SUMMARY
Broadly neutralizing antibodies (bNAbs) represent an alternative to drug therapy for the treatment of HIV-1 infection. Immunotherapy with single bNAbs often leads to emergence of escape variants, suggesting a potential benefit of combination bNAb therapy. Here a trispecific bNAb reduces viremia 100- to 1000-fold in viremic SHIV-infected macaques. After treatment discontinuation, viremia rebounds transiently and returns to low levels, through CD8-mediated immune control. These viruses remained sensitive to the trispecific antibody, despite loss of sensitivity to one of the parental bNAbs. Similarly, the trispecific bNAb suppresses the emergence of resistance in viruses derived from HIV-1 infected subjects, in contrast to parental bNAbs. Trispecific HIV-1 antibodies therefore mediates potent antiviral activity in vivo and may minimize the potential for immune escape.
Graphical Abstract

eTOC blurb:
Immunotherapy with monoclonal bNAbs leads to emergence of escape variants. Pegu et al. show that trispecific bNAb treatment reduces viremia up to 1000-fold in vivo and limits selection of resistant HIV-1 variants in cell culture. Thus, this antibody has therapeutic potential and may minimize immune escape.
INTRODUCTION
Broadly neutralizing antibodies (bNAbs) against the HIV-1 envelope glycoprotein (Env) exert protection against infection through multiple antiviral mechanisms (Pegu et al., 2017; Sok and Burton, 2018). In contrast to most commonly prescribed anti-retroviral drugs, including reverse transcriptase inhibitors and integrase inhibitors, antibodies can block viral entry and potentially engage other immune effector functions through Fc receptors (Asokan et al., 2020a; Bournazos et al., 2014; Bournazos and Ravetch, 2017; Lu et al., 2016; Lu et al., 2018; Richardson et al., 2018). Another feature of mAbs is their relatively long half-life, typically ranging from 2-4 weeks in humans with opportunities to further enhance both effector function and half life through antibody engineering (Asokan et al., 2020a; Bournazos et al., 2014; Cohen et al., 2019; Gaudinski et al., 2018; Ledgerwood et al., 2015; Mankarious et al., 1988). The antiviral activity of bNAbs in vivo suggests therapeutic potential for maintaining virus suppression induced by cART drugs and killing of virally infected T cells. For these reasons, bNAbs have been advanced in preclinical and clinical investigation for the prevention, treatment, and potential cure of HIV-1 infection (Caskey et al., 2019; Gama and Koup, 2018).
Despite the potency and breadth of single monoclonal bNAbs, resistant virus can rapidly emerge through selection or mutational escape (Caskey et al., 2015; Caskey et al., 2017; Lynch et al., 2015). To minimize the possibility of resistance and viral escape in vivo, combinations of bNAbs have been explored (Bar-On et al., 2018; Cohen et al., 2019; Mendoza et al., 2018). One clinical study reported that a combination of two bNAbs, directed to the CD4 binding site and V3 associated carbohydrate, suppressed viremia effectively in those with sensitive circulating virus, with effects that persisted after completion of therapy (Mendoza et al., 2018). Viral escape from one of the antibodies has been attributed to differences in persistence of the bNAbs, resulting in periods of monotherapy with declining levels of the bNAb that persists longer in vivo, a situation fostering selection of resistance. Therefore, it has been suggested that an optimal combination would require at a minimum three bNAbs targeting three different specificities to provide broad coverage, and matched pharmacokinetic profiles to maximize breadth of coverage and limit viral escape (Kong et al., 2015; Wagh et al., 2016). As an alternative to using such bNAb combinations, we developed a trispecific bNAb platform that can combine three different bNAb specificities in a single molecule (Xu et al., 2017). We previously showed that these trispecific bNAbs display greater breadth and potency than individual monoclonal bNAbs and can provide protection against challenge with a mixture of two SHIVs, each resistant to one of the bNAb specificities represented in the trispecific bNAb. Here, we show that a potent trispecific bNAb protects against viral replication in infected animals and show that it suppresses the generation of resistant virus in primary human T cell cultures.
RESULTS
Potent suppression of viremia and modulation of viral reservoir in vivo
We assessed whether the trispecific bNAb could exert an anti-viral effect on lentiviral infection in vivo. For this purpose, we tested the ability of N6/PGDM1400-10E8v4 to suppress plasma viremia in Indian rhesus macaques infected with SHIVBG505. N6/PGDM1400-10E8v4 is a trispecific bNAb comprising of the variable domains from three different bNAbs: (1) N6 that targets the CD4 binding site, (2) PGDM1400 that targets the V2 glycan site and (3) 10E8v4 that targets the membrane proximal external region (MPER) on the HIV-1 envelope glycoprotein. This trispecific bNAb neutralized SHIVBG505 more potently than the parental mAbs from which it was derived, with PGDM1400 the most potent followed by N6 and 10E8v4 (Table S1). It was comparable in potency to a related trispecific, VRC01/PGDM1400-10E8v4 described previously (Xu et al., 2017), with slightly greater breadth and potency against a large 208 isolate panel and a modestly shorter half life in vivo (Xu et al., 2017). Because only two injections at 1 week intervals were deemed feasible because of the expected monkey anti-human Ab response, N6/PGDM1400-10E8v4 was chosen for this study because of its potency profile. Naïve rhesus macaques (n=6) were challenged with SHIVBG505 intrarectally and followed through six weeks of infection when N6/PGDM1400-10E8v4 was administered intravenously at a dose of 20 mg/kg, with a repeat dose one week later. Following treatment, viral loads fell sharply in all animals, ranging from a 100- to 1000-fold decline (Fig. 1A, right panel). In contrast, untreated historical control animals challenged intrarectally with SHIVBG505 and matched for peak acute viremia, showed no such decline in viremia at these time points after challenge (Fig. 1A, left panel). There was a significant difference (p=0.002) in the slopes of plasma viral decay when comparing the plasma viral decline during the first two weeks after trispecific bNAb infusion to the plasma viral decline in the control animals (week 5/6 to week 14) post SHIVBG505 challenge (Fig. S1A–B). The circulating antibody levels dipped to between 1~10 μg/ml coincident with the emergence of anti-drug antibodies after two weeks (ADA, Figure S1C) in all animals. No further trispecific bNAb was administered after these two doses and as the levels declined, plasma viral load increased, returning transiently to near pre-treatment levels (Fig. 1B).
Figure 1. Effect of trispecific antibody treatment on plasma viral load in SHIVBG505 infected animals.

(A) Plasma viral load in untreated (control) and trispecific antibody treated (treatment) SHIVBG505 infected animals. Trispecific antibody (20 mg/kg iv) was given at days 42 and 49 post intrarectal challenge with SHIVBG505. Antibody infusion times are indicated by the vertical dashed lines at day 42 and 49 post SHIV challenge. (B) Plasma viral load (red line, left Y-axis) and trispecific antibody levels (black line, right Y-axis) for each individual animal. Antibody infusions are indicated by the vertical dashed lines at day 42 and 49.
To determine whether trispecific bNAb treatment affected viral replication in lymphoid tissues in vivo, axillary and inguinal lymph node biopsies were obtained before and after treatment. The number of free virions and viral RNA containing cells in the LN were quantified using RNAscope technology (Hsu et al., 2018), and compared before and after trispecific bNAb infusion (Figs. 2A–D, S1D–G). Similar to the sharp decline in plasma viremia, we saw a significant drop (p=0.005, Kruskal-Wallis test) in the number of viral RNA containing cells one week after the first trispecific bNAb infusion (Fig. 2E, left panel), confirming a significant reduction in viral load in lymphoid tissue. Ten weeks later, the number of infected cells remained low in lymph nodes, possibly due to the natural process of immunological control that is common in SHIV infected NHPs. The trispecific bNAb infusion did not significantly change the number of cell free virions trapped onto the follicular dendritic cell (FDC) network immediately after infusion, but they were significantly reduced (p=0.03, Kruskal-Wallis test) at the 16-week time point (Fig. 2E, right panel). These results indicated that the trispecific bNAb significantly reduced both plasma viremia and viral replication in lymphoid tissues, demonstrating potent in vivo anti-viral activity in this model.
Figure 2. SHIV RNA detection by in situ confocal staining.

(A) Representative confocal image of whole lymph node showing SHIV RNA (green), CD20 (orange), and cellular nucleic acids (blue). Scale bar: 500 μm (B) Magnified area from (A) that shows an extra-follicular cell infected with SHIV (top) and a further magnified image (bottom) that shows the virally infected cell associated with T cell markers of CD3 (cyan) and CD4 (red). (C) Magnified area from (A) that shows an intra-follicular cell that is infected with SHIV (top) and a further magnified image (bottom) that shows the virally infected cells associated with T cell markers of CD3 (cyan) and CD4 (red). (D) Magnified area from (A) that shows the intra-follicular area filled with free virus and a further magnified image (bottom) shows the free virus associated with the FDC network (pink). Scale bars: 20 μm (top) and 10 μm (bottom). Additional detailed, merged images are shown in figure S4. (E) RNAscope quantification of SHIV gag mRNA containing cells and free virions in lymph nodes obtained from SHIV-infected animals pre- and post-infusion of the trispecific antibody. Indicated p values were calculated using a nonparametric Kruskal-Wallis test.
