Abstract
Respiratory tract colonization with Scedosporium apiospermum in patients with chronic suppurative lung disease is a significant concern for lung transplantation candidates, since Scedosporium infections occurring posttransplantation are usually untreatable. Up to 10% of patients with cystic fibrosis attending our respiratory medicine unit have had Scedosporium organisms isolated from sputum samples. We therefore developed a molecular typing method to examine these isolates. Typing by PCR amplification of ribosomal intergenic spacer sequences demonstrated 20 different types from 52 isolates collected from the respiratory medicine unit and elsewhere in Australia. A single common type was isolated from 11 respiratory medicine unit inpatients. Two other types were isolated from more than one source: one from two respiratory medicine unit inpatients and one from two epidemiologically linked nonhuman sources. Multiple isolates were obtained from nine patients. This method demonstrated persistent carriage of isolates of the same type in one patient for 7 months. Two patients showed carriage of isolates with multiple typing patterns within a 3-month period. The high rate of isolation and the predominance of isolates with a single typing pattern from respiratory medicine unit patients may suggest transmission to patients from a source in the unit. There was no epidemiological evidence of direct patient-to-patient spread, and Scedosporium organisms were not isolated from dust, soil, or air samples from the unit. The source and route of transmission have yet to be determined.
Scedosporium apiospermum (perfect state, Pseudallescheria boydii) is a saprophytic mold with a worldwide distribution and a reservoir in soil. The most commonly described clinical problem associated with this fungus is Madura foot, a cutaneous mycetoma that follows spore implantation. S. apiospermum has also been reported to cause pneumonia in previously immunocompetent hosts after near-drowning incidents in water contaminated with soil or animal manure (1). Immunocompromised patients, including those with hematological malignancy or undergoing bone marrow or solid-organ transplantation, may suffer fatal pulmonary, sinus, and disseminated infections (4, 5, 10, 13). Patients with cavitating or chronic suppurative lung diseases may develop clinical problems similar to those caused by Aspergillus fumigatus, the most common being respiratory tract colonization; fungal balls (7) and allergic bronchopulmonary disease (9) have also been reported. S. apiospermum is usually resistant in vitro and in vivo to commonly available antifungal agents, such as itraconazole and amphotericin. There are, however, reports of successful treatment with both systemic miconazole and ketoconazole (2, 16), in some cases combined with surgery (3). Scedosporium respiratory tract colonization is regarded as a relative but not absolute contraindication to lung transplantation (11). Most centers consider such colonized lung transplant candidates on an individual basis because of these potential therapeutic difficulties.
We have recently noticed an increase in S. apiospermum isolates from patients with cystic fibrosis and bronchiectasis attending our respiratory medicine unit (RMU). As colonization has serious implications for the future management of some of these patients, we developed a typing method involving PCR amplification of ribosomal intergenic spacer sequences (IGS PCR) to elucidate the epidemiology of S. apiospermum. This is the first reported typing method for this fungus.
MATERIALS AND METHODS
Fungal strains.
Consecutive clinical isolates of S. apiospermum identified by the Western Australian Centre for Pathology and Medical Research Mycology laboratory over a 17-month period from July 1997 to November 1998 were included in this study. These isolates were obtained from patients attending the RMU or other departments of Sir Charles Gairdner Hospital, a tertiary referral center, and other isolates referred from elsewhere in Western Australia. Five additional isolates were obtained from centers in other Australian states. S. apiospermum isolates were identified by macroscopic and microscopic characteristics. Fungi were subcultured from primary isolation agar plates onto Sabouraud agar plus 50 mg of chloramphenicol (Oxoid; supplied by Acorn Biological Pty. Limited, Victoria Park East, Australia) per liter to confirm purity. Hyphal fragments were then suspended in sterile water and stored at room temperature.
DNA extraction.
