes of diarrhoea in children: a reanalysis of the GEMS case-control study. Lancet. 2016;388:1291–1301.:S0140-6736(16)31529-X. doi: 10.1016/S0140-6736(16)31529-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Cho MS, Ahn TY, Joh K, Kwon OS, Jheong WH, et al. A novel marker for the species-specific detection and quantitation of Shigella sonnei by targeting a methylase gene. J Microbiol Biotechnol. 2012;22:1113–1117. doi: 10.4014/jmb.1111.11006. [DOI] [PubMed] [Google Scholar]
- 83.Sansonetti PJ, Kopecko DJ, Formal SB. Shigella sonnei plasmids: evidence that a large plasmid is necessary for virulence. Infect Immun. 1981;34:75–83. doi: 10.1128/iai.34.1.75-83.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Feng L, Perepelov AV, Zhao G, Shevelev SD, Wang Q, et al. Structural and genetic evidence that the Escherichia coli O148 O antigen is the precursor of the Shigella dysenteriae type 1 O antigen and identification of a glucosyltransferase gene. Microbiology (Reading) 2007;153:139–147. doi: 10.1099/mic.0.2006/001107-0. [DOI] [PubMed] [Google Scholar]
- 85.Göhmann S, Manning PA, Alpert CA, Walker MJ, Timmis KN. Lipopolysaccharide O-antigen biosynthesis in Shigella dysenteriae serotype 1: analysis of the plasmid-carried rfp determinant. Microb Pathog. 1994;16:53–64. doi: 10.1006/mpat.1994.1005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.