The stability of the trispecific bNAb in vivo was assessed by measuring the neutralization breadth and potency of plasma from treated animals (Table S2–3). Low-level neutralization of autologous virus only was observed prior to treatment, as well as 10 weeks after treatment when plasma trispecific levels were undetectable. In contrast, broad and potent neutralization activity was detected in plasma at 9 days post-treatment demonstrating that the trispecific bNAb retained its broad neutralization activity in vivo.
To assess the impact of trispecific bNAb treatment on the viral quasispecies in these SHIV infected animals, we performed single genome amplification (SGA) to analyze the env sequences of the plasma virus populations present before and after the trispecific bNAb infusions in each individual SHIVBG505 infected animal (Table S4). The pre-infusion sequences for all animals were largely homogenous and similar to the parental SHIVBG505 sequence (Fig. S2). After bNAb treatment, at 9 to 10 weeks after SHIV challenge, when the viral load rebounded after trispecific bNAb levels decayed in plasma, the dominant viral sequence in three animals had mutations that removed the N160 glycan that is important for PGDM1400 neutralization (Fig. 3A, S2). Env pseudoviruses constructed from these env sequences demonstrated a loss of sensitivity to PGDM1400 but not to the trispecific bNAb (Fig. 3B, S2). These results show that infusion of a trispecific bNAb did not select for trispecific resistant mutants in SHIV infected animals.
Figure 3. Viral envelope mutations selected by trispecific antibody.
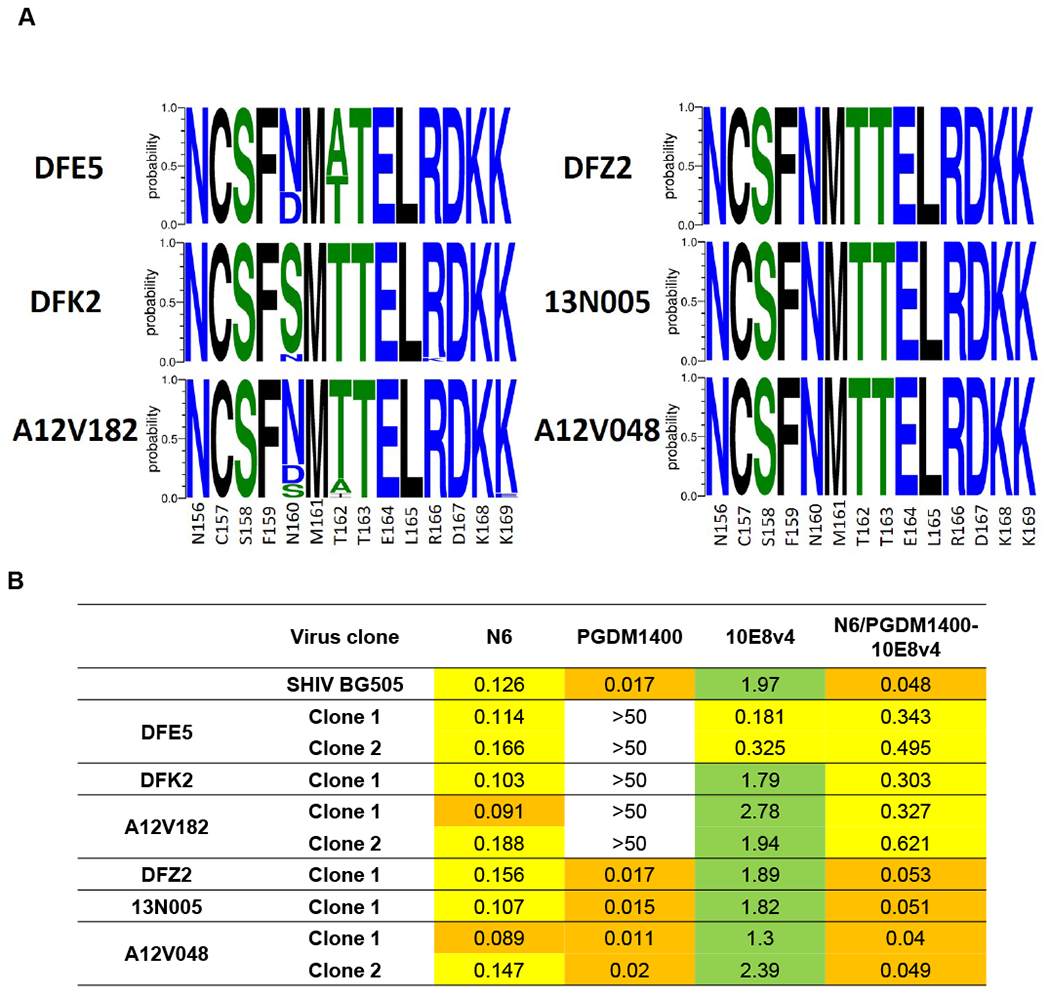
(A) Logo plots of SHIVBG505 Env V2 glycan region sequences obtained via Env-SGA in SHIVBG505 infected animals after trispecific antibody infusion. (B) Neutralization sensitivity to the parental and trispecific antibodies of pseudotyped viruses encoding the dominant plasma SHIV Env sequences obtained via Env-SGA post trispecific antibody infusion from each animal.
Long term follow-up of N6/PGDM1400-10E8v4 treated animals revealed relatively stable long term control of viremia in 5 out of the 6 animals compared to matched historical control animals infected with same challenge SHIVBG505 stock, although due to limited animal numbers this difference was not statistically significant (Fig. 1H). To determine the immune basis for this effect, we evaluated SHIV specific cellular immune responses in PBMC and lymph nodes. More than one year after treatment, both SHIV-specific CD4+ and CD8+ T cells were detectable in all animals (Figure S3A–B). To determine the cellular basis of immune control, animals were administered an anti-CD8+β mAb to deplete CD8+ T cells. This treatment led to varying levels of CD8+ T cell depletion in each animal which led to corresponding levels of transient increases in plasma viremia in each animal, suggesting that viral control following trispecific bNAb treatment was mediated by CD8+ T cell effector functions (Figure S3C).
Potent suppression of viral replication in human lymphocytes ex vivo
A desirable feature of trispecific bNAbs is their potential to prevent the emergence of resistant strains often seen with single bNAbs. We further addressed this question using CD4+ T cells from viremic HIV-1 infected donors in an ex vivo setting. Purified CD4+ T cells from five different viremic, untreated subjects were activated and co-cultured with uninfected CD4+ T cells from healthy donors in the presence of the trispecific or individual corresponding parental monoclonal bNAbs for a period of 2-4 weeks. The trispecific bNAb consistently suppressed the outgrowth of virus from all five donors (Fig. 4A–B). In contrast, virus replication emerged in the presence of parental bNAbs at levels that varied by donor. In three out of the five donors (donors 3-5), N6 led to similar viral suppression as was observed with the trispecific bNAb, whereas for the remaining two donors (donors 1 and 2), there was minimal suppression observed with N6 in comparison to potent suppression, as assessed by HIV-1 gag protein and RNA levels in the culture supernatants, observed with the trispecific bNAb. For the other parental monoclonal bNAbs, minimal or transient suppression was observed, and viral replication increased with time, suggesting the outgrowth of resistant variants.
Figure 4. Effect of monoclonal and trispecific antibodies on replication of HIV-1 from infected donor CD4+ T cells.

Activated CD4+ T cells from five viremic HIV-1 infected donors were cultured in the presence of the indicated antibodies for 2-3 weeks and viral replication was quantified by measuring HIV-1 p24 protein and gag RNA levels in the culture supernatants. (A) HIV-1 p24 protein levels (% of control, where control is denoted as 100% for each time point) in the culture supernatants were quantified by measuring HIV-1 p24 protein levels in the culture supernatant over time by ELISA. The levels shown are means of the levels measured in triplicate wells for each antibody from one experiment performed for each donor. (B) HIV-1 gag RNA levels in the culture supernatants were quantified at the end of the incubation using real time qPCR. Viral load (copies/ml) for each donor were as follows: donor 1: 4740, donor 2: 4503, donor 3: 3488, donor 4: 60,910, donor 5: 1810.