Hyphal fragments from water storage were subcultured onto Sabouraud agar to confirm purity. Spores were then inoculated from agar into 5 ml of Sabouraud broth (Oxoid) in a 5-cm sterile petri dish and incubated at 36°C until a mycelial mat had formed, at 3 to 5 days. A portion of the mat weighing approximately 10 to 20 mg was removed with sterile forceps, blotted with filter paper to remove excess liquid, and then suspended in 600 μl of filter-sterilized sorbitol buffer (1 M sorbitol, 14 mM beta-mercaptoethanol, 100 mM EDTA) containing 400 U of lyticase (catalog no. L4025; Sigma) for 1 h at 35°C, with occasional vortexing. The resultant fungal spheroplasts were precipitated by centrifugation at 15,000 × g for 5 min, and the supernatant was discarded. Spheroplasts were resuspended in 200 μl of proteinase K plus buffer ATL (Qiagen, Clifton Hill, Victoria, Australia) and incubated overnight in a water bath at 55°C. DNA was extracted using the Qiagen tissue kit protocol and eluted in 200 μl of buffer AE. DNA extracts were stored at −25°C. The DNA concentration was measured spectrophotometrically (Genequant; Pharmacia Biotech, Cambridge, England). The DNA yield was approximately 200 μg/ml.
PCR protocol.
The primers used span the ribosomal gene complex IGS region by targeting the conserved 5′ end of the 18S ribosomal gene and the 3′ end of the 26S ribosomal gene. Primer sequences were as follows: forward primer IGSL, 5′-TAG TAC GAG AGG AAC CGT-3′, and reverse primer IGSR, 5′-GCA TAT GAC TAC TGG CAG-3′. They correspond to bases 120 to 137 and 2285 to 2302 of the Aspergillus nidulans sequence (EMBL accession number Z27114) (14). Each PCR mixture contained 1 U of Taq polymerase (Amplitaq; Perkin-Elmer, Knoxfield, Australia), PCR buffer [67 mM Tris-HCl (pH 8.8), 16.6 mM (NH4)2SO4, 0.45% Triton X-100, 0.2 mg of gelatin per ml] (Biotech and Fisher Biotec, Belmont, Australia), 3 mM MgCl2 (Biotech), a 200 μM concentration of each deoxynucleoside triphosphate (Boehringer Mannheim and Roche Diagnostics, Castle View, Australia), a 0.5 μM concentration of each primer, and 4 μl of a 1-in-10 dilution of DNA extract (approximately 80 ng of DNA). Each reaction mixture was made up to 20 μl with deionized water treated with diethyl pyrocarbonate (catalog no. D5758; Sigma) and autoclaved. Reactions were carried out in a Perkin-Elmer GeneAmp PCR System 9700 thermocycler. Cycling conditions were 1 cycle of 94°C for 5 min and 45 cycles of denaturing at 94°C for 30 s, annealing at 45°C for 1 min, and extension at 72°C for 1 min, followed by one final extension step of 72°C for 7 min. All DNA extracts were assayed in duplicate, and each run included a negative control of deionized water treated with diethyl pyrocarbonate and autoclaved. Twenty microliters of PCR product was mixed with 4 μl of DNA loading dye, and 20 μl of the mixture was run on a 2.8% agarose gel in Tris acetate buffer and counterstained with ethidium bromide at 60 V for 3 h. Banding patterns were compared visually, with reference to a 123-bp fragment ladder (Gibco BRL, Life Technologies Pty. Limited, Mulgrave, Australia). All PCRs were repeated to confirm reproducibility of banding patterns between runs. All isolates were subcultured, and DNA extraction and PCR were repeated to confirm strain stability.
RMU setting and environmental sampling.
The RMU consists of two wards containing a total of 54 beds, a bronchoscopy suite, and associated offices and occupies a separate block on the hospital site. This block was constructed approximately 30 years ago as a tuberculosis isolation unit. RMU outpatients are seen at a different site in the main hospital block. Dust was collected from various environments on the RMU using a sterile cotton swab moistened with sterile distilled water. The locations were the main ward corridor, the bronchoscopy suite, the cystic fibrosis patients' lounge, the physiotherapy room, the ward kitchen, and the ward offices. Swabs were spread onto Sabouraud agar plus chloramphenicol (50 mg/liter) and onto rose bengal agar plus chloramphenicol (10 mg/liter) (Oxoid) and incubated at 26 and 36°C for 1 week. Surface soil was collected from around the roots of the two potted plants kept on the RMU. Approximately 10 mg of soil was mixed with 10 ml of sterile distilled water and agitated briefly, and particulate matter was allowed to precipitate for 5 min. Five microliters of suspension was then cultured onto Sabouraud and rose bengal media as described above. Air was sampled from within and immediately outside the RMU using an RCS air sampler (Biotest AG, Dreieich, Germany). Nutrient agar test strips from the RCS sampler were incubated at 36°C for 1 week. Fungi were identified using standard laboratory procedures.