DISCUSSION
We previously showed that the trispecific bNAb mediates broader in vitro neutralization and provides broader protection in NHPs challenged with a mixture of two SHIVs compared to individual parental bNAbs (Xu et al., 2017). Those results demonstrated the utility of trispecific bNAbs in preventing HIV-1 infection and their potential use as prophylactic agents. Here, we report that in viremic SHIV infected NHPs, two infusions of the trispecific bNAb treatment reduced plasma viremia 100- to 1000-fold and limited the evolution of mutations that could lead to escape from trispecific bNAb neutralization. In addition, the trispecific bNAb showed the most consistent suppression of viral replication in primary cultures of infected cells from viremic HIV-1 infected donors, in contrast to variable suppression observed by the individual parental bNAbs.
Unlike small molecule drugs, antibodies are known to actively engage the immune system and mediate various effector mechanisms via their Fc regions (Lu et al., 2018). It has been recently reported that the Fc-mediated effector functions contributes to the in vivo anti-viral effect of anti-HIV-1 antibodies (Asokan et al., 2020b; Wang et al., 2020). We also find that trispecific bNAbs like their parental monoclonal bNAbs can mediate anti-viral effects through multiple mechanisms, including direct viral neutralization, by engaging innate effector cells through Fc/Fc receptor interactions, and by fixing complement (Figures S4), suggesting that similar responses may contribute to the potent anti-viral effect observed for the trispecific bNAb.
The anti-viral effect observed in this model is substantial,with reductions in plasma viral load as high as 3 log10. While we did not directly compare the thereputic effect to single bNAbs, previously published studies with monoclonal bNAbs in this model demonstrated transient reductions of 1-1.5 log10 (Asokan et al., 2020b; Bolton et al., 2016; Julg et al., 2017). We observed long term viral control in most of the treated animals compared to historical control animals. In this regard, we demonstrate potent CD8+ T cell immunity post treatment in this study, as has been observed in other NHP and early clinical studies of bNAbs (Niessl et al., 2020; Nishimura et al., 2021; Nishimura et al., 2017). We note however, that virological control in the SHIV model likely occurs more frequently than in humans. Should similar trispecific bNAb treatment be successful in humans, the inherent long half-life may allow for infrequent administration and reduced dependence on chronic drug treatment and its attendant side effects. Infusion of the trispecific bNAb also led to a rapid decline in cell-associated viral RNA in lymphoid tissues, suggesting the potential to impact tissue reservoirs. In addition, we observed that the cell-free virions in the lymph nodes did not decrease as quickly as the cell-associated viral RNA in these tissues after infusion of the trispecific bNAb. The majority of cell-free virions observed in the lymph nodes were trapped on the follicular dendritic cell network, consistent with previous publications (Dave et al., 2018; Keele et al., 2008b). These trapped virions represent a very stable reservoir of infectious virus that has been shown to persist in the presence of neutralizing antibodies and cART regimens. Overall, this study is the first proof of concept demonstration that a trispecific bNAb can be used to treat lentiviral infection.
The development of resistance has been a recurrent problem for HIV-1 drug therapy, even with current highly effective cART regimens (Beyrer and Pozniak, 2017; Garbelli et al., 2017). To combat such multi-drug resistant variants, newer anti-viral drugs are constantly under development, by targeting novel mechanisms or prolonging half-life (Cambou and Landovitz, 2020). bNAbs are similarly limited in terms of efficacy due to rapid selection of resistance variants when administered alone to HIV-1 infected patients. Even when combinations of two bNAbs have been administered, development of resistance has been observed, due at least in part to differential pharmacokinetic decay profiles of the bNAbs, resulting in periods of monotherapy (Bar-On et al., 2018). It has been proposed that an ideal bNAb combination would require 3-4 different bNAbs to maximize coverage and minimize potential viral escape (Wagh et al., 2016). Due to varying pharmacokinetic and pharmacodynamic properties of each bNAb, such combination bNAb therapy will entail additional complexity, requiring manufacture of multiple antibodies and design of optimal formulations and dosing regimens, which is simplified by the use of a single biologic therapy. Very little viral replication was observed in long-term cultures of HIV+ donor CD4+ T cells in presence of the trispecific bNAb in contrast to parental monoclonal bNAbs, suggesting lack of selection for resistant viruses to the trispecific antibodies in this ex vivo model, although clinical trials in infected individuals will be needed to assess if any trispecific bNAb resistant viruses develop de novo. In addition, the lack of viral escape was also observed in the NHP SHIV model further reinforcing the advantage of having a single molecule targeting multiple specificities. The impact of multiple rounds of antibody administration on resistance development in humans must be addressed future clinical trials since the xenogeneic anti-human antibody response limits long-term evaluation of efficacy in the NHP model. Ongoing clinical experience with individual anti-Env monoclonal bNAbs suggest they maintain consistent pharmacokinetics after multiple infusions and thus, do not elicit funtionally important anti-human Ab responses (Mayer et al., 2017). Taken together, these findings also suggest a lower likelihood of developing viral resistance to the trispecific bNAb relative to the individual bNAbs from which it was derived.
A related trispecific bNAb, VRC01/PGDM1400-10E8v4 (also called SAR441236), is currently under evaluation for safety and efficacy in humans . Previous clinical experience with a related bispecific platform suggests that the safety and pharmacokinetic profile of the trispecific bNAb studied here should be similar to that of natural antibodies (Raghu et al., 2018). In summary, the in vivo antiviral effect demonstrated here, together with the protection against acquisition of infection in an NHP challenge model, suggest the potential of trispecific bnAbs for both treatment and prevention of HIV-1 infection. A recent phase 2b trial demonstrated that the bNAb VRC01 can reduce the risk of HIV- 1 infection, but only for the subset of viruses that are highly neutralization sensitive – with an IC80 value of < 1.0 μg/ml (Corey et al., 2021). This further highlights the utility of a bNAb molecule that can provide broad and highly potent neutralization. Overall, trispecific bNAbs represent a promising class of immunotherapeutic proteins against HIV-1 infection.
Limitations of the Study
There are several limitations of this study. Long term virological control has been observed in this SHIV model more commonly than for human HIV-1 infection. Our study measured longitudinal plasma viremia in each animal before and after treatment with the trispecific bNAb, but we used historical controls to assess the natural history of viremia in our SHIV challenge model. We also did not include a group of animals treated with a combination of the three parent bNAbs, so our concusions are limited to the in vivo effect of this one trispecific bNAb. Likewise in the ex vivo assays measuring outgrowth of viruses from HIV+ donor CD4+ T cells in presence of bNAbs, we compared the trispecific bNAb to individual parental bNAbs, but we did not include a mixture of the three parental bNAbs. Lastly, although we saw a rapid decline in cell-associated viral RNA in lymphoid tissues after trispecific bNAb infusion, the lack of similar data from untreated control animals limits the implication of this finding.
STAR★Methods
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by John R. Mascola (jmascola@mail.nih.gov).
Materials Availability
All new reagents are available by MTA for non-commercial research.
Data and Code Availability
The published article includes all data generated or analyzed during this study.
This study did not generate new code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
NHP studies
All animals were housed and cared for in accordance with Guide for Care and Use of Laboratory Animals Report number NIH 82-53 (Department of Health and Human Services, Bethesda, Maryland, USA, 1985) in a biosafety level 2 National Institute of Allergy and Infectious Diseases (NIAID) facility. All animal procedures and experiments were performed according to protocols approved by the Institutional Animal Care and Use Committee of the National Institute of Allergy and Infectious Diseases (NIH). Naïve Rhesus macaques were intrarectally challenged with SHIVBG505. In brief, animals were inoculated intrarectally with one milliliter of a 1:8 dilution of challenge stock. This corresponds to an animal infectious dose of approximately 5 based on the reported AID50 titer which was 1 milliliter of 1:120 dilution of the challenge stock (Li et al., 2021). The infection was confirmed by measuring plasma viremia weekly and allowed to progress for 6 weeks. At 6- and 7-weeks post challenge, the animals were administered low-endotoxin trispecific antibody preparations (<1 EU/mg) intravenously at 20 mg of Ab/kg of body weight. A control group of 4 animals were left untreated and viremia was followed weekly till week 12 post challenge.
Human subjects.
PBMCs were obtained from viremic HIV-1 infected donors enrolled in the VRC 601 trial (Lynch et al., 2015). VRC 601 was a single-site, phase 1, open-label, dose escalation study examining the safety and pharmacokinetics of the human mAh VRC-HIVMAB060-00-AB (VRC01) in HIV-infected adults. The study was conducted at the NIH Clinical Center by the VRC Clinical Trials Program, NIAID, NIH (ClinicalTrials.gov NCT01950325). The protocol was reviewed and approved by the NIAID Institutional Review Board. U.S. Department of Health and Human Services guidelines for conducting clinical research were followed. All subjects gave written informed consent before participation.
METHOD DETAILS
Generation of the trispecific and monoclonal antibodies.
Trispecific antibodies were produced by transient transfection of 4 expression plasmids into Expi293 cells using ExpiFectamine™ 293 Transfection Kit (Thermo Fisher Scientific) according to manufacturer’s protocol as previously described (Xu et al., 2017). Briefly, four expression plasmids for mAb arm light, heavy chains, and for CODV light, heavy chains respectively were co-transfected into Expi293 cells using ExpiFectamine™ 293 Transfection Kit (Thermo Fisher Scientific) according to manufacturer’s protocol. 25% (w/w) of each plasmid was diluted into Opti-MEM, mixed with pre-diluted ExpiFectamine reagent for 20-30 minutes at room temperature (RT), and added into Expi293 cells (2.5x106 cells/ml). An optimization of transfection to determine the best ratio of plasmids was often used to produce the trispecific antibody with good yield and purity. 4-5 days after transfection, the supernatant from transfected cells was collected and filtered through 0.45 μm filter unit (Nalgene). The trispecific antibody in the supernatant was purified using a 3-step procedure. First, protein A affinity purification was used, and the bound Ab was eluted using IgG elution buffer (Thermo Fisher Scientific). Second, the eluted product was dialyzed against PBS (pH7.4) overnight with 2 changes of PBS buffer. Any precipitate was cleared by filtration through 0.45 μm filter unit (Nalgene) before next step. Third, SEC purification (Hiload 16/600 Superdex 200pg, or Hiload 26/600 Superdex 200pg, GE Healthcare) was used to remove aggregates and different species in the preparation. The fractions were analyzed on reduced and non-reduced SDS-PAGE to identify the fractions that contained the monomeric trispecific antibody before combining them. The purified antibody was then aliquoted and kept at −80°C for long term storage. Similarly, monoclonal antibodies were produced as previously described (Pegu et al., 2014; Rudicell et al., 2014).
Antibody binding to cell surface antigen.
HIV-1 BaL infected CEM-NKr-CCR5 cell line, CEM-BaL, was used to assess binding of antibodies to cell surface expressed HIV-1 envelope glycoprotein. CEM-BaL was maintained in R10 media: RPMI1640 (Thermo Fisher Scientific), 1% Pen/Strep (Thermo Fisher Scientific), and 10% FBS (Sigma). Cells were counted and 5 x 105 cells were added to each FACS tube for staining. Cells were stained with 5 μg of each antibody for 15 minutes at room temperature. Following the incubation, cells were washed twice with PBS (Thermo Fisher Scientific) by centrifugation at 300 g for 3 minutes. Cells were stained with goat anti-human IgG (H+L) Alexa Fluor 647 (Thermo Fisher Scientific) and the aqua-blue live/dead fixable dead cell stain kit (Thermo Fisher Scientific) for 15 minutes at room temperature. Following the incubation, cells were washed twice with PBS by centrifugation at 300 g for 3 minutes. Cells were fixed with 1% PFA (Electron Microscopy Sciences) in PBS and data on binding of the different antibodies to the cell surface was acquired on a LSRFortessa X-50 flow cytometer (BD biosciences). Data were analyzed using FlowJo software (BD biosciences).
Antibody-Dependent Cell Cytoxicity (ADCC).
CEM-BaL cells were used as targets and human KHYG-1 NK cells expressing human CD16 (Alpert et al., 2012) were used as effector cells in this ADCC assay. The target cells were counted and stained with PKH26 red fluorescent cell linker membrane labeling dye (Sigma) in diluent C (Sigma) for 10 minutes followed by a wash in PBS by spinning at 300 g for 3 minutes. These labeled target cells were then resuspended in R10 media and added to a 96-well round bottom plate at a density of 1 x 105 cells/well. Antibody concentrations beginning at a final well concentration of 250 nM were diluted 5-fold in R10 for a total of 6 dilutions to 0.08 nM and added to corresponding wells. The effector cells were counted, resuspended in R10 media, and added to the plate at a density of 9 x 105 cells/well. The culture plate was incubated at 37°C for 6 hours. After the incubation, the culture plate was aspirated and washed twice with PBS. Cells were stained with the aqua live/dead fixable dead cell stain kit (Thermoe Fisher Scientific) and Annexin V, Alexa Flour 647 conjugate (Thermo Fisher Scientific) in annexin binding buffer (Thermo Fisher Scientific) for 15 minutes. The culture plate was then washed twice with annexin binding buffer and then the cells were fixed with 1% PFA in annexin binding buffer. The data quantifying the number of dead target cells was acquired on a LSRFortessa X-50 flow cytometer and analyzed using FlowJo software.
Antibody-Dependent Cell Phagocytosis (ADCP).
The ADCP assay was performed as previously described (Ackerman et al., 2011), with minor modifications. Briefly, 1 μm red fluorescent neutrAvidin biotin-binding microspheres (Thermo Fisher Scientific) were incubated with a biotinylated BG505 DS-SOSIP HIV-1 env trimer (Kwon et al., 2015) overnight at 4 °C. After overnight incubation and washes in PBS/BSA, trimer coated beads were co-cultured with bNAbs diluted 4-fold in PBS/BSA starting at a concentration of 39 ng/ml for a total of 6 dilutions for 2 hours in a 37°C tissue culture incubator. After incubation, 2 x 104 THP-1 cells were added for 4 hours at 37°C / 5% CO2. Cells were then fixed in 1% PFA and read for levels of phagocytosis activity by flow cytometry. The phagocytosis score was calculated based on flow cytometry analysis parameters as: (frequency of bead+ THP-1 cells) x (Geo. MFI of bead fluorescence within the bead+ THP-1 cell population/ Geo. MFI of bead fluorescence from the negative control condition containing THP-1 cells and beads without bNAbs).
Antibody-Dependent Complement Mediated Lysis (ADCML).
The ADCML assay was performed as previously described (Miller-Novak et al., 2018), with minor modifications. Replication competent HIV-1 BaL, grown in human PBMCs, was diluted in RPMI1640 media to a final volume of 1.25 ng/mL and added to a 96-well round-bottom tissue culture plate. Antibodies were diluted in RPMI1640 media for a final concentration of 30 nM and added to corresponding wells. The concentration of 30 nM was selected based on a previous titration done with a related bNAb, VRC07-523LS (Figure S4G, H). Normal human complement serum (Quidel) was diluted to 20% by final volume and added to each well of the plate. The plate was incubated for 24 hours in 37°C tissue culture incubator. After incubation, samples were diluted at fixed ratios in RPMI1640 media and the lysis was quantified by measuring HIV-1 p24 levels in the supernatant using HIV-1 p24 ELISA kit (ABL). Percent lysis was calculated as follows: (i) blank well values were subtracted from all sample well values, (ii) negative-control, non-specific control, and spontaneous complement activation control values were averaged and subtracted, and (iii) the values were divided by the average 100% lysis positive-control value and multiplied by 100.
SHIVBG505 challenge stock.
A SHIV.BG505.332N.375Y.dCT challenge stock grown in primary activated rhesus CD4 T cells was prepared as described (Li et al., 2016). This challenge stock was shown to reliably infect Indian rhesus macaques by intrarectal (Pauthner et al., 2019) and intravaginal (Arunachalam et al., 2020) routes of administration. The deep sequencing of the challenge stock has also been done and has been deposited in to the appropriate databases (Li et al., 2021).
Plasma viral load quantification.
Plasma viremia was quantitated using a PCR-based method to quantify SIV gag RNA levels with a detection limit of 15 copies/ml as described previously (Bolton et al., 2015).
Quantification of trispecific antibody levels.
Trispecific antibody levels were measured using quantitative ELISA based methods in which a resurfaced core gpl20 (RSC3)(Wu et al., 2010) coated microtiter plates were used to capture the administered antibodies followed by detection using a HRP-conjugated anti-human IgG antibody (Jackson Immunoresearch).
HIV-1 neutralization assays.
Neutralization of replication-competent SHIV challenge stocks or single–round-of-entry Env-pseudoviruses (cross-clade) was evaluated in vitro by using Tzm-bl target cells and a luciferase reporter assay as described (Li et al., 2005; Montefiori, 2009; Pegu et al., 2015). Briefly, HIV-1 Env pseudoviruses were generated by transfection in 293T cells of Env expression plasmids with full-length, Env-defective HIV genome SG3dEnv. To assess neutralization sensitivity of replication competent SHIVBG505, we used the same SHIVBG505 challenge stock that were used for in vivo infection. SHIVBG505 or HIV-1 pseudoviruses were incubated with the antibody or plasma for 30 min at 37°C before Tzm-bl cells were added. The protease inhibitor indinavir was added to assays with SHIV stocks to a final concentration of 1 μM to limit infection of target cells to a single round of viral replication. Luciferase expression was quantified 48 h after infection upon cell lysis and the addition of luciferin substrate (Promega).