RESULTS
Patient characteristics.
Fifty-two isolates from 31 sources (29 patients and two sea turtles [Chelonia mydas]) were studied (Table 1). The number of isolates from individual sources ranged from one to nine. Sources were divided into four epidemiological groups (Table 1): group 1, RMU inpatients; group 2, RMU outpatients; group 3, other patients from Western Australia; and group 4, human and nonhuman sources from other states of Australia. The origins and quantities of isolates were as follows: respiratory samples, 47; ear swabs, 3; unknown sites (human), 2; and nonhuman sources, 2. No Scedosporium organisms were isolated from the environmental samples.
TABLE 1.
Clinical features of sources of S. apiospermum isolates and IGS PCR types obtained
| Source no. | Age (yr) and sexa | Diagnosis | No. of isolatesd | Origin(s) | Type(s)e |
|---|---|---|---|---|---|
| Group 1 | |||||
| 1 | 30, M | Cystic fibrosis | 1 | Sputum | A |
| 2 | 19, M | Cystic fibrosis | 2 | Sputum | B |
| 3 | 24, F | Cystic fibrosis | 3 | Sputum | B, C |
| 4 | 20, F | Cystic fibrosis | 1 | Sputum | B |
| 5 | 35, M | Cystic fibrosis | 1 | Sputum | D |
| 6 | 57, F | Bronchiectasis | 2 | Sputum | B |
| 7 | 78, F | Bronchiectasis | 9 | BWb, sputum | C |
| 8 | 58, F | Bronchiectasis | 1 | Sputum | B |
| 9 | 64, F | Mycobacterium avium intracellulare pulmonary infection | 2 | Sputum | B, E |
| 10 | 68, M | Immunocompromised (fibrosing alveolitis), pneumonia | 3 | BW | B |
| 11 | 70, M | Carcinoma of lung | 1 | BW | B |
| 12 | 90, F | Carcinoma of lung | 1 | BW | B |
| 13 | 62, F | Carcinoma of lung | 1 | BW | B |
| 14 | 66, F | Carcinoma of lung | 1 | BW | B |
| Group 2 | |||||
| 15 | 57, M | Bronchiectasis | 1 | Sputum | F |
| 16 | 45, F | Asthma | 2 | Sputum | G |
| 17 | 50, F | Bronchiectasis | 1 | Sputum | H |
| Group 3 | |||||
| 18 | 47, M | Immunocompromised (renal transplant) pneumonia | 4 | BW, sputum | I |
| 19 | 68, F | Immunocompromised pneumonia | 1 | Sputum | B |
| 20 | 90, F | Pneumonia | 1 | Sputum | J |
| 21 | 58, F | Interstitial lung disease | 1 | BW | K |
| 22 | 87, M | Unknown | 1 | Sputum | L |
| 23 | 53, M | Bronchiectasis | 3 | Sputum | M |
| 24 | 84, M | Otitis externa | 1 | Ear swab | N |
| 25 | 43, F | Otitis externa | 1 | Ear swab | O |
| 26 | 28, M | Otitis externa | 1 | Ear swab | P |
| Group 4 | |||||
| From human sources | |||||
| 27 | NAc | NA | 1 | Unknown | Q |
| 28 | NA | NA | 1 | Unknown | R |
| 29 | NA | NA | 1 | BW | S |
| From C. mydas | |||||
| 30 | NA | NA | 1 | Cloaca | T |
| 31 | NA | NA | 1 | Eggshell | T |
M, male; F, female.
BW, bronchial washings.
NA, not available.
The total number of isolates was 52.
The number of types was 20.
PCR specifications.