Measurement of Anti-Drug Antibody Responses.
Anti-drug antibody (ADA) responses were evaluated as follows. Plasma from macaques that had been administered the trispecific bNAb were diluted with PBS containing 5% skim milk, 2% BSA and 0.05% Tween 20. Five-fold serial dilutions ranging from 1:50 to 1:781250 of these plasmas were then added in duplicate wells to 96-well ELISA plates coated with 2 μg/ml of the trispecific bNAb. The plate was incubated for 1 hour at room temperature followed by a PBS-T (PBS with 0.05% Tween-20) wash. Bound monkey IgGs were then probed with a horseradish peroxidase (HRP)-conjugated anti-monkey IgG, Fc-specific (Southern Biotech) for 30 minutes at room temperature. The plate was then washed and SureBlue TMB (Kirkegaard & Perry Laboratories, Gaithersburg, MD) substrate was added. Once color was developed (typically 15 to 20 min), stopping buffer (IN H2SO4) was added and the optical density at 450 nm was read. Endpoint titer was calculated by determining the lowest dilution that had optical density greater than five-fold of that in the background wells.
Detection of viral RNA in lymph node tissues.
Biopsied lymph nodes were removed of fat and fixed overnight in 4% Paraformaldehyde (PFA) (Electron Microscopy Science) at room temperature (RT) followed by a paraffin block embedding. Tissue sections (5 μm thickness) were mounted on SuperFrost slides (Thermo Fisher Scientific) and subjected to the RNAscope assay (ACD). Briefly, sections were baked for 1 hour at 60°C, deparaffinized using xylene (twice, 5 minutes each) and ethanol baths (twice, 2 minutes each), and subjected to antigen retrieval step at 100°C for 15 minutes and proteinase K treatment at 40°C for 15 minutes. Slides were hybridized with SHIVBG505 pooled probe spanning gag-pol (20ZZ, targeting the region 2184-3342) and vif-vpu-nef (34ZZ, targeting the region 2-751 nt) at 40°C for 2 hours before being subjected to amplification steps as per RNAscope® Multiplex Fluorescent V2 assay protocol, followed by with TSA-FITC (Perkin Elmer, 1:1500) hybridization step. Following RNAscope, visualization of B (CD20+) and T (CD3+, CD4+) cells and FDC network was achieved by incubation for 1 h with blocking solution (0.1 M Tris, 0.3% Triton X-100, 1% BSA), follow by an overnight incubation with primary, nonconjugated antibodies directed to CD3 (Dako, clone F7.2.38) and FDC (Sigma, clone CNA.42). Next day, slides were washed with PBS at RT for 15 minutes, and then stained for 2 hours at RT with fluorescently conjugated Donkey anti-mouse IgGla (AlexaFlour594 conjugate, Thermo Fisher Scientific) and goat anti-mouse IgGM (AlexaFlour546 conjugate, Thermo Fisher Scientific) to detect CD3 and FDC, respectively. After three washing steps, slides were blocked with 10% normal mouse/goat serum for 1 hour at RT, and incubated with eFlour660 conjugated anti-CD20 (eBiosciences, clone L26) and AlexaFlour700 conjugated anti-CD4 (R&D Systems, cat. FAB8165G, goat polyclonal). After an additional wash step, the slides were counterstained for nuclear staining with Syto40 (Thermo Fisher Scientific, 1:100000) for 40 minutes at RT and mounted with Prolong Gold Antifade (Thermo Fisher Scientific) and cured for over 24 hours in the dark. Tissues were imaged using a Nikon C2 confocal microscope with 40X objective (1.40 NA) and images were analysized in Imaris version 9.5.0 (Bitplane), a 3D image visualization and analysis software. SHIV gag RNA+ cells and free virions were quantified using the Spots wizard tool within Imaris. 7 μm was assigned as the diameter for the virus-producing cells for the Spots wizard tool based on previously established averaged diameter of 7.5 μm for cells (Deleage et al., 2016; Kuznetsov et al., 2003) and was the average diameter calculated from randomly selected SHIV gag RNA+ cells. 2 μm was assigned as the diameter for the free virions that accounts for the established size of virus particle diameter of 1-1.27 μm (Deleage et al., 2016; Kuznetsov et al., 2003) and reduce the detection of background fluorescence. Additonally, to minimize operator bias and error, the detected spots were independently verified by two individuals and were manually deleted if they were background artifacts and not detected in at least three confocal optical slices. Background artifacts were determined when the vRNA signal overlapped with extremely saturated nuclear syto40 fluoresence which were indicative of artifacts native to the tissue. Data were normalized by area (mm2) to account for differences in tissue size.
Single-genome amplification/sequencing of SHIV env:
Virions were pelleted from plasma and virion-associated RNA extracted as previously described (Li et al., 2016). This viral RNA was then used for single-genome amplification and sequencing of SHIV Env as previously described (O’Brien et al., 2019). Briefly, to generate env cDNA, reverse transcription of viral RNA was performed using Superscript III reverse transcriptase according to the manufacturer’s directions (Thermo Fisher Scientific) and a gene specific primer (5’-TGTAATAAATCCCTTCCAGTCCCCCC-3’). The env gene was then amplified by nested PCR using Platinum Taq DNA High Fidelity polymerase (Thermo Fisher Scientific) for both reactions according to the manufacturer’s protocol. Template cDNA was serially diluted until PCR-positive wells, scored based on gel electrophoresis, constituted less than 30% of the total number of reactions, as previously described (Keele et al., 2008a; Keele et al., 2009). Correctly sized amplicons were sequenced directly using Big Dye Terminator technology (Applied Biosystems). Both DNA strands were sequenced and overlapping sequence fragments for each amplicon were assembled and edited using the Sequencher 5.0 program (Gene Codes). To confirm PCR amplification from a single template, chromatograms were manually examined for multiple peaks. Sequences with mixed bases were excluded from further analysis.
Measurement of SHIV specific cellular immune responses:
Cryopreserved PBMC and lymph node cells were thawed and rested overnight in a 37C/5% CO2 incubator. The next morning, cells were stimulated with HIV-1 BG505 Env peptide pool (comprised of 156 individual peptides as 15mers overlapping by 11 aa in 100% DMSO, JPT Peptides) and SIVmac239 Gag peptide pool (comprised of 125 individual peptides as 15mers overlapping by 11 aa in 100% DMSO, NIH AIDS Reagent Program) at a final concentration of 2 μg/ml in the presence of monensin and costimulatory antibodies anti-CD28 and anti-CD49d (clones CD28.2 and 9F10, BD Biosciences) for 6 hours. Negative controls received an equal concentration of DMSO instead of peptides (final concentration of 0.5% and co-stimulatory antibodies. Intracellular cytokine staining was performed as described (Donaldson et al., 2019). The following monoclonal antibodies were used: CD3 APC-Cy7 (clone SP34.2, BD Biosciences), CD4 PE-Cy5.5 (clone S3.5, Invitrogen), CD8 BV570 (clone RPA-T8, Biolegend), CD45RA PE-Cy5 (clone 5H9, BD Biosciences), CCR7 BV650 (clone G043H7, Biolegend), CXCR3 BV711 (clone 1C6/CXCR3, BD Biosciences), PD-1 BUV737 (clone EH12.1, BD Biosciences), ICOS Pe-Cy7 (clone C398.4A, Biolegend), CD69 ECD (cloneTP1.55.3, Beckman Coulter), IFN-γ Ax700 (clone B27, Biolegend), IL-2 BV750 (clone MQ1-17H12, BD Biosciences), IL-4 BB700 (clone MP4-25D2, BD Biosciences), TNF-FITC (clone Mab11, BD Biosciences), ICOS Pe-Cy7 (clone C398.4A, Biolegend), CD69 ECD (cloneTP1.55.3, Beckman Coulter), IFN-γ Ax700 (clone B27, Biolegend), IL-2 BV750 (clone MQ1-17H12, BD Biosciences), IL-4 BB700 (clone MP4-25D2, BD Biosciences), and CD154 BV785 (clone 24-31, BioLegend). Aqua live/dead fixable dead cell stain kit (Thermo Fisher Scientific) was used to exclude dead cells. All antibodies were previously titrated to determine the optimal concentration. Samples were acquired on an BD FAC Symphony flow cytometer and analyzed using FlowJo version 9.9.6 (Treestar, Inc., Ashland, OR).
Depletion of CD8+ T lymphocytes:
After approximately one-year post SHIV challenge, the trispecific antibody treatment animals were injected intravenously with anti-CD8β mAb CD8b255R1 (National Institutes of Health Nonhuman Primate Reagent Resource Program) (50mg/kg). Whole blood was obtained and stained with fluorescently conjugated antibodies at different time points before and after infusion to monitor absolute counts of lymphocyte subsets in the blood using fluorescent bead containing trucount tubes (BD biosciences) as previously described (Pegu et al., 2015). In brief, the following antibodies were used to monitor the various lymphocyte subsets – anti-NHP CD45 FITC (clone D058-1283, BD Biosciences), anti-CD20 Pacific Blue (clone 2H7, BioLegend), anti-CD14 BV510 (M5E2, BioLegend), anti-CD69 BV605 (clone FN50, BioLegend), anti-CD16 BV711 (clone 3G8, BioLegend), anti-CD8 BV785 (clone RPA-T8, BioLegend), anti-CD4 Alexa Flour 700 (clone OKT4, BioLegend), anti-CD3 APC-Cy7 (clone SP34-2, BD Biosciences), anti-HLA-DR PE-Cy5.5 (clone TU36, ThermoFisher) and anti-CD159a PE-Cy7 (clone REA110, Milenyi Biotec). In addition, plasma viremia was quantified for up to 6 months post CD8+ T cell depletion.
Viral resistance assay:
For each individual assay, CD4+ T cells were negatively selected from frozen PBMC samples obtained from viremic HIV-1 infected donors using the CD4+ T cell isolation kit from (Miltenyi Biotec) as per manufacturer’s instructions. After isolation, the cells were activated in cRPMI media (RPMI+10% FBS+1% Pen-Strep) containing IL-2 (10 U/ml, Sigma) and PHA-P (5 μg/ml, Sigma) overnight. Additionally, uninfected CD4+ T cells from three different naïve donors were activated separately overnight using the same activation media. The following day, all cell cultures were counted then combined into a single bulk culture at a ratio of 1 HIV-1 infected donor:4 uninfected “feeder” cells, with equal parts from each of the three naïve donors. The bulk culture was maintained in cRPMI+IL2 at a density of 1 million cells/ml for several days until the HIV-1 p24 level in the supernatant was above 100 pg/ml. When the virus level in the culture reached this level, the cells were split and transferred to a 96-well culture plate with approximately 50,000 cells per well. The antibodies were diluted in cRPMI+IL2 media and added to each respective well at a concentration of 75 μg/ml, or no antibody for control. Each condition was run in triplicate wells. Half of the supernatant was removed three times a week for analysis and replaced with fresh media and antibody. Additionally, 25,000 activated CD4 “feeder” T cells were added to each well once a week to sustain the conditions necessary for further virus growth. The supernatant samples were run on HIV-1 p24 antigen capture ELISA plates (ABL) per the manufacturer’s instructions to quantify the levels of p24 protein in each well. The viral RNA was quantified in the supernatant in each well at the end of the culture by real time RT-PCR as previously described (Hataye et al., 2019).
HIV trispecific Ab cell surface binding and potent Fc effector functions:
Apart from their ability to neutralize virus directly, antibodies can engage immune mediators through their Fc effector domains to promote lysis of virus and virally infected cells (Lu et al., 2018; Parsons et al., 2018). We observed that two different trispecific bNAbs, VRC01/PGDM1400-10E8v4 and N6/PGDM1400-10E8v4, can bind to cell surface expressed HIV Env, and have ADCC (Antibody-Dependent Cell Cytotoxicity), ADCP (Antibody-Dependent Cell Phagocytosis), and ADCML (Antibody-Dependent Complement-Mediated Lysis) effector activities (Figures S4).
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical analysis comparing slopes of viral decay in infected animals was performed using an unpaired T-test. Comparison of SHIV gag mRNA containing cells and free virions in lymph nodes of infected animals was performed using a nonparametric Kruskal-Wallis test.
Supplementary Material
Key resources table
| REAGENT or | SOURCE | IDENTIFIE |
|---|---|---|
| Antibodies | ||
| VRC01 | (Wu et al., 2010) | AB_2491019 |
| VRC07-523LS | (Rudicell et al., 2014) | N/A |
| PGDM1400 | (Sok et al., 2014) | N/A |
| 10E8v4 | (Kwon et al., 2016) | N/A |
| N6 | (Huang et al., 2016) | N/A |
| VRC01/PGDM1400-10E8v4 | (Xu et al., 2017) | N/A |
| N6/PGDM1400-10E8v4 | (Li et al., 2021; Xu et al., 2017) | N/A |
| anti-CD8β mAb CD8b255R1 | NIH Nonhuman Primate Reagent Resource Program | AB_2716321 |
| Bacterial and virus strains | ||
| SHIVBG505 | N/A | |
| HIV-1 BaL | NIH AIDS reagent program | N/A |
| Biological samples | ||
| Human PBMCs | VRC 601 trial volunteers (NCT01950325) | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| HIV-1 BG505 Env peptide pool | JPT peptides | N/A |
| SIVmac239 Gag peptide pool | NIH AIDS reagent program | N/A |
| biotinylated BG505 DS-SOSIP HIV-1 env trimer | (Kwon et al., 2015) | N/A |
| Primer:HIV-l Env; forward: TGTAATAAATCCCTTC CAGTCCCCCC | (O’Brien et al., 2019) | N/A |
| Critical commercial assays | ||
| HIV-1 p24 ELISA kit | ABL | Cat# 5447 |
| Experimental models: Cell lines | ||
| Expi293F cells | ThermoFisher Scientific Inc | Cat# A14525 |
| CEM-NKr-CCR5 cell line | NIH AIDS reagent program | CVCL_X623 |
| KHYG-1 NK cells expressing human CD16 | Alpert et al 2012 | N/A |
| THP-1 cells | ATCC | CVCL_0006 |
| Tzm-bl cells | NIH AIDS reagent program | CVCL_B478 |
| Experimental models: Organisms/strains | ||
| Indian origin rhesus macaque | This paper | N/A |
| Oligonucleotides | ||
| SHIVBG505 pooled probe spanning gag-pol (20ZZ, targeting the region 2184-3342) and vif-vpu-nef (34ZZ, targeting the region 2-751 nt) | This paper | N/A |
| Software and algorithms | ||
| Imaris version 9.5.0 | Bitplane | SCR_007370 |
| GraphPad Prism Software | GraphPad Prism Software, Inc. | SCR_002798 |
| FlowJo software | BD biosciences | SCR_008520 |
| Sequencher 5.0 program | Gene Codes | SCR_001528 |
| Other | ||
| Sequence data, analyses, and resources related to single genome amplification/sequencing of SHIV env | This paper | N/A |
HIGHLIGHTS.
Trispecific bNAb reduce viremia 100- to 1000-fold in viremic SHIV-infected macaques.
After treatment completion in these macaques, long term viral control is CD8-mediated.
Trispecific but not parental bNAbs suppress the emergence of resistant virus in culture.
ACKNOWLEDGEMENTS
We thank Wenge Ding for preparation of SHIVBG505 challenge stocks; Carla Lawendowski for excellent program management; Alida Taylor, Hana Bao, Saran Bao, Jumugal Noor and Diana Scorpio from the VRC Translational Research Program for animal research support; and Brenda Hartman for assistance with graphics. The anti-CD8β mAh CD8b255R1 used in these studies was provided by the NIH Nonhuman Primate Reagent Resource (P40 OD028116, U24 AI126683). We thank the VRC 601 trial volunteers for their contribution and commitment to HIV research. We also acknowledge the contributions of the VRC 601 Study Team including Grace Chen, Pamela Costner, Ingelise Gordon and Julie Ledgerwood. Support for this work was provided in part by the Intramural Research Program of the Vaccine Research Center, National Institute of Allergy and Infectious Diseases, NIH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interests
L.X., Z-y.Y., G.J.N., R.R.W., E.R., J.R.M., R.A.K., N.A.D-R., T.Z. and A.P. are inventors on patent application WO 2017/074878 submitted by Sanofi, The United States of America, as represented by the Secretary, Department of Health and Human Services, and the National Institutes of Health, that discloses the use of anti-HIV antibodies.The other authors declare no competing interests.
REFERENCES
- NCT03705169:Pharmacokinetics of SAR441236; https://clinicaltrials.gov/ct2/show/NCT03705169.