PCR was optimized with respect to amount of DNA, concentrations of Taq polymerase, deoxynucleoside triphosphates, primers and magnesium, and cycling conditions. The parameters described resulted in the brightest bands, but the banding patterns were preserved over a >10-fold range of DNA concentrations, a range of magnesium chloride concentrations of 2.5 to 3.5 mM, and an annealing temperature down to 40°C (data not shown). Band patterns were reproducible, with the same set of bands being obtained by performing the PCR on different occasions with the same DNA extract.
Types of S. apiospermum.
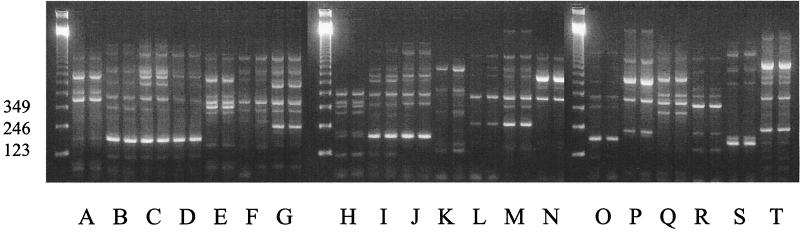
Analysis was restricted to the anamorph of the species, S. apiospermum. All isolates were typeable. This IGS PCR technique allowed the differentiation of 20 distinct types of S. apiospermum. In the absence of a reference collection of S. apiospermum strains with corresponding molecular characterizations, a single band difference was taken to indicate a distinct IGS PCR type (Fig. 1). Repeating the PCR with the same DNA extract generated the same banding pattern. Each IGS PCR type was stable on subculture (data not shown). Types B, C, and T were isolated from more than one source. Type B was isolated from 17 samples taken from 12 patients. Eleven of these patients were RMU inpatients. Type C was isolated from two RMU inpatients, and type T was isolated from two sea turtles from Queensland.
FIG. 1.
IGS PCR typing patterns of S. apiospermum. Twenty different types (A through T) are shown in duplicate. Molecular size standards (in base pairs) are shown in the left-hand lane.
Length and stability of carriage.
S. apiospermum was isolated on more than one occasion from each of nine patients. Seven of these patients had chronic suppurative lung disease. A single IGS PCR type was isolated from five of these patients, four of which were type B. One patient had isolates of type C from nine samples over a period of 7 months. Isolates with two IGS PCR types were isolated from each of two patients with chronic lung disease within a period of less than 3 months. Two immunocompromised patients with pneumonia had isolates with a single IGS PCR type on multiple bronchial washings and sputum samples. It is unknown whether pneumonia in either patient was caused by S. apiospermum, since no histological evidence of invasive fungal infection was obtained.
DISCUSSION
This is the first report of a typing method for S. apiospermum, a fungus that is emerging as a significant pathogen in immunocompromised patients. The only previously reported typing method for Scedosporium spp. has been a randomly amplified polymorphic DNA method (15) for the closely related (6) species Scedosporium inflatum. IGS PCR typing is technically straightforward and reproducible. Similar methods have been applied to numerous bacteria, including Burkholderia cepacia (8), and, more recently, to the fungus A. fumigatus (14).
This study has shown how IGS PCR typing may help to elucidate the epidemiology of S. apiospermum and to define the natural history of human colonization. The ecology of S. apiospermum bears some similarity to that of A. fumigatus. Both these environmental organisms are potential colonizers of patients with chronic suppurative lung disease. The natural history of S. apiospermum colonization appears to be similar to the more fully described relationship of A. fumigatus with these patients. We have demonstrated both isolation of S. apiospermum organisms with the same IGS PCR type over a prolonged period of time and carriage of multiple isolates of different IGS PCR types over a period of a few weeks in patients with chronic suppurative lung disease. Due to the lack of a reference collection with molecular characterization correlated to epidemiological information, we cannot conclude at this point whether this represents carriage of multiple strains or the prolonged carriage of the same strain. These two observations are, however, analogous to the results obtained for A. fumigatus typed by randomly amplified polymorphic DNA analysis (17) and DNA fingerprinting (12) for patients with cystic fibrosis. Scedosporium sp. colonization is, however, potentially more serious than Aspergillus sp. colonization in patients with cystic fibrosis, since the latter becomes a contraindication to lung transplantation only if the patient has additionally either an aspergilloma with pleural involvement or allergic bronchopulmonary aspergillosis with a large steroid requirement. In contrast, Scedosporium sp.-colonized lung transplant candidates may be excluded per se because of the therapeutic difficulties involved when invasive disease develops after transplantation.