- Ackerman ME, Moldt B, Wyatt RT, Dugast AS, McAndrew E, Tsoukas S, Jost S, Berger CT, Sciaranghella G, Liu Q, et al. (2011). A robust, high-throughput assay to determine the phagocytic activity of clinical antibody samples. J Immunol Methods 366, 8–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alpert MD, Heyer LN, Williams DE, Harvey JD, Greenough T, Allhorn M, and Evans DT (2012). A novel assay for antibody-dependent cell-mediated cytotoxicity against HIV-1- or SIV-infected cells reveals incomplete overlap with antibodies measured by neutralization and binding assays. J Virol 86, 12039–12052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arunachalam PS, Charles TP, Joag V, Bollimpelli VS, Scott MKD, Wimmers F, Burton SL, Labranche CC, Petitdemange C, Gangadhara S, et al. (2020). T cell-inducing vaccine durably prevents mucosal SHIV infection even with lower neutralizing antibody titers. Nat Med 26, 932–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asokan M, Dias J, Liu C, Maximova A, Ernste K, Pegu A, McKee K, Shi W, Chen X, Almasri C, et al. (2020a). Fc-mediated effector function contributes to the in vivo antiviral effect of an HIV neutralizing antibody. Proc Natl Acad Sci U S A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asokan M, Dias J, Liu C, Maximova A, Ernste K, Pegu A, McKee K, Shi W, Chen X, Almasri C, et al. (2020b). Fc-mediated effector function contributes to the in vivo antiviral effect of an HIV neutralizing antibody. Proc Natl Acad Sci U S A 117, 18754–18763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-On Y, Gruell H, Schoofs T, Pai JA, Nogueira L, Butler AL, Millard K, Lehmann C, Suarez I, Oliveira TY, et al. (2018). Safety and antiviral activity of combination HIV-1 broadly neutralizing antibodies in viremic individuals. Nat Med 24, 1701–1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyrer C, and Pozniak A (2017). HIV Drug Resistance - An Emerging Threat to Epidemic Control. N Engl J Med 377, 1605–1607. [DOI] [PubMed] [Google Scholar]
- Bolton DL, Pegu A, Wang K, McGinnis K, Nason M, Foulds K, Letukas V, Schmidt SD, Chen X, Todd JP, et al. (2016). Human Immunodeficiency Virus Type 1 Monoclonal Antibodies Suppress Acute Simian-Human Immunodeficiency Virus Viremia and Limit Seeding of Cell-Associated Viral Reservoirs. J Virol 90, 1321–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bournazos S, Klein F, Pietzsch J, Seaman MS, Nussenzweig MC, and Ravetch JV (2014). Broadly neutralizing anti-HIV-1 antibodies require Fc effector functions for in vivo activity. Cell 158, 1243–1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bournazos S, and Ravetch JV (2017). Anti-retroviral antibody FcgammaR-mediated effector functions. Immunol Rev 275, 285–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cambou MC, and Landovitz RJ (2020). Novel Antiretroviral Agents. Curr HIV/AIDS Rep 17, 118–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caskey M, Klein F, Lorenzi JC, Seaman MS, West AP Jr., Buckley N, Kremer G, Nogueira L, Braunschweig M, Scheid JF, et al. (2015). Viraemia suppressed in HIV-1-infected humans by broadly neutralizing antibody 3BNC117. Nature 522, 487–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caskey M, Klein F, and Nussenzweig MC (2019). Broadly neutralizing anti-HIV-1 monoclonal antibodies in the clinic. Nat Med 25, 547–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caskey M, Schoofs T, Gruell H, Settler A, Karagounis T, Kreider EF, Murrell B, Pfeifer N, Nogueira L, Oliveira TY, et al. (2017). Antibody 10–1074 suppresses viremia in HIV-1-infected individuals. Nat Med 23, 185–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen YZ, Butler AL, Millard K, Witmer-Pack M, Levin R, Unson-O’Brien C, Patel R, Shimeliovich I, Lorenzi JCC, Horowitz J, et al. (2019). Safety, pharmacokinetics, and immunogenicity of the combination of the broadly neutralizing anti-HIV-1 antibodies 3BNC117 and 10-1074 in healthy adults: A randomized, phase 1 study. PLoS One 14, e0219142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corey L, Gilbert PB, Juraska M, Montefiori DC, Morris L, Karuna ST, Edupuganti S, Mgodi NM, deCamp AC, Rudnicki E, et al. (2021). Two Randomized Trials of Neutralizing Antibodies to Prevent HIV-1 Acquisition. New England Journal of Medicine in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dave RS, Jain P, and Byrareddy SN (2018). Follicular Dendritic Cells of Lymph Nodes as Human Immunodeficiency Virus/Simian Immunodeficiency Virus Reservoirs and Insights on Cervical Lymph Node. Front Immunol 9, 805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deleage C, Wietgrefe SW, Del Prete G, Morcock DR, Hao XP, Piatak M Jr., Bess J, Anderson JL, Perkey KE, Reilly C, et al. (2016). Defining HIV and SIV Reservoirs in Lymphoid Tissues. Pathog Immun 1, 68–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson MM, Kao SF, and Foulds KE (2019). OMIP-052: An 18-Color Panel for Measuring Th1, Th2, Th17, and Tfh Responses in Rhesus Macaques. Cytometry A 95, 261–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gama L, and Koup RA (2018). New-Generation High-Potency and Designer Antibodies: Role in HIV-1 Treatment. Annu Rev Med 69, 409–419. [DOI] [PubMed] [Google Scholar]
- Garbelli A, Riva V, Crespan E, and Maga G (2017). How to win the HIV-1 drug resistance hurdle race: running faster or jumping higher? Biochem J 474, 1559–1577. [DOI] [PubMed] [Google Scholar]
- Gaudinski MR, Coates EE, Houser KV, Chen GL, Yamshchikov G, Saunders JG, Holman LA, Gordon I, Plummer S, Hendel CS, et al. (2018). Safety and pharmacokinetics of the Fc-modified HIV-1 human monoclonal antibody VRC01LS: A Phase 1 open-label clinical trial in healthy adults. PLoS Med 15, e1002493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hataye JM, Casazza JP, Best K, Liang CJ, Immonen TT, Ambrozak DR, Darko S, Henry AR, Laboune F, Maldarelli F, et al. (2019). Principles Governing Establishment versus Collapse of HIV-1 Cellular Spread. Cell Host Microbe 26, 748–763 e720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu DC, Sunyakumthorn P, Wegner M, Schuetz A, Silsorn D, Estes JD, Deleage C, Tomusange K, Lakhashe SK, Ruprecht RM, et al. (2018). Central Nervous System Inflammation and Infection during Early, Nonaccelerated Simian-Human Immunodeficiency Virus Infection in Rhesus Macaques. J Virol 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Kang BH, Ishida E, Zhou T, Griesman T, Sheng Z, Wu F, Doria-Rose NA, Zhang B, McKee K, et al. (2016). Identification of a CD4-Binding-Site Antibody to HIV that Evolved Near-Pan Neutralization Breadth. Immunity 45, 1108–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julg B, Pegu A, Abbink P, Liu J, Brinkman A, Molloy K, Mojta S, Chandrashekar A, Callow K, Wang K, et al. (2017). Virological Control by the CD4-Binding Site Antibody N6 in Simian-Human Immunodeficiency Virus-Infected Rhesus Monkeys. J Virol 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keele BF, Giorgi EE, Salazar-Gonzalez JF, Decker JM, Pham KT, Salazar MG, Sun C, Grayson T, Wang S, Li H, et al. (2008a). Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proc Natl Acad Sci U S A 105, 7552–7557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keele BF, Li H, Learn GH, Hraber P, Giorgi EE, Grayson T, Sun C, Chen Y, Yeh WW, Letvin NL, et al. (2009). Low-dose rectal inoculation of rhesus macaques by SIVsmE660 or SIVmac251 recapitulates human mucosal infection by HIV-1. J Exp Med 206, 1117–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keele BF, Tazi L, Gartner S, Liu Y, Burgon TB, Estes JD, Thacker TC, Crandall KA, McArthur JC, and Burton GF (2008b). Characterization of the follicular dendritic cell reservoir of human immunodeficiency virus type 1. J Virol 82, 5548–5561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong R, Louder MK, Wagh K, Bailer RT, deCamp A, Greene K, Gao H, Taft JD, Gazumyan A, Liu C, et al. (2015). Improving neutralization potency and breadth by combining broadly reactive HIV-1 antibodies targeting major neutralization epitopes. J Virol 89, 2659–2671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuznetsov YG, Victoria JG, Robinson WE Jr., and McPherson A (2003). Atomic force microscopy investigation of human immunodeficiency virus (HIV) and HIV-infected lymphocytes. J Virol 77, 11896–11909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon YD, Georgiev IS, Ofek G, Zhang B, Asokan M, Bailer RT, Bao A, Caruso W, Chen X, Choe M, et al. (2016). Optimization of the Solubility of HIV-1-Neutralizing Antibody 10E8 through Somatic Variation and Structure-Based Design. J Virol 90, 5899–5914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon YD, Pancera M, Acharya P, Georgiev IS, Crooks ET, Gorman J, Joyce MG, Guttman M, Ma X, Narpala S, et al. (2015). Crystal structure, conformational