S. apiospermum isolates of one IGS PCR type predominated among RMU inpatients. This could suggest that transmission might have occurred on the RMU. Two other explanations should also be considered. First, isolates of the B type might have a tropism for patients with chronic suppurative lung disease. Against this possibility are the observations that (i) isolates with multiple types (six types) were isolated from patients with cystic fibrosis and bronchiectasis, (ii) type B isolates were recovered only from the subset of these patients who were epidemiologically linked with the RMU, and (iii) type B isolates were isolated from respiratory specimens of many RMU patients with other diagnoses. The second possible explanation is that isolates of type B are more common than those of other types. The observation that a type B isolate was recovered from one patient with no apparent connection with the RMU, but who lived in the same metropolitan area, is the only circumstantial evidence in support of this hypothesis.
A means of S. apiospermum transmission was not determined. An environmental reservoir was not discovered, since environmental sampling did not yield Scedosporium sp. isolates. There was no evidence of person-to-person transmission, since, with the exception of the young adults with cystic fibrosis who attended the ward together (and who also met outside the hospital), there was no evidence of direct patient interaction. For the majority of RMU patients from whom isolates with IGS PCR type B were recovered, the inpatient periods did not overlap. Isolates with type C were recovered from two patients who were frequent RMU inpatients and who were admitted on one occasion at the same time. The significance of this is uncertain.
This is the first report of molecular typing of a collection of clinical isolates of S. apiospermum. We have demonstrated that S. apiospermum isolates of the same IGS PCR type can be recovered from some patients with chronic suppurative lung disease over a prolonged period of time and that multiple isolates of different IGS PCR types can be recovered over relatively short periods of time from the same patient. The predominance of a particular IGS PCR type associated with the RMU could suggest that a common source infects patients with chronic suppurative lung disease and other susceptible patients, but the lack of other molecular characterization permits only tentative conclusions. Molecular typing of this species by another method is therefore required to help establish the epidemiology of this species. Due to the significance of this isolate for potential lung transplantation candidates, understanding the epidemiology of this fungus should help in its future management.
ACKNOWLEDGMENTS
Tom Riley provided financial support for this project.
The staff of the Molecular Diagnostics Laboratory and Gracy Cherian of the Mycology Laboratory, PathCentre, gave technical assistance. Infection control nurses Helen Cadwallader and Anne Dyson assisted with environmental sampling and tracing admission records. David Ellis of the Mycology Laboratory, Women and Children's Hospital, Adelaide, kindly provided fungal isolates from elsewhere in Australia.
REFERENCES
- 1.Dworzack D L, Clark R B, Borkowski W J, Smith D L, Dykstra M, Pugsley M P, Horowitz E A, Connolly T L, McKinney D L, Hostetler M K, Fitzgibbons J F, Galant M. Pseudallescheria boydii brain abscess: association with near drowning and efficacy of high-dose, prolonged miconazole therapy in patients with multiple abscesses. Medicine. 1989;68:218–224. [PubMed] [Google Scholar]
- 2.Galgiani J N, Stevens D A, Graybill J R, Stevens D L, Tillinghast A J, Levine H B. Pseudallescheria boydii infections treated with ketoconazole. Chest. 1984;86:219–224. doi: 10.1378/chest.86.2.219. [DOI] [PubMed] [Google Scholar]
- 3.Garcia-Arata M I, Otero M J, Zomeno M, de la Figuera M A, de las Cuevas M C, Lopez-Brea M. Scedosporium apiospermum pneumonia after autologous bone marrow transplantation. Eur J Clin Microbiol Infect Dis. 1996;15:600–603. doi: 10.1007/BF01709371. [DOI] [PubMed] [Google Scholar]
- 4.Grigg A P, Phillips P, Durham S, Shepherd J D. Recurrent Pseudallescheria boydii sinusitis in acute leukemia. Scand J Infect Dis. 1993;25:263–267. doi: 10.3109/00365549309008495. [DOI] [PubMed] [Google Scholar]
- 5.Gumbart C H. Pseudallescheria boydii infection after bone marrow transplantation. Ann Intern Med. 1983;99:193–194. doi: 10.7326/0003-4819-99-2-193. [DOI] [PubMed] [Google Scholar]
- 6.Issakainen J, Jalava J, Eerola E, Campbell C K. Relatedness of Pseudallescheria, Scedosporium and Graphium pro parte based on SSU rRNA sequences. J Med Vet Mycol. 1997;35:389–398. [PubMed] [Google Scholar]
- 7.Kathuria S K, Rippon J. Nonaspergillus aspergilloma. Am J Clin Pathol. 1982;78:870–873. doi: 10.1093/ajcp/78.6.870. [DOI] [PubMed] [Google Scholar]
- 8.Kostman J R, Edlind T D, LiPuma J, Stull T L. Molecular epidemiology of Pseudomonas cepacia determined by polymerase chain reaction ribotyping. J Clin Microbiol. 1992;30:2084–2087. doi: 10.1128/jcm.30.8.2084-2087.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lake F R, Tribe A E, McAleer R, Froudist J, Thompson P J. Mixed allergic bronchopulmonary fungal disease due to Pseudallescheria boydii and Aspergillus. Thorax. 1990;45:489–492. doi: 10.1136/thx.45.6.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lopez F A, Crowley R S, Wastila L, Valantine H A, Remington J S. Scedosporium apiospermum (Pseudallescheria boydii) infection in a heart transplant recipient: a case of mistaken identity. J Heart Lung Transplant. 1998;17:321–324. [PubMed] [Google Scholar]
- 11.Maurer J R, Frost A E, Glanville A R, Estene M, Higenbottam T. International guidelines for the selection of lung transplant candidates. Am J Respir Crit Care Med. 1998;158:335–339. doi: 10.1164/ajrccm.158.1.15812. [DOI] [PubMed] [Google Scholar]
- 12.Neuvéglise C, Sarfati J, Debeaupuuis J P, Vu Thien H, Just J, Tournier G, Latgé J-P. Longitudinal study of Aspergillus fumigatus strains isolated from cystic fibrosis patients. Eur J Clin Microbiol Infect Dis. 1997;16:747–750. doi: 10.1007/BF01709257. [DOI] [PubMed] [Google Scholar]
- 13.Paradowski L J. Saprophytic fungal infections and lung transplantation—revisited. J Heart Lung Transplant. 1997;16:524–531. [PubMed] [Google Scholar]
- 14.Radford S A, Johnson E M, Leeming J P, Millar M R, Cornish J M, Foot A B M, Warnock D W. Molecular epidemiological study of Aspergillus fumigatus in a bone marrow transplantation unit by PCR amplification of ribosomal intergenic spacer sequences. J Clin Microbiol. 1998;36:1294–1299. doi: 10.1128/jcm.36.5.1294-1299.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.San Millán R, Quindós G, Garaizar J, Salesa R, Guarro J, Pontón J. Characterization of Scedosporium prolificans clinical isolates by randomly amplified polymorphic DNA analysis. J Clin Microbiol. 1997;35:2270–2274. doi: 10.1128/jcm.35.9.2270-2274.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tekavec J, Mlinaric-Missoni E, Babic-Vazic V. Pulmonary tuberculosis associated with invasive pseudallescheriasis. Chest. 1997;111:508–511. doi: 10.1378/chest.111.2.508. [DOI] [PubMed] [Google Scholar]
- 17.Verweij P E, Meis J F G M, Sarfati J, Hoogkamp-Korstanje J A A, Latgé J-P, Melchers W J G. Genotypic characterization of sequential Aspergillus fumigatus isolates from patients with cystic fibrosis. J Clin Microbiol. 1996;34:2595–2597. doi: 10.1128/jcm.34.10.2595-2597.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]