fixation and entry-related interactions of mature ligand-free HIV-1 Env. Nat Struct Mol Biol 22, 522–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ledgerwood JE, Coates EE, Yamshchikov G, Saunders JG, Holman L, Enama ME, DeZure A, Lynch RM, Gordon I, Plummer S, et al. (2015). Safety, pharmacokinetics and neutralization of the broadly neutralizing HIV-1 human monoclonal antibody VRC01 in healthy adults. Clin Exp Immunol 182, 289–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Wang S, Kong R, Ding W, Lee FH, Parker Z, Kim E, Learn GH, Hahn P, Policicchio B, et al. (2016). Envelope residue 375 substitutions in simian-human immunodeficiency viruses enhance CD4 binding and replication in rhesus macaques. Proc Natl Acad Sci U S A 113, E3413–3422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Wang S, Lee F-H, Roark RS, Murphy AI, Smith J, Zhao C, Rando J, Chohan N, Ding Y, et al. (2021). New SHIVs and Improved Design Strategy for Modeling HIV-1 Transmission, Immunopathogenesis, Prevention and Cure. bioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Gao F, Mascola JR, Stamatatos L, Polonis VR, Koutsoukos M, Voss G, Goepfert P, Gilbert P, Greene KM, et al. (2005). Human immunodeficiency virus type 1 env clones from acute and early subtype B infections for standardized assessments of vaccine-elicited neutralizing antibodies. J Virol 79, 10108–10125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu CL, Murakowski DK, Bournazos S, Schoofs T, Sarkar D, Halper-Stromberg A, Horwitz JA, Nogueira L, Golijanin J, Gazumyan A, et al. (2016). Enhanced clearance of HIV-1-infected cells by broadly neutralizing antibodies against HIV-1 in vivo. Science 352, 1001–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu LL, Suscovich TJ, Fortune SM, and Alter G (2018). Beyond binding: antibody effector functions in infectious diseases. Nat Rev Immunol 18, 46–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch RM, Boritz E, Coates EE, DeZure A, Madden P, Costner P, Enama ME, Plummer S, Holman L, Hendel CS, et al. (2015). Virologic effects of broadly neutralizing antibody VRC01 administration during chronic HIV-1 infection. Sci Transl Med 7, 319ra206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mankarious S, Lee M, Fischer S, Pyun KH, Ochs HD, Oxelius VA, and Wedgwood RJ (1988). The half-lives of IgG subclasses and specific antibodies in patients with primary immunodeficiency who are receiving intravenously administered immunoglobulin. J Lab Clin Med 112, 634–640. [PubMed] [Google Scholar]
- Mayer KH, Seaton KE, Huang Y, Grunenberg N, Isaacs A, Allen M, Ledgerwood JE, Frank I, Sobieszczyk ME, Baden LR, et al. (2017). Safety, pharmacokinetics, and immunological activities of multiple intravenous or subcutaneous doses of an anti-HIV monoclonal antibody, VRC01, administered to HIV-uninfected adults: Results of a phase 1 randomized trial. PLoS Med 14, e1002435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendoza P, Gruell H, Nogueira L, Pai JA, Butler AL, Millard K, Lehmann C, Suarez I, Oliveira TY, Lorenzi JCC, et al. (2018). Combination therapy with anti-HIV-1 antibodies maintains viral suppression. Nature 561, 479–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller-Novak LK, Das J, Musich TA, Demberg T, Weiner JA, Venzon DJ, Mohanram V, Vargas-Inchaustegui DA, Tuero I, Ackerman ME, et al. (2018). Analysis of Complement-Mediated Lysis of Simian Immunodeficiency Virus (SIV) and SIV-Infected Cells Reveals Sex Differences in Vaccine-Induced Immune Responses in Rhesus Macaques. J Virol 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montefiori DC (2009). Measuring HIV neutralization in a luciferase reporter gene assay. Methods Mol Biol 485, 395–405. [DOI] [PubMed] [Google Scholar]
- Niessl J, Baxter AE, Mendoza P, Jankovic M, Cohen YZ, Butler AL, Lu CL, Dube M, Shimeliovich I, Gruell H, et al. (2020). Combination anti-HIV-1 antibody therapy is associated with increased virus-specific T cell immunity. Nat Med 26, 222–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura Y, Donau OK, Dias J, Ferrando-Martinez S, Jesteadt E, Sadjadpour R, Gautam R, Buckler-White A, Geleziunas R, Koup RA, et al. (2021). Immunotherapy during the acute SHIV infection of macaques confers long-term suppression of viremia. J Exp Med 218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura Y, Gautam R, Chun TW, Sadjadpour R, Foulds KE, Shingai M, Klein F, Gazumyan A, Golijanin J, Donaldson M, et al. (2017). Early antibody therapy can induce long-lasting immunity to SHIV. Nature 543, 559–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien SP, Swanstrom AE, Pegu A, Ko SY, Immonen TT, Del Prete GQ, Fennessey CM, Gorman J, Foulds KE, Schmidt SD, et al. (2019). Rational design and in vivo selection of SHIVs encoding transmitted/founder subtype C HIV-1 envelopes. PLoS Pathog 15, e1007632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons MS, Chung AW, and Kent SJ (2018). Importance of Fc-mediated functions of anti-HIV-1 broadly neutralizing antibodies. Retrovirology 15, 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauthner MG, Nkolola JP, Havenar-Daughton C, Murrell B, Reiss SM, Bastidas R, Prevost J, Nedellec R, von Bredow B, Abbink P, et al. (2019). Vaccine-Induced Protection from Homologous Tier 2 SHIV Challenge in Nonhuman Primates Depends on Serum-Neutralizing Antibody Titers. Immunity 50, 241–252 e246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pegu A, Asokan M, Wu L, Wang K, Hataye J, Casazza JP, Guo X, Shi W, Georgiev I, Zhou T, et al. (2015). Activation and lysis of human CD4 cells latently infected with HIV-1. Nat Commun 6, 8447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pegu A, Hessell AJ, Mascola JR, and Haigwood NL (2017). Use of broadly neutralizing antibodies for HIV-1 prevention. Immunol Rev 275, 296–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pegu A, Yang ZY, Boyington JC, Wu L, Ko SY, Schmidt SD, McKee K, Kong WP, Shi W, Chen X, et al. (2014). Neutralizing antibodies to HIV-1 envelope protect more effectively in vivo than those to the CD4 receptor. Sci Transl Med 6, 243ra288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghu G, Richeldi L, Crestani B, Wung P, Bejuit R, Esperet C, Antoni C, and Soubrane C (2018). SAR156597 in idiopathic pulmonary fibrosis: a phase 2 placebo-controlled study (DRI11772). Eur Respir J 52. [DOI] [PubMed] [Google Scholar]
- Richardson SI, Chung AW, Natarajan H, Mabvakure B, Mkhize NN, Garrett N, Abdool Karim S, Moore PL, Ackerman ME, Alter G, and Morris L (2018). HIV-specific Fc effector function early in infection predicts the development of broadly neutralizing antibodies. PLoS Pathog 14, e1006987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudicell RS, Kwon YD, Ko SY, Pegu A, Louder MK, Georgiev IS, Wu X, Zhu J, Boyington JC, Chen X, et al. (2014). Enhanced potency of a broadly neutralizing HIV-1 antibody in vitro improves protection against lentiviral infection in vivo. J Virol 88, 12669–12682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sok D, and Burton DR (2018). Recent progress in broadly neutralizing antibodies to HIV. Nat Immunol 19, 1179–1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sok D, van Gils MJ, Pauthner M, Julien JP, Saye-Francisco KL, Hsueh J, Briney B, Lee JH, Le KM, Lee PS, et al. (2014). Recombinant HIV envelope trimer selects for quaternary-dependent antibodies targeting the trimer apex. Proc Natl Acad Sci U S A 111, 17624–17629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagh K, Bhattacharya T, Williamson C, Robles A, Bayne M, Garrity J, Rist M, Rademeyer C, Yoon H, Lapedes A, et al. (2016). Optimal Combinations of Broadly Neutralizing Antibodies for Prevention and Treatment of HIV-1 Clade C Infection. PLoS Pathog 12, e1005520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Gajjar MR, Yu J, Padte NN, Gettie A, Blanchard JL, Russell-Lodrigue K, Liao LE, Perelson AS, Huang Y, and Ho DD (2020). Quantifying the contribution of Fc-mediated effector functions to the antiviral activity of anti-HIV-1 IgG1 antibodies in vivo. Proc Natl Acad Sci U S A 117, 18002–18009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Yang ZY, Li Y, Hogerkorp CM, Schief WR, Seaman MS, Zhou T, Schmidt SD, Wu L, Xu L, et al. (2010). Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science 329, 856–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Pegu A, Rao E, Doria-Rose N, Beninga J, McKee K, Lord DM, Wei RR, Deng G, Louder M, et al. (2017). Trispecific broadly neutralizing HIV antibodies mediate potent SHIV protection in macaques. Science 358, 85–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The published article includes all data generated or analyzed during this study.
This study did not generate new code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.


